Abstract
The worldwide obesity epidemic has been mainly attributed to lifestyle changes. However, who becomes obese in an obesity-prone environment is largely determined by genetic factors. In the last twenty years, important progress has been made in the elucidation of the genetic architecture of obesity. In parallel with successful gene identifications, the number of gene-environment interaction (GEI) studies has grown rapidly. This paper reviews the growing body of evidence supporting gene-environment interactions in the field of obesity. Heritability, monogenic and polygenic obesity studies provide converging evidence that obesity-predisposing genes interact with a variety of environmental, lifestyle and treatment exposures. However, some skepticism remains regarding the validity of these studies based on several issues, which include statistical modelling, confounding, low replication rate, underpowered analyzes, biological assumptions and measurement precision. What follows in this review includes (1) an introduction to the study of GEI, (2) the evidence of GEI in the field of obesity, (3) an outline of the biological mechanisms that may explain these interaction effects, (4) methodological challenges associated with GEI studies and potential solutions, and (5) future directions of GEI research. Thus far, this growing body of evidence has provided a deeper understanding of GEI influencing obesity and may have tremendous applications in the emerging field of personalized medicine and individualized lifestyle recommendations.
Introduction
Over the past three decades, the prevalence of obesity has reached epidemic proportions throughout the world (1). This recent epidemic cannot be explained by sudden changes in the human population gene pool and has been mainly attributed to lifestyle modifications (2). Over-nutrition and decline in physical activity are the two “usual suspects”, but additional factors (reduced gut microflora diversity, sleep debt, endocrine disruptors, reduction in variability of ambient temperatures) have emerged as significant contributors to the escalating prevalence of obesity (3). If obesity is a multifactorial disorder that requires environmental influences to manifest, some individuals are more susceptible than others to weight gain in an obesity-prone environment, and who becomes obese at the individual level is largely determined by genetic factors (4). Technological and methodological breakthroughs in the last twenty years have led to important progress in the elucidation of the genetic architecture of obesity (5). The first two genes (LEP and MKKS) associated with a Mendelian non-syndromic or syndromic form of obesity were identified in 1997 and 2000 (6, 7). Seven years later, the first common variant (located in the intron 1 of the FTO gene) reproducibly associated with polygenic obesity was identified (8, 9). At the time we are writing, over 40 monogenic obesity loci (with or without syndromic features) and 130 polygenic obesity loci have been described, and this list is destined to grow over the coming years (5). In parallel with successful gene identification efforts, the number of studies on gene-environment interactions has grown rapidly (10). In the first segment of this review, we summarize the findings supporting gene-environment interaction in obesity from heritability, monogenic and polygenic studies and provide a biological hypothesis to explain these statistical interactions. The final section will outline methodological challenges associated with GEI studies, provide potential solutions to these issues based on existing evidence and highlight future directions of GEI research.
Definitions
The genetic etiology of obesity can be classified into two categories. First, Mendelian (or monogenic) obesity describes individuals who carry a rare gene variant with a dramatic impact on adiposity (11). These variants are associated with a high lifetime risk of disease and exhibit a near one-to-one relationship between genotype and phenotype (12–14). Monogenic obesity can be classified as syndromic or non-syndromic. Syndromic obesity refers to Mendelian obesity that co-occurs with a distinct set of clinical phenotypes, such as mental retardation, dysmorphic features and organ-specific developmental abnormalities (15). Syndromic forms of obesity result from chromosomal abnormalities or point mutations, which can be autosomal or X-linked disorders (16). Non-syndromic forms are caused by pathogenic mutations or structural variations in genes involved in the leptin/melanocortin pathway, and are mainly characterized by hyperphagic obesity (17). Homozygous/compound heterozygous carriers of pathogenic mutations in genes from the leptin/melanocortin pathway are exceedingly rare and lead to a fully penetrant form of early-onset extreme obesity. In contrast, heterozygous incomplete penetrant mutations in the same pathway account for a greater proportion of oligogenic obesity cases in population (5). Heterozygous loss-of-function mutation carriers in MC4R display mild obese phenotypes, which can be moderated by environmental factors that increase the risk of obesity (18, 19). Second, other cases of obesity can be attributed to the concerted presence of DNA variation in several genes (each with a relatively small effect), known as polygenic obesity (12). With respect to body weight regulation, recent simulations estimated that hundreds of variants with small to modest effect may account for the genetic architecture of complex traits such as obesity (20).
The concept of gene-environment interaction in the context of human diseases is not recent and has been discussed since proposed by J.B. Haldane in 1946 (21). The statistical definition of an interaction between two or more risk factors is simply the coefficient of the product term of the risk factors, also known as effect modification or effect modulation. Interaction is thus measured in terms of departure from a multiplicative or an additive model (22, 23). Alternatively, biological interaction between two factors is defined as their co-participation in the same causal mechanism to disease development (24). For statistical evidence of gene-environment interaction to be convincing, it is typically necessary to replicate the findings in additional samples and / or support the evidence with plausible underlying biological mechanisms (22).
The importance of gene-environment interactions in obesity: evidence from heritability, monogenic and polygenic studies
Heritability estimates are influenced by the environment
Early indications of the shared influence of genetics and the environment in shaping obesity originated from heritability studies involving environmental exposures among twins. Heritability is the proportion of total phenotypic variability caused by genetic variance in a population. Large pedigree, twin and adoption studies allow the calculation of heritability and they all evidence a strong genetic component in human obesity (25–27). Before the first obesity gene identification reports, scientists considered the possibility that heritability, a global estimate of genetic predisposition to obesity, may be modulated by specific environments (28). Specific environmental exposures known to mediate heritability estimates include biological, socio-economic factors and lifestyle factors.
In utero factors have been proposed to modulate offspring’s future risk for obesity (29). The higher estimates of heritability for BMI observed in mother-offspring pairs in comparison with father-offspring pairs suggest a possible modification effect of maternal in utero environment on the offspring’s genetic predisposition to obesity (30). Maternal weight gain during pregnancy may interact with genetic factors to render the offspring more susceptible to develop obesity in young adulthood (31).
Genetic influence on BMI may also interact with sex and age. Sex-specific genetic effects on BMI have been observed in adolescents as well as in adults (32, 33). Heritability of obesity also varies with age. A previous study of over 12,000 twin pairs reported a heritability of 4–9% for BMI at birth, which increased to more than 50% at 5 months of age (34). Heritability estimates increase from infancy to childhood (35), from childhood to pre-adolescence (36), from preadolescence to adolescence (37), and reach a plateau during adolescence and adulthood, and then slightly decrease in late adulthood (38). Longitudinal BMI change from adolescence to young adulthood and from young adulthood to adulthood is a heritable trait, but genetic variants for change in BMI partially overlap with those affecting the level of BMI (39, 40). Moreover, heritability estimates of obesity increase with the severity of obesity status (41).
The investigation of socio-economic factors and lifestyle behaviours has revealed many additional conditions that impact heritability estimates. One may presume that the emergence of a society characterized by food abundance and physical inactivity may increase the impact of environment (and therefore decrease the impact of genes) in the determination of the obese phenotypes. Counter intuitively, the proportion of variability in BMI attributable to genetic variation is increased among people born after the establishment of a modern ‘obesogenic’ environment (42–44). These results are congruent with the seminal work by Claude Bouchard and colleagues showing that the BMI response to long-term overfeeding in young adult male twins is mainly influenced by genetic factors (28). Twin studies have shown that a high level of physical activity can substantially reduce the influence of genetic factors on BMI in both young and older adults (45, 46). PT. Williams studied the parental contribution to offspring’s BMI in 47,691 adult runners and showed that vigorous physical activity (running distance ≥ 9 km/day) decreased the parental contribution to BMI, by 48–58 %, in comparison with runners with moderate physical activity (running distance < 3 km/day)(47). Socioeconomic research indicates that higher educational status is associated with decreased risk of obesity (48), but heritability estimates for BMI in late childhood/adolescence are positively correlated with the level of education of parents (49). Sleep duration is negatively associated with obesity (50). In a twin study, the heritability of BMI (h2 = 70%) in short-sleepers (< 7 hours/day) was more than twice the heritability of BMI (h2 = 32%) when sleep duration was longer (≥ 9 h/day) (51). Weight gain is a well-known adverse effect of antipsychotic medication (52), but a considerable degree of inter-individual variability has been described in literature (53). Two pilot twin/sibs comparison studies have reported heritability estimates of 60–80% for body weight gain in response to antipsychotics in adolescents and adults (54, 55). Weight loss in response to vigorous exercise, diet restriction or bariatric surgery is also highly variable which suggests a heritable component (56–58).
A recent analysis of the Framingham Heart Study analysed how the heritability of BMI was influenced by historical period, life course and physical activity (59). These authors reported that: 1) the heritability estimates of BMI were considerably larger after the mid 1980’s compared to the 3 preceding decades; 2) the genetic influence on BMI appears to decrease across the lifespan, with the greatest genetic influence observed during reproductive ages across historical period and 3) the heritability of BMI was considerably smaller among physically active individuals aged 21–50 years, but not among those >50 years old (59).
Obesity predisposing gene variants interact with the environment
Although heritability studies provided early evidence for the genetic contribution to obesity, recent efforts have focused on the identification of specific gene variants that impact obesity risk. Our knowledge about the genetic architecture of Mendelian (syndromic and non-syndromic) and polygenic forms of obesity has greatly expanded in the last 20 years (17). It is noteworthy that even some forms of Mendelian (syndromic and non-syndromic) obesity can display a somewhat variable phenotype (60–63). This can be attributed not only to genetic heterogeneity, gene-gene interactions and inheritance model (64, 65), but interactions with environmental factors should be considered as one of the causes for the variability in obese phenotypes (23). Since the rapid increase in obesity prevalence over the last few decades indicates a strong environmental influence on BMI (e.g. physical activity, diet, educational status, age, sex) (3), many researchers have worked on the identification of specific environmental factors that interact with monogenic and polygenic obesity predisposing genes. The existing evidence regarding the study of obesity indicates that lifestyle factors can significantly modify the impact of obesity predisposing gene variants.
Obesity predisposing gene variants interact with non-modifiable biological factors
Obesity predisposing gene variants interact with pregnancy and in utero factors
Pre-pregnancy maternal obesity and excessive weight gain during pregnancy are both associated with increased birth weight, higher rate of macrosomia in the offspring (66, 67) and higher risk of adiposity in offspring during childhood, adolescence and adulthood (29, 68, 69). Recently, a morbidly obese female patient with a rare homozygous LEPR mutation was reported to gain 110 lbs during pregnancy, far beyond the 11–40 lbs gestational weight gain range recommended by the Institute of Medicine, and gave birth to a baby with macrosomia (70). These data suggest that a Mendelian predisposition for obesity increases gestational weight gain and offspring’s birth weight.
However, no such effect on gestational weight gain was observed for a polygenic gene score composed of four common obesity-predisposing common variants in or near FTO, MC4R, TMEM18 and GNPDA2 (71). Studies with gene scores including more SNPs are needed to further investigate this hypothesis.
Prenatal exposure to maternal cigarette smoking was found to interact with genetic variation in OPRM1 to modulate fat intake in offspring (72). Among 956 adolescents, the T allele in OPRM1 was associated with lower fat intake but only in those without prenatal exposure to cigarette smoke (72). DNA methylation was significantly reduced within several CpGs across OPRM1 among adolescents exposed to prenatal maternal cigarette smoking compared to those not exposed (72).
Obesity predisposing gene variants interact with sex
Females are generally more likely to develop morbid obesity than males (73) and these discrepancies may be explained in part by sex-specific genetic effects. In line with this hypothesis, pathogenic monogenic mutations in MC4R have an effect on BMI about twice as strong in females as in males (18, 74).
Seven out of 14 polygenic loci convincingly associated with waist-to-hip ratio displayed sexual dimorphism, all with a stronger effect on the phenotype in women than in men (75, 76). A recent genome-wide interaction meta-analysis did not report any sex-specific for variants associated with BMI, but found 44 loci with sex-specific effects on waist-to-hip ratio adjusted for BMI (28 of the 44 loci displayed larger effects in women than men) (77).
Obesity predisposing gene variants interact with age
The syndrome of Prader-Willi has two distinct phenotypic stages. In infancy, it is characterized by poor suck, feeding problems and failure to thrive, followed by hyperphagia in later childhood that leads to excessive weight gain (78). Rare deletions in the region p11.2 of the chromosome 16 have been associated with a highly penetrant mendelian form of obesity with additional developmental features (79). These individuals generally have early feeding and growth difficulties, and start to gain excessive weight around 5–6 years of age. As a result, an incomplete penetrance for childhood obesity but a complete penetrance for adult obesity has been observed for the carriers of the chromosome 16p11.2 deletion (79, 80). The longitudinal study of adult MC4R mutation carriers shows an increasing age-dependent penetrance (18).
The life-course analysis of the intronic FTO gene variant and BMI in longitudinal studies indicates that this polygenic obesity-predisposing variant is negatively associated with BMI during infancy (age: 0–2.5 years) but positively associated with BMI from the age of 4 years, with an age-dependent increase during childhood, adolescence and young adulthood (36, 81–84). Most of the effect of the FTO intron 1 SNP on BMI gain occurs during this period, and no appreciable effect of FTO on BMI increase is observed during adulthood and agedness (83, 85–88). Studying the association of an obesity gene score from multiple markers in longitudinal cohorts provided similar results: the genetic predisposition score displayed a moderately positive association with birth weight, and more strongly associated with BMI gain during early infancy and childhood, but no association with BMI change during adulthood was observed (89–91). A negative genotype × age interaction between the PCSK1 rs6232 SNP and obesity traits was observed in two independent studies, and a recent meta-analysis of up to 331,175 individuals confirmed this result and identified a similar interaction between age and the PCSK1 rs6235 SNP on obesity (92–94). A genome-wide interaction meta-analysis also identified 15 BMI loci with age specific effects, 11 of which showed greater effect sizes in younger (<50 years) compared to older (≥ 50 years) adults (77).
Obesity predisposing gene variants interact with lifestyle factors
Obesity predisposing gene variants interact with an obesity-prone environment
The promotion and globalization of societal changes leading to an imbalance between calorie intake and calorie expenditure partly explain the current obesity epidemic, but interactions between genes and this obesity-prone environment also contribute to the development of obesity. Dudley et al. reported a significant cohort effect on the prevalence of obesity in Prader-Willi syndrome (62). Prevalence of obesity was higher in patients born after 1990 than before (62). A generation-dependent penetrance of MC4R pathogenic monogenic mutations on obesity was also found in multigenerational pedigrees, with the effect of mutations on the obesity phenotype being amplified by the emergence of an “obesogenic” environment (18, 95).
This trend is supported by a recent analysis of the Framingham Heart Study (FHS), which demonstrated that risk allele carriage in FTO rs9939609 was associated with a greater increase in BMI among individuals born after 1942 compared to those who were born before 1942 (96). The FTO intron 1 variant is weakly associated with BMI in South Asian Indian populations, but its effect on weight is stronger in urban compared to rural dwellers (97, 98). A lack of association of FTO with obesity-related traits was also observed in a Gambian rural population (99). The authors speculate that the impact of genetic variance in FTO rs9939609 on BMI may be marginal in lean populations where excess food is scarce, compared to populations where food is abundant (99). Lastly, the growing influence of obesity predisposing genes in ‘obesogenic’ environments has also been supported by the positive interaction between birth year and the impact of 32 obesity predisposing genes (100). Together, these data suggest a stronger influence of genetic factors on obesity in obesity-prone environments.
Obesity predisposing gene variants interact with physical activity
Recent data indicate that genetic predisposition to obesity can be blunted in part through physical activity. Over twenty independent studies reported an interaction between the FTO obesity risk genotype and physical activity on BMI variation or obesity in children, adolescents and adults (47, 101–122). An interaction between FTO intron 1 variant and the level of physical activity on obesity was recently confirmed in a meta-analysis of 218,166 adults where physical activity attenuated the odds of obesity by 27% conferred by the variant (123). No such interaction was found in 19 268 children and adolescents (123). We recently studied more accurate surrogates of physical activity and adiposity in a multi-ethnic study of 17 423 participants recruited in 17 low-, middle- and high-income countries and we observed that the effect of FTO rs1421085 on the variation of body adiposity index was reduced by 56% in the higher versus lower metabolic equivalent score tertiles (121). Similar results were obtained for a genetic predisposition score combining the information of 12 obesity-associated SNPs, and a high level of physical activity was associated with a 40% reduction in the genetic predisposition to obesity in adults (N=20 430), as well as for BMI level and BMI change across time (110). Physical activity was also found to attenuate the effect of a 28 SNP obesity gene score on BMI among a sample of East Asians and Europeans (124). We did not evidence any significant interaction between the quantitative level of physical activity and a 14 SNP obesity gene score on BMI or body adiposity index in an international multi-ethnic study of 17 423 participants (121). Our data, consistent with the conclusions of a recent meta-analysis in participants of European ancestry living in North America and Europe, suggest that the benefits of being physically active may be optimal in genetically predisposed people living in the more sedentary countries (121, 125).
A number of recent studies have analysed the interaction between sedentary behaviours and genetic risk for BMI, independent of physical activity level (111, 126, 127). The initial report by Qi et al demonstrated that prolonged television watching accentuated the impact of a 32 SNP genetic risk score on BMI (111), and a second study of an adolescent sample reported that screen time increased the impact of two SNPs (FLJ35779, GNPDA2), although these interactions were ethnic specific and of nominal significance (126). The most recent study of this interaction analyzed how total sitting time impacted the association between FTO rs9939609 and BMI among the Framingham Heart Study (FHS) and the Women’s Health Initiative Study (WHI), but the results were not significant (127).
Obesity predisposing gene variants interact with diet
Rouskas et al. reported that the penetrance of MC4R loss-of-function heterozygous mutations on obesity is exceptionally low (6.3 %) in the Greek population, in comparison with those observed in other European countries (60–100%)(128). A possible explanation of this ‘Greek paradox’ may be a protective effect of the Mediterranean diet against MC4R deficiency-induced obesity (128).
Several studies have characterized the impact of diet patterns on genetic predisposition to obesity. Independent cross-sectional and longitudinal samples of Caucasians and Latin Americans, suggest that a high daily energy intake, high fat intake or high saturated fat intake can amplify the effect of the FTO genotype on obesity risk in children, adolescents and adults (105, 114, 129–132). Higher intake of fried foods has also been shown to increase the impact of a 32 SNP gene score (and an FTO variant individually) on BMI over follow-up (133). These interactions were replicated in an independent cohort of 21 421 women (133). A recent 25 year follow-up study in Australia reported an interaction between rs9939609 in FTO and diet on BMI change (134). The prudent/healthy diet was associated with a greater BMI change among AA compared to TT genotypes. This interaction was observed at 17 years of follow-up, but was restricted to females (134). Increased intake of sugar-sweetened beverages has also been shown to increase the impact of a 32 SNP genetic risk score on BMI (135). This interaction effect was also observed for incident obesity cases and replicated in an independent cohort (N=21 740) (135).
Despite the many studies demonstrating that diet patterns can moderate the genetic risk for obesity, two recent meta-analyses did not detect significant interactions between diet patterns and obesity-associated gene variants (136, 137). Data from 177 330 adults (87% Whites, 10% Asian, 3% African American) did not indicate any significant interactions between the FTO variant and dietary intake of total energy, fat, protein or carbohydrate on BMI(137). A second meta-analysis of 68 317 Europeans did not detect an interaction between a 32 SNP genetic risk score and a multifactorial diet score on BMI (136). The diet score moderated the impact of two SNPs on BMI (LRRN6C and MTIF3), although these effects were nominal and the impact of these risk variants appeared to be greater among those consuming healthier diets (136). The authors speculate that the broad diet assessment in their analysis may have masked interactions of varying directions and magnitudes that were identified in previous studies (136).
The Apolipoprotein A-II (APOA2) −265 T>C promoter functional polymorphism appears to interact with high-saturated fat to increase BMI and obesity risk in several independent populations (Mediterranean, Asian, Caucasian, Hispanic and Carribean) (138, 139). High saturated fat intake was associated with significant increases in the genetic risk for obesity across populations (139). Specifically, the C allele homozygotes with high saturated fat intake displayed a 1.84 (95% CI, 1.38–2.47) odds of obesity compared to a 0.81 (95% CI, 0.59–1.11) odds in those with low saturated fat intake (139). A separate analysis of 1 225 obese adults demonstrated the C allele homozygotes with a high saturated fat intake (> 20.7 g/day) had higher waist circumference values than individuals with any other genotype in the high saturated fat intake group (140). While the underlying biological mechanism explaining this association is not fully understood, the APOA2 −265 T>C SNP has been associated with obesity risk eating behaviours such as meal skipping, and dietary modulation of plasma ghrelin (140).
The Apolipoprotein A5 protein influences plasma triglyceride concentrations in humans and regulators of the APOA5 gene (peroxisome proliferator-activated receptors, insulin, thyroid hormone) have been implicated in obesity risk (141, 142). In a weight loss study of 606 men with hyperlipidemia, C allele carriers of the −1131 T>C variant in the APOA5 gene displayed significantly greater BMI reduction while on a fat restriction diet (143). Additional evidence from the Framingham risk study suggests that carriers of the mutant C allele may have a lower risk of obesity compared to T allele homozygotes when consuming a diet high in monounsaturated fats (144). This interaction was also tested in a Mediterranean sample and greater fat intake was associated with obesity among T allele homozygotes while no association was observed among carriers of the mutant C allele (145). These studies suggest that the C allele in APOA5 may have a protective effect against obesity among individuals consuming a high fat diet.
Several studies have examined the interaction between diet patterns and PPARG Pro12Ala polymorphism with regards to obesity (146), (147). The risk allele 12Ala has been linked to increased obesity risk through meta-analysis, although some heterogeneous effects of this mutation have also been observed in interaction studies (148–150). Lamri et al analysed the interaction between PPARG Pro12Ala polymorphism and fat intake, and observed that AlaAla individuals displayed greater BMI values than Pro carriers among high fat consumers (151). In contrast, a study of 720 French Canadians found that higher amounts of saturated or total fat consumption were associated with greater waist circumference in Pro allele homozygotes but not in 12Ala carriers (147). Similar results were observed from studies analysing BMI. An investigation of the Health Nurses’ Study demonstrated that high total fat intake was associated with greater BMI among participants homozygous for the Pro allele but not among 12Ala carriers (152). This study also reported that monounsaturated fat intake was associated with decreased BMI among 12Ala allele carriers and this interaction was replicated in an independent weight loss study (153). Additional interaction studies of PPARG related to body composition have shown sex-specific effects (154, 155) and weight change analyses have reported inconsistent results (153, 156–159).
A more recent analysis found an interaction between an eight SNP obesity gene score and mono and polyunsaturated fatty acid intake (160). Among 2 346 children with low unsaturated fat intake, the gene score was associated with increased body fat mass index yet no association was present among unexposed children (160).
Obesity predisposing gene variants interact with psychosocial stress
A recent genome-wide interaction analysis identified a significant interaction between psychosocial stress and five SNPs within the Early B-cell Factor 1 (EBF1) gene and hip circumference (161). The interaction reached genome-wide significance among the subset of 2460 Whites in the Multi-Ethnic Study of Atherosclerosis (MESA) but was not significant among Chinese Americans, Blacks or Hispanics. This study reported that the impact of risk allele carriage in EBF1 on hip circumference was greater among participants with a greater chronic stress burden (161). The authors also replicated the interaction between psychosocial stress and three of the original five SNPs (rs17056278 C>G, rs17056298 C>G, and rs17056318 T>C) in EBF1 in the Framingham Offspring cohort(161). A subsequent analysis by the same research group replicated the EBF1 × psychosocial stress interaction on obesity (waist circumference or BMI) in the Family Heart Study Whites and at trend level in the Duke Caregiver study (162). The direction of the interaction effect was consistent across each of the studies: chronic psychosocial stress amplified the effect of EBF1 variation on BMI (162).
Obesity predisposing gene variants interact with educational status
Epidemiological studies have shown an association between a low level of education and higher risk of overweight and obesity (48). A significant negative association between BMI and educational status was found in non-carriers of MC4R mutations but not in MC4R loss-of function mutation carriers issued from the same pedigrees (18). These results show that a high level of education has no protective effect on obesity risk in presence of MC4R pathogenic mutations.
On the contrary, a significant gene × education interaction has been found in the intron 1 variant in FTO, the significant effect of the SNP on BMI and obesity risk restricted to subjects with no university education (163). This finding is supported by a recent study of European children (N=16 228) indicating that favourable socioeconomic status is protective against obesity, yet this effect was only observed in participants with the low risk genotype TT in FTO rs9939609 (164).
Obesity predisposing gene variants interact with smoking status
A meta-analysis of nine European study samples (N=24 198) demonstrated that smoking status moderated the association between genetic variation at the CHRNA5-CHRNA3-CHRNB4 locus (rs1051730) and BMI (165). While there was no evidence of association between variation at rs1051730 and BMI in never smokers, each additional risk allele (T) was associated with a BMI decrease of 0.16 and 0.33 kg/m2 among former and current smokers, respectively (165). A separate study of 14 131 Pakistani adults reported another gene × smoking interaction: the minor allele (T) in FLJ33534 was associated with lower BMI in current smokers and positively associated with BMI among adults who had never smoked (166). A number of gene × smoking interactions were identified when African Americans and Caucasians were analysed separately, but no significant interactions were observed in the overall sample from the Southern Community Cohort Study (167). Four nominally significant gene × smoking interactions were reported in a recent study of 16 157 Pakistani adults, with current smoking status amplifying the effect of PTBP2 rs11165643, HIP1 rs1167827 and GRID1 rs7899106 SNPs, and decreasing the effect of C6orf106 rs205262 SNP (120).
Obesity predisposing gene variants interact with alcohol consumption
Among a sample of 3 522 East Asians, increased alcohol consumption was associated with an increase in the effect of a 29 SNP GRS on BMI (122). Increased alcohol intake was also reported to increase the impact of PPARGC1A rs4619879 among African Americans, but this interaction was not significant in Caucasians or in the combined sample (167).
Obesity predisposing gene variants interact with disease status/response to treatment
Obesity predisposing gene variants interact with specific health conditions
Beyerlein et al suggest that pre-existent overweight may double the effect of an obesity genetic predisposition score on body fat mass in 4 837 European children (168). This association is supported by an independent study of 7 225 children of European ancestry which found that previously identified obesity predisposing loci had a greater impact on BMI among obese children compared to their non-obese counterparts (169). Similar results were observed in 1 930 adults of European descent (170). If true, it signifies that obesity predisposing genes may have an even more detrimental effect on weight gain once overweight/obesity is established. Depression predicts subsequent development of obesity (171) and depression status has been shown to amplify the effect of FTO SNPs on BMI (172). Moreover, obesity has an important role in the etiology of polycystic ovary syndrome (173) and FTO intronic SNP has larger effects on BMI in patients with polycystic ovary syndrome than in subject from the general population (174, 175).
Obesity predisposing gene variants interact with lifestyle modifications
A strict, fat-reduced, and carbohydrate-modified diet leads to a long-term marked weight reduction in adolescents with Prader-Willi syndrome who are already overweight (176). Importantly, if diagnosis is made at an early age and intensive diet management starts early, reasonable weight control is achieved in non-obese patients with Prader-Willi syndrome (177, 178). Regular exercise training has beneficial effects on body composition and weight loss in Prader-Willi syndrome patients (179, 180), especially as they tend to be less physically active than obese non-syndromic individuals (181). MC4R or POMC monogenic patients respond as well as non-monogenic obese patients to hypocaloric dietary or multidisciplinary (exercise, behavior, nutrition therapy) interventions (182, 183) but MC4R monogenic patients fail to maintain weight loss after intervention (183).
The obesity risk variant rs9939609, an allele in FTO, does not modify the weight loss response to lifestyle intervention (184–186) or caloric restriction (187, 188), but is associated with lower additional weight loss and higher risk of weight regain during the weight maintenance phase that follows the caloric restriction program (188). Carriers of the FTO intron 1 obesity risk variant experience a higher rate of dropout when they are submitted to a high-fat/low carbohydrate (in comparison with a low-fat / high carbohydrate) hypocaloric diet (189), but they achieve better weight maintenance than wild-type individuals during a 3-year intervention with a Mediterranean diet (190). FTO variation has also been shown to interact with protein intake to moderate the response to weight-loss interventions (191, 192). A two-year randomized control trial (RCT) found that higher protein intake was associated with improved weight loss and body composition among carriers of the FTO rs1558902 risk allele compared to non-carriers (191). Another analysis of the same trial (N >700) showed that individuals with the FTO rs9939609 risk (A) allele achieved more favourable changes in food cravings and appetite when consuming a higher-protein weight-loss diet (192). Carriers of the FTO obesity risk alleles also lose less weight in response to exercise training (193, 194). Among five childhood obesity susceptibility loci identified in a French-German genome-wide association studies (GWAS) meta-analysis (195), only one (SDCCAG8) was associated with differential weight loss after lifestyle intervention in 401 children and adolescents (196). Eight out of 15 obesity predisposing gene variants recently identified by GWAS showed trends of association with weight loss or weight regain during lifestyle intervention in 3 356 adults of the Diabetes Prevention Program (197).
The PLIN gene has received increasing support for its role in obesity risk and insulin resistance, and genetic variation at the perilipin locus has been shown to interact with diet behaviours (198–200). Two separate studies have shown that A carriers of the 11 482 G>A SNP at the perilipin locus lost less weight in response to weight loss interventions compared to non-carriers (199, 201).
Obesity predisposing gene variants interact with therapeutic treatment
As most obese persons are resistant to the weight-reducing effects of leptin, administration of recombinant leptin to obese subjects does not generally result in significant weight loss (202). However, patients with congenital leptin deficiency markedly reverse obesity and associated phenotypic abnormalities when they are treated with daily injections of recombinant human leptin (203, 204). Leptin administration reduces energy intake, fat mass, hyperinsulinemia, and hyperlipidemia, restores normal pubertal development, endocrine and immune function and improves neurocognitive performances (205). Although patients with complete leptin deficiency are extremely rare, leptin supplementation may eventually help a far greater number of obese patients with partial leptin deficiency (heterozygous for a loss-of-function mutation in the LEP gene) based on the observation that leptin therapy induces more significant weight loss in subjects with low leptin levels (206, 207).
The guanine nucleotide binding protein beta polypeptide 3 (GNB3) C825T functional gene variant predicts that obese individuals will benefit more from the anti-obesity drug sibutramine treatment. Sibutramine is a serotonin and norepinephrine reuptake inhibitor and given that GNB3 variance is associated with an altered response to G protein subunit activation (α 2-adrenergic activation) (208), there is biological evidence to support this interaction. Two independent studies showed that the carriers of the 825 T allele lose more weight in response to sibutramine administration than C allele homozygotes (209, 210). Lastly, obesity predisposing gene variants in FTO and MC4R are associated with more weight gain in response to antipsychotic treatments (211–215).
Obesity predisposing gene variants interact with bariatric surgery
Bariatric surgery is the most effective long-term treatment for severe obesity (216). Laparoscopic adjustable gastric banding did not result in a long-term weight reduction in a 18-years old patient with complete MC4R deficiency (217), and was associated with an high risk of conversion to bypass operations in individuals with partial MC4R deficiency (218). On the contrary, three studies confirmed that Roux-en-Y gastric bypass surgery was an efficient strategy to lose weight in MC4R mutation carriers (219–221). These results suggest that diversionary operations, which are more invasive but efficiently improve the neuro-hormonal control of satiety than gastric banding procedures, be recommended in the context of non-syndromic monogenic forms of hyperphagic obesity.
FTO risk allele carriers lose less weight than common allele homozygous individuals after banding surgery (222, 223), but experience a similar level or more weight reduction after gastric bypass surgery (222, 224).
Biological processes underlying statistical gene-environment interactions
Epigenetic changes are believed to be one of the primary mechanisms explaining interactions between environmental exposures and genetic variation (225, 226). Other potential mechanisms propose that the transcription changes induced by environmental exposures may vary across genotypes (225). Epigenetics is defined as heritable changes in gene function that cannot be explained by changes in the deoxyribonucleic acid (DNA) sequence, and the three main types of epigenetic modification in mammals include DNA methylation, histone modification and non-coding RNA (226, 227). Of these mechanisms, DNA methylation has received the most attention in human studies, and in mammals this process mainly occurs at CpG dinucleotides (228). Specifically, covalent bonding of a methyl group to the cytosine base creates a barrier that inhibits transcription factors from binding to the DNA helix (228, 229). CpG DNA methylation at gene promoters is typically associated with gene silencing, whereas CpG methylation in gene bodies is linked to gene activity (228). Given that epigenetic differences have been linked to obesity status, as well as genetic variation and a variety of pre and postnatal environmental factors, these processes likely represent a plausible mechanism of gene-environment interactions.
The emergence of new approaches to study epigenetic variation, such as epigenome-wide association studies (EWAS), has led to the identification of methylation patterns associated with obesity (230). Increased BMI among adults was found to be positively associated with increased methylation at the hypoxia-inducible factor 3 alpha (HIF-3α) locus in blood cells and adipose tissue (231). This finding was confirmed in two replication cohorts in the initial analysis (231), as well as two additional independent studies (232, 233). A separate EWAS of an African American sample identified an association between methylation at 37 CpG sites and BMI, which were replicated in two cohorts of European ancestry (232). Analyzing whole-genome DNA methylation and expression data in human adipose tissue from 96 males and 94 females revealed that DNA methylation and expression of 2 825 genes was correlated with BMI (234).
Existing evidence also supports the association between environmental exposures on DNA methylation patterns. Monozygotic twins, who are epigenetically indistinguishable at birth, exhibited drastically different overall content and genomic distribution of DNA methylation and histone acetylation in later life (235). Moreover, methylation and expression of 1050 genes have been found to vary with age (234, 236). The epigenetic divergence that occurs with aging likely reflects the accumulation of environmental exposures that influence methylation patterns. Prenatal factors including maternal BMI and variations in maternal methyl donor intake during pregnancy have been linked to methylation changes in the offspring (237), and multiple studies have shown that maternal vitamin B12, folate and cobalamin levels during pregnancy are associated with offspring adiposity (238, 239). Folate, vitamin B12 and choline are methyl donors and involved in the synthesis of methionine, the precursor of the universal donor of methyl groups needed for DNA methylation (S-adenosylmethionine). As a result, disregulation in any of these components can alter the epigenomic regulation of gene expression (240). With respect to postnatal determinants of DNA methylation, exercise interventions have been shown to alter the DNA methylation of 2817 genes in skeletal muscle and 7663 genes in adipose tissue (18 of which were obesity candidate genes) (241). The effect of exercise on DNA methylation appeared to be tissue specific, with the majority of genes in skeletal muscle displaying decreased DNA methylation (242), and the majority of genes in adipose tissue showed increased DNA methylation (243). These changes mirrored the patterns observed for gene expression: most of the genes showing concurrent changes in DNA methylation and expression displayed increased expression in skeletal muscle and decreased expression in adipose tissue (241). A recent review of 25 studies (16 observational 9 interventional) found that both acute and chronic exercise significantly influenced DNA methylation, and these changes occurred in a tissue- and gene-specific manner (244). DNA methylation changes have also been observed in response to high-fat intake (245–248), and after weight loss interventions the methylation profiles of adipose and muscle tissue among those formerly obese became more similar to lean individuals (249–252). These methylation changes involved a number of known obesity-associated loci, including LEPR, STAB1, ZNF608, HMGA1, MSRA, TUB, NRXN3, FTO, MC4R and BDNF (250, 251). Both exercise and diet have long been recognized as central determinants of body weight regulation in epidemiological studies (253, 254). The epigenetic changes induced by these exposures likely explain a portion of their association with BMI as well as their interaction with obesity predisposing gene variants (102, 125).
Although environmental factors have the potential to influence the epigenetic environment, it is estimated that approximately 20–40% of epigenetic variation can be attributed to genetic differences (226, 255, 256). Early evidence demonstrated that the risk allele of FTO promotes increased methylation of sites within intron 1 of the FTO gene, as well as greater methylation of additional genes (257). Other evidence identified 28 obesity-associated SNPs that were associated with differential methylation at 107 proximal CpG sites (258). A recent study of Trim28 haploinsufficiency used findings from mice and humans to demonstrate the variation in obesity phenotypes that can be induced through epigenetic changes (259). These authors also reported that FTO expression was decreased among Trim28_Low obese children compared to Trim28_Low lean individuals (259).
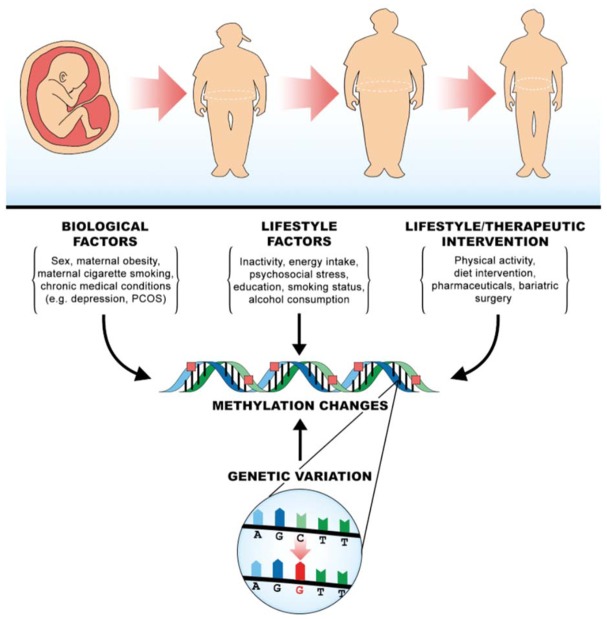
These epigenetic findings support an emerging mechanistic model to explain gene-environment interactions in obesity. Existing evidence indicates that obesity, pre and postnatal environmental factors and multiple obesity associated gene variants are linked with epigenetic patterns (Figure 1). Given that gene variants, such as those in FTO, and environmental factors both play a role in the methylation of obesity genes, the balance between these two effects likely impact the manifestation of genetic obesity. This biological model provides support for many of the statistical interactions reported to date, and further integration of genomic, epigenomic and transcriptomic data with gene-environment interaction studies will aid in uncovering these biological mechanisms.
Figure 1.
Biological model to explain gene-environment interactions in obesity.
Methodological considerations in gene-environment interaction studies of obesity: recent developments and future options
Despite the rapid growth of GEI studies in the field of obesity, there has been increasing scrutiny regarding the validity of these studies based on several issues including statistical modelling (260, 261), confounding (260, 262), a low replication rate (263–265), underpowered analyzes (266), lack of biological assumptions (267, 268) and measurement precision (269) (Table 1). The relevance of testing interactions between individual genetic variants and specific environmental exposures has also been questioned (260). Based on these concerns, some leaders of opinion have suggested that a large proportion of significant gene × environment interaction (GEI) findings are in fact false positives (23, 260). This scepticism has been adopted by multiple journals, which have implemented stringent criteria for candidate gene and interaction studies considered for review (270, 271).
Table 1.
Summary of methodological issues and solutions for gene-environment interaction studies in obesity.
| Methodological Issue | Suggested Solution | Reference (Lead author |
|---|---|---|
| Modelling the G × E cross-product terms | Include an additional coefficient to model non-linear genetic effects (β 4G2), and a second to account for non-linear interaction effects (β 5G2 × E) | Aliev, Bavav Genet, 2014 |
| Comparing biological frameworks (e.g. diathesis-stress model vs. differential susceptibility framework) | Adjust the parameters in the regression equation to compare alternate theoretical frameworks | Belsky, Psychol Bull, 2009 Widamen, Psychol Methods, 2012 |
| Selection of interaction scale (e.g. additive vs. multiplicative) | Consider the application of the interaction test a priori. Additive scales have been recommended for identifying heterogeneous effects across subgroups in public health settings, while multiplicative scales are suggested for studying disease etiology | Ottman, Prev Med, 1996 |
| Confounding of the G × E interaction term | Include all covariate × gene and covariate × environment interaction terms | Keller, Biol Psychiatry, 2014 |
| Shared heritability between the outcome and covariates | Avoid the inclusion of heritable covariates that are associated with the gene variant being tested | Aschard, Am J Hum Genet, 2015 |
| Correlation between the gene variant under study and the interacting environmental factor | Directly analyze the relationship between the interacting gene variant and environmental exposure to ensure that they are not correlated | VanderWeele, Am J Epidemiol, 2013 |
| Variations in gene expression/silencing, and changing the heritability of BMI throughout development | Use a repeated measures design or include a G × E × Time term if the sample size is sufficient | Liu, Environ Health, 2012 |
| Changing heritability of BMI throughout development | Use existing gene × age interactions to identify variants with differential effects across the lifespan | Elks, Front Endocrinol, 2012 Winkler, PLoS Genet, 2015 |
| Measurement error associated with the environmental exposure and outcome | Consider more accurate measurement tools or repeated measures in favour of large sample sizes with less accurate measures | Wong, Int J Epidemiol, 2003 |
This portion of the review will focus on issues in GEI studies related to (1) statistical modelling of interaction terms, (2) modelling of confounding variables, (3) timing of environmental exposure across the life span and (4) measurement of predictor and outcome variables. The final section will provide suggestions to these issues based on existing evidence and will outline future directions of GEI research.
Statistical modelling issues in gene-environment interaction research
Using a multiple linear regression model with the inclusion of a cross-product term signifying the product of environmental (E) and genetic (G) variables is the most common method to assess interactions (107, 114, 117, 125). Coding genetic polymorphisms is either performed to create a binary variable (under a recessive or dominant model) or a three-category variable based on an additive model, with the latter often used when the true functional model of a given marker is not known (261). A recent analysis demonstrated that modelling gene-environment interactions with a simple cross-product term (G × E) often produces misleading results when assuming an additive model (261). First, the simple cross-product model always forces the regression lines to be ordered (0, 1, 2 and never 0, 2, 1). While this assumption may be intuitive from a biological perspective, this approach will always predict an ordered effect of genotypic differences even when the data do not reflect this assumption (265). Second, the differences in slopes between the adjacent regression lines are always assumed to be the same (261). There is no rationale for this assumption and in practice, sampling error alone would be expected to create uneven differences between regression slopes (272). Alternatively, non-linear gene-environment interaction effects may be present, which could not be estimated accurately with only a cross-product interaction term (261, 273). Third, this model constrains all three regression lines to cross at the same point when interaction effects are present (261). This implies that there is a certain level of environmental exposure that confers the same level of risk/disease for all three different genotypes. There is no statistical or biological evidence to justify this assumption, especially since the specific genetic model is often not established for the genetic marker of interest (260). These simulations indicate that the models using only the cross-product term are more vulnerable to Type 1 and Type 2 errors (261). In all cases, including two additional coefficients, one to model non-linear genetic effects (β 4G2) and another to account for non-linear interaction effects (β 5G2 × E), represented the interaction (or lack of interaction) more accurately. Many authors recommend this model for genetic variants following an additive or unknown genetic model, and emphasize that failure of an interaction to match a plausible biological interaction likely indicates a false positive result (261, 274). In summary, re-conceptualizing interaction models to account for nonlinear effects removes the constraints of traditional regression techniques and provides a more accurate representations of gene-environment interaction effects (261).
Other authors contend that traditional GEI analyses neglect to test the a priori hypotheses that form the basis of these studies (275). The implicit framework adopted in traditional GEI analyses is the diathesis-stress model of environmental action (276), which specifies that certain individuals are more vulnerable to the adverse consequences of some exposures than others (277). As an extension of this limitation, exploratory approaches also fail to test or compare the competing predictions from alternative theoretical frameworks such as the more recent differential-susceptibility framework (278). This theory posits that some individuals are more susceptible to not only negative exposures, but to positive environmental influences as well (279). Based on this characterization some authors have proposed that gene variants classically referred to as ‘vulnerability genes’ be reclassified as ‘plasticity genes’ to correspond with the differential susceptibility framework (280). This theory has been proposed by several authors (278, 281), and has been applied to the study of GEI (280, 282, 283). In response to these competing frameworks, many statistical criteria were developed to distinguish differential-susceptibility interactions from those representing diathesis-stress theory (279, 284), and Widamen’s confirmatory method appears to be the most efficient (285). This technique directly evaluates alternative theoretical frameworks by aligning the analyses with each hypothesis (285). Specifically, this method systematically adjusts the parameters included in the regression equation to compare different theoretical frameworks, and specifies where the regression lines (representing each genetic subgroup) will cross relative to the value of the environmental exposure (285). With respect to the frameworks discussed above, the diathesis-stress theory models an ordinal interaction whereby the predicted outcome value for the genetically vulnerable subgroup is always less than that of the genetically low-risk group. The differential-susceptibility framework predicts that the risk of the outcome for the genetically malleable group can be higher or lower than the genetically non-malleable group depending on the level of the environmental exposure (275). It is important to note that Widaman’s confirmatory approach can be used for dominant/recessive or additive genetic models, and can be applied to other forms of statistical interaction involving competing hypotheses about the nature of the interaction (275, 286). Integration of this technique into interaction studies where the theoretical framework is uncertain may help to improve the accuracy and replication rates of interaction studies. This model is supported by biological evidence from a recent study, which demonstrated that TRIM28 knockout mice are alternatively lean or obese depending on subtle environmental changes (259). Other consequences of this framework have important implications for variants identified in GWAS. If a ‘plasticity gene’ displays opposite effects in two exposure groups, the main effect of this variant may not be identified in GWAS and this interaction effect could be missed. Another concern for GWAS is the possibility that interaction effects may only exist among subgroups with very rare exposures. Analyzing both ‘vulnerability’ and ‘plasticity’ gene frameworks may be a method to prevent these issues and identify additional gene-environment interactions in future studies.
Another consideration when analyzing interaction effects is the selection of either an additive or multiplicative interaction scale (287–289). This decision has important implications given that different scales can lead to different conclusions and consequently, different public health recommendations (290). An additive interaction exists when the combined genetic and environmental risk is significantly greater than would be expected if their effects were additive, whereas a multiplicative interaction describes a joint genetic and environmental risk that is greater than expected from multiplying their effects (23, 291). Some authors argue that the selection of measurement scale is less crucial when the underlying biological processes are not known, and both scales can be appropriate in certain situations (292–294). If the pathophysiology consists of a multistage process, such as cancer initiation and promotion stages, two factors that act at the same stage will generally fit an additive model and those acting at different stages will typically fit a multiplicative model (295, 296). It has also been suggested that if the main objective is to study public health impact, an additive scale is better suited to identify heterogeneous effects across subgroups, while the multiplicative scale is more appropriate for studying disease etiology (295).
Confounding issues in gene-environment interaction research
Several modelling strategies have been proposed to address the impact of confounding in gene-environment interaction studies (260, 262). Variables with the potential to offer alternative explanations of an interaction are typically entered into the regression equation as covariates to control for their potential confounding effects (265). While this method controls the influence of confounding on the main effect of the genotype and environment, it does not adjust for potential confounding of the interaction term (260). An alternative method has been proposed whereby all covariate × gene and covariate × environment interaction terms are included in the model that tests the gene × environment interaction of interest (260). If significant covariate interactions are observed, the validity of any gene × environment interactions may be compromised by the covariate and warrant additional analysis. Although there are potential objections to this modelling technique, the justifications of this approach are outlined by Keller (260). First, even though over-fitting the model may prevent accurate estimations of the many covariate interactions, the purpose of including covariate interactions is to control for their effects on the gene × environment interaction rather than producing accurate parameter estimates. Second, multicolinearity between the many interaction terms may diminish the strength of the main gene × environment interaction. This however is the purpose of this procedure and if inclusion of the covariates weakens the main interaction, then the covariates may be significantly influencing the interaction. Lastly, it is reassuring to recognize that the gene × environment interaction term is only marginally affected if there is no ‘true’ relationship between the covariate and the gene × environment interaction (260). One caveat to this approach is that shared heritability between the covariates and the outcome can introduce bias and increase the risk of false-positive results (297). Therefore, including heritable covariates in the model should be avoided if they are associated with the gene variant being tested (297).
Confounding issues can be further complicated if the interacting genetic variant and environmental exposure of interest are correlated (297, 298). Under these circumstances, simulations have demonstrated that uncontrolled confounding will bias the estimates of the main genetic effect and the gene-environment interaction even if the genetic and environmental factor are not associated with the outcome (299). If the genetic variant and environmental factors are independent, this is no longer an issue as long as unmeasured environmental confounders are not related to genetic factor. The issue of gene-environment dependence has been highlighted in extreme cases where the genetic variants are associated with both the environmental factor and the outcome. For example, variants on 15q25 have been linked to both smoking behaviours and lung cancer (300–302). As a result, some authors suggest directly analyzing the relationship between the interacting genetic variant and environmental exposure (299).
Considering time of exposure in gene-environment interaction research
Given that gene expression and silencing varies significantly throughout development, it may be important to consider time of exposure when modelling exposures that can have differential effects throughout the life cycle (303). Evidence from toxicology research indicates that many environmental exposures display distinct dose response curves that vary based on the developmental stage at which exposure occurs (304, 305). The identification of these developmental windows suggests a need to include time of exposure as a third interacting factor when analysing gene-environment interactions (268). However, the inclusion of a three-way interaction term dramatically increases the necessary sample size (260, 306) and this information is rarely available. Simulation studies indicate that the sample size required to detect three-way interactions is four-fold the sample size necessary to detect a two-way interaction of the same magnitude (307). Another statistical method to address this issue involves considering environmental exposure as a time-varying factor to analyze the lag effects of gene × time-varying environment interactions (268). Yet, the repeated measurements needed to measure lag effects are often not feasible due to the cost of repeated measurement in large studies. This constraint explains the high prevalence of cross-sectional case-control designs to study gene-environment interactions (268). The challenge of measuring variations in the impact of environmental exposures is compounded by changes in the heritability of the outcome across time. A meta-regression of heritability studies of BMI found that the genetic contribution to BMI varies with age: heritability was positively associated with age among child studies and negatively associated with age among studies of adults (308). A recent genome-wide interaction meta-analysis identified 15 BMI loci that interacted with age, 11 of which had a greater effect impact in younger (<50 years) compared to older (≥ 50 years) adults (77). Failure to address the time-varying effects of environmental exposures and heritability may account for some of the challenges with replicating gene-environment interactions (23, 260, 309).
Measurement issues in gene-environment interactions research
The measurement of the exposure and outcome also represent important considerations for gene-environment interaction research. Major determinants of power include allele frequency, genetic effect size, the magnitude of interaction effect, risk allele frequency, degree of genetic misclassification and measurement error associated with the exposure and outcome (269, 306, 310). Although the trade-off between precision and feasibility is common to most study designs, the large samples required to study interaction effects make this balance particularly important. Currently, the most notable gene-environment interactions in obesity have measured diet patterns or physical activity as environmental exposures (102, 107, 311, 312). The gold standard criterion measure for these exposures are a 7-day weighed diary and doubly labelled water, respectively. Unfortunately, the large number of participants required for these studies have restricted the measurement of these exposures to less precise instruments. The error associated with exposure measurement generally attenuates the estimate of the true effect size (313, 314). Similar problems occur when the outcome used is an indirect measure for the true outcome of interest. In gene-environment analyses of obesity, BMI is commonly used as the outcome (102, 125, 315, 316), which further contributes to this error given that BMI fails to distinguish between fat and fat-free mass (317).
Simulations have characterized how varying different determinants of power can impact the required sample size of gene-environment interaction studies (269, 306). As an example of these analyses (269), genetic misclassification was fixed at 2.5% to be consistent with prior empirical studies (318, 319) and the magnitude of effect for the common allele was also constant. With a correlation between the true and observed exposure and outcome of 0.6 and 0.7, respectively, a sample size of just over 9 500 is needed to detect an interaction at a significance of 10−4 with 95% power (17). However, the correlations of 0.6 and 0.7 between the true and observed exposure and outcome are unusually high for gene-environment interactions in obesity due to the cost of precise measurement tools (125, 310, 320). With more typical correlations of 0.3 and 0.4, the required sample size can increase to over 100 000 participants with all other variables held fixed (269). If precise instruments are not available to mitigate this error, performing repeated measurements is a useful strategy on condition that the error in repeated measures is not correlated (313). As an example, performing two independent repeated measures using a tool with a validity coefficient of 0.6 increases the overall validity coefficient to almost 0.8. With all other variables being fixed, this reduces the necessary sample size by more than a factor of six (269).
The value of improving measurement accuracy as opposed to increasing sample size can be reinforced with the example of physical activity measurement, a common exposure analyzed in gene-environment interactions of obesity (117, 125, 269). Physical activity is usually assessed by questionnaire, and even comprehensive instruments that address occupational and leisure activity rarely correlate with objective measures of energy expenditure above 0.3 (321). The physical activity assessment used in the EPIC-Norfolk study displayed an overall correlation of 0.44 with objective measures, although this fell to 0.28 after adjustment for age and sex (322). The error associated with measuring this exposure is compounded by the moderate correlation (0.5) of BMI with body fat percentage as measured by dual-energy x-ray absorptiometry (323). Using the EPIC-Norfolk questionnaire with BMI as an outcome would require almost 90 000 participants to detect an interaction that doubled the effect of a genetic variant, when the variant is present in 20% of the population (269). Since a doubling of genetic risk from an environmental exposure is at the upper limit of interaction effect estimates reported for common variants and exposures (324–326), some authors speculate that the majority of published interaction studies are underpowered and report “lucky” true-positives or false-positive results (23). A recent study by our group provides an empirical example of how measurement precision can influence statistical power. We analyzed physical activity × FTO interactions on BMI using two measures of physical activity: a three-level categorical variable and a quantitative estimate of energy expenditure. The categorical data was available in 99% of the sample while the quantitative energy expenditure data was only available in 63% of the sample. Despite this disparity, similar interactions were detected using both measures, which may suggest that the added precision of the energy expenditure data compensated for the decrease in sample size (121).
Given the sample size requirements imposed by this type of data, more direct measurement techniques have been proposed. Objective measures such as heart rate monitors carry increased precision while maintaining feasibility in moderately sized epidemiological studies (327). Heart rate monitor data have demonstrated a correlation with the gold standard of energy expenditure methods (doubly labelled water) of 0.73 (328). Two repeated measures can increase this correlation to over 0.88. Substituting this method of exposure measurement for questionnaire methods would decrease the necessary sample size to 9453, a decrease by a factor of 10 (269). Therefore, the gain in precision associated with more accurate measurements of exposure may be less resource intensive than accruing large sample sizes. The power implications of using precise measurement techniques suggest that smaller studies with more accurate measures of exposure and outcome may be better suited to detect gene-environment interactions than large sample sizes with imprecise measurement (269). The issue of measurement imprecision has long been debated in the nutrition field and ‘deep phenotyping’ strategies (measuring metabolic markers such as circulating plasma lipids as a surrogate of a high-fat diet) may be worthwhile alternatives to traditional self-report measures (329–331). Other assessments that may mitigate the concerns associated with traditional diet measures include ad libitum energy intake tests or analyzing the dietary information of food consumed in regulated settings such as cafeterias or restaurants (332).
The issue of direct and indirect measurement of genetic variants also has important implications for statistical power. In many current GWAS and GEI studies, the true susceptibility loci involved in the disease etiology is not known (or unavailable) and the linkage disequilibrium (LD) between marker alleles and the actual disease loci is used to study associations between gene variants and the phenotype under study (333, 334). Since this is an indirect approach, the effect estimate will be underestimated if the LD between the two variants is incomplete (r2< 1) (335). Previous studies have demonstrated that the sample size requirements of GEI studies can be strongly influenced by the marker allele frequency, disease allele frequency, the LD between these loci, as well as the main genetic and environmental effect, the prevalence/impact of the environmental exposure and the magnitude of the interaction (334, 335).
Future directions for gene-environment interactions and obesity
Given that specific environments can greatly impact the magnitude of genetic predisposition to obesity, the systematic study of gene-environment interactions constitutes an important field of investigation in order to inform public health strategies to prevent and manage obesity and other complex diseases. Gene-environment interaction studies in the context of various forms of obesity (monogenic, polygenic) and in diverse experimental designs (observational, interventional) (22) may lead to a better understanding of the protective or detrimental environmental exposures that modify the impact of certain genetic variants. Existing interactions need to be studied in additional obesity-prone (e.g. response to smoking cessation, response to insulin therapy in diabetic subjects) or obesity-protective (e.g. lifestyle intervention, response to the anti-obesity drug orlistat administration or to bariatric surgery) conditions. Gene-environment interaction studies are complementary to observational epidemiology, interventional study or clinical trials, and will certainly help to elaborate efficient strategies to reverse the obesity epidemic.
Currently, GWAS for obesity-related traits have focused on the marginal gene effect ignoring gene-environment interaction entirely (336). Gene-environment interactions are nevertheless frequent in obesity, and statistical models that do not properly account for gene-environment interactions may attenuate the marginal effect size and reduce the power to detect true associations (23, 337). Applying a joint test for a main genotype effect and gene-environment interaction may increase the power to identify an individual SNP associated with a disease outcome (338–340). As many completed GWAS for obesity have been conducted on samples with large amounts of existing environmental data, performing gene-environment-wide interaction studies (GEWIS) in these existing datasets is a cost-effective strategy to find additional obesity-associated gene variants that interact with specific environments but have been missed by conventional GWAS (341). Since large sample sizes and meta-analytical approaches are required to reliably detect SNPs with subtle gene-environment interaction patterns (342), GEWIS for obesity have been initiated in the context of large international obesity consortiums like GIANT (343). Although these methods show promise, recent simulations indicate that this technique is more appropriate for analyzing interactions between genetic risk scores rather than individual SNPs, due to the reduced power when analyzing the small effect sizes of individual SNPs (344). As a potential solution, Marigorta and Gibson suggest selecting participants who are at a high-risk for obesity based on environmental exposure (344). This strategy has the potential to identify environmental exposures that can modulate the impact of specific variants associated with obesity (344). However, challenges associated with GEWIS include identifying adequately sized cohorts with appropriate genetic and phenotypic data, as well as issues with statistical power. As a novel alternative to these techniques, variance prioritization was developed as a method to model genetic associations with genetic variance, without requiring knowledge of the interacting variables (345). Since the main effects of gene variants involved in interactions are typically associated with a large degree of variance, this strategy exploits this pattern to rank and prioritize variance estimates to identify gene variants associated with a large degree of variance in a quantitative trait (345).
Bayesian methods have also been developed to integrate variations in multiple SNPs within a given gene/region, and examine how an environmental exposure moderates the risk of these genetic profiles (346). This method was applied to the Environment and Genetics of Lung Cancer Etiology (EAGLE) study and detected a smoking × genetic profile interaction that was not detected by conventional interaction tests (346). Artificial neural networks have been applied to interaction analyses and simulations suggest that this technique may be particularly valuable for detecting nonlinear penetrance and interaction effects (347). Other analytical approaches have been developed to test interactions while addressing the common concern of statistical power. These techniques, termed “cocktail methods,” involve a three-step approach to testing genome-wide gene-environment interactions while preserving the type 1 error rate and increasing power by 30–40% under certain circumstances (348). These three steps include screening, multiple testing and G×E testing, and current simulations of this technique have been applied to binary environmental variables, although this approach is applicable to continuous environmental data (348). While early analysis of these novel techniques has been positive, further real data application of these methods will reveal the generalizability of these approaches.
Recent GWAS for obesity have collected phenotypic information in individuals living in a broad range of environments. While successful, this approach may fail to identify potential gene variants associated with obesity-related traits in a context dependent manner. Gene identification efforts may therefore be targeted in populations that display homogeneous environment and lifestyle factors across time and across the community, as observed in the Plain people (349). Performing genetic association studies for adiposity change in response to a standard major environment modification (antipsychotic drug use, smoking cessation, intensive caloric restriction, anti-obesity drug therapy, obesity surgery) is another valuable way to control the environmental exposure, lower sources of heterogeneity and provide a more comprehensive molecular basis for genetic predisposition to obesity.
In order to refine the search for interaction variants, statistical GEI tests could be combined with methylation quantitative trait loci (meQTL), expression quantitative trait loci (eQTL), and protein quantitative trait loci (pQTL) to focus on SNPs with a plausible biological mechanism for interaction (258). Specifically, a joint test could be applied to (1) identify genetic variants that statistically interact with a given environmental exposure (e.g. physical activity level) to modulate an outcome (e.g. BMI), (2) test if the same genetic variants are also eQTL, meQTL and/or pQTL for a given locus and (3) determine if the methylation, expression and protein level of the same locus is modulated by the same environmental factor (243). A similar test could be applied to analyse the interaction between an individual SNP and multiple environmental factors. Since methylation is influenced by several environmental exposures (physical activity (243), diet (248), sleep (350)) identifying SNPs that redundantly interact with multiple exposures may be a method to exploit this pattern. The ‘Identifying REdundant Gene-environment InteractionS’ (REGIS) method may increase the probability of detecting ‘true’ and replicable gene-environment interactions. Another avenue for future research is to study gene-environment interactions jointly in mouse and human studies (351). The development of the clustered regularly interspaced short palindromic repeat (CRISPR) system for gene targeting and editing creates a new opportunity to study ‘humanized’ genetically modified mice carrying human mutations (352, 353). Combining this biological data from animal studies with statistical evidence of interaction form human epidemiological studies is also likely to improve the validity of gene-environment interaction studies (23).
Conclusion
Heritability, syndromic, monogenic and polygenic obesity studies provide converging evidence that obesity predisposing genetic factors strongly interact with environment, from birth to agedness and in a wide range of situations. A prolific period of discovery is foreseen in this fast-moving field, especially with the many methodological innovations that attempt to address the ‘missing heritability’ of obesity. To effectively tackle this knowledge gap, prospective studies need to incorporate current methodological evidence to optimize the validity of emerging evidence. Emerging epigenetic studies have demonstrated that obesity, genetic variants and environmental exposures can influence DNA methylation, which provides a mechanistic model to support the statistical interactions from genetic epidemiology. A comprehensive understanding of gene-environment interactions in obesity may lead to tremendous applications in the emerging field of personalized medicine and individualized lifestyle recommendations. Evidence from interaction studies suggests that specific subgroups of individuals may have an increased risk to develop obesity in specific environments but may also benefit more from lifestyle interventions, a treatment or a surgical procedures (354). This information will help determine if population-wide or personalized subgroup interventions are the best suited to fight the worldwide obesity epidemic (355, 356).
Summary statement.
Current research has identified several environmental exposures that can moderate the impact of genetic risk factors on obesity. This paper reviews these studies (gene-environment interactions) in the obesity field and outlines the methodological challenges of these investigations.
Acknowledgments
We would like to thank Michelle Turcotte for her technical assistance with figure design and creation.
FUNDING
D. M. holds a Canada Research Chair in Genetics of Obesity.
Abbreviations
- EWAS
(epigenome-wide association studies), genome-wide analysis of epigenetic modifications associated with disease phenotypes:
- SNP
(single nucleotide polymorphism), a single nucleotide variation that is common in >1% of the population;
- eQTL
(expression quantitative trait locus), a locus that influences the expression levels of mRNAs or proteins;
- GEWIS
(genome-wide interaction studies), simultaneous testing of genetic variants theoretically covering the whole genome in interaction with an environmental factor;
- meQTL
(methylation quantitative trait locus), a locus linked to different patterns of DNA methylation;
- pQTL
(protein quantitative trait locus), a locus associated with variations in protein levels;
- QTL
(quantitative trait locus), region of DNA containing or linked to genes that underlie a quantitative trait.
Footnotes
DECLARATION OF INTEREST
The authors declare no financial interests.
References
- 1.Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 2011;377(9765):557–67. doi: 10.1016/S0140-6736(10)62037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 3.McAllister EJ, Dhurandhar NV, Keith SW, Aronne LJ, Barger J, Baskin M, et al. Ten putative contributors to the obesity epidemic. Critical reviews in food science and nutrition. 2009;49(10):868–913. doi: 10.1080/10408390903372599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levin BE. Factors promoting and ameliorating the development of obesity. Physiology & behavior. 2005;86(5):633–9. doi: 10.1016/j.physbeh.2005.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Choquet H, Meyre D. Molecular basis of obesity: current status and future prospects. Curr Genomics. 2011;12(3):154–68. doi: 10.2174/138920211795677921. Epub 2011/11/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, et al. Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature. 1997;387(6636):903–8. doi: 10.1038/43185. [DOI] [PubMed] [Google Scholar]
- 7.Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, et al. Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet. 2000;26(1):67–70. doi: 10.1038/79201. [DOI] [PubMed] [Google Scholar]
- 8.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–6. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 10.Khoury MJ, Wacholder S. Invited commentary: from genome-wide association studies to gene-environment-wide interaction studies--challenges and opportunities. American journal of epidemiology. 2009;169(2):227–30. doi: 10.1093/aje/kwn351. discussion 34–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blakemore AI, Froguel P. Investigation of Mendelian forms of obesity holds out the prospect of personalized medicine. Ann N Y Acad Sci. 2010;1214:180–9. doi: 10.1111/j.1749-6632.2010.05880.x. Epub 2010/12/24. [DOI] [PubMed] [Google Scholar]
- 12.Hinney A, Vogel CI, Hebebrand J. From monogenic to polygenic obesity: recent advances. Eur Child Adolesc Psychiatry. 2010;19(3):297–310. doi: 10.1007/s00787-010-0096-6. Epub 2010/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoury MJ, Beaty TH, Cohen BH. Scope and strategies of genetic epidemiology: analysis of articles published in Genetic Epidemiology, 1984–1991. Genet Epidemiol. 1993;10(5):321–9. doi: 10.1002/gepi.1370100505. Epub 1993/01/01. [DOI] [PubMed] [Google Scholar]
- 14.Narod SA. Modifiers of risk of hereditary breast and ovarian cancer. Nat Rev Cancer. 2002;2(2):113–23. doi: 10.1038/nrc726. Epub 2003/03/15. [DOI] [PubMed] [Google Scholar]
- 15.Ichihara S, Yamada Y. Genetic factors for human obesity. Cell Mol Life Sci. 2008;65(7–8):1086–98. doi: 10.1007/s00018-007-7453-8. Epub 2007/12/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung WK, Leibel RL. Molecular physiology of syndromic obesities in humans. Trends Endocrinol Metab. 2005;16(6):267–72. doi: 10.1016/j.tem.2005.06.009. Epub 2005/07/12. [DOI] [PubMed] [Google Scholar]
- 17.Choquet H, Meyre D. Molecular Basis of Obesity: Current Status and Future Prospects. Current genomics. 2011 May;12(3):154–68. doi: 10.2174/138920211795677921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stutzmann F, Tan K, Vatin V, Dina C, Jouret B, Tichet J, et al. Prevalence of melanocortin-4 receptor deficiency in Europeans and their age-dependent penetrance in multigenerational pedigrees. Diabetes. 2008;57(9):2511–8. doi: 10.2337/db08-0153. Epub 2008/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanikova DSM, Buzga M, Skopkova M, Ticha L, Petrasova M, et al. Age of obesity onset in MC4R mutation carriers. Endocrine regulations. 2014;49(3):137–40. doi: 10.4149/endo_2015_03_137. [DOI] [PubMed] [Google Scholar]
- 20.Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat Genet. 2012;44(5):483–9. doi: 10.1038/ng.2232. Epub 2012/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haldane JB. The interaction of nature and nurture. Ann Eugen. 1946;13(3):197–205. doi: 10.1111/j.1469-1809.1946.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 22.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6(4):287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 23.Franks PW, Pearson E, Florez JC. Gene-environment and gene-treatment interactions in type 2 diabetes: progress, pitfalls, and prospects. Diabetes Care. 2013;36(5):1413–21. doi: 10.2337/dc12-2211. Epub 2013/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rothman KJ, Greenland S, Walker AM. Concepts of interaction. American journal of epidemiology. 1980;112(4):467–70. doi: 10.1093/oxfordjournals.aje.a113015. [DOI] [PubMed] [Google Scholar]
- 25.Bayoumi RA, Al-Yahyaee SA, Albarwani SA, Rizvi SG, Al-Hadabi S, Al-Ubaidi FF, et al. Heritability of determinants of the metabolic syndrome among healthy Arabs of the Oman family study. Obesity (Silver Spring) 2007;15(3):551–6. doi: 10.1038/oby.2007.555. [DOI] [PubMed] [Google Scholar]
- 26.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. Jama. 1986;256(1):51–4. [PubMed] [Google Scholar]
- 27.Stunkard AJ, Sorensen TI, Hanis C, Teasdale TW, Chakraborty R, Schull WJ, et al. An adoption study of human obesity. N Engl J Med. 1986;314(4):193–8. doi: 10.1056/NEJM198601233140401. [DOI] [PubMed] [Google Scholar]
- 28.Bouchard C, Tremblay A, Despres JP, Nadeau A, Lupien PJ, Theriault G, et al. The response to long-term overfeeding in identical twins. N Engl J Med. 1990;322(21):1477–82. doi: 10.1056/NEJM199005243222101. [DOI] [PubMed] [Google Scholar]
- 29.Whitaker RC. Predicting preschooler obesity at birth: the role of maternal obesity in early pregnancy. Pediatrics. 2004;114(1):e29–36. doi: 10.1542/peds.114.1.e29. [DOI] [PubMed] [Google Scholar]
- 30.Murrin CM, Kelly GE, Tremblay RE, Kelleher CC. Body mass index and height over three generations: evidence from the Lifeways cross-generational cohort study. BMC public health. 2012;12(1):81. doi: 10.1186/1471-2458-12-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawlor DA, Lichtenstein P, Fraser A, Langstrom N. Does maternal weight gain in pregnancy have long-term effects on offspring adiposity? A sibling study in a prospective cohort of 146,894 men from 136,050 families. The American journal of clinical nutrition. 2011;94(1):142–8. doi: 10.3945/ajcn.110.009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietilainen KH, Kaprio J, Rissanen A, Winter T, Rimpela A, Viken RJ, et al. Distribution and heritability of BMI in Finnish adolescents aged 16y and 17y: a study of 4884 twins and 2509 singletons. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1999;23(2):107–15. doi: 10.1038/sj.ijo.0800767. [DOI] [PubMed] [Google Scholar]
- 33.Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, et al. Sex differences in heritability of BMI: a comparative study of results from twin studies in eight countries. Twin Res. 2003;6(5):409–21. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- 34.Dubois L, Ohm Kyvik K, Girard M, Tatone-Tokuda F, Perusse D, Hjelmborg J, et al. Genetic and environmental contributions to weight, height, and BMI from birth to 19 years of age: an international study of over 12,000 twin pairs. PLoS One. 2012;7(2):e30153. doi: 10.1371/journal.pone.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demerath EW, Choh AC, Czerwinski SA, Lee M, Sun SS, Chumlea WC, et al. Genetic and environmental influences on infant weight and weight change: the Fels Longitudinal Study. Am J Hum Biol. 2007;19(5):692–702. doi: 10.1002/ajhb.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haworth CM, Carnell S, Meaburn EL, Davis OS, Plomin R, Wardle J. Increasing heritability of BMI and stronger associations with the FTO gene over childhood. Obesity (Silver Spring) 2008;16(12):2663–8. doi: 10.1038/oby.2008.434. [DOI] [PubMed] [Google Scholar]
- 37.Lajunen HR, Kaprio J, Keski-Rahkonen A, Rose RJ, Pulkkinen L, Rissanen A, et al. Genetic and environmental effects on body mass index during adolescence: a prospective study among Finnish twins. Int J Obes (Lond) 2009;33(5):559–67. doi: 10.1038/ijo.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan C, Guo B, Warner C, Fowler T, Barrett T, Boomsma D, et al. Heritability of body mass index in pre-adolescence, young adulthood and late adulthood. European journal of epidemiology. 2012 doi: 10.1007/s10654-012-9678-6. [DOI] [PubMed] [Google Scholar]
- 39.North KE, Graff M, Adair LS, Lange EM, Lange LA, Guo G, et al. Genetic Epidemiology of BMI and Body Mass Change From Adolescence to Young Adulthood. Obesity (Silver Spring) 2009 doi: 10.1038/oby.2009.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hjelmborg JB, Fagnani C, Silventoinen K, McGue M, Korkeila M, Christensen K, et al. Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity (Silver Spring) 2008;16(4):847–52. doi: 10.1038/oby.2007.135. [DOI] [PubMed] [Google Scholar]
- 41.Allison DB, Faith MS, Nathan JS. Risch’s lambda values for human obesity. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 1996;20(11):990–9. [PubMed] [Google Scholar]
- 42.Rokholm B, Silventoinen K, Angquist L, Skytthe A, Kyvik KO, Sorensen TI. Increased genetic variance of BMI with a higher prevalence of obesity. PLoS One. 2011;6(6):e20816. doi: 10.1371/journal.pone.0020816. Epub 2011/07/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wardle J, Carnell S, Haworth CM, Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. The American journal of clinical nutrition. 2008;87(2):398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 44.Rokholm B, Silventoinen K, Tynelius P, Gamborg M, Sorensen TI, Rasmussen F. Increasing genetic variance of body mass index during the Swedish obesity epidemic. PLoS One. 2011;6(11):e27135. doi: 10.1371/journal.pone.0027135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mustelin L, Silventoinen K, Pietilainen K, Rissanen A, Kaprio J. Physical activity reduces the influence of genetic effects on BMI and waist circumference: a study in young adult twins. Int J Obes (Lond) 2009;33(1):29–36. doi: 10.1038/ijo.2008.258. [DOI] [PubMed] [Google Scholar]
- 46.McCaffery JM, Papandonatos GD, Bond DS, Lyons MJ, Wing RR. Gene X environment interaction of vigorous exercise and body mass index among male Vietnam-era twins. The American journal of clinical nutrition. 2009;89(4):1011–8. doi: 10.3945/ajcn.2008.27170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams PT. Attenuating effect of vigorous physical activity on the risk for inherited obesity: a study of 47,691 runners. PLoS One. 2012;7(2):e31436. doi: 10.1371/journal.pone.0031436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roskam AJ, Kunst AE, Van Oyen H, Demarest S, Klumbiene J, Regidor E, et al. Comparative appraisal of educational inequalities in overweight and obesity among adults in 19 European countries. Int J Epidemiol. 2010;39(2):392–404. doi: 10.1093/ije/dyp329. [DOI] [PubMed] [Google Scholar]
- 49.Lajunen HR, Kaprio J, Rose RJ, Pulkkinen L, Silventoinen K. Genetic and environmental influences on BMI from late childhood to adolescence are modified by parental education. Obesity (Silver Spring) 2012;20(3):583–9. doi: 10.1038/oby.2011.304. [DOI] [PubMed] [Google Scholar]
- 50.Cappuccio FP, Taggart FM, Kandala NB, Currie A, Peile E, Stranges S, et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep. 2008;31(5):619–26. doi: 10.1093/sleep/31.5.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson NF, Harden KP, Buchwald D, Vitiello MV, Pack AI, Weigle DS, et al. Sleep duration and body mass index in twins: a gene-environment interaction. Sleep. 2012;35(5):1–7. doi: 10.5665/sleep.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maher AR, Maglione M, Bagley S, Suttorp M, Hu JH, Ewing B, et al. Efficacy and comparative effectiveness of atypical antipsychotic medications for off-label uses in adults: a systematic review and meta-analysis. Jama. 2011;306(12):1359–69. doi: 10.1001/jama.2011.1360. [DOI] [PubMed] [Google Scholar]
- 53.Correll CU, Malhotra AK. Pharmacogenetics of antipsychotic-induced weight gain. Psychopharmacology. 2004;174(4):477–89. doi: 10.1007/s00213-004-1949-9. [DOI] [PubMed] [Google Scholar]
- 54.Theisen FM, Gebhardt S, Haberhausen M, Heinzel-Gutenbrunner M, Wehmeier PM, Krieg JC, et al. Clozapine-induced weight gain: a study in monozygotic twins and same-sex sib pairs. Psychiatric genetics. 2005;15(4):285–9. doi: 10.1097/00041444-200512000-00011. [DOI] [PubMed] [Google Scholar]
- 55.Gebhardt S, Theisen FM, Haberhausen M, Heinzel-Gutenbrunner M, Wehmeier PM, Krieg JC, et al. Body weight gain induced by atypical antipsychotics: an extension of the monozygotic twin and sib pair study. J Clin Pharm Ther. 2010;35(2):207–11. doi: 10.1111/j.1365-2710.2009.01084.x. [DOI] [PubMed] [Google Scholar]
- 56.Hainer V, Stunkard AJ, Kunesova M, Parizkova J, Stich V, Allison DB. Intrapair resemblance in very low calorie diet-induced weight loss in female obese identical twins. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2000;24(8):1051–7. doi: 10.1038/sj.ijo.0801358. [DOI] [PubMed] [Google Scholar]
- 57.Bouchard C, Tremblay A, Despres JP, Theriault G, Nadeau A, Lupien PJ, et al. The response to exercise with constant energy intake in identical twins. Obes Res. 1994;2(5):400–10. doi: 10.1002/j.1550-8528.1994.tb00087.x. [DOI] [PubMed] [Google Scholar]
- 58.Hatoum IJ, Greenawalt DM, Cotsapas C, Reitman ML, Daly MJ, Kaplan LM. Heritability of the weight loss response to gastric bypass surgery. J Clin Endocrinol Metab. 2011;96(10):E1630–3. doi: 10.1210/jc.2011-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo G, Liu H, Wang L, Shen H, Hu W. The Genome-Wide Influence on Human BMI Depends on Physical Activity, Life Course, and Historical Period. Demography. 2015;52(5):1651–70. doi: 10.1007/s13524-015-0421-2. Epub 2015/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaisse C, Clement K, Durand E, Hercberg S, Guy-Grand B, Froguel P. Melanocortin-4 receptor mutations are a frequent and heterogeneous cause of morbid obesity. J Clin Invest. 2000;106(2):253–62. doi: 10.1172/JCI9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore SJ, Green JS, Fan Y, Bhogal AK, Dicks E, Fernandez BA, et al. Clinical and genetic epidemiology of Bardet-Biedl syndrome in Newfoundland: a 22-year prospective, population-based, cohort study. American journal of medical genetics Part A. 2005;132(4):352–60. doi: 10.1002/ajmg.a.30406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudley O, McManus B, Vogels A, Whittington J, Muscatelli F. Cross-cultural comparisons of obesity and growth in Prader-Willi syndrome. Journal of intellectual disability research : JIDR. 2008;52(Pt 5):426–36. doi: 10.1111/j.1365-2788.2008.01044.x. [DOI] [PubMed] [Google Scholar]
- 63.Bachmann-Gagescu R, Mefford HC, Cowan C, Glew GM, Hing AV, Wallace S, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genetics in medicine : official journal of the American College of Medical Genetics. 2010;12(10):641–7. doi: 10.1097/GIM.0b013e3181ef4286. [DOI] [PubMed] [Google Scholar]
- 64.Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O’Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348(12):1085–95. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 65.Carmi R, Elbedour K, Stone EM, Sheffield VC. Phenotypic differences among patients with Bardet-Biedl syndrome linked to three different chromosome loci. American journal of medical genetics. 1995;59(2):199–203. doi: 10.1002/ajmg.1320590216. [DOI] [PubMed] [Google Scholar]
- 66.Sebire NJ, Jolly M, Harris JP, Wadsworth J, Joffe M, Beard RW, et al. Maternal obesity and pregnancy outcome: a study of 287,213 pregnancies in London. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2001;25(8):1175–82. doi: 10.1038/sj.ijo.0801670. [DOI] [PubMed] [Google Scholar]
- 67.Crozier SR, Inskip HM, Godfrey KM, Cooper C, Harvey NC, Cole ZA, et al. Weight gain in pregnancy and childhood body composition: findings from the Southampton Women’s Survey. The American journal of clinical nutrition. 2010;91(6):1745–51. doi: 10.3945/ajcn.2009.29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oken E, Rifas-Shiman SL, Field AE, Frazier AL, Gillman MW. Maternal gestational weight gain and offspring weight in adolescence. Obstet Gynecol. 2008;112(5):999–1006. doi: 10.1097/AOG.0b013e31818a5d50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds RM, Osmond C, Phillips DI, Godfrey KM. Maternal BMI, Parity, and Pregnancy Weight Gain: Influences on Offspring Adiposity in Young Adulthood. J Clin Endocrinol Metab. 2010 doi: 10.1210/jc.2010-0697. [DOI] [PubMed] [Google Scholar]
- 70.Nizard J, Dommergue M, Clement K. Pregnancy in a woman with a leptin-receptor mutation. N Engl J Med. 2012;366(11):1064–5. doi: 10.1056/NEJMc1200116. [DOI] [PubMed] [Google Scholar]
- 71.Lawlor DA, Fraser A, Macdonald-Wallis C, Nelson SM, Palmer TM, Davey Smith G, et al. Maternal and offspring adiposity-related genetic variants and gestational weight gain. The American journal of clinical nutrition. 2011;94(1):149–55. doi: 10.3945/ajcn.110.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee KW, Abrahamowicz M, Leonard GT, Richer L, Perron M, Veillette S, et al. Prenatal exposure to cigarette smoke interacts with to modulate dietary preference for fat. J Psychiatry Neurosci. 2014;39(4):130263. doi: 10.1503/jpn.130263. Epub 2014/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999–2004. Jama. 2006;295(13):1549–55. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 74.Dempfle A, Hinney A, Heinzel-Gutenbrunner M, Raab M, Geller F, Gudermann T, et al. Large quantitative effect of melanocortin-4 receptor gene mutations on body mass index. J Med Genet. 2004;41(10):795–800. doi: 10.1136/jmg.2004.018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5(6):e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015;11(10):e1005378. doi: 10.1371/journal.pgen.1005378. Epub 2015/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Butler JV, Whittington JE, Holland AJ, McAllister CJ, Goldstone AP. The transition between the phenotypes of Prader-Willi syndrome during infancy and early childhood. Dev Med Child Neurol. 2010;52(6):e88–93. doi: 10.1111/j.1469-8749.2009.03530.x. [DOI] [PubMed] [Google Scholar]
- 79.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463(7281):671–5. doi: 10.1038/nature08727. Epub 2010/02/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Y, Zhu H, Miller DT, Gusella JF, Platt OS, Wu BL, et al. Age- and gender-dependent obesity in individuals with 16p11.2 deletion. Journal of genetics and genomics = Yi chuan xue bao. 2011;38(9):403–9. doi: 10.1016/j.jgg.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 81.Kaakinen M, Laara E, Pouta A, Hartikainen AL, Laitinen J, Tammelin TH, et al. Life-Course Analysis of a Fat Mass and Obesity-Associated (FTO) Gene Variant and Body Mass Index in the Northern Finland Birth Cohort 1966 Using Structural Equation Modeling. American journal of epidemiology. 2010 doi: 10.1093/aje/kwq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CN, et al. Association between Common Variation at the FTO Locus and Changes in Body Mass Index from Infancy to Late Childhood: The Complex Nature of Genetic Association through Growth and Development. PLoS Genet. 2011;7(2):e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hertel JK, Johansson S, Sonestedt E, Jonsson A, Lie RT, Platou CG, et al. FTO, Type 2 Diabetes, and Weight Gain Throughout Adult Life: A Meta-Analysis of 41,504 Subjects From the Scandinavian HUNT, MDC, and MPP Studies. Diabetes. 2011 doi: 10.2337/db10-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rzehak P, Scherag A, Grallert H, Sausenthaler S, Koletzko S, Bauer CP, et al. Associations between BMI and the FTO gene are age dependent: results from the GINI and LISA birth cohort studies up to age 6 years. Obes Facts. 2010;3(3):173–80. doi: 10.1159/000314612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qi L, Kang K, Zhang C, van Dam RM, Kraft P, Hunter D, et al. Fat mass-and obesity-associated (FTO) gene variant is associated with obesity: longitudinal analyses in two cohort studies and functional test. Diabetes. 2008;57(11):3145–51. doi: 10.2337/db08-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wangensteen T, Egeland T, Akselsen H, Holmen J, Undlien D, Retterstol L. FTO genotype and weight gain in obese and normal weight adults from a Norwegian population based cohort (the HUNT study) Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2010;118(9):649–52. doi: 10.1055/s-0030-1249636. [DOI] [PubMed] [Google Scholar]
- 87.Vimaleswaran KS, Angquist L, Hansen RD, van der AD, Bouatia-Naji N, Holst C, et al. Association Between FTO Variant and Change in Body Weight and Its Interaction With Dietary Factors: The DiOGenes Study. Obesity (Silver Spring) 2012 doi: 10.1038/oby.2012.49. [DOI] [PubMed] [Google Scholar]
- 88.Jacobsson JA, Almen MS, Benedict C, Hedberg LA, Michaelsson K, Brooks S, et al. Detailed analysis of variants in FTO in association with body composition in a cohort of 70-year-olds suggests a weakened effect among elderly. PLoS One. 2011;6(5):e20158. doi: 10.1371/journal.pone.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elks CE, Loos RJ, Sharp SJ, Langenberg C, Ring SM, Timpson NJ, et al. Genetic markers of adult obesity risk are associated with greater early infancy weight gain and growth. PLoS Med. 2010;7(5):e1000284. doi: 10.1371/journal.pmed.1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kilpelainen TO, den Hoed M, Ong KK, Grontved A, Brage S, Jameson K, et al. Obesity-susceptibility loci have a limited influence on birth weight: a meta-analysis of up to 28,219 individuals. The American journal of clinical nutrition. 2011;93(4):851–60. doi: 10.3945/ajcn.110.000828. [DOI] [PubMed] [Google Scholar]
- 91.Elks CE, Loos RJ, Hardy R, Wills AK, Wong A, Wareham NJ, et al. Adult obesity susceptibility variants are associated with greater childhood weight gain and a faster tempo of growth: the 1946 British Birth Cohort Study. The American journal of clinical nutrition. 2012 doi: 10.3945/ajcn.111.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kilpelainen TO, Bingham SA, Khaw KT, Wareham NJ, Loos RJ. Association of variants in the PCSK1 gene with obesity in the EPIC-Norfolk study. Hum Mol Genet. 2009;18(18):3496–501. doi: 10.1093/hmg/ddp280. Epub 2009/06/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Choquet H, Kasberger J, Hamidovic A, Jorgenson E. Contribution of Common PCSK1 Genetic Variants to Obesity in 8,359 Subjects from Multi-Ethnic American Population. PLoS One. 2013;8(2):e57857. doi: 10.1371/journal.pone.0057857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nead KT, Li A, Wehner MR, Neupane B, Gustafsson S, Butterworth A, et al. Contribution of common non-synonymous variants in PCSK1 to body mass index variation and risk of obesity: a systematic review and meta-analysis with evidence from up to 331 175 individuals. Hum Mol Genet. 2015;24(12):3582–94. doi: 10.1093/hmg/ddv097. Epub 2015/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wangensteen T, Kolsgaard ML, Mattingsdal M, Joner G, Tonstad S, Undlien D, et al. Mutations in the melanocortin 4 receptor (MC4R) gene in obese patients in Norway. Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2009;117(6):266–73. doi: 10.1055/s-0028-1102942. [DOI] [PubMed] [Google Scholar]
- 96.Rosenquist JN, Lehrer SF, O’Malley AJ, Zaslavsky AM, Smoller JW, Christakis NA. Cohort of birth modifies the association between FTO genotype and BMI. Proc Natl Acad Sci U S A. 2015;112(2):354–9. doi: 10.1073/pnas.1411893111. Epub 2014/12/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Taylor AE, Sandeep MN, Janipalli CS, Giambartolomei C, Evans DM, Kranthi Kumar MV, et al. Associations of FTO and MC4R Variants with Obesity Traits in Indians and the Role of Rural/Urban Environment as a Possible Effect Modifier. J Obes. 2011;2011:307542. doi: 10.1155/2011/307542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vasan SK, Fall T, Neville MJ, Antonisamy B, Fall CH, Geethanjali FS, et al. Associations of Variants in FTO and Near MC4R With Obesity Traits in South Asian Indians. Obesity (Silver Spring) 2012 doi: 10.1038/oby.2012.64. [DOI] [PubMed] [Google Scholar]
- 99.Hennig BJ, Fulford AJ, Sirugo G, Rayco-Solon P, Hattersley AT, Frayling TM, et al. FTO gene variation and measures of body mass in an African population. BMC Med Genet. 2009;10:21. doi: 10.1186/1471-2350-10-21. Epub 2009/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Demerath EW, Choh AC, Johnson W, Curran JE, Lee M, Bellis C, et al. The positive association of obesity variants with adulthood adiposity strengthens over an 80-year period: a gene-by-birth year interaction. Human Heredity. 2013;75(2–4):175–85. doi: 10.1159/000351742. Epub 2013/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Richardson AS, North KE, Graff M, Young KM, Mohlke KL, Lange LA, et al. Moderate to vigorous physical activity interactions with genetic variants and body mass index in a large US ethnically diverse cohort. Pediatr Obes. 2013 doi: 10.1111/j.2047-6310.2013.00152.x. Epub 2013/03/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. Epub 2011/11/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90(2):425–8. doi: 10.3945/ajcn.2009.27652. Epub 2009/06/26. [DOI] [PubMed] [Google Scholar]
- 104.Demerath EW, Lutsey PL, Monda KL, Linda Kao WH, Bressler J, Pankow JS, et al. Interaction of FTO and physical activity level on adiposity in African-American and European-American adults: the ARIC study. Obesity (Silver Spring) 2011;19(9):1866–72. doi: 10.1038/oby.2011.131. Epub 2011/05/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sonestedt E, Gullberg B, Ericson U, Wirfalt E, Hedblad B, Orho-Melander M. Association between fat intake, physical activity and mortality depending on genetic variation in FTO. Int J Obes (Lond) 2011;35(8):1041–9. doi: 10.1038/ijo.2010.263. Epub 2010/12/24. [DOI] [PubMed] [Google Scholar]
- 106.Ruiz JR, Labayen I, Ortega FB, Legry V, Moreno LA, Dallongeville J, et al. Attenuation of the effect of the FTO rs9939609 polymorphism on total and central body fat by physical activity in adolescents: the HELENA study. Arch Pediatr Adolesc Med. 2010;164(4):328–33. doi: 10.1001/archpediatrics.2010.29. Epub 2010/04/07. [DOI] [PubMed] [Google Scholar]
- 107.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 108.Moore SC, Gunter MJ, Daniel CR, Reddy KS, George PS, Yurgalevitch S, et al. Common genetic variants and central adiposity among Asian-Indians. Obesity (Silver Spring) 2012;20(9):1902–8. doi: 10.1038/oby.2011.238. Epub 2011/07/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee HJ, Kim IK, Kang JH, Ahn Y, Han BG, Lee JY, et al. Effects of common FTO gene variants associated with BMI on dietary intake and physical activity in Koreans. Clin Chim Acta. 2010;411(21–22):1716–22. doi: 10.1016/j.cca.2010.07.010. Epub 2010/07/24. [DOI] [PubMed] [Google Scholar]
- 110.Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS Med. 2010;7(8) doi: 10.1371/journal.pmed.1000332. Epub 2010/09/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qi Q, Li Y, Chomistek AK, Kang JH, Curhan GC, Pasquale LR, et al. Television watching, leisure time physical activity, and the genetic predisposition in relation to body mass index in women and men. Circulation. 2012;126(15):1821–7. doi: 10.1161/CIRCULATIONAHA.112.098061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O’Connell JR, et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med. 2008;168(16):1791–7. doi: 10.1001/archinte.168.16.1791. Epub 2008/09/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rankinen T, Rice T, Teran-Garcia M, Rao DC, Bouchard C. FTO genotype is associated with exercise training-induced changes in body composition. Obesity (Silver Spring) 2010;18(2):322–6. doi: 10.1038/oby.2009.205. Epub 2009/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ahmad T, Lee IM, Pare G, Chasman DI, Rose L, Ridker PM, et al. Lifestyle Interaction With Fat Mass and Obesity-Associated (FTO) Genotype and Risk of Obesity in Apparently Healthy U.S. Women. Diabetes Care. 2011;34(3):675–80. doi: 10.2337/dc10-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xi B, Shen Y, Zhang M, Liu X, Zhao X, Wu L, et al. The common rs9939609 variant of the fat mass and obesity-associated gene is associated with obesity risk in children and adolescents of Beijing, China. BMC Med Genet. 2010;11:107. doi: 10.1186/1471-2350-11-107. Epub 2010/07/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mitchell JA, Church TS, Rankinen T, Earnest CP, Sui X, Blair SN. FTO genotype and the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring) 2010;18(3):641–3. doi: 10.1038/oby.2009.311. Epub 2009/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Corella D, Ortega-Azorin C, Sorli JV, Covas MI, Carrasco P, Salas-Salvado J, et al. Statistical and biological gene-lifestyle interactions of MC4R and FTO with diet and physical activity on obesity: new effects on alcohol consumption. PLoS One. 2012;7(12):e52344. doi: 10.1371/journal.pone.0052344. Epub 2013/01/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scott RA, Bailey ME, Moran CN, Wilson RH, Fuku N, Tanaka M, et al. FTO genotype and adiposity in children: physical activity levels influence the effect of the risk genotype in adolescent males. European journal of human genetics : EJHG. 2010;18(12):1339–43. doi: 10.1038/ejhg.2010.131. Epub 2010/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cauchi S, Stutzmann F, Cavalcanti-Proenca C, Durand E, Pouta A, Hartikainen AL, et al. Combined effects of MC4R and FTO common genetic variants on obesity in European general populations. J Mol Med (Berl) 2009;87(5):537–46. doi: 10.1007/s00109-009-0451-6. Epub 2009/03/04. [DOI] [PubMed] [Google Scholar]
- 120.Ahmad S, Zhao W, Renstrom F, Rasheed A, Samuel M, Zaidi M, et al. Physical activity, smoking, and genetic predisposition to obesity in people from Pakistan: the PROMIS study. BMC Med Genet. 2015;16(1):114. doi: 10.1186/s12881-015-0259-x. Epub 2015/12/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reddon H, Gerstein HC, Engert JC, Mohan V, Bosch J, Desai D, et al. Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study. Sci Rep. 2016;6:18672. doi: 10.1038/srep18672. Epub 2016/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakamura S, Narimatsu H, Sato H, Sho R, Otani K, Kawasaki R, et al. Gene-environment interactions in obesity: implication for future applications in preventive medicine. J Hum Genet. 2015 doi: 10.1038/jhg.2015.148. Epub 2015/12/15. [DOI] [PubMed] [Google Scholar]
- 123.Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu J, Loos RJ, Lu L, Zong G, Gan W, Ye X, et al. Associations of genetic risk score with obesity and related traits and the modifying effect of physical activity in a Chinese Han population. PLoS One. 2014;9(3):e91442. doi: 10.1371/journal.pone.0091442. Epub 2014/03/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D, et al. Gene × physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS Genet. 2013;9(7):e1003607. doi: 10.1371/journal.pgen.1003607. Epub 2013/08/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Graff M, North KE, Richardson AS, Young KM, Mohlke KL, Lange LA, et al. Screen time behaviours may interact with obesity genes, independent of physical activity, to influence adolescent BMI in an ethnically diverse cohort. Pediatr Obes. 2013;8(6):e74–9. doi: 10.1111/j.2047-6310.2013.00195.x. Epub 2013/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klimentidis YC, Arora A, Chougule A, Zhou J, Raichlen DA. FTO association and interaction with time spent sitting. Int J Obes (Lond) 2015 doi: 10.1038/ijo.2015.190. Epub 2015/09/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rouskas K, Meyre D, Stutzmann F, Paletas K, Papazoglou D, Vatin V, et al. Loss-of-Function Mutations in MC4R Are Very Rare in the Greek Severely Obese Adult Population. Obesity (Silver Spring) 2012 doi: 10.1038/oby.2012.77. [DOI] [PubMed] [Google Scholar]
- 129.Corella D, Arnett DK, Tucker KL, Kabagambe EK, Tsai M, Parnell LD, et al. A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI. J Nutr. 2011;141(12):2219–25. doi: 10.3945/jn.111.143826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Phillips CM, Kesse-Guyot E, McManus R, Hercberg S, Lairon D, Planells R, et al. High dietary saturated fat intake accentuates obesity risk associated with the fat mass and obesity-associated gene in adults. J Nutr. 2012;142(5):824–31. doi: 10.3945/jn.111.153460. Epub 2012/03/30. [DOI] [PubMed] [Google Scholar]
- 131.Moleres A, Ochoa MC, Rendo-Urteaga T, Martinez-Gonzalez MA, Azcona San Julian MC, Martinez JA, et al. Dietary fatty acid distribution modifies obesity risk linked to the rs9939609 polymorphism of the fat mass and obesity-associated gene in a Spanish case-control study of children. The British journal of nutrition. 2012;107(4):533–8. doi: 10.1017/S0007114511003424. Epub 2011/07/30. [DOI] [PubMed] [Google Scholar]
- 132.Lappalainen T, Lindstrom J, Paananen J, Eriksson JG, Karhunen L, Tuomilehto J, et al. Association of the fat mass and obesity-associated (FTO) gene variant (rs9939609) with dietary intake in the Finnish Diabetes Prevention Study. The British journal of nutrition. 2012;108(10):1859–65. doi: 10.1017/S0007114511007410. Epub 2012/01/24. [DOI] [PubMed] [Google Scholar]
- 133.Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK, et al. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. Bmj. 2014;348:g1610. doi: 10.1136/bmj.g1610. Epub 2014/03/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pennell CE, Ang QW, Warrington NM, Oddy WH, Beilin LJ, Newnham JP, et al. Gene-environment interactions between the fat mass and obesity-associated (FTO) gene and nutrition modify the association between FTO and obesity in childhood and adolescence. J Paediatr Child Health. 2014;50:56. [Google Scholar]
- 135.Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, et al. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367(15):1387–96. doi: 10.1056/NEJMoa1203039. Epub 2012/09/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nettleton JA, Follis JL, Ngwa JS, Smith CE, Ahmad S, Tanaka T, et al. Gene × dietary pattern interactions in obesity: analysis of up to 68 317 adults of European ancestry. Hum Mol Genet. 2015;24(16):4728–38. doi: 10.1093/hmg/ddv186. Epub 2015/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Qi Q, Kilpelainen TO, Downer MK, Tanaka T, Smith CE, Sluijs I, et al. FTO genetic variants, dietary intake and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014;23(25):6961–72. doi: 10.1093/hmg/ddu411. Epub 2014/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Corella D, Tai ES, Sorli JV, Chew SK, Coltell O, Sotos-Prieto M, et al. Association between the APOA2 promoter polymorphism and body weight in Mediterranean and Asian populations: replication of a gene-saturated fat interaction. Int J Obes (Lond) 2011;35(5):666–75. doi: 10.1038/ijo.2010.187. Epub 2010/10/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Corella D, Peloso G, Arnett DK, Demissie S, Cupples LA, Tucker K, et al. APOA2, dietary fat, and body mass index: replication of a gene-diet interaction in 3 independent populations. Arch Intern Med. 2009;169(20):1897–906. doi: 10.1001/archinternmed.2009.343. Epub 2009/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith CE, Ordovas JM, Sanchez-Moreno C, Lee YC, Garaulet M. Apolipoprotein A-II polymorphism: relationships to behavioural and hormonal mediators of obesity. Int J Obes (Lond) 2012;36(1):130–6. doi: 10.1038/ijo.2011.24. Epub 2011/03/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Prieur X, Coste H, Rodriguez JC. The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-alpha and contains a novel farnesoid X-activated receptor response element. J Biol Chem. 2003;278(28):25468–80. doi: 10.1074/jbc.M301302200. Epub 2003/04/24. [DOI] [PubMed] [Google Scholar]
- 142.Prieur X, Huby T, Coste H, Schaap FG, Chapman MJ, Rodriguez JC. Thyroid hormone regulates the hypotriglyceridemic gene APOA5. J Biol Chem. 2005;280(30):27533–43. doi: 10.1074/jbc.M503139200. Epub 2005/06/09. [DOI] [PubMed] [Google Scholar]
- 143.Aberle J, Evans D, Beil FU, Seedorf U. A polymorphism in the apolipoprotein A5 gene is associated with weight loss after short-term diet. Clin Genet. 2005;68(2):152–4. doi: 10.1111/j.1399-0004.2005.00463.x. Epub 2005/07/06. [DOI] [PubMed] [Google Scholar]
- 144.Corella D, Lai CQ, Demissie S, Cupples LA, Manning AK, Tucker KL, et al. APOA5 gene variation modulates the effects of dietary fat intake on body mass index and obesity risk in the Framingham Heart Study. J Mol Med (Berl) 2007;85(2):119–28. doi: 10.1007/s00109-006-0147-0. Epub 2007/01/11. [DOI] [PubMed] [Google Scholar]
- 145.Sanchez-Moreno C, Ordovas JM, Smith CE, Baraza JC, Lee YC, Garaulet M. APOA5 gene variation interacts with dietary fat intake to modulate obesity and circulating triglycerides in a Mediterranean population. J Nutr. 2011;141(3):380–5. doi: 10.3945/jn.110.130344. Epub 2011/01/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marti A, Corbalan MS, Martinez-Gonzalez MA, Forga L, Martinez JA. CHO intake alters obesity risk associated with Pro12Ala polymorphism of PPARgamma gene. J Physiol Biochem. 2002;58(4):219–20. doi: 10.1007/BF03179859. Epub 2003/05/15. [DOI] [PubMed] [Google Scholar]
- 147.Robitaille J, Despres JP, Perusse L, Vohl MC. The PPAR-gamma P12A polymorphism modulates the relationship between dietary fat intake and components of the metabolic syndrome: results from the Quebec Family Study. Clin Genet. 2003;63(2):109–16. doi: 10.1034/j.1399-0004.2003.00026.x. Epub 2003/03/13. [DOI] [PubMed] [Google Scholar]
- 148.Papoutsakis C. Gene-Diet Interactions and Obesity Indices. Current Nutrition Reports. 2012;1(3):142–52. [Google Scholar]
- 149.Galbete C, Toledo E, Martinez-Gonzalez MA, Martinez JA, Guillen-Grima F, Marti A. Pro12Ala variant of the PPARG2 gene increases body mass index: An updated meta-analysis encompassing 49,092 subjects. Obesity (Silver Spring) 2013;21(7):1486–95. doi: 10.1002/oby.20150. Epub 2013/05/15. [DOI] [PubMed] [Google Scholar]
- 150.Galbete C, Toledo J, Martinez-Gonzalez MA, Martinez JA, Guillen-Grima F, Marti A. Lifestyle factors modify obesity risk linked to PPARG2 and FTO variants in an elderly population: a cross-sectional analysis in the SUN Project. Genes & nutrition. 2013;8(1):61–7. doi: 10.1007/s12263-012-0296-4. Epub 2012/04/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lamri A, Abi Khalil C, Jaziri R, Velho G, Lantieri O, Vol S, et al. Dietary fat intake and polymorphisms at the PPARG locus modulate BMI and type 2 diabetes risk in the D.E.S.I.R. prospective study. Int J Obes (Lond) 2012;36(2):218–24. doi: 10.1038/ijo.2011.91. Epub 2011/05/05. [DOI] [PubMed] [Google Scholar]
- 152.Memisoglu A, Hu FB, Hankinson SE, Manson JE, De Vivo I, Willett WC, et al. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12(22):2923–9. doi: 10.1093/hmg/ddg318. Epub 2003/09/25. [DOI] [PubMed] [Google Scholar]
- 153.Garaulet M, Smith CE, Hernandez-Gonzalez T, Lee YC, Ordovas JM. PPARgamma Pro12Ala interacts with fat intake for obesity and weight loss in a behavioural treatment based on the Mediterranean diet. Mol Nutr Food Res. 2011;55(12):1771–9. doi: 10.1002/mnfr.201100437. Epub 2011/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anderson AL, Harris TB, Houston DK, Tylavsky FA, Lee JS, Sellmeyer DE, et al. Relationships of dietary patterns with body composition in older adults differ by gender and PPAR-gamma Pro12Ala genotype. Eur J Nutr. 2010;49(7):385–94. doi: 10.1007/s00394-010-0096-9. Epub 2010/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dedoussis GV, Manios Y, Kourlaba G, Kanoni S, Lagou V, Butler J, et al. An age-dependent diet-modified effect of the PPARgamma Pro12Ala polymorphism in children. Metabolism. 2011;60(4):467–73. doi: 10.1016/j.metabol.2010.04.007. Epub 2010/06/29. [DOI] [PubMed] [Google Scholar]
- 156.Lindi VI, Uusitupa MI, Lindstrom J, Louheranta A, Eriksson JG, Valle TT, et al. Association of the Pro12Ala polymorphism in the PPAR-gamma2 gene with 3-year incidence of type 2 diabetes and body weight change in the Finnish Diabetes Prevention Study. Diabetes. 2002;51(8):2581–6. doi: 10.2337/diabetes.51.8.2581. Epub 2002/07/30. [DOI] [PubMed] [Google Scholar]
- 157.Franks PW, Jablonski KA, Delahanty L, Hanson RL, Kahn SE, Altshuler D, et al. The Pro12Ala variant at the peroxisome proliferator-activated receptor gamma gene and change in obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2007;50(12):2451–60. doi: 10.1007/s00125-007-0826-6. Epub 2007/09/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Adamo KB, Dent R, Langefeld CD, Cox M, Williams K, Carrick KM, et al. Peroxisome proliferator-activated receptor gamma 2 and acyl-CoA synthetase 5 polymorphisms influence diet response. Obesity (Silver Spring) 2007;15(5):1068–75. doi: 10.1038/oby.2007.630. Epub 2007/05/15. [DOI] [PubMed] [Google Scholar]
- 159.Nicklas BJ, van Rossum EF, Berman DM, Ryan AS, Dennis KE, Shuldiner AR. Genetic variation in the peroxisome proliferator-activated receptor-gamma2 gene (Pro12Ala) affects metabolic responses to weight loss and subsequent weight regain. Diabetes. 2001;50(9):2172–6. doi: 10.2337/diabetes.50.9.2172. Epub 2001/08/28. [DOI] [PubMed] [Google Scholar]
- 160.Riedel C, von Kries R, Fenske N, Strauch K, Ness AR, Beyerlein A. Interactions of genetic and environmental risk factors with respect to body fat mass in children: results from the ALSPAC study. Obesity (Silver Spring) 2013;21(6):1238–42. doi: 10.1002/oby.20196. Epub 2013/05/15. [DOI] [PubMed] [Google Scholar]
- 161.Singh A, Babyak MA, Nolan DK, Brummett BH, Jiang R, Siegler IC, et al. Gene by stress genome-wide interaction analysis and path analysis identify EBF1 as a cardiovascular and metabolic risk gene. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.189. Epub 2014/10/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Singh A, Babyak MA, Brummett BH, Jiang R, Watkins LL, Barefoot JC, et al. Computing a Synthetic Chronic Psychosocial Stress Measurement in Multiple Datasets and its Application in the Replication of G × E Interactions of the EBF1 Gene. Genet Epidemiol. 2015;39(6):489–97. doi: 10.1002/gepi.21910. Epub 2015/07/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Corella D, Carrasco P, Sorli JV, Coltell O, Ortega-Azorin C, Guillen M, et al. Education modulates the association of the FTO rs9939609 polymorphism with body mass index and obesity risk in the Mediterranean population. Nutr Metab Cardiovasc Dis. 2012;22(8):651–8. doi: 10.1016/j.numecd.2010.10.006. Epub 2010/12/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Foraita R, Gunther F, Gwozdz W, Reisch LA, Russo P, Lauria F, et al. Does the FTO gene interact with the socioeconomic status on the obesity development among young European children? Results from the IDEFICS study. Int J Obes (Lond) 2015;39(1):1–6. doi: 10.1038/ijo.2014.156. Epub 2014/08/20. [DOI] [PubMed] [Google Scholar]
- 165.Freathy RM, Kazeem GR, Morris RW, Johnson PC, Paternoster L, Ebrahim S, et al. Genetic variation at CHRNA5-CHRNA3-CHRNB4 interacts with smoking status to influence body mass index. Int J Epidemiol. 2011;40(6):1617–28. doi: 10.1093/ije/dyr077. Epub 2011/05/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ahmad S, Zhao W, Renstrom F, Rasheed A, Zaidi M, Samuel M, et al. A novel interaction between the FLJ33534 locus and smoking in obesity: a genome-wide study of 14 131 Pakistani adults. Int J Obes (Lond) 2016;40(1):186–90. doi: 10.1038/ijo.2015.152. Epub 2015/08/19. [DOI] [PubMed] [Google Scholar]
- 167.Edwards TL, Velez Edwards DR, Villegas R, Cohen SS, Buchowski MS, Fowke JH, et al. HTR1B, ADIPOR1, PPARGC1A, and CYP19A1 and obesity in a cohort of Caucasians and African Americans: an evaluation of gene-environment interactions and candidate genes. Am J Epidemiol. 2012;175(1):11–21. doi: 10.1093/aje/kwr272. Epub 2011/11/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beyerlein A, von Kries R, Ness AR, Ong KK. Genetic markers of obesity risk: stronger associations with body composition in overweight compared to normal-weight children. PLoS One. 2011;6(4):e19057. doi: 10.1371/journal.pone.0019057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mitchell JA, Hakonarson H, Rebbeck TR, Grant SF. Obesity-susceptibility loci and the tails of the pediatric BMI distribution. Obesity (Silver Spring) 2013;21(6):1256–60. doi: 10.1002/oby.20319. Epub 2013/02/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Williams PT. Quantile-specific penetrance of genes affecting lipoproteins, adiposity and height. PLoS One. 2012;7(1):e28764. doi: 10.1371/journal.pone.0028764. Epub 2012/01/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Luppino FS, de Wit LM, Bouvy PF, Stijnen T, Cuijpers P, Penninx BW, et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Archives of general psychiatry. 2010;67(3):220–9. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 172.Rivera M, Cohen-Woods S, Kapur K, Breen G, Ng MY, Butler AW, et al. Depressive disorder moderates the effect of the FTO gene on body mass index. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.45. [DOI] [PubMed] [Google Scholar]
- 173.Barber TM, McCarthy MI, Wass JA, Franks S. Obesity and polycystic ovary syndrome. Clinical endocrinology. 2006;65(2):137–45. doi: 10.1111/j.1365-2265.2006.02587.x. [DOI] [PubMed] [Google Scholar]
- 174.Kowalska I, Malecki MT, Straczkowski M, Skupien J, Karczewska-Kupczewska M, Nikolajuk A, et al. The FTO gene modifies weight, fat mass and insulin sensitivity in women with polycystic ovary syndrome, where its role may be larger than in other phenotypes. Diabetes & metabolism. 2009;35(4):328–31. doi: 10.1016/j.diabet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 175.Tan S, Scherag A, Janssen OE, Hahn S, Lahner H, Dietz T, et al. Large effects on body mass index and insulin resistance of fat mass and obesity associated gene (FTO) variants in patients with polycystic ovary syndrome (PCOS) BMC Med Genet. 2010;11:12. doi: 10.1186/1471-2350-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bonfig W, Dokoupil K, Schmidt H. A special, strict, fat-reduced, and carbohydrate-modified diet leads to marked weight reduction even in overweight adolescents with Prader-Willi syndrome (PWS) TheScientificWorldJournal. 2009;9:934–9. doi: 10.1100/tsw.2009.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wigren M, Hansen S. Prader-Willi syndrome: clinical picture, psychosocial support and current management. Child: care, health and development. 2003;29(6):449–56. doi: 10.1046/j.1365-2214.2003.00364.x. [DOI] [PubMed] [Google Scholar]
- 178.Vogels A, Fryns JP. Age at diagnosis, body mass index and physical morbidity in children and adults with the Prader-Willi syndrome. Genet Couns. 2004;15(4):397–404. [PubMed] [Google Scholar]
- 179.Silverthorn KH, Hornak JE. Beneficial effects of exercise on aerobic capacity and body composition in adults with Prader-Willi syndrome. Am J Ment Retard. 1993;97(6):654–8. [PubMed] [Google Scholar]
- 180.Eiholzer U, Nordmann Y, l’Allemand D, Schlumpf M, Schmid S, Kromeyer-Hauschild K. Improving body composition and physical activity in Prader-Willi Syndrome. J Pediatr. 2003;142(1):73–8. doi: 10.1067/mpd.2003.mpd0334. [DOI] [PubMed] [Google Scholar]
- 181.Butler MG, Theodoro MF, Bittel DC, Donnelly JE. Energy expenditure and physical activity in Prader-Willi syndrome: comparison with obese subjects. American journal of medical genetics Part A. 2007;143(5):449–59. doi: 10.1002/ajmg.a.31507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Santoro N, Perrone L, Cirillo G, Raimondo P, Amato A, Coppola F, et al. Weight loss in obese children carrying the proopiomelanocortin R236G variant. J Endocrinol Invest. 2006;29(3):226–30. doi: 10.1007/BF03345544. [DOI] [PubMed] [Google Scholar]
- 183.Reinehr T, Hebebrand J, Friedel S, Toschke AM, Brumm H, Biebermann H, et al. Lifestyle intervention in obese children with variations in the melanocortin 4 receptor gene. Obesity (Silver Spring) 2009;17(2):382–9. doi: 10.1038/oby.2008.422. [DOI] [PubMed] [Google Scholar]
- 184.Muller TD, Hinney A, Scherag A, Nguyen TT, Schreiner F, Schafer H, et al. ‘Fat mass and obesity associated’ gene (FTO): no significant association of variant rs9939609 with weight loss in a lifestyle intervention and lipid metabolism markers in German obese children and adolescents. BMC Med Genet. 2008;9:85. doi: 10.1186/1471-2350-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Franks PW, Jablonski KA, Delahanty LM, McAteer JB, Kahn SE, Knowler WC, et al. Assessing gene-treatment interactions at the FTO and INSIG2 loci on obesity-related traits in the Diabetes Prevention Program. Diabetologia. 2008;51(12):2214–23. doi: 10.1007/s00125-008-1158-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lappalainen TJ, Tolppanen AM, Kolehmainen M, Schwab U, Lindstrom J, Tuomilehto J, et al. The common variant in the FTO gene did not modify the effect of lifestyle changes on body weight: the Finnish Diabetes Prevention Study. Obesity (Silver Spring) 2009;17(4):832–6. doi: 10.1038/oby.2008.618. [DOI] [PubMed] [Google Scholar]
- 187.Matsuo T, Nakata Y, Murotake Y, Hotta K, Tanaka K. Effects of FTO Genotype on Weight Loss and Metabolic Risk Factors in Response to Calorie Restriction Among Japanese Women. Obesity (Silver Spring) 2011 doi: 10.1038/oby.2011.322. [DOI] [PubMed] [Google Scholar]
- 188.Woehning A, Schultz JH, Roeder E, Moeltner A, Isermann B, Nawroth PP, et al. The A-allele of the common FTO gene variant rs9939609 complicates weight maintenance in severe obese patients. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.14. [DOI] [PubMed] [Google Scholar]
- 189.Grau K, Hansen T, Holst C, Astrup A, Saris WH, Arner P, et al. Macronutrient-specific effect of FTO rs9939609 in response to a 10-week randomized hypo-energetic diet among obese Europeans. Int J Obes (Lond) 2009;33(11):1227–34. doi: 10.1038/ijo.2009.159. [DOI] [PubMed] [Google Scholar]
- 190.Razquin C, Martinez JA, Martinez-Gonzalez MA, Bes-Rastrollo M, Fernandez-Crehuet J, Marti A. A 3-year intervention with a Mediterranean diet modified the association between the rs9939609 gene variant in FTO and body weight changes. Int J Obes (Lond) 2010;34(2):266–72. doi: 10.1038/ijo.2009.233. [DOI] [PubMed] [Google Scholar]
- 191.Zhang X, Qi Q, Zhang C, Smith SR, Hu FB, Sacks FM, et al. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial. Diabetes. 2012;61(11):3005–11. doi: 10.2337/db11-1799. Epub 2012/08/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Huang T, Qi Q, Li Y, Hu FB, Bray GA, Sacks FM, et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr. 2014;99(5):1126–30. doi: 10.3945/ajcn.113.082164. Epub 2014/03/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mitchell JA, Church TS, Rankinen T, Earnest CP, Sui X, Blair SN. FTO genotype and the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring) 18(3):641–3. doi: 10.1038/oby.2009.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Rankinen T, Rice T, Teran-Garcia M, Rao DC, Bouchard C. FTO genotype is associated with exercise training-induced changes in body composition. Obesity (Silver Spring) 2010;18(2):322–6. doi: 10.1038/oby.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6(4):e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Scherag A, Kleber M, Boes T, Kolbe AL, Ruth A, Grallert H, et al. SDCCAG8 obesity alleles and reduced weight loss after a lifestyle intervention in overweight children and adolescents. Obesity (Silver Spring) 2012;20(2):466–70. doi: 10.1038/oby.2011.339. [DOI] [PubMed] [Google Scholar]
- 197.Delahanty LM, Pan Q, Jablonski KA, Watson KE, McCaffery JM, Shuldiner A, et al. Genetic predictors of weight loss and weight regain after intensive lifestyle modification, metformin treatment, or standard care in the Diabetes Prevention Program. Diabetes Care. 2012;35(2):363–6. doi: 10.2337/dc11-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tai ES, Ordovas JM. The role of perilipin in human obesity and insulin resistance. Curr Opin Lipidol. 2007;18(2):152–6. doi: 10.1097/MOL.0b013e328086aeab. Epub 2007/03/14. [DOI] [PubMed] [Google Scholar]
- 199.Corella D, Qi L, Sorli JV, Godoy D, Portoles O, Coltell O, et al. Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. The Journal of clinical endocrinology and metabolism. 2005;90(9):5121–6. doi: 10.1210/jc.2005-0576. Epub 2005/06/30. [DOI] [PubMed] [Google Scholar]
- 200.Jang Y, Kim OY, Lee JH, Koh SJ, Chae JS, Kim JY, et al. Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 2006;30(11):1601–8. doi: 10.1038/sj.ijo.0803312. Epub 2006/04/06. [DOI] [PubMed] [Google Scholar]
- 201.Ruiz JR, Larrarte E, Margareto J, Ares R, Alkorta P, Labayen I. Preliminary findings on the role of PLIN1 polymorphisms on body composition and energy metabolism response to energy restriction in obese women. The British journal of nutrition. 2011;106(4):486–90. doi: 10.1017/S0007114511000432. Epub 2011/03/12. [DOI] [PubMed] [Google Scholar]
- 202.Zelissen PM, Stenlof K, Lean ME, Fogteloo J, Keulen ET, Wilding J, et al. Effect of three treatment schedules of recombinant methionyl human leptin on body weight in obese adults: a randomized, placebo-controlled trial. Diabetes, obesity & metabolism. 2005;7(6):755–61. doi: 10.1111/j.1463-1326.2005.00468.x. [DOI] [PubMed] [Google Scholar]
- 203.Farooqi IS, Matarese G, Lord GM, Keogh JM, Lawrence E, Agwu C, et al. Beneficial effects of leptin on obesity, T cell hyporesponsiveness, and neuroendocrine/metabolic dysfunction of human congenital leptin deficiency. J Clin Invest. 2002;110(8):1093–103. doi: 10.1172/JCI15693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Licinio J, Caglayan S, Ozata M, Yildiz BO, de Miranda PB, O’Kirwan F, et al. Phenotypic effects of leptin replacement on morbid obesity, diabetes mellitus, hypogonadism, and behavior in leptin-deficient adults. Proc Natl Acad Sci U S A. 2004;101(13):4531–6. doi: 10.1073/pnas.0308767101. Epub 2004/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Paz-Filho G, Wong ML, Licinio J. Ten years of leptin replacement therapy. Obesity reviews : an official journal of the International Association for the Study of Obesity. doi: 10.1111/j.1467-789X.2010.00840.x. [DOI] [PubMed] [Google Scholar]
- 206.Heymsfield SB, Greenberg AS, Fujioka K, Dixon RM, Kushner R, Hunt T, et al. Recombinant leptin for weight loss in obese and lean adults: a randomized, controlled, dose-escalation trial. Jama. 1999;282(16):1568–75. doi: 10.1001/jama.282.16.1568. [DOI] [PubMed] [Google Scholar]
- 207.Farooqi IS, Keogh JM, Kamath S, Jones S, Gibson WT, Trussell R, et al. Partial leptin deficiency and human adiposity. Nature. 2001;414(6859):34–5. doi: 10.1038/35102112. [DOI] [PubMed] [Google Scholar]
- 208.Baumgart D, Naber C, Haude M, Oldenburg O, Erbel R, Heusch G, et al. G protein beta3 subunit 825T allele and enhanced coronary vasoconstriction on alpha(2)-adrenoceptor activation. Circ Res. 1999;85(10):965–9. doi: 10.1161/01.res.85.10.965. Epub 1999/11/13. [DOI] [PubMed] [Google Scholar]
- 209.Hsiao DJ, Wu LS, Huang SY, Lin E. Weight loss and body fat reduction under sibutramine therapy in obesity with the C825T polymorphism in the GNB3 gene. Pharmacogenetics and genomics. 2009;19(9):730–3. doi: 10.1097/FPC.0b013e3283307cf1. [DOI] [PubMed] [Google Scholar]
- 210.Grudell AB, Sweetser S, Camilleri M, Eckert DJ, Vazquez-Roque MI, Carlson PJ, et al. A controlled pharmacogenetic trial of sibutramine on weight loss and body composition in obese or overweight adults. Gastroenterology. 2008;135(4):1142–54. doi: 10.1053/j.gastro.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tiwari HK, Patki A, Lieberman J, Stroup TS, Allison DB, Leibel RL, et al. Association of allelic variation in genes mediating aspects of energy homeostasis with weight gain during administration of antipsychotic drugs (CATIE Study) Frontiers in genetics. 2011;2(56) doi: 10.3389/fgene.2011.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chowdhury NI, Tiwari AK, Souza RP, Zai CC, Shaikh SA, Chen S, et al. Genetic association study between antipsychotic-induced weight gain and the melanocortin-4 receptor gene. The pharmacogenomics journal. 2012 doi: 10.1038/tpj.2011.66. [DOI] [PubMed] [Google Scholar]
- 213.Roffeei SN, Mohamed Z, Reynolds GP, Said MA, Hatim A, Mohamed EH, et al. Association of FTO, LEPR and MTHFR gene polymorphisms with metabolic syndrome in schizophrenia patients receiving antipsychotics. Pharmacogenomics. 2014;15(4):477–85. doi: 10.2217/pgs.13.220. Epub 2014/03/15. [DOI] [PubMed] [Google Scholar]
- 214.Czerwensky F, Leucht S, Steimer W. Association of the common MC4R rs17782313 polymorphism with antipsychotic-related weight gain. J Clin Psychopharmacol. 2013;33(1):74–9. doi: 10.1097/JCP.0b013e31827772db. Epub 2013/01/02. [DOI] [PubMed] [Google Scholar]
- 215.Czerwensky F, Leucht S, Steimer W. MC4R rs489693: a clinical risk factor for second generation antipsychotic-related weight gain? Int J Neuropsychopharmacol. 2013;16(9):2103–9. doi: 10.1017/S1461145713000849. Epub 2013/08/08. [DOI] [PubMed] [Google Scholar]
- 216.Sjostrom L, Lindroos AK, Peltonen M, Torgerson J, Bouchard C, Carlsson B, et al. Lifestyle, diabetes, and cardiovascular risk factors 10 years after bariatric surgery. N Engl J Med. 2004;351(26):2683–93. doi: 10.1056/NEJMoa035622. [DOI] [PubMed] [Google Scholar]
- 217.Aslan IR, Ranadive SA, Ersoy BA, Rogers SJ, Lustig RH, Vaisse C. Bariatric surgery in a patient with complete MC4R deficiency. Int J Obes (Lond) 2011;35(3):457–61. doi: 10.1038/ijo.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stutzmann F, Keller R, Labrune Y, Durand E, Calvacanti-Proenca C, Potoczna N, et al. Variability of the effect of bariatric surgery according to the genotype of the MC4R. Diabetes & metabolism. 2010;36(1):A23–A4. [Google Scholar]
- 219.Aslan IR, Campos GM, Calton MA, Evans DS, Merriman RB, Vaisse C. Weight Loss after Roux-en-Y Gastric Bypass in Obese Patients Heterozygous for MC4R Mutations. Obes Surg. 2011 doi: 10.1007/s11695-010-0295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hatoum IJ, Stylopoulos N, Vanhoose AM, Boyd KL, Yin DP, Ellacott KL, et al. Melanocortin-4 Receptor Signaling Is Required for Weight Loss after Gastric Bypass Surgery. J Clin Endocrinol Metab. 2012 doi: 10.1210/jc.2011-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Censani M, Conroy R, Deng L, Oberfield SE, McMahon DJ, Zitsman JL, et al. Weight loss after bariatric surgery in morbidly obese adolescents with MC4R mutations. Obesity (Silver Spring) 2014;22(1):225–31. doi: 10.1002/oby.20511. Epub 2013/06/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sarzynski MA, Jacobson P, Rankinen T, Carlsson B, Sjostrom L, Bouchard C, et al. Associations of markers in 11 obesity candidate genes with maximal weight loss and weight regain in the SOS bariatric surgery cases. Int J Obes (Lond) 2011;35(5):676–83. doi: 10.1038/ijo.2010.166. [DOI] [PubMed] [Google Scholar]
- 223.Bandstein M, Schultes B, Ernst B, Thurnheer M, Schioth HB, Benedict C. The Role of FTO and Vitamin D for the Weight Loss Effect of Roux-en-Y Gastric Bypass Surgery in Obese Patients. Obes Surg. 2015 doi: 10.1007/s11695-015-1644-4. Epub 2015/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liou TH, Chen HH, Wang W, Wu SF, Lee YC, Yang WS, et al. ESR1, FTO, and UCP2 genes interact with bariatric surgery affecting weight loss and glycemic control in severely obese patients. Obes Surg. 2011;21(11):1758–65. doi: 10.1007/s11695-011-0457-3. [DOI] [PubMed] [Google Scholar]
- 225.Franks PW, Ling C. Epigenetics and obesity: the devil is in the details. BMC Med. 2010;8:88. doi: 10.1186/1741-7015-8-88. Epub 2010/12/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.van Dijk SJ, Molloy PL, Varinli H, Morrison JL, Muhlhausler BS. Epigenetics and human obesity. Int J Obes (Lond) 2015;39(1):85–97. doi: 10.1038/ijo.2014.34. Epub 2014/02/26. [DOI] [PubMed] [Google Scholar]
- 227.Feil RFM. Epigenetics and the environment: emerging patterns and implications. Nature Reviews Genetics. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 228.Youngson NA, Morris MJ. What obesity research tells us about epigenetic mechanisms. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110337. doi: 10.1098/rstb.2011.0337. Epub 2012/11/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–16. doi: 10.1038/ng.520. Epub 2010/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–41. doi: 10.1038/nrg3000. Epub 2011/07/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aissi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet. 2014;383(9933):1990–8. doi: 10.1016/S0140-6736(13)62674-4. Epub 2014/03/19. [DOI] [PubMed] [Google Scholar]
- 232.Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, et al. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet. 2015;24(15):4464–79. doi: 10.1093/hmg/ddv161. Epub 2015/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pan H, Lin X, Wu Y, Chen L, Teh AL, Soh SE, et al. HIF3A association with adiposity: the story begins before birth. Epigenomics. 2015;7(6):937–50. doi: 10.2217/epi.15.45. Epub 2015/05/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ronn T, Volkov P, Gillberg L, Kokosar M, Perfilyev A, Jacobsen AL, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum Mol Genet. 2015;24(13):3792–813. doi: 10.1093/hmg/ddv124. Epub 2015/04/12. [DOI] [PubMed] [Google Scholar]
- 235.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102(30):10604–9. doi: 10.1073/pnas.0500398102. Epub 2005/07/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ronn T, Poulsen P, Hansson O, Holmkvist J, Almgren P, Nilsson P, et al. Age influences DNA methylation and gene expression of COX7A1 in human skeletal muscle. Diabetologia. 2008;51(7):1159–68. doi: 10.1007/s00125-008-1018-8. Epub 2008/05/20. [DOI] [PubMed] [Google Scholar]
- 237.Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun. 2014;5:5592. doi: 10.1038/ncomms6592. Epub 2014/11/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yajnik CS, Deshmukh US. Fetal programming: maternal nutrition and role of one-carbon metabolism. Rev Endocr Metab Disord. 2012;13(2):121–7. doi: 10.1007/s11154-012-9214-8. Epub 2012/03/15. [DOI] [PubMed] [Google Scholar]
- 239.Yajnik CS, Deshpande SS, Jackson AA, Refsum H, Rao S, Fisher DJ, et al. Vitamin B12 and folate concentrations during pregnancy and insulin resistance in the offspring: the Pune Maternal Nutrition Study. Diabetologia. 2008;51(1):29–38. doi: 10.1007/s00125-007-0793-y. Epub 2007/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Gueant JL, Namour F, Gueant-Rodriguez RM, Daval JL. Folate and fetal programming: a play in epigenomics? Trends Endocrinol Metab. 2013;24(6):279–89. doi: 10.1016/j.tem.2013.01.010. Epub 2013/03/12. [DOI] [PubMed] [Google Scholar]
- 241.Ling C, Ronn T. Epigenetic adaptation to regular exercise in humans. Drug Discov Today. 2014;19(7):1015–8. doi: 10.1016/j.drudis.2014.03.006. Epub 2014/03/19. [DOI] [PubMed] [Google Scholar]
- 242.Nitert MD, Dayeh T, Volkov P, Elgzyri T, Hall E, Nilsson E, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–32. doi: 10.2337/db11-1653. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ronn T, Volkov P, Davegardh C, Dayeh T, Hall E, Olsson AH, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. Epub 2013/07/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiol (Oxf) 2015;213(1):39–59. doi: 10.1111/apha.12414. Epub 2014/10/28. [DOI] [PubMed] [Google Scholar]
- 245.Gillberg L, Jacobsen SC, Ronn T, Brons C, Vaag A. PPARGC1A DNA methylation in subcutaneous adipose tissue in low birth weight subjects--impact of 5 days of high-fat overfeeding. Metabolism. 2014;63(2):263–71. doi: 10.1016/j.metabol.2013.10.003. Epub 2013/11/23. [DOI] [PubMed] [Google Scholar]
- 246.Jacobsen SC, Brons C, Bork-Jensen J, Ribel-Madsen R, Yang B, Lara E, et al. Effects of short-term high-fat overfeeding on genome-wide DNA methylation in the skeletal muscle of healthy young men. Diabetologia. 2012;55(12):3341–9. doi: 10.1007/s00125-012-2717-8. Epub 2012/09/11. [DOI] [PubMed] [Google Scholar]
- 247.Jacobsen SC, Gillberg L, Bork-Jensen J, Ribel-Madsen R, Lara E, Calvanese V, et al. Young men with low birthweight exhibit decreased plasticity of genome-wide muscle DNA methylation by high-fat overfeeding. Diabetologia. 2014;57(6):1154–8. doi: 10.1007/s00125-014-3198-8. Epub 2014/02/27. [DOI] [PubMed] [Google Scholar]
- 248.Voisin S, Almen MS, Moschonis G, Chrousos GP, Manios Y, Schioth HB. Dietary fat quality impacts genome-wide DNA methylation patterns in a cross-sectional study of Greek preadolescents. European journal of human genetics : EJHG. 2015;23(5):654–62. doi: 10.1038/ejhg.2014.139. Epub 2014/07/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Barres R, Kirchner H, Rasmussen M, Yan J, Kantor FR, Krook A, et al. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Rep. 2013;3(4):1020–7. doi: 10.1016/j.celrep.2013.03.018. Epub 2013/04/16. [DOI] [PubMed] [Google Scholar]
- 250.Benton MC, Johnstone A, Eccles D, Harmon B, Hayes MT, Lea RA, et al. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 2015;16:8. doi: 10.1186/s13059-014-0569-x. Epub 2015/02/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Huang YT, Maccani JZ, Hawley NL, Wing RR, Kelsey KT, McCaffery JM. Epigenetic patterns in successful weight loss maintainers: a pilot study. Int J Obes (Lond) 2015;39(5):865–8. doi: 10.1038/ijo.2014.213. Epub 2014/12/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P, et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab. 2015;21(1):138–49. doi: 10.1016/j.cmet.2014.12.014. Epub 2015/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Speakman JR, O’Rahilly S. Fat: an evolving issue. Dis Model Mech. 2012;5(5):569–73. doi: 10.1242/dmm.010553. Epub 2012/08/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ladabaum U, Mannalithara A, Myer PA, Singh G. Obesity, Abdominal Obesity, Physical Activity, and Caloric Intake in U.S. Adults: 1988–2010. Am J Med. 2014 doi: 10.1016/j.amjmed.2014.02.026. Epub 2014/03/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res. 2014;24(7):1064–74. doi: 10.1101/gr.171439.113. Epub 2014/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93(5):876–90. doi: 10.1016/j.ajhg.2013.10.004. Epub 2013/11/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Bell CG, Finer S, Lindgren CM, Wilson GA, Rakyan VK, Teschendorff AE, et al. Integrated genetic and epigenetic analysis identifies haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One. 2010;5(11):e14040. doi: 10.1371/journal.pone.0014040. Epub 2010/12/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Voisin S, Almen MS, Zheleznyakova GY, Lundberg L, Zarei S, Castillo S, et al. Many obesity-associated SNPs strongly associate with DNA methylation changes at proximal promoters and enhancers. Genome Med. 2015;7(1):103. doi: 10.1186/s13073-015-0225-4. Epub 2015/10/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dalgaard K, Landgraf K, Heyne S, Lempradl A, Longinotto J, Gossens K, et al. Trim28 Haploinsufficiency Triggers Bi-stable Epigenetic Obesity. Cell. 2016;164(3):353–64. doi: 10.1016/j.cell.2015.12.025. Epub 2016/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Keller MC. Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol Psychiatry. 2014;75(1):18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Aliev F, Latendresse SJ, Bacanu S-A, Neale MC, Dick DM. Testing for measured gene-environment interaction: problems with the use of cross-product terms and a regression model reparameterization solution. Behav Genet. 2014;44(2):165–81. doi: 10.1007/s10519-014-9642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Vanderweele TJ, Ko Y-A, Mukherjee B. Environmental confounding in gene-environment interaction studies. Am J Epidemiol. 2013;178(1):144–52. doi: 10.1093/aje/kws439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Underwood PC, Chamarthi B, Williams JS, Sun B, Vaidya A, Raby BA, et al. Replication and meta-analysis of the gene-environment interaction between body mass index and the interleukin-6 promoter polymorphism with higher insulin resistance. Metabolism: Clinical & Experimental. 2012;61(5):667–71. doi: 10.1016/j.metabol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Munafo MR, Flint J. Replication and heterogeneity in gene × environment interaction studies. Int J Neuropsychopharmacol. 2009;12(6):727–9. doi: 10.1017/S1461145709000479. Epub 2009/05/30. [DOI] [PubMed] [Google Scholar]
- 265.Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168(10):1041–9. doi: 10.1176/appi.ajp.2011.11020191. Epub 2011/09/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Franks PW, Nettleton JA. Invited commentary: Gene X lifestyle interactions and complex disease traits--inferring cause and effect from observational data, sine qua non. Am J Epidemiol. 2010;172(9):992–7. doi: 10.1093/aje/kwq280. discussion 8–9. Epub 2010/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Fleming JL, Huang TH, Toland AE. The role of parental and grandparental epigenetic alterations in familial cancer risk. Cancer Res. 2008;68(22):9116–21. doi: 10.1158/0008-5472.CAN-08-2184. Epub 2008/11/18. [DOI] [PubMed] [Google Scholar]
- 268.Liu CY, Maity A, Lin X, Wright RO, Christiani DC. Design and analysis issues in gene and environment studies. Environ Health. 2012;11:93. doi: 10.1186/1476-069X-11-93. Epub 2012/12/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wong MY, Day NE, Luan JA, Chan KP, Wareham NJ. The detection of gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol. 2003;32(1):51–7. doi: 10.1093/ije/dyg002. Epub 2003/04/12. [DOI] [PubMed] [Google Scholar]
- 270.Hewitt JK. Editorial policy on candidate gene association and candidate gene-by-environment interaction studies of complex traits. Behav Genet. 2012;42(1):1–2. doi: 10.1007/s10519-011-9504-z. Epub 2011/09/20. [DOI] [PubMed] [Google Scholar]
- 271.Johnston CLB, Matthys W. Editorial policy for candidate gene studies. J Abnorm Child Psychol. 2013;41:511–4. [Google Scholar]
- 272.Pinelli M, Scala G, Amato R, Cocozza S, Miele G. Simulating gene-gene and gene-environment interactions in complex diseases: Gene-Environment iNteraction Simulator 2. BMC Bioinformatics. 2012;13:132. doi: 10.1186/1471-2105-13-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Van Hulle CA, Lahey BB, Rathouz PJ. Operating characteristics of alternative statistical methods for detecting gene-by-measured environment interaction in the presence of gene-environment correlation in twin and sibling studies. Behav Genet. 2013;43(1):71–84. doi: 10.1007/s10519-012-9568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Dick DM, Agrawal A, Keller MC, Adkins A, Aliev F, Monroe S, et al. Candidate gene-environment interaction research: reflections and recommendations. Perspect Psychol Sci. 2015;10(1):37–59. doi: 10.1177/1745691614556682. Epub 2015/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Belsky J, Pluess M, Widaman KF. Confirmatory and competitive evaluation of alternative gene-environment interaction hypotheses. J Child Psychol Psychiatry. 2013;54(10):1135–43. doi: 10.1111/jcpp.12075. Epub 2013/04/27. [DOI] [PubMed] [Google Scholar]
- 276.Zubin J, Spring B. Vulnerability--a new view of schizophrenia. J Abnorm Psychol. 1977;86(2):103–26. doi: 10.1037//0021-843x.86.2.103. Epub 1977/04/01. [DOI] [PubMed] [Google Scholar]
- 277.Burmeister M, McInnis MG, Zollner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9(7):527–40. doi: 10.1038/nrg2381. Epub 2008/06/19. [DOI] [PubMed] [Google Scholar]
- 278.Belsky J, Pluess M. Beyond diathesis stress: differential susceptibility to environmental influences. Psychol Bull. 2009;135(6):885–908. doi: 10.1037/a0017376. Epub 2009/11/04. [DOI] [PubMed] [Google Scholar]
- 279.Belsky J, Bakermans-Kranenburg MJ, van IJzendoorn MH. For better and for worse: Differential Susceptibility to environmental influences. Current Directions in Psychological Science. 2007;16:300–4. [Google Scholar]
- 280.Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Mol Psychiatry. 2009;14(8):746–54. doi: 10.1038/mp.2009.44. Epub 2009/05/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Ellis BJ, Boyce WT, Belsky J, Bakermans-Kranenburg MJ, van Ijzendoorn MH. Differential susceptibility to the environment: an evolutionary--neurodevelopmental theory. Dev Psychopathol. 2011;23(1):7–28. doi: 10.1017/S0954579410000611. Epub 2011/01/26. [DOI] [PubMed] [Google Scholar]
- 282.Bakermans-Kranenburg MJ, van Ijzendoorn MH. Gene-environment interaction of the dopamine D4 receptor (DRD4) and observed maternal insensitivity predicting externalizing behavior in preschoolers. Dev Psychobiol. 2006;48(5):406–9. doi: 10.1002/dev.20152. Epub 2006/06/14. [DOI] [PubMed] [Google Scholar]
- 283.Belsky J, Pluess M. Beyond risk, resilience, and dysregulation: phenotypic plasticity and human development. Dev Psychopathol. 2013;25(4 Pt 2):1243–61. doi: 10.1017/S095457941300059X. Epub 2013/12/18. [DOI] [PubMed] [Google Scholar]
- 284.Roisman GI, Newman DA, Fraley RC, Haltigan JD, Groh AM, Haydon KC. Distinguishing differential susceptibility from diathesis-stress: recommendations for evaluating interaction effects. Dev Psychopathol. 2012;24(2):389–409. doi: 10.1017/S0954579412000065. Epub 2012/05/09. [DOI] [PubMed] [Google Scholar]
- 285.Widaman KF, Helm JL, Castro-Schilo L, Pluess M, Stallings MC, Belsky J. Distinguishing ordinal and disordinal interactions. Psychol Methods. 2012;17(4):615–22. doi: 10.1037/a0030003. Epub 2012/09/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Brown GL, Mangelsdorf SC, Neff C. Father involvement, paternal sensitivity, and father-child attachment security in the first 3 years. J Fam Psychol. 2012;26(3):421–30. doi: 10.1037/a0027836. Epub 2012/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Blot WJ, Day NE. Synergism and interaction: are they equivalent? Am J Epidemiol. 1979;110(1):99–100. doi: 10.1093/oxfordjournals.aje.a112793. Epub 1979/07/01. [DOI] [PubMed] [Google Scholar]
- 288.Saracci R. Interaction and synergism. Am J Epidemiol. 1980;112(4):465–6. doi: 10.1093/oxfordjournals.aje.a113014. Epub 1980/10/01. [DOI] [PubMed] [Google Scholar]
- 289.VanderWeele TJ, Robins JM. The identification of synergism in the sufficient-component-cause framework. Epidemiology. 2007;18(3):329–39. doi: 10.1097/01.ede.0000260218.66432.88. Epub 2007/04/17. [DOI] [PubMed] [Google Scholar]
- 290.Vandenbroucke JP. New public health and old rhetoric. Bmj. 1994;308(6935):994–5. doi: 10.1136/bmj.308.6935.994. Epub 1994/04/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ottman R. Gene-environment interaction: definitions and study designs. Prev Med. 1996;25(6):764–70. doi: 10.1006/pmed.1996.0117. Epub 1996/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Koopman JS. Causal models and sources of interaction. Am J Epidemiol. 1977;106(6):439–44. doi: 10.1093/oxfordjournals.aje.a112489. Epub 1977/12/01. [DOI] [PubMed] [Google Scholar]
- 293.Kupper LL, Hogan MD. Interaction in epidemiologic studies. Am J Epidemiol. 1978;108(6):447–53. doi: 10.1093/oxfordjournals.aje.a112643. Epub 1978/12/01. [DOI] [PubMed] [Google Scholar]
- 294.Walter SD, Holford TR. Additive, multiplicative, and other models for disease risks. Am J Epidemiol. 1978;108(5):341–6. doi: 10.1093/oxfordjournals.aje.a112629. Epub 1978/11/01. [DOI] [PubMed] [Google Scholar]
- 295.Rothman KJ, Greenland S, Walker AM. Concepts of interaction. Am J Epidemiol. 1980;112(4):467–70. doi: 10.1093/oxfordjournals.aje.a113015. Epub 1980/10/01. [DOI] [PubMed] [Google Scholar]
- 296.Siemiatycki J, Thomas DC. Biological models and statistical interactions: an example from multistage carcinogenesis. Int J Epidemiol. 1981;10(4):383–7. doi: 10.1093/ije/10.4.383. Epub 1981/12/01. [DOI] [PubMed] [Google Scholar]
- 297.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96(2):329–39. doi: 10.1016/j.ajhg.2014.12.021. Epub 2015/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Dudbridge F, Fletcher O. Gene-environment dependence creates spurious gene-environment interaction. Am J Hum Genet. 2014;95(3):301–7. doi: 10.1016/j.ajhg.2014.07.014. Epub 2014/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Vanderweele TJ, Ko YA, Mukherjee B. Environmental confounding in gene-environment interaction studies. Am J Epidemiol. 2013;178(1):144–52. doi: 10.1093/aje/kws439. Epub 2013/07/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–22. doi: 10.1038/ng.109. Epub 2008/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–7. doi: 10.1038/nature06885. Epub 2008/04/04. [DOI] [PubMed] [Google Scholar]
- 302.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–42. doi: 10.1038/nature06846. Epub 2008/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Nederhof E, Bouma EMC, Riese H, Laceulle OM, Ormel J, Oldehinkel AJ. Evidence for plasticity genotypes in a gene-gene-environment interaction: the TRAILS study. Genes, Brain, & Behavior. 2010;9(8):968–73. doi: 10.1111/j.1601-183X.2010.00637.x. [DOI] [PubMed] [Google Scholar]
- 304.Woodruff TJ. Bridging epidemiology and model organisms to increase understanding of endocrine disrupting chemicals and human health effects. J Steroid Biochem Mol Biol. 2011;127(1–2):108–17. doi: 10.1016/j.jsbmb.2010.11.007. Epub 2010/11/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Buck Louis GM, Schisterman EF, Sweeney AM, Wilcosky TC, Gore-Langton RE, Lynch CD, et al. Designing prospective cohort studies for assessing reproductive and developmental toxicity during sensitive windows of human reproduction and development--the LIFE Study. Paediatr Perinat Epidemiol. 2011;25(5):413–24. doi: 10.1111/j.1365-3016.2011.01205.x. Epub 2011/08/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ege MJ, Strachan DP. Comparisons of power of statistical methods for gene-environment interaction analyses. Eur J Epidemiol. 2013;28(10):785–97. doi: 10.1007/s10654-013-9837-4. [DOI] [PubMed] [Google Scholar]
- 307.Heo M, Leon AC. Sample sizes required to detect two-way and three-way interactions involving slope differences in mixed-effects linear models. J Biopharm Stat. 2010;20(4):787–802. doi: 10.1080/10543401003618819. Epub 2010/05/25. [DOI] [PubMed] [Google Scholar]
- 308.Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. Epub 2012/05/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Joseph PG, Pare G, Anand SS. Exploring gene-environment relationships in cardiovascular disease. The Canadian journal of cardiology. 2013;29(1):37–45. doi: 10.1016/j.cjca.2012.10.009. Epub 2012/12/25. [DOI] [PubMed] [Google Scholar]
- 310.Luan JA, Wong MY, Day NE, Wareham NJ. Sample size determination for studies of gene-environment interaction. Int J Epidemiol. 2001;30(5):1035–40. doi: 10.1093/ije/30.5.1035. Epub 2001/11/02. [DOI] [PubMed] [Google Scholar]
- 311.Corella D, Arnett DK, Tucker KL, Kabagambe EK, Tsai M, Parnell LD, et al. A high intake of saturated fatty acids strengthens the association between the fat mass and obesity-associated gene and BMI. J Nutr. 2011;141(12):2219–25. doi: 10.3945/jn.111.143826. Epub 2011/11/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90(5):1418–25. doi: 10.3945/ajcn.2009.27958. Epub 2009/09/04. [DOI] [PubMed] [Google Scholar]
- 313.Armstrong BKWE, Saracci R. Principles of Exposure Measurement in Epidemiology. Oxford: Oxford University Press; 1994. [Google Scholar]
- 314.Wegener JW, Meyrer H, Rupp J, Nawrath H. Barnidipine block of L-type Ca(2+) channel currents in rat ventricular cardiomyocytes. British journal of pharmacology. 2000;130(8):2015–23. doi: 10.1038/sj.bjp.0703514. Epub 2000/08/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Manco M, Panunzi S, Macfarlane DP, Golay A, Melander O, Konrad T, et al. One-hour plasma glucose identifies insulin resistance and beta-cell dysfunction in individuals with normal glucose tolerance: cross-sectional data from the Relationship between Insulin Sensitivity and Cardiovascular Risk (RISC) study. Diabetes Care. 2010;33(9):2090–7. doi: 10.2337/dc09-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Whitlock G, Lewington S, Sherliker P, Clarke R, Emberson J, Halsey J, et al. Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet. 2009;373(9669):1083–96. doi: 10.1016/S0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Muller MJ, Bosy-Westphal A, Krawczak M. Genetic studies of common types of obesity: a critique of the current use of phenotypes. Obes Rev. 2010;11(8):612–8. doi: 10.1111/j.1467-789X.2010.00734.x. Epub 2010/03/30. [DOI] [PubMed] [Google Scholar]
- 318.Pastinen T, Raitio M, Lindroos K, Tainola P, Peltonen L, Syvanen AC. A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res. 2000;10(7):1031–42. doi: 10.1101/gr.10.7.1031. Epub 2000/07/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Prince JA, Feuk L, Howell WM, Jobs M, Emahazion T, Blennow K, et al. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res. 2001;11(1):152–62. doi: 10.1101/gr.150201. Epub 2001/01/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Peters T, Brage S, Westgate K, Franks PW, Gradmark A, Diaz MJT, et al. Validity of a short questionnaire to assess physical activity in 10 European countries. Eur J Epidemiol. 2012;27(1):15–25. doi: 10.1007/s10654-011-9625-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Welk GJ, Kim Y, Stanfill B, Osthus DA, Calabro MA, Nusser SM, et al. Validity of 24-h Physical Activity Recall: Physical Activity Measurement Survey. Med Sci Sports Exerc. 2014;46(10):2014–24. doi: 10.1249/MSS.0000000000000314. Epub 2014/02/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wareham NJ, Jakes RW, Rennie KL, Mitchell J, Hennings S, Day NE. Validity and repeatability of the EPIC-Norfolk Physical Activity Questionnaire. Int J Epidemiol. 2002;31(1):168–74. doi: 10.1093/ije/31.1.168. Epub 2002/03/27. [DOI] [PubMed] [Google Scholar]
- 323.Vinknes KJ, Elshorbagy AK, Drevon CA, Gjesdal CG, Tell GS, Nygard O, et al. Evaluation of the body adiposity index in a Caucasian population: the Hordaland health study. Am J Epidemiol. 2013;177(6):586–92. doi: 10.1093/aje/kws271. Epub 2013/02/28. [DOI] [PubMed] [Google Scholar]
- 324.Wang K, Li WD, Zhang CK, Wang Z, Glessner JT, Grant SF, et al. A genome-wide association study on obesity and obesity-related traits. PLoS One. 2011;6(4):e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–11. doi: 10.1038/ng.1087. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Franks PW, Mesa JL, Harding AH, Wareham NJ. Gene-lifestyle interaction on risk of type 2 diabetes. Nutr Metab Cardiovasc Dis. 2007;17(2):104–24. doi: 10.1016/j.numecd.2006.04.001. Epub 2006/10/03. [DOI] [PubMed] [Google Scholar]
- 327.Livingstone MB, Prentice AM, Coward WA, Ceesay SM, Strain JJ, McKenna PG, et al. Simultaneous measurement of free-living energy expenditure by the doubly labeled water method and heart-rate monitoring. Am J Clin Nutr. 1990;52(1):59–65. doi: 10.1093/ajcn/52.1.59. Epub 1990/07/01. [DOI] [PubMed] [Google Scholar]
- 328.Wareham NJ, Wong MY, Day NE. Glucose intolerance and physical inactivity: the relative importance of low habitual energy expenditure and cardiorespiratory fitness. Am J Epidemiol. 2000;152(2):132–9. doi: 10.1093/aje/152.2.132. Epub 2000/07/26. [DOI] [PubMed] [Google Scholar]
- 329.Dhurandhar NV, Schoeller D, Brown AW, Heymsfield SB, Thomas D, Sorensen TI, et al. Energy balance measurement: when something is not better than nothing. Int J Obes (Lond) 2015;39(7):1109–13. doi: 10.1038/ijo.2014.199. Epub 2014/11/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Delude CM. Deep phenotyping: The details of disease. Nature. 2015;527(7576):S14–5. doi: 10.1038/527S14a. Epub 2015/11/05. [DOI] [PubMed] [Google Scholar]
- 331.Hodson L, Skeaff CM, Fielding BA. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog Lipid Res. 2008;47(5):348–80. doi: 10.1016/j.plipres.2008.03.003. Epub 2008/04/26. [DOI] [PubMed] [Google Scholar]
- 332.Cameron JD, Goldfield GS, Riou ME, Finlayson GS, Blundell JE, Doucet E. Energy depletion by diet or aerobic exercise alone: impact of energy deficit modality on appetite parameters. Am J Clin Nutr. 2016 doi: 10.3945/ajcn.115.115584. Epub 2016/02/19. [DOI] [PubMed] [Google Scholar]
- 333.Ottman R. An epidemiologic approach to gene-environment interaction. Genet Epidemiol. 1990;7(3):177–85. doi: 10.1002/gepi.1370070302. Epub 1990/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev Genet. 2004;5(2):89–100. doi: 10.1038/nrg1270. Epub 2004/01/22. [DOI] [PubMed] [Google Scholar]
- 335.Hein R, Beckmann L, Chang-Claude J. Sample size requirements for indirect association studies of gene-environment interactions (G × E) Genet Epidemiol. 2008;32(3):235–45. doi: 10.1002/gepi.20298. Epub 2008/01/01. [DOI] [PubMed] [Google Scholar]
- 336.Williamson E, Ponsonby AL, Carlin J, Dwyer T. Effect of including environmental data in investigations of gene-disease associations in the presence of qualitative interactions. Genet Epidemiol. 2010;34(6):552–60. doi: 10.1002/gepi.20511. [DOI] [PubMed] [Google Scholar]
- 337.Khoury MJ, Adams MJ, Jr, Flanders WD. An epidemiologic approach to ecogenetics. Am J Hum Genet. 1988;42(1):89–95. [PMC free article] [PubMed] [Google Scholar]
- 338.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–9. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 339.Dai JY, Logsdon BA, Huang Y, Hsu L, Reiner AP, Prentice RL, et al. Simultaneously testing for marginal genetic association and gene-environment interaction. Am J Epidemiol. 2012;176(2):164–73. doi: 10.1093/aje/kwr521. Epub 2012/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Manning AK, LaValley M, Liu CT, Rice K, An P, Liu Y, et al. Meta-analysis of gene-environment interaction: joint estimation of SNP and SNP × environment regression coefficients. Genet Epidemiol. 2011;35(1):11–8. doi: 10.1002/gepi.20546. Epub 2010/12/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. American journal of epidemiology. 2009;169(2):219–26. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Aschard H, Hancock DB, London SJ, Kraft P. Genome-wide meta-analysis of joint tests for genetic and gene-environment interaction effects. Hum Hered. 2011;70(4):292–300. doi: 10.1159/000323318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. doi: 10.1038/nature14132. Epub 2015/02/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Marigorta UM, Gibson G. A simulation study of gene-by-environment interactions in GWAS implies ample hidden effects. Front Genet. 2014;5:225. doi: 10.3389/fgene.2014.00225. Epub 2014/08/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Pare G, Cook NR, Ridker PM, Chasman DI. On the use of variance per genotype as a tool to identify quantitative trait interaction effects: a report from the Women’s Genome Health Study. PLoS Genet. 2010;6(6):e1000981. doi: 10.1371/journal.pgen.1000981. Epub 2010/06/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Yu K, Wacholder S, Wheeler W, Wang Z, Caporaso N, Landi MT, et al. A flexible Bayesian model for studying gene-environment interaction. PLoS Genet. 2012;8(1):e1002482. doi: 10.1371/journal.pgen.1002482. Epub 2012/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Gunther F, Pigeot I, Bammann K. Artificial neural networks modeling gene-environment interaction. BMC Genetics. 2012;13:37. doi: 10.1186/1471-2156-13-37. Epub 2012/05/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Hsu L, Jiao S, Dai JY, Hutter C, Peters U, Kooperberg C. Powerful cocktail methods for detecting genome-wide gene-environment interaction. Genet Epidemiol. 2012;36(3):183–94. doi: 10.1002/gepi.21610. Epub 2012/06/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61(4):233–47. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 350.Cedernaes J, Osler ME, Voisin S, Broman JE, Vogel H, Dickson SL, et al. Acute Sleep Loss Induces Tissue-Specific Epigenetic and Transcriptional Alterations to Circadian Clock Genes in Men. The Journal of clinical endocrinology and metabolism. 2015;100(9):E1255–61. doi: 10.1210/JC.2015-2284. Epub 2015/07/15. [DOI] [PubMed] [Google Scholar]
- 351.Sonestedt E, Lyssenko V, Ericson U, Gullberg B, Wirfalt E, Groop L, et al. Genetic variation in the glucose-dependent insulinotropic polypeptide receptor modifies the association between carbohydrate and fat intake and risk of type 2 diabetes in the Malmo Diet and Cancer cohort. The Journal of clinical endocrinology and metabolism. 2012;97(5):E810–8. doi: 10.1210/jc.2011-2444. Epub 2012/03/09. [DOI] [PubMed] [Google Scholar]
- 352.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. doi: 10.1038/nbt.2842. Epub 2014/03/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Qin W, Kutny PM, Maser RS, Dion SL, Lamont JD, Zhang Y, et al. Generating Mouse Models Using CRISPR-Cas9-Mediated Genome Editing. Current protocols in mouse biology. 2016;6(1):39–66. doi: 10.1002/9780470942390.mo150178. Epub 2016/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Blakemore AI, Froguel P. Investigation of Mendelian forms of obesity holds out the prospect of personalized medicine. Annals of the New York Academy of Sciences. 2010;1214:180–9. doi: 10.1111/j.1749-6632.2010.05880.x. [DOI] [PubMed] [Google Scholar]
- 355.Doyle YG, Furey A, Flowers J. Sick individuals and sick populations: 20 years later. Journal of epidemiology and community health. 2006;60(5):396–8. doi: 10.1136/jech.2005.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Bloss CS, Jeste DV, Schork NJ. Genomics for disease treatment and prevention. Psychiatr Clin North Am. 2011;34(1):147–66. doi: 10.1016/j.psc.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]