Abstract
Protein kinase RIOK-1 is a non-ribosomal factor essential for rRNA cleavage and ribosome small subunit maturation. It is encoded in all eukaryotic organisms. The RIOK-1 encoding gene of Caenorhabditis elegans (Ce-riok-1) is expressed in the neuronal and reproductive systems in larvae and adults of this free-living nematode, and it supports larval growth and development of the adult gonad. In spite of its recognised roles in model organisms such as C. elegans, little is known about the function of this molecule in parasitic nematodes. In a previous study, we characterised the structure, transcriptional profiles and in vivo transcriptional expression patterns of the Ss-riok-1 of human and canine parasitic nematode Strongyloides stercoralis. Here, we extend previous work to undertake functional studies, using transgenesis to assess the roles of Ss-RIOK-1 in the development of S. stercoralis. The results revealed that recombinant Ss-RIOK-1 with D282A mutation at its catalytic site lost its kinase phosphorylation activity in vitro. Both wild-type and mutant Ss-RIOK-1s were expressed in the cytoplasm of neurons and some hypodermal cells in the wild-type strain (UPD) of S. stercoralis. Larvae expressing the dominant negative mutant Ss-RIOK-1 that lost the catalytic activity had a decreased mobility and a severe defect in development to the infective L3 stage. Our findings demonstrated that Ss-RIOK-1 is essential for the development and survival of free-living larvae of S. stercoralis, and that catalytic activity is essential for its function in the parasitic nematode.
Keywords: Strongyloides stercoralis, Protein kinases, RIOK-1, Development, Transgenesis
1. Introduction
RIOK-1 protein kinase belongs to the RIO atypical protein kinase family, which contains three members: RIOK-1, RIOK-2 and RIOK-3 (Angermayr and Bandlow, 1997; LaRonde-LeBlanc and Wlodawer, 2005a; LaRonde, 2014). From archaea to human, all organisms have at least RIOK-1 and RIOK-2 (LaRonde-LeBlanc and Wlodawer, 2005b). RIOK-1s possess a two-lobed structure, which is a common property of eukaryotic protein kinases (ePKs). However, they lack the substrate-binding loop that is also common to ePKs (LaRonde-LeBlanc and Wlodawer, 2004, 2005a). Nevertheless, the findings in archaea, yeast cells and human cells indicated that RIOK-1 has the ability to hydrolyze ATP (Angermayr et al., 2002; Widmann et al., 2012). Based on its ability to phosphorylate common kinase substrates in vitro, RIOK-1 was recognised as a Ser/Thr protein kinase (Angermayr et al., 2002). However, RIOK-1 can also phosphorylate the RNA polymerase I (POL-I) subunit 43 at Tyr 73, causing this subunit to dissociate from the Pol I complex, thus down-regulating the polymerase activity (Iacovella et al., 2015). This finding highlights the importance of yeast RIOK-1 in regulating Pol I activity and chromosome stability in the nucleus with a cell cycle-dependent mechanism (Iacovella et al., 2015).
RIOK-1 is also a non-ribosomal factor essential for ribosomal small subunit maturation in the cytoplasm (Vanrobays et al., 2001). A lack of RIOK-1 disrupts ribosomal biogenesis, causing 20S pre-rRNA and immature small subunits to accumulate in the cytoplasm, and delay cell cycle progression (Angermayr et al., 2002). RIOK-1 interacts with pre-40S ribosomal small subunit after it is transported from the nucleus (Widmann et al., 2012). During this process, nearly all non-ribosome factors dissociate from the pre-ribosome particles except for NOB-1 (Nin one binding protein) and PNO-1 (Partner of NOB-1) (Widmann et al., 2012; Turowski et al., 2014). RIOK-1 supports NOB-1 to cleave 20S pre-rRNA by hydrolyzing ATP to complete the final step in maturation of the pre-ribosome small subunit (Turowski et al., 2014). The function of RIOK-1 in cell biology requires the capacity to bind ATP and catalyse its hydrolysis (Turowski et al., 2014; Widmann et al., 2012). Although RIOK-1 lacks a loop for substrate binding, it can still be associated into different protein complexes that have the ability to interact with potential substrates and phosphorylate those (Guderian et al., 2011; Read et al., 2013).
Knockout of riok-1 causes a growth defect in yeast cells, and the reversal of this defect requires both the catalytic activity as well as the ATP-binding capability of RIOK-1, indicating that both of these functions of RIOK-1 are essential for the growth of yeast cells (Soudet et al., 2010; Ferreira-Cerca et al., 2014). In addition, the C-terminus is essential for RIOK-1 to bind to the pre-ribosome small subunit (Ferreira-Cerca et al., 2014) and be transported into the nucleus to regulate the activity of POL-I and promote the stability of nuclear rDNA (Iacovella et al., 2015).
The function of RIO protein kinase in multicellular organisms is less well understood than the functions in single cells. In Caenorhabditis elegans, a high throughput RNA interference (RNAi) screen revealed that both RIOK-1 and RIOK-2 are essential for development and reproduction in this nematode (Fraser et al., 2000; Rual et al., 2004; Sonnichsen et al., 2005). Furthermore, RIOK-1 for C. elegans (Ce-RIOK-1) modulates the Ras signalling pathway, and is localised in neurons and tissues of the male and female reproductive systems, including somatic gonad, spermathaeca, uterus and vulva cells (Weinberg et al., 2014). Consistent with the localization in the reproductive tract of C. elegans, knock-down of Ce-riok-1 by RNAi causes a defect in germ cell proliferation (Mendes et al., 2015). RNAi knockdown also causes enlarged germ cell nuclei, abnormal oocyte maturation in the proximal gonad and an endomitotic oocyte (EMO) phenotype (Mendes et al., 2015), proposed to be caused by excess DNA duplication and the ectopic activation of the MAPK pathway in the gonad. In addition, RNAi knockdown of Ce-riok-1 causes strong early larval arrest, indicating that it is involved in embryogenesis and early larval development (Mendes et al., 2015).
In spite of the crucial role of RIOK-1 in C. elegans, very little is understood about its function in parasitic nematodes. Our previous work showed that the Strongyloides stercoralis genome encodes a RIOK-1 kinase, Ss-riok-1, that is transcribed throughout development in S. stercoralis, with the highest transcription being in the infective L3 (iL3). Ss-riok-1 is expressed in neurons of post free-living (PFL) L1s and L2s (Yuan et al., 2014). However, whether Ss-RIOK-1 is essential for larval survival or development in this parasitic nematode as it is in C. elegans, is still unknown.
In the present study, a GFP-fused, kinase-dead mutant form of Ss-RIOK-1 was expressed in S. stercoralis under the regulation of its specific transcriptional regulatory region, and sub-cellular localization of the recombinant protein was determined. The influence of expression of mutant RIOK-1 on development of PFL larvae was then assessed. The results support the conclusions that Ss-RIOK-1 is an essential kinase for the survival of S. stercoralis larvae and that its catalytic activity is necessary for Ss-RIOK-1 to function in this capacity.
2. Materials and methods
2.1. Ethics statement
The S. stercoralis (UPD strain) was maintained in prednisolone-treated Beagles, in accordance with a protocol (permit No. SYXK-0029) approved by the Animal Ethics and Animal Experimentation Committee of Hubei Province, China. The care and maintenance of dogs were in strict accordance with the regulations for the Administration of Affairs Concerning Experimental Animals of PR China.
2.2. Parasite maintenance and culture
The UPD strain of S. stercoralis was maintained in prednisolone-treated dogs and cultured as previously described (Schad et al., 1984; Lok, 2007). Faeces were collected and mixed with charcoal and cultured at 22 °C. Free-living adult S. stercoralis for microinjection were isolated from charcoal coprocultures using Baermann funnels (Bowman and Lynn, 1995), incubated for 2 days at 22 °C and then placed on Nematode Growth Medium (NGM) agar plates seeded with Escherichia coli OP50. After microinjection of parental free-living females, larval progeny were screened for GFP expression, and positive worms were transferred onto new NGM plates with E. coli OP50.
2.3. RIOK-1 constructs for prokaryotic expression and transgenesis
To make catalytically inactive mutant RIOK-1, the coding sequence of Ss-riok-1 (GenBank Accession No. KJ701282.1) was cloned into pMD-19T for sequencing, and a pair of primers, Ss-riok-1 282DAF and Ss-riok-1 282DAR (Supplementary Table S1), that contain the mutation site which alters the Asp282 to Ala was designed based on the cDNA sequence of Ss-riok-1 (GenBank Accession No. KJ701282). Ss-riok-1 cloning plasmid (100 ng) was used as template and mutant Ss-riok-1 was amplified by pfu DNA polymerase (Thermo Scientific, USA) under the following cycle conditions: 98 °C for 3 min; then 18 cycles of 98 °C for 30 s, 60 °C for 45 s, 72 °C for 10 min; final extension at 72 °C for 10 min. Then, 1 μL of Dpn I enzyme was added to the PCR compounds at 37 °C for 30 min. Ten microliter of PCR product were used to transform E. coli DH5α competent cells and the mutant Ss-riok-1 construct was sequenced.
The GFP-coding sequence for prokaryotic expression was amplified from the vector which was made by inserting the GFP-coding sequence into a pGEX6p-1 vector (Kaelin et al., 1992) with primers gfp-F and gfp-R (Supplementary Table S1). The PCR cycling conditions were 98 °C for 3 min; 25 cycles of 98 °C for 30 s, 55 °C for 30 s, 72 °C for 2 min; final extension at 72 °C for 10 min. gfp-fused wild-type Ss-riok-1 or mutant Ss-riok-1 D282A was amplified with primer pairs Ss-rio1-bamHI and Ss-rio1-xhoI (Supplementary Table S1) and then cloned into a pET-32a (LaVallie et al., 1993) expression vector using the ClonExpress™ II kit (Vazyme, USA). Positive clones were detected by PCR amplification from a bacteria-containing recombinant plasmid and sent for sequencing. The GFP-coding sequence was also cloned into the pET-32a vector.
To insert the wild-type and mutant Ss-riok-1 coding sequences into the previous Ss-riok-1 promoter transgene construct pRP1 (Yuan et al., 2014), the terminal codon of gfp was mutated to a restriction site BspE I. Two pairs of site mutation primers were designed as pRP1-bspeIF1 and pRP1-bspeIR1 as well as pRP1-bspeIF2 and pRP1-bspeIR2 (Supplementary Table S1). Both wild-type and mutant coding sequences (CDS) of Ss-riok-1 were amplified first by primer pairs Ss-rio1BspeF and Ss-rio1EcorR (Supplementary Table S1) and then inserted into pRP1 between the BspE I and EcoR I digestion sites using the ClonExpress™ II kit (Vazyme, USA). The constructs with CDS of wild-type Ss-riok-1 and mutant Ss-riok-1 were designed as pRP7 and pRP8, respectively. Each of the transgene constructs was extracted from E. coli cells by a TIAN pure Midi Plasmid Kit (Tiangen Biotech, China) and then was diluted to 23 ng/μL and stored at −20 °C until use.
The rescue constructs were made by mixing pRP8 and pRP7 at concentration ratios (pRP8 to pRP7) of 3:1, 2:1 and 1:1, keeping the total plasmid concentration in the mixture constant at 23 ng/μL.
2.4. In vitro kinase assay
The recombinant prokaryotic expression constructs were then used to transform E. coli Transetta strain (Transgene, China) cells for protein expression. The bacterial cells were diluted by 1:100 into new Luria–Bertani (LB)/Amp+medium, after 3 h of growth at 37 °C. The bacteria were induced with IPTG (1:4000), grown at 20 °C, mixed at 150 rpm/min overnight and then concentrated by centrifugation at 8160 g/min for 2 min. The bacteria were resuspended in a buffer containing 300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, 10 mM Tris base, pH 8.0, individually passed through a 0.45 mm filter, and loaded onto 3 mL of Ni–NTA Resin affinity columns (Transgene, China). Finally, the recombinant proteins were eluted with gradient imidazole solution. All the samples were checked by SDS–PAGE and the specific proteins were concentrated by Ultra-50 KD centrifugal filter devices (Millipore, Germany).
To assess the autophosphorylation activity of the fusion protein, 10 μg of GFP-fused wild-type Ss-RIOK-1, GFP-fused mutant Ss-RIOK-1 D282A or GFP were mixed with 50 mM Tris–Cl pH 7.5, 10 mM MgCl2, 1 mM cold ATP, 50 mM NaCl, 8 μCi ATP[γ32P] individually in 20 μL reaction mixtures. Reaction mixtures were incubated at 37 °C for 1 h and samples were loaded on SDS–PAGE. The radioactive signals were checked by autoradiography.
2.5. Parasite transformation
Strongyloides stercoralis were transformed with pRP7 or pRP8 constructs by gonadal microinjection as previously described (Lok, 2007). Transformed free-living females were transferred to new NGM plates layered with E. coli OP50. Each transformed female was mated with two wild-type males. All the transformed worms were cultured at 22 °C.
As a control, 10 non-transformed free-living females and 20 non-transformed free-living males were transferred to a new NGM plate with E. coli OP50 cultured at 22 °C overnight for egg laying. Then these adults were picked from the plate, and their eggs and hatching progeny were incubated at 22 °C for different time intervals.
2.6. Screening S. stercoralis larvae for transgene expression
Transgenic F1 larvae were identified based on GFP expression using an Olympus SZX16 stereomicroscope with coaxial epifluorescence. Tissues and cells with GFP expression were assessed in detail using an Olympus BX51 compound microscope equipped with Differential Interference Contrast (DIC) optics and epifluorescence (Olympus America Inc., Center Valley, Pennsylvania, USA) as previously described. Briefly, larvae were transferred to 2% agarose pads containing 100 mM levamisole on microscope slides. Images of parasites were captured with a Pixera Penguin 150 CL camera with Viewfinder 3.0.1 (Pixera Corporation, Japan) software.
To assess larval motility, worms were transferred to new NGM plates with E. coli OP50. Plates with larvae were placed on an Olympus SZX16 microscope, and videos of larval locomotion were captured with an Olympus DP73 camera with Olympus cell sens-standard1.8.1 (Olympus, Japan) software.
2.7. Morphometrics
Total body lengths of F1 transgenic and control S. stercoralis larvae were measured at 48, 72 and 96 h after microinjection of parental free-living females using Image J software (http://rsb.info.nih.gov/ij/). Total lengths of pharynxes of F1 transgenic and control S. stercoralis larvae were also measured at 96 h after microinjection of parental free-living females. Body length and the ratio of pharynx length to body length were measured in live transgenic larvae. A minimum of 33 larvae were measured, with the exception of non-transformed larvae (n = 8) at 48 h, pRP8 group (n = 14) at 72 h and (n = 18) at 96 h, because only limited larvae were alive at 72 h and 96 h in this group. The values for body length and the ratio of pharynx length to body length were plotted in Prism (GraphPad Software, Inc., La Jolla CA, USA). One-way ANOVA with Bonferroni’s Multiple Comparison Test was used to analyse data in the three groups at each time point. For developmental stage classification, larvae were assigned to one of three classes of development based on their status at 96 h after microinjection of parental free-living females. The “iL3” larvae were those that had completed the L2–L3 moult and had the characteristic forked tail. “Moulting larvae” were those with morphological characteristics of the iL3, but had retained the loosened L2 cuticle. Larvae in an earlier developmental phase were called “young larvae” (Supplementary Fig. S1). Dead transgenic larvae from each group were also counted into the three stages according to the same standard. The percentage composition of larvae by developmental class for each experimental group was calculated as a test statistic. Transformations were repeated at least three times, with free-living adults collected from different coprocultures. All larvae in the non-transformed group (n = 68), pRP7 group (n = 37) and pRP8 group (n = 46), mutant/wild-type 3:1 group (n = 65), mutant/wild-type 2:1 group (n = 47), mutant/wild-type 1:1 group (n = 47) were collected and assessed. The data on iL3 proportion and survival rate of larvae were subjected to the “normalise” algorithm in Prism (GraphPad Software, Inc.) and then were analysed by one-way ANOVA with Bonferroni’s Multiple Comparison Test. A statistical probability (P) of less than 0.05 was the criterion for significance.
3. Results
3.1. An aspartate in the active site is essential for kinase activity of Ss-RIOK-1
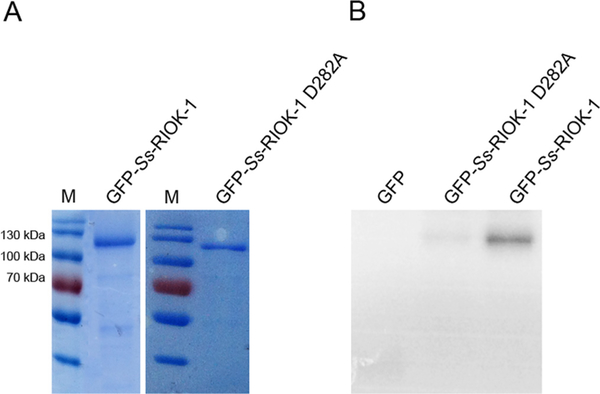
To ascertain the structural requirements for catalytic activity of Ss-RIOK-1, a single site mutation, D282A, was introduced into Ss-RIOK-1. This site was selected because although Ss-RIOK-1 belongs to the atypical kinase family, it retains the aspartate in the active site that is conserved through ePKs. Our previous study (Yuan et al., 2014) revealed that recombinant GST-Ss-RIOK-1 has both autophosphorylation activity and the ability to phosphorylate the common kinase substrate myelin basic protein (MBP). Therefore, to confirm that the D282A mutation ablated catalytic activity of the recombinant Ss-RIOK-1::GFP expressed from our transgene construct, we assessed the kinase activities of GFP-fused wild-type and mutant Ss-RIOK-1 (Fig. 1A) using an autophosphorylation assay. The assay clearly revealed that GFP-fused wild-type Ss-RIOK-1 underwent autophosphorylation, while the GFP-fused D282A mutant did not (Fig. 1B). Purified GFP protein was used as a negative control in this experiment, and it had no autophosphorylation activity.
Fig. 1.
In vitro kinase assay of GFP-fused wild-type Strongyloides stercoralis (Ss)-RIOK-1 and Ss-RIOK-1 D282A mutant. (A) Purified, bacterially expressed GFP-fused wild-type Ss-RIOK-1 and purified GFP-fused D282A mutant Ss-RIOK-1 as compared with molecular weight standards (M). (B) Autoradiograph comparing autophosphorylation activity of GFP-tag only (GFP), GFP-fused mutant (GFP-Ss-RIOK-1 D282A) and wild-type (GFP-Ss-RIOK-1) Ss-RIOK-1s after incubation with ATP[γ32P].
3.2. Ss-RIOK-1 predominantly localises in the cytoplasm of cells
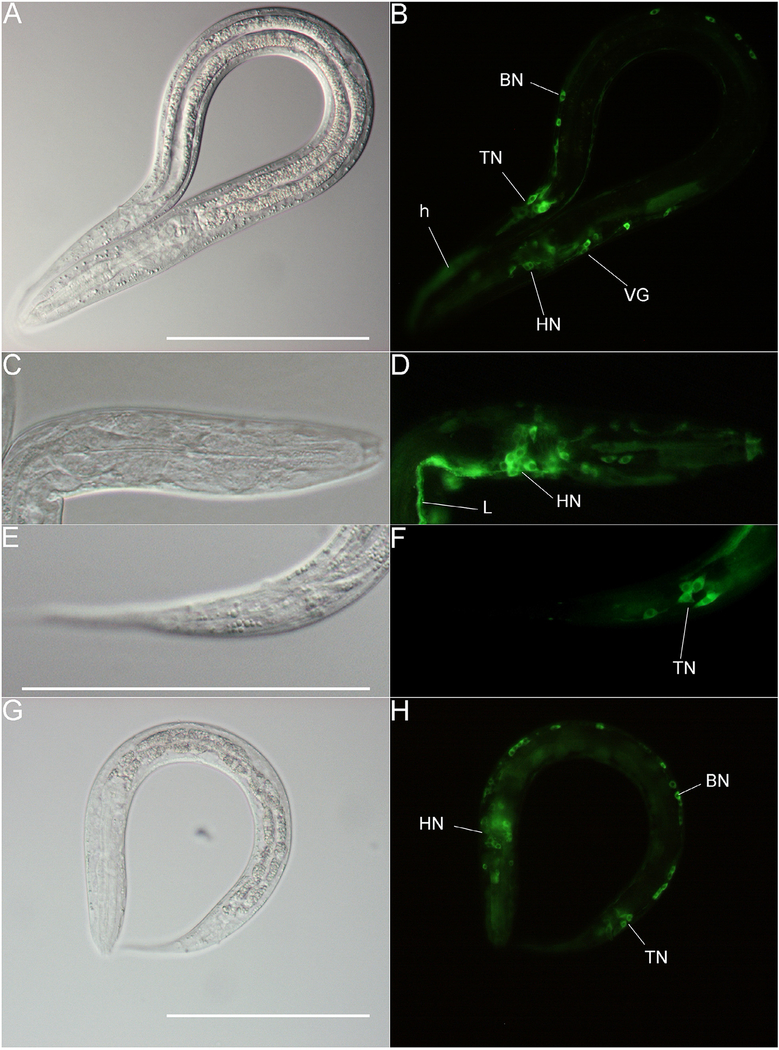
We showed previously that a putative Ss-riok-1 promoter drives gfp expression in neurons of PFL L2s (Yuan et al., 2014). When the coding sequence of Ss-riok-1 was fused downstream of gfp, the expressed fusion protein (Ss-RIOK-1::GFP) predominated in the cytoplasm of neurons and hypodermal cells of PFL L1-L2 (Fig. 2A–F). The D282A mutation in RIOK-1, which disrupted catalytic activity, did not interfere with this pattern of subcellular localization (Fig. 2G, H). These observations served to confirm that wild-type and mutant Ss-RIOK-1s were transcribed and expressed from their respective transgene constructs in S. stercoralis. Neurons expressing Ss-RIOK-1::GFP included the cell bodies of head neurons and their axons, tail neurons and some motor neurons of the body wall (Fig. 2B, D, F). Neurons expressing the D282A mutant Ss-RIOK-1::GFP showed no obvious morphological abnormalities (Fig. 2G, H). The expression patterns of GFP-Ss-RIOK-1 shifted to a predominantly hypodermal one when larvae developed to iL3s (Supplementary Fig. S2).
Fig. 2.
Subcellular localization of GFP-fused wild-type and mutant Strongyloides stercoralis (Ss)-RIOK-1. Differential Interference Contrast (DIC) (A, C, E) and fluorescence images (B, D, F) of wild-type GFP fused Ss-RIOK-1 transgenic post free-living L2s. DIC (G) and fluorescence images (H) of GFP-fused mutant Ss-RIOK-1 transgenic PFL L2s. GFP is expressed in the hypodermis (h), head neurons (HN), body motor neurons (BN), tail neurons (TN), ventral ganglion (VG) and longitudinal nerve tracts (L). Scale bar = 100 μm.
3.3. Expression of mutant Ss-RIOK-1 caused a lethal phenotype in transgenic larvae
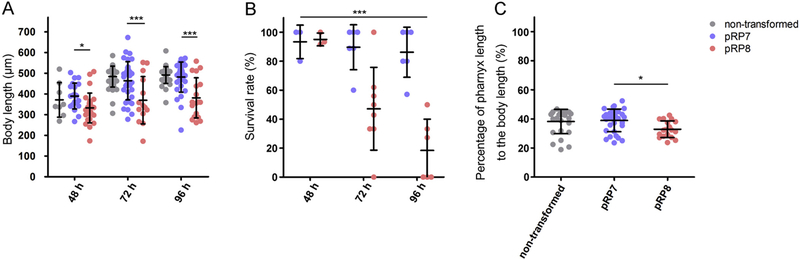
In contrast to its lack of effect on the anatomical expression pattern and on subcellular localization, expression of the D282A mutant Ss-RIOK-2::GFP strongly suppressed larval development in transgenic S. stercoralis, ending with the death of the larvae at the PFL L1–L2 stages. F1 larvae expressing either the mutant or wild-type GFP fusion proteins had developed to PFL L1–L2 at 48 h after transformation. The nerve ring and lateral ganglia encircling the pharynx, the sites of Ss-RIOK-1 expression, were well organised (Fig. 2D, H) in PFL L1s, but the mean total body length of PFL L1s from the mutant (pRP8) Ss-RIOK-1 expression group was significantly shorter than that of the wild-type (pRP7) Ss-RIOK-1 expression group (Fig. 3A). We did not observe obvious defects in morphology or motility in either transgenic or non-transgenic PFL L1s (data not shown). In contrast to the findings at 48 h after transformation, a strong lethal phenotype became evident among larvae expressing the D282A mutant Ss-RIOK-1::GFP at 72 and 96 h after transformation, when development of the larvae expressing the wild-type Ss-RIOK-1::GFP proceeded to the PFL L2 and beyond. Only 47% of larvae were alive from the GFP-tagged mutant RIOK-1 expression group compared with 89% in the GFP tagged wild-type RIOK-1 group at 72 h after injection. The survival rate dropped to 18% at 96 h after injection in the GFP tagged mutant RIOK-1 expression group, but maintained 86% in the GFP tagged wild-type RIOK-1 expression group (Fig. 3B). Larvae expressing the mutant RIOK-1 which did not survive past the 48 h observation time point corresponded to development to the PFL L2 stage, as indicated by body length measurements of dead larvae expressing the GFP tagged mutant RIOK-1 (Supplementary Fig. S3).
Fig. 3.
Expression of GFP-fused mutant Strongyloides stercoralis (Ss)-RIOK-1 in vivo caused arrest/lethal phenotype in post free-living larval stages. Length of transgenic larvae at different time points following transformation (A). Larval lengths measured from live parasites in the mutant Ss-RIOK-1 expression group (pRP8), wild-type Ss-RIOK-1 expression group (pRP7) and non-transformed control group at 48 h, 72 h and 96 h after transformation. Survival rates of transgenic larvae expressing different constructs at different time points following transformation (B). Pharynx length as a percentage of total body length in transgenic and wild-type larvae sampled 96 h after transformation (C). Data are means from three or more than three biological replicates of the experiment. Error bars indicate S.D. Asterisks denote statistically significant differences of body length and pharynx to body length ratio were revealed by one-way ANOVA with Bonferroni’s Multiple Comparison Test. *P < 0.05, **P < 0.01, ***P < 0.001.
The body lengths of the few live larvae expressing the mutant fusion protein were significantly (P < 0.05 at 48 h, and P < 0.001 at 72 h and 96 h) shorter than those expressing the wild-type fusion protein and those in the non-transformed control group at the same age (Fig. 3A). By contrast, there was no significant difference in body length between larvae expressing the wild-type fusion protein and those from the non-transformed control group. In addition, larvae expressing the wild-type fusion protein exhibited a pharynx/body ratio similar to that of larvae from the non-transformed group at 96 h after injection (Fig. 3C). As evidenced by their notched tails, larvae expressing the wild-type fusion protein fully developed to iL3s. As expected, the pharynx/body ratio of mutant fusion protein expression larvae was significantly lower than the two other groups (Fig. 3C).
3.4. The D282A mutant phenotype could be rescued by co-expression of wild-type Ss-RIOK-1::GFP fusion proteins
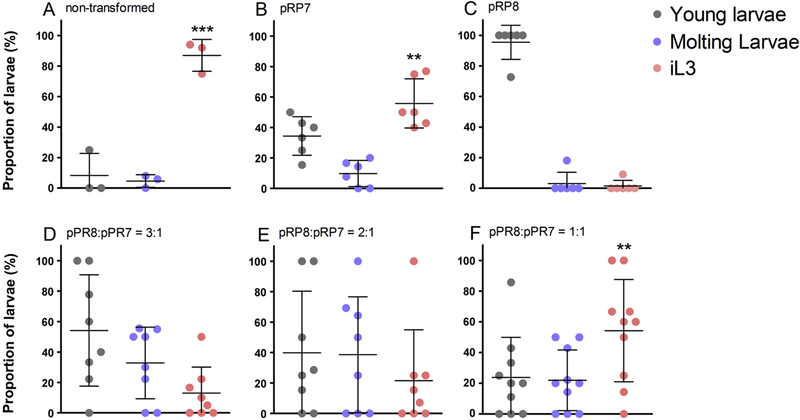
RIOK-1 is known to interact with ribosome subunits and function in the late maturation of the small pre-ribosome subunit in the cytoplasm of yeast and human cells (Vanrobays et al., 2001; Widmann et al., 2012; Turowski et al., 2014). In designing our mutant RIOK-1::GFP expressing construct, we hypothesised that the catalytically inactive RIOK-1 would be incorporated into the ribosome particle but would be incapable of supporting ribosome subunit maturation. By virtue of its expression, likely from a multicopy episomal transgene array, this fusion protein was considered likely to disrupt the function of endogenous Ss-RIOK-1 by interfering with its interaction with the ribosome particle, thus exerting a dominant negative effect. To test this hypothesis, and confirm a gene-specific effect of the D282A mutant fusion protein, we investigated the capacity of the wild-type fusion protein to rescue the growth stunting and lethality phenotypes we observed in worms expressing the mutant fusion protein. To this end, we co-transformed S. stercoralis free-living female adults with plasmids pRP7 and pRP8 at different concentration ratios, and compared resulting phenotypes with untransformed controls and controls transformed with constructs encoding either the mutant or wild-type fusion protein alone. Ninety-six hours after transformation with the construct mixtures, the populations of transgenic larvae were separated into three phenotypic classes (Supplementary Fig. S1). Proportions of larvae that developed to iL3s increased in response to the increasing relative concentration of the pRP7 (wild-type) construct in the co-transforming plasmid mixture administered to parental free-living females (Fig. 4D–F).
Fig. 4.
The arrest/lethal phenotype resulting from GFP-fused D282A mutant Strongyloides stercoralis (Ss)-RIOK-1 expression was rescued by co-expression of GFP-fused wild-type Ss-RIOK-1. Percentages of larvae classified into three developmental classes, young larvae, moulting larvae and infective L3s (iL3), from groups transformed with plasmid mixtures containing varying ratios of the mutant construct (pRP8) to the wild-type construct (pRP7), with the mutant or wild-type construct alone and the non-injection group. All the transgenic larvae were collected and classified into the three groups, including dead worms. Counts were done at 96 h after transformation. Error bars indicate S.D. Asterisks denote the proportion of iL3s in these groups were statistically much higher than that in pRP8 group, which was revealed by one-way ANOVA analysis with Bonferroni’s Multiple Comparison Test. *P < 0.05, **P < 0.01, ***P < 0.001.
Notably, the iL3 proportion from the group co-transformed with the highest relative concentrations of the wild-type fusion protein, which had 54% of iL3s at 96 h (Fig. 4F), was not statistically different from the proportion of iL3s in the group that was transformed with wild-type encoding the construct alone, which had 55% of iL3s at 96 h (Fig.4B). Thus, increasing the concentration of GFP-fused wild-type Ss-RIOK-1 encoding construct rescued completely the dominant negative phenotype conferred by expression of the GFP-fused mutant Ss-RIOK-1. Dead worms could be observed in every developing stage from each group expressing different plasmid mixtures; however, the dead larvae only accounted for a small proportion of each group when compared with larvae expressing the mutant fusion protein alone, in which the majority of larvae died. In the context of this experiment, however, only one larva from the group expressing the mutant fusion protein (transformed with pRP8) alone progressed to the iL3 stage, and two were in the process of moulting.
4. Discussion
Based on deep bioinformatic analyses of the genomic and transcriptomic datasets from some parasitic nematode species (Campbell et al., 2011; Breugelmans et al., 2014), the RIO atypical kinase, RIOK-1, is considered to be a potential target for new drugs to control parasitic nematode diseases which affect one-quarter of the world’s population (Miguel and Kremer, 2004). However, little was known about the degree to which RIOK-1 participates in regulating the development of parasitic nematodes prior to this investigation. Previously we have found that Ss-riok-1 was transcribed in each developmental stage of S. stercoralis (see Yuan et al., 2014). In the present study, we employed reverse genetic methods involving expression of GFP-tagged wild-type and catalytically inactive Ss-RIOK-1 to study aspects of the function of this protein kinase in the development and survival of free-living stages of S. stercoralis. However, the parasitic stages were not investigated here due to the difficulty in collecting enough transgenic larvae to establish the parasitic life cycle in the host.
Under the control of its own transcriptional regulatory region, GFP-fused RIOK-1 was expressed in neurons of the PFL L1-L2 stage and in the hypodermis of iL3s. The neuronal localization that we observed for Ss-RIOK-1 is similar to localization of its homologue in C. elegans (see Weinberg et al., 2014; Mendes et al., 2015). However, the shift in expression patterns during development from L2 to L3 had not been observed in continuously developing C. elegans. This could be due to the greater similarity of S. stercoralis iL3s to dauer larvae of C. elegans than to its continuously developing L3s. Another difference in the localizations of Ss-RIOK-1 and Ce-RIOK-1 is the absence of the parasite kinase from the gonads of PFL S. stercoralis, which could also be explained by the similarity of early PFL larval development by S. stercoralis to dauer entry in C. elegans where development of the gonad is arrested at an early stage (Narbonne and Roy, 2006).
Ss-RIOK-1 was found to localise in the cytoplasm in larval cells, which is consistent with the general functions of homologs in yeast and human cells as essential factors in ribosomal biogenesis (Vanrobays et al., 2003; Widmann et al., 2012; Turowski et al., 2014). However, considering its essential function in ribosome maturation in other systems, Ss-RIOK-1, similar to its C. elegans homolog (Weinberg et al., 2014; Mendes et al., 2015), was expressed in fewer tissues than expected. A possible explanation for this could be that Ss-RIOK-1 is expressed in a broader range of tissues but at levels that are below those that can be detected by visual observation of GFP reporter expression. Such a scenario has been reported in yeast (Angermayr and Bandlow, 1997). In addition, we cannot exclude that Ss-RIOK-1 might localise to the nucleus in a cell cycle-dependent manner as does its homologue in yeast. Such transient nuclear localization would be difficult to observe during live worm imaging (Iacovella et al., 2015).
RIOK-1 is essential for growth of yeast and C. elegans (Fraser et al., 2000; Ashrafi et al., 2003; Rual et al., 2004; Sonnichsen et al., 2005; Ferreira-Cerca et al., 2014; Turowski et al., 2014). Our data indicated that it is also crucial for the development and survival of PFL larval stages of S. stercoralis. We adopted the approach of interrogating Ss-RIOK-1 function by expressing a GFP-fused catalytically inactive mutant form of the kinase as a putatively dominant negative transgene. This approach was based on evidence that the catalytic activity is essential for RIOK-1 to function in ribosome maturation as well as maintaining rDNA stability and promoting its segregation (Widmann et al., 2012; Ferreira-Cerca et al., 2014; Iacovella et al., 2015). This measure was necessary because most animal-parasitic nematodes, including S. stercoralis, either lack or have diminished sensitivity to RNAi (Geldhof et al., 2007; Maule et al., 2011), and methods for direct gene disruption or editing are presently in their infancy for these parasites (Lok et al., 2016). A similar, dominant negative approach using transgenic worms succeeded in interrupting the function of a target gene (Ss-daf-16) to produce informative phenotypes (Castelletto et al., 2009). Expression of GFP-fused kinase ‘dead’ Ss-RIOK-1 caused severe developmental arrest and, ultimately, death at the developmental interval from PFL L2 to iL3, corresponding to the observation time points between 72 and 96 h following transformation. The data indicate a progressive lethal phenotype together with suppression of development during this time interval, which corresponds with a rise in abundance of Ss-riok-1 transcripts during development from PFL L1 to iL3 (Yuan et al., 2014). This observation suggests that the expression of Ss-RIOK-1 is regulated, and is more crucial in parasitic stages, as the transcript abundance in these stages is higher than that in the PFL stages. The developmental arrest/lethal phenotype that we observed in S. stercoralis expressing the mutant construct is consistent with the lethal phenotype caused by RNAi silencing of riok-1 in C. elegans (see Fraser et al., 2000; Simmer et al., 2003; Sonnichsen et al., 2005; Mendes et al., 2015), which suggests that RIOK-1 is required for early larval development in both C. elegans and S. stercoralis. Similar phenotypes, including the arrest of larval growth and cell cycle arrest of germ cell, have been reported for the ribosomal factor mutant strain of C. elegans (see Kudron and Reinke, 2008). RNAi-based silencing of Ce-riok-1 by feeding early L1s also interferes with the proliferation of germ cells and maturation of oocytes, and causes an EMO phenotype (Mendes et al., 2015). Because the expression of Ss-RIOK-1 was not observed in the gonadal primordium of S. stercoralis larvae, we were not able to establish unequivocally whether Ss-RIOK-1 functions in the germ line and in gonad development in S. stercoralis.
The expression of even wild-type Ss-RIOK-1-GFP may be toxic to transformed S. stercoralis, which might explain the observation of more GFP-expressing embryos that could not hatch during the attempt to inject the parental worms with higher concentrations of plasmid pRP7. Consequently, in our rescue experiments, the total DNA concentrations in mixtures of wild-type and mutant constructs were maintained at the same levels as that at which the constructs were injected individually, but with the increasing proportions of GFP-fused wild-type Ss-RIOK-1. Results of the rescue experiment indicate that wild-type Ss-RIOK-1-GFP can completely rescue the arrest/lethal phenotype caused by the mutant Ss-RIOK-1-GFP. Dose dependency of the effect underscores the specificity of the rescue. The loss of catalytic activity might also affect the function of Ss-RIOK-1 in other protein complexes, as its homologue is associated with protein Arginine Methyl transferase 5 (PRMT-5) and the mammalian target of rapamycin complex 2 (mTor-complex-2) (Guderian et al., 2011; Read et al., 2013; Marjon et al., 2016) in human cells. Taken together, these findings suggest a more complex mechanism and significant function for RIOK-1 than previously appreciated in regulating cellular processes generally, and in enabling growth and survival of parasitic nematode larvae specifically.
In conclusion, the present study provides evidence of a fundamental role for Ss-RIOK-1 in supporting the development and survival of larval S. stercoralis. It reveals that Ss-RIOK-1 localises predominantly in the cytoplasm of the parasite cells and that its catalytic activity is necessary for the development and survival of this parasitic nematode. It must be noted that RIOK-1 is present both in the host and parasitic nematodes, and this may present significant challenges in development of selective inhibitors of the parasite kinase. However, specific inhibitors designed to compete with binding of ATP to parasite RIOK-1s, thus blocking its catalytic activity, inhibitors that could block the interaction of RIOK-1 with other protein complexes or the activation of RIOK-1 from its upstream regulator would be expected to suppress the development of parasitic nematodes, ultimately causing their death in the complex milieu of the host. These multiple points of attack should facilitate the design of such RIOK-1 inhibitors and could constitute a new avenue to an anthelmintic treatment.
Supplementary Material
Acknowledgements
Special thanks to Fan Liu and Mudassar Nizaz Mughal for assistance with taking care of experimental dogs and to Dr. Bang Shen for his helpful discussions regarding the rescue experiment. This work was supported by “Fundamental Research Funds for the Central Universities”, China (Program No. 2662015PY180) and “the National Key Basic Research Program (973 program) of China” (Grant No. 2015CB150300) to MH, by a grant from The National Institute of Health (NIH), USA (AI-50688) to JBL and by funds from Huazhong Agricultural University Scientific & Technology Self-innovation Foundation (Program No. 2015RC005), the National Health and Medical Research Council (NHMRC) and the Australian Research Council (ARC) of Australia to RBG.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ijpara.2017.06.005.
References
- Angermayr M, Bandlow W, 1997. The type of basal promoter determines the regulated or constitutive mode of transcription in the common control region of the yeast gene pair GCY1/RIO1. J. Biol. Chem 272, 31630–31635. [DOI] [PubMed] [Google Scholar]
- Angermayr M, Roidl A, Bandlow W, 2002. Yeast Rio1p is the founding member of a novel subfamily of protein serine kinases involved in the control of cell cycle progression. Mol. Microbiol 44, 309–324. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G, 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272. [DOI] [PubMed] [Google Scholar]
- Bowman DD, Lynn RC, 1995. Georgi’s Parasitology for Veterinarians. W B Saunders Co, Saint Louis, Missouri, U.S.A. [Google Scholar]
- Breugelmans B, Jex AR, Korhonen PK, Mangiola S, Young ND, Sternberg PW, Boag PR, Hofmann A, Gasser RB, 2014. Bioinformatic exploration of RIO protein kinases of parasitic and free-living nematodes. Int. J. Parasitol 44, 827–836. [DOI] [PubMed] [Google Scholar]
- Campbell BE, Boag PR, Hofmann A, Cantacessi C, Wang CK, Taylor P, Hu M, Sindhu ZU, Loukas A, Sternberg PW, Gasser RB, 2011. Atypical (RIO) protein kinases from Haemonchus contortus–promise as new targets for nematocidal drugs. Biotechnol. Adv 29, 338–350. [DOI] [PubMed] [Google Scholar]
- Castelletto ML, Massey HC Jr., Lok JB, 2009. Morphogenesis of Strongyloides stercoralis infective larvae requires the DAF-16 ortholog FKTF-1. PLoS Pathog. 5, e1000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Kiburu I, Thomson E, LaRonde N, Hurt E, 2014. Dominant Rio1 kinase/ATPase catalytic mutant induces trapping of late pre-40S biogenesis factors in 80S-like ribosomes. Nucleic Acids Res. 42, 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J, 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Geldhof P, Visser A, Clark D, Saunders G, Britton C, Gilleard J, Berriman M, Knox D, 2007. RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitology 134, 609–619. [DOI] [PubMed] [Google Scholar]
- Guderian G, Peter C, Wiesner J, Sickmann A, Schulze-Osthoff K, Fischer U, Grimmler M, 2011. RioK1, a new interactor of protein arginine methyltransferase 5 (PRMT5), competes with pICln for binding and modulates PRMT5 complex composition and substrate specificity. J. Biol. Chem 286, 1976–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovella MG, Golfieri C, Massari LF, Busnelli S, Pagliuca C, Dal Maschio M, Infantino V, Visintin R, Mechtler K, Ferreira-Cerca S, 2015. Rio1 promotes rDNA stability and downregulates RNA polymerase I to ensure rDNA segregation. Nat. Commun 6, 6643. [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr., Krek W, Sellers WR, DeCaprio JA, Ajchenbaum F, Fuchs CS, Chittenden T, Li Y, Farnham PJ, Blanar MA, et al. , 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70, 351–364. [DOI] [PubMed] [Google Scholar]
- Kudron MM, Reinke V, 2008. C. elegans nucleostemin is required for larval growth and germline stem cell division. PLoS Genet. 4, e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A, 2004. Crystal structure of A. fulgidus Rio2 defines a new family of serine protein kinases. Structure 12, 1585–1594. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A, 2005a. A family portrait of the RIO kinases. J. Biol. Chem 280, 37297–37300. [DOI] [PubMed] [Google Scholar]
- LaRonde-LeBlanc N, Wlodawer A, 2005b. The RIO kinases: an atypical protein kinase family required for ribosome biogenesis and cell cycle progression. Biochim. Biophys. Acta 1754, 14–24. [DOI] [PubMed] [Google Scholar]
- LaRonde NA, 2014. The ancient microbial RIO kinases. J. Biol. Chem 289, 9488–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie ER, DiBlasio EA, Kovacic S, Grant KL, Schendel PF, McCoy JM, 1993. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Biotechnology (N Y) 11, 187–193. [DOI] [PubMed] [Google Scholar]
- Lok J, Shao H, Massey H, Li X, 2016. Transgenesis in Strongyloides and related parasitic nematodes: historical perspectives, current functional genomic applications and progress towards gene disruption and editing. Parasitology, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok JB, 2007. In: Strongyloides stercoralis: a model for translational research on parasitic nematode biology. WormBook, pp. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjon K, Cameron MJ, Quang P, Clasquin MF, Mandley E, Kunii K, McVay M, Choe S, Kernytsky A, Gross S, 2016. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 15, 574–587. [DOI] [PubMed] [Google Scholar]
- Maule AG, McVeigh P, Dalzell JJ, Atkinson L, Mousley A, Marks NJ, 2011. An eye on RNAi in nematode parasites. Trends Parasitol. 27, 505–513. [DOI] [PubMed] [Google Scholar]
- Mendes TK, Novakovic S, Raymant G, Bertram SE, Esmaillie R, Nadarajan S, Breugelmans B, Hofmann A, Gasser RB, Colaiacovo MP, Boag PR, 2015. Investigating the role of RIO protein kinases in Caenorhabditis elegans. PLoS One 10, e0117444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel E, Kremer M, 2004. Worms: identifying impacts on education and health in the presence of treatment externalities. Econometrica 72, 159–217. [Google Scholar]
- Narbonne P, Roy R, 2006. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development 133, 611–619. [DOI] [PubMed] [Google Scholar]
- Read RD, Fenton TR, Gomez GG, Wykosky J, Vandenberg SR, Babic I, Iwanami A, Yang H, Cavenee WK, Mischel PS, Furnari FB, Thomas JB, 2013. A kinome-wide RNAi screen in Drosophila glia reveals that the RIO kinases mediate cell proliferation and survival through TORC2-Akt signaling in glioblastoma. PLoS Genet. 9, e1003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M, 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeomebased RNAi library. Genome Res. 14, 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schad GA, Hellman ME, Muncey DW, 1984. Strongyloides stercoralis: hyperinfection in immunosuppressed dogs. Exp. Parasitol 57, 287–296. [DOI] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, Fraser AG, Ahringer J, Plasterk RH, 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1, E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ, 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469. [DOI] [PubMed] [Google Scholar]
- Soudet J, Gélugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A, 2010. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 29, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D, 2014. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res. 42, 12189–12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M, 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol 23, 2083–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobays E, Gleizes PE, Bousquet-Antonelli C, Noaillac-Depeyre J, Caizergues-Ferrer M, Gelugne JP, 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Schulze E, Fatouros C, Schmidt E, Baumeister R, Brummer T, 2014. Expression pattern and first functional characterization of riok-1 in Caenorhabditis elegans. Gene Expr. Patterns 15, 124–134. [DOI] [PubMed] [Google Scholar]
- Widmann B, Wandrey F, Badertscher L, Wyler E, Pfannstiel J, Zemp I, Kutay U, 2012. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 23, 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Lok JB, Stoltzfus JD, Gasser RB, Fang F, Lei WQ, Fang R, Zhou YQ, Zhao JL, Hu M, 2014. Toward understanding the functional role of Ss-riok-1, a RIO protein kinase-encoding gene of Strongyloides stercoralis. PLoS Negl Trop Dis 8, e3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.