Abstract
Background
Pain is a subjective experience derived from complex interactions among biological, environmental, and psychosocial pathways. Sex differences in pain sensitivity and chronic pain prevalence are well established. However, the molecular basis underlying these sex dimorphisms are poorly understood particularly with regard to the role of the peripheral nervous system. Here we sought to identify shared and distinct gene networks functioning in the peripheral nervous systems that may contribute to sex differences of pain in rats after nerve injury.
Results
We performed RNA-seq on dorsal root ganglia following chronic constriction injury of the sciatic nerve in male and female rats. Analysis from paired naive and injured tissues showed that 1513 genes were differentially expressed between sexes. Genes which facilitated synaptic transmission in naïve and injured females did not show increased expression in males.
Conclusions
Appreciating sex-related gene expression differences and similarities in neuropathic pain models may help to improve the translational relevance to clinical populations and efficacy of clinical trials of this major health issue.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5512-9) contains supplementary material, which is available to authorized users.
Keywords: Dorsal root ganglion, Sex differences, RNA-seq, Gene expression, Peripheral nervous system, Nerve injury
Background
Once established chronic pain is often resistant to existing treatments and associated with adverse health outcomes such as decreased quality of life [1], alterations in mood [2] and sleep patterns [3], and disability [4]. Sex differences in the susceptibility to most chronic pain conditions are well established [5] and experimental pain studies consistently demonstrate greater pain sensitivity, increased pain ratings, and decreased tolerance to a variety of pain modalities in women versus men [5–9]. However, the mechanisms underpinning these sex dimorphisms are poorly understood.
While the inclusion of women in clinical trials has increased dramatically during the past 20 years, a strong bias towards the exclusive use of male animals in preclinical studies of neuropathic pain has persisted [10, 11]. Arguably, the limited use of females in mechanistic studies of neuropathic pain has obfuscated our understanding, management, and treatment of neuropathic pain in either sex [10, 11]. The few studies that have used rodent models of chronic pain to examine sex-specific differences have primarily focused on the hormonal and neuroimmune effects on pain modulation in the central nervous system (CNS) [12]. For example, activated immune cells (e.g., microglia) in the spinal cord dorsal horn release inflammatory mediators (e.g., cytokines) in response to tissue damage which promote neuronal excitability [13]. Although the role of microglia-neuronal signaling pathways in pain pathophysiology had been characterized using predominantly male mice, recent evidence demonstrated that female and male mice use distinct immune cell types to modulate pain behaviors [14, 15].
Chronic pain following peripheral nerve injury is associated with profound changes in gene expression that alter synaptic plasticity and neuroimmune interactions. The majority of studies have examined gene expression changes exclusively in males [10, 11, 16]. One notable exception is a 2006 study by LaCroix-Fralish and colleagues [17] in which microarrays were used to measure the gene expression in the rat dorsal horn after spinal nerve ligation in both male and female rats. Despite the potential for gene expression to yield insight into the molecular underpinnings of chronic pain, a transcriptome-wide assessment of gene expression in the peripheral nervous system (PNS) after nerve injury in male and female rats has not been reported.
Here we sought to identify the similarities and differences in gene regulation between male and female rats in the PNS following peripheral nerve injury. We compare large-scale transcriptome analyses of dorsal root ganglia (DRGs) from male and female rats, in both naive and a peripheral nerve injury model, and identified differential expression of mediators of neurochemical mechanisms. Our RNA-seq suggests that data sexually dimorphic roles of sensory neurons and glia in the DRG may be an important consideration in clinical drug trials designed to evaluate treatments for chronic pain.
Results
RNA-seq transcriptome profiles of DRGs from gonadally intact, adult male and female rats were measured from naïve animals and animals at 14 days after sciatic chronic constriction injury (CCI) (Fig. 1). Basic biological influences that regulate responses to nociceptive stimuli and the generation of chronic pain have largely been investigated using neuropathic pain models in rodents. In an effort to isolate the pain behaviors from other experimental procedures used to create the model the majority of these prior studies used sham procedures as a comparison group. In sham procedures, all experimental procedures are performed with the exception of nerve injury from mechanical manipulation or administration of an active compound. In the present study, we use naïve animals as a comparison group so that we may capture all changes that are associated with clinical neuropathic pain conditions (e.g., skin incision, damage to nerve terminals, deep muscle tissue with associated inflammatory response).
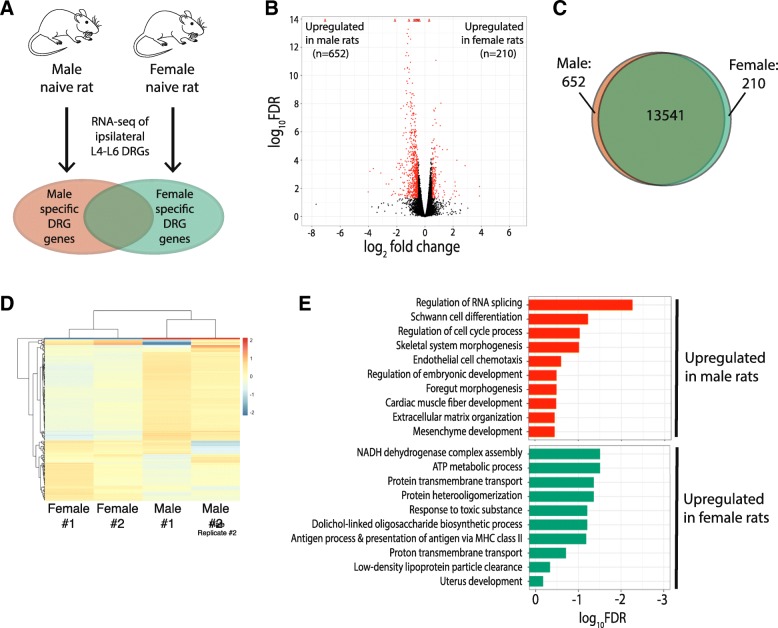
Fig. 1.

Schematic diagram of experimental procedures. Male and female rats were randomly assigned to the naïve group or receive CCI. Total RNA was isolated from the L4-L6 DRG of naïve rats and on day 14 after CCI to the sciatic nerve. Libraries were constructed after poly(A) selection and sequenced. RNA-seq was performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG), defined as genes expressed after CCI versus naïve with a |log2fold change (FC)| > 0.5 and an FDR < 0.05 were compared between male and female rats. DRG = dorsal root ganglion; CCI = chronic constriction injury. Line art of rat in this figure and herein were drawn by KES
Differentially regulated genes in DRGs of naïve male and female rats
To identify sex-specific differences of gene expression in DRG under physiological conditions, we performed RNA-seq and measured all poly(A)-containing transcripts expressed in the L4-L6 DRG of naïve rats (Fig. 2a). Expression levels of 14,403 genes were evaluated and the majority (13,541 genes; 94.0%) showed no significant difference in expression level between males and females (Fig. 2b). Of the 862 genes with expression levels that differed significantly between males and females, 652 (75.6%) showed increased in expression in males versus females and 210 (24.4%) showed increased expression in females versus males (Fig. 2c-d). Lists of these genes with increased and decreased expression in the DRGs between male and female rats are provided as Additional file 1 and Additional file 2. These male and female gene lists were then subject to gene ontology (GO) analysis using Metascape. GO pathways associated with genes that had relatively increased expression in the male naive DRG include Regulation of RNA splicing (GO:0043484), Schwann cell differentiation (GO:0014037), and Regulation of cell cycle process (GO:0010564) (Fig. 2e). GO pathways associated with genes that had relatively increased expression in the female naive DRG were different from that in males and include NADH dehydrogenase complex assembly (GO:0010257), ATP metabolic process (GO:0046034), and Antigen process and presentation of antigen via MHC class II (GO: 0002495).
Fig. 2.
Sex differences of gene expression in DRGs from naïve rats. a Schematic diagram of experiment. RNA-seq performed on ipsilateral L4-L6 DRGs from naïve male and female rats. Differentially expressed genes (DEG) in one sex are identified by a |log2fold change (FC)| > 0.5 and an FDR < 0.05. b Volcano plot of differential gene expression in naïve rats between male and female rats. Significant genes are designated in red. Fold change represents the ratio of gene expression in female to male rats. c Venn diagram representing the number of genes expressed in males (orange), females (green), and in both sexes (overlap). d Heatmap that show the log2FC of the 200 most variable genes between naïve male and naive female rats. e Gene ontology pathways associated with increased expression of genes in male (top; orange) and female (bottom; green) naïve DRG. DRG dorsal root ganglion
Differentially regulated genes in DRGs after nerve injury in male rats
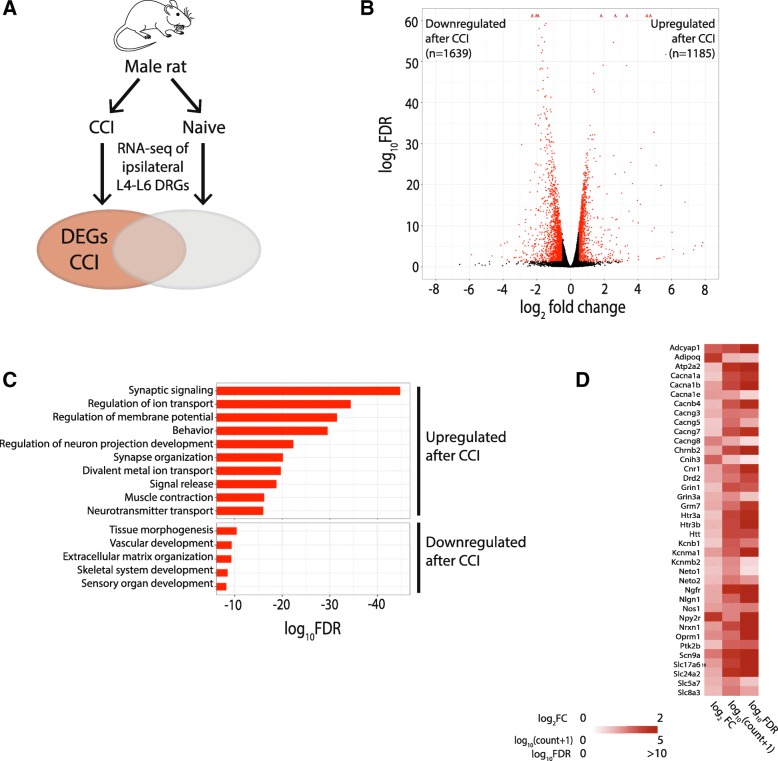
To determine the effects of peripheral nerve injury on gene expression in the DRGs of male rats, we evaluated RNA-seq data obtained 14 days after CCI and compared these expression profiles to those from naive males (Fig. 3a). Compared to naive DRGs, DRGs from male CCI rats differentially expressed 2824 genes (19.5%) (Fig. 3b). Of these 2824 differential genes, 1185 genes (42.0%) were upregulated after injury and 1639 genes (58.0%) were downregulated after injury (Fig. 3b). Despite the use of naïve rats as the comparison group, we found that a large number of upregulated genes after CCI (e.g., Reg3b, Vgf, Atf3, Cacna2d1, Gal, Npy, Gap43) are the same as those previously reported to be upregulated in the DRG after nerve injury which used sham-operated rats as the comparison group [16, 18]. The top 25 upregulated and downregulated genes after CCI ranked by the false discovery rate (FDR) are listed in Table 1 and Table 2, respectively. GO analysis of the upregulated transcripts confirmed significant enrichment among nerve injury and pain related processes (Fig. 3c). A total of 38 genes were present in the three most significant GO biological processes and include voltage-gated ion channels (e.g., Cacna1a, Cacna1b, Cacna1e, Scn9a), ligand-gated ion channels (e.g., Chrnb2, Grin1, Htr3a), and G-protein coupled receptors (e.g., Cnr1, Npy2r, Oprm1) (Fig. 3d). Among the 652 genes that showed increased expression in DRGs of naïve male rats compared to female naïve rats, 549 genes (84.2%) were downregulated following CCI whereas only 7 genes (1.1%) were upregulated following CCI.
Fig. 3.
Differential gene expression between CCI and naïve groups in male rats. a Schematic diagram of experiment. Male rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) are defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an adjusted p-value < 0.05. b Volcano plot showing RNA-seq data of DRGs from naïve male rats and male rats following CCI. DEGs are designated in red. Triangles represent genes with extremely high log10FDR values. c Gene ontology pathways associated with increased (top) and decreased (bottom) differential expression in CCI versus naive. d Heatmap that shows the log2FC, log10(gene count + psuedocount of 1), and the –log10FDR for the upregulated genes enriched in the top 3 GO terms shown in c). DRG dorsal root ganglia, CCI chronic constriction injury, FC fold change, FDR false discovery rate
Table 1.
Top 25 differentially upregulated genes following CCI in males
| Ensembl ID | Symbol | Full gene name | log2 Fold Change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000003745 | Atf3 | Activating transcription factor 3 | 2.59 | 0.11 | 1.10E-121 |
| ENSRNOG00000004805 | Stac2 | SH3 and cysteine rich domain 2 | 4.58 | 0.20 | 3.87E-111 |
| ENSRNOG00000006151 | Reg3b | Regenerating family member 3 beta | 4.74 | 0.23 | 1.76E-89 |
| ENSRNOG00000010079 | Car3 | Carbonic anhydrase 3 | 3.32 | 0.17 | 5.73E-80 |
| ENSRNOG00000033531 | Cacna2d1 | Calcium voltage-gated channel auxiliary subunit alpha2delta 1 | 1.80 | 0.10 | 1.89E-71 |
| ENSRNOG00000015156 | Gal | Galanin and GMAP prepropeptide | 2.54 | 0.16 | 2.02E-55 |
| ENSRNOG00000018808 | Vip | Vasoactive intestinal peptide | 5.65 | 0.36 | 1.79E-52 |
| ENSRNOG00000010803 | Gabra5 | Gamma-aminobutyric acid type A receptor alpha 5 subunit | 1.93 | 0.13 | 7.30E-50 |
| ENSRNOG00000056493 | Mybpc1 | Myosin binding protein C, slow type | 3.33 | 0.22 | 8.95E-50 |
| ENSRNOG00000017333 | Syt4 | Synaptotagmin 4 | 1.37 | 0.09 | 6.47E-48 |
| ENSRNOG00000049882 | Adcyap1 | Adenylate cyclase activating polypeptide 1 | 1.34 | 0.09 | 6.37E-44 |
| ENSRNOG00000013654 | Cbln2 | Cerebellin 2 precursor | 1.42 | 0.10 | 2.33E-43 |
| ENSRNOG00000006639 | Scn9a | Sodium voltage-gated channel alpha subunit 9 | 1.16 | 0.09 | 3.63E-35 |
| ENSRNOG00000010478 | Serpina3n | Serine (or cysteine) peptidase inhibitor, clade A, member 3 N | 4.95 | 0.40 | 1.71E-33 |
| ENSRNOG00000015220 | Crtac1 | Cartilage acidic protein 1 | 1.43 | 0.12 | 7.35E-33 |
| ENSRNOG00000007324 | Plxna2 | Plexin A2 | 1.30 | 0.10 | 7.35E-33 |
| ENSRNOG00000004874 | Flrt3 | Fibronectin leucine rich transmembrane protein 3 | 1.18 | 0.10 | 9.78E-33 |
| ENSRNOG00000004067 | Nrcam | Neuronal cell adhesion molecule | 1.05 | 0.09 | 7.15E-32 |
| ENSRNOG00000047466 | Bdnf | Brain-derived neurotrophic factor | 1.34 | 0.11 | 8.21E-32 |
| ENSRNOG00000024832 | Gpr158 | G protein-coupled receptor 158 | 1.26 | 0.10 | 8.77E-32 |
| ENSRNOG00000053334 | Stmn4 | Stathmin 4 | 1.45 | 0.12 | 1.46E-29 |
| ENSRNOG00000018598 | Ankrd1 | Ankyrin repeat domain 1 | 4.19 | 0.36 | 1.47E-29 |
| ENSRNOG00000007354 | Trpa1 | Transient receptor potential cation channel, subfamily A, member 1 | 1.15 | 0.10 | 3.17E-29 |
| ENSRNOG00000017445 | Tubb2b | Tubulin, beta 2B class IIb | 1.00 | 0.09 | 1.83E-28 |
| ENSRNOG00000011696 | Lifr | Leukemia inhibitory factor receptor alpha | 0.93 | 0.08 | 3.32E-28 |
Table 2.
Top 25 differentially downregulated genes following CCI in males
| Ensembl ID | Symbol | Full gene name | log2 Fold Change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000016516 | Mbp | Myelin basic protein | −2.03 | 0.09 | 1.20E-104 |
| ENSRNOG00000018642 | Leng8 | Leukocyte receptor cluster member 8 | −2.32 | 0.12 | 2.45E-80 |
| ENSRNOG00000012906 | Bcas1 | Breast carcinoma amplified sequence 1 | −1.93 | 0.10 | 3.11E-74 |
| ENSRNOG00000020942 | Plekha4 | Pleckstrin homology domain containing A4 | −1.46 | 0.09 | 4.65E-60 |
| ENSRNOG00000026704 | Drp2 | Dystrophin related protein 2 | −1.53 | 0.09 | 1.37E-59 |
| ENSRNOG00000010626 | Sphk1 | Sphingosine kinase 1 | −1.86 | 0.11 | 8.00E-59 |
| ENSRNOG00000025768 | Clk1 | CDC-like kinase 1 | −1.71 | 0.10 | 1.23E-56 |
| ENSRNOG00000018369 | Prx | Periaxin | −1.54 | 0.10 | 1.45E-55 |
| ENSRNOG00000013387 | Tpcn2 | Two pore segment channel 2 | −1.91 | 0.12 | 1.16E-54 |
| ENSRNOG00000030266 | Plekhg2 | Pleckstrin homology and RhoGEF domain containing G2 | −1.67 | 0.11 | 1.58E-53 |
| ENSRNOG00000008782 | Pnisr | PNN interacting serine and arginine rich protein | −1.72 | 0.11 | 6.57E-53 |
| ENSRNOG00000048139 | Ncmap | Noncompact myelin associated protein | −1.65 | 0.11 | 7.10E-51 |
| ENSRNOG00000061731 | Plxnb3 | Plexin B3 | −1.76 | 0.12 | 3.68E-49 |
| ENSRNOG00000003809 | Sat1 | Spermidine/spermine N1-acetyl transferase 1 | −1.36 | 0.09 | 3.78E-49 |
| ENSRNOG00000020763 | Snrnp70 | Small nuclear ribonucleoprotein U1 subunit 70 | −1.66 | 0.11 | 5.41E-49 |
| ENSRNOG00000017309 | Rsrp1 | Arginine and serine rich protein 1 | −1.81 | 0.12 | 9.01E-49 |
| ENSRNOG00000060614 | Pxdn | Peroxidasin homolog (Drosophila) | −1.35 | 0.09 | 7.10E-47 |
| ENSRNOG00000018748 | Slc16a11 | Solute carrier family 16, member 11 | −1.62 | 0.11 | 3.90E-46 |
| ENSRNOG00000020525 | Col5a3 | Collagen type V alpha 3 chain | −1.63 | 0.11 | 4.46E-44 |
| ENSRNOG00000007657 | Col27a1 | Collagen type XXVII alpha 1 chain | −1.93 | 0.13 | 1.02E-43 |
| ENSRNOG00000029535 | Nrbp2 | Nuclear receptor binding protein 2 | −1.46 | 0.10 | 2.00E-42 |
| ENSRNOG00000009951 | Aif1l | Allograft inflammatory factor 1-like | −1.52 | 0.11 | 3.17E-42 |
| ENSRNOG00000011921 | Dusp4 | Dual specificity phosphatase 4 | −1.52 | 0.11 | 4.30E-42 |
| ENSRNOG00000001254 | Col6a2 | Collagen type VI alpha 2 chain | −1.16 | 0.08 | 4.72E-42 |
| ENSRNOG00000006209 | Mbd6 | Methyl-CpG binding domain protein 6 | −1.60 | 0.12 | 1.12E-39 |
Differentially regulated genes in DRGs after nerve injury in female rats
To determine the effects of peripheral nerve injury on gene expression in female rats, we evaluated RNA-seq data at 14 days after CCI, and compared the expression profiles to those from the naive females (Fig. 4a). In comparison to naive DRGs, 817 (5.7%) genes were differentially regulated after nerve injury (Fig. 4b). Of these 817 differential genes, 471 genes (57.6%) were upregulated after injury and 346 genes (42.3%) were downregulated. The top 25 upregulated and downregulated genes after CCI ranked by DR are listed in Table 3 and Table 4, respectively. GO analysis identified that 2 of the top enriched GO biological processes in the differentially expressed genes in females are shared between males and females following nerve injury (i.e., Behavior (GO:0007610), Regulation of Ion Transport (GO:0043269); Fig. 4c). However, even though males and females share enrichment in these biological processes, just 20 and 23% of these genes are common to both males and females (Additional file 3). A total of 130 genes show enrichment in the “Positive regulation of nervous system development” GO biological process and include genes that encode for cytokines (e.g., Il6, Il1r1), chemokines (e.g., Cxcl13, Cxcl9), and enzymes (e.g., Doux2, Gch1, Tnik) (Fig. 4d). Among the 210 genes with increased expression in naive DRGs of female rats compared to the naïve male rats, 30 (14.2%) were downregulated and 4 (1.9%) were upregulated following CCI.
Fig. 4.
Differential gene expression between CCI and naïve groups in female rats. a Schematic diagram of experiment. Female rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) are defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an FDR < 0.05. b Volcano plot showing RNA-seq data of DRGs from naïve female rats and male rats following CCI. DEGs are designated in red. Triangles represent genes with extremely high log10FDR and/or log2FC values. c Gene ontology pathways associated with increased (top) and decreased (bottom) differential expression in CCI versus naïve. d Heatmap that shows the log2FC, log10(gene count + psuedocount of 1), and the –log10FDR for the upregulated genes enriched in the top 2 GO terms shown in c). DRG dorsal root ganglia, CCI chronic constriction injury, FC fold change, FDR false discovery rate
Table 3.
Top 25 differentially upregulated genes following CCI in females
| Ensembl ID | Symbol | Full gene name | log2 Fold Change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000003745 | Atf3 | Activating transcription factor 3 | 3.38 | 0.12 | 5.16E-171 |
| ENSRNOG00000004805 | Stac2 | SH3 and cysteine rich domain 2 | 5.12 | 0.20 | 2.98E-135 |
| ENSRNOG00000006151 | Reg3b | Regenerating family member 3 beta | 5.91 | 0.25 | 2.92E-122 |
| ENSRNOG00000018808 | Vip | Vasoactive intestinal peptide | 5.63 | 0.26 | 8.71E-98 |
| ENSRNOG00000033531 | Cacna2d1 | Calcium voltage-gated channel auxiliary subunit alpha2delta 1 | 1.93 | 0.10 | 1.21E-74 |
| ENSRNOG00000015156 | Gal | Galanin and GMAP prepropeptide | 3.10 | 0.17 | 1.42E-67 |
| ENSRNOG00000001416 | Vgf | VGF nerve growth factor inducible | 1.98 | 0.12 | 1.87E-58 |
| ENSRNOG00000049882 | Adcyap1 | Adenylate cyclase activating polypeptide 1 | 1.87 | 0.12 | 2.61E-51 |
| ENSRNOG00000020136 | Tgm1 | Transglutaminase 1 | 5.11 | 0.34 | 4.71E-47 |
| ENSRNOG00000004874 | Flrt3 | Fibronectin leucine rich transmembrane protein 3 | 1.66 | 0.11 | 1.82E-44 |
| ENSRNOG00000010478 | Serpina3n | Serine (or cysteine) peptidase inhibitor, clade A, member 3 N | 8.15 | 0.60 | 2.29E-39 |
| ENSRNOG00000017333 | Syt4 | Synaptotagmin 4 | 1.26 | 0.09 | 3.28E-37 |
| ENSRNOG00000000065 | Pde6b | Phosphodiesterase 6B | 4.01 | 0.31 | 1.22E-34 |
| ENSRNOG00000003031 | Atp2b4 | ATPase plasma membrane Ca2+ transporting 4 | 1.36 | 0.11 | 3.05E-33 |
| ENSRNOG00000018598 | Ankrd1 | Ankyrin repeat domain 1 | 4.56 | 0.37 | 8.96E-33 |
| ENSRNOG00000019447 | Ecel1 | Endothelin converting enzyme-like 1 | 3.05 | 0.25 | 6.10E-32 |
| ENSRNOG00000001476 | Cldn4 | Claudin 4 | 6.36 | 0.53 | 1.11E-30 |
| ENSRNOG00000017445 | Tubb2b | Tubulin, beta 2B class IIb | 1.16 | 0.10 | 7.43E-28 |
| ENSRNOG00000012422 | Tnik | TRAF2 and NCK interacting kinase | 1.35 | 0.12 | 3.79E-27 |
| ENSRNOG00000013521 | Dhfr | Dihydrofolate reductase | 1.25 | 0.11 | 1.04E-26 |
| ENSRNOG00000005615 | Gadd45a | Growth arrest and DNA-damage-inducible, alpha | 1.22 | 0.11 | 5.32E-25 |
| ENSRNOG00000013496 | Crisp3 | Cysteine-rich secretory protein 3 | 6.65 | 0.61 | 5.43E-25 |
| ENSRNOG00000047466 | Bdnf | Brain-derived neurotrophic factor | 1.27 | 0.12 | 1.89E-24 |
| ENSRNOG00000010803 | Gabra5 | Gamma-aminobutyric acid type A receptor alpha 5 subunit | 1.58 | 0.15 | 2.36E-24 |
| ENSRNOG00000001528 | Gap43 | Growth associated protein 43 | 1.02 | 0.10 | 1.01E-23 |
Table 4.
Top 25 differentially downregulated genes following CCI in females
| Ensembl ID | Symbol | Full gene name | log2 Fold Change | Standard error | FDR |
|---|---|---|---|---|---|
| ENSRNOG00000047098 | Hbb-b1 | Hemoglobin, beta adult major chain | −3.41 | 0.16 | 1.87E-100 |
| ENSRNOG00000061299 | LOC100134871 | Beta globin minor gene | −3.22 | 0.15 | 5.47E-94 |
| ENSRNOG00000045989 | Hba-a1 | Hemoglobin alpha, adult chain 1 | −3.02 | 0.23 | 8.70E-37 |
| ENSRNOG00000000167 | Alas2 | 5′-aminolevulinate synthase 2 | −2.73 | 0.24 | 2.67E-26 |
| ENSRNOG00000020951 | Slc4a1 | Solute carrier family 4 member 1 | −3.09 | 0.31 | 3.26E-20 |
| ENSRNOG00000047321 | Hba2 | Hemoglobin, alpha 2 | −3.23 | 0.34 | 3.87E-19 |
| ENSRNOG00000058105 | Hbb | Hemoglobin subunit beta | −3.14 | 0.36 | 3.24E-16 |
| ENSRNOG00000029886 | Hba1 | Hemoglobin, alpha 1 | −3.15 | 0.36 | 1.68E-15 |
| ENSRNOG00000032002 | Hapln1 | Hyaluronan and proteoglycan link protein 1 | −0.91 | 0.13 | 5.84E-10 |
| ENSRNOG00000024330 | Ngp | Neutrophilic granule protein | −3.22 | 0.48 | 1.97E-09 |
| ENSRNOG00000000814 | Fabp7 | Fatty acid binding protein 7 | −0.73 | 0.11 | 2.23E-09 |
| ENSRNOG00000028707 | Np4 | Defensin NP-4 precursor | −3.14 | 0.47 | 2.35E-09 |
| ENSRNOG00000018958 | Mt3 | Metallothionein 3 | −0.77 | 0.12 | 9.01E-09 |
| ENSRNOG00000048139 | Ncmap | Noncompact myelin associated protein | −0.78 | 0.12 | 1.33E-08 |
| ENSRNOG00000006471 | Pvalb | Parvalbumin | −0.84 | 0.13 | 2.91E-08 |
| ENSRNOG00000017619 | Aldh1a1 | Aldehyde dehydrogenase 1 family, member A1 | −0.98 | 0.16 | 4.22E-08 |
| ENSRNOG00000003809 | Sat1 | Spermidine/spermine N1-acetyl transferase 1 | −0.74 | 0.12 | 8.26E-08 |
| ENSRNOG00000010402 | Hspb2 | Heat shock protein family B (small) member 2 | −1.01 | 0.17 | 1.31E-07 |
| ENSRNOG00000011917 | Cd79b | CD79b molecule | −3.28 | 0.55 | 1.36E-07 |
| ENSRNOG00000016752 | Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 | −0.73 | 0.12 | 1.98E-07 |
| ENSRNOG00000019183 | Alox15 | Arachidonate 15-lipoxygenase | −2.34 | 0.41 | 5.15E-07 |
| ENSRNOG00000013228 | Scrg1 | Stimulator of chondrogenesis 1 | −1.18 | 0.21 | 7.18E-07 |
| ENSRNOG00000010031 | Vtn | Vitronectin | −0.67 | 0.12 | 7.18E-07 |
| ENSRNOG00000010408 | Polr2k | RNA polymerase II subunit K | −0.93 | 0.17 | 1.26E-06 |
| ENSRNOG00000019660 | Spib | Spi-B transcription factor | −2.73 | 0.49 | 1.35E-06 |
Comparison of differentially regulated genes in DRGs after nerve injury between males and females
To identify sex-specific transcriptional changes that occur following peripheral nerve injury, we compared the lists of differentially expressed genes after CCI between male and female rats (Fig. 5a). We first visualized the extent of similarity/dissimilarity in the transcriptional profiles of the individual samples using the first two principal components from principal component analysis of all genes (Fig. 5b). The first two principal components accounted for a total of 79% of the total variance among the samples and produced distinct clusters of the samples by treatment condition (i.e., naive, CCI) and sex. This segregation of samples was confirmed by hierarchical clustering of the top 200 genes that showed the greatest variability across all groups (Fig. 5c). Of the 3127 genes that were differentially regulated between males and females, only 514 genes (16.4%) were common to both males and females regardless of direction of transcriptional changes (Fig. 5d).
Fig. 5.
Comparison of differential gene expression between CCI and naïve groups in male and female rats. a Schematic diagram of experiment. Male and female rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an FDR < 0.05. b Principal component analysis of male and female rats from the naïve and CCI groups. c Heatmap that shows the log2FC of the 200 most differentially expressed genes between the CCI and Naïve groups in female and male rats. d Venn diagram shows the number of genes that are differentially expressed in males (orange) and females (green). Log2FC expression between the CCI and naive males (x-axis) and females (y-axis). Threshold of |log2FC| > 0.5(dashed lines) with an FDR < 0.05 designates DEGs in female rats only (green), male rats only (orange), and in both male and female rats (brown). DRG dorsal root ganglia, CCI chronic constriction injury, FC fold change, FDR false discovery rate
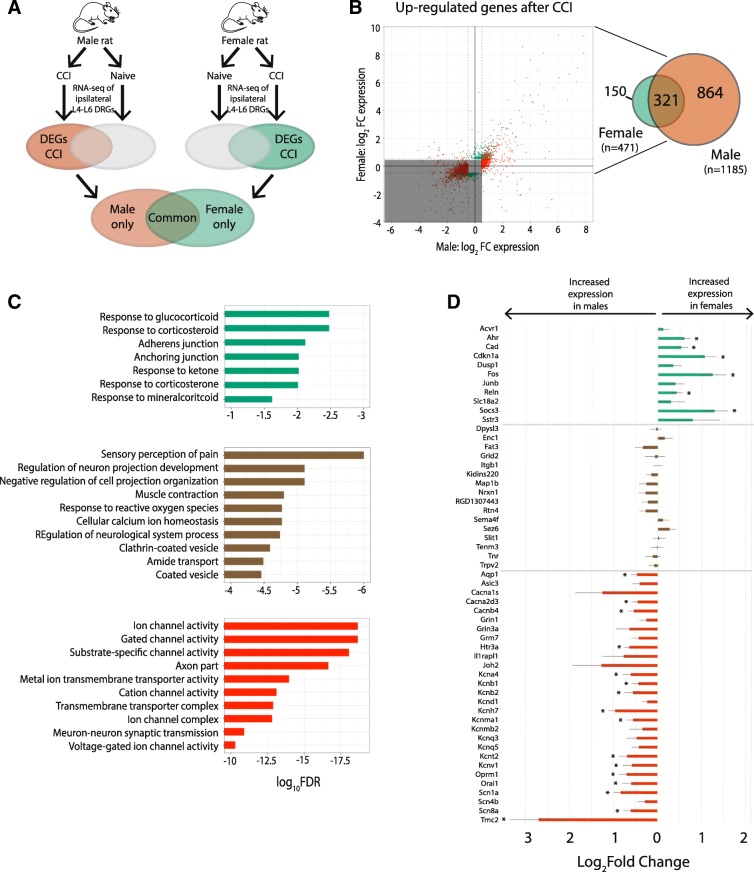
We then divided all upregulated genes that had an FDR < 0.05 and |log2FC| > 0.5 into the following 5 groups: 1) genes upregulated in both males and females, 2) genes upregulated in females with no change of gene expression in males, 3) genes upregulated in males with no change of gene expression in females, 4) genes upregulated in females and downregulated in males, and 5) genes upregulated in males and downregulated in females (Fig. 6b). Of the 1185 genes that were upregulated in males, 321 (27.1%) were also upregulated in females. We used GeneMANIA to identify functional pathways enriched in this gene set (Fig. 6c). The top functional pathways identified in this set of upregulated genes were Sensory Perception of Pain, Regulation of Neuron Projection Development, and Negative Regulation of Cell Projection Organization.
Fig. 6.
Gene ontology in differentially expressed genes that are upregulated after CCI. a Schematic diagram of experiment. Male and female rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an FDR < 0.05. b Log2FC expression between the CCI and naive males (x-axis) and females (y-axis) for DEGs upregulated in CCI versus naive. Threshold of |log2FC| > 0.5 (dashed lines) with an FDR < 0.05 designates DEGs in female rats only (green), male rats only (orange), and in both male and female rats (brown). Venn diagram shows the numbers of DEGs identified in each of these groups. c Functional pathway analysis lists the top gene ontology pathways with the FDR for each term in female rats (top; green), in both male and female rats (middle; brown) and in male rats (bottom; orange). d For each of the top-ranking pathways, the log2FC expression values for DEGs common to several of these pathways are plotted. Genes that significantly differ between males and females with an FDR < 0.05 are indicated with an asterisk. Positive values of the log2FC indicate increased expression in females compared to males and negative values indicate increased expression in males versus females. DRG dorsal root ganglia, CCI chronic constriction injury, FDR false discovery rate, FC fold change
A total of 146 genes were specifically upregulated in females after CCI and had no significant change in gene expression in males. The functional pathways identified from this set of genes include response to glucocorticoid (GO:0051384) and response to corticosteroid (GO:0051412) (Fig. 6c). Some examples of genes associated with these pathways include Ahr, Cad, Cdkn1a, Fos, Reln, and Socs3 (Fig. 6d).
A total of 859 genes were specifically upregulated in males after CCI and had no expression change in females. The functional pathways identified from this set of genes include gated channel activity (GO:002836), ion cannel activity (GO:0022839), substrate-specific channel activity (GO:0022838), and metal ion transmembrane transporter activity (GO:0046873) (Fig. 6c). Examples of genes associated with these pathways include potassium channels (i.e., Kcna4, Kcnb1, Kcnh7, Kcnma1, Kcnq5, Kcnt2, Kcnv1), calcium channels (i.e., Cacna2d3, Cacnb4), Oprm1, and Scn8a (Fig. 6d). A complete list of genes found to be significantly differentially expressed between males and females after CCI is provided in Additional file 4. Four genes were regulated in opposite directions after injury. Adamts4, Cyp2s1, Top2a, and Cenpf were upregulated in female rats and downregulated in male rats after CCI compared with naive controls. Actg2, Casq1, Pla2g2a, Acta2, and Clec1b were upregulated in males and downregulated in females.
We divided the genes downregulated after CCI that had an FDR < 0.05 and |log2FC| > 0.5 into the following 3 groups: 1) genes downregulated in both males and females, 2) genes downregulated in females with no change of gene expression in males, 3) genes downregulated in males with no change of gene expression in females (Additional file 5B). No significant functional pathways were enriched using the genes common to both males and females or in genes significantly downregulated only in females. Genes downregulated only in females showed significant enrichment in the Mitochondrial membrane part GO cellular component (Additional file 5C). Genes downregulated only in males showed significant enrichment in pathways enriched for extracellular matrix-related pathways (Additional file 5C).
Additional file 6 shows the results of biologic validation of 10 genes by qPCR using a separate cohort of animals. We first identified an appropriate endogenous control gene by evaluating 6 candidate reference genes that showed the most stable gene expression among naïve and injured DRG of both sexes. The NormFinder algorithm calculated the lowest stability score for Hmbs which is consistent with stable expression among conditions (Table 5). Due to missing data from one sample Ywhaz could not be included in this analysis. The relative changes in gene expression derived by qPCR were in agreement with those detected by RNA-seq.
Table 5.
Stability values of candidate reference genes in the DRG as determined by NormFinder
| Ranking Order | Gene or pair of genes | Stability |
|---|---|---|
| 1 | Hmbs | 0.13 |
| 2 | Hmbs & Actb | 0.16 |
| 3 | Hmbs & G6pd | 0.18 |
| 4 | Actb & G6pd | 0.19 |
| 5 | Actb | 0.25 |
| 6 | Sdha & Hmbs | 0.29 |
| 7 | G6pd | 0.30 |
| 8 | Sdha & Actb | 0.37 |
| 9 | Sdha & G6 ph | 0.38 |
| 10 | Sdha | 0.55 |
| 11 | Polr2 | 0.95 |
Discussion
Few studies have addressed sex-specific alterations in gene expression in neuropathic pain models, and as a result, the molecular pathways underlying sex differences in neuropathic pain are poorly understood. In addition, gene expression data are available for several types of neuronal tissue from rats, mice, and humans; however, to our knowledge, RNA-seq derived whole transcriptome analysis of the DRG, which contain the cell bodies of primary sensory neurons, has not been performed in these organisms. In the present study, we use RNA-seq to compare the gene expression profiles in lumbar DRGs between male and female rats under both naïve and nerve injury conditions. While additional biological replicates would allow additional genes with more subtle gene expression changes to be identified, our results highlight vast differences in the regulation of genes in PNS that are associated with pain-relevant pathways between females and males after injury. However, both sexes upregulate similar groups of genes associated with neuron regrowth pathways.
Sex differences in gene expression in the naïve DRG
Our gene expression profiles from naive DRGs of male and female rats provide important insights into the normal expression of PNS genes in an unperturbed organism. Remarkably, only 6% of all genes expressed in the naïve DRGs were differentially expressed by sex. Furthermore, these genes that were differentially expressed between sexes did not enrich for pathways directly implicated in either nerve injury or nociception. However, genes that had increased expression in naïve females did show enrichment in immune-related pathways. Increased expression in females of immune-related genes is not unexpected since the connection between sex hormones and immune function is tightly linked. While comprehensive mechanisms for the sexual dimorphism in nociceptive pain have yet to be determined, current evidence supports an important role of sex hormones in pain modulation through the innate and adaptive immune systems [12, 19]. Overall, females mount a stronger humoral, cellular, and inflammatory response than males which is largely attributed to differences in circulating levels of sex hormones especially the decreased levels of androgens [20, 21]. The effects of circulating androgens are known to dampen proinflammatory cytokine production through release of IL-10 by Th1 cells [22] in males. Inflammatory cytokines modulate neuronal excitability through changes in the composition, density, and spatial distribution of ion channels and receptors in the neuronal membrane [23, 24]. Therefore, males may experience less pain as a result of a damped pro-inflammatory immune response after a pain-initiating stimulus. Indeed, evidence from experimental pain studies of pain-free individuals demonstrates a more pronounced temporal summation and greater pain sensitivity in females versus males [5].
Gene expression that showed similar changes after nerve injury between males and females
Genes that were similarly upregulated in the DRG of both sexes after nerve injury were related to neuron regeneration, pain, and intercellular signaling. Following nerve injury, neuronal pathways involved in the regeneration of damaged axons are activated in attempts to recover lost motor, sensory, and autonomic functions. These regenerative mechanisms involve sprouting from the end of the damaged axon toward the denervated segments of the cell, collateral branching of the undamaged neurons around the injury, and long-term reorganization of neuronal circuits in the CNS in response to aberrant peripheral signals [25]. Changes in gene expression and redistribution of receptors and ion channels in the neuronal membrane [26] may partially compensate for incomplete reinnervation; however, altered sensation and loss of fine motor control may produce functional deficits as well as maladaptive alterations (e.g., enhanced spinal reflexes, neuropathic pain).
The few existing studies of sex differences in nerve regeneration suggest that regeneration of myelinated neurons occurs more rapidly in females than males [27–29]. However, Tong and colleagues recently found no sex differences in axon regeneration of axotomized neurons [30]. Our findings support the latter study since after CCI no significant sex-specific transcriptional differences exist in genes known to be involved in axon regeneration and neuron projection development. Discrepancies in neuron regeneration timing may be attributed to differences of nerve injury models, measures of regeneration, and timing of assessments.
Gene expression profiles after injury differ between males and females
After nerve injury, male and female rats showed transcriptional changes in genes that share common GO biological processes (i.e., Behavior, Regulation of ion transport). However, less than one fourth of the genes that were enriched for each of these common processes are shared between males and females. For example, Akap9 (A-kinase anchor protein 9) is a member of the Regulation of Ion Transport GO biologic process and, in our study, is significantly upregulated in females but not in males after CCI. Akap9 interacts with members in signaling transduction pathways (e.g., protein kinase A, serine/threonine kinase protein kinase N, protein phosphatase 1) [31] and loss-of-function mutations are associated with Long QT syndromes [32]. The role of Akap9 in peripheral nerves has not been reported; however, its link to regulating membrane potential suggests Akap9 could contribute to a sex-specific pain mechanism by increasing neuron excitability after nerve injury in females.
Oprm1 is also represented in the Regulation of Ion Transport GO biologic process, but in contrast to Akap9, Oprm1 is significantly upregulated after injury in males but not in females. Importantly, Oprm1 encodes the μ-opioid receptor (MOR) which is targeted by the agonist morphine to produce profound analgesia. Zubieta and colleagues [33] found that, in relation to women, men experienced increased activation of the endogenous analgesic mechanisms in several brain regions during infusion of hypertonic saline into the masseter muscle as demonstrated by increased binding of radiolabeled carfentanil to MORs. Further, several preclinical studies report that morphine produces greater analgesia in male animals in the setting of chronic inflammatory pain [34, 35]. In the present DRG-focused study, the higher Oprm1 expression in males after injury unveiled a possible peripheral mechanism that contributes to sex differences in morphine analgesia and endogenous analgesic mechanisms. Consistent with this finding, we previously demonstrated a relative decrease in efficacy of a peripherally-acting MOR agonist DALDA (dermorphin [D-Arg2, Lys4 (1–4) amide) to inhibit neuropathic pain-related behavior after systemic administration in female rats versus male rats [36].
Another important DRG transcript we found upregulated after CCI in both male and female rats was Csf1 (colony-stimulating factor 1). Csf1 is transported from the DRG to the spinal cord dorsal horn where it binds to its receptor (i.e., Csf1r) located on microglia whose activation promotes the CNS changes necessary for mechanical hypersensitivity [37]. Our results are consistent with a recent study [37] in which Csf1 was reported to be expressed de novo in injured sensory neurons following peripheral nerve injury. Importantly, we found Csf1 expression was 1.7 times higher in females than in males. Further studies will be required to examine if and how sex-specific regulation of Csf1 factors into differences between males and females in their susceptibility to chronic pain syndromes.
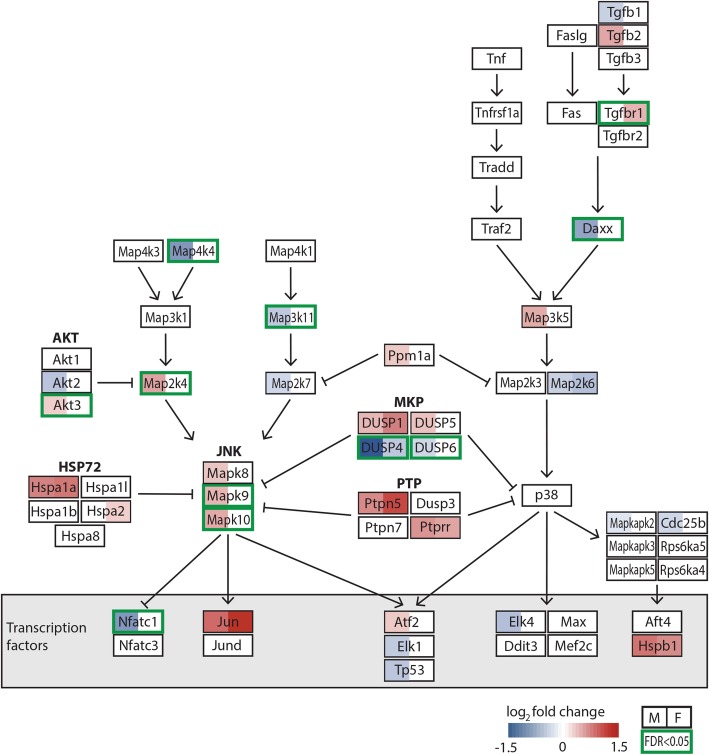
We also identify many growth factors, hormones, cytokines, and neurotransmitters that are differentially up- and downregulated following CCI in males and/or females. Once these molecules bind their cognate receptors on cell membranes, they induce kinase and phosphatase activity in signal transduction cascades which can ultimately modify the expression of specific transcription factors and their respective target genes. Indeed, many of the sex-specific expression differences we identified were in genes that produce key proteins in the mitogen-activated protein kinase (MAPK) pathway (Fig. 7). Alterations of the MAPK pathways are associated with the development of pain hypersensitivity (for review see [38, 39]). Prior studies have attributed MAPK signaling within DRG cells to pain phenotypes ([40–42]). However, sex-based differences were not considered in these studies. For example, a study that examined CCI in male rats found that Jun (c-Jun, V-Jun avian sarcoma virus 17 oncogene homolog) is upregulated in the DRG after CCI and associated with increased expression of proteins that contribute to mechanisms involved in neuropathic pain (e.g., neuropeptide Y, vasoactive intestinal peptide) [43]. While our results are consistent with this study, we found that Jun expression was 1.4 times greater in females than males. We also found that expression of Fos (FBJ murine osteosarcoma viral oncogene homolog) was also significantly upregulated in females after CCI, but downregulated in males. Since c-Jun and c-fos combine to form the AP-1 early response transcription factor, sex-specific differences in the activity of AP-1 may contribute to the differential gene expression observed after CCI.
Fig. 7.
Modified KEGG map of the JNK and p38 MAP kinase signaling pathway showing transcriptional changes. Rectangles represent key genes within the pathway. Coloration in the left half and right half of each rectangle indicates the log2FC in gene expression following CCI versus naïve for male (left) and female (right). Only gene expression changes with an FDR < 0.05 are shown. Red shades indicated increased gene expression after CCI compared with naïve and blue shades indicated decreased gene expression as shown on the color key. Green frames identify genes that show differential gene expression between female and male DRGs with an FDR < 0.05. DRG dorsal root ganglion, CCI chronic constriction injury, FDR false discovery rate, FC fold change
While we were unable to resolve cell type specificity of gene expression from bulk DRG, recent single cell expression studies have identified subpopulations of neuronal and non-neuronal cells within male DRG [44, 45]. Ultimately, understanding how sex-specific differences in PNS gene expression arise at a single cell level could help facilitate drugs regimens that limit off-target effects. While our study was limited to understanding differential gene expression during the maintenance phase of nerve-injury induced chronic pain, earlier time points should yield valuable insights into transcriptional changes that occur during the developmental phase. In addition, future studies are needed to examine the functional implications of differentially expressed genes between males and females in the DRG after CCI.
Conclusions
Our findings build upon existing knowledge of differential pain sensitivity and susceptibility to chronic pain and highlight the profound differences in peripheral mechanisms between males and females after nerve injury which has not been well studied. First, baseline differences in immune function in the PNS may predispose females to developing chronic pain conditions. Second, functional pathways relevant to both sexes that become altered after injury may have important underlying differences in gene expression. Failures of preclinical treatments to translate into humans may be at least partially due to sex-specific alterations in gene expression and existing biases for exclusion of females in preclinical studies. Increased attention to sex differences in preclinical pain studies may improve the translational relevance to clinical populations.
Methods
Animal models
Adult male and female Sprague Dawley rats (RRID: RGD_5508397; Harlan Bioproducts for Science, Indianapolis, IN) were randomly assigned to undergo CCI surgery or no treatment (i.e., naive control). All surgical procedures were performed by the same individual to avoid variation in technique. All animals: Animals were allowed to acclimate for a minimum of 48 h before any experimental procedures, housed 2–3 per cage, and given access ad libitum to food and water. CCI surgery group: CCI surgery to the sciatic nerve was performed as previously described [46]. Under 2% isoflurane, a small incision was made at the level of the mid-thigh. The sciatic nerve was exposed by blunt dissection through the biceps femoris. The nerve trunk proximal to the distal branching point was loosely ligated with four 4–0 silk sutures placed approximately 0.5 mm apart until the epineuria was slightly compressed and minor twitching of the relevant muscles was observed. The muscle layer was closed with 4–0 silk suture and the wound closed with metal clips. On postoperative day 14, naive and CCI rats were euthanized by overdose of isoflurane and the ipsilateral L4-L6 DRGs were quickly dissected, immediately submerged in liquid nitrogen, and stored at − 80 °C until RNA extraction. The ipsilateral L4-L6 DRGs from a single rat were pooled and define each sample. Of note, we did not identify the stage of estrus in the female rats. Existing literature suggests that freely cycling rodents do not exhibit increased variability of study outcomes including pain behaviors [10, 47–50]. In addition, during the 14-day period between surgery and tissue harvest, each rat passes through each stage of the estrus cycle multiple times during which any differences due to the stage of estrus would average out. Veterinary care and animal housing is provided by Johns Hopkins University Animal Services and the Division of Comparative Medicine.
Behavior testing
Mechanical hypersensitivity was measured using von Frey monofilaments as previously described [51]. Animals were assessed at baseline and on day 14 after CCI (Additional file 7). The animals were placed in individual plexiglass cages on top of a wire mesh and allowed to acclimate for 1 h. Tactile stimulation to the midplantar surface of each hind paw was performed using calibrated monofilaments with gradually increasing stiffness (i.e., 0.37, 0.61, 1.23, 2.0, 4.0, 5.93, 10.0, 13.5 g). Each monofilament was applied for 4–6 s in the area between the footpads on the plantar surface of the hind paw. Monofilaments with increasing force were applied until a positive response was observed (e.g., abrupt paw withdrawal, shaking, licking). When a positive response was observed, the monofilament with the next lower force was then applied. The test continued 1) for 5 stimulations after a positive test was observed or 2) the upper or lower range of the von Frey monofilament set was obtained. The paw withdrawal threshold was determined as previously described. If a rat did not achieve at least a 50% reduction in baseline paw withdrawal threshold after 48 h or on day 14, then this animal was not used (n = 3). In addition to one animal of each sex that failed to show adequate reduction of withdrawal thresholds, one naïve animal was remove from analysis due to responses at a much lower mechanical threshold than normal.
RNA isolation
Total RNA was extracted from pooled ipsilateral lumbar DRGs (L4–6) from one rat using the Qiagen RNeasy Mini Prep Kit (Qiagen, Valencia, CA; #74104) with on-column DNase digestion (Qiagen; #79254) according to manufacturer’s instructions. RNA concentration was measured using the Nanodrop ND-2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and RNA integrity was assessed using RNA Nano Eukaryote chips in an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Library construction and sequencing
Five hundred nanograms of total RNA per sample was used to construct sequencing libraries (n = 1 rat/sample and run in independent biological duplicates per group per sex). Strand-specific RNA libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs; # E7420S) after poly(A) selection by the NEBNext poly(A) mRNA Isolation Module (New England Biolabs; #X7490) according to manufacturer’s instructions. Samples were barcoded using the recommended NEBNext Multiplex Oligos (New England Biolabs; #E7490). Size range and quality of libraries were verified on the Agilent 2100 Bioanalyzer (Agilent Technologies; Palo Alto, CA). RNA-seq libraries were quantified by qPCR using the KAPA library quantification kit (KAPA Biosystems). Each library was normalized to 2 nM and pooled in equimolar concentrations. Paired-end sequencing was performed on an Illumina HiSeq2500 (Illumina, San Diego, CA). Two independent biological replicates of 1 rat per group per sex were run for a total of 8 libraries. Libraries were sequenced to an average depth of 47.6 million reads per sample in two batches. Samples from each batch were multiplexed and resequenced in a single HiSeq lane to evaluate for batch effects. Data was preprocessed using the identical pipeline and Pearson correlation coefficients were calculated (Additional file 8). No correction for batch effects was performed.
Data analysis
Sequencing reads were aligned to annotated RefSeq genes in the rat reference genome (rn6) using HISAT2 [52]. Aligned reads were filtered to remove ribosomal RNA and visualized using the Integrative Genomics Viewer [53]. A gene count matrix that contained raw transcript counts for each annotated gene were generated using the featureCounts function of the Subread package in R [54] against the Ensemble rn6 transcriptome. We filtered this matrix by removing genes with zero counts across all samples and relied on the automatic and independent filtering used by DESeq2 to determine the most appropriate threshold for removing genes with low counts [55].
To identify genes that were differentially regulated following nerve injury, transcript counts were normalized and log2 transformed using the default normalization procedures in DESeq2 [55]. The differential expression analysis was first performed separately for each sex using default parameters. This analysis identified differentially expressed genes between the naive and CCI groups within males or females. The interaction of sex on differential gene expression after injury was evaluated by the interaction term included in the design matrix within DESeq2. All downstream analyses on RNA-seq data were performed on data obtained from DESeq2. An adjusted p-value (i.e., FDR) < 0.05 and an absolute log2 fold change > 0.5 were used to define differentially expressed transcripts between naive and injured animals. DESeq2 adjusts for multiple testing by implementing the procedures of Benjamini and Hochberg [55]. Genes with differential expression between groups were then included in pathway analysis to infer their functional roles and relationships.
Gene ontology analysis for enriched biological processes in each set of differentially enriched genes identified by DESeq2 was performed using Metascape [http://metascape.org] [56] with a minimum enrichment of 1.0 and a p-value cutoff of 0.05. Metascape compiles data monthly from publicly available resources (e.g. NCBI, Reactome, GO, KEGG) to provide comprehensive analysis of a list of genes. All significant differentially enriched genes identified by DESeq2 were used to construct network-based functional associations using the GeneMANIA algorithm [57] as a plug-in within Cytoscape version 3.4.0 [58]. Raw and processed sequencing data has been deposited in the NCBI GEO database under accession #GEO100122.
Validation of RNA-seq by qPCR
Total RNA extracted from DRGs of a separate cohort of rats was used to confirm the relationship of gene expression trends between sexes in selected genes by qPCR. As previously described, first-strand cDNA synthesis from 500 ng total RNA in a 20 μl reaction was performed using random hexamer primers and the SuperScript III Reverse Transcriptase (ThermoFisher Scientific) according to manufacturer’s instructions. cDNA was diluted 1:4 with nuclease-free water and stored at − 20 °C. mRNA sequence for each gene was retrieved from NCBI. Forward and reverse primers for each gene were designed using the PrimerQuest Tool (IDT, Coralville, Iowa) to span one or more introns. Primers were obtained through Integrated DNA Technologies (IDT, Coralville, Iowa) and sequences are provided in Additional file 9.
Each 20 μl qPCR reaction consisted of 10ul 2X Power SYBR Green Master Mix (Thermo Fisher Scientific, Waltham, MA), 200 nM each forward and reverse primer, and 2 μl diluted cDNA. PCR of each target was performed using the 7900HT Fast Real-Time PCR system (Applied Biosystems, Waltham, MA) with the following thermocycling conditions: initial denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s. Two biological replicates were assayed for each group and each biological replicate was run in triplicate for each target gene. Nuclease-free water was included in each plate as a no-template control.
PCR efficiencies of each primer set were determined using the slope of standard curves constructed with Cq values obtained from 5-fold serial dilutions of pooled cDNA from DRGs of each group (e.g., male naïve, female naïve, male CCI, female CCI). The efficiency was calculated using they formula: E = 10–1/slope. Dissociation curve analysis was used to identify amplification of non-specific products including primer dimers.
Identification of an endogenous control
To identify a stable endogenous control gene for normalization of the target genes in qPCR we selected six candidate reference genes (i.e., Sdha, Hmbs, Polr2a, G6pd, Ywhaz, Actb) that showed little variation in the RNA-seq data. The expression stability of each candidate reference gene across groups was analyzed using the NormFinder function in R (https://www.moma.dk/normfinder-software) [59]. For each candidate gene, NormFinder (RRID: SCR_003387) calculates a stability value for each gene based on the genes expression variation among samples within the same group and variation between different group. This stability value enables candidate genes to be ranked according to their expression stability among different experimental conditions. The gene or pair of genes with the lowest stability value (i.e., highest expression stability) was selected as the endogenous control for relative gene expression calculations of each target gene.
Statistical analysis
Differential gene expression analysis
Several pairwise comparisons were performed. First, we compared gene expression in the DRGs from naive rats in male and females to identify transcripts that are expressed at significantly higher or lower levels than in the opposite sex. Then, we identified transcripts that were differentially regulated following injury compared with the naive tissue within the same sex (i.e., male CCI versus male naive tissue, female CCI versus female naïve tissue). Last, we compared genes that were differentially regulated following injury between males and females.
Quantitative real-time PCR
Default settings were used to define quantification cycle (Cq) values using SDS software version 2.4.1 (Applied Biosystems, Waltham, MA). The Cq values were averaged over three technical replicates. If the standard deviation of this average was > 0.20, the outlying replicate was removed and the Cq was averaged over the two remaining technical replicates. The 2−ΔΔCT method was used to convert Cq values into relative gene expression for each gene [60].
Additional files
List of genes with increased expression in male versus female rats in the naive DRG. (XLSX 75 kb)
List of genes with increased expression in female versus male rats in the naive DRG. (XLSX 43 kb)
Overlap of differentially expressed genes after CCI between males and females in common functional pathways. (TIF 932 kb)
Complete list of genes that are significantly differentially regulated between males and females. (XLSX 164 kb)
Co-expression networks of differentially expressed genes. A) Schematic diagram of experiment. Male and female rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an adjusted p-value < 0.05. B) Log2FC expression between the CCI and naive males (x-axis) and females (y-axis) for DEGs downregulated in CCI versus naive. Threshold of |log2FC| > 0.5 (dashed lines) with an adjusted p-value< 0.05 designates DEGs in female rats only (green), male rats only (red), and in both male and female rats (brown). Venn diagram shows the numbers of DEGs identified in each of these groups. C-D) Co-expression network for differentially expressed genes in C) female rats only D) and male rats only with decreased gene expression (top) obtained from GeneMANIA. Functional pathway analysis lists the top 5 gene ontology pathways (bottom) with the FDR for each term Co-expression networks of differentially expressed genes. DRG = dorsal root ganglia; CCI = chronic constriction injury; FDR = false discovery rate. DRG = dorsal root ganglia; CCI = chronic constriction injury; FDR = false discovery rate; FC = fold change. (TIF 817 kb)
qPCR validation of RNA-seq data. Quantitative PCR was used to confirm the relationship of gene expression after nerve injury of DRGs from male and female rats: A) Increased relative expression in females only was confirmed in Ahr, Cdkn1a and Tnfaip6, B) Increased relative expression in males only was confirmed in Faim2, Phyhipl, and Trpm6, C) Increased expression in both sexes after injury confirmed in Csf1 and Oprm1, and D) no change in relative expression in Sec62, Stk38l. Values represent the mean standard deviation log2Expression of 2 biological replicates. (TIF 1039 kb)
Paw withdrawal thresholds to mechanical stimulation. (TIF 218 kb)
Scatterplots of the log2FPKM of the initially sequenced batch versus the log2FPKM of the resequenced batch for each sample resequenced. (TIF 506 kb)
List of primers used for qPCR validation. (XLSX 13 kb)
Acknowledgements
We thank members of the Taverna and Guan labs for critical reading of this manuscript and Rakel Tryggvadóttir and Colin Callahan for their technical assistance. The abstract for this study has previously been published as part of the American Pain Society conference proceedings.
Funding
This study was supported by the Blaustein Pain Research and Education Endowment (K.E.S., S.D.T.) and National Institutes of Health grants R01GM106024 (S.D.T.), R01GM118760 (S.D.T.), F32NR015728 (K.E.S.), R01NS070814 (Y.G.), and R21NS099879 (Y.G.), and R01HG006282 (H.J.). The funding agencies had no role in the design of the study, sample collection, analysis or interpretation of data, or in writing the manuscript.
Availability of data and materials
The RNA-seq data are available in the Gene Expression Omnibus repository under accession # GSE100122 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100122).
Abbreviations
- ATP
Adenosine Triphosphate
- CCI
Chronic Constriction Injury
- CNS
Central Nervous System
- DALDA
Dermorphin [D-Arg2, Lys4 (1–4) amide
- DRG
Dorsal Root Ganglion
- FC
Fold Change
- FDR
False Discovery Rate
- GO
Gene Ontology
- IL
Interleukin
- MAPK
Mitogen-activated Protein Kinase
- MHC
Major Histocompatibility Complex
- MOR
Morphine Opioid Receptor
- NADH
Nicotinamide adenine dinucleotide
- PCR
Polymerase Chain Reaction
- PNS
Peripheral Nervous System
- RNA
Ribonucleic Acid
Authors’ contributions
KES, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article; WZ, Analysis and interpretation of data, Drafting or revising the article; ZJ, Analysis and interpretation of data; Drafting or revising the article; SH, Acquisition of data and assisting with experiments and data analysis; ZC, Acquisition of data and assisting with experiments and data analysis; HJ, Analysis and interpretation of data, Drafting or revising the article; YG, Conception and design, Analysis and interpretation of data, Drafting or revising the article; SDT, Conception and design, Analysis and interpretation of data, Drafting or revising the article. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures involving animals were reviewed and approved by the Johns Hopkins Animal Care and use Committee (Approval #RA18M84) and were performed in accordance with NIH Guide for the Care and Use of Laboratory Animals.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kimberly E. Stephens, Email: kstephe3@jhmi.edu
Weiqiang Zhou, Email: kenandzhou@hotmail.com.
Zhicheng Ji, Email: zhichengji@gmail.com.
Zhiyong Chen, Email: zchen56@jhmi.edu.
Shaoqiu He, Email: she11@jhmi.edu.
Hongkai Ji, Email: hji@jhu.edu.
Yun Guan, Email: yguan1@jhmi.edu.
Sean D. Taverna, Email: staverna@jhmi.edu
References
- 1.Pinto-Meza A, Fernandez A, Fullana MA, Haro JM, Palao D, Luciano JV, Serrano-Blanco A. Impact of mental disorders and chronic physical conditions in health-related quality of life among primary care patients: results from an epidemiological study. Qual Life Res. 2009;18(8):1011–1018. doi: 10.1007/s11136-009-9522-y. [DOI] [PubMed] [Google Scholar]
- 2.Schaefer C, Mann R, Sadosky A, Daniel S, Parsons B, Nieshoff E, Tuchman M, Nalamachu S, Anschel A, Stacey BR. Burden of illness associated with peripheral and central neuropathic pain among adults seeking treatment in the United States: a patient-centered evaluation. Pain Med. 2014;15(12):2105–2119. doi: 10.1111/pme.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheatle MD, Foster S, Pinkett A, Lesneski M, Qu D, Dhingra L. Assessing and managing sleep disturbance in patients with chronic pain. Anesthesiol Clin. 2016;34(2):379–393. doi: 10.1016/j.anclin.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Global Burden of Disease Study C Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2015;386(9995):743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fillingim RB, King CD, Ribeiro-Dasilva MC, Rahim-Williams B, Riley JL., 3rd Sex, gender, and pain: a review of recent clinical and experimental findings. J Pain. 2009;10(5):447–485. doi: 10.1016/j.jpain.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartley EJ, Fillingim RB. Sex differences in pain: a brief review of clinical and experimental findings. Br J Anaesth. 2013;111(1):52–58. doi: 10.1093/bja/aet127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouhassira D, Lanteri-Minet M, Attal N, Laurent B, Touboul C. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain. 2008;136(3):380–387. doi: 10.1016/j.pain.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Torrance N, Smith BH, Bennett MI, Lee AJ. The epidemiology of chronic pain of predominantly neuropathic origin. Results from a general population survey. J Pain. 2006;7(4):281–289. doi: 10.1016/j.jpain.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Mogil JS. Sex differences in pain and pain inhibition: multiple explanations of a controversial phenomenon. Nat Rev Neurosci. 2012;13(12):859–866. doi: 10.1038/nrn3360. [DOI] [PubMed] [Google Scholar]
- 10.Mogil JS, Chanda ML. The case for the inclusion of female subjects in basic science studies of pain. Pain. 2005;117(1–2):1–5. doi: 10.1016/j.pain.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci Biobehav Rev. 2011;35(3):565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosen S, Ham B, Mogil JS. Sex differences in neuroimmunity and pain. J Neurosci Res. 2017;95(1–2):500–508. doi: 10.1002/jnr.23831. [DOI] [PubMed] [Google Scholar]
- 13.Grace PM, Hutchinson MR, Maier SF, Watkins LR. Pathological pain and the neuroimmune interface. Nat Rev Immunol. 2014;14(4):217–231. doi: 10.1038/nri3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorge RE, LaCroix-Fralish ML, Tuttle AH, Sotocinal SG, Austin JS, Ritchie J, Chanda ML, Graham AC, Topham L, Beggs S, et al. Spinal cord toll-like receptor 4 mediates inflammatory and neuropathic hypersensitivity in male but not female mice. J Neurosci. 2011;31(43):15450–15454. doi: 10.1523/JNEUROSCI.3859-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorge RE, Mapplebeck JC, Rosen S, Beggs S, Taves S, Alexander JK, Martin LJ, Austin JS, Sotocinal SG, Chen D, et al. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci. 2015;18(8):1081–1083. doi: 10.1038/nn.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaCroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152(8):1888–1898. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 17.LaCroix-Fralish ML, Tawfik VL, Spratt KF, DeLeo JA. Sex differences in lumbar spinal cord gene expression following experimental lumbar radiculopathy. J Mol Neurosci. 2006;30(3):283–295. doi: 10.1385/JMN:30:3:283. [DOI] [PubMed] [Google Scholar]
- 18.Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133(9):2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorge RE, Totsch SK. Sex differences in pain. J Neurosci Res. 2017;95(6):1271–1281. doi: 10.1002/jnr.23841. [DOI] [PubMed] [Google Scholar]
- 20.Gaillard RC, Spinedi E. Sex- and stress-steroids interactions and the immune system: evidence for a neuroendocrine-immunological sexual dimorphism. Domest Anim Endocrinol. 1998;15(5):345–352. doi: 10.1016/s0739-7240(98)00028-9. [DOI] [PubMed] [Google Scholar]
- 21.Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum Reprod Update. 2005;11(4):411–423. doi: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- 22.Bebo BF, Jr, Schuster JC, Vandenbark AA, Offner H. Androgens alter the cytokine profile and reduce encephalitogenicity of myelin-reactive T cells. J Immunol. 1999;162(1):35–40. [PubMed] [Google Scholar]
- 23.von Banchet GS, Kiehl M, Schaible HG. Acute and long-term effects of IL-6 on cultured dorsal root ganglion neurones from adult rat. J Neurochem. 2005;94(1):238–248. doi: 10.1111/j.1471-4159.2005.03185.x. [DOI] [PubMed] [Google Scholar]
- 24.Inoue A, Ikoma K, Morioka N, Kumagai K, Hashimoto T, Hide I, Nakata Y. Interleukin-1beta induces substance P release from primary afferent neurons through the cyclooxygenase-2 system. J Neurochem. 1999;73(5):2206–2213. [PubMed] [Google Scholar]
- 25.Hanz S, Fainzilber M. Retrograde signaling in injured nerve--the axon reaction revisited. J Neurochem. 2006;99(1):13–19. doi: 10.1111/j.1471-4159.2006.04089.x. [DOI] [PubMed] [Google Scholar]
- 26.Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci. 2002;5(Suppl):1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 27.Kovacic U, Sketelj J, Bajrovic FF. Sex-related differences in recovery of cutaneous nociception after end-to-side nerve repair in the rat. J Plast Reconstr Aesthet Surg. 2009;62(6):806–813. doi: 10.1016/j.bjps.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 28.Kovacic U, Sketelj J, Bajrovic FF. Sex-related difference in collateral sprouting of nociceptive axons after peripheral nerve injury in the rat. Exp Neurol. 2003;184(1):479–488. doi: 10.1016/s0014-4886(03)00269-3. [DOI] [PubMed] [Google Scholar]
- 29.Kovacic U, Zele T, Osredkar J, Sketelj J, Bajrovic FF. Sex-related differences in the regeneration of sensory axons and recovery of nociception after peripheral nerve crush in the rat. Exp Neurol. 2004;189(1):94–104. doi: 10.1016/j.expneurol.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 30.Tong LL, Ding YQ, Jing HB, Li XY, Qi JG. Differential motor and sensory functional recovery in male but not female adult rats is associated with remyelination rather than axon regeneration after sciatic nerve crush. Neuroreport. 2015;26(7):429–437. doi: 10.1097/WNR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J Biol Chem. 1999;274(24):17267–17274. doi: 10.1074/jbc.274.24.17267. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Marquardt ML, Tester DJ, Sampson KJ, Ackerman MJ, Kass RS. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc Natl Acad Sci U S A. 2007;104(52):20990–20995. doi: 10.1073/pnas.0710527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zubieta JK, Smith YR, Bueller JA, Xu Y, Kilbourn MR, Jewett DM, Meyer CR, Koeppe RA, Stohler CS. Mu-opioid receptor-mediated antinociceptive responses differ in men and women. J Neurosci. 2002;22(12):5100–5107. doi: 10.1523/JNEUROSCI.22-12-05100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loyd DR, Murphy AZ. Sex differences in the anatomical and functional organization of the periaqueductal gray-rostral ventromedial medullary pathway in the rat: a potential circuit mediating the sexually dimorphic actions of morphine. J Comp Neurol. 2006;496(5):723–738. doi: 10.1002/cne.20962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Traub RJ, Murphy AZ. Persistent pain model reveals sex difference in morphine potency. Am J Physiol Regul Integr Comp Physiol. 2006;291(2):R300–R306. doi: 10.1152/ajpregu.00022.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiwari V, Yang F, He SQ, Shechter R, Zhang C, Shu B, Zhang T, Tiwari V, Wang Y, Dong X, et al. Activation of peripheral mu-opioid receptors by Dermorphin [D-Arg2, Lys4] (1-4) amide leads to modality-preferred inhibition of neuropathic pain. Anesthesiology. 2016;124(3):706–720. doi: 10.1097/ALN.0000000000000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan Z, Kuhn JA, Wang X, Colquitt B, Solorzano C, Vaman S, Guan AK, Evans-Reinsch Z, Braz J, Devor M, et al. Injured sensory neuron-derived CSF1 induces microglial proliferation and DAP12-dependent pain. Nat Neurosci. 2016;19(1):94–101. doi: 10.1038/nn.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji RR, Gereau RW, Malcangio M, Strichartz GR. MAP kinase and pain. Brain Res Rev. 2009;60(1):135–148. doi: 10.1016/j.brainresrev.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obata K, Noguchi K. MAPK activation in nociceptive neurons and pain hypersensitivity. Life Sci. 2004;74(21):2643–2653. doi: 10.1016/j.lfs.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 40.Montague K, Simeoli R, Valente J, Malcangio M. A novel interaction between CX3CR1 and CCR2 signalling in monocytes constitutes an underlying mechanism for persistent vincristine-induced pain. J Neuroinflammation. 2018;15(1):101. doi: 10.1186/s12974-018-1116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Li C, Deng Z, Du E, Xu C. Long non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience. 2018;374:214–222. doi: 10.1016/j.neuroscience.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 42.Sun W, Zhang L, Li R. Overexpression of miR-206 ameliorates chronic constriction injury-induced neuropathic pain in rats via the MEK/ERK pathway by targeting brain-derived neurotrophic factor. Neurosci Lett. 2017;646:68–74. doi: 10.1016/j.neulet.2016.12.047. [DOI] [PubMed] [Google Scholar]
- 43.Son SJ, Lee KM, Jeon SM, Park ES, Park KM, Cho HJ. Activation of transcription factor c-Jun in dorsal root ganglia induces VIP and NPY upregulation and contributes to the pathogenesis of neuropathic pain. Exp Neurol. 2007;204(1):467–472. doi: 10.1016/j.expneurol.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 45.Li CL, Li KC, Wu D, Chen Y, Luo H, Zhao JR, Wang SS, Sun MM, Lu YJ, Zhong YQ, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26(1):83–102. doi: 10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 47.Prendergast BJ, Onishi KG, Zucker I. Female mice liberated for inclusion in neuroscience and biomedical research. Neurosci Biobehav Rev. 2014;40:1–5. doi: 10.1016/j.neubiorev.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Klein SL, Schiebinger L, Stefanick ML, Cahill L, Danska J, de Vries GJ, Kibbe MR, McCarthy MM, Mogil JS, Woodruff TK, et al. Opinion: sex inclusion in basic research drives discovery. Proc Natl Acad Sci U S A. 2015;112(17):5257–5258. doi: 10.1073/pnas.1502843112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vacca V, Marinelli S, Pieroni L, Urbani A, Luvisetto S, Pavone F. 17beta-estradiol counteracts neuropathic pain: a behavioural, immunohistochemical, and proteomic investigation on sex-related differences in mice. Sci Rep. 2016;6:18980. doi: 10.1038/srep18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vacca V, Marinelli S, Pieroni L, Urbani A, Luvisetto S, Pavone F. Higher pain perception and lack of recovery from neuropathic pain in females: a behavioural, immunohistochemical, and proteomic investigation on sex-related differences in mice. Pain. 2014;155(2):388–402. doi: 10.1016/j.pain.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 51.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 52.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tripathi S, Pohl MO, Zhou Y, Rodriguez-Frandsen A, Wang G, Stein DA, Moulton HM, DeJesus P, Che J, Mulder LC, et al. Meta- and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montojo J, Zuberi K, Rodriguez H, Kazi F, Wright G, Donaldson SL, Morris Q, Bader GD. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics. 2010;26(22):2927–2928. doi: 10.1093/bioinformatics/btq562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 60.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes with increased expression in male versus female rats in the naive DRG. (XLSX 75 kb)
List of genes with increased expression in female versus male rats in the naive DRG. (XLSX 43 kb)
Overlap of differentially expressed genes after CCI between males and females in common functional pathways. (TIF 932 kb)
Complete list of genes that are significantly differentially regulated between males and females. (XLSX 164 kb)
Co-expression networks of differentially expressed genes. A) Schematic diagram of experiment. Male and female rats were randomly assigned to the naïve group or receive CCI. RNA-seq performed on ipsilateral L4-L6 DRGs from each animal. Differentially expressed genes (DEG) defined as genes expressed after CCI versus naïve with a |log2FC| > 0.5 and an adjusted p-value < 0.05. B) Log2FC expression between the CCI and naive males (x-axis) and females (y-axis) for DEGs downregulated in CCI versus naive. Threshold of |log2FC| > 0.5 (dashed lines) with an adjusted p-value< 0.05 designates DEGs in female rats only (green), male rats only (red), and in both male and female rats (brown). Venn diagram shows the numbers of DEGs identified in each of these groups. C-D) Co-expression network for differentially expressed genes in C) female rats only D) and male rats only with decreased gene expression (top) obtained from GeneMANIA. Functional pathway analysis lists the top 5 gene ontology pathways (bottom) with the FDR for each term Co-expression networks of differentially expressed genes. DRG = dorsal root ganglia; CCI = chronic constriction injury; FDR = false discovery rate. DRG = dorsal root ganglia; CCI = chronic constriction injury; FDR = false discovery rate; FC = fold change. (TIF 817 kb)
qPCR validation of RNA-seq data. Quantitative PCR was used to confirm the relationship of gene expression after nerve injury of DRGs from male and female rats: A) Increased relative expression in females only was confirmed in Ahr, Cdkn1a and Tnfaip6, B) Increased relative expression in males only was confirmed in Faim2, Phyhipl, and Trpm6, C) Increased expression in both sexes after injury confirmed in Csf1 and Oprm1, and D) no change in relative expression in Sec62, Stk38l. Values represent the mean standard deviation log2Expression of 2 biological replicates. (TIF 1039 kb)
Paw withdrawal thresholds to mechanical stimulation. (TIF 218 kb)
Scatterplots of the log2FPKM of the initially sequenced batch versus the log2FPKM of the resequenced batch for each sample resequenced. (TIF 506 kb)
List of primers used for qPCR validation. (XLSX 13 kb)
Data Availability Statement
The RNA-seq data are available in the Gene Expression Omnibus repository under accession # GSE100122 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100122).