Abstract
We report on a girl, born to first-cousin Lebanese parents, with severe intellectual disability, congenital hip luxation, cardiac malformation, short stature, facial dysmorphic features including microcephaly, sparse hair, bilateral epicanthal folds, ataxia, seizures, and elevated lactate and pyruvate levels in serum. Whole exome sequencing was carried out on the patient's DNA. Potentially causal homozygous variants in the MED25 (p.Ile173Thr) and COQ8A (p.Arg512Trp) genes were found. The potential pathogenicity of these variants, and the possibility that the 2 variants could synergistically act to produce the phenotype reported, is discussed.
Key Words: Cerebellar ataxia, Dysmorphology, Whole exome sequencing
Syndromic genetic disorders are often clinically heterogeneous and display a wide phenotypic variability. With the advent of next-generation sequencing technologies, it is now possible to identify causal mutations in 2 or more genes that may synergistically influence the phenotype of such complex disorders [Mégarbané, 2018]. Such digenic mutations have been shown to be present in genes that influence the same phenotype, as in the case of congenital hypogonadotropic hypogonadism [Méndez et al., 2015] and Alport syndrome [Mencarelli et al., 2015]. On the other hand, biallelic variants in 2 different genes causing the co-occurrence of 2 different clinical phenotypes have also been reported in some families [Pedurupillay et al., 2016; Umair et al., 2017].
Here, we report a Lebanese consanguineous family where a girl presented with severe psychomotor delay, atrial septal defect, hip luxation, facial dysmorphic features, and seizures. Whole exome sequencing (WES) led to the identification of 2 novel homozygous missense variants in the COQ8A and MED25 genes. The contribution of each of these variants to the phenotype is discussed.
Clinical Report
The girl is the only child of consanguineous Lebanese parents, born at term by a C-section after a normal pregnancy following in vitro fertilization. Her birth weight was 3,150 g (35th centile), length 50 cm (35th centile), and OFC was 37 cm (75th centile). At birth, she had a bilateral congenital hip luxation, treated by a cast, and an ostium secundum. The parents noticed a developmental delay at around the age of 6 months. She started to sit at the age of 1 year/2 months and to walk with assistance at the age of 3 years. At 4 years/9 months, she had her first episode of seizures. An EEG showed the presence of a focal discharge in the left temporal area. Seizures were medically well controlled.
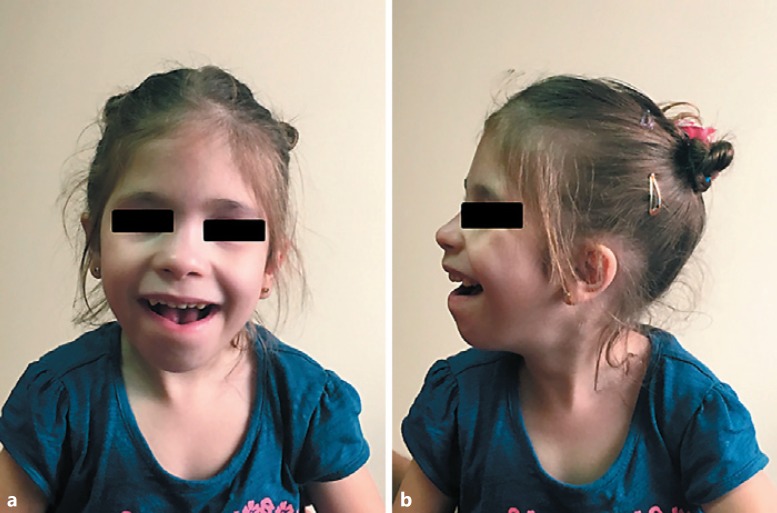
She was referred to our institute at the age of 5 years. She was unable to speak a few words, but she could understand simple orders. Her height was 100 cm (5th centile), weight 17.2 kg (50th centile), and OFC was 46.5 cm (<3rd centile). Dysmporphic facial features included microcephaly, sparse hair, a high forehead, bilateral epicanthal folds, broad nasal tip, flat and short philtrum, thin upper lip, and a large mouth (Fig. 1a, b). Her skin was normal without scars or hyperelasticity. Neurological examination revealed difficulty maintaining a normal upright posture, an unsteady gait, weakness in fine motor skills, normal muscle tone and strength, normal reflexes, flat feet, and occasional stereotypic movements of the hands. Total body X-rays showed no abnormalities. Atrial septal defect was detected on the echocardiogram. Brain MRI at the age of 1 year showed cerebral atrophy mainly in the frontotemporal region. Clinical examination of the parents was normal.
Fig. 1.
Frontal (a) and lateral (b) view of the patient. Note the microcephaly, high forehead, bilateral epicanthal folds, flat and short philtrum, thin upper lip, and a large mouth.
Materials and Methods
Genomic DNA was isolated from the patient's blood sample using standard techniques.
Whole Exome Sequencing
Approximately 37 Mb (214,405 exons) of the consensus coding sequences were enriched from fragmented genomic DNA by more than 340,000 probes designed against the human genome (Nextera Rapid Capture Exome, Illumina Inc., San Diego, CA, USA) and the generated library sequenced on an Illumina NextSeq or HiSeq 4000 platform (Illumina) to an average coverage depth 70–100×. Data was processed using bcl2fastq software 2.17.1.14 (Illumina) and fed to an in-house pipeline based on the 1000 Genomes Project data and Genome Analysis Toolkit (GATK) best practice recommendations. Variants were called using Burrows-Wheeler Alignment software with the Maximal Exact Matches algorithm and GATK HaplotypeCaller, freebayes, and samtools. Variants were annotated using Annovar Alamut version 2.4.5 (Interactive Biosoftware, Rouen, France) and in-house ad hoc bioinformatics tools and compared to the in-house mutation database (CentoMD®), the Human Gene Mutation Database (HGMD®), ClinVar, and an ethnic database containing over 750 exomes belonging to Arab individuals from the Arabian Peninsula. Variants were evaluated based on the mode of inheritance and compatibility with the clinical phenotype. All clinical features were translated to the Human Phenotype Ontology (HPO) and filtered against the HPO-annotated variant list. Finally, clinical information was reevaluated and the data filtered again based on medical differential diagnostic assumptions.
Results
The results of complete blood counts, serum electrolytes, blood glucose levels, triglycerides, cholesterol, amino acid studies of plasma and urine, urinary screening for organic acids, urinalysis, thyroid, liver and renal function tests, CPK, Ca, and P were all unremarkable. Serum levels were increased for both lactate (59 mg/dL) and pyruvate (1.23 mg/dL), while serum coenzyme Q10 (CoQ10) was reported in the normal range. Muscular biopsy was not performed. High-resolution R-banding showed a normal 46,XX karyotype.
WES detected a homozygous missense variant in the COQ8A (ADCK3) gene. This NM_020247.4:c.1534C>T (p.Arg512Trp) variant is extremely rare in the gnomAD database: frequency 0.0001026 (29/282,644 alleles), with no described homozygotes. It is predicted to be probably damaging by PolyPhen-2, deleterious by PROVEAN, and disease causing by MutationTaster. Following the recommendations of the American College of Medical Genetics and Genomics, this variant is also classified as a variant of uncertain significance, class 3 [Richards et al., 2015].
Another unreported homozygous variant was found in the MED25 gene (NM_030973.3:c.518T>C; p.Ile173Thr). This variant has also never before been reported in association with a disease and is present at extremely low allele frequencies in the gnomAD Browser: 9/251,742 (frequency 3.575e−6). No homozygotes were present in this database for this variant. It is predicted to be benign by PolyPhen-2, to be damaging according to PROVEAN, and disease causing according to MutationTaster. It is classified as a variant of uncertain significance, class 3, according to the recommendations of American College of Medical Genetics and Genomics [Richards et al., 2015].
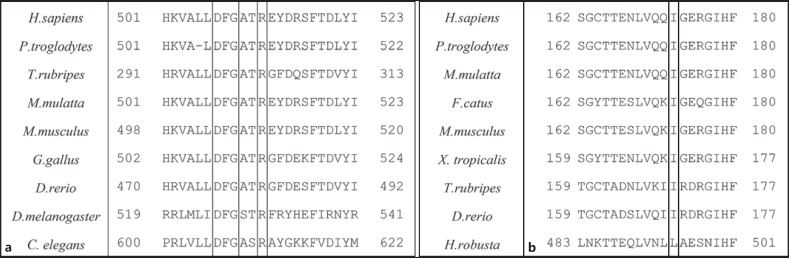
Both variants were confirmed by Sanger sequencing and found in heterozygous state in both parents. In addition, both variants were found to be affecting highly conserved residues in the protein (Fig. 2).
Fig. 2.
Multiple alignment of COQ8A (a) and MED25 (b) protein sequences using HomoloGene showing the conserved nature of the Arg512 and Ile173 residues. The DFG motif in the COQ8A sequences is also specified.
Discussion
Here, we report on a female patient of consanguineous parents with severe neurodevelopmental delay, seizures, ataxia, absence of speech, intellectual disability, stereotypic hand movements, and microcephaly.
The MECP2 and FOXG1 genes were sequenced, but no pathogenic mutations were found. A WES was performed, and 2 homozygous variants were found in 2 different genes in the patient.
The first variant was found in the COQ8A gene, the homolog of yeast Coq8, involved in the synthesis of CoQ10, or ubiquinone, an electron carrier critical for electron transfer by the mitochondrial inner membrane respiratory chain, therefore, essential for proper functioning of the mitochondrial respiratory chain [Lagier-Tourenne et al., 2008]. Pathogenic variants in the COQ8A gene are known to cause autosomal recessive primary CoQ10 deficiency type 4, also known as autosomal recessive spinocerebellar ataxia type 9 (OMIM 612016). This disorder is characterized by childhood onset of cerebellar ataxia and exercise intolerance, but some affected individuals develop seizures and have mild mental impairment, indicating variable severity. Our patient displayed ataxia, seizures, and increased lactate levels in serum that are characteristic of this condition (Table 1). Nevertheless, she did not have pes cavus or cerebellar hypoplasia on the MRI, and CoQ10 levels were normal, which is different from primary CoQ10 deficiency type 4 syndrome; although, it is possible that CoQ10 levels may be reduced in the muscle biopsy which was not performed. Moreover, another particularity in our patient is the early onset of signs of abnormal gait as well as the severe intellectual disability. These discrepancies between our patient and previously reported cases may be explained by the fact that the homozygous COQ8A variant in our patient is situated within the protein kinase domain, which is different from all other cases, but also secondary to the effect of a modifying gene. Interestingly, the p.Arg512Trp variant reported here is located very close to both a previously described p.Tyr514Cys mutation in a compound heterozygous patient [Lagier-Tourenne et al., 2008] as well as a conserved DFG motif that is known to play an important role in regulating kinase activity.
Table 1.
Clinical comparison between the present case and other reported patients with MED25 and COQ8A mutations
| Present case | Basel-Vanagaite et al., 2015 | Figueiredo et al., 2015 | COQ8A-related disorder | |
|---|---|---|---|---|
| Congenital hip luxation | + | – | NA | – |
| Developmental delay | + | + | + | + |
| Absent/delayed speech | + | + | + | – |
| Short stature | + | – | NA | – |
| Seizures | + | + | NA | + |
| Ataxia | + | – | – | + |
| Hypotonia | + | + | NA | – |
| Microcephaly | + | + | NA | – |
| Tall forehead | – | – | + | – |
| Sparse hair | + | + | – | – |
| Sparse eyebrows | – | + | – | – |
| Nevus flammeus | – | + | – | – |
| Ptosis | – | + | – | – |
| Hypertelorism | + | + | – | – |
| Epicanthus | + | + | – | – |
| Cataract | – | + | NA | – |
| Microcornea | – | + | NA | – |
| Strabismus | + | + | NA | – |
| Downslanting palpebral |
||||
| fissures | – | + | – | – |
| Broad nasal tip | + | – | + | – |
| Short philtrum | + | + | + | – |
| Large mouth | + | – | – | – |
| Cleft palate | – | + | – | – |
| Prominent chin | – | + | – | |
| Wide cupid bow | – | + | – | |
| Thin upper lip | + | + | + | – |
| Pes cavus | – | – | NA | + |
| Increased serum lactate | + | – | NA | + |
| Decreased coenzyme Q10 | – | – | NA | + |
| Brain malformations | – | + | NA | + |
| Heart malformations | + | + | NA | – |
| Abnormal genitalia | – | + | NA | – |
NA, information not available; +, present; –, absent.
The second variant identified by WES sequencing in our patient is a missense p.Ile173Thr variant in the MED25 gene. This gene encodes a component of the transcriptional coactivator complex termed the “Mediator complex” involved in the regulated transcription of nearly all RNA polymerase II-dependent genes. The encoded protein plays a role in chromatin modification and in preinitiation complex assembly. MED25 was first implicated in the pathogenesis of autosomal recessive axonal Charcot-Marie-Tooth disease (ARCMT2B) [Leal et al., 2001, 2009]. However, recent studies suggest that the mutation implicated in CMT2B is likely to be a rare benign variant rather than pathogenic [Figueiredo et al., 2015].
Beside the ARCMT2B mutation, mutations in MED25 have been described in 2 other diseases: Basel-Vanagaite-Smirin-Yosef syndrome (OMIM 616449) and syndromic intellectual disability [Basel-Vanagaite et al., 2015; Figueiredo et al., 2015]. Our patient shares many clinical features with the patients mentioned in these studies (Table 1). Interestingly, the novel p.Ile173Thr variant identified in this study, in MED25, is located in the same functional domain, the von Willebrand-associated domain, as the p.Arg140Trp and p.Tyr39Cys mutations reported in the patients with Basel-Vanagaite-Smirin-Yosef syndrome and syndromic intellectual disability. The von Willebrand-associated domain is known to direct the incorporation of the protein into the Mediator complex [Sela et al., 2013]. In addition, homology studies clearly show the conserved nature of the residue across species. Even in distantly related species, such as annelids, the Ileu residue was noticed to be replaced only by another non-polar residue, like leucine (Fig. 2). Therefore, it is highly likely that a change to the polar threonine residue at this position would not be tolerated. Together with the extremely rare allele frequency in the gnomAD database and the predicted damaging effect of the variant, this variant is likely to be influencing the phenotype of our patient. We thus propose that our patient's phenotype is the combined result of 2 variants, the COQ8A variant being the “highly penetrant mendelizing variant”, and MED25 being the phenotype-modifying variant [Lupski et al., 2011].
Since the reported clinical phenotypes for mutations in both COQ8A and MED25 show a large degree of overlap, it is difficult to assign individual clinical features in our patient to effects of either of the mutations. For instance, intellectual disability in our patient could be due to a severe manifestation of autosomal recessive spinocerebellar ataxia type 9, or due to syndromic intellectual disability caused by the MED25 mutation. Functional studies in model organisms would help reveal the individual contribution of these variants to the overall phenotype.
Statement of Ethics
Informed consent for genetic analysis was obtained from the family in compliance with national ethics regulation.
Disclosure Statement
The authors have no conflicts of interest to disclose.
Acknowledgment
We are thankful to the family for their cooperation.
References
- 1.Basel-Vanagaite L, Smirin-Yosef P, Essakow JL, Tzur S, Lagovsky I. Homozygous MED25 mutation implicated in eye-intellectual disability syndrome. Hum Genet. 2015;134:577–587. doi: 10.1007/s00439-015-1541-x. [DOI] [PubMed] [Google Scholar]
- 2.Figueiredo T, Melo US, Pessoa AL, Nobrega PR, Kitajima JP. Homozygous missense mutation in MED25 segregates with syndromic intellectual disability in a large consanguineous family. J Med Genet. 2015;52:123–127. doi: 10.1136/jmedgenet-2014-102793. [DOI] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C, Tazir M, López LC, Quinzii CM, Assoum M. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal A, Morera B, Del Valle G, Heuss D, Kayser C. A second locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 19q13.3. Am J Hum Genet. 2001;68:269–274. doi: 10.1086/316934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leal A, Huehne K, Bauer F, Sticht H, Berger P. Identification of the variant Ala335Val of MED25 as responsible for CMT2B2: molecular data, functional studies of the SH3 recognition motif and correlation between wild-type MED25 and PMP22 RNA levels in CMT1A animal models. Neurogenetics. 2009;10:375–376. doi: 10.1007/s10048-009-0213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mégarbané A. Clinical genetics revisited: effect of new techniques (next-generation sequencing, comparative genomic hybridization) on previous diagnoses. Middle East J Med Genet. 2018;7:1–6. [Google Scholar]
- 8.Mencarelli MA, Heidet L, Storey H, van Geel M, Knebelmann B. Evidence of digenic inheritance in Alport syndrome. J Med Genet. 2015;52:163–174. doi: 10.1136/jmedgenet-2014-102822. [DOI] [PubMed] [Google Scholar]
- 9.Méndez JP, Zenteno JC, Coronel A, Soriano-Ursúa MA, Valencia-Villalvazo EY. Triallelic digenic mutation in the prokineticin 2 and GNRH receptor genes in two brothers with normosmic congenital hypogonadotropic hypogonadism. Endocr Res. 2015;40:166–171. doi: 10.3109/07435800.2014.982327. [DOI] [PubMed] [Google Scholar]
- 10.Pedurupillay CR, Landsend EC, Vigeland MD, Ansar M, Frengen E. Segregation of incomplete achromatopsia and alopecia due to PDE6H and LPAR6 variants in a consanguineous family from Pakistan. Genes (Basel) 2016;7:E41. doi: 10.3390/genes7080041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, Bick D, Das S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sela D, Conkright JJ, Chen L, Gilmore J, Washburn MP. Role for human mediator subunit MED25 in recruitment of mediator to promoters by endoplasmic reticulum stress-responsive transcription factor ATF6α. J Biol Chem. 2013;288:26179–26187. doi: 10.1074/jbc.M113.496968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umair M, Seidel H, Ahmed I, Ullah A, Haack TB. Ellis-van Creveld syndrome and profound deafness resulted by sequence variants in the EVC/EVC2 and TMC1 genes. J Genet. 2017;96:1005–1014. doi: 10.1007/s12041-017-0868-6. [DOI] [PubMed] [Google Scholar]