Abstract
The gap between domain experts and natural language processing expertise is a barrier to extracting understanding from clinical text. We describe a prototype tool for interactive review and revision of natural language processing models of binary concepts extracted from clinical notes. We evaluated our prototype in a user study involving 9 physicians, who used our tool to build and revise models for 2 colonoscopy quality variables. We report changes in performance relative to the quantity of feedback. Using initial training sets as small as 10 documents, expert review led to final F1scores for the “appendiceal-orifice” variable between 0.78 and 0.91 (with improvements ranging from 13.26% to 29.90%). F1for “biopsy” ranged between 0.88 and 0.94 (−1.52% to 11.74% improvements). The average System Usability Scale score was 70.56. Subjective feedback also suggests possible design improvements.
Keywords: natural language processing (NLP), electronic health records, machine learning, user-computer interface, medical informatics
INTRODUCTION
Although electronic health records have long been recognized as vital sources of information for decision support systems and data-driven quality measures,1 extraction of information from unstructured clinical notes presents many challenges.2 Despite recent advances in natural language processing (NLP) techniques, the process is often expensive and time-consuming and requires expert construction of gold standard training corpora.3 Current tools also lack provisions for domain experts to inspect NLP outcomes and make corrections that might improve the results.4
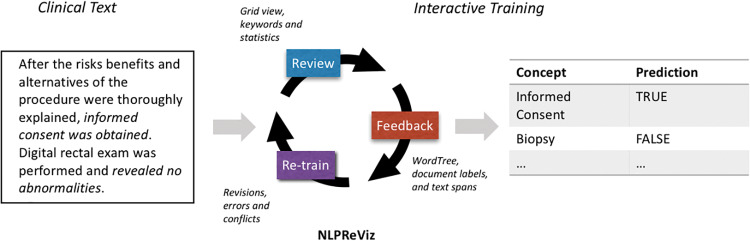
We present NLPReViz, a prototype tool designed for clinicians and clinical researchers to train, review, and revise NLP models (Figure 1 ). Our work is informed by recent work in interactive machine learning, information visualization, and NLP.
Figure 1.
An interactive machine learning cycle begins with the review step, with output from the learning model displayed to the user. User feedback is used by the system to improve upon the machine learning models by providing labels for documents that were previously not part of the training set, or by correcting any misclassified documents. After retraining, a new model is created and the tool shows prediction changes and provides guidance for resolving potentially contradictory feedback items.
Interactive machine learning (also known as “human-in-the-loop”) systems provide users with outputs sufficient for eliciting feedback, forming a closed loop capable of building continuously improving predictive models.5,6 This approach has been used for interactive document clustering,7 document retrieval,8 image segmentation,9 and music composition,10 and in medical informatics, including for subspace clustering11 and literature review.12
These systems require effective displays to present outputs and elicit user feedback. We have adapted elements from systems such as Word Tree,13 Jigsaw,14 and others. Word Tree shows a graphical overview of common word sequences and the contexts in which they are found. Visual analytics tools such as Jigsaw present overviews of document collections in support of expert analysis of large text corpora.
Our work is complementary to recent efforts in the development of usable NLP tools. D’Avolio et al.15 describe a prototype system called the Automated Retrieval Console (ARC) that combines existing tools for creating text annotations (Knowtator)16 and deriving NLP features (cTAKES)17 within a common user interface that can be used to configure the machine learning algorithms and export their results. RapTAT extends this effort to demonstrate how interactive annotation can be used to reduce the time required to create an annotated corpus.18
Our work complements these efforts, focusing not only on the annotation stage, but also on facilitating expert review of NLP results and supporting the elements of an interactive machine learning system for clinical text. We have extended ideas from Kulesza et al.,19 incorporating Zaidan et al.’s20 rationale framework through multiple interactive means of providing feedback, thus turning user input into additional training data suitable for retraining models. A preliminary evaluation with 9 clinicians reviewing colonoscopy notes suggests that this approach might enable rapid and inexpensive construction of high-quality NLP models.
MATERIALS AND METHODS
Interface design
The design of our tool was informed by prior work on interactive learning systems:5,6,19
- Review displays support interpretation of NLP results both within and across documents.
- R1: Document displays highlight NLP results and, where possible, show evidence for the results extracted from the text.
- R2: Overview displays support comparisons between documents and identification of frequent words or phrases associated with NLP results.
- Feedback mechanisms provide usable and efficient means of updating NLP models.
- R3: Interaction tools support selection of text as evidence for selected interpretations.
- R4: Conflicting or inconsistent feedback should be identified and presented to the user for appropriate resolution.
- Retrain involves results of model revisions that should be apparent to users.
- R5: Displays should help users understand changes in predictions and other model revisions.
Figure 2 shows the user interface for NLPReViz. A video demo can be found at vimeo.com/trivedigaurav/emr-demo. We built a prototype, evaluated it with a think-aloud study, and revised it based on the participants’ feedback.21 Our tool is available for download along with source code and documentation at NLPReViz.github.io.
Figure 2.
User interface: (A) The grid view shows the extracted variables in columns and individual documents in rows, providing an overview of NLP results. Below the grid, we have statistics on the active variable, with (B) the distribution of classifications for the selected variable and (C) the list of top indicators for that variable aggregated across all documents in the dataset. (D) Indicators from the active report are shown on the right. (E) The document view shows the full text of the patient reports, with the indicator terms highlighted. (F) Feedback can be sent using the control bar on the top or a right-click context menu. (G) The Word Tree view provides the ability to search and explore word sequence patterns found across the documents in the corpus, and to provide feedback that will be used to retrain NLP models. In this figure, we built the tree by searching for the word “biopsy” and then drilled down upon the node “hot.” The word tree now contains all the sentences in the dataset with the phrase “hot biopsy.” This allows the user to get an idea of all the scenarios in which “hot biopsy” has been used. Hovering over different nodes in the tree will highlight specific paths in the tree with the selected term. (H) The retrain view lists user-provided feedback, including any potential inconsistencies, and specifies changes in variable assignments due to retraining. In the example above, the user has selected a text span documenting “informed consent” in a report. However, the user also labeled the report incorrectly, possibly in error. NLPReViz points this out as conflicting feedback.
Learning with rationales
We use “bag-of-words” and Support Vector Machine classifiers with linear kernels to predict binary classifications for concept variables extracted from documents. Our model for incorporating user feedback adapts a framework proposed by Zaidan et al.,20 in which the domain experts supply not only the correct label but also a span of text that serves as a rationale for their labeling decision. Rationales are turned into pseudo-examples providing additional training data.20,22 Rationales have been shown to be effective for predicting sentiments of movie reviews, for example.22 We adapted this approach for use on clinical text by constructing one merged pseudo-example per document from the annotations received against them (see Supplementary Appendix: Annotator Rationale Method for an example). Rationales are constructed from user interactions with the tool and are used to retrain the Support Vector Machine models.
Evaluation
Our evaluation addressed 2 key questions: (1) Can clinicians successfully use NLPReViz to provide feedback for improving NLP models? and (2) Can this feedback be effective with a small set of initial training data?
This study was approved as exempt by the University of Pittsburgh’s Institutional Review Board under project PRO15020008.
Dataset
We used a reduced dataset of colonoscopy reports prepared by Harkema et al.23 along with their gold standard label set. Participants worked with 2 variables: “biopsy” and “appendiceal-orifice.” A document was marked “true” for the biopsy variable if the report indicated that a sample of tissue was tested through a biopsy procedure. The appendiceal-orifice variable indicates whether that region of the colon was reached and was explicitly noted during the colonoscopy. Our dataset consisted of 453 documents, split into 2 parts, two-thirds for a development set to conduct the user study and one-third held out as a test set to evaluate system performance.
Participants
We identified a convenience sample of participants with MD degrees and knowledge of colonoscopy procedures. Participants were given a $50 gift card for 90 min of participation via web conferencing. One participant (p9) experienced technical difficulties, resulting in a shorter study time.
To address the question of sensitivity to the size of the initial training set, we used 2 splits to build initial training models. The first group of 4 participants (p1–p4) started with models built on 10 annotated documents. Initial models for the second group (p5–p8 and p9) were based on 30 annotated documents. The same 173 documents were used in the test set for both groups.
Protocol
Each session began with a participant background questionnaire, followed by a 15-min walkthrough of the interface and an introduction to the annotation guidelines used to prepare our gold standard labels.23 Participants were given up to 1 h to annotate and build models, roughly divided between the 2 variables. We reminded them to retrain at regular intervals, particularly if they provided >10 consecutive feedback items without retraining. After finishing both variables, participants completed the System Usability Scale (SUS)24 and discussed reactions to the tool.
We evaluated performance of the models on the test set using the harmonic mean of recall and precision, F1score at each retraining step. We calculated Cohen’s κ statistic25 to measure agreement of the complete set of each participant’s feedback items with the gold standard labels. To compare user feedback to a possibly optimal set of labels, we simulated feedback actions using gold standard labels. Ten random feedback items (without rationales) were added at each step, ranging from 10 to 280 items. This was repeated 50 times to compute an average.
RESULTS
Nine physicians participated in the study (Table 1). The average SUS score was 70.56 out of 100. An SUS score of 68 is considered average usability.24
Table 1.
Descriptions of study participants
| Degree | Position | Years in position | Role | NLP experience? | SUS score | |
|---|---|---|---|---|---|---|
| Group 1: Initial model trained with 10 documents | ||||||
| p1 | MD PhD | Assistant Professor | <5 | Clinician, Researcher | Yes; current and past projects | 75 |
| p2 | MD | Faculty Researcher | 5–10 | Clinician, Researcher | No; using in future project | 55 |
| p3 | MD | Faculty Researcher | <5 | Clinician | No | 90 |
| p4 | MD | Fellow | <5 | Clinician | No | 77.5 |
| Group 2: Initial model trained with 30 documents | ||||||
| p5 | MD | Resident Physician | <5 | Clinician | No | 67.5 |
| p6 | MD | Fellow | <5 | Clinician, Researcher | No | 77.5 |
| p7 | MD | Resident Physician | <5 | Clinician | No | 85 |
| p8 | MD | Resident Physician | <5 | Clinician | No | 52.5 |
| p9a | MD MAS | Physician | 5–10 | Clinician, Researcher | Currently involved in a project | 55 |
We obtained the System Usability Scale scores24 using a post-study questionnaire.
ap9 faced technical difficulties during the study, which may have influenced perceived usability.
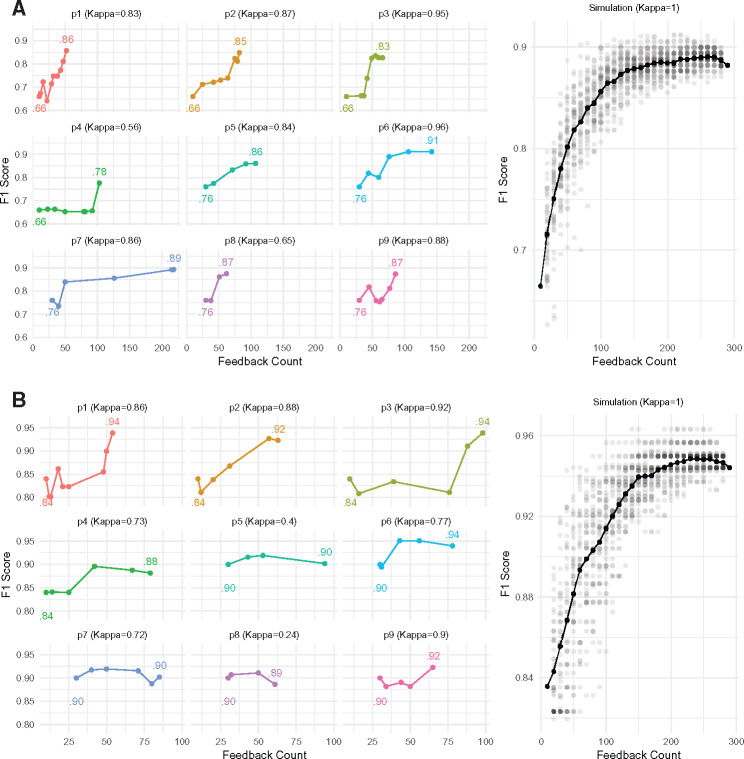
The changes in F1scores on the test set (relative to gold standard labels) for the 9 participants are shown in Figure 3 , along with their Cohen’s κ scores indicating agreement of feedback with the gold standard labels. Scores are plotted against the cumulative number of records affected by user feedback actions after each retraining step. Performance improved in 17 of 18 tasks, with improvement as high as 29.90% (Figure 3 and Supplementary Appendix). Differences in participants’ approaches to annotation and retraining are summarized in Table 2.
Figure 3.
(A) Appendiceal-orifice; (B) biopsy. Plots showing the variations in F1-scores for the 2 variables as the participants provided feedback. These results are shown for the test dataset only. Participants p1–p4 started with an initial training set of 10 documents, while p5 onward used a model trained on 30 training documents. Differences in the spacing of the points in each graph reflect differences in feedback rates across participants. Kappa scores next to the participant IDs indicate how their feedback compared to the gold standard labels. The simulation plots on the right show comparisons with feedback based on gold standard labels. The solid line shows the average of 50 simulation runs. These simulations include only document-level feedback from the gold standard labels, without any rationales.
Table 2.
Activity patterns of the individual participants for both variables combined
| Docs opened | Total feedback | Unique feedback | Unseen feedback | Model count | Error count | Type of feedback |
|||
|---|---|---|---|---|---|---|---|---|---|
| Word Tree (docs) | Span | Label | |||||||
| p1 | 129 | 86 | 66 | 14 | 17 | 2 | 1 (19) | 16 | 51 |
| p2 | 117 | 125 | 112 | 1 | 12 | 6 | 4 (40) | 24 | 61 |
| p3 | 71 | 144 | 88 | 5 | 12 | 3 | 2 (16) | 28 | 98 |
| p4 | 94 | 162 | 93 | 4 | 12 | 0 | 6 (43) | 26 | 106 |
| p5 | 104 | 141 | 133 | 14 | 7 | 1 | 1 (18) | 6 | 117 |
| p6 | 170 | 301 | 230 | 29 | 6 | 2 | 3 (44) | 58 | 181 |
| p7 | 50 | 243 | 202 | 141 | 6 | 2 | 8 (190) | 21 | 32 |
| p8 | 54 | 63 | 55 | 0 | 6 | 1 | 0 (0) | 23 | 40 |
| p9 | 68 | 91 | 81 | 0 | 10 | 2 | 0 (0) | 34 | 57 |
| Indicates documents viewed; we do not assume agreement in case of no feedback | Number of feedback items provided | Unique document labels inferred from feedback | Documents labeled without viewing them first (when using Word Tree) | Number of training iterations | Conflicts and overrides in provided feedback | Feedback items provided using the different feedback input mechanisms: the Word Tree view (along with documents affected), highlighting spans or assigning a label to the document | |||
Participants p1–p4 started with an initial model trained on 10 documents, while p5–p9 started with 30 documents.
Open-ended subjective feedback was positive. Others commented about the learnability of the tool: “I thought it was very easy to use and straightforward,” “The process was very easy with a little bit of guidance,” and “May need some initial training – may be complex for somebody who hasn’t done [annotations] before.” Further comments were related to incremental design enhancements discussed in the following section.
DISCUSSION
Our study suggests that physicians can use NLPReViz to provide effective feedback for building NLP models with relatively low levels of effort. We observed notable improvements for most users in a short time span (30 min), starting with small training sets (as few as 10 documents). We found improvements in F1scores across all users for “appendiceal-orifice,” though results were more mixed for “biopsy.” Examination of less successful efforts indicated that some participants found the biopsy annotation guidelines to be challenging. These difficulties were associated with the lower kappa scores (eg, p8, “biopsy”) between the user-provided labels and gold standard labels. Favorable performance of models based on participant feedback relative to results of simulations using gold standard labels (Figure 3) suggests that NLPReViz can be used to elicit feedback suitable for improving NLP models.
Due to the small sample size, we were unable to statistically compare the relative improvements in performance across the size of the training set. Difficulties in sampling also confound the results somewhat, as the later participants (p5–p9), who used the models based on larger training sets, are also those who were generally less experienced with colonoscopy reports. Other questions for future investigation include identifying forms of feedback that are useful under different circumstances. Subjective feedback also suggested several possible improvements to our design. One particularly interesting suggestion was to indicate that a phrase was irrelevant to a classification of a document. For example, the phrase “hot biopsy forceps” usually described the tool used to remove polyps and not a biopsy procedure.
Our learning system can be extended to use n-grams features with the same interface. The main limitation of our prototype lies in the use of binary variables. This approach allows us to represent complex concepts like “patient has no family history of colon cancer” and also categorical variables by decomposing them into multiple variables. However, more sophisticated NLP models will require extending the interaction techniques.
Initial success with small training sets also suggests the possibility of exploring “zero-training” cold start26 and developing annotation guidelines de novo, perhaps with preannotation techniques like those used in RapTAT.18 Future work involves developing extensions for easy integration of our system with other tools, including documenting our representational state transfer (REST) calls so that the front end can be supported by a different learning system, for example. We are also interested in exploring alternative training mechanisms necessary for a moving NLPReViz toward a fully active learning approach, providing real-time model updates and prioritized items with every feedback instance. Finally, the use of our tool to reduce annotation expense presents another question: Without a gold standard annotated test set, how will users know when resulting models are “good enough”?
CONCLUSION
Interactive tools are needed to close the gap between domain expertise and NLP skills to ease the extraction of computable understanding from clinical texts. We present NLPReViz, an interactive tool for finding patterns of interest, reviewing text, and revising models. Our user interface design complements the rationale-based learning system that we adopted to incorporate user feedback. It allows domain experts without machine learning experience to build models and give feedback to improve them iteratively. Our user study supports the viability of our approach by demonstrating notable improvements in performance metrics in a short time span with minimal initial training. Further work will be needed to apply these strategies to a broader range of NLP problems.
Funding
This research was supported by National Institutes of Health grant 5R01LM010964. GT also acknowledges a graduate student research fellowship from the Department of Biomedical Informatics at the University of Pittsburgh.
Contributors
All authors made contributions to the conception of the work. GT led the development of the interactive tool and the evaluation studies under the supervision of HH. PP developed the NLP back-end system with RH and JW. GT, HH, and RH drafted the manuscript, with critical revisions by PP and WWC.
Competing interests
The authors have no competing interests to declare.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank our user study participants. We would also like to thank Dr Ateev Mehrotra for providing the colonoscopy reports dataset.
References
- 1. Dick RS, Steen EB, Detmer DE, et al. The Computer-based Patient Record: an Essential Technology for Health Care. National Academy Press: Washington, DC; 1997. [PubMed] [Google Scholar]
- 2. Malmasi S, Sandor NL, Hosomura N, Goldberg M, Skentzos S, Turchin A. Canary: An NLP platform for clinicians and researchers. Appl Clin Inform. 2017;2:447–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friedman C, Johnson SB. Natural language processing in biomedicine. In: Shortliffe EH, Cimino JJ, ed. Biomedical Informatics: Computer Applications in Health Care and Biomedicine, Health Informatics, Chapter 8. 3rd ed. New York, NY: Springer; 2006:312–43. [Google Scholar]
- 4. Holzinger A, Stocker C, Ofner B, et al. Combining HCI, natural language processing, and knowledge discovery-potential of IBM content analytics as an assistive technology in the biomedical field. Human-Computer Interaction and Knowledge Discovery in Complex, Unstructured, Big Data. Springer: Berlin, Heidelberg; 2013:13–24. [Google Scholar]
- 5. Amershi S, Fogarty J, Kapoor A, et al. Effective end-user interaction with machine learning. Proc Twenty-Fifth AAAI Conf Artificial Intell; 2011. https://www.microsoft.com/en-us/research/publication/effective-end-user-interaction-machine-learning/. [Google Scholar]
- 6. Amershi S, Cakmak M, Knox WB, et al. Power to the people: the role of humans in interactive machine learning. AI Magazine. 2014;354:105–20. [Google Scholar]
- 7. Chau DH, Kittur A, Hong JI, et al. Apolo: making sense of large network data by combining rich user interaction and machine learning. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. New York, NY: ACM; 2011:167–176. [Google Scholar]
- 8. Heimerl F, Koch S, Bosch H, et al. Visual classifier training for text document retrieval. IEEE Transactions on Visualization and Computer Graphics. 2012;1812:2839–48. [DOI] [PubMed] [Google Scholar]
- 9. Fails J, Olsen D Jr. Interactive machine learning. In Proceedings of the 8th International Conference on Intelligent User Interfaces. ACM; 2003:39–45. [Google Scholar]
- 10. Fiebrink R, Cook PR, Trueman D. Human model evaluation in interactive supervised learning. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. ACM; 2011:147–56. [Google Scholar]
- 11. Hund M, Böhm D, Sturm W, et al. Visual analytics for concept exploration in subspaces of patient groups. Brain Inform. 2016;34:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wallace BC, Small K, Brodley CE, et al. Deploying an interactive machine learning system in an evidence-based practice center: abstrackr. In Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. ACM; 2012:819–24. [Google Scholar]
- 13. Wattenberg M, Viegas FB. The Word Tree, an interactive visual concordance. IEEE Transact Visualization Comput Graphics. 2008;146:1221–28. [DOI] [PubMed] [Google Scholar]
- 14. Stasko J, Görg C, Liu Z. Jigsaw: Supporting investigative analysis through interactive visualization. Inform Visualization. 2008;72:118–32. [Google Scholar]
- 15. D’Avolio LW, Nguyen TM, Goryachev S, et al. Automated concept-level information extraction to reduce the need for custom software and rules development. J Am Med Inform Assoc. 2011;185:607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ogren PV. Knowtator: a protégé plug-in for annotated corpus construction. In Proceedings of the 2006 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology. Association for Computational Linguistics: Morristown, NJ; 273–5. [Google Scholar]
- 17. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;175:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gobbel GT, Garvin J, Reeves R, et al. Assisted annotation of medical free text using RapTAT. J Am Med Inform Assoc. 2014;215:833–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kulesza T, Burnett M, Wong WK, Stumpf S. Principles of explanatory debugging to personalize interactive machine learning. Proceedings of the 20th International Conference on Intelligent User Interfaces. ACM; 2015;2015:126–37. [Google Scholar]
- 20. Zaidan OF, Eisner J, Piatko C. Using “annotator rationales” to improve machine learning for text categorization. In Human Language Technologies 2007: The Conference of the North American Chapter of the Association for Computational Linguistics; Proceedings of the Main Conference. In NAACL-HLT. Association for Computational Linguistics: Rochester, NY; 2007: 260–7. [Google Scholar]
- 21. Trivedi G, Pham P, Chapman W, et al. An interactive tool for natural language processing on clinical text. arXiv e-prints; 2017. https://arxiv.org/abs/1707.01890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yessenalina A, Yue Y, Cardie C. Multi-level structured models for document-level sentiment classification. In Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics: Stroudsburg, PA; 1046–56. [Google Scholar]
- 23. Harkema H, Chapman WW, Saul M, et al. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inform Assoc. 2011;18(Suppl):150–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooke J. SUS: a quick and dirty usability scale. In Jordan PW, Weerdmeester B, Thomas A, Mclelland IL, ed. Usability Evaluation in Industry. Taylor and Francis: London; 1996. [Google Scholar]
- 25. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Measurement. 1960;201:37–46. [Google Scholar]
- 26. Holzinger A. Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Inform. 2016;32:119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.