Short abstract
Objective
The aim of this study was to measure the extracellular matrix protein Spondin-2 (SPON2) in hepatocellular carcinoma (HCC) tissues and to determine its potential value as a prognostic indicator by assessing its correlation with clinicopathological variables and survival.
Methods
SPON2 mRNA expression was assessed in 20 matched pairs of HCC and non-cancerous liver tissues by quantitative reverse transcription-polymerase chain reaction analysis. SPON2 protein expression was determined in 107 matched pairs of HCC and normal liver tissue by immunohistochemical staining of tissue microarrays.
Results
Analysis of patient tissues and Oncomine datasets showed that SPON2 mRNA and SPON2 protein expression were both significantly upregulated in HCC tissues, compared with non-cancerous liver tissue; moreover, both correlated significantly with tumor size. Kaplan-Meier analysis revealed that HCC patients who showed high levels of cytoplasmic SPON2 protein had poorer survival following curative resection, compared with HCC patients who exhibited low protein expression levels. Multivariate Cox regression analysis showed that tumor thrombus and SPON2 protein expression both independently correlated with reduced survival in HCC patients.
Conclusion
Upregulated expression of SPON2 protein in tumor tissue could be an effective prognostic indicator for patients with HCC.
Keywords: Hepatocellular carcinoma, immunohistochemistry, prognosis, Spondin-2, tissue microarrays, tumor thrombus
Introduction
In 2012, there were an estimated 782,500 new cases of liver cancer and more than 745,500 deaths due to the disease worldwide; more than 50% of these deaths occurred in China.1 Most primary liver cancers (70%–90%) are hepatocellular carcinoma (HCC); notably, HCC is the third leading cause of cancer death in China, and was responsible for more than 400,000 deaths in 2015.2 Although multiple risk factors for HCC have been identified, including chemical and viral exposure, it remains a particularly lethal disease because it is typically asymptomatic prior to reaching the terminal stage. Serum alpha-fetoprotein (AFP) is commonly used as a biomarker for HCC; however, it demonstrates low sensitivity and specificity. Moreover, its level varies with the tumor type and patient population. There is thus a critical need to identify alternative diagnostic and prognostic markers for HCC.
Spondin-2 (SPON2) is a secreted extracellular matrix protein and a member of the Mindin/F-Spondin family.3 SPON2 has multiple functions in a variety of crucial cellular processes, including development of neurons, recruitment of inflammatory cells, and activation of the innate immune response.4,5 Recently, SPON2 overexpression was found in HCC,6–8 colorectal carcinoma,9–11 gastric cancer,12,13 Barrett’s adenocarcinoma,14 prostate cancer,15–20 ovarian cancer,21,22 pancreatic cancer,23 pulmonary adenocarcinoma,24 and breast cancer.25 In addition, SPON2 has been proposed as a new serum and histological diagnostic biomarker, as well as an independent prognostic indicator for colorectal carcinoma,10,11 gastric cancer,13 prostate cancer,17,19,20 and ovarian cancer.21,22
SPON2 expression is elevated in HCC tumor tissues, compared with matched non-cancerous liver tissue.6–8 Zhang et al.8 reported that HCC patients with upregulated SPON2 expression had better overall survival (OS). However, an inverse relationship between SPON2 expression and OS has been found in other inflammatory tumors, such as colorectal carcinoma10,11 and gastric cancer.13 Therefore, the relationship between SPON2 expression and prognosis in HCC remains unclear. The purpose of the present study was to assess expression of SPON2 mRNA and SPON2 protein in HCC and determine its association with clinicopathological features and patient prognosis.
Materials and methods
Patients and tissue samples
Two sets of patient samples were evaluated. We obtained HCC and matched non-cancerous liver tissue from 20 HCC patients undergoing curative resection at Affiliated Hospital of Nantong University (Jiangsu, China) from June 2013 to June 2014. These samples were used to analyze SPON2 mRNA expression by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). We also obtained matched tumor and normal liver tissue samples from 107 HCC patients undergoing curative resection at the same hospital between January 2004 and December 2009; these tissues were used to construct tissue microarrays (TMA) for immunohistochemical (IHC) analysis of SPON2 protein. All patients had pathologically confirmed HCC; none had undergone chemotherapy, radiation therapy, or immunotherapy prior to curative surgical resection. Post-surgical follow-ups were completed by December 2014 (median follow-up 39 months, range 1–85 months). Clinicopathological data were collected from medical records during the inpatient stay and included sex, age, differentiation grade, tumor diameter, Child-Pugh stage, hepatocirrhosis status, hepatitis B virus (HBV) infection, tumor thrombus, AFP level, Barcelona Clinic Liver Cancer (BCLC) stage, envelope invasion, and tumor satellite lesions. All patients provided written informed consent before surgery, and the study protocol was approved by the Human Research Ethics Committee of Affiliated Hospital of Nantong University.
RNA extraction and qRT-PCR
Total RNA was extracted from the 20 paired HCC and matched non-cancerous liver tissue samples by using TRIzol reagent (Invitrogen, Karlsruhe, Germany). One-step qRT-PCR analysis was performed on an Applied Biosystems 7500 Real-Time PCR System by using the LightCycler FastStart DNA Master SYBR Green I Kit (Roche Diagnostics, Tokyo, Japan). The SPON2 primers were as follows: forward 5′-AAGAACCAGTACGTCAGTAACGG-3′ and reverse 5′-CACAAACGAGACCAGCGAGT-3′ (201-bp). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified to normalize the SPON2 mRNA levels (GAPDH forward primer 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse primer 5′-AGGGGCCATCCACATCTTC-3′). The reverse transcription conditions were 42°C for 60 minutes and 70°C for 5 minutes. The PCR conditions were 10 minutes at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 30 s at 72°C. All reactions were performed in quadruplicate.
TMA-IHC
TMA construction and IHC staining were performed as previously described.26,27 The anti-SPON2 primary antibody was from Affinity (#DF4682, China). Immunostaining levels were evaluated independently by two investigators who were blinded to the sample identities. The staining intensity was scored as: 0 (−, no staining), 1 (+, weak staining), 2 (++, moderate staining), or 3 (+++, strong staining). The proportion of cells staining positive for SPON2 was also recorded. The product of the intensity score and percentage positive cells was recorded as the final SPON2 staining score (range 0–300). For analysis, samples were classified as “low” or “high” SPON2 protein expression based on a cutoff point of 100, which was identified by using X-tile software (Rimm Laboratory, Yale University, New Haven, CT, USA) as described previously.28 Scores of 0 to 100 and 101 to 300 were considered low and high expression, respectively.
Bioinformatic analysis of the Oncomine database
Datasets of SPON2 mRNA expression in normal and diseased liver tissues were obtained from the Oncomine database (http://www.oncomine.org/resource/login.html) as described elsewhere.11 “SPON2,” “Hepatocellular Carcinoma,” “mRNA,” and “Cancer vs Normal Analysis” filters were applied as the search terms to compare SPON2 mRNA expression level between HCC and other liver tissues. “Survival Status” was applied as an additional filter to evaluate the association between expression and survival. “SPON2,” “Hepatocellular Carcinoma,” and “mRNA” filters were applied to evaluate the correlation between expression and clinicopathological parameters. SPON2 mRNA expression levels were further classified into greater than or less than the median groups according to the log2 median-centered intensity value obtained with Microsoft Excel.
Statistical analysis
Statistical analysis was performed by using SPSS version 24.0 software (IBM Corp., Armonk, NY, USA) and Prism 6.01 software (GraphPad, La Jolla, CA, USA). Student’s t-test was used to evaluate differences in SPON2 mRNA expression levels. The relationships between SPON2 mRNA or SPON2 protein expression and clinicopathological parameters were analyzed by using χ2 tests. Kaplan-Meier curves and the log-rank test were used to evaluate survival. Features verified to have statistically significant prognostic value in a univariate Cox regression model were then entered into a multivariate Cox regression model. Differences were regarded as statistically significant at P<0.05.
Results
Upregulation of SPON2 mRNA expression in HCC tissues
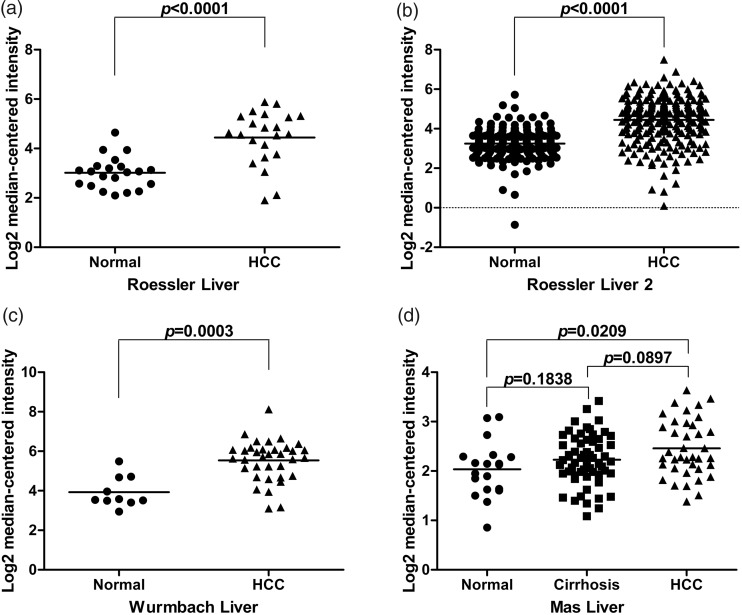
To assess whether SPON2 is differentially expressed in HCC and normal liver tissues at the mRNA level, we analyzed datasets obtained from the Oncomine database. SPON2 mRNA levels were significantly higher in HCC tissues, compared with non-cancerous liver tissues, in the Roessler Liver dataset (n=22, P<0.0001; Figure 1a), Roessler Liver 2 dataset (n=225, P<0.0001; Figure 1b), and Wurmbach Liver dataset (n=35, P=0.0003; Figure 1c). The Mas Liver dataset (Figure 1d) showed no difference in SPON2 mRNA between normal and cirrhotic liver samples (n=58, P=0.1838) or between cirrhotic and HCC samples (n=38, P=0.0897); however, SPON2 mRNA was significantly upregulated in HCC tissues, compared with normal liver (n=38, P=0.0209). These analyses of a total of 320 HCC samples and 270 non-cancerous liver control samples indicated that SPON2 mRNA expression is increased in HCC. To confirm this result, we performed qRT-PCR analysis of SPON2 mRNA expression in tissues obtained from 20 HCC patients in our local population; SPON2 mRNA was significantly higher in HCC tissues, compared with matched non-cancerous liver tissues (n=20, P<0.0001; Figure 2a).
Figure 1.
Mining of the Oncomine database indicates that SPON2 mRNA expression is upregulated in HCC tissues. (a–c) SPON2 mRNA expression in normal liver and HCC tissues, based on the Roessler Liver dataset (a), Roessler Liver 2 dataset (b), and Wurmbach Liver dataset (c). (d) SPON2 mRNA expression in normal liver, cirrhotic liver, and HCC tissues, based on the Mas Liver dataset.
Figure 2.
Expression of SPON2 in HCC and normal liver tissue. (a) qRT-PCR analysis of SPON2 mRNA expression in 20 paired HCC and matched non-cancerous liver tissues. (b) TMA-IHC analysis of SPON2 protein expression in 107 paired HCC and matched non-cancerous liver tissues.
Upregulation of SPON2 protein expression in HCC tissues
Next, we analyzed SPON2 protein expression by IHC staining of a TMA composed of 107 paired HCC and matched non-cancerous liver tissues. As shown in Figure 2b, SPON2 protein was primarily detected in the cytoplasm of HCC cells. Notably, SPON2 protein expression was detected in a significantly greater number of samples from HCC tissues (56/107, 52.336%) than from matched non-cancerous liver tissues (21/107, 19.626%; P<0.0001), Therefore, upregulated expression of SPON2 appears to be associated with development of HCC.
Association between upregulated SPON2 expression and clinicopathological parameters in HCC patients
To assess the association between SPON2 mRNA expression and clinicopathological parameters of HCC patients, we analyzed the Wurmbach Liver, Jia Liver, and Chiang Liver datasets in the Oncomine database. Each dataset was divided into high and low SPON2 expression groups based on the median SPON2 mRNA level. We found that the SPON2 mRNA level was significantly associated with tumor size in the Wurmbach Liver dataset (χ2=4.309, P=0.038) and with age in both the Jia Liver (χ2=5.069, P=0.014) and Chiang Liver (χ2=5.199, P=0.023) datasets (Table 1). Analysis of the TMA-IHC data revealed associations between the SPON2 protein level and differentiation grade (χ2=12.914, P=0.002), tumor diameter (χ2=4.809, P=0.028), and Child-Pugh stage (χ2=10.178, P=0.001) (Table 2). However, other clinical features, including sex, age, hepatocirrhosis status, HBV infection, tumor thrombus, AFP level, BCLC stage, envelope invasion, and tumor satellite lesions, were not significantly correlated with SPON2 protein expression. Taken together, these findings demonstrate that the increase in SPON2 expression in HCC was related to tumor size.
Table 1.
Association between SPON2 mRNA expression and clinicopathological parameters in HCC patients based on Oncomine datasets.
| Dataseta | Clinicopathological parameters | n |
SPON2 expression |
χ2 | P value | |
|---|---|---|---|---|---|---|
| Below-median | Above-median | |||||
| Wurmbach Liver | ||||||
| Size (cm) | ||||||
| ≤5 | 23 | 11 | 12 | 4.309 | 0.038 | |
| >5 | 10 | 1 | 9 | |||
| Chiang Liver | ||||||
| Age (years) | ||||||
| ≤50 | 5 | 0 | 5 | 5.199 | 0.023 | |
| >50 | 76 | 40 | 36 | |||
| Jia Liver | ||||||
| Age (years) | ||||||
| ≤50 | 121 | 51 | 70 | 5.069 | 0.014 | |
| >50 | 117 | 68 | 49 | |||
The analysis was performed by using datasets from the Oncomine cancer gene expression microarray database (https://www.oncomine.org/resource/login.html).
Table 2.
Association between SPON2 protein expression and clinicopathological parameters in HCC patients based on TMA-IHC analysis.
| Clinicopathological parameters | n | Low expression | High expression | χ2 | P value |
|---|---|---|---|---|---|
| Sex | |||||
| Male | 75 | 35 | 40 | 0.100 | 0.752 |
| Female | 32 | 16 | 16 | ||
| Age (years) | |||||
| ≤50 | 45 | 22 | 23 | 0.047 | 0.829 |
| >50 | 62 | 29 | 33 | ||
| Grade of differentiation | |||||
| Low | 39 | 26 | 13 | 12.914 | 0.002* |
| Middle | 35 | 17 | 18 | ||
| High | 33 | 8 | 25 | ||
| Tumor diameter (cm) | |||||
| ≤5 | 49 | 29 | 20 | 4.809 | 0.028* |
| >5 | 58 | 22 | 36 | ||
| Child-Pugh stage | |||||
| A | 38 | 26 | 12 | 10.178 | 0.001* |
| B or C | 69 | 25 | 44 | ||
| Hepatocirrhosis | |||||
| Absent | 24 | 11 | 13 | 0.042 | 0.838 |
| Present | 83 | 40 | 43 | ||
| HBV infection | |||||
| Absent | 41 | 21 | 20 | 0.337 | 0.562 |
| Present | 66 | 30 | 36 | ||
| Tumor thrombus | |||||
| Absent | 47 | 25 | 22 | 1.027 | 0.311 |
| Present | 60 | 26 | 34 | ||
| AFP (ng/mL) | |||||
| ≤20 | 52 | 27 | 25 | 0.736 | 0.391 |
| >20 | 55 | 24 | 31 | ||
| BCLC stage | |||||
| A | 24 | 14 | 10 | 1.412 | 0.235 |
| B, C, or D | 83 | 37 | 46 | ||
| Envelope | |||||
| Absent | 63 | 26 | 37 | 2.511 | 0.113 |
| Present | 44 | 25 | 19 | ||
| Tumor satellite | |||||
| Absent | 58 | 26 | 32 | 0.408 | 0.523 |
| Present | 49 | 25 | 24 |
*P<0.05; Serum Alpha-Fetoprotein (AFP); Hepatitis B Virus (HBV); Barcelona Clinic Liver Cancer (BCLC).
Association between upregulated SPON2 expression and OS in HCC
To determine whether SPON2 mRNA expression correlated with OS in HCC patients, we performed a Kaplan-Meier survival analysis. Using the Hoshida Liver Statistics dataset (n=118, including 80 completed and 38 censored cases) in the Oncomine database, no relationship could be detected between SPON2 mRNA expression and prognosis (log-rank test=1.399, P=0.2368; Figure 3a). However, when the analysis was performed using the TMA-IHC dataset, high SPON2 protein expression in the cytoplasm of HCC cells correlated significantly with poor prognosis (log-rank test=4.381, P=0.036; Figure 3b). These results suggest that the level of SPON2 protein in tumor tissue could be a prognostic indicator in HCC.
Figure 3.
Kaplan-Meier survival analysis of SPON2 expression in HCC patients. Correlation between prognosis of HCC patients and tumor expression of (a) SPON2 mRNA, based on the Oncomine Hoshida Liver Statistics dataset, and (b) SPON2 protein, based on the TMA-IHC dataset.
SPON2 protein expression level as a prognostic predictor in HCC
We performed univariate and multivariate Cox regression analyses of SPON2 protein expression and clinicopathological parameters in the TMA-IHC dataset to identify potential predictors of OS and disease-free survival (DFS) in HCC patients. Univariate analyses indicated that tumor thrombus, BCLC stage, and SPON2 protein expression were significantly associated with OS and DFS in HCC patients (Table 3). When these factors were included in a multivariate Cox regression model, tumor thrombus and SPON2 protein expression remained independent prognostic predictors for HCC (Table 3).
Table 3.
Univariate and multivariate analysis of overall survival and disease-free survival in HCC patients.
| Characteristics |
Overall Survival |
Disease-Free Survival |
||||||
|---|---|---|---|---|---|---|---|---|
|
Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
|||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Sex | ||||||||
| Male vs. female | 1.037 (0.499–2.151) | 0.932 | 0.890 (0.428–1.847) | 0.754 | ||||
| Age (year) | ||||||||
| ≤50 vs. >50 | 0.811 (0.367–1.791) | 0.604 | 0.592 (0.275–1.275) | 0.180 | ||||
| Grade of differentiation | ||||||||
| Low/Middle/High | 0.764 (0.372–1.573) | 0.466 | 0.581 (0.290–1.166) | 0.127 | ||||
| Tumor diameter (cm) | ||||||||
| ≤5 vs. >5 | 1.639 (0.460–5.836) | 0.446 | 2.967 (0.742–11.856) | 0.124 | ||||
| Child-Pugh stage | ||||||||
| A vs. B or C | 0.874 (0.170–4.503) | 0.872 | 1.180 (0.370–3.766) | 0.780 | ||||
| Hepatocirrhosis | ||||||||
| − vs. + | 1.896 (0.789–4.556) | 0.153 | 2.264 (0.950–5.397) | 0.065 | ||||
| HBV infection | ||||||||
| − vs. + | 1.541 (0.778–3.052) | 0.215 | 1.359 (0.700–2.638) | 0.365 | ||||
| Tumor thrombus | ||||||||
| − vs. + | 2.400 (1.219–4.726) | 0.011* | 2.372 (1.264–4.452) | 0.007* | 2.123 (1.086–4.153) | 0.028* | 2.079 (1.113–3.884) | 0.022* |
| AFP (ng/mL) | ||||||||
| ≤20 vs. >20 | 1.369 (0.717–2.613) | 0.341 | 1.735 (0.905–3.324) | 0.097 | ||||
| BCLC stage | ||||||||
| A vs. B, C, or D | 0.367 (0.154–0.879) | 0.024* | 1.053 (0.515–0.252) | 0.069 | 0.398 (0.167–0.951) | 0.038* | 0.681 (0.340–1.363) | 0.278 |
| Envelope | ||||||||
| − vs. + | 0.637 (0.189–2.150) | 0.467 | 0.394 (0.103–1.506) | 0.173 | ||||
| Tumor satellite | ||||||||
| − vs. + | 1.829 (0.924–3.622) | 0.083 | 1.797 (0.920–3.510) | 0.086 | ||||
| SPON2 expression | ||||||||
| Low vs. High | 4.519 (2.142–9.533) | <0.001* | 3.093 (1.659–5.766) | <0.001* | 4.330 (2.056–9.121) | <0.001* | 3.175 (1.692–5.960) | <0.001* |
*P<0.05; Serum Alpha-Fetoprotein (AFP); Hepatitis B Virus (HBV); Barcelona Clinic Liver Cancer (BCLC); Hazard Ratio (HR); Confidence Interval (CI).
Discussion
In this study, we evaluated a total of 320 cancer samples and 270 normal liver controls from four independent datasets in the Oncomine database; we determined that SPON2 mRNA expression is upregulated in HCC, compared with normal liver tissue. We verified these results by qRT-PCR analysis of 20 paired HCC and matched non-cancerous tissue samples from our local patient population. Furthermore, we examined SPON2 protein expression levels by TMA-IHC analysis of 107 HCC and matched non-cancerous liver tissues, which demonstrated that SPON2 protein levels were significantly elevated in HCC tissues, compared with normal liver tissue; this result is consistent with previous findings.7,8 Collectively, our results support an association between increased SPON2 expression and HCC carcinogenesis and/or malignancy.
Recently, Zhang et al.8 reported that SPON2 expression in HCC tissues was closely correlated with vascular invasion and tumor-node-metastasis stage. In the current study, we found that SPON2 protein expression correlated with differentiation grade, tumor diameter, and Child-Pugh stage. Tumor size is an important predictor of vascular invasion. SPON2 is thought to promote viability, migration, invasion, and colony formation in colorectal carcinoma cells.10 However, overexpression of SPON2 inhibits the migration and invasion abilities of HCC cell lines.7,8 These conflicting findings suggest that the role of SPON2 in cell migration and invasion is complex.
Tumors with limited invasive abilities are typically associated with better prognosis. Elevated SPON2 expression has been established as a prognostic biomarker for some inflammatory tumors, such as gastric cancer and colorectal carcinoma.9–11,13 However, the utility of SPON2 as a prognostic biomarker in HCC is unclear. In the current study, Kaplan-Meier analysis revealed that HCC patients with higher SPON2 protein expression had significantly worse OS, compared with patients with lower tumor protein expression; this suggested that SPON2 protein could be a useful prognostic biomarker for HCC. Consistent with this, SPON2 protein expression level and tumor thrombus correlated with OS in univariate analysis and were independent predictors of poorer OS in multivariate analysis.
One limitation of our study is that it was a retrospective observational study; thus, the results may not be representative of other HCC populations. Zhang et al.8 reported that increased SPON2 expression in HCC patients was predictive of good survival. They speculated that SPON2 could prevent HCC progression by suppressing metastasis and facilitating M1-like macrophage recruitment to the tumor microenvironment. In contrast, Schmid et al.10 found that SPON2 expression induced liver metastasis in xenografted mice. Therefore, the results of the current study must be verified in prospective studies with larger patient cohorts. Analysis of SuperPaths (http://pathcards.genecards.org) suggests that SPON2 may interact with various cellular pathways, such as the ERK, Rho Family GTPase, and MAPK signaling pathways (Jaccard similarity scores of 0.61, 0.61, and 0.58, respectively). It will be particularly useful to investigate whether and how these pathways might play a role in regulating SPON2 expression and function in HCC.
In conclusion, this study indicated that upregulated expression of SPON2 protein in HCC tissues is significantly correlated with reduced OS and DFS following curative resection. Measurement of the SPON2 protein level in tumor tissue may therefore be a valuable prognostic indicator for HCC.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
National Natural Science Foundation of China (No. 81672409).
China Postdoctoral Science Foundation (2016M590489, 2017T100393).
Postdoctoral Science Foundation of Jiangsu Province (1601101C).
Scientific and Technological Innovation and Demonstration Project of Nantong City (MS32016018, MS12017001-6, MS12017007-5).
Jiangsu Provincial Medical Youth Talent (QNRC2016700).
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–132. [DOI] [PubMed] [Google Scholar]
- 3.Clark HF, Gurney AL, Abaya E, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res 2003; 13: 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YW, Li H, Zhang J, et al. The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat Immunol 2004; 5: 88–97. [DOI] [PubMed] [Google Scholar]
- 5.Jia W, Li H, He YW. The extracellular matrix protein mindin serves as an integrin ligand and is critical for inflammatory cell recruitment. Blood 2005; 106: 3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo JH, Ren B, Keryanov S, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 2006; 44: 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao CH, Yeh SC, Huang YH, et al. Positive regulation of spondin 2 by thyroid hormone is associated with cell migration and invasion. Endocr Relat Cancer 2010; 17: 99–111. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YL, Li Q, Yang XM, et al. SPON2 promotes M1-like macrophage recruitment and inhibits hepatocellular carcinoma metastasis by distinct integrin-Rho GTPase-hippo pathways. Cancer Res 2018; 78: 2305–2317. [DOI] [PubMed] [Google Scholar]
- 9.Chandrasinghe P, Stebbing J, Warusavitarne J. The MACC1-SPON2 axis: a new biomarker and therapeutic target in colorectal cancer. Oncogene 2017; 36: 1474–1475. [DOI] [PubMed] [Google Scholar]
- 10.Schmid F, Wang Q, Huska MR, et al. SPON2, a newly identified target gene of MACC1, drives colorectal cancer metastasis in mice and is prognostic for colorectal cancer patient survival. Oncogene 2016; 35: 5942–5952. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q, Wang XQ, Wang J, et al. Upregulation of spondin-2 predicts poor survival of colorectal carcinoma patients. Oncotarget 2015; 6: 15095–15110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajkumar T, Vijayalakshmi N, Gopal G, et al. Identification and validation of genes involved in gastric tumorigenesis. Cancer Cell Int 2010; 10: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin C, Lin JR, Ma L, et al. Elevated spondin-2 expression correlates with progression and prognosis in gastric cancer. Oncotarget 2017; 8: 10416–10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Razvi MH, Peng D, Dar AA, et al. Transcriptional oncogenomic hot spots in Barrett's adenocarcinomas: serial analysis of gene expression. Genes Chromosomes Cancer 2007; 46: 914–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbieri CE. Evolution of novel biomarkers for detection of prostate cancer. J Urol 2013; 190: 1970–1971. [DOI] [PubMed] [Google Scholar]
- 16.Edwards S, Campbell C, Flohr P, et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br J Cancer 2005; 92: 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JW, Kim ST, Turner AR, et al. Identification of new differentially methylated genes that have potential functional consequences in prostate cancer. PLoS One 2012; 7: e48455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romanuik TL, Ueda T, Le N, et al. Novel biomarkers for prostate cancer including noncoding transcripts. Am J Pathol 2009; 175: 2264–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian X, Li C, Pang B, et al. Spondin-2 (SPON2), a more prostate-cancer-specific diagnostic biomarker. PLoS One 2012; 7: e37225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucarelli G, Rutigliano M, Bettocchi C, et al. Spondin-2, a secreted extracellular matrix protein, is a novel diagnostic biomarker for prostate cancer. J Urol 2013; 190: 2271–2277. [DOI] [PubMed] [Google Scholar]
- 21.Anderson GL, McIntosh M, Wu L, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst 2010; 102: 26–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon I, Liu Y, Krall KL, et al. Evaluation of the novel serum markers B7-H4, Spondin 2, and DcR3 for diagnosis and early detection of ovarian cancer. Gynecol Oncol 2007; 106: 112–118. [DOI] [PubMed] [Google Scholar]
- 23.Badea L, Herlea V, Dima SO, et al. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology 2008; 55: 2016–2027. [PubMed] [Google Scholar]
- 24.Yuan X, Bian T, Liu J, et al. Spondin2 is a new prognostic biomarker for lung adenocarcinoma. Oncotarget 2017; 8: 59324–59332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellsworth RE, Seebach J, Field LA, et al. A gene expression signature that defines breast cancer metastases. Clin Exp Metastasis 2009; 26: 205–213. [DOI] [PubMed] [Google Scholar]
- 26.Wang F, Feng Y, Li P, et al. RASSF10 is an epigenetically inactivated tumor suppressor and independent prognostic factor in hepatocellular carcinoma. Oncotarget 2016; 7: 4279–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei H, Lian S, Zhang S, et al. High expression of ROR2 in cancer cell correlates with unfavorable prognosis in colorectal cancer. Biochem Biophys Res Commun 2014; 453: 703–709. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Wang W, Li P, et al. High TREM2 expression correlates with poor prognosis in gastric cancer. Hum Pathol 2018; 72: 91–99. [DOI] [PubMed] [Google Scholar]