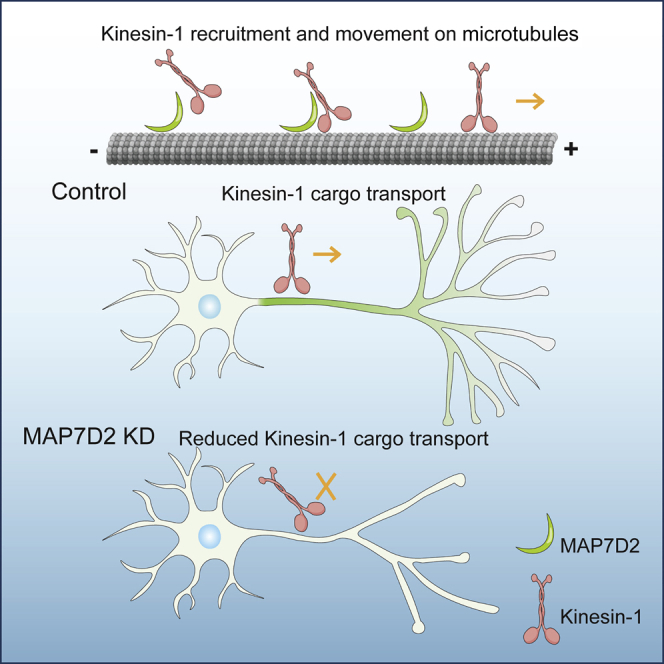
Summary
The motor protein kinesin-1 plays an important role in polarized sorting of transport vesicles to the axon. However, the mechanism by which the axonal entry of kinesin-1-dependent cargo transport is regulated remains unclear. Microtubule-associated protein MAP7 (ensconsin in Drosophila) is an essential kinesin-1 cofactor and promotes kinesin-1 recruitment to microtubules. Here, we found that MAP7 family member MAP7D2 concentrates at the proximal axon, where it overlaps with the axon initial segment and interacts with kinesin-1. Depletion of MAP7D2 results in reduced axonal cargo entry and defects in axon development and neuronal migration. We propose a model in which MAP7D2 in the proximal axon locally promotes kinesin-1-mediated cargo entry into the axon.
Keywords: microtubule, microtubule-associated protein, kinesin, axon initial segment, transport
Graphical Abstract
Highlights
-
•
The microtubule-binding domain of MAP7D2 targets the proximal axon
-
•
MAP7D2 is required for axon development
-
•
MAP7D2 associates with all three kinesin-1 family members
-
•
MAP7D2 promotes kinesin-1-mediated cargo transport in axons
Kinesin-1 is well known for transporting vesicles and organelles into to the axon. Pan et al. show that MAP7D2 concentrates at the proximal axon, regulates kinesin-1 activity, and locally promotes kinesin-1-mediated cargo trafficking into axons. Specific microtubule-associated proteins (MAPs) that locally control motor activities is an emerging concept.
Introduction
The microtubule cytoskeleton and proper coordination over intracellular organelle transport are critically important for the development and function of neurons. Microtubules serve as tracks for long-distance cargo trafficking and have an intrinsic polarity that is recognized by molecular motors to transport cargo in a directional manner. Plus-end-directed kinesin-1 motors selectively drive transport vesicles into axons and are required for polarized cargo sorting at the axon initial segment (AIS) (Farías et al., 2015, Kapitein et al., 2010). However, the mechanism by which the axonal entry of kinesin-1-dependent cargo transport is regulated remains unclear.
Several regulatory mechanisms have been proposed to contribute to polarized cargo trafficking. For instance, the preference of kinesin-1 for stable microtubules may contribute to sorting and trafficking of axonal cargo (Farías et al., 2015). By using optical nanoscopy, it was recently shown that within the mixed dendritic microtubule arrays stable microtubules are mostly oriented plus-end inward and guide kinesin-1 motors out of dendrites (Tas et al., 2017). This particular microtubule organization favors kinesin-1 transport toward the axon; however, additional sorting mechanisms may play a role at the proximal axon.
The microtubule-associated protein MAP7 (ensconsin in Drosophila) was described as an essential kinesin-1 cofactor by promoting kinesin-1 recruitment to microtubules (Barlan et al., 2013, Sung et al., 2008). MAP7 is required for all known kinesin-1-dependent processes in polarized Drosophila oocytes (Metivier et al., 2018, Sung et al., 2008) and essential for kinesin-1-mediated myonuclear positioning in mammalian myotubes (Metzger et al., 2012). Moreover, MAP7 cooperates with kinesin-1 to transport cargo (Barlan et al., 2013) and triggers centrosome separation during interphase (Gallaud et al., 2014). The mammalian MAP7 family members include MAP7 (ensconsin, E-MAP-115), MAP7D1, MAP7D2, and MAP7D3. All family members share a highly conserved N-terminal coiled-coil motif, which interacts with microtubules and a conserved C-terminal MAP7 coiled-coil domain that is presumed to bind to kinesin-1. MAP7 and MAP7D1 are broadly expressed and have been described to play a role in neuronal developmental processes, including in axonal development in cultured neurons (Koizumi et al., 2017, Tymanskyj et al., 2017). MAP7D2 on the other hand is predominantly expressed in brain tissues (Niida and Yachie, 2011); however, little is known about its localization and function in neuronal cells.
In this study, we show that MAP7D2 interacts with all three kinesin-1 family members and accumulates in the proximal axon through its N-terminal microtubule-binding domain. Depletion of MAP7D2 results in reduced axonal cargo entry and defects in axon formation and outgrowth during early stages of neuronal development. These data indicate that MAP7D2 is a local kinesin-1 regulator that promotes cargo entry into the axon.
Results
MAP7D2 Localizes to the Proximal Axon
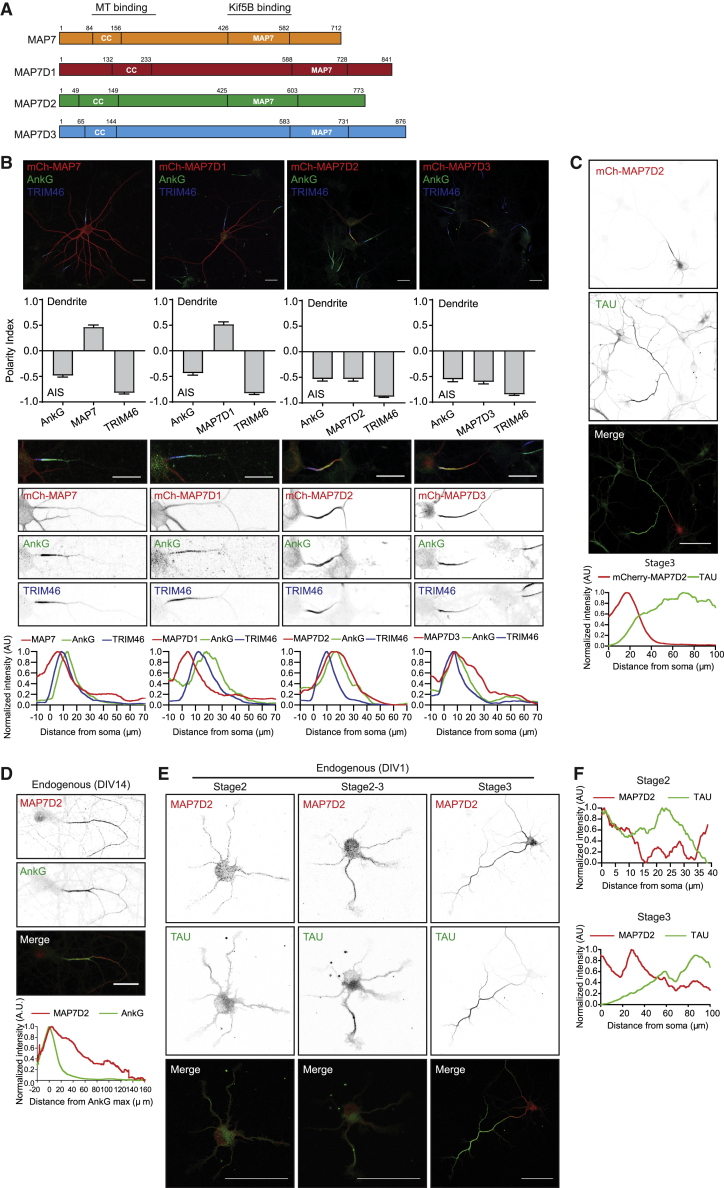
To study the subcellular distributions of MAP7 family members in neurons, we first expressed mCherry-tagged MAP7, MAP7D1, MAP7D2, and MAP7D3 in primary cultured hippocampal neurons (Figure 1A). Whereas MAP7 and MAP7D1 are mainly present in the somatodendritic compartment, MAP7D2 and MAP7D3 localize to the proximal axon overlapping with the AIS markers TRIM46 and AnkyrinG (AnkG) (Figure 1B). MAP7D2 is not abundant in other parts of the axon, evident by the lack of Tau colocalization (Figure 1C). Moreover, by labeling neurons with an antibody against MAP7 confirmed the dendrite localization (Figure S1A), evident by the intensity of MAP7 decreasing in the TRIM46 positive axon and the polarity index being biased to dendrites (Figures S1B and S1C). These data suggest that MAP7 family members have a distinct distribution in neurons.
Figure 1.
MAP7D2 Is Enriched in Proximal Axon
(A) Schematic domain structure of human MAP7 family members. Numbers represent amino acids.
(B) DIV15 neurons expressing mCherry-tagged MAP7 proteins and co-stained for AnkG (green) and TRIM46 (blue). Bar graph shows the polarity index of MAP7 proteins together with AnkG and TRIM46 (n > 10 neurons in each group). Bottom panels are zooms of the proximal axons and line scans for the normalized intensity of each channel from soma to axon.
(C) DIV3 neurons expressing mCherry-MAP7D2 and stained for TAU (green). Line graphs of each channel are shown.
(D) DIV14 neurons stained with endogenous MAP7D2 (red) and AnkG (green). Line graph shows that MAP7D2 fluorescence aligns with AnkG maximum intensity (n = 21).
(E and F) DIV1 neurons stained for endogenous MAP7D2 (red) and TAU (green) (E). Line scans for stages 2 and 3 show the normalized fluorescent intensity from soma to axon (F).
Scale bars: 20 μm in (B) and (D) and 50 μm in (C) and (E).
Since MAP7D3 is only expressed in non-brain tissues and MAP7D2 is specifically present in brain tissues (Niida and Yachie, 2011, Uhlén et al., 2015, Zhang et al., 2014), we decided to further investigate the neuronal function of MAP7D2. To study the localization of endogenous MAP7D2, we performed immunofluorescence labeling of cultured neurons. In agreement with the exogenous mCherry-MAP7D2 distribution, antibodies against endogenous MAP7D2 label the proximal axon overlapping with AnkG (Figure 1D) but also extend into the axon. The MAP7D2 antibody is highly specific, as it cannot recognize the overexpression of the other MAP7 proteins (Figure S1D). We did not detect any endogenous MAP7D3 in the proximal axon by labeling neurons with a MAP7D3-specific antibody (Figure S1E), and MAP7D3 is only present at microtubules in WT HeLa cells but not in MAP7D3 KO HeLa cells, while MAP7D2 is both absent in WT or MAP7D3 KO HeLa cells (Figures S1F and S1G), again suggesting that MAP7D3 is only expressed in non-brain tissues where MAP7D2 is not expressed. Taken together, these data indicate that MAP7D2 is exclusively expressed in the proximal axon of hippocampal neuron.
We next determined the localization in DIV1 (1 day in vitro) neuron cultures, which contain both stage 2 (unpolarized) and stage 3 (polarized) cells. MAP7D2 is found in the cell body and throughout the neurites in unpolarized stage 2 cells but strongly accumulates at the proximal axon in polarized stage 3 neurons (Figures 1E and 1F). These data indicate that MAP7D2 localizes to the proximal axon directly after neuronal polarization.
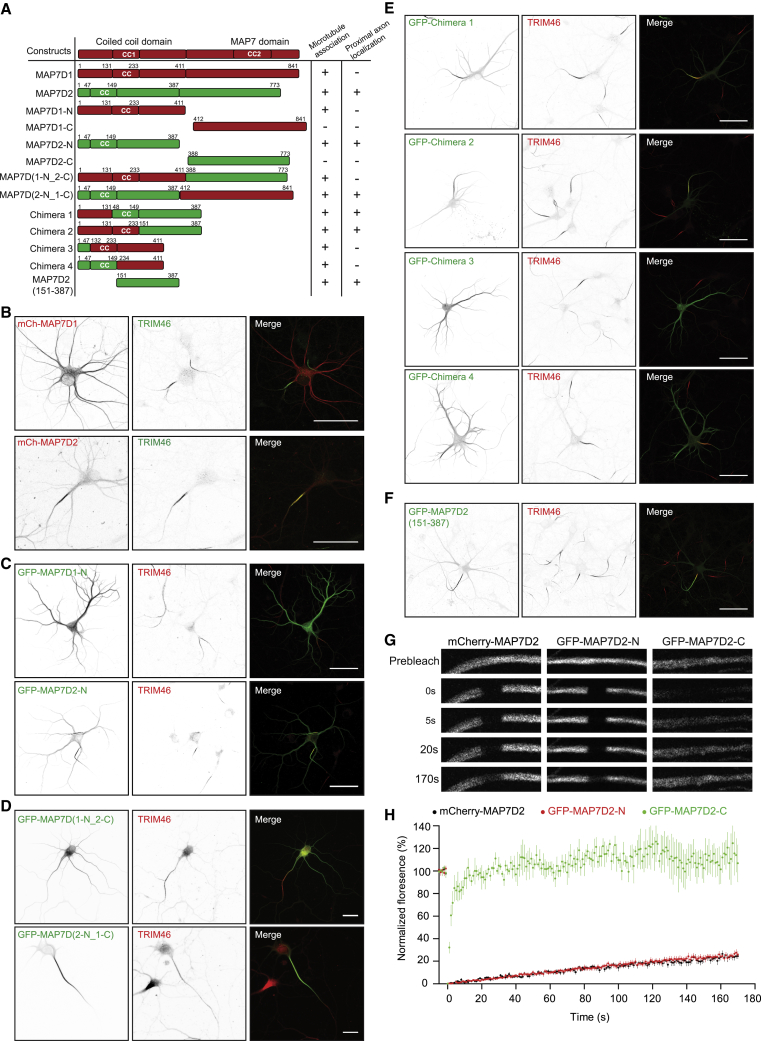
MAP7D2 Localizes to Proximal Axon through Its Microtubule-Binding Domain
To determine which domain is required for the proximal axon localization, we generated MAP7D1 and MAP7D2 truncation constructs (Figure 2A). The C-terminal domains of both MAP7D1 (MAP7D1-C) as well as MAP7D2 (MAP7D2-C) are diffusely localized throughout the neuron and accumulate at axon tips (Figure S2A). In contrast the N-terminal microtubule-binding region of MAP7D2 (MAP7D2-N) accumulates at the proximal axon and the N-terminal domain of MAP7D1 (MAP7D1-N) localizes to dendrites similar to the full-length proteins (Figures 2B and 2C). To map the minimal regions required for the axon-specific localization, we generated chimeric proteins by swapping domains of the N-terminal parts of MAP7D1 and MAP7D2 (Figure 2A). All N-terminal containing truncations and chimeras of MAP7D1 and MAP7D2 showed microtubule binding (Figures S2B–S2E). Chimeras containing the MAP7D2 region downstream of the coiled-coil motif (chimeras 1 and 2) localized to the proximal axon, while constructs containing the similar region of MAP7D1 (chimeras 3 and 4) were enriched in the dendrites (Figure 2E). GFP- and hemagglutinin (HA)-tagged constructs containing the 151–387 region of MAP7D2 strongly associated with microtubules in COS7 cells and showed proximal axon localization in neurons (Figures 2F and S2F).
Figure 2.
MAP7D2 Stably Localizes to Proximal Axon through Its Domain Amino Acid 151–387
(A) Overview of MAP7D1 and MAP7D2 truncations and chimeras.
(B) DIV9 neurons expressing mCherry-tagged MAP7D1 and MAP7D2, respectively, and co-stained for TRIM46 (green).
(C–F) DIV4 or DIV9 neurons expressing with GFP-tagged N-MAP7D1, N-MAP7D (C) -, indicated chimeras in (A) (D and E), or MAP7D2 amino acid (aa) 151–387 (F) and co-stained for TRIM46 (red).
(G and H) FRAP images (G) and quantifications (H) of mCherry-MAP7D2, GFP-N-MAP7D2, and GFP-C-MAP7D2 fluorescence recovery in the axons of DIV9 neurons. n = 4–7 neurons. Error bars represent SEM.
Scale bars: 20 μm in (D) and 50 μm in (B), (C), (E), and (F).
To determine the dynamics of MAP7D2 in cells, we performed fluorescence recovery after photobleaching (FRAP) experiments in the proximal axon using N-terminal, C-terminal, or full-length MAP7D2. The MAP7D2 C-terminus quickly and completely recovers, while full-length and N-terminal MAP7D2 only showed ∼20% recovery after 3 min (Figures 2G and 2H). In dendrites, MAP7 and MAP7D1 show very similar recovery profiles (∼20%–30% recovery after 3 min) (Figures S2G and S2H). Together these data indicate that MAP7D2 stably localizes to the proximal axon through its N-terminal microtubule-binding domain.
TRIM46 Is Required for Proximal Axon Localization of MAP7D2
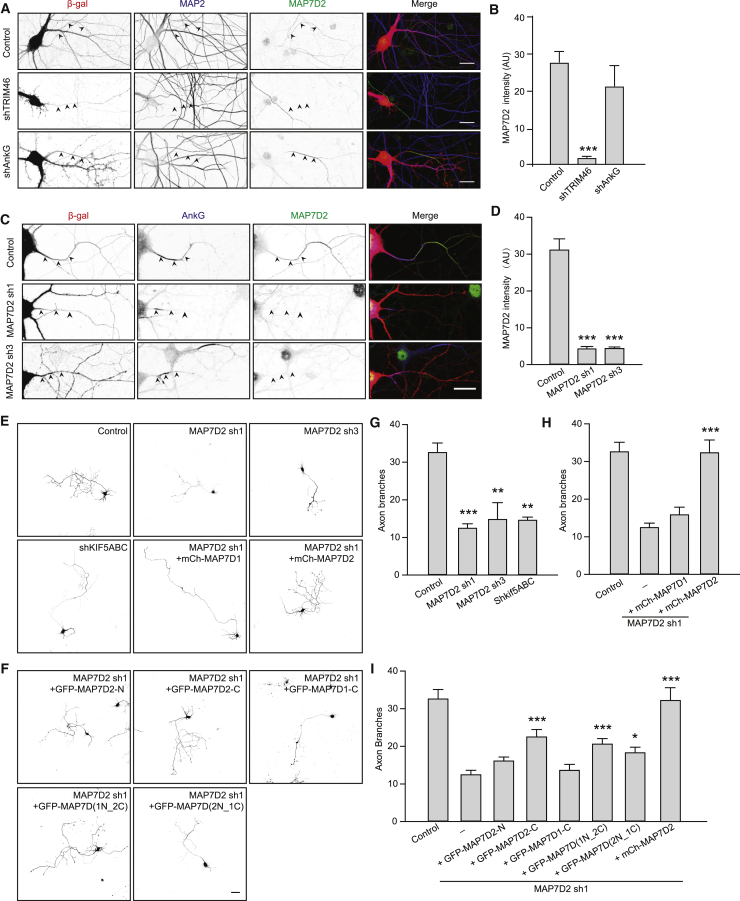
Since MAP7D2 localizes to the proximal axon, we next determined the underlying mechanism of this specific localization. Previously several proteins were shown to localize to the AIS and to be important for this structure, including the microtubule binding protein TRIM46 and AIS scaffolding protein AnkG. TRIM46 forms parallel microtubule bundles in the proximal axon and plays key role in axon specification (van Beuningen et al., 2015). AnkG is the major AIS scaffold protein required for AIS assembly, which forms a barrier between axonal and somatodendritic compartment for neuronal polarity maintenance (Winckler et al., 1999). We first depleted TRIM46 or AnkG with short hairpin RNAs (shRNAs) and determined the MAP7D2 localization at the proximal axon. MAP7D2 staining was still present in AnkG knockdown neurons but largely decreased from the proximal axon in TRIM46-depleted cells (Figures 3A and 3B), suggesting that MAP7D2’s localization to the proximal axon depends on TRIM46 but not AnkG. To address whether MAP7D2 interacts with TRIM46, we co-expressed them in COS7 cells where they co-localize on microtubule bundles. However, when we expressed a truncated TRIM46 protein lacking the microtubule binding domain (TRIM46 ΔCOS) (van Beuningen et al., 2015) together with MAP7D2, the colocalization is lost (Figure S3A). This suggests that MAP7D2 does not directly interact with TRIM46.
Figure 3.
MAP7D2 Is Important for Axon Development
(A) DIV14 control, TRIM46-depleted, or AnkG-depleted neurons co-transfected with “β”-gal (red) and stained for MAP2 (blue) and MAP7D2 (green). Arrowheads point to the proximal axon.
(B) Bar graph shows the average intensity of MAP7D2 in each condition. n = 9–10 neurons.
(C) DIV14 control or MAP7D2-depleted neurons co-transfected with “β”-gal fill and stained for AnkG (blue), MAP7D2 (green).
(D) Bar graph shows the average intensity in the specified condition. n = 15–20 neurons
(E and F) Representative images of DIV4 neurons of indicated conditions, together with GFP (E) or mCherry fill (F).
(G–I) Quantifications of the numbers of axon branches of indicated conditions. n = 19–123 neurons.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Unpaired t test. Error bars represent SEM. Statistics in (G) were from control, and statistics in (H) and (I) were from MAP7D2 sh1. Scale bars: 20 μm in (A) and (C) and 50 μm in (E) and (F).
To test the effect of MAP7D2 depletion on TRIM46 and AnkG localization, we generated several independent shRNAs to deplete MAP7D2 expression and validated the knockdown efficiency by staining transfected neurons with MAP7D2 antibody. Compared to control cells, the MAP7D2 staining was reduced by ∼85% in neurons transfected with either shRNA1 or shRNA3 (Figures 3C and 3D). However, the MAP7D2 knockdown only slightly reduced the levels of both TRIM46 as well as AnkG (Figures S3B–S3D), which may be caused by the reduced diameter of the AIS segment upon MAP7D2 depletion (Figures S3E and S3F). MAP7D3 could not be detected upon MAP7D2 depletion and neither did we find changes in MAP7 localization, arguing against a potential compensatory mechanism by other MAP7 proteins (Figure S4).
MAP7D2 Is Important for Neuronal Migrations and Axon Development
Since MAP7D2 localizes to the proximal axon, we tested whether MAP7D2 is important for axon development. We found that developing DIV4 neurons depleted of MAP7D2 show reduced axon branching compared to control neurons (Figures 3E and 3G). The axonal phenotype is rescued by re-expression of MAP7D2 but not MAP7D1 (Figures 3E and 3H), potentially due to their difference in cellular localization. In addition to axon branching, we tested whether MAP7D2 is important for axon formation. Low concentration of the microtubule stabilizing drug taxol has been shown to induce the formation of multiple axons, somewhat mimicking the process of axon initiation (Witte et al., 2008). Indeed, after taxol treatment, control neurons form 5 axons on average. Neurons depleted of MAP7D2 do not form multiple axons after taxol addition, indicating that MAP7D2 is important for taxol-induced axon formation (Figures S5A and S5B). To confirm these results, we electroporated hippocampal neurons with shRNAs targeting MAP7D2 or control before plating and analyzed axon formation in non-taxol treated neurons at DIV3 by using Tau and TRIM46 antibodies to mark the axon (Figure S5C). Compared to control cells, there was a small but significantly reduction in the number of Tau or TRIM46 positive neurons after MAP7D2 knockdown. Conversely, the overexpression of MAP7D2 leads to a slight increased number of axons (Figures S5D and S5E). Therefore, MAP7D2 is important for axon branching and potentially also for axon formation.
Neurons in the developing neocortex migrate from the ventricular zone to the different layers in the cortical plate. To test whether MAP7D2 is involved in this process, we performed ex vivo electroporation on mouse E14.5 embryos and cultured brain slices for 4 days to allow GFP-labeled cells in ventricular zone to migrate. Whereas control neurons efficiently migrated to the upper layers of the cortical plate, upon MAP7D2 knockdown neurons accumulated in the ventricular zone. Importantly, this migration defect could be rescued by re-expressing MAP7D2, showing that MAP7D2 is important for neuronal migration (Figures S5F–S5H).
MAP7D2 Activity Requires Proximal Axon Localization and Kinesin Binding
To address the functional differences between MAP7D1 and MAP7D2, we expressed truncation as well as chimeric constructs (Figures 2A–2D and S2A), as it was recently shown in Drosophila that MAP7 C-terminal kinesin-binding domain was sufficient to rescue several of the mutant phenotype (Metivier et al., 2018). Interestingly, we also observed a partial rescue by overexpressing of MAP7D2-C; however, MAP7D1-C could not reverse the knockdown phenotype. Moreover, even when we fused MAP7D1-C to the AIS targeting MAP7D2-N (chimera MAP7D(2N_1C)), it only partially rescues the MAP7D2 knockdown phenotype (Figures 3F and 3I), suggesting that MAP7D2 C-terminus plays a specific role in axonal branching. We next tested whether the axonal branching phenotype of MAP7D2 can be phenocopied by knocking down kinesin-1. To address this, we depleted all three kinesin-1 family members (KIF5A, KIF5B, and KIF5C) and observed a similar axonal branching phenotype as for MAP7D2 (Figures 3E and 3G). Taken together, these data suggest that both the proximal axon localization as well as specific features of the MAP7D2 C-terminus are important determinants for axonal branching.
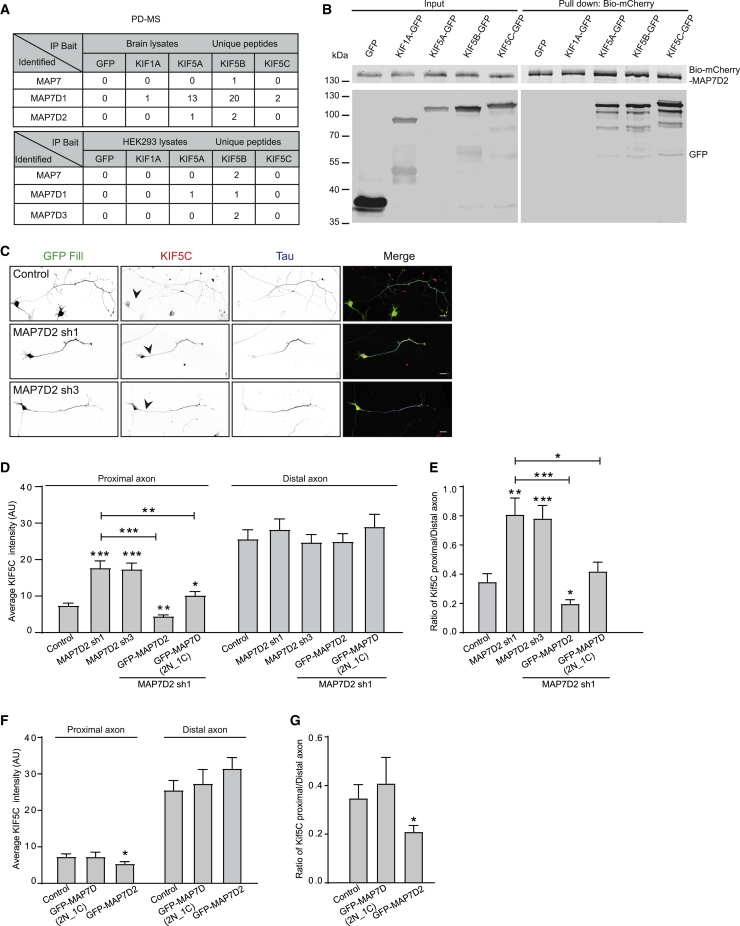
All Three Kinesin-1 Family Members Interact with MAP7D2
Previous studies have reported the association between the first coil coil of kinesin-1 and MAP7 (Metzger et al., 2012). To study the interaction in an unbiased manner, we performed pull-down experiments followed by mass spectrometry. Here, GFP-tagged motor domain and coiled-coil region (MDC) of KIF5A-MDC (1–566) and KIF5B-MDC (1–807) were expressed in HEK293 cells, isolated with beads coupled with GFP antibodies, and incubated with rat brain extracts, and isolated proteins were analyzed by mass spectrometry. In these pull-down experiments, we used GFP, KIF1A-MDC (1–500), and the KIF5C-MD (1–401) lacking the first coil coil as controls. Both KIF5A-MDC and KIF5B-MDC pulled down MAP7 family members from human embryonic kidney (HEK) cells and brain extracts (Figure 4A). More MAP7D1 was pulled down from brain lysates, which can be explained by the fact that dendritic-enriched MAP7D1 in neurons is much more abundant than axonal MAP7D2. The differences in MAP7 protein pull-down between brain tissue lysates versus HEK293 cell lysates again indicates that MAP7D2 is exclusively expressed in brain tissues, while MAP7D3 is only expressed in non-brain tissues. These mass spectrometry results were confirmed by more direct pull-down experiments in HEK cells coexpressing MAP7D2 and kinesin-1 constructs (Figure 4B), indicating that all three kinesin-1 family members interact with MAP7D2.
Figure 4.
MAP7D2 Interacts with Kinesin-1 Family and Affects Kinesin-1 Distribution in the Proximal Axon
(A) Indicated baits incubated with rat brain and HEK293 cell extracts identified proteins by mass spectrometry. PD-MS means pull-down-based mass spectrometry.
(B) Western blot analysis of biotinylated (bio) pull-down from extracts of HEK293 cells transfected with indicated constructs, probed for GFP and mCherry.
(C) Representative images of neurons with control, MAP7D2 sh1, or sh3 and fixed at DIV3 and stained for KIF5C and the axonal marker Tau. Arrowheads point to the proximal axon.
(D and E) Quantifications of KIF5C intensity (D) or ratio (E) at the beginning and the end of axon in indicated conditions. n = 15–32 neurons.
(F and G) Quantifications of KIF5C intensity (F) or ratio (G) at the beginning and the end of axon in specified conditions. n = 19–27 neurons.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Unpaired t test. Error bars represent SEM. Scale bars: 20 μm in (C).
MAP7D2 Regulates Kinesin-1 Activity
MAP7 has been shown to stimulate kinesin-1 activity by promoting microtubule landing and activity (Sung et al., 2008). To determine whether MAP7D2 affects kinesin-1 activity, we co-expressed a truncated constitutively active KIF5 (KIF5C-MDC) together with a full-length MAP7D2 or MAP7D2-N that lack of kinesin-1 association domain in COS7 cells. KIF5C-MDC was strongly recruited to microtubules in the presence of MAP7D2 but not MAP7D2-N (Figure S6A). The lack of overlap between the proteins probably reflects that kinesin-1 is recruited to the microtubules by MAP7D2 and then walks away. The microtubule binding but non-walking KIF5A(G235A) (Rigor-KIF5A) was found to preferentially bind to proximal axonal microtubules (Farías et al., 2015). To investigate whether the recruitment of Rigor-KIF5A is affected by MAP7D2, we expressed it in control or MAP7D2-depleted neuron. We observed a significant decrease of the Rigor-KIF5A in the proximal axon in MAP7D2-depleted neuron (Figures S6B and S6C), suggesting that MAP7D2 is important for kinesin-1 recruitment to microtubules in the proximal axon.
To further address MAP7D2′s role on kinesin-1, we analyzed endogenous KIF5C upon MAP7D2 depletion in DIV3 neurons. In control neurons, KIF5C is largely absent from the proximal axon where endogenous MAP7D2 is localized (Figures 4C–4E). Upon MAP7D2 depletion, however, there is a significant increase in the proximal axon localized kinesin-1, which could be rescued by re-expression of the full-length MAP7D2 and only partially by MAP7D(2N_1C) (Figures 4D and 4E). In addition, overexpression of MAP7D2 leads to a significant reduction in proximal localized kinesin-1, whereas MAP7D(2N_1C) did not (Figures 4F and 4G). Together this suggests that MAP7D2 stimulates kinesin-1 activity in the proximal axon.
MAP7D2 Is Required for Kinesin-1-Based Cargo Transport
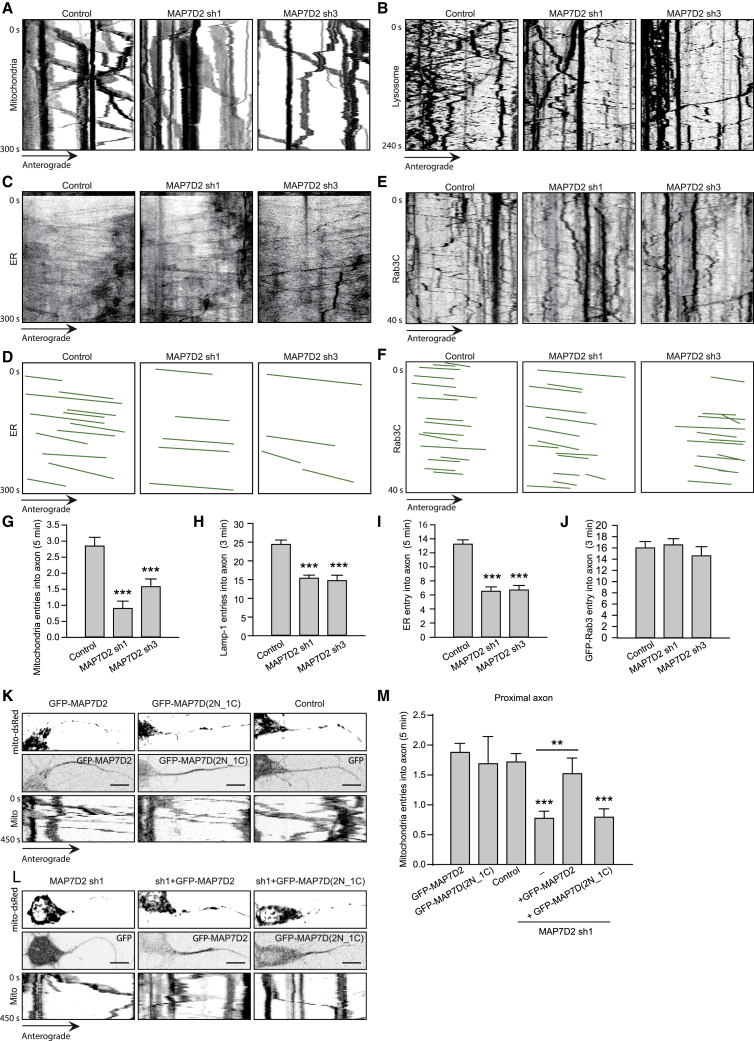
The axonal transport of mitochondria, endoplasmic reticulum (ER) and lysosomes strongly depends on kinesin-1 activity (Farías et al., 2017, van Spronsen et al., 2013, Woźniak et al., 2009). To investigate whether kinesin-1-mediated cargo trafficking is affected by MAP7D2, we performed live-cell imaging experiments using the cargos indicated above. We observed a marked decrease in the number of mitochondria entering the axon in MAP7D2-depleted neurons (Figures 5A and 5G). Also, the transport events in the distal axon were reduced (Figures S6D and S6E), which may well be due to reduced activation of kinesin-1 in the axon (Figures 4C–4E). Similar results were found for the axon entry of lysosomes and the ER (Figures 5B–5D, 5H, and 5I). Imaging of Rab3 vesicles, which are mainly transported by kinesin-3 family members (Kevenaar et al., 2016), did not reveal differences between control and MAP7D2 knockdown cells (Figures 5E, 5F, and 5J). Finally, we were able to fully rescue the axonal entry of mitochondria upon MAP7D2 depletion by re-expression of the full-length protein, but not the MAP7D(2N_1C) (Figures 5K–5M). Together these data indicate that MAP7D2 is required for kinesin-1-mediated cargo transport to enter the axon.
Figure 5.
MAP7D2 Promotes Kinesin-1-Based Cargo Transport
(A–C) Kymographs showing live-cell imaging of mitochondria, lysosome, or ER (after FRAP) in control or MAP7D2-depleted neurons co-transfected with mito-DsRed (A), LAMP1-GFP (B), or GFP-SBP-Rtn4A (C).
(D) Drawing of the anterograde ER movements along the proximal axon in (C).
(E) Kymographs showing live-cell imaging of Rab3 vesicles in control or MAP7D2-depleted neurons.
(F) Drawing of the anterograde Rab3 vesicles movement along the proximal axon in (D).
(G–J) Quantifications of mitochondria (G), lysosome (H), ER (I), or Rab3 vesicles (J) entry into axon as described for (A)–(F). n = 12–44 neurons.
(K–M) Stills and kymographs for live-cell imaging of mitochondria in neurons co-transfected with mito-DsRed and indicated constructs (K and L). Quantifications for mitochondria entry into axon within 5 min upon specified conditions (M). n = 12–21 neurons. Unpaired t test. Scale bars: 10 μm.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Mann-Whitney rank-sum test. Error bars represent SEM.
Discussion
In this study, we analyzed the proteins of the MAP7 family in rat hippocampus neurons. Mammals have 4 MAP7 family members, of which MAP7, MAP7D1, and MAP7D2 are expressed in neuronal tissues. We found that these proteins have very specific localization patterns; whereas MAP7 and MAP7D1 localize to the somatodendritic compartment, MAP7D2 specifically accumulates to the proximal axon, largely overlapping with the AIS, and also extends some into the axon. This is markedly different to sensory dorsal root ganglion (DRG) neurons, which do not possess dendrites nor a classic AIS, where MAP7 was shown to localize to axon branchpoints (Tymanskyj et al., 2017, Tymanskyj et al., 2018). We found that MAP7D2 is tightly associated with proximal axon via an N-terminal microtubule binding fragment, which we mapped to a 235-amino-acid domain. The proximal axon localization of MAP7D2 depends on TRIM46, which bundles microtubules in the AIS in a parallel fashion (van Beuningen et al., 2015). We speculate that MAP7D2 may recognize a modification and/or a change in the microtubule lattice induced by TRIM46 at the AIS. Additionally, as MAP7 microtubule binding and subcellular localization was shown to be influenced by phosphorylation (Ramkumar et al., 2018), this might also play a role in targeting MAP7D2 to the proximal axon.
Like for other MAP7 proteins, we show that MAP7D2 can interact with all three KIF5 (kinesin-1) isoforms but not with KIF1A (kinesin-3). Accordingly, depletion of MAP7D2 leads to a reduction of kinesin-1-dependent cargo to enter the axon, whereas kinesin-3-dependent RAB3 vesicles were unaffected. Moreover, we observed a decreased recruitment of rigor KIF5A to the proximal axon, whereas endogenous KIF5C is increased in the proximal axon, suggesting that the MAP7D2 is important for both kinesin-1 recruitment and processivity in the proximal axon. These data lead to a model where the kinesin-1 activator MAP7D2 is localized to the proximal axon to stimulate kinesin-1-dependent cargo entry into the axon. We also observed reduced mitochondria transport in the distal axon upon MAP7D2 depletion, which may well be caused by the reduced activation of axonal kinesin-1, although we cannot exclude that there are low remaining levels of MAP7D2 in the distal segment promoting kinesin-1 transport.
A recent study addressed the functioning of the MAP7 family in both cultured cells as well as with purified proteins in vitro and found that there is a high degree of redundancy (Hooikaas et al., 2018). In neurons, we find that the expression of both MAP7D1 as well as MAP7D2 C-terminus end up at the tips of the axon branches, suggesting that they bind to kinesin-1 and are co-transported. The MAP7D2 C-terminus can partially rescue the axonal branching phenotype. When the MAP7D1 C-terminus is attached to the AIS binding domain of MAP7D2, it hardly rescues the axon branching phenotype. Similarly, full-length MAP7D2 was able to rescue the MAP7D2 knockdown-induced kinesin-1 accumulation at the proximal axon and the reduced axonal entries of mitochondria, whereas the chimera construct could not or only partially rescue the defects. This suggests that MAP7D2 has a unique function in regulating kinesin-1 activity in the proximal axon to regulate branching.
The idea that spatially confined microtubule-associated proteins form a “MAP code” that locally control microtubule motor activities is an emerging concept. In neurons, for example, doublecortin stimulates kinesin-3 to target dendrites (Lipka et al., 2016), whereas MAP2 and SEPT9 were found to prevents kinesin-1 cargo to enter the dendrite (Gumy et al., 2017, Karasmanis et al., 2018). For axons, the protein Tau is suggested to prevent kinesin-1 landing and/or activity on axonal microtubules (Seitz et al., 2002, Vershinin et al., 2007). In this study, we found that MAP7D2 promotes kinesin-1 transport into the axon of hippocampal neurons. It was shown that MAP7 and Tau compete for microtubule binding in vitro (Monroy et al., 2018). Whether the same applies for Tau and MAP7D2 remains to be seen, although we did not observe clear proximal relocalization of Tau upon MAP7D2 depletion. Since mutations in kinesin-1 family member KIF5A are associated with several neurological diseases (Crimella et al., 2012, Nicolas et al., 2018), in the future it will be interesting to address the functions of MAP7 family members in these diseases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-MAP7D2 | Atlas Antibodies | Cat# HPA051508; RRID: AB_2681513) |
| Mouse polyclonal anti-MAP7 | Abnova Corporation | Cat# H00009053-B01P; RRID: AB_10714227 |
| Rabbit polyclonal anti-MAP7D3 | Atlas Antibodies | Cat# HPA035598; RRID: AB_10671108 |
| Mouse monoclonal anti-AnkG | UC Davis/NIH NeuroMab Facility | Cat# 75-146; RRID: AB_10673030 |
| Rabbit polyclonal anti-TRIM46 | From C.C. Hoogenraad, (van Beuningen et al., 2015) | N/A |
| Mouse monoclonal anti-Tau-1 | Millipore | Cat# MAB3420; RRID: AB_94855) |
| Rat monoclonal anti-HA | Roche | Cat# 11867423001; RRID: AB_390918 |
| Rabbit polyclonal anti-Kinesin 5C | Abcam | Cat# ab5630; RRID:AB_304999 |
| Rabbit polyclonal anti-GFP | Abcam | Cat# ab290; RRID:AB_303395 |
| Mouse monoclonal anti-mCherry | Clontech Laboratories | Cat# 632543; RRID: AB_2307319 |
| Goat Anti-Rabbit IgG Antibody, IRDye 680LT Conjugated | LI-COR Biosciences | Cat# 827-11081; RRID: AB_10795015 |
| Goat Anti-Mouse IgG Antibody, IRDye 680LT Conjugated | LI-COR Biosciences | Cat# 827-11080; RRID: AB_10795014 |
| Goat Anti-Rabbit IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat# 827-08365; RRID: AB_10796098 |
| Goat Anti-Mouse IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat# 827-08364; RRID: AB_10793856 |
| anti-mouse Alexa488 | Life Technologies | Cat# A11029; RRID: AB_2534088 |
| anti-rabbit Alexa488 | Life Technologies | Cat# A11034; RRID: AB_2576217 |
| anti-rat Alexa488 | Life Technologies | Cat# A11006; RRID: AB_2534074 |
| anti-mouse Alexa568 | Life Technologies | Cat# A11031; RRID: AB_144696 |
| anti-rabbit Alexa568 | Life Technologies | Cat# A11036; RRID: AB_10563566 |
| anti-rat Alexa568 | Life Technologies | Cat# A-11077; RRID: AB_2534121 |
| anti-mouse Alexa647 | Life Technologies | Cat# A21236; RRID: AB_2535805 |
| anti-rabbit Alexa647 | Life Technologies | Cat# A21245; RRID: AB_2535813 |
| anti-mouse Alexa405 | Life Technologies | Cat# A-31553; RRID: AB_221604 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Paclitaxel | Sigma-Aldrich | T7402 |
| Fugene6 | Promega | E2691 |
| Lipofectamine 2000 | Thermofisher | 11668019 |
| Vectashield mounting medium | Vectorlabs | H-1000 |
| Critical Commercial Assays | ||
| Rat Neuron Nucleofector kit | Amaxa | VVPG-1003 |
| Experimental Models: Cell Lines | ||
| African Green Monkey SV40-transformed kidney ATCC CRL-1651 fibroblast (COS-7) | ATCC | CRL-1651 |
| Human embryonic kidney 239T (HEK293T) | ATCC | CRL-3216 |
| HeLa (Kyoto) | (Hooikaas et al., 2018) | N/A |
| HeLa MAP7D3 CRISPR/Cas9 knockout lines | (Hooikaas et al., 2018) | N/A |
| Experimental Models: Organisms/Strains | ||
| Rat (Wistar) | Janvier | N/A |
| Mouse (C57 BL/6JRj) | Janvier | N/A |
| Oligonucleotides | ||
| pSuper-rat MAP7D2 shRNA #1 targeting sequence: ggaacctcctatgagtaaa | This paper | N/A |
| pSuper-rat MAP7D2 shRNA #3 targeting sequence: ctgaagaagttcaatctat | This paper | N/A |
| pSuper-mouse MAP7D2 shRNA targeting sequence: gagacaaagattagccaaa | This paper | N/A |
| pSuper-rat KIF5A shRNA targeting sequence: gagacatcttcaaccacat | This paper | N/A |
| pSuper-rat KIF5B shRNA targeting sequence: tggagggtaaacttcatga | This paper | N/A |
| pSuper-rat KIF5C shRNA targeting sequence: tgagatctacttggacaaa | This paper | N/A |
| pSuper-rat TRIM46 shRNAs | (van Beuningen et al., 2015) | N/A |
| pSuper-rat AnkG shRNAs | (Kuijpers et al., 2016) | N/A |
| Recombinant DNA | ||
| pGW1-GFP | (Kapitein et al., 2010) | N/A |
| Bio-mCherry-MAP7 | (Hooikaas et al., 2018) | N/A |
| Bio-mCherry-MAP7D1 | (Hooikaas et al., 2018) | N/A |
| Bio-mCherry-MAP7D2 | (Hooikaas et al., 2018) | N/A |
| Bio-mCherry-MAP7D3 | (Hooikaas et al., 2018) | N/A |
| GFP-MAP7D1-N | This paper | N/A |
| GFP-MAP7D1-C | This paper | N/A |
| GFP-MAP7D2-N | This paper | N/A |
| GFP-MAP7D2-C | This paper | N/A |
| GFP-MAP7D(1N_2C) | This paper | N/A |
| GFP-MAP7D(2N_1C) | This paper | N/A |
| GFP-MAP7D2 | This paper | N/A |
| GFP-chimera 1 | This paper | N/A |
| GFP-chimera 2 | This paper | N/A |
| GFP-chimera 3 | This paper | N/A |
| GFP-chimera 4 | This paper | N/A |
| GFP-MAP7D2 (151-387) | This paper | N/A |
| HA-MAP7D2 (151-387) | This paper | N/A |
| MARCKS-GFP | (De Paola et al., 2003) | N/A |
| pβactin-GFP-FRB | (Kapitein et al., 2010) | N/A |
| pβactin-HA-β-galactosidase | (Hoogenraad et al., 2005) | N/A |
| KIF1A (1-500)-GFP-FRB | (Lipka et al., 2016) | N/A |
| KIF5A (1-566)-GFP-FRB | (Lipka et al., 2016) | N/A |
| KIF5B (1-807)-GFP-FRB | (Kapitein et al., 2010) | N/A |
| KIF5C (1-401)-GFP-FRB | (Lipka et al., 2016) | N/A |
| KIF5B (1-560)-GFP-FRB | (Gumy et al., 2017) | N/A |
| KIF5C (1-560)-GFP-FRB | This paper | N/A |
| GFP-TRIM46 | (van Beuningen et al., 2015) | N/A |
| GFP-TRIM46 ΔCOS | (van Beuningen et al., 2015) | N/A |
| Mito-dsRed | (van Spronsen et al., 2013) | N/A |
| LAMP1-GFP | (Farías et al., 2017) | N/A |
| GFP-SBP-Rtn4A | Kind gift from Dr. Ginny Farías | N/A |
| GFP-Rab3C | (Kevenaar et al., 2016) | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| NeuronJ | (Meijering et al., 2004) | N/A |
| Kymoreslicewide | Github | https://github.com/ekatrukha/KymoResliceWide |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Casper Hoogenraad (c.hoogenraad@uu.nl).
Experimental Model and Subject Details
Animals
All animal experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the Federal Government of the Netherlands, and were approved by the Animal Ethical Review Committee (DEC) of Utrecht University.
Heterologous cell and Hippocampal neuron cultures
HEK293 cells, HeLa cells and African Green Monkey SV40-transformed kidney fibroblast (COS7) cells were cultured in a 10 cm plate with 10 mL 50% DMEM/ 50% Ham’s F10 medium containing 10% FBS (GIBCO) and 1% Penicillin/streptomycin in incubator with 37°C and 5% CO2. WT and MAP7D3 knockout HeLa cells lines were already described (Hooikaas et al., 2018).
Hippocampus were dissected out from embryonic 18 Janvier WISTAR rat brain and dissociated with Trypsin in 37°C water bath. 100K/well neurons were plated in glass coverslips pre-coated with Poly-L-lysine (37.5 μg/ml, Sigma) and Laminin (1.25 μg/ml, Sigma). Neurobasal medium (Invitrogen) containing 2% B27 (Invitrogen), 0.5 mM glutamine (Invitogen), 15.6 μM glutamate (Sigam) and 1% penicillin/streptomycin (Invitrogen) was used for neuron culture.
Ex vivo electroporation and organotypic brain slice cultures
Pregnant C57BL/6JRj mice were sacrificed by cervical dislocation, and brains from E14.5 embryos were electroporated with 1.5μl DNA mixture containing a MARCKS-GFP together with shRNA target to MAP7D2 or pSuper empty vector (control), which was dissolved in MQ with 0.05% FastGreen FCF Dye (Sigma). The DNA mix was injected in the lateral ventricles of the embryonic brains using borosilicate glass micro-pipettes (World Precision Instruments) and a PLI-100A Picoliter Microinjector (Warner Instruments). Embryonic brains (region in the motor cortex) were electroporated using platinum plated electrodes (Nepagene) with an ECM 830 Electro Square Porator (Harvard Apparatus) set to 3 unipolar pulses of 100ms at 30V with 100ms intervals. Embryonic brains were then isolated and collected in ice-cold cHBSS, embedded in 3% SeaPlaque GTG Agarose (Lonza) in cHBSS and sectioned coronally into 300 μm thick slices using a VT1000 S Vibratome (Leica). Slices were collected on Poly-L-lysine and Laminin-coated culture membrane inserts (Falcon), placed on top of slice culture medium (70% v/v Basal Eagle Medium, 26% v/v cHBSS, 20mM D-glucose, 1mM L-glutamine, 0.1 mg/mL penicillin/streptomycin) and cultured 4 days prior to fixation.
Method Details
Antibody and reagents
The following antibodies were used for the immunofluorescence staining: mouse anti-AnkG (Neuromab, 75-146); rabbit anti-TRIM46 (van Beuningen et al., 2015); rabbit anti-MAP7D2 (Atlas Antibodies, HPA051508); mouse anti-Tau (Chemicon, MAB3420); rat anti-HA (Roche, 1-867-423); mouse anti-MAP7 (MaxPab, B01P); Alexa Fluor 405-, Alexa Fluor 488-, Alexa Fluor 568-, Alexa Fluor 647-conjugated secondary antibodies (Life Technologies). The following antibodies were used for western blot: mouse anti-mCherry (Clonetech, 632543); rabbit anti-GFP (Abcam, ab290); IRDye 680LT- and IRDye 800CW-conjugated secondary antibodies (LI-COR Biosciences).
Other reagents used in this study include: Taxol (Sigma, T7402), Lipofectamine2000 (Thermofiser, 11668019), Vectashield mounting medium (Vectorlabs, H-1000).
DNA and shRNA constructs
All full-length MAP7 family protein constructs were cloned by a PCR-based strategy into a Bio-mCherry-C1 vector, except for MAP7D1 which has been cloned into a Bio-mCherry-C3 vector. MAP7 constructs were generated from HeLa cDNA, MAP7D1 was cloned based on cDNA from IMAGE clone IRATp970A04109D (Source Bioscience), MAP7D2 was cloned based on cDNA of IMAGE clone IRAKp961B22199Q (Source Bioscience) and MAP7D3 was cloned based on cDNA of IMAGE clone IRAKp961K1163Q (Source Bioscience). All chimeras and truncation constructs of MAP7D1 and MAP7D2 were cloned into GW1 vector (Kapitein et al., 2010). MARCKS-GFP for ex vivo experiment was described before (De Paola et al., 2003). The kinesin constructs: pβactin-GFP-FRB, KIF1A (1-500)-GFP, KIF5A (1-566)-GFP, KIF5B (1-807)-GFP, KIF5B (1-560)-GFP and KIF5C (1-401)-GFP (Gumy et al., 2017, Kapitein et al., 2010, Kevenaar et al., 2016, Lipka et al., 2016). KIF5C (1-560) was generated by using full length rat KIF5C cDNA (NM_001107730.1) as PCR template and was cloned into pβactin-GFP-FRB vector. Plasmids for live imaging experiments: Mito-dsRed (van Spronsen et al., 2013), GFP-SBP-Rtn4A was cloned into GFP C1 vector from human RTN4A-GFP (ADDGENE #61807) by Dr. Ginny Farías, LAMP1-GFP was cloned into GFP N1 vector from Rat LAMP1-RFP (ADDGENE #1817) by Dr. Ginny Farías (Farías et al., 2017), GFP-Rab3C (Kevenaar et al., 2016). The following shRNAs were used for knockdown experiments: Rat MAP7D2 shRNA #1 (5′-ggaacctcctatgagtaaa-3′), MAP7D2 shRNA #3 (5′-ctgaagaagttcaatctat-3′), KIF5A shRNA (5′-gagacatcttcaaccacat-3′), KIF5B shRNA (5′-tggagggtaaacttcatga-3′), KIF5C shRNA (5′-tgagatctacttggacaaa-3′), mouse MAP7D2 shRNA (5′-gagacaaagattagccaaa-3′). ShRNAs for TRIM46 and AnkG have been described previously (Kuijpers et al., 2016, van Beuningen et al., 2015).
Transfections and immunofluorescence
HEK293, HeLa, or COS7 cells were diluted in a ratio 1:3 24 hours before transfection. MaxPEI was used for transfection based on manufacture’s protocol and incubated for 24 hours to 36 hours before pull-down experiments.
For the DIV0 hippocampus neuron transfection, the Amaxa Rat Neuron Nucleofector kit (Lonza) was used based on manufacture’s protocol. Briefly, 120K/well neurons were transfected with 2 μg DNA and cultured in Neurobasal medium (Invitrogen) containing 2% B27 (Invitrogen), 0.5 mM glutamine (Invitrogen), 15.6 μM glutamate (Sigam) and 1% penicillin/streptomycin (Invitrogen), after 3 days’ incubation, neurons were fixed for immunofluorescence and imaging.
For neurons older than DIV0, Lipofectamin 2000 (Invitrogen) was used for transfection via manufacture’s protocol. Briefly, 1.8 μg DNA and 3.3 μL Lipofectamin 2000 reagent were mixed in 200 μL Neurobasal medium for 30 minutes. The neuronal growth medium was transferred to a new 12 well plate, and the original plate was filled with fresh Neurobasal medium supplemented with 0.5 mM glutamine. The DNA mix was added into neurons for 45 minutes, then neurons were washed with pre-warmed Neurobasal medium and placed to the new plate with growth medium. Neurons were imaged after 1 day (all the overexpression experiments) or 3 days (all knockdown experiments) transfection.
For immunofluorescence experiments, cells were fixed with 4% PFA (paraformaldehyde) for 10-15 minutes, washed 3 times with PBS (phosphate buffer saline), and incubated with primary antibodies in GDB buffer (0.2% BSA, 0.8 M NaCl, 0.5% Triton X-100, 30 mM phosphate buffer) overnight at 4°C. Cells were then washed with PBS for 3 times, incubated with secondary antibodies in GDB buffer for 45 minutes at room temperature, followed by washing 3 times with PBS and mounting with Vectashield mounting medium (Vector Laboratories).
Fixed cell imaging
After immunofluorescence staining, neurons were imaged using a Nikon Eclipse 80i upright microscope or a Zeiss LSM700 confocal laser scanning microscope. A Plan Fluor 40x NA 1.30 oil objective was used for upright microscope. For the confocal microscope, a Plan Apochromat 20x NA 0.8 dry objective, 40x NA 1.3 oil and 63x NA 1.4 oil objectives were used.
Live cell imaging and Photo bleaching experiments
All the live imaging was performed on a Nikon Eclipse TE2000E inverted microscope equipped with Evolve 512 EMCCD camera (Roper Scientific), spinning disk confocal (Roper Scientific), incubate chamber (Tokai Hit) and MetaMorph 7.7.5 software (Molecular Device) was used for all the live imaging experiments. In order to visualize mitochondria, ER, lysosome and Rab3 vesicles, neurons were transfected with Mito-dsRed, GFP-SBP-Rtn4A, Lamp1-GFP and GFP-Rab3C respectively as described previously. Neurons were maintained in 37°C /5% CO2 and imaged with 1 frame/s for 5 minutes using a Plan Fluor 40x NA 1.3 oil objective for the mitochondria and ER transport, 1 frame/s for 3 minutes using a Plan Apo VC 60x NA 1.3 oil objective for the lysosome transport and 4 frames/s for 40 s by using a Plan Apo VC 100x NA 1.4 oil objective for the Rab3 vesicle transport. For image acquisition of ER movement, fluorescence recovery after photobleaching (FRAP) experiment was performed at the beginning of axons by using the ILas2 system (Roper Scientific). For the mitochondria imaging upon MAP7D2 rescue and distal axon transport experiment, multi-position acquisition setting was used for imaging 3 neurons at the same time, in total for 15-30 minutes, and quantifications were normalized to 5 minutes. For FRAP experiments with all MAP7 family members, movies were taken for 3 minutes at 1 frame/s and indicated areas were bleached at 10 s.
GFP pull-down and mass spectrometry
GFP pull-down experiments were performed using GFP-trap magnetic beads (ChromoTek) against either HEK293 cell lysates or rat brain lysates. HEK293 cells were transfected with different kinesin constructs tagged with GFP at the C terminus or GFP-FRB empty construct as control for 2 days and then lysed by incubating cells for 30 minutes on ice in lysis buffer (20 mM Tris-HCl pH8.0, 150 mM NaCl, 1% Triton X-100 and 1x protease inhibitor cocktail), cell lysates were centrifuged at 13,000 rpm for 10 minutes and the supernatants were incubated with beads for 40 minutes at 4°C. Rat brains were lysed with the same lysis buffer, centrifuged at 16,000 g for 20 minutes and the supernatants were incubated with the beads already bound to kinesins with GFP tagged constructs for 2 hours. After incubation, beads were washed 3 times with wash buffer (25 mM Tris-HCl pH8.0, 100 mM NaCl, 0.1% NP40). For protein elution, beads were boiled with NuPAGE LDS 4 sample buffer (Invitrogen), centrifuged, and supernatants were run on a 4%–12% NuPAGE tris-acetate gel (Invitrogen). Gels were stained with Colloidal Blue kit (Invitrogen), and samples were cut from SDS-PAGE gel lanes for mass spectrometry as described before (Kevenaar et al., 2016).
Biotin-streptavidin pull-down and western blot
Streptavidin pull-down assays were performed by using Dynabeads M-280 streptavidin beads (Invitrogen). HEK293 cells were transfected with bio-mCherry-MAP7D2 and BirA together with GFP-FRB or GFP-FRB tagged KIF1A-MDC (1-500), KIF5A-MDC (1-566), KIF5B-MDC (1-560) or KIF5C-MDC (1-560). After 24-48 hours, cells were harvested in ice-cold PBS and lysed with lysis buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100 and 1x protease inhibitor cocktail). Cell lysates were centrifuged at 13,000 rpm for 7 minutes and the supernatants were incubated with streptavidin beads which were already blocked by 0.2% Chicken Egg Albumine (Sigma). After incubating for 40 minutes at room teraperature, beads were washed 3-5 times with wash buffer (50mM Tris pH7.5, 150 mM NaCl, 0.5% Triton X-100 and 1x protease inhibitor cocktail). Samples were eluted with SDS/DTT sample buffer and boiled for subsequent western blot assay.
For the western blot assays, samples were loaded into 10% SDS-PAGE gels and transferred to nitrocellulose membrane. Membranes were blocked with 2% BSA (bovine serum albumin) in PBS/0.05% Tween 20. Primary antibodies were diluted in blocking buffer and incubated with the membranes overnight at 4°C, washed 3 times with PBS/0.05% Tween 20 and incubated with secondary IRDye 680LT or IRDye 800LT antibodies for 45 minutes at room temperature. Membranes were then washed 3 times with PBS/0.05% Tween 20 and scanned on Odyssey Infrared Imaging system (LI-COR Biosciences).
Slice immunofluorescence and imaging
Mice brain slices from 4 days organotypic cultures were fixed with 4% paraformaldehyde in PBS. Fixed slices were then permeabilized and blocked in 10% Normal Goat Serum (NGS)/0.2% Triton X-100/PBS. The signal was increased by staining with an anti-GFP antibody overnight, followed by secondary Alexa 488 antibody staining. Slices were washed 4 times for 15 min in PBS in between. Slices were mounted using Vectashield mounting medium (Vector Laboratories) containing DAPI. Z stack acquisitions were taken using a LSM700 (Zeiss) confocal microscope equipped with a Plan-Apochromat 20x NA 0.8 objective with a 0.5x magnification.
Quantification and Statistical Analysis
All statistical details including the definitions of n, numbers of n and statistical tests performed can be found in each figure and figure legends. Sigma Plot or GraphPad was used for graphs and statistics. Unpaired t-test or Mann-Whitney U test was performed for statistics and p < 0.05 was considered significant.
Image processing and analysis were performed using ImageJ, MATLAB, and Adobe Photoshop. Phylogenetic analysis Conserved Coiled coil motif within MAP7 family proteins was defined by using Pfam and Coiled Coils Prediction (Prabi), while MAP7 domain was defined using Pfam.
Protein localizations in neurons
To study protein localization in neurons, images stained for different axonal markers were obtained by confocal microscope, tracings were made along axons by using segmented lines in ImageJ and average intensity profiles were normalized by MATLAB program as previously described (van Beuningen et al., 2015). For the endogenous MAP7D2 and AnkG distributions, the relative alignment of intensity profiles were made according to the position of maximum intensity in the AnKG staining.
Analysis of polarity index
MAP7 family members were expressed in DIV9 neurons, fixed at DIV10, stained for AnkG and TRIM46. Average intensity of 20 μm in the AIS or dendrite was measured. Rigor-KIF5A-GFP was expressed in DIV1 neurons together with control or MAP7D2 shRNAs, and fixed at DIV4, stained for TRIM46 for marker of the axon, average intensity of 10 μm in the AIS or dendrite was measured, at least 2 dendrites were included and background was subtracted. Polarity index was calculated with the formula: PI = (Id-Ia)/(Id+Ia). Id corresponds to mean dendrite intensity, while Ia is the mean AIS intensity. PI > 0 indicates the polarization is biased toward dendrite and PI < 0 to the AIS.
Analysis of endogenous protein
To validate the efficiency of MAP7D2 shRNAs, neurons transfected with control or shRNAs were stained with MAP7D2 antibody. The staining intensity was calculated by measuring the average intensity of the proximal axons (50 μm) and subtracting the intensity of areas adjacent to the axons with the same length, leaving a final intensity value per axon. For TRIM46 and AnkG level analysis, the average intensity of 20 μm in the AIS in the transfected neuron was normalized to the same region of non-transfected neighboring neuron. For the MAP7D2 shRNA specificity experiment, MAP7D3 (average intensity at the proximal axon of around 20 μm) and MAP7 (average soma intensity) were measured.
Analysis of FRAP experiments
For the FRAP analysis, the mean intensity of bleached areas were correct by subtracting background with the same ROI region frame by frame and calculated as I(t) = I_(bleached region) (t) – I_background (t). The normalization of recovery R_norm (t) was calculated according to the formula:
I (0) corresponds to the mean intensity of regions directly after bleaching, < I(Δt) > -intensity averaged over 5 frames before bleaching. To correct bleaching due to imaging, the non-bleached region next to the bleached region was account and calculated:
I_control corresponds to the mean intensity of the non-bleached region and time interval Δt denotes 5 initial frames before bleaching. The curves were made by averaging each frame the recovery rate.
Quantification of ex vivo neuronal migration
The degree of neuronal migration was quantified as described previously (Hand et al., 2005). Briefly, the positions of the cell bodies of GFP-positive neurons within the cortex were measured by ImageJ plugin, the Analyze Particle. The relative position of neurons at the regions of interest was recorded in terms of distance between the ventricle and the pial surface. The position information, including neurons across the entire Z series in the electroporated neocortial region together with the top (pial surface) and bottom (ventricle) boundaries, were imported to Excel. Based on the relative position of each neuron and the total numbers of neurons counted, the radial cell distribution along the radial axis was obtained by further data processing using an Excel macro and expressed as percentage of migration.
Quantification of axon branching
To quantify the axon branches of each neuron, an ImageJ plugin, NeuronJ (Meijering et al., 2004) was used for tracing the morphology of axon.
Quantification of axon formation by electroporation or Taxol inducement
For the analysis of axon formation by electroporation, neurons were stained with either TAU or TRIM46 as markers for polarization. For the Taxol inducement of axon formation, DIV3 neurons were transfected with Psuperb, MAP7D2 shRNA together with GFP as described before, after 1 day, a concentration of 20 nM Taxol was added to neurons for maintaining 48 hours. Neurons were stained with TRIM46 as an axon marker, imaged on the confocal microscope, and images were analyzed using ImageJ software.
Quantifications of organelle and vesicle movement
To quantify organelle dynamics, kymographs were made using Kymoreslicewide plugins in ImageJ on the proximal or distal axons. For the ER movement, FRAP was used to remove the background.
Acknowledgments
We thank Dr. Ginny Farías for sharing Rigor-KIF5A, ER, and lysosome constructs. The work was supported by Chinese Scholarship Council scholarship (CSC; to Y.C. and X.P.) and the Netherlands Organization for Scientific Research (NWO-ALW-VICI 865.10.010; to C.C.H.), the Netherlands Organization for Health Research and Development (ZonMW-TOP 912.16.058; to C.C.H.), and the European Research Council (ERC) (ERC-consolidator 617050 to C.C.H.).
Author Contributions
X.P. designed and performed the experiments, analyzed results, and wrote the manuscript; Y.C. performed pull-down and Rigor KIF5A experiments; R.S. performed the mass spectrometry experiment; L.W. and S.P. performed ex vivo electroporation and organotypic slice cultures; P.J.H., M.M., and L.W. provided critical reagents; A.A. gave advice and edited the manuscript; M.H. and C.C.H. designed experiments, supervised the research, and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: February 19, 2019
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.celrep.2019.01.084.
Contributor Information
Martin Harterink, Email: m.harterink@uu.nl.
Casper C. Hoogenraad, Email: c.hoogenraad@uu.nl.
Supplemental Information
References
- Barlan K., Lu W., Gelfand V.I. The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 2013;23:317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimella C., Baschirotto C., Arnoldi A., Tonelli A., Tenderini E., Airoldi G., Martinuzzi A., Trabacca A., Losito L., Scarlato M. Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2. Clin. Genet. 2012;82:157–164. doi: 10.1111/j.1399-0004.2011.01717.x. [DOI] [PubMed] [Google Scholar]
- De Paola V., Arber S., Caroni P. AMPA receptors regulate dynamic equilibrium of presynaptic terminals in mature hippocampal networks. Nat. Neurosci. 2003;6:491–500. doi: 10.1038/nn1046. [DOI] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., Britt D.J., Guo X., Bonifacino J.S. Sorting of dendritic and axonal vesicles at the pre-axonal exclusion zone. Cell Rep. 2015;13:1221–1232. doi: 10.1016/j.celrep.2015.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., Bonifacino J.S. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 2017;114:E2955–E2964. doi: 10.1073/pnas.1616363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaud E., Caous R., Pascal A., Bazile F., Gagné J.-P., Huet S., Poirier G.G., Chrétien D., Richard-Parpaillon L., Giet R. Ensconsin/Map7 promotes microtubule growth and centrosome separation in Drosophila neural stem cells. J. Cell Biol. 2014;204:1111–1121. doi: 10.1083/jcb.201311094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy L.F., Katrukha E.A., Grigoriev I., Jaarsma D., Kapitein L.C., Akhmanova A., Hoogenraad C.C. MAP2 defines a pre-axonal filtering zone to regulate KIF1- versus KIF5-dependent cargo transport in sensory neurons. Neuron. 2017;94:347–362. doi: 10.1016/j.neuron.2017.03.046. [DOI] [PubMed] [Google Scholar]
- Hand R., Bortone D., Mattar P., Nguyen L., Heng J.I., Guerrier S., Boutt E., Peters E., Barnes A.P., Parras C. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Milstein A.D., Ethell I.M., Henkemeyer M., Sheng M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat. Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Hooikaas P.J., Martin M., Kuijntjes G.-J., Peeters C.A.E., Katrukha E.A., Ferrari L., Stucchi R., Verhagen D.G.F., van Riel W.E., Altelaar A.F.M. MAP7 family proteins are microtubule-tethered allosteric activators of kinesin-1. bioRxiv. 2018 [Google Scholar]
- Kapitein L.C., Schlager M.A., Kuijpers M., Wulf P.S., van Spronsen M., MacKintosh F.C., Hoogenraad C.C. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr. Biol. 2010;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Karasmanis E.P., Phan C.-T., Angelis D., Kesisova I.A., Hoogenraad C.C., McKenney R.J., Spiliotis E.T. Polarity of neuronal membrane traffic requires sorting of kinesin motor cargo during entry into dendrites by a microtubule-associated septin. Dev. Cell. 2018;46:204–218. doi: 10.1016/j.devcel.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar J.T., Bianchi S., van Spronsen M., Olieric N., Lipka J., Frias C.P., Mikhaylova M., Harterink M., Keijzer N., Wulf P.S. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 2016;26:849–861. doi: 10.1016/j.cub.2016.01.048. [DOI] [PubMed] [Google Scholar]
- Koizumi H., Fujioka H., Togashi K., Thompson J., Yates J.R., 3rd, Gleeson J.G., Emoto K. DCLK1 phosphorylates the microtubule-associated protein MAP7D1 to promote axon elongation in cortical neurons. Dev. Neurobiol. 2017;77:493–510. doi: 10.1002/dneu.22428. [DOI] [PubMed] [Google Scholar]
- Kuijpers M., van de Willige D., Freal A., Chazeau A., Franker M.A., Hofenk J., Rodrigues R.J.C., Kapitein L.C., Akhmanova A., Jaarsma D., Hoogenraad C.C. Dynein regulator NDEL1 controls polarized cargo transport at the axon initial segment. Neuron. 2016;89:461–471. doi: 10.1016/j.neuron.2016.01.022. [DOI] [PubMed] [Google Scholar]
- Lipka J., Kapitein L.C., Jaworski J., Hoogenraad C.C. Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 2016;35:302–318. doi: 10.15252/embj.201592929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E., Jacob M., Sarria J.C., Steiner P., Hirling H., Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Metivier M., Monroy B., Gallaud E., Caous R., Pascal A., Richard-Parpaillon L., Guichet A., Ori-McKenney K., Giet R. The Kinesin-1 binding domain of Ensconsin/MAP7 promotes Kinesin-1 activation in vivo. bioRxiv. 2018 [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E.S., Richardson B.E., Gomes E.R., Baylies M.K. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy B.Y., Sawyer D.L., Ackermann B.E., Borden M.M., Tan T.C., Ori-McKenney K.M. Competition between microtubule-associated proteins directs motor transport. Nat. Commun. 2018;9:1487. doi: 10.1038/s41467-018-03909-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A., Kenna K.P., Renton A.E., Ticozzi N., Faghri F., Chia R., Dominov J.A., Kenna B.J., Nalls M.A., Keagle P., ITALSGEN Consortium. Genomic Translation for ALS Care (GTAC) Consortium. ALS Sequencing Consortium. NYGC ALS Consortium. Answer ALS Foundation. Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Consortium. SLAGEN Consortium. French ALS Consortium. Project MinE ALS Sequencing Consortium Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97:1268–1283. doi: 10.1016/j.neuron.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niida Y., Yachie A. MAP7D2 is a brain expressing X-linked maternal imprinted gene in humans. Nature Precedings. 2011 http://precedings.nature.com/documents/6684/version/1 hdl:10101/npre.2011.6684.1. [Google Scholar]
- Ramkumar A., Jong B.Y., Ori-McKenney K.M. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev. Dyn. 2018;247:138–155. doi: 10.1002/dvdy.24599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz A., Kojima H., Oiwa K., Mandelkow E.-M., Song Y.-H., Mandelkow E. Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J. 2002;21:4896–4905. doi: 10.1093/emboj/cdf503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.-H., Telley I.A., Papadaki P., Ephrussi A., Surrey T., Rørth P. Drosophila ensconsin promotes productive recruitment of Kinesin-1 to microtubules. Dev. Cell. 2008;15:866–876. doi: 10.1016/j.devcel.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Tas R.P., Chazeau A., Cloin B.M.C., Lambers M.L.A., Hoogenraad C.C., Kapitein L.C. Differentiation between oppositely oriented microtubules controls polarized neuronal transport. Neuron. 2017;96:1264–1271.e5. doi: 10.1016/j.neuron.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymanskyj S.R., Yang B., Falnikar A., Lepore A.C., Ma L. MAP7 regulates axon collateral branch development in dorsal root ganglion neurons. J. Neurosci. 2017;37:1648–1661. doi: 10.1523/JNEUROSCI.3260-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymanskyj S.R., Yang B.H., Verhey K.J., Ma L. MAP7 regulates axon morphogenesis by recruiting kinesin-1 to microtubules and modulating organelle transport. eLife. 2018;7:e36374. doi: 10.7554/eLife.36374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- van Beuningen S.F.B., Will L., Harterink M., Chazeau A., van Battum E.Y., Frias C.P., Franker M.A.M., Katrukha E.A., Stucchi R., Vocking K. TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron. 2015;88:1208–1226. doi: 10.1016/j.neuron.2015.11.012. [DOI] [PubMed] [Google Scholar]
- van Spronsen M., Mikhaylova M., Lipka J., Schlager M.A., van den Heuvel D.J., Kuijpers M., Wulf P.S., Keijzer N., Demmers J., Kapitein L.C. TRAK/Milton motor-adaptor proteins steer mitochondrial trafficking to axons and dendrites. Neuron. 2013;77:485–502. doi: 10.1016/j.neuron.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Vershinin M., Carter B.C., Razafsky D.S., King S.J., Gross S.P. Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. USA. 2007;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winckler B., Forscher P., Mellman I. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 1999;397:698–701. doi: 10.1038/17806. [DOI] [PubMed] [Google Scholar]
- Witte H., Neukirchen D., Bradke F. Microtubule stabilization specifies initial neuronal polarization. J. Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woźniak M.J., Bola B., Brownhill K., Yang Y.-C., Levakova V., Allan V.J. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J. Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.