Abstract.
With recent advances in the field of deep learning, the use of convolutional neural networks (CNNs) in medical imaging has become very encouraging. The aim of our paper is to propose a patch-based CNN method for automated mass detection in full-field digital mammograms (FFDM). In addition to evaluating CNNs pretrained with the ImageNet dataset, we investigate the use of transfer learning for a particular domain adaptation. First, the CNN is trained using a large public database of digitized mammograms (CBIS-DDSM dataset), and then the model is transferred and tested onto the smaller database of digital mammograms (INbreast dataset). We evaluate three widely used CNNs (VGG16, ResNet50, InceptionV3) and show that the InceptionV3 obtains the best performance for classifying the mass and nonmass breast region for CBIS-DDSM. We further show the benefit of domain adaptation between the CBIS-DDSM (digitized) and INbreast (digital) datasets using the InceptionV3 CNN. Mass detection evaluation follows a fivefold cross-validation strategy using free-response operating characteristic curves. Results show that the transfer learning from CBIS-DDSM obtains a substantially higher performance with the best true positive rate (TPR) of at 1.67 false positives per image (FPI), compared with transfer learning from ImageNet with TPR of at 2.1 FPI. In addition, the proposed framework improves upon mass detection results described in the literature on the INbreast database, in terms of both TPR and FPI.
Keywords: mass detection, mammograms, convolution neural networks, transfer learning, computer aided detection, breast image analysis
1. Introduction
Breast cancer is the most common form of cancer in the female population. In the USA, it is estimated that of women will be diagnosed with breast cancer at some point during their lifetime.1 Moreover, it has been demonstrated that the breast cancer survival rate is strongly dependent on the stage at which cancer is diagnosed. Although digital breast tomosynthesis is gradually being adopted, x-ray mammography is still the gold standard imaging modality used for breast cancer screening due to its fast acquisition and cost-effectiveness. However, for certain population groups (young women or women with dense breasts), it has been shown to have a reduced sensitivity, which may result in more missed cancers.2
In the past decade, research in breast image analysis has mainly focused on the development of computer-aided detection or diagnosis (CAD) systems to assist radiologists in the diagnosis. Traditionally, mammography CAD systems relied on hand-engineered features, which showed limited accuracy in complex scenarios. More recently, with the advent of deep learning methods, CAD systems learn automatically which image features are more relevant to be used to perform a diagnosis, boosting the performance of these systems. The term “deep learning” can be defined as any one of a set of methods that learn data representations using multiple levels of representation.3 They are obtained by composing simple but nonlinear models that transform the representation from one level (starting with the raw input) into increasing levels of representation. Deep learning strategies have gained a lot of interest in various fields, including object detection,3–7 image recognition,5–11 natural language processing,12,13 speech recognition,14,15 etc.
Although several authors have proposed the use of traditional machine learning and content-based image retrieval techniques to classify masses and microcalcifications,16,17 the exploitation of deep learning frameworks in the field of breast imaging has been limited, as only a small number of datasets are publicly available (e.g., DDSM,18 INbreast19). In this sense, one should mention the early paper of Kozegar et al.,20 who used an iterative breast segmentation approach to subsequently classify the regions using traditional feature selection and machine learning paradigms. Later, Dhungel et al.21 proposed a multiscale deep belief network classifier, followed by a cascade of region-based convolutional neural networks (R-CNN) and cascades of random forest classifiers for mass detection, while Carneiro et al.22 proposed the use of CNN models pretrained using a computer vision database (ImageNet) for classifying benign and malignant lesions in the DDSM and INbreast datasets.
More recently, Lotter et al.23 trained a CNN patch-based classifier to classify lesions in the DDSM dataset and subsequently developed a scanning model to provide full mammogram classification, achieving an area under receiver operating curve () of 0.92 on the DDSM dataset. In the same year, Dhungel at al.24 used a deep learning methodology to develop an approach for mass detection, segmentation, and classification in mammograms and tested the approach on the INbreast dataset. Detection results had a true positive rate (TPR) of at five false positives per image (FPI) on testing data.
In another work, breast abnormalities (masses, microcalcifications) were simultaneously detected using a faster R-CNN model and a CNN-based classifier25 obtaining a TPR of 0.93 at 0.56 FPI for mass mammograms using a subset of the INbreast database. Recently, Ribli et al.26 used fast R-CNN for the classification and detection of malignant and benign lesions with a TPR of 0.90 at 0.3 FPI, using a subset of the INbreast database with lesions. Regarding the use of private mammography datasets, Becker et al.27 developed a multipurpose image analysis software to detect and classify abnormalities, obtaining an of 0.79 on the testing set. In other works, Kooi et al.28 used a larger private database of images to provide a comparison between traditional mammography CAD systems relying on hand-crafted features and the CNN methods. It was shown that the CNN model trained on a patch level with a large database outperformed state-of-the-art CAD systems and equivalued (less experienced) radiologists with an of 0.88.
Generally, the training process for supervised deep CNNs requires a large number of annotated samples to avoid overfitting to the training dataset. This issue is often addressed by researchers using transfer learning (also known as domain adaptation). Here, the aim is to fine-tune a pretrained model (trained on a larger database) on a smaller dataset.29 Transfer learning is considered to be an efficient methodology, in which the knowledge from one image domain can be transferred to another image domain. Azizpour et al.30 suggested that the success of any transfer learning approach highly depends on the extent of similarity between the databases on which a CNN is pretrained and the database to which the image features are transferred. Tajbakhsh et. al.31 debated if the use of pretrained deep CNNs with sufficient fine-tuning could eliminate the need for training a deep CNN from scratch. The authors also analyzed the influence of the choice of the training samples on the performance of CNNs and concluded that there is no set rule to say if a shallow tuning or deep tuning is beneficial and that the optimal method is dependent on the type of application.
In the direction of an automated CAD system, the techniques for mass-like lesion detection and classification follow a two stage pipeline with candidate detector and latter classifying the masses.24,28,32 Recently Chougrad et al.33 focused on the classification of breast masses and demonstrated that an increased performance could be achieved using transfer learning from natural images to mammograms. The authors compared the performance of three CNNs for the classification of breast masses into malignant and benign, showing that better classification could be obtained using transfer learning from the natural images (ImageNet). In this work, we have developed an automated framework for detecting masses in full mammograms. Here, we use the concept of transfer learning to enhance the performance of the automated framework. Note that, in contrast to Chougrad et al.,33 we are dealing with the problem of mass detection instead of classification and have analyzed different CNNs for classifying mass and nonmass regions instead of classifying masses into benign and malignant.
In this work, the first step is to analyze the performance of three popular deep CNN architectures (VGG16, ResNet50, InceptionV3) in terms of mass and nonmass classification on a large public dataset of digitized mammogram (CBIS-DDSM). Second, the best performing CNN is used to classify mass and nonmass regions in another small public dataset (INbreast). Here, a study is performed for mass detection in mammograms, comparing the results when the transfer learning is performed between the images of similar domains (i.e., digitized and digital mammograms) against the results obtained when the transfer learning is performed between the images of different domains (mammograms and natural images). The classification results are evaluated using the testing accuracy, while the detection results are evaluated using the free-response operating characteristic (FROC)34 analysis.
The paper is structured as follows: Sec. 2 provides the details of the datasets used and CNN architectures, followed by the methodology for training and testing the CNN models for classification and detection of masses. Section 3 provides the details of the experiments performed in this work; Sec. 4 presents the results and discussion, and the paper finishes with Sec. 5, where conclusions and future work are stated.
2. Methodology
In this section, we describe the datasets used, the sampling procedure for generating input patches, the CNN architectures, and the strategy used for training the CNN, followed by the strategy used for detection of masses in mammograms.
A fully automated framework for mass detection is developed (see Fig. 1); it is initialized by extracting small regions of the image (referred to as patches) to be used for training the CNN. The model obtained after the CNN training is first used to classify the unseen testing patches as mass and nonmass patches (with different probabilities). The patches are then recombined to reconstruct the whole mammogram and subsequently the classification probabilities (of each patch) are used to obtain the mass probability map (MPM) for the mammogram and obtain the probable mass region defined by a bounding box.
Fig. 1.
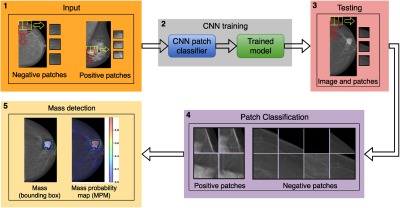
The proposed framework for automated detection of masses in mammograms, where the first block shows the patch extraction strategy using sliding window for negative (nonmass candidates) and positive (mass candidates) mammograms, followed by CNN training in block 2 to obtain a trained model. The third block shows the patch extraction from a test image followed by patch classification using the trained model (shown in block 4). The block 5 shows the MPM and the detection on the original image (with the green bounding box).
2.1. Datasets
2.1.1. CBIS-DDSM
The DDSM18 database contains digitized images from scanned mammography films compressed with lossless JPEG encoding. In this work, we have used a version of the database, i.e., CBIS-DDSM,35 containing a subset of the original DDSM images in the standard DICOM format. The database was downloaded on October 10, 2017, from the CBIS-DDSM website36 containing 3061 mammograms of 1597 cases. In total, there are 1698 masses in 1592 images from 891 cases, which include both cranio-caudal (CC) and medio-lateral oblique (MLO) views for most of the screened breasts. The CBIS-DDSM database contains pixelwise annotations for the regions of interest (RoI), e.g., masses and calcifications, as well as lesion’s pathology, i.e., benign or malignant.
The CBIS-DDSM database is composed of digitized film-screen mammography images, which implies a nonhomogeneous intensity distribution of the background (nonbreast area). Therefore, a segmentation step using Otsu segmentation37 is used to differentiate between the breast area and the background. Following the standard training and testing split of the data as suggested by Lee et al.,35 the images are first divided into training and testing sets with 1231 and 361 images, respectively. Further, the training set is subdivided into the training and validation images with 985 and 246 images, respectively.
2.1.2. INbreast
The INbreast dataset is composed of digital mammograms acquired using a Siemens MammoNovation mammography system (Siemens Healthineers, Erlangen, Germany). The images were acquired from 115 cases with CC and MLO breast views, leading to a total of 410 images available in DICOM format. From these, a total of 116 masses can be found in 107 mammograms from 50 cases. In this work, we have not considered the cases with follow-up studies (different acquisition times) as different cases, thus resulting in a total of 108 cases. Regarding preprocessing of these full-field digital mammograms (FFDM), global thresholding is performed to segment the breast region from the background and all right breasts are mirrored horizontally to keep the same orientation.
The dataset contains pixel-level mass annotations and histological information about the type of cancers. The dataset also contains some mammograms with multiple masses. We have found that in four mammograms, the lesions are very close and the bounding-boxes overlap, so we consider them as a single lesion. Thus, the total number of masses in this paper is considered to be 112 instead of 116. A fivefold cross validation is used to analyze the performance on the whole dataset. The dataset is divided into training (60%), validation (20%), and testing (20%) sets on the case level per fold. The distribution is performed in an stratified manner to ascertain equal ratios of normal and abnormal cases.
2.2. Input Patch Extraction
In this work, a sliding window approach is used to scan the whole breast and extract all the possible patches from the image (see Fig. 1). The total number of patches is controlled by the stride (), which also defines the minimum overlap between two consecutive patches. All the patches are then classified based on the annotations provided in the dataset. For example, a patch is labeled as positive (mass candidate) if the central pixel of the patch lies inside the mass (verified using the corresponding RoI annotation); otherwise, it is assigned a negative (no mass) label.
Since in the CBIS-DDSM database normal images (without any abnormalities) are not available, an equal number of positive and negative patches are extracted from mass images considering that all the positive patches are extracted first and then an equal number of negative patches are randomly selected (excluding the border area patches due to high contrast difference). This provides a balanced dataset for training the CNN.
On the other hand, the INbreast dataset contains mammograms with and without masses, so positive patches are extracted only from the mammograms with masses. To maintain a balance between positive and negative samples for the CNN training, the negative patches are extracted from the mammograms without masses using the following formulation:
| (1) |
where is the number of positive patches and is the total number of nonmass mammograms in the training or validation set and is the required number of patches to be randomly selected from each of the nonmass mammograms. Table 1 provides the details of the patches extracted from the two datasets.
Table 1.
Data description: pos refers to positives (masses) and neg to negatives (nonmasses).
| Dataset | Splits | Cases | Images | Patches | Stride | |||
|---|---|---|---|---|---|---|---|---|
| Pos | Neg | Pos (mass) | Neg | Pos | Neg | |||
| CBIS-DDSM | Train | 553 | — | 985 (1055) | — | 25979 | 25979 | 56 |
| Validation | 138 | — | 246 (263) | — | 6210 | 6210 | 56 | |
| Test | 201 | — | 361 (378) | — | 8694 | 8694 | 56 | |
| INbreast | Train | 30 | 35 | 66 (68) | 191 | 2020 | 2101 | 56 |
| Validation | 10 | 11 | 20 (21) | 61 | 539 | 549 | 56 | |
| Test | 10 | 12 | 21 (23) | 51 | 882 | 918 | 56 | |
2.3. CNN Architectures
For patch classification, we evaluated three popular, widely used CNN architectures (VGG16, ResNet50, and InceptionV3) that have already proven to be excellent for image classifications using the ImageNet dataset, which we use for transfer learning from natural images to digitized and digital mammograms.
2.3.1. VGG16
The VGG38 network is the contribution from the Visual Geometry Group, University of Oxford, and consists of very small convolutional filters () with a depth of 16 to 19 weight layers, resulting in a simple architecture. In this work, the VGG16 is used; it consists of 13 convolutional layers and 2 fully connected or dense layers, followed by an output dense layer with a softmax activation function. There are also five max pool layers in the network.
2.3.2. ResNet50
The ResNet5039 architecture consists of convolutional layers, pooling layers, and multiple residual layers, each containing several bottleneck blocks: a stack of three convolutional layers followed by batch normalization (BN) layers. The ResNet50 structure has four residual layers, each comprising 3, 4, 6, and 3 bottleneck blocks from bottom to top, followed by a dense layer and the output layer with softmax activation function. In total, there are 179 layers in the ResNet50 architecture.
2.3.3. InceptionV3
The InceptionV340 model has been developed by Google and is also known as GoogleNet. The computational cost and memory requirement of the Inception network is much lower than VGG and ResNet50, which makes it a prominent network to be used in Big Data scenarios. The Inception network consists of a collection of Inception modules, each of which uses sets of kernels to represent larger kernels in a computationally efficient manner. The network implemented here has five convolutional layers, each followed by a BN layer, 2 pooling layers, and 11 inception modules.
2.4. CNN Training
The CNNs described above are initially trained on the ImageNet dataset with input dimensions , where the three dimensions represent red, green, and blue color channels. Since extracted patches from mammograms contain only one channel (gray level), each patch () has been replicated onto the three-color channels to make the input patches compatible with the input of the pretrained CNNs. To train a CNN, preprocessing or intensity normalization is an important step. In this work, as part of preprocessing, a zero mean normalization is applied based on global contrast normalization (GCN), as described by Chougrad et al.33
For CNN training, the dataset is split into training and validation sets. The training set is used to train the network and update its weights, while the validation set is used to measure how well the trained CNN is performing after each epoch. An epoch here describes the number of times the algorithm processes the entire dataset. Further, data augmentation is used to generate more samples from already existing training data. In this work, the negative and positive patches are augmented on-the-fly using horizontal flipping, rotation of up to 30 deg, and rescaling by a factor chosen between 0.75 and 1.25, as commonly used in the literature.24,25,31,33
We first analyze the performance of the different CNNs for classifying mass and nonmass region in the CBIS-DDSM dataset. The optimizer used is Adam41 and the batch size is 128 (for a GPU of 12 GB). Early stopping is used on validation loss and is set to 10 epochs. For the random weight initialization, the CNNs are trained for 100 epochs (maximum) using a learning rate of . Further, the extent of transfer learning is analyzed by transferring the domain from natural images to DDSMs. This is carried out using the pretrained ImageNet weights to initialize the CNNs and fine-tune the CNN for 100 epochs (maximum) using a learning rate of . A higher learning rate is used while training the models initialized using randoms weights because training the CNN from scratch would require more time to learn the features pertaining to the images being analyzed. By contrast, when the CNN is initialized using pretrained weights (where the model has already been trained on millions of images), the features learned during initial training are sensitive to the extent of training, so a smaller learning rate is used to preserve pretrained features when fine-tuned.
Computations were performed on a Linux workstation with 12 CPU cores and a NVIDIA TitanX Pascal GPU with 12GB memory using Keras-2 library with Tensorflow as the backend.
2.5. Mass Detection on INbreast
The best performing CNN model is subsequently fine-tuned to transfer the feature domain from DDSM to FFDMs in the INbreast dataset. After fine-tuning the CNN weights using the INbreast training and validation dataset (using a learning rate of ), mass detection is performed in a fully automated manner without any human intervention. This is achieved using the following steps (see blocks 3 to 5 in Fig. 1):
-
Step 1.
First, all the possible patches are extracted from each image using the sliding window approach.
-
Step 2.
The patches are analyzed using the trained CNN to obtain the mass probability of the given patch. Patches are then used to reconstruct the image and generate the MPM using the linear interpolation of the predicted probabilities.
-
Step 3.
The MPM is then thresholded at different probability levels. This step results in the creation of different regions (each region represents a probable mass) in the mammogram such that each pixel in those regions has the probability greater than the threshold value.
-
Step 4.
A bounding box is created to enclose each probable region using connected component analysis. A mass is considered detected if the intersection over union (IoU) between the bounding box and the annotated ground truth is greater than 0.2, as suggested in earlier works.20,24,42,43
2.6. Evaluation Metric
The evaluation metrics used in this work are (a) the testing accuracy of the model, (b) the area under the receiver operating curve , and (c) FROC curve. The FROC curve is used to evaluate the performance of the detection tool on the INbreast dataset and is plotted between the fraction of correctly identified lesions as TPR and the number of FPI for all decision thresholds. The TPR is evaluated as , where and refer to the mean and standard deviation, respectively.
3. Experimental Results
This section presents the different training and transfer learning experiments performed to evaluate the CNN models. Note that, in all cases, the original resolution of the processed DICOM mammograms is used. A patch-level dataset is generated containing patches of size extracted from the original mammograms and is used as the input for the CNNs. We first transfer the domain of convolutional features from natural images to DDSMs. This is achieved by training the CNNs on the CBIS-DDSM dataset. Later, the trained CBIS-DDSM network is fine-tuned on the INbreast dataset containing fully digital mammograms.
In all experiments, the input patches for training the CNN are generated using a stride of . The stride value is selected to obtain a trade-off between the computational requirements and the number of training samples. In the following experiments, a total of patches for CBIS-DDSM and patches for the INbreast dataset are used (see Table 1).
Experiment #1: The training for each of the three CNNs previously described, i.e., VGG16, ResNet50, and InceptionV3, is performed on the CBIS-DDSM dataset using the pretrained weights obtained from the ImageNet database. This initialization is compared against the randomly initialized CNNs for classifying masses. To demonstrate the potential of transfer learning for mass classification, the CNN training was repeated multiple times (owing to the randomness of the training procedure).
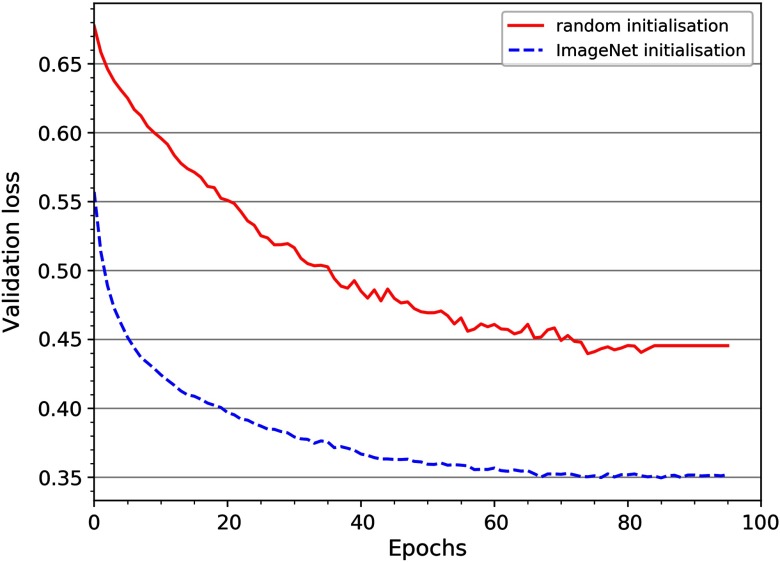
Table 2 compares the results between the random and ImageNet weight initialization. Note that, in all cases, the initialization with ImageNet weights obtained a better accuracy compared with random initialization, and InceptionV3 CNN obtained the highest testing accuracy , and of . Moreover, as shown in Fig. 2, the randomly initialized CNN required a larger number of epochs to converge than the pretrained InceptionV3, demonstrating the benefits of pretraining on ImageNet.
Table 2.
Classification performance (testing accuracy) for mass and nonmass regions in CBIS-DDSM dataset for VGG16, ResNet50, and InceptionV3, where and refer to the mean and standard deviation, respectively, for five independent training results.
| Model | Pretrained | Time per epoch (s) | Testing accuracy () | Az |
|---|---|---|---|---|
| VGG16 | No | 518 | ||
| Yes | 465 | |||
| Resnet50 | No | 483 | ||
| Yes | 438 | |||
| InceptionV3 | No | 338 | ||
| Yes | 310 |
Fig. 2.
Validation loss for the random and ImageNet initialization of InceptionV3.
The obtained results show that the difference in performance (testing accuracy) of the pretrained InceptionV3 with pretrained ResNet50 and VGG16, respectively, was statistically significant (). Also, for each CNN, the difference in performance between the random and ImageNet initialization was found to be statistically significant (). For the rest of the paper, all experiments are performed using the pretrained InceptionV3 CNN model, which provides the best results on the CBIS-DDSM dataset.
Experiment #2: Since both the INbreast and CBIS-DDSM are mammography datasets, with the only difference being the mode of acquisition (scanned films and fully digital mammograms), the feature space of the CNN for one is very likely to be relevant to the other dataset. So, in this experiment, we fine-tune (using learning rate) the best model obtained from Exp #1 on the INbreast dataset, i.e., the model pretrained on ImageNet dataset and fine-tuned on CBIS-DDSM. Here, the fivefold cross-validation strategy is used to analyze the performance of the network on the whole INbreast dataset.
Table 3 shows the impact of transfer learning on InceptionV3 CNN. The results indicate that, using the transfer learning between the images of similar domains (), the testing accuracy is improved to compared with that obtained with the database of natural images ().
Table 3.
Testing accuracy for classifying mass and nonmass regions in INbreast dataset, where and refer to the mean and standard deviation, respectively, for fivefold cross validation.
| Model | Pretrained weight | Fine-tuning on INbreast | Testing accuracy () | Training cascade |
|---|---|---|---|---|
| InceptionV3 | ImageNet | Yes | ||
| InceptionV3 | CBIS-DDSM | Yes |
Experiment #3: In the third experiment, we use the best model obtained from Exp , i.e., , to detect the masses in full mammograms in an automated manner without any human intervention. Here, the full mammogram is divided into small patches using the sliding window approach with a stride of . The trained model is then used to classify these patches into mass and nonmass regions and generate the MPM images (see Fig. 1). The mass detection is then performed following the methodology described in steps 3 and 4 in Sec. 2.5.
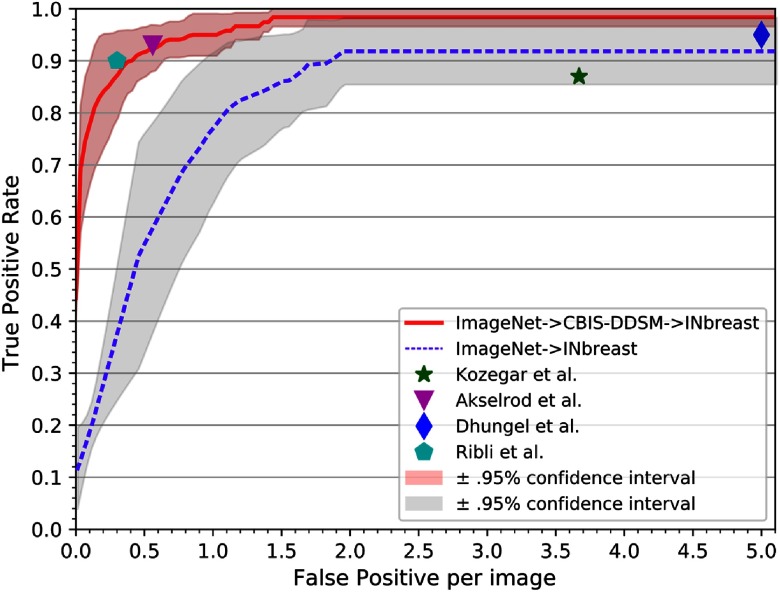
Mass detection is performed on the INbreast dataset using a fivefold cross validation strategy to analyze the entire dataset. The detection performance on the full INbreast dataset is analyzed using FROC curves, as shown in Fig. 3, where the upper and lower bounds are presented in 95% confidence interval. It is observed that for the same evaluation measure of IoU , the performance of CNN is substantially higher when the transfer learning is performed between the images of similar domains (i.e., ) with at 1.67 FPI (Fig. 3), compared withthat obtained when using database of natural images () with at 2.1 FPI (Fig. 3).
Fig. 3.
FROC curve for mass detection on INbreast using transfer learning: testing performance of InceptionV3 pretrained on CBIS-DDSM and fine-tuned on INbreast dataset is plotted using fivefold cross-validation strategy. The operating points from the literature are shown for direct comparison with the proposed framework.
Figure 4 illustrates examples of mass detection on a few testing images (unseen during training) performed using the best model obtained using fine-tuning.
Fig. 4.
Mass detection examples in INbreast using the strategy. (a)–(h) Correct detections are illustrated, (i)–(j) FP cases, and (k)–(l) missed mass cases. Blue contours represent the ground truth (masses), green bounding boxes correspond to the detection of the mass (TP), and red squares show the FP.
Figures 4(a)–4(h) show examples of correctly detected masses in CC and MLO views with variable lesion sizes and contrasts. In addition, Figs. 4(i) and 4(j) show examples of false positive (FP) detections (red squares), where dense tissue areas mimic the appearance of lesion-like structures and a false positive in the pectoral region. Note that the proposed method is unable to detect only 2 masses (very small size) out of the total of 112 lesions within the INbreast dataset. The two undetected masses are shown in Figs. 4(k) and 4(l).
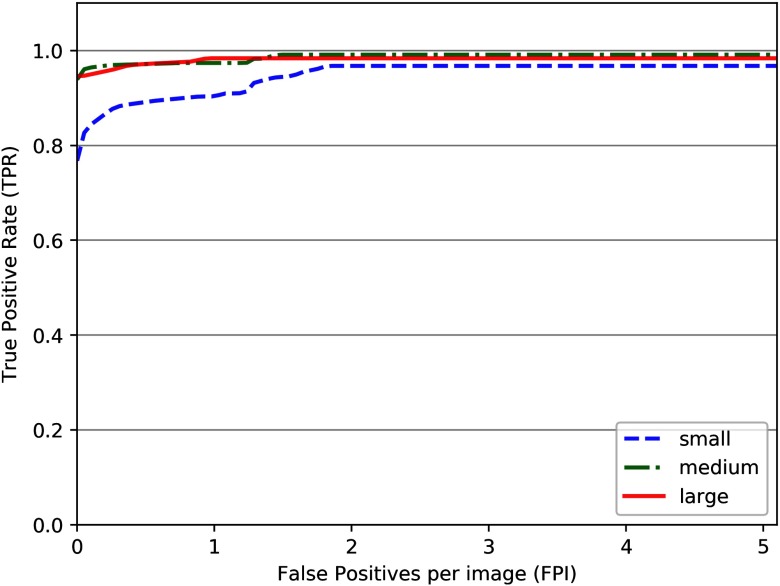
To analyze the performance across different mass sizes, we have divided the lesions into three categories (following radiological criteria), i.e., small lesions (area ), medium size lesions (), and large lesions (area ), and analyzed the performance of the proposed detection framework. This is shown in Fig. 5. The results show that the small lesions have a TPR of 0.89 at 0.5 FPI, while the medium and large lesions have the same TPR of 0.97 at 0.5 FPI. Consequently, the detection performance is inferior for small lesions below .
Fig. 5.
FROC curve showing the performance of the proposed framework on INbreast dataset with different lesion sizes.
4. Discussion
In this paper, we developed an end-to-end mass detection framework using a CNN-based patch classification approach. To generalize the applicability of the proposed framework, we analyzed three different CNN architectures and employed two public datasets containing digitized and digital mammograms.
The interesting aspect of the transfer learning is to reuse the CNN model pretrained for a completely different problem and obtain better results using less complex algorithms. In this regard, first, we examined the benefit of transfer learning between two entirely different image domains, i.e., natural images and mammograms. In this context, we compared the performance of CNNs with randomly initialized weights versus pretrained (ImageNet) weight initialization for the purpose of mass classification in mammograms. As shown in Table 2, despite the differences in the two image domains, the pretrained CNNs performed substantially better than the randomly initialized CNNs. These results gave confidence on the applicability of transfer learning in the context of mammograms. This also supported the fact that the pretrained CNN is able to efficiently use the information of universal features and patterns learned from the ImageNet.
In CNN training, the use of a smaller stride did not increase the variability in the data, so we empirically found a good stride value to perform training (). During the testing, mass probabilities were calculated on each patch and then used to obtain the MPM for the whole mammogram. To analyze the performance of network with respect to the stride used, we tried varying patch strides while testing. This step demanded a trade-off between the accuracy and the computational cost. Very large strides resulted in a poorer localized predictions, whereas very small strides required very high computational cost. For the testing process, we extracted the patches using strides of . We also tried a higher detection threshold (), which resulted in at 1.7 FPI.
The proposed framework produces the best TPR of at 1.67 FPI and a TPR of at 0.5 FPI. The detection performance of the proposed framework is superior in terms of TPR when compared with other state-of-the-art methods using the INbreast dataset (Table 4 and in Fig. 3) on various other operating points.
Table 4.
Comparison between this work and results published in the literature using INbreast dataset, where and refer to the mean and standard deviation, respectively, for fivefold cross validation.
For the purpose of preprocessing, two different approaches were investigated: (1) we scaled the image intensities between 0-255 before extracting the patches and (2) we applied GCN normalization to obtain the zero mean over the input patches. Both the approaches showed different impact on the fine-tuning process, with the GCN approach showing higher performance compared with the scaling approach. Thus, the results in Exp#2 and 3 were performed using GCN preprocessing. Further, we investigated different stride values, which resulted in a smaller or larger number of patches than those presented in Sec. 3. Increasing the stride also increases the similarity in the input data (owing to higher overlap), and vice-versa. It was observed that CNNs performed better when trained with patches with more variability in spite of a small amount of input data compared with the number of CNN parameters to be trained. This behavior could be explained by the use of data augmentation at every epoch during training, which increases the size of the data by increasing variations in the input data.
There are some important things to note about training the CNN: (1) we tried to fine-tune the CNNs by training only the last few layers (also referred to as shallow tuning), as discussed in the literature,31,33 with no significant improvement in the classification results on the CBIS-DDSM and INbreast dataset.44 So, we finally fine-tuned the CNNs by training all the layers at a small learning rate. (2) It was also observed that the random weight initialization took a larger number of epochs to converge than initializing using ImageNet weights.
5. Conclusions
In this work, a transfer learning approach is used for automated mass detection in mammograms. For this purpose, widely used CNN models are analyzed for the detection of breast masses using two public mammogram databases (CBIS-DDSM and INbreast). The methodology presented uses regions of an image (patches) to train the CNNs. The results of training the CNN on CBIS-DDSM demonstrated that the feature domain of the CNN can be well adapted from natural images to classify masses in mammograms. Thereafter, it has been shown that the performance of CNN (in terms of mass detection) can be substantially enhanced using the transfer learning from the images of similar domain (i.e., mammograms), compared with the images of different domains (natural images). The automated framework developed in the work (using InceptionV3) has shown to obtain the best results based on TPR and FPI, outperforming current state-of-the-art approaches using the same INbreast dataset.
In this work, the patch classification is based on the classification of the central pixel. In future work, analysis of whether training the CNN using the volume (i.e., no. of pixels) of tumour within each patch could increase the accuracy of the prediction will be conducted. Further, the developed methodology will be extended for the segmentation of masses in mammograms. Also, the impact of domain adaptation using different FFDMs datasets (i.e., from different vendors) will be investigated. Finally, future work will also focus on the use of transfer learning for image domain adaptation from 2-D mammography to 3-D breast tomosynthesis.
Acknowledgments
This work is partially supported by SMARTER project funded by Ministry of Economy and Competitiveness of Spain, under project reference DPI2015-68442-R.A. is funded by the support of the Secretariat of Universities and Research, Ministry of Economy and Knowledge, Government of Catalonia Ref. ECO/1794/2015 FIDGR-2016. The authors gratefully acknowledge the support of the NVIDIA Corporation with the donation of the Titan X Pascal GPU used for this research.
Biographies
Richa Agarwal is a PhD student in the ViCOROB group at University of Girona, Spain. She received her BTech degree (2011) in India. In 2016, she graduated from Bourgoune University, France, with a masters in computer vision. In the last 3 years, she has been working in the field of medical imaging. Her research interests include image segmentation and registration, and the use of deep learning methods for breast cancer screening.
Oliver Diaz is a postoctoral researcher at the ViCOROB, University of Girona, Spain. He received his PhD (2013) from the Centre for Vision, Speech, and Signal Processing (University of Surrey, United Kingdom). Currently, he is working on the Spanish project SMARTER, where he combines the design of image processing algorithms with the analysis of the physics involved during image acquisition. He also works in close collaboration with the University Hospital Parc Taul (Spain).
Xavier Lladó is an associate professor and the head of the Department of Computer Architecture and Technology at University of Girona. He obtained his BS degree (1999) and PhD (2004) from University of Girona. His research interests are in the field of image processing computer vision and especially in medical image analysis. He has published more than 200 papers in journals and for conferences. He has been a member of the program committees of several conferences and a senior member of the IEEE.
Moi Hoon Yap is a reader (associate professor) in computer vision at Manchester Metropolitan University and a Royal Society Industry Fellow with Image Metrics Ltd. She received her PhD in computer science from Loughborough University in 2009. After her PhD, she worked as postdoctoral research assistant in the Centre for Visual Computing at the University of Bradford. She serves as an associate editor for the Journal of Open Research Software and as a reviewer for IEEE Transactions/Journals.
Robert Martí is currently an associate professor at the ViCOROB research group. He received both BSc and MSc degrees in computer science (1997 to 1999) from the University of Girona. In 2003, he obtained his PhD from the School of Information Systems (now School of Computing Sciences) at the University of East Anglia. His research interests are in the fields of image analysis, pattern recognition, and computer vision, especially focusing on image registration, feature extraction, and classification.
Disclosures
No conflicts of interests, financial or otherwise, are declared by the authors.
References
- 1.Stewart B., Wild C., Eds., “World cancer report,” International Agency for Research on Cancer, World Health Organization; (2014). [Google Scholar]
- 2.Ries L. A., et al. , “SEER cancer statistics review, 1975-2003,” 1975-2003, National Cancer Institute, Bethesda, Maryland (2006).
- 3.LeCun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521(7553), 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 4.Birdwell R. L., et al. , “Mammographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection,” Radiology 219(1), 192–202 (2001). 10.1148/radiology.219.1.r01ap16192 [DOI] [PubMed] [Google Scholar]
- 5.Girshick R., et al. , “Rich feature hierarchies for accurate object detection and semantic segmentation,” in Proc. Conf. Comput. Vision and Pattern Recognit., IEEE, pp. 580–587 (2014). 10.1109/CVPR.2014.81 [DOI] [Google Scholar]
- 6.Szegedy C., Toshev A., Erhan D., “Deep neural networks for object detection,” in Adv. Neural Inf. Process. Syst., pp. 2553–2561 (2013). [Google Scholar]
- 7.Ren S., et al. , “Faster R-CNN: towards real-time object detection with region proposal networks,” in Adv. Neural Inf. Process. Syst., pp. 91–99 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Krizhevsky A., Sutskever I., Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Adv. Neural Inf. Process. Syst., pp. 1097–1105 (2012). [Google Scholar]
- 9.Szegedy C., et al. , “Going deeper with convolutions,” in Proc. Conf. Comput. Vision and Pattern Recognit., IEEE, pp. 1–9 (2015). 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- 10.Farabet C., et al. , “Learning hierarchical features for scene labeling,” IEEE Trans. Pattern Anal. Mach. Intell. 35(8), 1915–1929 (2013). 10.1109/TPAMI.2012.231 [DOI] [PubMed] [Google Scholar]
- 11.Tompson J. J., et al. , “Joint training of a convolutional network and a graphical model for human pose estimation,” in Adv. Neural Inf. Process. Syst., pp. 1799–1807 (2014). [Google Scholar]
- 12.Collobert R., et al. , “Natural language processing (almost) from scratch,” J. Mach. Learn. Res. 12(Aug), 2493–2537 (2011). [Google Scholar]
- 13.Bordes A., Chopra S., Weston J., “Question answering with subgraph embeddings,” in Proc. Conf. Empirical Methods in Nat. Lang. Process., pp. 615–620 (2014). 10.3115/v1/D14-1067 [DOI] [Google Scholar]
- 14.Mikolov T., et al. , “Strategies for training large scale neural network language models,” in IEEE Workshop Automatic Speech Recognit. and Understanding (ASRU), IEEE, pp. 196–201 (2011). 10.1109/ASRU.2011.6163930 [DOI] [Google Scholar]
- 15.Hinton G., et al. , “Deep neural networks for acoustic modeling in speech recognition: The shared views of four research groups,” IEEE Signal Process. Mag. 29(6), 82–97 (2012). 10.1109/MSP.2012.2205597 [DOI] [Google Scholar]
- 16.Giger M. L., Karssemeijer N., Schnabel J. A., “Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer,” Annu. Rev. Biomed. Eng. 15, 327–357 (2013). 10.1146/annurev-bioeng-071812-152416 [DOI] [PubMed] [Google Scholar]
- 17.Oliver A., et al. , “A review of automatic mass detection and segmentation in mammographic images,” Med. Image Anal. 14(2), 87–110 (2010). 10.1016/j.media.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 18.Heath M., et al. , “The digital database for screening mammography,” in Proc. of the 5th Int. Workshop on Digital Mammography 431–434 (2000). [Google Scholar]
- 19.Moreira I. C., et al. , “INbreast: toward a full-field digital mammographic database,” Acad. Radiol. 19(2), 236–248 (2012). 10.1016/j.acra.2011.09.014 [DOI] [PubMed] [Google Scholar]
- 20.Kozegar E., et al. , “Assessment of a novel mass detection algorithm in mammograms,” J. Cancer Res. Ther. 9(4), 592–600 (2013). 10.4103/0973-1482.126453 [DOI] [PubMed] [Google Scholar]
- 21.Dhungel N., Carneiro G., Bradley A. P., “Automated mass detection in mammograms using cascaded deep learning and random forests,” in Int. Conf. Digital Image Comput. Tech. and Appl. (DICTA), IEEE, pp. 1–8 (2015). 10.1109/DICTA.2015.7371234 [DOI] [Google Scholar]
- 22.Carneiro G., Nascimento J., Bradley A. P., “Unregistered multiview mammogram analysis with pre-trained deep learning models,” Lect. Notes Comput. Sci. 9351, 652–660 (2015). 10.1007/978-3-319-24574-4 [DOI] [Google Scholar]
- 23.Lotter W., Sorensen G., Cox D., “A multi-scale CNN and curriculum learning strategy for mammogram classification,” in Third Int. Workshop Deep Learn. Med. Image Anal. and Multimodal Learn. Clin. Decis. Support, Springer, pp. 169–177 (2017). 10.1007/978-3-319-67558-9_20 [DOI] [Google Scholar]
- 24.Dhungel N., Carneiro G., Bradley A. P., “A deep learning approach for the analysis of masses in mammograms with minimal user intervention,” Med. Image Anal. 37, 114–128 (2017). 10.1016/j.media.2017.01.009 [DOI] [PubMed] [Google Scholar]
- 25.Akselrod-Ballin A., et al. , “Deep learning for automatic detection of abnormal findings in breast mammography,” Lect. Notes Comput. Sci. 10553, 321–329 (2017). 10.1007/978-3-319-67558-9 [DOI] [Google Scholar]
- 26.Ribli D., et al. , “Detecting and classifying lesions in mammograms with deep learning,” Sci. Rep. 8(1), 4165 (2018). 10.1038/s41598-018-22437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A., et al. , “Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer,” Invest. Radiol. 52, 434–440 (2017). 10.1097/RLI.0000000000000358 [DOI] [PubMed] [Google Scholar]
- 28.Kooi T., et al. , “Large scale deep learning for computer aided detection of mammographic lesions,” Med. Image Anal. 35, 303–312 (2017). 10.1016/j.media.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 29.Yosinski J., et al. , “How transferable are features in deep neural networks?” in Adv. Neural Inf. Process. Syst., pp. 3320–3328 (2014). [Google Scholar]
- 30.Azizpour H., et al. , “From generic to specific deep representations for visual recognition,” in IEEE Conf. Comput. Vision and Pattern Recognit. Workshops (CVPRW), Boston, Massachusetts, IEEE; (2015). 10.1109/CVPRW.2015.7301270 [DOI] [Google Scholar]
- 31.Tajbakhsh N., et al. , “Convolutional neural networks for medical image analysis: Full training or fine tuning?” IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016). 10.1109/TMI.2016.2535302 [DOI] [PubMed] [Google Scholar]
- 32.Litjens G., et al. , “A survey on deep learning in medical image analysis,” Med. Image Anal. 42, 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 33.Chougrad H., Zouaki H., Alheyane O., “Deep convolutional neural networks for breast cancer screening,” Comput. Methods Prog. Biomed. 157, 19–30 (2018). 10.1016/j.cmpb.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 34.Bunch P. C., et al. , “A free response approach to the measurement and characterization of radiographic observer performance,” Proc. SPIE 0127, 124–135 (1977). 10.1117/12.955926 [DOI] [Google Scholar]
- 35.Lee R. S., et al. , “A curated mammography data set for use in computer-aided detection and diagnosis research,” Sci. Data 4, 170177 (2017). 10.1038/sdata.2017.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark K., et al. , “The cancer imaging archive (TCIA): maintaining and operating a public information repository,” J. Digital Imaging 26(6), 1045–1057 (2013). 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otsu N., “A threshold selection method from gray-level histograms,” IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979). 10.1109/TSMC.1979.4310076 [DOI] [Google Scholar]
- 38.Simonyan K., Zisserman A., “Very deep convolutional networks for large-scale image recognition,” in Int. Conf. Learn. Represent. (ICLR) (2015). [Google Scholar]
- 39.He K., et al. , “Deep residual learning for image recognition,” in Proc. Conf. Comput. Vision and Pattern Recognit., IEEE, pp. 770–778 (2016). 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 40.Szegedy C., et al. , “Rethinking the inception architecture for computer vision,” in Proc. Conf. Comput. Vision and Pattern Recognit., IEEE, pp. 2818–2826 (2016). [Google Scholar]
- 41.Kingma D. P., Ba J. L., “Adam: A method for stochastic optimization,” in Proc. 3rd Int. Conf. Learn. Represent. (ICLR), ACM; (2014). [Google Scholar]
- 42.te Brake G. M., Karssemeijer N., Hendriks J. H., “An automatic method to discriminate malignant masses from normal tissue in digital mammograms,” Phys. Med. Biol. 45(10), 2843–2857 (2000). 10.1088/0031-9155/45/10/308 [DOI] [PubMed] [Google Scholar]
- 43.Campanini R., et al. , “A novel featureless approach to mass detection in digital mammograms based on support vector machines,” Phys. Med. Biol. 49(6), 961–975 (2004). 10.1088/0031-9155/49/6/007 [DOI] [PubMed] [Google Scholar]
- 44.Agarwal R., et al. , “Mass detection in mammograms using pre-trained deep learning models,” Proc. SPIE 10718, 107181F (2018). 10.1117/12.2317681 [DOI] [Google Scholar]