Abstract
We propose Segment Convolutional Neural Networks (Seg-CNNs) for classifying relations from clinical notes. Seg-CNNs use only word-embedding features without manual feature engineering. Unlike typical CNN models, relations between 2 concepts are identified by simultaneously learning separate representations for text segments in a sentence: preceding, concept1, middle, concept2, and succeeding. We evaluate Seg-CNN on the i2b2/VA relation classification challenge dataset. We show that Seg-CNN achieves a state-of-the-art micro-average F-measure of 0.742 for overall evaluation, 0.686 for classifying medical problem–treatment relations, 0.820 for medical problem–test relations, and 0.702 for medical problem–medical problem relations. We demonstrate the benefits of learning segment-level representations. We show that medical domain word embeddings help improve relation classification. Seg-CNNs can be trained quickly for the i2b2/VA dataset on a graphics processing unit (GPU) platform. These results support the use of CNNs computed over segments of text for classifying medical relations, as they show state-of-the-art performance while requiring no manual feature engineering.
Keywords: natural language processing, medical relation classification, convolutional neural network, machine learning
INTRODUCTION AND RELATED WORK
It is now well established that automated extraction of knowledge from biomedical literature or clinical notes involves accurately identifying not only the conceptual entities, but also the varied relationships among those concepts.1–4 The task generally involves annotating unstructured text with named entities and classifying the relations between these annotated entities. Relation identification has received increasing attention over the past decade, and is critical in applications including clinical decision-making, clinical trial screening, and pharmacovigilance.5–12
Some of the advances in the state-of-the-art clinical natural language processing (NLP) systems for classifying medical relations were documented in the 2010 i2b2/VA challenge workshop, which attracted international teams to address shared tasks on identifying the possible relations between medical problems and treatments, between medical problems and tests, and between pairs of medical problems.13 All participating systems in the 2010 i2b2/VA challenge utilized heavy feature engineering for their machine learning models13; many also harvested features from existing NLP pipelines such as cTakes,14 MetaMap,15 and GeniaTagger.16 All systems combined lexical, syntactic, and semantic features. Some teams complemented their machine learning systems with annotated and/or unannotated external data.17–25 Others supplemented their machine learning systems with rules that capture linguistic patterns of relations.23,25,26 One of the top-performing teams17 performed a follow-up study by employing a composite kernel–based model that consists of concept kernels, connection kernels, and tree kernels in order to map lexical, semantic, and syntactic features onto higher-dimensional space.27 They reported an improvement of 0.01 micro-averaged F-measure (0.731–0.742) on their overall challenge scores.
Unfortunately, systems that use human-engineered features often do not generalize well to new datasets.3,28 Recent studies on applying convolutional neural networks (CNNs) to clinical datasets aimed to automatically learn feature representations to reduce the need for engineered features and have achieved some success on specific tasks, such as medical image analysis.29 Most recently, Sahu et al.30 applied CNN to i2b2/VA relation classification and learned a single sentence-level representation for each relation, making use of embedding, semantic, and syntactic features; however, the top challenge participating systems still maintain state-of-the-art performance.17,19 Their sentence-CNN learns a relation representation for the entire sentence but does not explicitly distinguish the segments that form the relations preceding, concept1, middle, concept2, and succeeding. This is inconsistent with the observation that the 5 segments of text have different roles in determining the relation class.31,32 Thus the motivating question for this study is whether we can design CNNs with only word-embedding features and no manual feature engineering to effectively classify the relations among medical concepts as stated in the clinical narratives. Our system learns one representation for each segment, uses only embedding features, attains an F-measure matching the state-of-the-art system, and performs modestly better than the challenge participating systems.
METHODS AND MATERIALS
Dataset
This work utilized the corpus and target relations from the 2010 i2b2/VA challenge,13 which include relations from the following 3 categories: medical problem–treatment (TrP) relations, medical problem–test (TeP) relations, and medical problem–medical problem (PP) relations. Each category contains a list of possible relations. For example, the PP relation category includes problems that are related to each other (PIP) and that have no relation (None). The supplementary material shows detailed relation descriptions and statistics. For the i2b2/VA relation classification task, the named entities are given, so there is no need to run named entity recognition. The relation challenge data are publicly available through i2b2/VA at https://www.i2b2.org/NLP/Relations/.
Word embeddings
The word embeddings are meaningful real-valued vectors where semantically similar words usually have close embedding vectors. The word embeddings learned by neural networks often capture linguistic regularities and patterns that are useful in language modeling.33 Thus using word-embedding vectors trained from an unsupervised neural language model as features is a popular approach in NLP, especially CNN-based methods.30,34–36 We applied word2vec33 to learn word embeddings from different corpora using the continuous bag-of-words method. We experimented with both the general domain New York Times corpus37 containing 1.9 million documents and the Medical Information Mart for Intensive Care (MIMIC)-III clinical notes corpus38 that contains 2 million clinical notes. Earlier studies aggregated (max- or mean-aggregation) embedding vectors for feature generation,36 which we adopted as baseline models, as shown in Figure 1.
Figure 1.
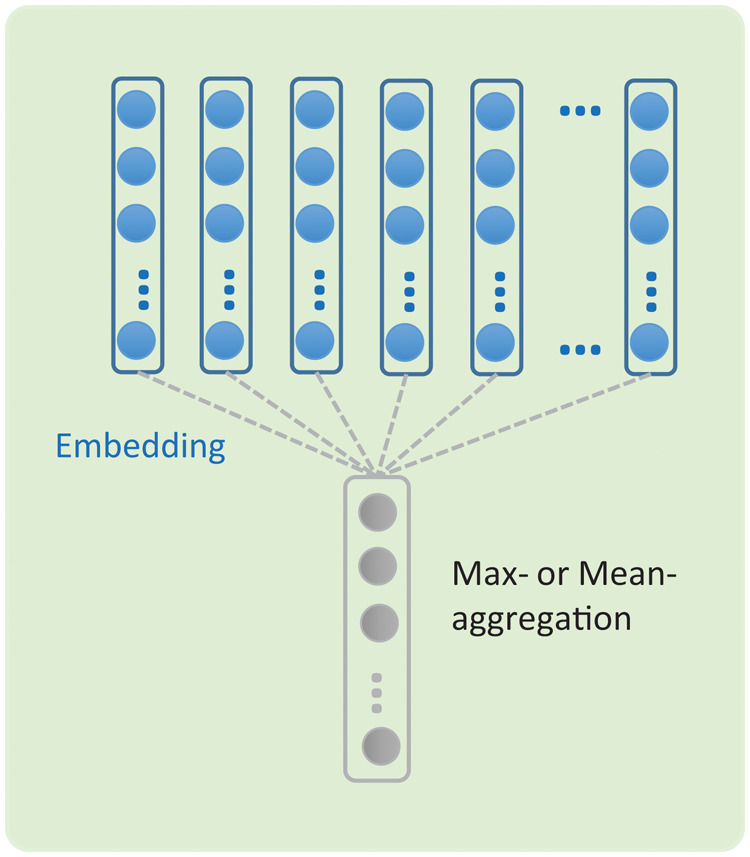
A simple embedding aggregation model.
Sentence-CNN for relation classification
Previously, CNNs have been applied to modeling and classifying sentences and short text.34,39 Relation classification needs finer detail, because one sentence may contain multiple distinct mentions of relations, each with its own concept text and context. One way to represent context is to record the relative positions of individual words to the 2 medical concepts being related.30,40 This approach was used by Sahu et al.30 on i2b2/VA relations, which we reimplemented as a comparison model. Our reimplementation augments the embedding vector of each word by appending 2 integers that indicate its position relative to concept1 and concept2, denoted by and , respectively. For example, in the sentence “Her [neuroimaging studies] revealed evidence of [lumbar stenosis],” “Her” is at −1 distance and “revealed” is at +1 distance away from “neuroimaging studies” (concept1), hence their values are −1 and +1, respectively. For all words in concept1 (“neuroimaging” and “studies”), values are set to 0. We pass a sequence of [embedding; position] vectors to the convolution layer and then a max-pooling layer, termed as a convolution unit in Figure 2 (A). We then input mapped features to a softmax classifier in order to classify the relations.
Figure 2.
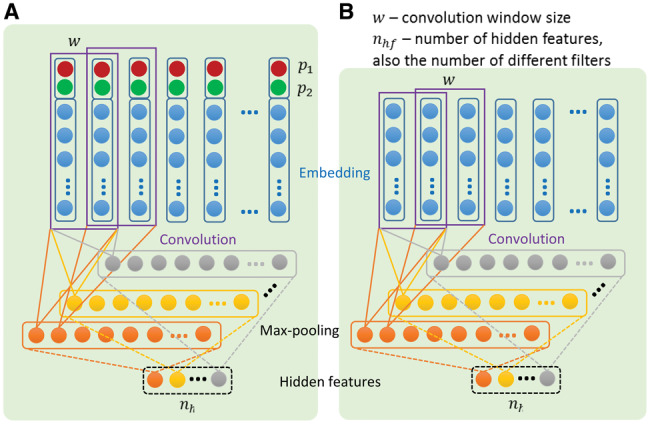
The convolution units for (A) Sentence-CNN model and (B) Seg-CNN model for relation classification. In the figure, w is the convolution window size and nhf is the number of hidden features, as well as the number of different filters. In (A), position features are appended to the embedding features.
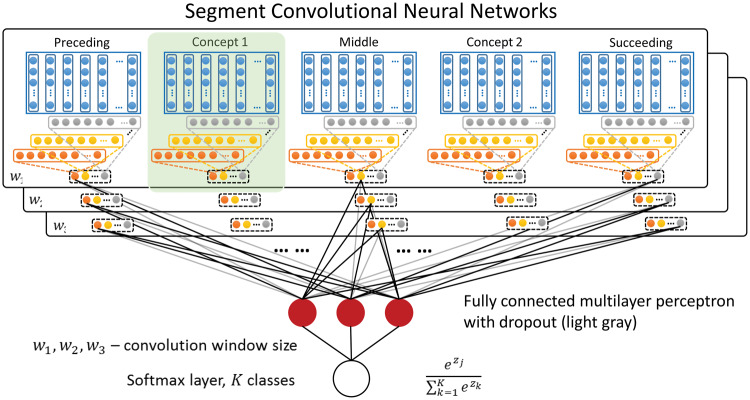
Seg-CNN for relation classification
Sentence-CNN learns a relation representation for the entire sentence but does not explicitly distinguish segments. This is inconsistent with the observation that the 5 segments of text have different roles in determining the relation class.31 We therefore propose Seg-CNN, which consists of multiple convolution units that process the preceding (tokenized words before the first concept), concept1 (tokenized words in the first concept), middle (tokenized words between the 2 concepts), concept2 (tokenized words in the second concept), or succeeding (tokenized words after the second concept) segment, respectively. Each convolution unit uses a sliding window (eg, of size or ) to process a segment and consists of a convolution layer, then a max-pooling layer, to produce multiple hidden features (see Figure 2 [B]). In the following description, let be the word-embedding dimension. A segment with length (number of words) is represented as a matrix , concatenating its word embeddings as columns.
In a convolution unit, one hidden feature is produced by one filter as follows (henceforth we use feature and filter interchangeably). Let be the convolution weight of the jth filter (, where hf stands for hidden features) with a window size of w. Let * denote the operation of element-wise matrix multiplication and the summation operation across matrix entries. Let be the convolution bias and the rectified linear unit activation function. Sliding the convolution window across a length- segment gives
| (1) |
where , comma (,) separates different dimensions, colon (:) denotes a span, and, in particular, a stand-alone colon indicates an entire span of a dimension. Note the difference between convolution in Figure 2 (B) and simple aggregation in Figure 1. The output of the convolutional layer varies in length depending on the number of words in the segment. We then apply a max-pooling operation to produce
| (2) |
as the resulting hidden feature of this filter. The intuition of max-pooling is to capture the most important feature, ie, the one with the highest value, for each feature map, effectively filtering out less informative compositions of words. Max-pooling also guarantees that the extracted features are independent of their location and the segment length.
Figure 3 shows how convolution units are constructed and organized in Seg-CNN. For a specific convolution unit of a segment, each filter can be considered as a linguistic feature detector that learns to recognize a specific feature over w-grams. With such filters, we have a hidden layer of feature vector for the segment s. With m different window sizes, we have a -dimensional vector g:
| (3) |
Figure 3.
Segment convolutional neural network (Seg-CNN). Concept and context text are divided into 5 segments: before the first concept (preceding), of the first concept (concept1), between the 2 concepts (middle), of the second concept (concept2), and after the second concept (succeeding). Each concept is processed by the convolution unit as shown in Figure 2 (B).
The vector g concatenates the hidden features for all segments of a relation. We input g to a fully connected layer (with weight W and bias b) to produce a size- vector , where is the number of relation classes. We then apply a softmax layer to compute the probability for the th class as in
| (4) |
Then the relation class is chosen as .
EXPERIMENTS AND RESULTS
The top systems from i2b2/VA challenge participants still represent the state of the art for this dataset.17,19 In order to fairly compare Seg-CNN with those systems, we used the same training and test datasets. To optimize the hyperparameters for our models, we randomly selected 10% of the training dataset as the validation set. We trained word embeddings on the New York Times and MIMIC-III corpora, respectively, with multiple embedding dimensions from 300 to 600. We chose [3–5] as convolution window sizes. When inspecting relation categories, we found that the PP relation category had a highly imbalanced class ratio (nearly 8 times more None labels than PIP labels). Following de Bruijn et al.,17 we down-sampled the training set to a PIP/None ratio of 1:4. In both sentence- and Seg-CNN models, we experimented with multiple numbers of hidden features (100, 150, and 200).
Some concepts are annotated on the head word (eg, single-word annotations), others include preceding and succeeding modifiers (eg, spanning >20 words). To overcome these annotation inconsistencies, we allowed the concept text to be padded, backward and forward, with neighboring words (experimenting with padding sizes from 3 to 10). Although padding introduces redundancy between concepts and context, the downstream fully connected layer acts as a feature selector. Optimal padding size, number of hidden features, and embedding dimensions were chosen based on validation set performance. For regularization on the CNN models, we used the 50% random dropout41 on the output of the max-pooling layer. Dropout randomly drops the values of a portion (50% in our experiment) of hidden units, thus preventing co-adaptation of these hidden units and reducing overfitting.42
For evaluation, we computed the same micro-averaged precision, recall, and F-measure as used in the challenge (see Table 1). Comparing the micro-averaged F-measure, Seg-CNN ranks first in all relation classification tasks compared with the challenge participating systems with heavily engineered features from the i2b2/VA challenge, even though Seg-CNN uses only word embeddings without feature engineering. Moreover, Seg-CNN outperforms all comparison models, including max- and mean-aggregation of embedding and sentence-CNN. This is consistent with our intuition on the benefits of learning separate feature representations for different segments. As the follow-up study by Zhu et al.27 that attained the state of the art only reported the overall evaluations – 0.755 (precision), 0.726 (recall), and 0.742 (F-measure) – we also report the overall metrics from Seg-CNN as 0.748 (precision), 0.736 (recall), and 0.742 (F-measure). Seg-CNN matches the state-of-the-art F-measure while using only word embedding and minimal feature engineering. Note that the performance shows considerable difference over the 3 categories of relations (TrP, TeP, and PP), which is true for both our CNN models and the challenge participating systems. This is likely due to multiple issues, including the number of labels to classify (6 labels for the TrP relation category and 3 labels for the TeP relation category) and the class imbalance (the highest imbalance for the PP relation category). The observation that Seg-CNN consistently performs modestly better than challenge participating systems across the 3 categories suggests that Seg-CNN is not less robust to these issues than the contrasting systems.
Table 1.
Performance of the CNN models with word embedding trained on the MIMIC-III corpus (when not explicitly noted) or on the general domain New York Times corpus (NYT)
| System | Medical problem–treatment relations |
Medical problem–test relations |
Medical problem–medical problem relations |
||||||
|---|---|---|---|---|---|---|---|---|---|
| R | P | F | R | P | F | R | P | F | |
| Seg-CNN | 0.685 | .687 | 0.686 | 0.804 | .836 | 0.820 | 0.704 | .700 | 0.702 |
| Sentence-CNN | 0.642 | .641 | 0.641 | 0.760 | .812 | 0.785 | 0.679 | .693 | 0.686 |
| Embedding max | 0.636 | .645 | 0.641 | 0.770 | .816 | 0.791 | 0.741 | .554 | 0.634 |
| Embedding mean | 0.632 | .618 | 0.625 | 0.770 | .825 | 0.796 | 0.786 | .533 | 0.635 |
| Seg-CNN (NYT) | 0.641 | .690 | 0.665 | 0.790 | .835 | 0.812 | 0.708 | .681 | 0.694 |
| Seg-CNN (NYT + MIMIC) | 0.653 | .706 | 0.678 | 0.788 | .848 | 0.817 | 0.710 | .689 | 0.700 |
| Roberts et al.19 | 0.686 | .672 | 0.679 | 0.833 | .798 | 0.815 | 0.726 | .664 | 0.694 |
| deBruijn et al.17 | 0.583 | .750 | 0.656 | 0.789 | .843 | 0.815 | 0.712 | .691 | 0.701 |
| Grouin et al.26 | 0.646 | .647 | 0.647 | 0.801 | .792 | 0.797 | 0.645 | .670 | 0.657 |
| Patrick et al.24 | 0.599 | .671 | 0.633 | 0.774 | .813 | 0.793 | 0.627 | .677 | 0.651 |
| Jonnalagadda et al.21 | 0.679 | .581 | 0.626 | 0.828 | .765 | 0.795 | 0.730 | .586 | 0.650 |
| Divita et al.18 | 0.582 | .704 | 0.637 | 0.782 | .794 | 0.788 | 0.534 | .710 | 0.610 |
| Solt et al.20 | 0.629 | .621 | 0.625 | 0.779 | .801 | 0.790 | 0.711 | .469 | 0.565 |
| Demner-Fushman et al.23 | 0.612 | .642 | 0.626 | 0.677 | .835 | 0.748 | 0.533 | .662 | 0.591 |
| Anick et al.22 | 0.619 | .596 | 0.608 | 0.787 | .744 | 0.765 | 0.502 | .631 | 0.559 |
| Cohen et al.25 | 0.578 | .606 | 0.591 | 0.781 | .750 | 0.765 | 0.492 | .627 | 0.552 |
Performance of i2b2/VA challenge participating systems are also included for comparison (gray). The Seg-CNN best performance is attained with the hyperparameter combinations (200 embedding dimension, 100 hidden features, pad size 7) for TrP relations, (500, 150, 4) for TeP relations, and (400, 100, 10) for PP relations. The comparison model Sentence-CNN attains best performance with (400 embedding dimension, 200 hidden features) for TrP relations, (500, 200) for TeP relations, and (300, 150) for PP relations. Seg-CNN using New York Times embedding has best-performance hyperparameters at (600, 200, 8) for TrP relations, (500, 200, 4) for TeP relations, and (500, 200, 10) for PP relations. Seg-CNN using embedding trained from the New York Times and MIMIC-III corpora has best-performance hyperparameters at (600, 200, 6) for TrP relations, (300, 150, 4) for TeP relations, and (600, 150, 9) for PP relations. Best micro-averaged F-measures are in bold.
We implemented our models using the Theano package43 and ran them on an NVidia Tesla GPU with cuDNN library enabled. We have made our codes available on a public repository (https://github.com/yuanluo/seg_cnn). Table 2 shows the training time required by the Seg-CNN and Sentence-CNN using medical word embeddings. The training times are within a reasonable 7-min window for all the model-task combinations.
Table 2.
Running time of the CNN models with word embedding trained on medical corpus
| System | Problem-treatment relations | Problem-test relations | Problem-problem relations |
|---|---|---|---|
| Seg-CNN | 120s | 217s | 413s |
| Sentence-CNN | 165s | 156s | 369s |
The model hyperparameters for corresponding models are the optimal ones listed in Table 1. The time is measured by number of seconds.
DISCUSSION
In order to evaluate the impact of the corpus used to train word embeddings, we report in Table 1 the performance of Seg-CNN using a general domain embedding. Comparing these results to Seg-CNN with medical word embeddings, we see about a 2% drop in micro-averaged F-measure. This drop is consistent with the distinct characteristics of clinical narratives, many of which are fragmented text abundant with acronyms (eg, CABG for coronary artery bypass grafting) and abbreviations (eg, s/p for status post). CNNs with general domain embeddings likely miss critical information carried by such words. For example, “The patient developed [medical problem] s/p [treatment]” usually indicates a treatment-cause-problem relation. A larger embedding corpus typically leads to better embedding33; however, in this work, word embeddings from general plus medical corpora did not outperform medical embeddings only. It is our future work to explore whether the difference between the New York Times corpus and the MIMIC-III corpus overshadows the benefits of additional corpora, and whether other embedding methods such as Skip-Gram33 could produce better embeddings.
The performance of Seg-CNN is better than that of Sentence-CNN and embedding aggregations. Our Sentence-CNN is similar to that of Sahu et al.,30 but does not use linguistic features such as part of speech, phrase chunking, etc. In addition, Sahu et al.,30 combined the i2b2/VA training and test datasets and performed cross-validation, and thus had considerably more training data. Although the performance of Sentence-CNN is lower than the performance of state-of-the-art i2b2/VA challenge participant models, Seg-CNN’s performance is slightly higher. This observation confirms the intuition on the benefits of learning individual representations for different segments. Seg-CNN’s improvement over the state-of-the-art systems was modest, indicating room for further improvement. There may still be merit in the linguistic features (as shown in Sahu et al.30) and domain-specific knowledge. The impact of domain-specific knowledge is also evident from the fact that Seg-CNN with medical embeddings outperformed Seg-CNN with general-domain embeddings. We plan to investigate whether tighter integration of linguistic features and domain knowledge into CNNs could result in further improvements for relation classification.
CONCLUSION
In this work, we showed that Seg-CNN achieved state-of-the-art performance on the i2b2/VA relation classification challenge datasets, without manual feature engineering. We also showed that Seg-CNN outperforms a Sentence-CNN model and embedding aggregation models, which is consistent with the intuition that learning individual representation for each of the preceding, concept1, middle, concept2, and succeeding segment can provide useful information in discerning relations between concepts. We evaluated the impact of word embeddings on the performance of Seg-CNN and showed that medical word embeddings can help improve relation classification. These results are not only encouraging, but also suggestive of future directions, such as effective use of embedding corpora and tighter integration of domain knowledge into CNN models.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank i2b2 National Center for Biomedical Computing, funded by U54LM008748, for creating the clinical records originally prepared for the i2b2/VA relation classification challenge. We would like to also thank the NVidia GPU grant program for providing the GPU used in our computation.
Funding
This work was supported by National Institutes of Health grants UL1TR001422, P50-HG007738, and 1R01MH106577-01A1 and the MIT-Philips collaborative research project.
Competing interests
The authors have no competing interests to declare.
Contributors
YL formulated the original problem, designed and implemented the Seg-CNN and comparison models, evaluated the systems’ performance, and wrote the first draft of the paper. YC helped with debugging the models, tuning the hyperparameters, and evaluating the systems. OU, PS, and JS formulated the original problem and provided helpful feedback and revisions to the paper.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
References
- 1. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394. [PMC free article] [PubMed] [Google Scholar]
- 2. Cimino JJ. In defense of the desiderata. J Biomed Inform. 2006;393:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y, Uzuner Ö, Szolovits P. Bridging semantics and syntax with graph algorithms—state-of-the-art of extracting biomedical relations. Briefings Bioinform. 2016;181:160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;366:462–77. [DOI] [PubMed] [Google Scholar]
- 5. Luo Y, Sohani AR, Hochberg EP, Szolovits P. Automatic lymphoma classification with sentence subgraph mining from pathology reports. J Am Med Inform Assoc. 2014;215:824–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo Y, Xin Y, Hochberg E, Joshi R, Uzuner O, Szolovits P. Subgraph augmented non-negative tensor factorization (SANTF) for modeling clinical narrative text. J Am Med Inform Assoc. 2015;225:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weng C, Wu X, Luo Z, Boland MR, Theodoratos D, Johnson SB. EliXR: an approach to eligibility criteria extraction and representation. J Am Med Inform Assoc. 2011;18(Suppl 1):i116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coulet A, Shah NH, Garten Y, Musen M, Altman RB. Using text to build semantic networks for pharmacogenomics. J Biomed Inform. 2010;436:1009–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinform. 2009;102:S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu M, Wu Y, Chen Y, et al. . Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc. 2012;19(e1):e28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harpaz R, Vilar S, DuMouchel W, et al. . Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inform Assoc. 2013;203:413–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luo Y, Thompson W, Herr T, et al. . Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf. 2017. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 13. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;185:552–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Savova GK, Masanz JJ, Ogren PV, et al. . Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;175:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Paper presented at the AMIA Symposium, Washington, DC; 2001. [PMC free article] [PubMed] [Google Scholar]
- 16. Tsuruoka Y, Tsujii Ji. Bidirectional inference with the easiest-first strategy for tagging sequence data. Paper presented at the Conference on Human Language Technology and Empirical Methods in Natural Language Processing, Vancouver, BC; 2005. [Google Scholar]
- 17. de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc. 2011;185:557–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Divita G, Treitler O, Kim Y, et al. . Salt Lake City VA’s challenge submissions. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 19. Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc. 2011;185:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Solt I, Szidarovszky FP, Tikk D. Concept, assertion and relation extraction at the 2010 i2b2 relation extraction challenge using parsing information and dictionaries. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 21. Jonnalagadda S, Cohen T, Wu S, Gonzalez G. Enhancing clinical concept extraction with distributional semantics. J Biomed Inform. 2012;451:129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Anick P, Hong P, Xue N, Anick D. I2B2 2010 challenge: machine learning for information extraction from patient records. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 23. Demner-Fushman D, Apostolova E, Doğan RI, et al. . NLM’s system description for the fourth i2b2/VA challenge. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 24. Patrick JD, Nguyen DHM, Wang Y, Li M. i2b2 Challenges in Clinical Natural Language Processing 2010. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 25. Cohen AM, Ambert K, Yang J, et al. . OHSU/portland VAMC team participation in the 2010 i2b2/VA challenge tasks. Paper presented at the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 26. Grouin C, Abacha A, Bernhard D, et al. . CARAMBA: concept, assertion, and relation annotation using machine-learning based approaches. Paper presented at the 2010 i2b2/ VA Workshop on Challenges in Natural Language Processing for Clinical Data Boston, MA; 2010. [Google Scholar]
- 27. Zhu X, Cherry C, Kiritchenko S, Martin J, De Bruijn B. Detecting concept relations in clinical text: Insights from a state-of-the-art model. J Biomed Inform. 2013;462:275–85. [DOI] [PubMed] [Google Scholar]
- 28. Björne J, Salakoski T. Generalizing biomedical event extraction. Paper presented at the BioNLP Shared Task Workshop, Portland, OR; 2011. [Google Scholar]
- 29. Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. Paper presented at the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA;2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sahu SK, Anand A, Oruganty K, Gattu M. Relation extraction from clinical texts using domain invariant convolutional neural network. arXiv preprint arXiv:160609370. 2016. [Google Scholar]
- 31. Uzuner O, Mailoa J, Ryan R, Sibanda T. Semantic relations for problem-oriented medical records. Artif Intell Med. 2010;502:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo Y. Recurrent neural networks for classifying relations in clinical notes. J Biomed Inform. 2017;72:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mikolov T, Dean J. Distributed representations of words and phrases and their compositionality. Adv Neural Inf Process Syst. 2013. [Google Scholar]
- 34. Kim Y. Convolutional neural networks for sentence classification. arXiv preprint arXiv:14085882. 2014. [Google Scholar]
- 35. Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P. Natural language processing (almost) from scratch. J Machine Learning Res. 2011;12:2493–537. [Google Scholar]
- 36. Nikfarjam A, Sarker A, O’Connor K, Ginn R, Gonzalez G. Pharmacovigilance from social media: mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J Am Med Inform Assoc. 2015;223:671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sandhaus E. The New York Times Annotated Corpus 2008; DVD. Accessed October 3, 2017. [Google Scholar]
- 38. Johnson AE, Pollard TJ, Shen L, et al. . MIMIC-III, a freely accessible critical care database. Scientific Data. 2016;3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalchbrenner N, Grefenstette E, Blunsom P. A convolutional neural network for modelling sentences. arXiv preprint arXiv:14042188. 2014. [Google Scholar]
- 40. Zeng D, Liu K, Lai S, Zhou G, Zhao J. Relation classification via convolutional deep neural network. Paper presented at COLING 2014, Dublin. [Google Scholar]
- 41. Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR. Improving neural networks by preventing co-adaptation of feature detectors. arXiv preprint arXiv:12070580. 2012. [Google Scholar]
- 42. Srivastava N, Hinton GE, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Machine Learning Res. 2014;151:1929–58. [Google Scholar]
- 43. Bergstra J, Breuleux O, Bastien F, et al. . Theano: A CPU and GPU math compiler in Python. Paper presented at the 9th Python in Science Conference 2010, Austin, TX. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.