ABSTRACT
Intraflagellar transport (IFT), which is essential for the formation and function of cilia in most organisms, is the trafficking of IFT trains (i.e. assemblies of IFT particles) that carry cargo within the cilium. Defects in IFT cause several human diseases. IFT trains contain the complexes IFT-A and IFT-B. To dissect the functions of these complexes, we studied a Chlamydomonas mutant that is null for the IFT-A protein IFT140. The mutation had no effect on IFT-B but destabilized IFT-A, preventing flagella assembly. Therefore, IFT-A assembly requires IFT140. Truncated IFT140, which lacks the N-terminal WD repeats of the protein, partially rescued IFT and supported formation of half-length flagella that contained normal levels of IFT-B but greatly reduced amounts of IFT-A. The axonemes of these flagella had normal ultrastructure and, as investigated by SDS-PAGE, normal composition. However, composition of the flagellar ‘membrane+matrix’ was abnormal. Analysis of the latter fraction by mass spectrometry revealed decreases in small GTPases, lipid-anchored proteins and cell signaling proteins. Thus, IFT-A is specialized for the import of membrane-associated proteins. Abnormal levels of the latter are likely to account for the multiple phenotypes of patients with defects in IFT140.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Flagella, Intraflagellar transport, BBSome, GTPases, Jeune asphyxiating thoracic dystrophy (JATD), Mainzer-Saldino syndrome (MSS)
Summary: Intraflagellar transport complex A is required for the efficient movement of groups of membrane and signal transduction proteins into flagella.
INTRODUCTION
Cilia are established and maintained by intraflagellar transport (IFT), which is the movement of “trains” of protein complexes (IFT particles) from the ciliary base to the tip (anterograde IFT) and from the tip back to the ciliary base (retrograde IFT) (Kozminski et al., 1993). IFT in the model organism Chlamydomonas reinhardtii occurs through the action of two separate motors: kinesin-2, and dynein-1b, which are responsible for anterograde and retrograde IFT, respectively. Main components of IFT trains include IFT complex A (IFT-A), IFT complex B (IFT-B) and the Bardet–Biedl syndrome protein complex (BBSome). IFT-B is generally considered to function in anterograde IFT including the movement of proteins into cilia, whereas IFT-A is generally considered to function in retrograde IFT including the movement of proteins out of cilia (Ishikawa and Marshall, 2011; Taschner et al., 2012); the BBSome is believed to function as a cargo adaptor for export of signaling proteins from the cilium (Eguether et al., 2014; Lechtreck, 2015; Lechtreck et al., 2009; Ye et al., 2018).
Interest in IFT function and its role in the development of human diseases has increased significantly in the last 15 years. Much of this research has focused on IFT-B, its interacting partners and its cargo. Comparatively little work has been done on IFT-A, which is organized into a ‘core’ consisting of IFT144, IFT140 and IFT122, and a ‘periphery’ consisting of IFT139, IFT121 and IFT43 (Behal et al., 2012; Mukhopadhyay at al., 2010). Except for IFT43 and IFT139, all IFT-A components are predicted to be composed of N-terminal WD-repeat domains and C-terminal tetratricopeptide repeat (TPR) domains, both of which can have diverse functions (Grove et al., 2008; Smith et al., 1999; Zeytuni and Zarivach, 2012). IFT139 is predicted to contain TPR domains throughout its length, whereas IFT43 is predicted to contain a single, low-probability coiled-coil domain (Behal et al., 2012; Cole, 2003; Taschner et al., 2012).
Initial studies on IFT-A were performed in Chlamydomonas using temperature-sensitive mutants of IFT139 and IFT144. At the permissive temperature, these mutants displayed lower levels of IFT-A in the flagella and bulging flagellar tips that contained IFT-B. At the restrictive temperature, these mutants resorbed their flagella (Piperno et al., 1998; Iomini et al., 2001). An observed decrease in retrograde IFT velocity coupled with the accumulation of IFT-B in the flagella implicated IFT-A as a key component of retrograde IFT (Piperno et al., 1998; Iomini et al., 2001). More recently, several studies have suggested a role for IFT-A in the ciliary localization of the membrane proteins smoothened (Smo), melanin-concentrating hormone receptor (Mchr1), Gpcr161, opsin, the GTPase Arl13b, inositol polyphosphate-5-phosphatase E (INPP5E), the somatostatin receptor 3 (SSTR3), the 5-HT6 receptor (HTR6), and a transient receptor potential cation channel (TRP) subfamily V (TRPV) ion channel (Brear et al., 2014; Crouse et al., 2014; Fu et al., 2016; Hirano et al., 2017; Lee et al., 2008; Liem et al., 2012; Mukhopadhyay et al., 2010, 2013; Sun et al., 2012). However, despite a potentially crucial role for IFT-A in the anterograde movement of ciliary proteins, a thorough global analysis of IFT-A function has yet to be performed.
We are taking advantage of Chlamydomonas to dissect the role of IFT-A in anterograde IFT. Previous studies using Chlamydomonas applied a truncation strategy to successfully analyze specific functions of IFT-B components in vivo (Brown et al., 2015; Hou and Witman, 2017; Kubo et al., 2016). Here, we use a similar strategy coupled with state-of-the-art light microscopy of living and fixed cells and mass spectrometry (MS) of isolated flagellar fractions to obtain a broad overview of the functions of IFT140 and its domains in vivo. IFT140 is of particular interest because, in humans, mutations in it cause multi-system ciliopathies including Jeune asphyxiating thoracic dystrophy (JATD) and Mainzer-Saldino syndrome (MSS, also known as conorenal syndrome). Hallmark features of JATD include shortened ribs and a narrowing of the chest often leading to asphyxia-related perinatal death. Other common features of JATD include progressive cystic nephropathy, retinal dystrophy, pancreatic cysts and liver disease (Schmidts et al., 2013). MSS is characterized by phalangeal abnormalities, chronic renal disease and retinal dystrophy (Perrault et al., 2012). IFT140 mutations are also associated with non-syndromic Leber congenital amaurosis (LCA) and retinitis pigmentosa (RP), which are both genetically heterogeneous retinal degenerative disorders (Hull et al., 2016; Jonassen et al., 2012; Khan et al., 2014; Xu et al., 2015).
The function of IFT140 has previously been investigated in Trypanosoma brucei. Depletion of IFT140 by RNA interference (RNAi) resulted in shortened flagella, accumulation of vesicles containing IFT-like material near the flagellar base and aberrant entry of proteins into the flagella of IFT140RNAi cells suggesting that depletion of IFT140 may alter the effectiveness of the transition zone (TZ) gate (Absalon et al., 2008). Recent studies in Caenorhabditis elegans, including mutants for IFT140 and other IFT-A proteins, concluded that IFT-A is necessary for the correct localization of pericentriolar matrix proteins of the Meckel-Gruber syndrome module (Scheidel and Blacque, 2018). A Chlamydomonas mutant that is likely to express a version of IFT140 lacking amino acid residues C-terminal to the TPR domains also has been reported (Zhu et al., 2017). This mutant shows some reduction of IFT-A proteins in the cell body and the flagella, and shows increased levels of IFT-B proteins in the flagella. The level of IFT motors were also increased in the mutant flagella. The mutant regenerated its flagella at a slow rate, leading to the conclusion that it is defective in mobilizing flagellar precursors to the base of the flagellum. However, the effect of the mutation on potential cargos of IFT-A was not explored. Another Chlamydomonas IFT140 mutant (ift140-3), reported by Lin et al., lacks cilia and retains normal transition zone protein localization (Lin et al., 2018).
Here, we report an IFT140 mutant allele (ift140-1; Picariello et al., 2015) that appears to be null for the protein. The complete loss of IFT140 destabilized IFT-A and prevented the formation of flagella but had no effect on the levels or localization of IFT-B in the cell body. Rescue of the mutant with a construct expressing the C-terminal TPR domains but lacking the N-terminal WD-repeat domains stabilized IFT-A and supported the formation of half-length flagella, but resulted in aberrant IFT and loss from the flagella of a specific subset of signal transduction components and membrane-associated proteins. These results reveal distinct functions for the N- and C-terminal regions of IFT140. Taken together, these data indicate an indispensable role for IFT140 in the stability of IFT-A, and a potential role for IFT-A in the anterograde trafficking of numerous ciliary signaling and membrane-associated proteins.
RESULTS
Generation of a Chlamydomonas IFT140 mutant
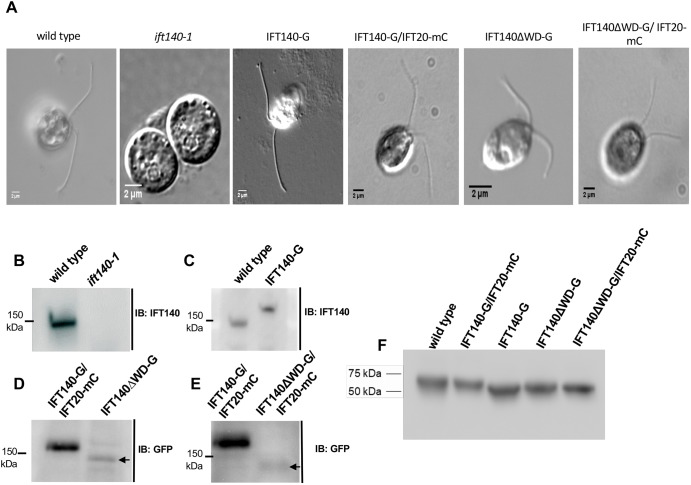
A collection of Chlamydomonas mutants with defects in motility was generated by Pete Lefebvre's group (University of Minnesota, St. Paul) by insertional mutagenesis using linearized pMN24 (containing the NIT1 gene); this collection was subsequently screened by Southern blotting to identify a strain with a restriction fragment length polymorphism (RFLP) in the gene encoding IFT140 (Hou, 2007). Analysis by PCR confirmed the insertion of at least some of the pMN24 sequence into exon 20 of IFT140, and Southern blot analysis revealed that a 1457-bp fragment, spanning IFT140 exons 20–24 in wild type, was missing from the mutant (Fig. 1A,B). This mutant allele has been designated ift140-1 (Picariello et al., 2015). ift140-1 cells are incapable of forming flagella (Fig. 2A ift140-1), and IFT140 was not detected on western blots of ift140-1 whole-cell extracts (WCEs) using an antibody directed against the C-terminus of the protein (Fig. 2B). Transformation of the ift140-1 mutant with full-length IFT140 tagged with super-folder GFP (sfGFP) fully rescued flagellar assembly (Fig. 2A IFT140-G, and Fig. 2C); so, if an N-terminal fragment of IFT140 is expressed in ift140-1, it does not have a readily detectable dominant-negative effect. Rescue of ift140-1 confirms that the phenotype is due to the mutation of IFT140.
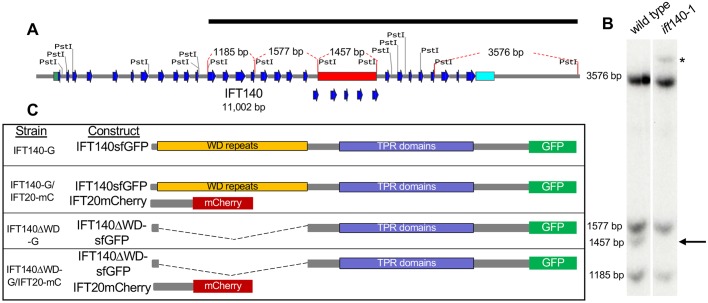
Fig. 1.
Identification of a Chlamydomonas reinhardtii IFT140 mutant. (A) Map of the IFT140 genomic sequence on chromosome 8. The 5′-UTR is highlighted in green, the 3′-UTR is highlighted in cyan, the blue arrows indicate exons 1–32, the red bar indicates the region of the gene (exons 20–24, 1457 bp) affected by the insertion of NIT1, and the black bar indicates the IFT140 genomic sequence used as the Southern blot probe. The fragment sizes indicated by red dashed lines on the gene map in A correspond to bands detected in B. (B) Southern blot from wild type and ift140-1 revealed loss of the 1457-bp band and appearance of a new band (asterisk) consistent with an insertion of an exogenous DNA sequence. (C) Schematic of the rescued cell lines and constructs used in these experiments. IFT140-G expresses full-length IFT140sfGFP in an ift140-1 background. IFT140-G/IFT20-mC expresses both full-length IFT140sfGFP and a full-length version of the IFT-B subunit IFT20 tagged with mCherry in an ift140-1 ift20-1 background. IFT140ΔWD-G cells express truncated IFT140sfGFP in an ift140-1 background. IFT140ΔWD-G/IFT20-mC cells express both truncated IFT140sfGFP and full-length IFT20mCherry in an ift140-1 ift20-1 background.
Fig. 2.
Characterization of the ift140-1 mutant and related strains. (A) Differential interference contrast images of the cell lines used in this study. ift140-1 cells are aflagellate. (B–E) Western blots of WCEs from the indicated cell lines demonstrating the level of expression of each IFT140 construct compared to wild-type or IFT140-G/IFT20-mC control cells. (F) βF1-ATPase loading control for blots shown in panels B–E.
IFT140 loss affects the stability of IFT-A but not the stability or the cytoplasmic localization of IFT-B
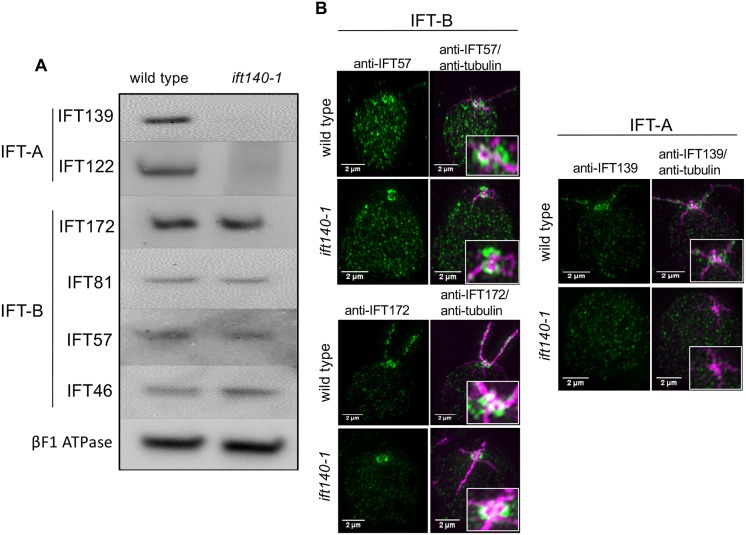
Western blots of WCE from wild-type and ift140-1 cells were probed for components of IFT-A and IFT-B. Loss of IFT140 severely affected the cytoplasmic protein levels of at least two other IFT-A components, IFT122 and IFT139 (Fig. 3A). To determine whether this decrease of IFT-A proteins reflected transcriptional control vs stability, quantitative PCR was performed with ift140 wild-type and ift140-1 cDNA templates utilizing primers directed towards the 3′-ends of IFT139 and IFT140. In ift140-1, the level of IFT139 transcripts was increased 6.8-fold relative to wild type, whereas IFT140 transcripts were undetectable. Therefore, the reduced levels of IFT-A proteins in the cytoplasm are likely to be the result of instability and resultant degradation of IFT-A in the absence of IFT140. In contrast, the IFT-B components IFT172, IFT81, IFT57 and IFT46 were not affected by the loss of IFT140 (Fig. 3A), similar to what has been reported for loss of the IFT-A subunit IFT43 (Zhu et al., 2017).
Fig. 3.
The absence of IFT140 affects the abundance and localization of IFT-A but not of IFT-B. (A) Western blot showing that loss of IFT140 in ift140-1 cells severely affected the levels of the IFT-A proteins IFT139 and IFT122. In contrast, levels of the IFT-B proteins examined remained unchanged. βF1 ATPase is the loading control. (B) SIM images of IFT components in wild-type and ift140-1 cells. In both cell types, IFT-B – as represented by IFT57 and IFT172 – localized in ring-shaped structures near the basal body. This suggests that IFT-B can localize properly in the peri-basal body area in the absence of IFT-A. IFT139 was not detected in ift140-1 cells by SIM, consistent with western blot results.
We assessed the localization of IFT-A and IFT-B by super-resolution structured-illumination microscopy (SIM). In wild-type cells, IFT-A and IFT-B form a pair of ring-like structures at the apical end of the cell, slightly distal to the basal body complex (Fig. 3B) (Brown et al., 2015). In the absence of IFT140, the IFT-B components IFT172 and IFT57 localize normally (Fig. 3B), suggesting that IFT-B is correctly recruited to the peri-basal body region, even in the absence of IFT-A. Consistent with our western blots, IFT139 is undetectable by SIM in ift140-1 cells (Fig. 3B), providing further evidence that IFT-A is destabilized in the absence of IFT140.
Generation of strains expressing various IFT140 constructs
To investigate the specific functions of IFT140, as well as the roles of its N-terminal region containing WD repeats and its C-terminal region containing TPR domains, we generated strains expressing modified versions of the protein. The schematic in Fig. 1C illustrates the constructs expressed in each of the strains. The strains were generated in the appropriate null mutant background, either ift140-1 for strains expressing only full-length or truncated IFT140-GFP (IFT140-G and IFT140ΔWD-G) or ift140-1 ift20-1 for strains expressing full-length or truncated IFT140-sfGFP plus the IFT-B subunit IFT20 tagged with mCherry (IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC). Strain IFT140-G served as the control for most experiments utilizing strain IFT140ΔWD-G; strain IFT140-G/IFT20-mC served as the control for experiments utilizing strain IFT140ΔWD-G/IFT20-mC. Full-length IFT140-sfGFP was expressed at wild-type levels (Fig. 2C), whereas the truncated protein was expressed at lower levels in several independently isolated strains (Fig. 2D,E). Quantitative PCR using cDNA and primers directed towards either IFT139 or IFT140 revealed respective increases of 3.5- or 7.5-fold, respectively, in the transcripts of these genes in IFT140ΔWD-G/IFT20-mC cells relative to IFT140-G/IFT20-mC cells. Thus, the reduction of IFT140 and IFT139 (see next subsection) proteins in these cells is not due to reduced transcription but, more likely, due to instability of the truncated IFT140. Despite the lower levels of truncated IFT140, IFT140ΔWD-G and IFT140ΔWD-G/IFT20-mC cells did form flagella (Fig. 2A).
Expression of IFT140 that lacks its N-terminal WD-repeat domains partially rescues the instability of IFT-A and supports the formation of half-length flagella
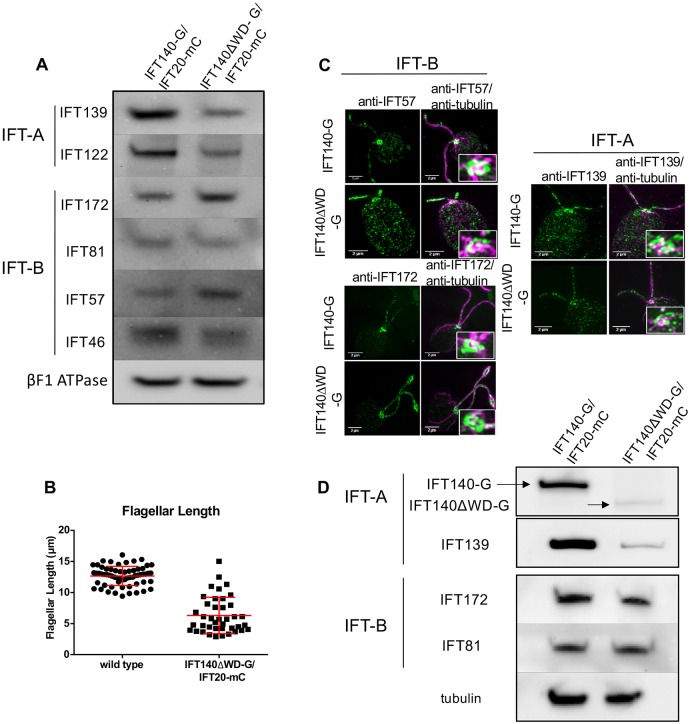
Expression of the IFT140ΔWD protein resulted in the partial recovery of IFT122 and IFT139 (Fig. 4A), and allowed ∼50% of cells to form flagella that averaged approximately one-half the length of control flagella (Fig. 4B). Therefore, the C-terminal TPR domains of IFT140 are sufficient to, at least partially, stabilize IFT-A. IFT-B levels in IFT140ΔWD-G/IFT20-mC cells are almost normal (Fig. 4A).
Fig. 4.
Expression of IFT140 lacking its N-terminal WD-repeats results in a partial recovery of IFT-A and formation of half-length flagella with reduced levels of IFT-A. (A) Western blots of WCE from IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC cells show a partial restoration of IFT-A proteins. Some IFT-B protein levels were slightly increased. βF1 ATPase is the loading control. (B) Flagellar-length measurements for wild-type and IFT140ΔWD-G/IFT20-mC cells. Wild-type flagella are ∼12.5 µm long (n=63 flagella) whereas IFT140ΔWD-G/IFT20-mC flagella averaged about 6.5-µm long (n=43 flagella). Approximately 50% of IFT140ΔWD-G/IFT20-mC cells were flagellated under normal culture conditions; cells without flagella were not included in the analysis. (C) SIM images of IFT140-G and IFT140ΔWD-G cells labelled with anti-tubulin and anti-IFT57, anti-IFT172 or anti-IFT139 antibodies. Expression of IFT140ΔWD-G led to partial recovery of IFT-A in the basal body region and in the flagella. Scale bars: 2 µm. (D) Western blots of isolated flagella of IFT140ΔWD-G/IFT20-mC and control cells. IFT140-G and IFT140ΔWD-G were detected with anti-GFP; IFT139, IFT172 and IFT81 were detected with antibodies against those proteins; and tubulin, which was the loading control, was detected with anti-acetylated tubulin.
The peri-basal body localization of IFT139 was partially restored in cells expressing IFT140ΔWD-G (Fig. 4C), consistent with the partial recovery of IFT-A protein levels seen in western blots (Fig. 4A). As in the complete absence of IFT140, the IFT-B subunits IFT57 and IFT172 localize normally to the peri-basal body region in these cells (Fig. 4C).
Western blots of isolated flagella of IFT140ΔWD-G/IFT20-mC and control cells indicated that the former had normal levels of IFT-B (as represented by IFT172 and IFT81) but substantially reduced levels of IFT-A (as represented by IFT139 and IFT140) (Fig. 4D).
IFT140 is necessary for the formation of flagella but not of the basal body or TZ
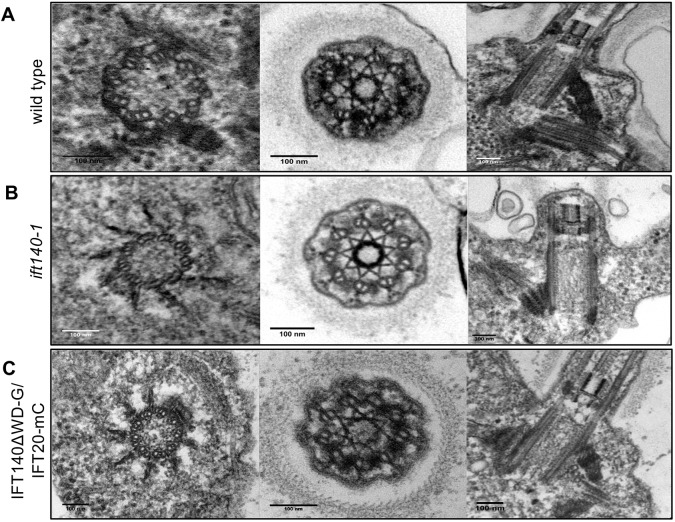
Neither complete loss of IFT140 nor the truncation of its N-terminus altered the ultrastructure of the basal body or the TZ (Fig. 5), in agreement with the results of Lin et al. (2018) that IFT140 is not needed for localization of the TZ protein NPHP4. Nevertheless, ift140-1 cells are incapable of forming flagella with any discernable axonemal ultrastructure (Figs 5B and 6B, arrow and enlargement). Instead, the cells form very short flagellar stubs that contain a small amount of electron-dense material. This suggests that some proteins can traverse the TZ and enter the flagellar stubs but are unable to form an axoneme.
Fig. 5.
TEM revealed normal basal body and transition zone architecture in ift140-1 and IFT140ΔWD-G/IFT20-mC cells. (A) Longitudinal and cross sections of basal bodies and the transition zone (TZ) in wild-type cells. (B,C) Basal bodies and TZs in ift140-1 cells (B) and IFT140ΔWD-G/IFT20-mC cells (C) show normal ultrastructure. Scale bars: 100 nm.
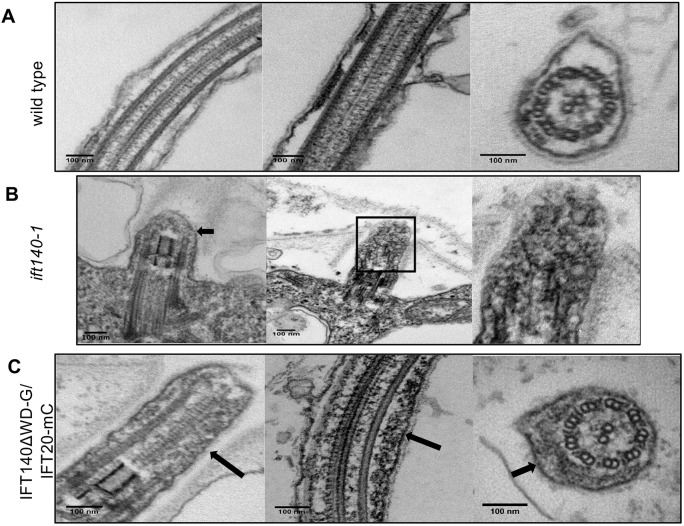
Fig. 6.
ift140-1 cells fail to form a flagellar axoneme, whereas IFT140ΔWD-G/IFT20-mC cells form an axoneme with apparently normal ultrastructure. (A) Longitudinal and cross sections of wild-type flagella. (B) Longitudinal section of a flagellar stub in ift140-1 cells. ift140-1 flagella barely extend past the transition zone (TZ) and lack an axoneme but contain amorphous accumulations of electron-dense material (arrows and magnification of boxed area). (C) Longitudinal and cross sections of flagella from IFT140ΔWD-G/IFT20-mC cells. IFT140ΔWD-G/IFT20-mC flagella form axonemes with apparently normal ultrastructure but still contain amorphous accumulations of electron-dense material (arrows).
In contrast to ift140-1, the approximately half-length flagella formed by IFT140ΔWD-G/IFT20-mC cells have normal axonemal ultrastructure (Fig. 6C), indicating that major structural components of the axoneme are delivered to the flagella, even in the absence of IFT140 WD-repeat domains and normal levels of IFT-A. Additionally, it suggests that the C-terminal region of IFT140 is sufficient to support formation of IFT trains and cargo transport. Accumulations of electron-dense material also were observed in IFT140ΔWD-G/IFT20-mC flagella (Fig. 6C, arrows). These accumulations suggest that retrograde IFT is defective in cells lacking the IFT140 WD repeats.
IFT frequency and velocity are decreased in IFT140ΔWD-G/IFT20-mC cells
To determine how loss of the IFT140 WD repeats affected IFT, we recorded IFT in IFT140-G/IFT20-mC (control) and IFT140ΔWD-G/IFT20-mC cells using live-cell total internal reflection fluorescence (TIRF) video microscopy and generated kymograms from the resulting videos (Fig. 7A,B). In IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC cells, almost all tracks of IFT140sfGFP and IFT20mCherry were coincident, demonstrating for the first time in living cells that IFT-A and IFT-B are present in the same IFT trains. Quantitative analysis of the kymograms revealed that the frequency of IFT trains moving in both anterograde and retrograde directions in IFT140ΔWD-G/IFT20-mC flagella was lower than in control flagella, and lower in long IFT140ΔWD-G/IFT20-mC flagella compared to short IFT140ΔWD-G/IFT20-mC flagella (Fig. 7C,D). Decreased IFT frequency also coincided with an increase in the number of static IFT particles, which was greater in long IFT140ΔWD-G/IFT20-mC flagella than in short flagella (Fig. 7B, horizontal lines). The decrease in IFT frequency in IFT140ΔWD-G/IFT20-mC flagella might stem from the progressive accumulation and stalling of IFT particles, which itself is indicative of a defect in retrograde IFT. Perhaps relevant to this, retrograde tracks were wider in long IFT140ΔWD-G/IFT20-mC flagella than in short IFT140ΔWD-G/IFT20-mC flagella, indicating that retrograde IFT trains are not only fewer but possibly larger in IFT140ΔWD-G/IFT20-mC flagella. In IFT140ΔWD-G/IFT20-mC cells, anterograde IFT velocities were 5% lower than normal, and were similar in both short and long flagella (Fig. 7E); despite showing statistical significance, this decrease might prove physiologically insignificant. However, average retrograde IFT velocity in IFT140ΔWD-G/IFT20-mC cells was decreased by 40% (Fig. 7F). In contrast to frequency, there was no difference in the distribution of velocities in long and short IFT140ΔWD-G/IFT20-mC flagella. Therefore, the lower IFT velocity in IFT140ΔWD-G/IFT20-mC cells does not appear to be a downstream consequence of cargo accumulation in the flagella but might reflect a fundamental inability of IFT140ΔWD-G/IFT20-mC cells to move retrograde IFT trains at normal speed.
Fig. 7.
IFT velocity and frequency are decreased in IFT140ΔWD-G/IFT20-mC flagella. (A) Kymographs showing anterograde and retrograde IFT for IFT140 and IFT20 in IFT140-G/IFT20-mC control cells. (B) Kymographs showing anterograde and retrograde IFT across a range of flagellar lengths in IFT140ΔWD-G/IFT20-mC cells. (C,D) Dot plots showing mean anterograde and retrograde IFT frequency for IFT140-G/IFT20-mC (n=43), IFT140ΔWD-G/IFT20-mC short flagella (n=17), and IFT140ΔWD-G/IFT20-mC long flagella (n=22). Overall, IFT frequency is decreased in IFT140ΔWD-G/IFT20-mC flagella and is lower in long than short IFT140ΔWD-G/IFT20-mC flagella. (E,F) Dot plots showing average anterograde IFT velocity for IFT140-G/IFT20-mC (n=157), IFT140ΔWD-G/IFT20-mC short flagella (n=157), and IFT140ΔWD-G/IFT20-mC long flagella (n=166) and average retrograde IFT velocity for IFT140-G/IFT20-mC (n=72), IFT140ΔWD-G/IFT20-mC short flagella (n=72), and IFT140ΔWD-G/IFT20-mC long flagella (n=72). Anterograde IFT velocity was only slightly lower in IFT140ΔWD-G/IFT20-mC flagella compared to IFT140-G/IFT20-mC flagella; however, retrograde IFT velocity was dramatically affected by the absence of the IFT140 WD repeats. The spread of velocity values is similar in IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC cells; however, velocity values are shifted downwards in IFT140ΔWD-G/IFT20-mC flagella regardless of their length. Data are presented as mean±s.d. *P≤0.01 according to a Wilcoxon rank-sum test. Short flagella were ≤3 µm; long flagella were >3 µm. Scale bars: 2 µm.
MS analysis of the ‘membrane+matrix’ fraction from isolated IFT140ΔWD-G/IFT20-mC flagella reveals altered levels of proteins in several classes
Isolated flagella of IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC cells were treated with non-ionic detergent to solubilize membrane proteins as well as soluble components of the flagellar cytoplasm; the resulting ‘membrane+matrix’ fraction was then separated from the axonemes by centrifugation. Analysis by SDS-PAGE and silver-staining revealed most changes to be in the ‘membrane+matrix’ fraction; there were few – if any – differences between the axonemal fractions (Fig. 8A). Therefore, we used MS coupled with label-free intensity-based absolute quantification (iBAQ) (Schwanhäusser et al., 2011) to compare the ‘membrane+matrix’ fractions from IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC flagella in two independent replicates. Details of all proteins identified in these experiments are in Table S12; however, obvious contaminants, including cell wall and some other cell body proteins, are excluded from the other tables and our analyses below. To eliminate potential low-level contaminants, we also excluded proteins that showed abundance values of <5.0 E+06 in either the control or experimental samples, unless they previously had been identified as flagellar proteins. Despite these precautions, we cannot exclude the possibility that some of the proteins in our analysis are contaminants originating from the cell body.
Fig. 8.
Effect of IFT140 WD repeat loss on the protein composition of flagella. (A) SDS-PAGE of axoneme and ‘membrane+matrix’ fractions from IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC flagella. Asterisks indicate differences between bands in the IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC ‘membrane+matrix’ fractions. (B) Proteins identified by MS of IFT140-G/IFT20-mC and IFT140ΔWD-G/IFT20-mC ‘membrane+matrix’ fractions were curated into separate groups and individual fold-change values (IFT140ΔWD-G/IFT20-mC:IFT140-G/IFT20-mC) based on iBAQ scores are shown in the graph. Red lines indicate a 2.0-fold increase and a 2.0-fold decrease. Green lines indicate median values for each protein group. IFT-A, GTPases, myristoylated proteins and geranylgeranylated proteins all have their median values decreased ≥2.0-fold, suggesting that recruitment of these protein classes is impaired in IFT140ΔWD-G/IFT20-mC flagella. The proteins included in each class are listed in Tables S1–S11.
Increases or decreases are presented as fold change, calculated using the proteome software Scaffold, which adds abundance values for control and experimental replicates and then calculates fold changes from these values. A fold change of 2.0 means that a protein is twice as abundant in the experimental sample as in the control sample; a fold change of 0.5 means the protein is half as abundant in the experimental sample as compared to the control sample.
Levels of tubulin showed no change; because IFT of tubulin is elevated in rapidly growing flagella (Craft et al., 2015), this suggests that IFT140ΔWD-G/IFT20-mC flagella have transitioned to the slow elongation phase (Rosenbaum et al., 1969; Wood et al., 2012), also referred to as ‘steady state’, despite averaging only half-length.
Fig. 8B shows increases or decreases of non-tubulin proteins in several protein groups; an individual protein may be included in more than one group. Tables S1–S11 give detailed data for the proteins in each group.
The 16 IFT-B proteins showed little or no change in IFT140ΔWD-G/IFT20-mC vs IFT140-G/IFT20-mC ‘membrane+matrix’ (Fig. 8B and Table S1). Therefore, even though IFT-B (and IFT-A) particles were prone to stalling in the IFT140ΔWD-G/IFT20-mC flagella, levels of IFT-B are relatively normal.
In contrast, all six IFT-A proteins were greatly decreased in the IFT140ΔWD-G/IFT20-mC sample, with levels ranging from 0.2–0.05 of those in the control sample (Fig. 8B and Table S1). The simplest explanation is that loss of the IFT140 WD domains has a severe effect on entry of IFT-A into flagella.
Trending in the opposite direction from the IFT-A proteins, all six structural polypeptides of the BBSome were highly elevated (3.4- to 6.1-fold) in the IFT140ΔWD-G/IFT20-mC sample (Fig. 8B and Table S1). An increase in BBSome proteins is consistent with a defect in retrograde IFT and, previously, has been observed in mutants with defects in dynein-1b (Lechtreck et al., 2013), as well as an IFT-B mutant that has reduced flagellar levels of IFT-A (Brown et al., 2015). Chlamydomonas BBS3B, the BBSome membrane recruitment factor, which is most similar to mammalian BBS3 (officially known as ARL6), did not show this pattern: the level of BBS3B was unchanged in IFT140ΔWD-G/IFT20-mC flagella. A second Chlamydomonas protein, BBS3A, which is less closely related to mammalian BBS3 and whose function is still unknown, was reduced to 0.2 of control (Table S12).
There was no difference in the amounts of the three kinesin-2 subunits in IFT140ΔWD-G/IFT20-mC vs control samples (Table S2), consistent with the very small difference in anterograde velocities in these strains (Fig. 7E). Similarly, levels of dynein-1b heavy chain, and of the intermediate chains D1bIC1 and D1bIC2, were essentially unchanged, whereas the light-intermediate chain D1bLIC was decreased to 0.3 of control. This might indicate impaired assembly or stability of dynein-1b, which could account for the reduced retrograde IFT velocity. D1bLIC has been implicated in dynein-1b cargo binding (Hou et al., 2004), so loss of this protein might contribute to the stalling of IFT particles in the IFT140ΔWD-G/IFT20-mC flagellum (Fig. 7).
Sixty-three of 67 proteins from the outer and inner dynein arms, radial spokes, and central microtubule complex were moderately increased in the IFT140ΔWD-G/IFT20-mC sample; the median fold increase for the group of all 67 proteins was 2.4 (Fig. 8B and Table S3). Similarly, the majority of the nexin-dynein regulatory complex (N-DRC) components were moderately increased, with the exception of DRC1, which showed a 38-fold increase. The low abundance of DRC1 relative to other N-DRC components in the control sample might have contributed to the large fold-change associated with this protein. The moderate increase associated with the other N-DRC components is informative because, in regenerating flagella, IFT of the N-DRC – which was investigated by focusing on its component DRC4 – is elevated ∼10-fold compared with its level in non-regenerating flagella (Wren et al., 2013). This provides additional evidence that the IFT140ΔWD-G/IFT20-mC flagella are not in a regenerating state. The most probable reason that all of these structural proteins are in the ‘membrane+matrix’ fraction is that they are the products of axonemal turnover (Song and Dentler, 2001) and, as such, are likely to be cargos of retrograde IFT (Qin et al., 2004). Their increase in the IFT140ΔWD-G/IFT20-mC flagella could reflect impaired retrograde IFT and/or instability of the axoneme.
As groups, GTPases, myristoylated proteins and geranylgeranylated proteins stand out as having median values that are decreased by ≥2-fold in the mutant relative to the control (Fig. 8B). Of 20 GTPases, 18 were decreased, with 13 being decreased to 0.5–0.1 of control (Table S4). Many of these are predicted to be myristoylated or geranylgeranylated (Tables S5 and S6). Of 30 proteins predicted to be myristoylated, 26 were decreased, with 19 being decreased to 0.5–0.006 of control; one protein of this group, protein phosphatase 2C, was moderately abundant in control flagella but could not be detected in mutant flagella. The only three proteins that were increased in this group were an AMP-regulated protein kinase/serine-threonine protein kinase (AMPK/STPK), phospholipase D (PLD) and ubiquitin-protein ligase (Phytozome accession number: Cre12.g548100.t1.1). PLD – and probably AMPK/STPK – continuously cycle through the flagellum, entering independently of IFT but exiting via the BBSome and IFT (Lechtreck et al., 2009; Lechtreck et al., 2013). Hence, impaired retrograde IFT undoubtedly explains the increase of PLD in the mutant flagella. Of 21 proteins predicted to be geranylgeranylated, 17 were decreased, with 14 proteins being decreased to 0.5–0.02 of control levels. One of these, ankyrin, was moderately abundant in control flagella but could not be detected in the mutant flagella. Most of the proteins in the GTPase group, and all of those in the myristoylated and geranylgeranylated groups are predicted to be peripheral membrane proteins.
The ATPases, kinases, phosphatases, transmembrane proteins, and channels showed median increases or decreases of less than 2.0-fold (Figure 8B). However, there was considerable variation within the groups, with some proteins being greatly increased and some greatly decreased (Tables S7-S11). For example, among the ATPases, FAP39 (EDP02029.1), a plasma membrane Ca2+-transporting ATPase (XP_001692057.1), and a calmodulin-binding Ca2+-transporting ATPase (EDP02541.1), all also predicted to be transmembrane proteins, were decreased to 0.2, 0.2, and 0.3 of control, respectively, while a putative plasma membrane-type proton ATPase, which might be a contaminant, was increased 10-fold. Among the kinases, the ODA5-associated flagellar adenylate kinase, which is predicted to be myristoylated, was decreased to 0.04 of control. In contrast, FAP42 (XP_001697065.1), which is an adenylate/guanylate kinase-like protein that is a component of the central microtubule complex (Mitchell et al., 2005), was increased nearly 3-fold, consistent with the general increase in axonemal components. Among transmembrane proteins, 16 of 23 showed decreased levels in the mutant, with about half, including the three ATPases mentioned above, being decreased to 0.5-0.2 of their control levels. The two predicted transmembrane proteins whose levels showed the highest increase in the mutant were FAP260 and FAP49, both PAS-domain containing proteins that may actually be axonemal proteins (Pazour et al., 2005). Therefore, in general, the levels of transmembrane proteins were decreased in the mutant. Only a few ion channel proteins were detected; of these, the transient receptor potential ion channel M-type (TRPM) ion channel, which is predicted to have transmembrane domains by the TMHMM server, v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/), was decreased to 0.003 of its control level.
Other proteins of potential interest not listed in Tables S1–S11 include kinesin-like calmodulin-binding protein (increased 16-fold), FAP265 (decreased to 0.2 of control) and Chlamydomonas microtubule plus-end binding protein 1 (EB1, hereafter referred to as CrEB1; decreased to 0.1 of control). POC7 was decreased to 0.6 of its wild-type level. Although this is less of a decrease compared to that of the other proteins singled out here, it is of potential interest because POC7 is homologous to mammalian PDE6D and UNC119, both of which have been implicated in the import of lipid-modified proteins into cilia.
DISCUSSION
IFT140 is necessary for assembly of IFT-A that, in turn, is necessary for flagellar formation
We have identified a so-far-unknown insertional mutation affecting the 1457-bp region of IFT140 that encodes a portion of the C-terminal TPR domains of IFT140. IFT140 transcripts were undetectable in the mutant. The mutation destabilized both peripheral and core IFT-A proteins and prevented cells from forming flagella. These results suggest that the mutant is null for IFT140, and show that IFT140 is required for IFT-A stability and the formation of flagella. Despite the severe effects loss of IFT140 has on IFT-A, IFT-B localization and protein levels remained unaffected, indicating that cellular levels of IFT-B are regulated independently of IFT-A and that IFT-B is recruited to the basal body region independently of IFT-A. The fact that, in ift140-1, IFT-B proteins are present in the cytoplasm at wild-type levels and localize normally to the peri-basal body region, but that flagella formation is impaired, raises the question of whether IFT-A is necessary for entry of IFT-B into the flagellum (see below under ‘Is IFT-A required for IFT trains to enter flagella?).
The TPR domains of ITF140 stabilize IFT-A assembly, whereas its WD repeats are required for fast retrograde IFT
The availability of the ift140-1 mutant enabled us to explore the function of the WD-repeat and TPR domains of IFT140 by transforming the mutant with a construct expressing a version of IFT140 lacking the WD-repeat domains. Our results revealed that the TPR domains are sufficient to restore IFT-A complex formation and to partially rescue flagellar assembly. This is consistent with the known functions of TPR domains, which can have roles in intra-complex interactions and the stabilization of protein complexes (Grove et al., 2008). Given the high level of predicted structural conservation in multiple IFT-A components (Taschner et al., 2012), the TPR domains of IFT144, IFT139, IFT122 and IFT121 are likely to also function in IFT-A assembly and stability.
Although IFT-A appeared to be stabilized by the truncated IFT140 containing only the TPR domains, loss of the N-terminal WD repeats severely affected the velocity of retrograde IFT. Two scenarios might account for the observed decrease in retrograde IFT velocity. First, the WD repeats might be required, directly or indirectly, for full activation of dynein-1b. It is possible that, in the absence of the IFT140 WD repeats, the retrograde IFT train cannot properly interact with dynein-1b, resulting in incomplete ‘untrapping’ of dynein-1b autoinhibition at the flagellar tip (Toropova et al., 2017; Jordan et al., 2018) and leading to decreased average retrograde IFT velocity. Second, the decreased level of IFT-A in these flagella might lead to retrograde IFT trains being formed with fewer IFT-A complexes. Previous work has suggested that dynein-1b binds to IFT-A for retrograde IFT (Pedersen et al., 2006; Williamson et al., 2012) and more-recent work has suggested that dynein-based transport velocity is dependent on the number of dyneins attached to the cargo (Urnavicius et al., 2018). With fewer IFT-A complexes per train to bind dynein-1b in IFT140ΔWD-G-20C cells, retrograde IFT trains would move at decreased velocity compared to control IFT trains.
We also observed that flagella of cells that express the truncated IFT140 accumulated static IFT particles in a length-dependent manner; this also could be due to weaker interactions of the IFT particles with the retrograde motors. Although retrograde IFT events are infrequent in these longer flagella, the resulting tracks tended to be wider than retrograde IFT tracks in shorter flagella, suggesting that the retrograde IFT trains are larger than normal. Albeit infrequent, movement of larger IFT trains to the flagellar base would result in some IFT-particle proteins being available for assembly into new anterograde IFT particles, precluding a complete redistribution of IFT particles from the cell body into the flagella, as occurs when the retrograde IFT motor is defective (Pazour et al., 1999), and enabling some flagellar assembly through anterograde IFT.
Is IFT-A required for IFT trains to enter flagella?
Ift140-1 cells, completely lacking IFT-A, are unable to form a flagellum, yet IFT140ΔWD-G/IFT20-mC cells expressing a small amount of IFT-A can form flagella. Thus, a threshold amount of IFT-A is necessary for flagellar formation. This raises the question of whether a minimum amount of IFT-A must be in an IFT train for the train to enter the organelle. In a recent cryo-electron tomographic study of IFT trains in Chlamydomonas flagella, Jordan et al. (2018) reported the apparent absence of IFT-A complexes in some trains of the mutant ift139-2, whose flagellar ‘membrane+matrix’ fraction contains low levels of IFT-A proteins but normal levels of IFT-B proteins. They concluded that IFT-A is not absolutely required for anterograde train formation. However, IFT-A proteins are greatly reduced in IFT140ΔWD-G/IFT20-mC cell bodies and are reduced in IFT140ΔWD-G/IFT20-mC flagella by an amount similar to that reported by Jordan and colleagues for the ift139-2 mutant, yet virtually all anterograde IFT trains in these flagella appear to contain both IFT-A and IFT-B by immunofluorescence microscopy (Fig. 7B). These disparate observations might reflect the difficulty of detecting, by cryo-electron tomography, small amounts of IFT-A that do not have a regular periodicity on a train. Alternatively, they might be explained if ift139-2 cell bodies contain significantly less IFT-A than IFT140ΔWD-G/IFT20-mC cell bodies. In any case, if all or nearly all IFT trains still contain IFT-A when the level of IFT-A is greatly reduced in the cell body, it would suggest that the cell has a quality-control machinery that allows trains to enter the flagellum only when they contain both IFT-B and IFT-A. If so, the reduced anterograde IFT frequency in IFT140ΔWD-G/IFT20-mC flagella (Fig. 7C) might be because trains are retained in the peri-basal body region until sufficient IFT-A associates with the train to license entry of the train into the flagellum – an event that is likely to be delayed by the greatly reduced levels of IFT-A in the cell body.
IFT-A specializes in import into flagella of membrane-associated proteins
Our ability to isolate flagella from cells expressing IFT140 that lacks its WD repeats allowed us to analyze the ‘membrane+matrix’ fraction of these flagella by MS, in order to obtain a broad, unbiased view of how the composition of this fraction was affected by loss of the WD repeats. We observed several changes that are indicative of a defect in retrograde IFT, including a large increase in BBSome proteins and a smaller increase in axonemal structural proteins that, in both cases, probably resulted from failure to efficiently return the proteins to the cell body. Consistent with our western blot results, IFT-B proteins were present at normal levels, whereas those of IFT-A proteins were greatly decreased. Because the level of IFT-A particles was reduced in IFT140ΔWD-G/IFT20-mC cell bodies, it is possible that the reduced number of IFT trains entering the flagella of this strain contained less IFT-A than IFT-B, and that most IFT-A particles that did enter were cycled back to the cell body relatively efficiently while return of IFT-B particles was impaired. This could result in coincidental accumulation of IFT-B to almost normal levels despite reduced import of both IFT-A and IFT-B.
The above abnormalities in IFT require caution in concluding that loss of any specific protein from the flagellum indicates that that protein is imported through direct interaction with the WD repeats of IFT140. Nevertheless, it is possible to attribute many of the changes in protein composition to defects in IFT-A, if not specifically to IFT140. Implicit in most of the following discussion is the assumption that differences in the amounts of proteins in the ‘membrane+matrix’ fraction of control vs experimental represent differences in trafficking of the proteins into and out of the flagellum. Changes in the expression of proteins in response to IFT perturbation could affect their levels in the flagellum but the end result of such changes would be captured in our data and would not change our conclusions on how mutation of IFT140 affects flagellar protein content.
We are, therefore, able to draw three conclusions. First, our results confirm that IFT-A has a main role in retrograde IFT, as evidenced by the accumulation of proteins in the flagellum, specifically axonemal structural proteins and BBSome proteins. This is in agreement with previous studies that suggested that IFT-A is important for retrograde IFT (Iomini et al., 2009; Jonassen et al., 2012; Liem et al., 2012; Tran et al., 2008).
Second, IFT-A has a main role in import of proteins into the flagellum, especially of proteins predicted to be membrane-associated, including many small GTPases, and myristoylated and geranylgeranylated proteins. Other recent studies have implicated IFT-A in the import of individual membrane-associated proteins such as Smo, Mchr1, Gpcr161, rhodopsin, Arl13b, INPP5E, SSTR3 and HTR6 (Badgandi et al., 2017; Brear et al., 2014; Fu et al., 2016; Hirano et al., 2017; Liem et al., 2012; Mukhopadhyay et al., 2010, 2013; Sun et al., 2012). Our results indicate that IFT-A has a crucial role in the import of a large number of such proteins.
Third, our results provide no indication that IFT-A has a main role in the import of axonemal structural proteins into the flagellum. The flagella of cells that express truncated IFT140 appeared to have ultrastructurally normal axonemes, and SDS-PAGE analysis revealed few, if any, changes in the axonemal fraction of these cells, despite the greatly reduced amount of IFT-A. Moreover, MS analysis revealed few, if any, decreases in the 67 axonemal components detected in the flagellar ‘membrane+matrix’ fraction. Therefore, import of most axonemal proteins is likely to be mediated by IFT-B. This is consistent with previous work showing that specific IFT-B subunits are involved in import of tubulin, outer arm dynein and other axonemal proteins into the flagellum (Craft et al., 2015; Hou and Witman, 2017; Hou et al., 2007; Ishikawa and Marshall, 2011; Kubo et al., 2016), and that loss of IFT-B precludes flagellar assembly (Brazelton et al., 2001; Cole et al., 1998; Jiang et al., 2017; Jonassen et al., 2008; Pazour et al., 2002; Taschner et al., 2012; Zhang et al., 2017). Considering all of the above, it appears that IFT-A is specialized for the import of membrane-associated proteins, whereas IFT-B is specialized for the import of axonemal structural proteins.
We have previously proposed that IFT-A is initially delivered to the apical plasma membrane of the cell and then recruited to IFT-B at the transition fibers (Brown et al., 2015). Consistent with this, evidence has been presented that IFT-A has a role in the recruitment of flagellar precursors to the base of the flagellum (Wu et al., 2017; Zhu et al., 2017). Since localization of IFT-A at the flagellar base was only partially restored in IFT140ΔWD-G/IFT20-mC cells, impaired cargo recruitment by IFT-A to the base of the flagellum might have contributed to the decrease in membrane proteins that we observed in the flagella of these cells. In any case, IFT-A appears to be specialized for trafficking of flagellar membrane proteins.
Loss of specific flagellar proteins is likely to explain the phenotype of cells expressing IFT140 that lacks its WD repeats
Among the proteins whose levels were severely decreased in the flagella of cells expressing truncated IFT140 are several that are known to affect IFT and/or ciliary assembly. These include Arl3, Arl13, CrEB1, Rab8, Ran and Ca2+-dependent protein kinase 1 (CDPK1). Thus, the decreases in these proteins is likely to contribute to the flagellar phenotype of these cells.
Arl3 is present in both cell body and cilium, where it has an important role in ciliogenesis (Cuvillier et al., 2000; Li et al., 2010). Its main function is probably the release of lipidated cargo proteins from their carrier proteins (Gotthardt et al., 2015; Stephen et al., 2018). Therefore, a decrease in ciliary Arl3 might contribute to the decreased amounts of these proteins in IFT140ΔWD-G/IFT20-mC flagella by preventing correct cargo unloading. Several proteins, including Arl3, have been shown to diffuse into and/or out of cilia, in addition to relying on IFT (Chien et al., 2017; Craft et al., 2015; Engel et al., 2012; Harris et al., 2018preprint; Kösling et al., 2018; Luo et al., 2017). Proteins relying solely on diffusion for exit from flagella should show no change in our analysis, so the 0.5-fold decrease in Arl3 might be a result of disrupting the IFT component of its trafficking. However, we cannot rule out the possibility that changes to the TZ or the flagellar lipid content in IFT140ΔWD-G/IFT20-mC cells disrupted the mechanisms that retain proteins, such as Arl3, in the flagellum.
The guanine exchange factor for Arl3 is Arl13 (Arl13b in vertebrates and ARL-13 in C. elegans), which is localized to cilia and converts Arl3-GDP entering the cilium to Arl3-GTP; the latter is then able to interact with carrier proteins to release their lipidated cargos (Gotthardt et al., 2015; Ivanova et al., 2017). Accordingly, studies have demonstrated a role for Arl13b/ARL-13 in the correct localization of several ciliary membrane proteins (Cevik et al., 2010; Humbert et al., 2012; Larkins et al., 2011; Li et al., 2012; Nozaki et al., 2017). Defects in Arl13b perturb ciliary length and ultrastructure (Caspary et al., 2007; Larkins et al., 2011), and Arl13b/ARL-13 might have a role in the regulation of IFT (Cevik et al., 2010; Nozaki et al., 2017). The decrease in Arl13 levels that we observed in the current study is similar to the findings that have been reported for Arl13b in mouse and human cells that have defects in IFT144 and IFT121 (Fu et al., 2016; Liem et al., 2012), indicating that the decrease is likely to be due to the more generalized effect on IFT-A, rather than the specific defect in IFT140. Although Arl13b has been reported to interact with IFT-B (Cevik et al., 2013), our results are consistent with the finding that it can interact with IFT-A independently of IFT-B (Fu et al., 2016).
In Chlamydomonas, CrEB1 is the only close homolog of human microtubule end-binding proteins 1 and 3 (EB1 and EB3; officially known as MAPRE1 and MAPRE3, respectively). In mammals, EB1 and EB3 are crucial for primary cilia assembly (Schrøder et al., 2011; Schrøder et al., 2007). Like EB1 and EB3, CrEB1 localizes to both the proximal region of the basal body and the flagellar tip (Pedersen et al., 2003). Decreased levels of CrEB1 in the flagella could affect IFT turnaround, contributing to the large accumulations of cargo that we observed in IFT140ΔWD-G/IFT20-mC flagella.
The small GTPase Rab8, in coordination with the BBSome, promotes ciliary membrane extension during ciliogenesis by initiating the formation of a vesicular structure near the centriole (Nachury et al., 2007). Rab8 is also implicated in the transport of several ciliary membrane proteins including rhodopsin, fibrocystin, polycystin 1 and polycystin 2 (Blacque et al., 2017; Follit et al., 2010; Ward et al., 2011). Thus, decreased levels of Rab8 might affect IFT140ΔWD-G-20C flagella in two ways: first, defective flagellar membrane formation due to decreased Rab8 could contribute to the shortened flagella observed in our analysis; second, decreased levels of Rab8 are likely to affect the transport of subsets of membrane proteins into the flagella.
Previous work has shown that a ciliary/cytoplasmic gradient of Ran is required for the entry of Kif17 into the cilia of mammalian cells (Dishinger et al., 2010) and it has been proposed that, similar to nuclear import, import of Kif17 and other proteins into the cilium is regulated by a Ran/importin system (Dishinger et al., 2010; Kee et al., 2012). Therefore, decreased ciliary Ran due to a defect in IFT140 probably renders cells unable to correctly establish a Ran-GTP/Ran-GDP gradient across the flagellar TZ, which would severely affect flagellar entry of any proteins relying on this system.
CDPK1 also was substantially decreased in IFT140ΔWD-G/IFT20-mC flagella. CDPK1 is involved in regulation of IFT entry and turnaround at the flagellar tip (Liang and Pan, 2013; Liang et al., 2014). Therefore, decreased levels of CDPK1 in the flagellum might affect IFT turnaround, contributing to the accumulation of cargo and the decreased retrograde IFT frequency in IFT140ΔWD-G/IFT20-mC flagella.
Absence of IFT140 WD repeats results in flagellar loss of proteins involved in Ca2+-signaling
Also decreased in flagella of cells that express IFT140 lacking the WD repeats were several transmembrane proteins that are involved in intraciliary ion regulation. Chlamydomonas TRPM, which is a homolog of the human TRP subfamily C (TRPC), was reduced to <0.01 of its control level and, thus, might be trafficked into the cilium through direct interaction with the WD repeats of IFT140. TRPC channels are non-selective cation channels that are Ca2+ permeable (Pablo et al., 2017). Four Ca2+-transporting ATPases [FAP39, XP_001692057.1, EDP02541 and EDP05262 (officially known as ACA1 and ACA2, respectively)], were also reduced in flagella of cells expressing truncated IFT140. Interestingly, Lee et al. (2008) reported that IFT140 is essential for expression of a ciliary TRPV ion channel in Drosophila melanogaster.
Intriguingly, previous work has shown that clearance of IFT accumulations from the trailing flagellum of Chlamydomonas cells gliding on a solid substrate is accompanied by and dependent upon transient elevations of intraflagellar Ca2+ (Collingridge et al., 2013; Shih et al., 2013). Therefore, disruption of Ca2+ signaling at flagellar adhesion sites due to loss of flagellar Ca2+ channels may contribute to the increased levels of static IFT particles observed in IFT140ΔWD-G/IFT20-mC flagella.
Loss of specific ciliary proteins caused by defective IFT140 probably explains the pathogenesis of IFT140-linked human disease
Impairment of retrograde IFT, such as we observed in cells expressing truncated IFT140, is known to cause defects in Hedgehog signaling (Qin et al., 2011; Tran et al., 2008) and to result in some of the human disease phenotypes associated with defects in IFT140. In addition, if mutations of IFT140 in human patients cause loss of some of the same ciliary proteins that were depleted in flagella of Chlamydomonas cells expressing truncated IFT140, it could explain the multiple phenotypes associated with JATD, MSS, LCA and RP. For example, Arl13b is crucial in the regulation of entry into cilia of Smo, a key player in Hedgehog signaling (Caspary et al., 2007; Larkins et al., 2011), and Arl3 interacts with Kif7, which regulates Hedgehog signaling (Gotthardt et al., 2015; Hanke-Gogokhia et al., 2016; Lai et al., 2011; Schwarz et al., 2017). Similarly, defective membrane biogenesis due to a decrease in ciliary ARL3, ARL13b or RAB8 could affect the renewal of photoreceptor outer segment disks, contributing to the retinal degeneration associated with RP and LCA (Hanke-Gogokhia et al., 2016; Yildiz and Khanna, 2012).
Among other proteins severely decreased in flagella of cells expressing IFT140 that lacks its WD repeats was the small ARF-related GTPase A1A (ArfA1A). Arf1, the mammalian homolog of Chlamydomonas ArfA1A, is known to function in controlling levels of phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] in the Golgi membrane (Godi et al., 1999) and could have a similar function in the ciliary membrane. Altered ciliary PtdIns(4,5)P2 levels affect ciliary membrane protein localization as well as Hedgehog signaling (Garcia-Gonzalo et al., 2015) and could accelerate cyst progression in MSS and JATD (Badgandi et al., 2017). Consistent with this, loss of IFT140 in mice results in defective Hedgehog signaling in highly cystic kidney tissue (Jonassen et al., 2012).
In summary, our work provides the first large scale analysis of changes in protein composition of cilia as a result of mutation of an IFT-A component. Defective anterograde trafficking of proteins – especially membrane-associated proteins – identified in our analysis are likely to explain why mutations of IFT140 in humans cause the skeletal, renal and hepatic phenotypes associated with JATD and MSS, as well as the ocular phenotypes associated with RP and LCA.
MATERIALS AND METHODS
Strains and cell culture
Chlamydomonas reinhardtii cells were grown in liquid TAP medium (Gorman and Levine, 1965), with a light/dark cycle of 14/10 h and with aeration by 5% CO2 (Witman, 1986). For some experiments cells were treated with autolysin to release cells from the mother cell wall. Wild-type strain 137c (nit1::nit2 mt+) is available from the Chlamydomonas Resource Center as CC-125. The IFT20-mCherry strain (ift20-1 IFT20-mCherry mt−) was generated by rescuing an ift20-1 null mutant with a construct expressing IFT20-mCherry (Lechtreck et al., 2009). The original ift140-1 strain (nit1-1 ac17 sr1 ift140-1::NIT1 mt+) was identified by screening (Hou, 2007) a collection of insertional mutants that had been generated by transforming strain A54-e18 (CC-2929; nit1-1 ac17 sr1 mt+) with the plasmid pMN24 linearized with EcoR1 (Nelson et al., 1994; Zhang and Lefebvre, 1997); this collection was generously provided to us by Trung Bui and Carolyn Silflow (University of Minnesota at St. Paul, MN) via William Dentler (University of Kansas, Lawrence, KS). Strains generated specifically for this study include IFT140-G (ift140-1::NIT1 IFT140-sfGFP mt+), IFT140-G/IFT20-mC (ift20-1 IFT20-mCherry ift140-1::NIT1 IFT140-sfGFP mt+), IFT140ΔWD-G (ift140-1::NIT1 IFT140Δ70-335-sfGFP mt+), and IFT140ΔWD-G/IFT20-mC (ift20-1 IFT20-mCherry ift140-1::NIT1 IFT140Δ70-335-sfGFP mt+).
Generation of strains and constructs
IFT140 was amplified in three fragments by PCR from the bacterial artificial chromosome 03K07 (available from the Chlamydomonas Resource Center) and ligated into pNEB193 (New England Biolabs, Ipswich, MA). Super-folder GFP (sfGFP) was ligated into the IFT140 sequence immediately upstream of the stop codon at a BamHI cut-site (inserted in-frame during amplification of the IFT140 sequence) to create pIFT140-sfGFP. The AphVIII- (Paromomycin-) resistance gene was treated with PacI and ligated into the PacI site in pIFT140-sfGFP to generate pIFT140-sfGFP-aphVIII. The IFT140-G strain was generated by transformation of the original ift140-1 strain with pIFT140-sfGFP-aphVIII. ift140-20mCh was generated by crossing the IFT20-mCherry strain with the ift140-1 strain. To generate strain IFT140-G/IFT20-mC expressing both IFT20-mCherry and IFT140-sfGFP, pIFT140-sfGFP-aphVIII was transformed into the ift140-20mCh strain. To construct a plasmid for generation of the IFT140ΔWD-G and IFT140ΔWD-G/IFT20-mC strains, pIFT140-sfGFP-aphVIII was used as a template for overlap-extension PCR. Briefly, primer pairs were designed to amplify regions flanking the sequence that encodes the IFT140 WD-repeat domains (L76-K325, 1123-3316 bp). The resulting PCR products were annealed and treated with the restriction endonucleases BspEI and EcoRI-HF. The endonuclease-treated fragment was then ligated into pIFT140-sfGFP-aphVIII that had been treated with the same restriction enzymes. The resulting plasmid, pIFT140ΔWD-sfGFP-aphVIII, was transformed into ift140-1 to generate the IFT140ΔWD-G strain. The same plasmid was used to transform the ift140-20C strain to yield the IFT140ΔWD-G/IFT20-mC strain. All transformations were carried out by electroporation as described previously (Brown et al., 2012). Selected transformants were then crossed at least once to strain 137c.
Antibodies
A recombinant protein consisting of IFT140 amino acids 1182–1384 was fused to maltose-binding protein and used to generate rabbit polyclonal antibodies (Lampire Biological Laboratories, Everett, PA). The resulting antiserum was affinity purified against a recombinant protein consisting of amino acids 1182–1384 fused to glutathione-S-transferase that had been expressed in BL21 gold (DE3) cells (Agilent Technologies, Santa Clara, CA). Additional antibodies used in this study were: mouse monoclonal anti-acetylated tubulin T6793 (Sigma-Aldrich, St Louis, MO); rabbit polyclonal anti-IFT122 (Behal et al., 2012), anti-β-F1-ATPase (Agrisera, Vännäs, Sweden); anti-GFP (AbCam, Cambridge, UK); guinea pig polyclonal anti-IFT46 (generously provided by Hongmin Qin; Hou et al., 2007); and mouse monoclonal anti-IFT172, anti-IFT139, anti-IFT81 and anti-IFT57 (all generously provided by Douglas Cole) (Behal et al., 2012; Cole et al., 1998).
Differential interference contrast imaging and flagellar length measurements
Cells were cultured as described above and harvested by centrifugation. The resulting cell pellet was resuspended in an appropriate volume of TAP growth medium and the cells were allowed to adhere to coverslips for 3 min. Coverslips were then immersed in −20°C methanol for 5 min, allowed to air dry for 30 min, and re-hydrated with immunofluorescence blocking buffer (IFB) (6% fish skin gelatin, 1% BSA and 0.05% Tween-20 in PBS pH 7.0) for 30 min at room temperature (RT). Excess IFB was wicked off and the cells were mounted in ProLong Antifade Gold (Invitrogen, Carlsbad, CA). Cells were imaged using a Nikon Ti-U microscope with a 60×1.49 NA oil immersion lens, a 1.4 NA oil immersion condenser and a 1.5× relay lens. Images were acquired using a Clara Interline camera (Andor, South Windsor, CT) and NIS-Elements software (Nikon Instruments, Inc., Melville, NY). Fiji (National Institutes of Health, Bethesda, MD) was used to measure flagella length, and to adjust brightness and contrast where applicable.
Immunofluorescence microscopy
Cells were cultured as described above, harvested by centrifugation and then treated with autolysin for 30 min prior to processing for immunofluorescence microscopy as follows: Live cells were allowed to adhere to polyethlyleineimine-coated coverslips for 3 min. Excess cells were wicked off and the coverslips immersed in −20°C methanol for 3 min. The coverslips were then air dried for 30 min and rehydrated with IFB for 30 min at RT. Cells were incubated in primary antibodies (anti-IFT172, at 1:5; anti-IFT139, at 1:5; and anti-IFT57, at 1:5) in IFB for 1 h at RT or at 4°C overnight, washed 4×15 min with IFB, incubated with secondary antibodies (Alexa Fluor-conjugated IgG, Invitrogen) in IFB for 1 h at RT, washed 4×15 min in IFB and mounted in ProLong Antifade Gold. Cells were imaged using a GE DeltaVision OMX structured-illumination super-resolution microscope (GE Healthcare Life Sciences, Pittsburgh, PA) with a 60× plan-apochromat 1.42 NA oil-immersion objective (Olympus, Waltham, MA) using immersion oil with a refractive index of 1.514. Images were captured on sCMOS cameras using the GE OMX software package. Images were reconstructed and registered using SoftWorx software (GE Healthcare Life Sciences) that was calibrated using the GE Healthcare target registration slide and a Weiner filter constant of 0.002. Where applicable Fiji was used to make brightness and or contrast adjustments to the images under comparison.
TIRF microscopy
Cells in ∼10 µl of culture medium were allowed to adhere to a cleaned glass coverslip for 5 min. The excess medium was removed and the coverslip inverted and placed on an 8-well slide (6 mm, Tekdon, Inc., Myakka City, FL, part no. #8-60) with each well containing 2 µl of 10 mM HEPES and 6.25 mM EGTA, pH 7.5. The cells were visualized using the GE DeltaVision OMX microscope described above in conjunction with the total internal reflection fluorescence (TIRF) illumination module using immersion oil with a refractive index of 1.514. Videos were recorded at 14 frames/s using sCMOS cameras at RT. Fiji with the multi-kymograph plug-in was used to generate kymographs from the acquired movies and was also used to adjust brightness and contrast of movies and kymographs where applicable.
Electron microscopy
For ultrastructural analysis, cells were harvested by centrifugation, gently resuspended in conditioned growth medium, then fixed in 2% glutaraldehyde for 15 min at RT. Cells were pelleted, the primary fixative was removed, and the cells were resuspended in secondary fixative (1% glutaraldehyde with 100 mM sodium cacodylate pH 7.2) for 2 h at RT. Cells were pelleted and washed 3×15 min each in 100 mM sodium cacodylate pH 7.2. 50 µl of the washed cell pellet was added to a microcentrifuge tube containing 50 µl of 2% agarose dissolved in 100 mM sodium cacodylate pH 7.2. The cell suspension was solidified on ice and was removed by cutting open the end of the microcentrifuge tube. The cell/agarose gel was then cut into ∼1 mm3 cubes, post-fixed with 1% OsO4 in 50 mM sodium cacodylate, washed 3× with ice-cold water, and stained overnight in fresh 1% uranyl acetate in water. Stained samples were dehydrated, embedded in Embed-812 (Electron Microscopy Sciences, Hatfield, PA), and thin-sectioned. Sections were examined under a Philips CM10 electron microscope and images acquired with a Gatan (Pleasanton, CA) digital camera.
SDS-PAGE and western blotting
For western blots of whole-cell extracts (WCEs), mid-log phase cells were pelleted by centrifugation and resuspended in 5×-denaturing sample buffer [50 mM Tris pH 8.0, 160 mM dithiothreitol (DTT), 5 mM EDTA, 50% sucrose, and 5% SDS], heated to 90°C for 5 min, then passed 6× through a 26-gauge needle. Protein concentration was determined in an RC/DC protein assay (BioRad Laboratories, Hercules, CA). Equal amounts of protein from control and test samples were subjected to SDS-PAGE and transferred to Immobilon P (EMD Millipore, Billerica, MA) membrane for subsequent analysis. Membranes were probed with the following antibodies at the indicated dilutions: anti-IFT140 (1:1000), anti-IFT122 (1:1000), anti-GFP (1:2000), anti- β-F1-ATPase (1:10,000), anti-IFT172 (1:5), anti-IFT139 (1:20), anti-IFT81 (1:400), anti-IFT57 (1:5) and anti-IFT46 (1:20,000).
Flagella isolation and MS
Flagella of IFT140-G/IFT20-mC (control) and IFT140ΔWD-G/IFT20-mC cells were harvested using dibucaine and fractionated into ‘membrane+matrix’ and axonemal fractions (Craige et al., 2013; Witman, 1986). Protein concentrations were determined as described above and equal amounts of the ‘membrane+matrix’ fractions from control and test cells were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. A fully resolved SDS-polyacrylamide gel was cut into five fractions and each fraction was processed separately as described. Gel bands were cut into 1×1 mm pieces and placed in 1.5-ml Eppendorf tubes with 1 ml of water for 30 min. The water was removed and 200 µl of 250 mM ammonium bicarbonate was added. For reduction, 20 µl of a 45 mM solution of DTT was added and the samples were incubated at 50°C for 30 min, then cooled to RT. For alkylation, 20 µl of a 100 mM iodoacetamide solution was added and allowed to react for 30 min. The gel slices were washed 2× with 1-ml water aliquots. The water was removed and 1 ml of 50:50 (50 mM ammonium bicarbonate:acetonitrile) was placed in each tube and samples were incubated at RT for 1 h. The solution was then removed and 200 µl of acetonitrile was added to each tube until the gel slices turned opaque white. The acetonitrile was removed and gel slices were further dried in a Speed Vac (Thermo-Fisher, Waltham, MA). Gel slices were rehydrated in 100 µl of 4 ng/µl sequencing-grade trypsin (Sigma-Aldrich, St. Louis, MO) in 0.01% ProteaseMAX Surfactant (Promega, Madison, WI): 50 mM ammonium bicarbonate. Additional bicarbonate buffer was added to ensure complete submersion of the gel slices. Samples were incubated at 37°C for 18 h. The supernatant of each sample was then removed and placed in a separate 1.5-ml tube. Gel slices were further extracted with 200 µl of 80:20 (acetonitrile: 1% formic acid). The extracts were combined with the supernatants of each sample. The samples were then completely dried in a Speed Vac.
Tryptic peptide digests were reconstituted in 20 µl of 5% acetonitrile containing 0.1% (v/v) trifluoroacetic acid and separated on a NanoACQUITY UPLC (Waters, Milford, MA). In brief, a 4.0-µl injection was loaded in 5% acetonitrile containing 0.1% formic acid at 4.0 µl/min for 4.0 min onto a 100-µm ID fused-silica pre-column packed with 2 cm of 5-µm (200 Å) Magic C18AQ resin (Bruker-Michrom, Auburn, CA). The sample was eluted using a gradient at 300 nl/min onto a 75-µm I.D. analytical column packed with 25 cm of 3 µm (100 Å) Magic C18AQ particles in a gravity-pulled tip. The solvents were A, water (0.1% formic acid); and B, acetonitrile (0.1% formic acid). A linear gradient was developed from 5% solvent A to 35% solvent B in 60 min. Ions were introduced by positive electrospray ionization via liquid junction into a Q Exactive hybrid mass spectrometer (Thermo-Fisher). Mass spectra were acquired over m/z 300-1750 at 70,000 resolution (m/z 200) and data-dependent acquisition selected the top 10 most abundant precursor ions for tandem MS by HCD fragmentation using an isolation width of 1.6 Da, collision energy of 27 and a resolution of 17,500.
Raw data files were peak processed with Proteome Discoverer (version 1.4, Thermo-Fisher) prior to database searching with Mascot Server (version 2.4) against the Chlamy Index of NCBInr database (PRJNIFT140-G/IFT20-mC260). Search parameters included trypsin specificity with two missed cleavages or no enzymatic specificity. The variable modifications of oxidized methionine, pyroglutamic acid for N-terminal glutamine, N-terminal acetylation of the protein, and a fixed modification for carbamidomethyl cysteine were considered. The mass tolerances were 10 ppm for the precursor and 0.05 Da for the fragments. Search results were then loaded into the Scaffold Viewer (Proteome Software, Inc., Portland, OR) for peptide/protein validation and label-free quantification using the iBAQ method (Schwanhäusser et al., 2011).
Three paired isolates of control and IFT140ΔWD-G/IFT20-mC ‘membrane+matrix’ were analyzed as above. However, one of the datasets had anomalies in the amounts of IFT and some cargo proteins in the control sample. These anomalies were not seen in the other samples of IFT140-G/IFT20-mC or in many samples of ‘membrane+matrix’ from wild-type that we have analyzed. As confidence in this dataset was low, it was dropped from further analysis. The low statistical power achievable with two independent replicates precluded the inclusion of a statistical analysis of the MS dataset. Known flagellar proteins were curated using the Chlamydomonas flagellar proteome (Pazour et al., 2005). Proteins were assigned to specific lipid modified subgroups based on the output of the GPS-Lipid prediction program (Xie et al., 2016).
Supplementary Material
Acknowledgements
We thank Drs Douglas Cole (University of Idaho) and Hongmin Qin (Texas A&M University) for antibodies; Dr Branch Craige, [University of Massachusetts Medical School (UMMS)] for help with differential interference contrast microscopy; Drs Gregory Hendricks and Lara Strittmatter of the UMMS Electron Microscopy Facility for expert assistance with electron microscopy; and Drs John Leszyk and Michelle Dubuke at the UMMS Proteomics and Mass Spectrometry Facility for their expert assistance with MS.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.P., J.M.B., Y.H., G.S., G.B.W.; Methodology: T.P., J.M.B., Y.H., G.B.W.; Validation: T.P., G.B.W.; Formal analysis: T.P., O.D.K., G.B.W.; Investigation: T.P., J.M.B., G.S., G.J.P.; Resources: T.P., D.A.C., K.L., G.B.W.; Data curation: T.P., J.M.B., Y.H., G.B.W.; Writing - original draft: T.P., G.B.W.; Writing - review & editing: T.P., J.M.B., O.D.K., K.L., G.J.P., G.B.W.; Visualization: T.P., J.M.B., G.B.W.; Supervision: J.M.B., G.B.W.; Project administration: G.B.W.; Funding acquisition: G.B.W. and G.J.P.
Funding
This work was supported by National Institutes of Health (NIH) [grant numbers: GM060992 and DK103632 to G.J.P., GM110413 to K.L., GM030626 and GM122574 to G.B.W.], and by the Robert W. Booth Endowment at UMMS to G.B.W. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.220749.supplemental
References
- Absalon S., Blisnick T., Bonhivers M., Kohl L., Cayet N., Toutirais G., Buisson J., Robinson D. and Bastin P. (2008). Flagellum elongation is required for correct structure, orientation and function of the flagellar pocket in Trypanosoma brucei. J. Cell Sci. 121, 3704-3716. 10.1242/jcs.035626 [DOI] [PubMed] [Google Scholar]
- Badgandi H. B., Hwang S.-H., Shimada I. S., Loriot E. and Mukhopadhyay S. (2017). Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 216, 743-760. 10.1083/jcb.201607095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behal R. H., Miller M. S., Qin H., Lucker B. F., Jones A. and Cole D. G. (2012). Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex a proteins. J. Biol. Chem. 287, 11689-11703. 10.1074/jbc.M111.287102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque O. E., Scheidel N. and Kuhns S. (2017). Rab GTPases in cilium formation and function. Small GTPases 3, 1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton W. J., Amundsen C. D., Silflow C. D. and Lefebvre P. A. (2001). The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11, 1591-1594. 10.1016/S0960-9822(01)00485-7 [DOI] [PubMed] [Google Scholar]
- Brear A. G., Yoon J., Wojtyniak M. and Sengupta P. (2014). Diverse cell type-specific mechanisms localize G protein-coupled receptors to Caenorhabditis elegans sensory cilia. Genetics 197, 667-684. 10.1534/genetics.114.161349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M., DiPetrillo C. G., Smith E. F. and Witman G. B. (2012). A FAP46 mutant provides new insights into the function and assembly of the C1d complex of the ciliary central apparatus. J. Cell Sci. 125, 3904-3913. 10.1242/jcs.107151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. M., Cochran D. A., Craige B., Kubo T. and Witman G. B. (2015). Assembly of IFT trains at the ciliary base depends on IFT74. Curr. Biol. 25, 1583-1593. 10.1016/j.cub.2015.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T., Larkins C. E. and Anderson K. V. (2007). The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell 12, 767-778. 10.1016/j.devcel.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Cevik S., Hori Y., Kaplan O. I., Kida K., Toivenon T., Foley-Fisher C., Cottell D., Katada T., Kontani K. and Blacque O. E. (2010). Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 188, 953-969. 10.1083/jcb.200908133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik S., Sanders A. A. W. M., Van Wijk E., Boldt K., Clarke L., van Reeuwijk J., Hori Y., Horn N., Hetterschijt L., Wdowicz A. et al. (2013). Active transport and diffusion barriers restrict joubert syndrome-associated arl13b/arl-13 to an inv-like ciliary membrane subdomain. PLoS Genet. 9, e1003977 10.1371/journal.pgen.1003977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien A., Shih S. M., Bower R., Tritschler D., Porter M. E. and Yildiz A. (2017). Dynamics of the IFT machinery at the ciliary tip. eLife Sci. 6, e28606 10.7554/eLife.28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G. (2003). The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435-442. 10.1034/j.1600-0854.2003.t01-1-00103.x [DOI] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C. and Rosenbaum J. L. (1998). Chlamydomonas Kinesin-II–dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge P., Brownlee C. and Wheeler G. L. (2013). Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr. Biol. 23, 2311-2318. 10.1016/j.cub.2013.09.059 [DOI] [PubMed] [Google Scholar]
- Craft J. M., Harris J. A., Hyman S., Kner P. and Lechtreck K. F. (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 208, 223-237. 10.1083/jcb.201409036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B., Brown J. M. and Witman G. B. (2013). Isolation of Chlamydomonas Flagella. Curr Protoc Cell Biol. 03, Unit -3.41.9 10.1002/0471143030.cb0341s59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse J. A., Lopes V. S., San Agustin J. T., Keady B. T., Williams D. S. and Pazour G. J. (2014). Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton 71, 302-310. 10.1002/cm.21173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvillier A., Redon F., Antoine J. C., Chardin P., DeVos T. and Merlin G. (2000). LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci. 113, 2065. [DOI] [PubMed] [Google Scholar]
- Dishinger J. F., Kee H. L., Jenkins P. M., Fan S., Hurd T. W., Hammond J. W., Truong Y. N.-T., Margolis B., Martens J. R. and Verhey K. J. (2010). Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and Ran-GTP. Nat. Cell Biol. 12, 703-710. 10.1038/ncb2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J. T., Keady B. T., Jonassen J. A., Liang Y., Francis R., Tobita K., Johnson C. A., Abdelhamed Z. A., Lo C. W. et al. (2014). IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment. Dev. Cell 31, 279-290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel B. D., Ishikawa H., Wemmer K. A., Geimer S., Wakabayashi K.-I., Hirono M., Craige B., Pazour G. J., Witman G. B., Kamiya R. et al. (2012). The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J. Cell Biol. 199, 151-167. 10.1083/jcb.201206068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit J. A., Li L., Vucica Y. and Pazour G. J. (2010). The cytoplasmic tail of fibrocystin contains a ciliary targeting sequence. J. Cell Biol. 188, 21-28. 10.1083/jcb.200910096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W., Wang L., Kim S., Li J. and Dynlacht B. D. (2016). Role for the IFT-A complex in selective transport to the primary cilium. Cell Rep. 17, 1505-1517. 10.1016/j.celrep.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R., Phua S. C., Roberson E. C., Garcia G., Abedin M., Schurmans S., Inoue T. and Reiter J. F. (2015). Phosphoinositides regulate ciliary protein trafficking to modulate Hedgehog signaling. Dev. Cell 34, 400-409. 10.1016/j.devcel.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A., Pertile P., Meyers R., Marra P., Di Tullio G., Iurisci C., Luini A., Corda D. and De Matteis M. A. (1999). ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P 2 on the Golgi complex. Nat. Cell Biol. 1, 280-287. 10.1038/12993 [DOI] [PubMed] [Google Scholar]
- Gorman D. S. and Levine R. P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665-1669. 10.1073/pnas.54.6.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K., Lokaj M., Koerner C., Falk N., Gießl A. and Wittinghofer A. (2015). A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. eLife Sci. 4, e11859 10.7554/eLife.11859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove T. Z., Cortajarena A. L. and Regan L. (2008). Ligand binding by repeat proteins: natural and designed. Curr. Opin. Struct. Biol. 18, 507-515. 10.1016/j.sbi.2008.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke-Gogokhia C., Wu Z., Gerstner C. D., Frederick J. M., Zhang H. and Baehr W. (2016). Arf-like protein 3 (ARL3) regulates protein trafficking and ciliogenesis in mouse photoreceptors. J. Biol. Chem. 291, 7142-7155. 10.1074/jbc.M115.710954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. A., Van De Weghe J. M., Kubo T., Witman G. B. and Lechtreck K. (2018). Diffusion rather than IFT provides most of the tubulin required for axonemal assembly. BioRxiv. 10.1101/286573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Katoh Y. and Nakayama K. (2017). Intraflagellar transport-A complex mediates ciliary entry and retrograde trafficking of ciliary G protein–coupled receptors. Mol. Biol. Cell 28, 429-439. 10.1091/mbc.e16-11-0813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. (2007). Identification and characterization of components of the intraflagellar transport (IFT) machinery. Ph.D. Dissertation. University of Massachusetts Medical School Worcester, MA. [Google Scholar]
- Hou Y. and Witman G. B. (2017). The N-terminus of IFT46 mediates intraflagellar transport of outer arm dynein and its cargo-adaptor ODA16. Mol. Biol. Cell 28, 2420-2433. 10.1091/mbc.e17-03-0172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Pazour G. J. and Witman G. B. (2004). A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell 15, 4382-4394. 10.1091/mbc.e04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Qin H., Follit J. A., Pazour G. J., Rosenbaum J. L. and Witman G. B. (2007). Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653-665. 10.1083/jcb.200608041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull S., Owen N., Islam F., Tracey-White D., Plagnol V., Holder G. E., Michaelides M., Carss K., Raymond F. L., Rozet J.-M. et al. (2016). Nonsyndromic retinal dystrophy due to bi-allelic mutations in the ciliary transport gene IFT140. Invest. Ophthalmol. Vis. Sci. 57, 1053-1062. 10.1167/iovs.15-17976 [DOI] [PubMed] [Google Scholar]
- Humbert M. C., Weihbrecht K., Searby C. C., Li Y., Pope R. M., Sheffield V. C. and Seo S. (2012). ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA 109, 19691-19696. 10.1073/pnas.1210916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Babaev-Khaimov V., Sassaroli M. and Piperno G. (2001). Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 153, 13-24. 10.1083/jcb.153.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Li L., Esparza J. M. and Dutcher S. K. (2009). Retrograde intraflagellar transport mutants identify complex a proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics 183, 885-896. 10.1534/genetics.109.101915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. and Marshall W. F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222-234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Ivanova A. A., Caspary T., Seyfried N. T., Duong D. M., West A. B., Liu Z. and Kahn R. A. (2017). Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J. Biol. Chem. 292, 11091-11108. 10.1074/jbc.M117.784025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Hernandez D., Hernandez C., Ding Z., Nan B., Aufderheide K. and Qin H. (2017). IFT57 stabilizes the assembled intraflagellar transport complex and mediates transport of motility-related flagellar cargo. J. Cell Sci. 130, 879-891. 10.1242/jcs.199117 [DOI] [PubMed] [Google Scholar]
- Jonassen J. A., San Agustin J., Follit J. A. and Pazour G. J. (2008). Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J. Cell Biol. 183, 377-384. 10.1083/jcb.200808137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen J. A., SanAgustin J., Baker S. P. and Pazour G. J. (2012). Disruption of IFT complex a causes cystic kidneys without mitotic spindle misorientation. J. Am. Soc. Nephrol. 23, 641-651. 10.1681/ASN.2011080829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Diener D. R., Stepanek L. and Pigino G. (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250-1255. 10.1038/s41556-018-0213-1 [DOI] [PubMed] [Google Scholar]
- Kee H. L., Dishinger J. F., Blasius T. L., Liu C.-J., Margolis B. and Verhey K. J. (2012). A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14, 431-437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. O., Bolz H. J. and Bergmann C. (2014). Early-onset severe retinal dystrophy as the initial presentation of IFT140-related skeletal ciliopathy. J. AAPOS 18, 203-205. 10.1016/j.jaapos.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Kösling S. K., Fansa E. K., Maffini S. and Wittinghofer A. (2018). Mechanism and dynamics of INPP5E transport into and inside the ciliary compartment. Biol. Chem. 399, 277 10.1515/hsz-2017-0226 [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P. and Rosenbaum J. L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519-5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Brown J. M., Bellve K., Craige B., Craft J. M., Fogarty K., Lechtreck K. F. and Witman G. B. (2016). Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J. Cell Sci. 129, 2106-2119. 10.1242/jcs.187120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. K., Gupta N., Wen X., Rangell L., Chih B., Peterson A. S., Bazan J. F., Li L. and Scales S. J. (2011). Functional characterization of putative cilia genes by high-content analysis. Mol. Biol. Cell 22, 1104-1119. 10.1091/mbc.e10-07-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins C. E., Aviles G. D. G., East M. P., Kahn R. A. and Caspary T. (2011). Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol. Biol. Cell 22, 4694-4703. 10.1091/mbc.e10-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K. F. (2015). IFT–cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40, 765-778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K.-F., Johnson E. C., Sakai T., Cochran D., Ballif B. A., Rush J., Pazour G. J., Ikebe M. and Witman G. B. (2009). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117-1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck K. F., Brown J. M., Sampaio J. L., Craft J. M., Shevchenko A., Evans J. E. and Witman G. B. (2013). Cycling of the signaling protein Phospholipase D through cilia requires the BBSome only for the export phase. J. Cell Biol. 201, 249-261. 10.1083/jcb.201207139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Sivan-Loukianova E., Eberl D. F. and Kernan M. J. (2008). An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr. Biol. 18, 1899-1906. 10.1016/j.cub.2008.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wei Q., Zhang Y., Ling K. and Hu J. (2010). The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 189, 1039-1051. 10.1083/jcb.200912001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ling K. and Hu J. (2012). The emerging role of Arf/Arl small gtpases in cilia and ciliopathies. J. Cell. Biochem. 113, 2201-2207. 10.1002/jcb.24116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y. and Pan J. (2013). Regulation of flagellar biogenesis by a calcium dependent protein kinase in Chlamydomonas reinhardtii. PLoS ONE 8, e69902 10.1371/journal.pone.0069902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Pang Y., Wu Q., Hu Z., Han X., Xu Y., Deng H. and Pan J. (2014). FLA8/KIF3B phosphorylation regulates Kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell 30, 585-597. 10.1016/j.devcel.2014.07.019 [DOI] [PubMed] [Google Scholar]
- Liem K. F. Jr, Ashe A., He M., Satir P., Moran J., Beier D., Wicking C. and Anderson K. V. (2012). The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J. Cell Biol. 197, 789-800. 10.1083/jcb.201110049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Guo S., and Dutcher S. K. (2018). RPGRIP1L helps to establish the ciliary gate for entry of proteins. J. Cell Sci. 131, jcs220905 10.1242/jcs.220905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Ruba A., Takao D., Zweifel L. P., Lim R. Y. H., Verhey K. J. and Yang W. (2017). Axonemal lumen dominates cytosolic protein diffusion inside the primary cilium. Sci. Rep. 7, 15793 10.1038/s41598-017-16103-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B. F., Pedersen L. B., Feely M., Rosenbaum J. L. and Mitchell D. R. (2005). ATP production in Chlamydomonas reinhardtii flagella by glycolytic enzymes. Mol. Biol. Cell. 16, 4509-4518. 10.1091/mbc.E05-04-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C. D., Lane W. S., Scales S. J. and Jackson P. K. (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Gene Dev. 24, 2180-2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S. J. and Jackson P. K. (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the sonic hedgehog pathway via camp signaling. Cell 152, 210-223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C. et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201-1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nelson J. A., Savereide P. B. and Lefebvre P. A. (1994). The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell. Biol. 14, 4011-4019. 10.1128/MCB.14.6.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S., Katoh Y., Terada M., Michisaka S., Funabashi T., Takahashi S., Kontani K. and Nakayama K. (2017). Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J. Cell Sci. 130, 563-576. 10.1242/jcs.197004 [DOI] [PubMed] [Google Scholar]
- Pablo J. L., DeCaen P. G. and Clapham D. E. (2017). Progress in ciliary ion channel physiology. J. Gen. Physiol. 149, 37-47. 10.1085/jgp.201611696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L. and Witman G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Baker S. A., Deane J. A., Cole D. G., Dickert B. L., Rosenbaum J. L., Witman G. B. and Besharse J. C. (2002). The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 157, 103-114. 10.1083/jcb.200107108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J. and Witman G. B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103-113. 10.1083/jcb.200504008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. B., Geimer S., Sloboda R. D. and Rosenbaum J. L. (2003). The microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 13, 1969-1974. 10.1016/j.cub.2003.10.058 [DOI] [PubMed] [Google Scholar]
- Pedersen L. B., Geimer S. and Rosenbaum J. L. (2006). Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr. Biol. 16, 450-459. 10.1016/j.cub.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Perrault I., Saunier S., Hanein S., Filhol E., Bizet A., Collins F., Salih M., Silva E., Baudouin V., Oud M. et al. (2012). Mainzer-Saldino syndrome is a ciliopathy caused by mutations in the IFT140 gene. Cilia 1, O28 10.1186/2046-2530-1-S1-O28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picariello T., Brown J. M., Swank G., Cochran D. A., Pazour G. J. and Witman G. B. (2015). IFT140 is required for anterograde IFT and flagellar formation in Chlamydomonas p. 4523 (Abstract #P153).
- Piperno G., Siuda E., Henderson S., Segil M., Vaananen H. and Sassaroli M. (1998). Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol 143, 1591-1601. 10.1083/jcb.143.6.1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Diener D. R., Geimer S., Cole D. G. and Rosenbaum J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Lin Y., Norman R. X., Ko H. W. and Eggenschwiler J. T. (2011). Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. USA 108, 1456-1461. 10.1073/pnas.1011410108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J. L., Moulder J. E. and Ringo D. L. (1969). Flagellar elongation and shortening in Chlamydomonas: the use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41, 600-619. 10.1083/jcb.41.2.600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidel N. and Blacque O. E. (2018). Intraflagellar transport complex A genes differentially regulate cilium formation and transition zone gating. Curr. Biol. 28, 3279-3287.e2. 10.1016/j.cub.2018.08.017 [DOI] [PubMed] [Google Scholar]
- Schmidts M., Frank V., Eisenberger T., Al Turki S., Bizet A. A., Antony D., Rix S., Decker C., Bachmann N., Bald M. et al. (2013). Combined NGS approaches identify mutations in the intraflagellar transport gene IFT140 in skeletal ciliopathies with early progressive kidney disease. Hum. Mutat. 34, 714-724. 10.1002/humu.22294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrøder J. M., Schneider L., Christensen S. T. and Pedersen L. B. (2007). EB1 Is required for primary cilia assembly in fibroblasts. Curr. Biol. 17, 1134-1139. 10.1016/j.cub.2007.05.055 [DOI] [PubMed] [Google Scholar]
- Schrøder J. M., Larsen J., Komarova Y., Akhmanova A., Thorsteinsson R. I., Grigoriev I., Manguso R., Christensen S. T., Pedersen S. F., Geimer S. et al. (2011). EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J. Cell Sci. 124, 2539-2551. 10.1242/jcs.085852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W. and Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337-342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Schwarz N., Lane A., Jovanovic K., Parfitt D. A., Aguila M., Thompson C. L., da Cruz L., Coffey P. J., Chapple J. P., Hardcastle A. J. et al. (2017). Arl3 and RP2 regulate the trafficking of ciliary tip kinesins. Hum. Mol. Genet. 26, 2480-2492. 10.1093/hmg/ddx143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S. M., Engel B. D., Kocabas F., Bilyard T., Gennerich A., Marshall W. F. and Yildiz A. (2013). Intraflagellar transport drives flagellar surface motility. eLife Sci. 2, e00744 10.7554/eLife.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Gaitatzes C., Saxena K. and Neer E. J. (1999). The WD repeat: a common architecture for diverse functions. Trends Biol. Sci. 24, 181-185. 10.1016/S0968-0004(99)01384-5 [DOI] [PubMed] [Google Scholar]
- Song L. and Dentler W. L. (2001). Flagellar protein dynamics in Chlamydomonas. J. Biol. Chem. 276, 29754-29763. 10.1074/jbc.M103184200 [DOI] [PubMed] [Google Scholar]
- Stephen L. A., Elmaghloob Y. and Ismail S. (2018). Maintaining protein composition in cilia. Biol. Chem. 399, 1 10.1515/hsz-2017-0168 [DOI] [PubMed] [Google Scholar]
- Sun X., Haley J., Bulgakov O. V., Cai X., McGinnis J. and Li T. (2012). Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia 1, 21-21 10.1186/2046-2530-1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Bhogaraju S. and Lorentzen E. (2012). Architecture and function of IFT complex proteins in ciliogenesis. Differentiation 83, S12-S22. 10.1016/j.diff.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K., Mladenov M. and Roberts A. J. (2017). Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding Elements. Nat. Struct. Mol. Biol. 24, 461-468. 10.1038/nsmb.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P. V., Haycraft C. J., Besschetnova T. Y., Turbe-Doan A., Stottmann R. W., Herron B. J., Chesebro A. L., Qiu H., Scherz P. J., Shah J. V. et al. (2008). THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 40, 403-410. 10.1038/ng.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L., Lau C. K., Elshenawy M. M., Morales-Rios E., Motz C., Yildiz A. and Carter A. P. (2018). Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202-206. 10.1038/nature25462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H. H., Brown-Glaberman U., Wang J., Morita Y., Alper S. L., Bedrick E. J., Gattone V. H., Deretic D. and Wandinger-Ness A. (2011). A conserved signal and GTPase complex are required for the ciliary transport of polycystin-1. Mol. Biol. Cell 22, 3289-3305. 10.1091/mbc.e11-01-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson S. M., Silva D. A., Richey E. and Qin H. (2012). Probing the role of IFT particle complex A and B in flagellar entry and exit of IFT-dynein in Chlamydomonas. Protoplasma 249, 851-856. 10.1007/s00709-011-0311-4 [DOI] [PubMed] [Google Scholar]
- Witman G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Meth. Enzymol. 134, 280-290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Wood C. R., Wang Z., Diener D., Zones J. M., Rosenbaum J. and Umen J. G. (2012). IFT proteins accumulate during cell division and localize to the cleavage furrow in Chlamydomonas. PLoS ONE 7, e30729 10.1371/journal.pone.0030729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren K. N., Craft J. M., Tritschler D., Schauer A., Patel D. K., Smith E. F., Porter M. E., Kner P. and Lechtreck K. F. (2013). A differential cargo-loading model of ciliary length regulation by IFT. Curr. Biol. 23, 2463-2471. 10.1016/j.cub.2013.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang Y., Sun B., McMahon A. P. and Wang Y. (2017). Hedgehog signaling: from basic biology to cancer therapy. Cell Chem. Biol. 24, 252-280. 10.1016/j.chembiol.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zheng Y., Li H., Luo X., He Z., Cao S., Shi Y., Zhao Q., Xue Y., Zuo Z. et al. (2016). GPS-Lipid: a robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 6, 28249 10.1038/srep28249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Yang L., Wang F., Li H., Wang X., Wang W., Ge Z., Wang K., Zhao L., Li H. et al. (2015). Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum. Genet. 134, 1069-1078. 10.1007/s00439-015-1586-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F., Nager A. R. and Nachury M. V. (2018). BBSome trains remove activated GPCRs from cilia by enabling passage through the transition zone. J. Cell Biol. 217, 1847-1868. 10.1083/jcb.201709041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz O. and Khanna H. (2012). CIliary signaling cascades in photoreceptors. Vision Res. 75, 112-116. 10.1016/j.visres.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N. and Zarivach R. (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397-405. 10.1016/j.str.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Zhang D. and Lefebvre P. A. (1997). Far1, a negative regulatory locus required for the repression of the nitrate reductase gene in Chlamydomonas reinhardtii. Genetics 146, 121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Taylor S. P., Nevarez L., Lachman R. S., Nickerson D. A., Bamshad M., University of Washington Center for Mendelian Genomics Consortium, Krakow D. and Cohn D. H. (2017). IFT52 mutations destabilize anterograde complex assembly, disrupt ciliogenesis and result in short rib polydactyly syndrome. Hum. Mol. Genet. 25, 4012-4020. 10.1093/hmg/ddw241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Zhu X., Wang L., Liang Y., Feng Q. and Pan J. (2017). Functional exploration of the IFT-A complex in intraflagellar transport and ciliogenesis. PLoS Genet. 13, e1006627 10.1371/journal.pgen.1006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.