Abstract
Objectives
We evaluated the performance and time to result for pathogen identification (ID) and antimicrobial susceptibility testing (AST) of the Accelerate Pheno™ system (AXDX) compared with standard of care (SOC) methods. We also assessed the hypothetical improvement in antibiotic utilization if AXDX had been implemented.
Methods
Clinical samples from patients with monomicrobial Gram-negative bacteraemia were tested and compared between AXDX and the SOC methods of the VERIGENE® and Bruker MALDI Biotyper® systems for ID and the VITEK® 2 system for AST. Additionally, charts were reviewed to calculate theoretical times to antibiotic de-escalation, escalation and active and optimal therapy
Results
ID mean time was 21 h for MALDI-TOF MS, 4.4 h for VERIGENE® and 3.7 h for AXDX. AST mean time was 35 h for VITEK® 2 and 9.0 h for AXDX. For ID, positive percentage agreement was 95.9% and negative percentage agreement was 99.9%. For AST, essential agreement was 94.5% and categorical agreement was 93.5%. If AXDX results had been available to inform patient care, 25% of patients could have been put on active therapy sooner, while 78% of patients who had therapy optimized during hospitalization could have had therapy optimized sooner. Additionally, AXDX could have reduced time to de-escalation (16 versus 31 h) and escalation (19 versus 31 h) compared with SOC.
Conclusions
By providing fast and reliable ID and AST results, AXDX has the potential to improve antimicrobial utilization and enhance antimicrobial stewardship.
Introduction
Given that mortality increases with each hour of delayed treatment, early recognition of sepsis and initiation of targeted antibiotic therapy for patients with Gram-negative rod (GNR) bacteraemia is crucial for optimal outcomes.1,2 Moreover, antimicrobial resistance in GNRs is known to be complex.3 This results in limited therapeutic options, which further increases the risk of morbidity and prolonged hospitalization, as well as mortality.1,4
Thus, there is a critical need for rapid and reliable diagnostics that facilitate the timely selection of appropriate antimicrobials. Integrating results from rapid laboratory methods for organism identification (ID) and antimicrobial susceptibility testing (AST), together with support from an antimicrobial stewardship programme, permits clinicians to streamline therapy decisions.3,5–7 This allows timely provision of life-saving treatment, along with de-escalation from broad-spectrum therapy,7 which can lead to decreased length of hospital stay, reduced hospital costs and improved patient outcomes.1,4,8–12
The Accelerate Pheno™ system (AXDX) is a new technology that quickly identifies the most common organisms in bloodstream infections (BSIs) using fluorescence in situ hybridization and morphokinetic cellular analysis to provide fast AST results.13 This technology has proven to be a useful tool to provide rapid and reliable ID and AST results,14–22 but questions remain regarding its potential clinical impact. Unlike other rapid diagnostics, AXDX provides phenotypic AST results including MICs, a format familiar to clinicians,23 potentially prompting antibiotic de-escalation ahead of an active stewardship intervention.
Our primary study objective was to compare the ID/AST performance and time to result (TTR) of AXDX with current laboratory standard of care (SOC) methods. Secondarily, we assessed the relationship between TTR and the projected times to antibiotic escalation, de-escalation and active and optimal therapy when using AXDX results for patients with GNR bacteraemia.
Materials and methods
Study design and population
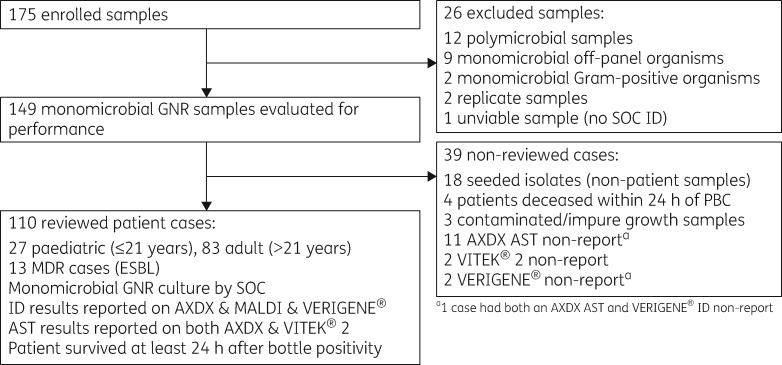
A prospective study evaluating ID/AST performance and TTR of AXDX compared with current SOC for samples of GNR bacteraemia was conducted at Indiana University Health Pathology Laboratory (Indianapolis, IN, USA). Additionally, an evaluation of whether the implementation of AXDX could theoretically lead to improvements in times to antibiotic escalation and de-escalation, as well as times to active and optimal therapy, was retrospectively assessed by clinical review. Patients seen at any of the Indiana University Health Hospitals, including Riley Hospital for Children, from September to December 2017 were eligible for inclusion if they had a monomicrobial GNR-positive blood culture (PBC). Exclusion criteria for the performance evaluation were polymicrobial samples, samples with off-panel organisms, or samples from patients with recurrent bacteraemia within 30 days. For the theoretical clinical review, exclusion criteria removed samples with an off-panel organism, contaminated/impure growth, presence of a concurrent infection site that showed at least one organism not isolated from blood, polymicrobial samples or samples from patients who died within 24 h of PBC (Figure 1).
Figure 1.
Flow diagram of study design and population inclusivity criteria.
Pathogen ID, AST and TTR
Patient blood samples were collected using aerobic, anaerobic or paediatric blood culture bottles and incubated in the BACTEC® FX blood culture monitoring system. Gram staining was performed after a blood culture bottle flagged positive, followed by sub-culturing to appropriate solid media based on Gram stain results. The AXDX system (Accelerate PhenoTest™ BC kit and software version 1.3.1.15) was run in parallel with SOC methods for pathogen ID [VERIGENE® (Luminex Corporation, Northbrook, IL, USA) and MALDI-TOF MS using the MALDI Biotyper® system (Bruker Daltonics, Fremont, CA, USA)] and AST [VITEK® 2 system: GN73 cards (bioMérieux, Durham, NC, USA)]. All potential ESBL-producing isolates were initially flagged on the VITEK® 2 system, followed by confirmatory testing using the CLSI ESBL disc diffusion test from isolated colonies.24 VERIGENE® results were reviewed for each confirmed ESBL-producing isolate, specifically for the presence of the CTX-M resistance gene. Timing calculations included adjustments to account for delays in setup and reporting times between SOC and AXDX.
Theoretical clinical data and outcomes
Demographic and clinical data were collected on eligible patients (Figure 1). Charts were retrospectively reviewed by an infectious diseases clinician or pharmacist to assess potential timing of changes in antimicrobial interventions that could have been made if AXDX AST results had been available. These included escalating therapy when appropriate, starting active or optimal therapy, and de-escalating unnecessary therapy. The review included adjudicated ID and AST results from the performance evaluation portion of the study and was conducted under the assumption that testing with AXDX was completed and results were acted on by the antimicrobial stewardship team 24 h/day. One hour was added to the AXDX AST report time to account for technologist review and reporting time. Patients with antimicrobial intervention occurring after the theoretical AXDX AST report time were assumed to have had that intervention time reduced by the difference between the theoretical AXDX AST report time and the actual SOC AST report time. If an intervention time was eligible for a reduction due to earlier AXDX AST, the reduction was limited such that the intervention would not occur sooner than the theoretical AXDX AST report time for that patient. Cases that did not fall within evidence-based antimicrobial stewardship programme guidelines5 were adjudicated by an infectious diseases physician. Active therapy was defined as the first antimicrobial dose to which the organism was susceptible by conventional antimicrobial testing. Optimal therapy was defined as the earliest optimal (‘institution preferred’) dose of antimicrobial from time of blood culture positivity. Times to active and optimal therapy were compared with times when AXDX ID and AST results were available. Escalation therapy was conversion to an antimicrobial with a broader spectrum of activity while de-escalation was conversion to an antimicrobial with a narrower spectrum of activity.
Statistical analysis
Positive percentage agreement (PPA) and negative percentage agreement (NPA) between AXDX and SOC ID results were calculated for on-panel target organisms. Essential agreement (EA), categorical agreement (CA), major errors (ME) and very major errors (VMEs) were also calculated between AXDX and SOC AST results. CA was defined as susceptible, intermediate or resistant results that matched between AXDX and SOC based on 2016 MIC breakpoints set by CLSI.24 EA was defined as agreement of MICs within ±1 doubling dilution between AXDX and SOC. A VME occurs when a sample is called resistant by SOC and susceptible by AXDX. An ME occurs when a sample is called susceptible by SOC and resistant by AXDX. Samples with VMEs or MEs were adjudicated by performing broth microdilution (BMD), which is the gold standard method for AST.
Statistical analyses were performed with SciPy,25 using the Python programming language.26 Statistical analyses comparing independent quantitative variables (e.g. TTR of patient results compared between AXDX and SOC methods) were performed using the two-sided Mann–Whitney U-test.27 Computed probability less than the significance level (α) of 0.05 was considered statistically significant. Where relevant, mean (μ) and standard deviation (σ) are noted.
Results
AXDX performance evaluation
After excluding 26 samples, 149 samples, including 18 seeded isolates (non-patient samples), 32 fresh paediatric (age ≤21 years) samples and 99 fresh adult (age >21 years) samples from patients with monomicrobial GNR bacteraemia, were available for performance evaluation (Figure 1). PPA was 95.9% and NPA was 99.9% for ID (Table 1). There were six false-negatives (three Enterobacter spp., one Proteus spp., one Citrobacter spp. and one Pseudomonas aeruginosa). The VERIGENE® system did not detect Enterobacter gergoviae or Klebsiella variicola in two different samples, which were detected by AXDX (identified as Enterobacter spp. and Klebsiella spp., respectively). Following discrepancy adjudication testing via BMD, EA was 94.5% and CA was 93.5%. SOC in AST was predominantly the VITEK® 2 system with confirmatory Etests for two cases (Table 2). Eleven percent (16/149) of samples were found to be ESBL producers. AXDX had an overall agreement rate of 93.7% for both EA and CA when comparing AST from the VITEK® 2 system with these specific isolates (data not shown).
Table 1.
ID performance of AXDX versus MALDI-TOF MS and VERIGENE® system
| Microbe | PPAa | NPAb |
|---|---|---|
| Escherichia coli | 64/64 (100%) | 83/83 (100%) |
| Klebsiella spp. | 32/32 (100%) | 115/116 (99.1%) |
| Enterobacter spp. | 16/19 (84.2%) | 130/130 (100%) |
| Proteus spp. | 4/5 (80%) | 144/144 (100%) |
| Citrobacter spp. | 4/5 (80%) | 144/144 (100%) |
| Serratia marcescens | 2/2 (100%) | 147/147 (100%) |
| Acinetobacter baumannii | 4/4 (100%) | 145/145 (100%) |
| P. aeruginosa | 16/17 (94.1%) | 131/131 (100%) |
| Total | 142/148 (95.9%) | 1039/1040 (99.9%) |
One indeterminate result (E. coli) was excluded from PPA calculation.
Three indeterminate results (one each for E. coli, Klebsiella spp. and P. aeruginosa) were excluded from NPA calculation.
Table 2.
AST performance of AXDX compared with standard methodsa, following adjudication
| Antibiotic | EA | CA | VME | ME | S | I | R |
|---|---|---|---|---|---|---|---|
| Ampicillin/sulbactam | 84/93 (90.3%) | 74/93 (79.6%) | 0 | 1 | 47 | 14 | 32 |
| Piperacillin/tazobactam | 94/103 (91.3%) | 97/103 (94.2%) | 1 | 0 | 86 | 2 | 15 |
| Cefepime | 122/134 (91.0%) | 117/134 (87.3%) | 0 | 1 | 105 | 7 | 22 |
| Ceftazidime | 113/132 (85.6%) | 111/132 (84.1%) | 0 | 2 | 97 | 3 | 32 |
| Ceftriaxone | 115/115 (100%) | 113/115 (98.3%) | 0 | 0 | 81 | 1 | 33 |
| Meropenem | 123/128 (96.1%) | 123/128 (96.1%) | 0 | 2 | 114 | 0 | 14 |
| Amikacin | 125/127 (98.4%) | 126/127 (99.2%) | 0 | 0 | 121 | 1 | 5 |
| Gentamicin | 125/131 (95.4%) | 130/131 (99.2%) | 0 | 0 | 111 | 1 | 19 |
| Tobramycin | 126/131 (96.2%) | 125/131 (95.4%) | 0 | 0 | 108 | 7 | 16 |
| Ciprofloxacin | 131/132 (99.2%) | 130/132 (98.5%) | 0 | 0 | 93 | 1 | 38 |
| Aztreonam | 1/1 (100%) | 1/1 (100%) | 0 | 0 | 0 | 0 | 1 |
| Total | 1159/1227 (94.5%) | 1147/1227 (93.5%) | 1 | 6 | 963 | 37 | 227 |
VITEK® 2 (n=147) and Etest (n=2).
Based on discrepancy adjudication via BMD, three initial AXDX VMEs were adjudicated to one VME against AXDX for piperacillin/tazobactam (Table 2), and one ME and one minor error against VITEK® 2 for ampicillin/sulbactam and cefepime, respectively. Sixteen initial AXDX MEs were adjudicated to six MEs against AXDX (Table 2) and five VMEs against VITEK® 2 (including two for ceftazidime and two for cefepime with ESBL Escherichia coli and one for aztreonam). This indicates that AXDX tended to overcall resistance while the VITEK® 2 system tended to overcall susceptibility for the discrepancy-adjudicated combinations. The remaining five initial AXDX MEs were adjudicated to minor errors (two for ampicillin/sulbactam, two for cefepime and one for ceftazidime).
Timing/workflow results
Despite highly variable organism growth time from collection to positive culture (Figure S1, available as Supplementary data at JAC Online), AXDX had a mean instrument run time of 1.3 h for ID from time of set-up, which was consistent among runs (σ=0.01 h), compared with 2.0 h for the VERIGENE® system, which was also consistent among runs (σ=0.4 h) (Table 3). Mean time for MALDI-TOF MS confirmatory testing was 21 h from time of positivity and was variable (σ=7.2 h). AXDX required a mean time of 6.6 h from time of set-up and 9.0 h from time of positivity for AST, compared with 9.2 and 35 h, respectively, for the VITEK® 2 system. (Table 3). AXDX instrument run time for AST was also consistent (σ=0.05 h) compared with the VITEK® 2 system (σ=1.4 h). AXDX decreased the initial time required to prepare PBCs with GNRs from up to 30 min to nearly 3 min, with the potential to save ∼45 min in total technologist time required for SOC methods.
Table 3.
Mean time to assay result by method (n=110)
| Assay | Method | Instrument run time (h)a | Time from positivity (h)a |
|---|---|---|---|
| ID | VERIGENE® | 2.0±0.4 | 4.4±1.7 |
| MALDI-TOF MS | NA | 21±7.2 | |
| AXDX | 1.3±0.01 | 3.7±1.7 | |
| AST | VITEK® 2 | 9.2±1.4 | 35±7.7 |
| AXDX | 6.6±0.05 | 9.0±1.7 |
NA, not applicable.
Times presented are mean (μ)±standard deviation (σ). n=110 for all results, P<0.001 between timing results for all listed methods in each assay group.
Microbiology results
A total of 149 pathogens were isolated, with E. coli and Klebsiella spp. being the most frequently isolated GNRs. As indicated earlier, 11% (16/149) of the isolates were ESBL producers. Of these 16, 1 (6.3%) did not carry the CTX-M resistance gene marker. Nine organisms not present on the VERIGENE® and AXDX ID panels made up 5.5% of total monomicrobial GNR cultures tested (prior to their exclusion in the analysed dataset) and included one each of the following organisms: Bacteroides fragilis, Salmonella spp., Shewanella spp., Dialister spp., Bacteroides thetaiotaomicron, Burkholderia gladioli, Pantoea calida, Achromobacter xylosoxidans and Aeromonas spp.
Patient characteristics
Following exclusions, 110 monomicrobial GNR patient samples, including 27 fresh paediatric (age ≤21 years) and 83 fresh adult (age >21 years) samples, were available for chart review (Figure 1). Demographic and other characteristics for these patients are summarized in Table 4. The sources of bacteraemia were most commonly genitourinary (38.2%), intra-abdominal (25.5%) and central vascular catheters (14.5%), followed by skin/soft tissue (3.6%), respiratory (2.7%), bone/joint (1.8%), cardiac (1.8%) and other sources (0.9%). Additionally, 10.9% of sources were unidentified.
Table 4.
Characteristics of case-reviewed patient populations
| Category | Description | Total cases (n = 110) | Paediatric cases (n = 27) | Adult cases (n = 83) |
|---|---|---|---|---|
| Pathogen | E. coli | 56 (51%) | 8 (30%) | 48 (58%) |
| Klebsiella spp. | 26 (24%) | 10 (37%) | 16 (19%) | |
| Enterobacter spp. | 11 (10%) | 6 (22%) | 5 (6%) | |
| P. aeruginosa | 9 (8%) | 3 (11%) | 6 (7%) | |
| Acinetobacter baumannii | 3 (3%) | – | 3 (4%) | |
| Citrobacter spp. | 3 (3%) | – | 3 (4%) | |
| Proteus spp. | 2 (2%) | – | 2 (2%) | |
| Gender | female | 50 (45%) | 13 (48%) | 37 (45%) |
| male | 60 (55%) | 14 (52%) | 46 (55%) | |
| Age | 1 month <1 year | 5 (5%) | 5 (19%) | – |
| ≥1 year ≤18 years | 19 (17%) | 19 (70%) | – | |
| >18 years ≤21 years | 3 (3%) | 3 (11%) | – | |
| >21 years <50 years | 14 (13%) | – | 14 (17%) | |
| ≥50 years <60 years | 21 (19%) | – | 21 (25%) | |
| ≥60 years <70 years | 20 (18%) | – | 20 (24%) | |
| ≥70 years <80 years | 18 (16%) | – | 18 (22%) | |
| ≥80 years | 10 (9%) | – | 10 (12%) | |
| Admission service | haematology/oncology | 13 (12%) | 8 (30%) | 5 (6%) |
| critical care | 10 (9%) | 2 (7%) | 8 (10%) | |
| transplant | 14 (13%) | 5 (19%) | 9 (11%) | |
| general wards | 73 (66%) | 12 (44%) | 61 (73%) | |
| Intensive care | ICU visit during therapy | 36 (33%) | 7 (26%) | 29 (35%) |
| no ICU visit | 74 (67%) | 20 (74%) | 54 (65%) |
Theoretical clinical data
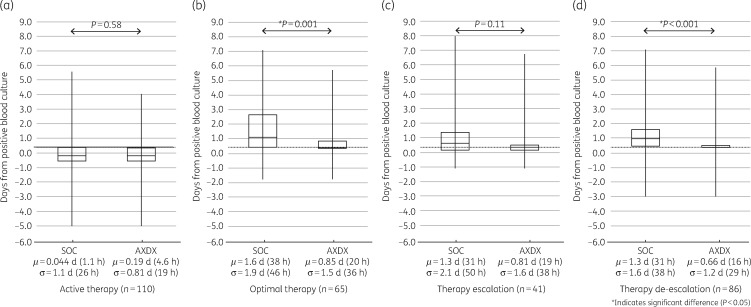
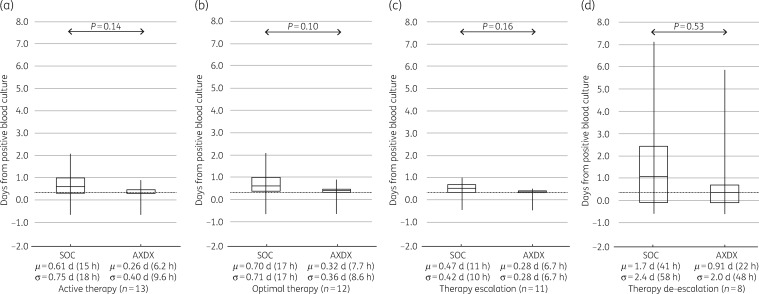
AXDX-derived AST results (minimum of 9.0 h after positivity) for all cases showed a theoretical mean time to de-escalation from time of positive culture of 0.66 days (16 h) compared with 1.3 days (31 h) for current SOC (P < 0.001) (Figure 2d). For cases involving ESBL isolates, theoretical mean time to de-escalation was 0.91 days (22 h) using AXDX results compared with 1.7 days (41 h) for SOC (P=0.53) (Figure 3d). With respect to theoretical escalation events from time of blood culture positivity, patients could have been escalated on average by 0.81 days (19 h) using AXDX results compared with 1.3 days (31 h) for SOC (P=0.11) (Figure 2c). For ESBL cases, mean AXDX escalation was 0.28 days (6.7 h) compared with 0.47 days (11 h) for SOC methods (P=0.16) (Figure 3c).
Figure 2.
All-patient therapy intervention times. SOC compared with theoretical AXDX for (a) active therapy, (b) optimal therapy when possible, (c) first escalation of therapy when required and (d) first de-escalation of therapy when possible. Box plots display median and IQR with tails indicating the minimum and maximum of observed values, and the notes below show the mean (μ) and standard deviation (σ). The grey dotted line represents mean AXDX AST time (9.0 h) for reference.
Figure 3.
ESBL-patient therapy intervention times. SOC compared with theoretical AXDX for (a) active therapy, (b) optimal therapy when possible, (c) first escalation of therapy when required and (d) first de-escalation of therapy when possible. Box plots display median and IQR with tails indicating the minimum and maximum of observed values, and the notes below show the mean (μ) and standard deviation (σ). The grey dotted line represents mean AXDX AST time (9.0 h) for reference.
Despite current SOC reporting (call to clinician after Gram stain), 25% (28/110) of the patients were not on active therapy at the time AXDX AST results were theoretically available (minimum 9.0 h after positivity), and therefore could have theoretically been acted upon sooner. Overall, mean time to active therapy with current SOC was 0.04 days (1 h), where AXDX could have been 0.19 days (4.6 h, P=0.58) (Figure 2a). Theoretical mean time to active therapy for ESBL isolates was 0.26 days (6 h) using AXDX results compared with 0.61 days (15 h) for SOC (P=0.14) (Figure 3a).
Fifty-nine percent of reviewed cases (65/110) had therapy optimized during hospitalization, and of these 78% (51/65) could have theoretically been put on optimal therapy sooner with AXDX AST results. Mean time to optimize therapy with current SOC was 1.6 days (38 h), where AXDX could have been 0.85 days (20 h, P=0.001) (Figure 2b). For ESBL isolates, theoretical mean time to optimization was 0.32 days (7.7 h) using AXDX results compared with 0.7 days (17 h) for SOC (P=0.10) (Figure 3b).
To evaluate potential clinical benefits of rapid AST results, a preliminary simple linear regression analysis was performed in the paediatric and adult populations to determine whether faster optimization of antimicrobial therapy (within 3 days of PBC) could lead to an overall decrease in total days of therapy. In the paediatric population, each day of reduction in the time to optimal patient therapy could correlate to a 2.0 day reduction in hospital-administered therapy time across all paediatric patients, and a 3.4 day reduction for patients on a general ward (Figure S2a). The adult population showed substantial variation in time to therapy optimization, ranging from ∼1 day prior to 7 days after PBC. The same linear regression across this population showed a limited effect of therapy optimization, with only a 0.7 day reduction across all adult patients. When focusing on adult patients with lower Charlson comorbidity index (CCI) scores, however, simple regression showed a 1.9 day reduction in total therapy for CCI ≤2 in this population (Figure S2b).
Discussion
The major findings of this study are that AXDX has the potential to significantly reduce time to pathogen ID and AST results for monomicrobial BSIs compared with SOC, and improve times to antibiotic de-escalation and active and optimal therapy.
Multiple studies have verified the accuracy and timing by which AXDX provides pathogen ID and AST;14–22 however, there are few data on the potential impact this novel assay has on clinical outcomes. Thus, our findings in this earliest cohort of investigations demonstrate the hypothetical impact of AXDX results on time to active and optimal therapy. Additionally, our results show the potential utility of AXDX to enhance antimicrobial stewardship by promoting faster antimicrobial de-escalation, which has been associated with improved clinical outcomes.28,29
Given the antimicrobial resistance crisis, there is a critical need for rapid and reliable diagnostics that permit the timely selection of antimicrobial therapy and enhanced antibiotic stewardship. During the empirical treatment period, studies show that many patients with both community- and healthcare-acquired bacteraemia often receive incorrect, inadequate or excessively broad-spectrum therapy.11,30–32 Inappropriate and widespread use of broad-spectrum antimicrobials not only contributes to patients enduring drug toxicity, increased lengths of stay, secondary infections and additional costs, but also increases the frequency of antimicrobial-resistant organisms.1,31,33 Poorly targeted therapy for a BSI may also increase a patient’s risk of developing sepsis after the primary hospitalization.34
Our results showed that AXDX has the potential to enhance the capacity for antimicrobial de-escalation, compared with current standard methods (16 compared with 31 h, P < 0.001). However, for more complex cases involving confirmed ESBL isolates, this effect did not reach significance (22 compared with 41 h, P=0.53). These findings may be explained by the low sample size (n=8), or by the fact that more-resistant organisms generally require escalation, coupled with the assumption that physicians may be more hesitant to de-escalate until clinical improvement is observed. Conversely, rapid AST was less likely to affect an escalation event across the general patient population than for patients with an ESBL-producing organism.
At the time of PBC detection, our study showed that 25% of patients could have been put on active therapy sooner had the AXDX AST results been made available. Interestingly, rapid AST did not have a statistically significant effect on active therapy across all patients (P=0.58). But for more resistant cases (i.e. ESBLs), there was a notable increase in the average time to active therapy (1.1 h for all patients compared with 15 h for ESBLs). Additionally, there was a notable decrease in variability of the AXDX results: σ=19 h for all patients compared with 9.6 h for ESBLs. While these results are based on a low sample size, the increased average time suggests that empirical therapy is less likely to be active against resistant organisms, and the reduced AXDX variability implies that rapid and consistently timed AST results such as those produced by AXDX would be useful when selecting an appropriate active treatment for resistant organisms.
For cases of non-ESBL and ESBL isolates, rapid AST would significantly improve time to optimal therapy. Our clinical review revealed that 78% of patients who received optimized therapy could have had therapy optimized earlier had AXDX AST results been available. Additionally, we found that each day required to optimize patient therapy in the paediatric population could correlate to a reduction of 2.0 and 3.4 days in hospital-administered therapy across all paediatric patients and in general ward paediatric patients, respectively. For adults, the effect of optimization and reduction in total days of antimicrobial therapy was only shown in less critically ill patients (CCI ≤2). Future prospective studies on more detailed populations are needed to develop more sophisticated models of antibiotic therapy versus CCI and/or paediatric severity of illness metrics. Overall, however, these data illustrate the potential of AXDX to enable treating physicians and stewards to optimize therapy for patients with microbial infections (including MDR organisms) sooner, to promote reduction in hospital-administered therapy and to reduce patient risk.
Clinical microbiology laboratories have utilized various rapid diagnostics for BSIs with GNRs, including VERIGENE®, BioFire, the FilmArray® Blood Culture Identification (BCID) Panel and MALDI-TOF MS. These diagnostic modalities have been shown to decrease mortality and shorten hospital stays, even more so if they are used in conjunction with a robust stewardship programme.10 Because these modalities are unable to completely replace traditional microbiology techniques, the complexity of laboratory workflow must increase to fully glean these clinical benefits. Furthermore, data supporting de-escalation are much less robust with molecular AST detection, reflecting the lack of confidence and understanding physicians have for molecular results.5,35,36
Similar to other studies,14–22 this study shows that AXDX provides reliable results for ID and AST that are comparable to other standard testing, such as molecular, proteomics-based and conventional phenotypic methods. There were, however, instances in which SOC methods for routine ID and AST were recommended by AXDX (11% of the time). This included AST non-reportable results (n=11, 7.3%) and ‘off-panel’ organisms not identified by AXDX (n=9, 5.5%). Consequently, like other rapid diagnostics, AXDX will not be able to replace traditional modalities for all ID and AST combinations, particularly for off-panel or polymicrobial cultures.16,18,23 However, upon evaluation of our workflow, AXDX substantially reduced overall ‘hands-on’ time for laboratory technologists, improving our workflow (decreasing setup and processing time 10-fold, from nearly 30 min for SOC to ∼3 min for AXDX), with the potential to save ∼45 min in total technologist time required for SOC methods.
This study has several limitations. First, the study site is a quaternary referral centre, with multiple state-wide satellite hospitals, caring for a varied patient mix including immunocompromised patients and a broad spread of resistance patterns, making these findings less generalizable to all healthcare settings. These findings will have the greatest impact in locales with similar practices and resources to our study site. Second, our study population was limited to GNR BSIs and did not address Gram-positive or yeast BSIs. While this does limit our study population, GNR BSIs have been shown to have increasing complexity of antimicrobial resistance,1,3–5,23 along with infectious disease physicians ranking this type of infection as one of the most unmet needs in pathogen diagnostics.37 Third, polymicrobial samples as indicated by Gram stain (∼10% of all PBCs) were excluded due to limitations on AXDX producing AST for all pathogens in these samples.16,19 Consequently, for this select subset, not all aspects of patient care that could have influenced clinical outcomes were captured. Furthermore, the lack of polymicrobial infections being tested (in addition to excluding Gram-positive and yeast BSIs) may have increased the performance of AXDX with respect to other studies. It should be noted that AXDX’s potential for clinical impact with Gram-positive bacteraemia has been shown in recent publications.16,18–20 Fourth, several assumptions (described in the Materials and methods section) regarding the potential clinical impact of decreased TTR, including how the theoretical data used were derived from retrospective chart review, limit our ability to infer the true result times and clinical impact. Fifth, optimal or ‘institution preferred’ therapy varies among different institutions, which may lead to these results being less generalizable to other healthcare settings. Sixth, the overall cost of implementing the AXDX technology was not assessed. Although AXDX can function as a standalone test for a large percentage of clinical isolates, it cannot remove the need for subculture of PBCs to ensure detection of other off-panel organisms (∼5.5%, not including polymicrobial samples).16,18,23 Finally, TTR and percentage agreement values were only compared with SOC modalities at the test site. Clinical studies comparing AXDX with rapid diagnostic tests implemented at other laboratories, specifically those that include molecular AST, are needed to evaluate the true impact of AXDX’s phenotypic AST results.
Conclusions
AXDX significantly reduces ID/AST TTR for PBC specimens, and thus can potentially aid in effective antimicrobial stewardship. Our findings highlight the potential clinical benefits of utilizing rapid phenotypic susceptibility testing to allow earlier modification of empirical therapy. Given the nature of this study, the true clinical significance of these time differences is unknown. However, given the strong evidence that each hour of delayed treatment in patients with GNR bacteraemia increases mortality,1,2 it stands to reason that these time differences could have a significant clinical impact.
Supplementary Material
Acknowledgements
We especially thank the following: Medical Technologist Mike Rayl, and all other medical laboratory scientists at the Indiana University Health Pathology laboratory who assisted with set up and data acquisition; Mu Shan, biostatistician at Indiana University School of Medicine, for data review and manuscript preparation; John Prichard and Christina Chantell from Accelerate Diagnostics, Inc. for technical support and manuscript preparation, respectively.
Funding
This study was supported by internal funding by the Department of Pathology and Laboratory Medicine through the Indiana University School of Medicine. Accelerate Diagnostics, Inc. provided the Accelerate Pheno™ system modules and half of the kits used in the study. Accelerate Diagnostics, Inc. was not involved in study design, data collection, or data interpretation.
Transparency declarations
N. W. S. has stock options and is an employee of Accelerate Diagnostics, Inc. and was involved in data management, figure design, and manuscript preparation. J. G. S. has received research grant support from Accelerate Diagnostics, Inc. All other authors: none to declare.
This article forms part of a Supplement sponsored by Accelerate Diagnostics, Inc.
References
- 1. Kang CI, Kim SH, Park WB. et al. Bloodstream infections caused by antibiotic-resistant Gram-negative bacilli: risk factors for mortality and impact of inappropriate initial antimicrobial therapy on outcome. Antimicrob Agents Chemother 2005; 49: 760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar A, Roberts D, Wood KE. et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34: 1589–96. [DOI] [PubMed] [Google Scholar]
- 3. Peleg AY, Hooper DC.. Hospital-acquired infections due to Gram-negative bacteria. N Engl J Med 2010; 362: 1804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perez KK, Olsen RJ, Musick WL. et al. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J Infect 2014; 69: 216–25. [DOI] [PubMed] [Google Scholar]
- 5. Barlam TF, Cosgrove SE, Abbo LM. et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62: e51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Messacar K, Parker SK, Todd JK. et al. Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J Clin Microbiol 2017; 55: 715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee R, Teng CB, Cunningham SA. et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis 2015; 61: 1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang AM, Newton D, Kunapuli A. et al. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013; 57: 1237–45. [DOI] [PubMed] [Google Scholar]
- 9. Perez KK, Olsen RJ, Musick WL. et al. Integrating rapid pathogen identification and antimicrobial stewardship significantly decreases hospital costs. Arch Pathol Lab Med 2013; 137: 1247–54. [DOI] [PubMed] [Google Scholar]
- 10. Timbrook TT, Morton JB, McConeghy KW. et al. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 2017; 64: 15–23. [DOI] [PubMed] [Google Scholar]
- 11. Bauer KA, West JE, Balada-Llasat JM. et al. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis 2010; 51: 1074–80. [DOI] [PubMed] [Google Scholar]
- 12. Zhang D, Micek ST, Kollef MH.. Time to appropriate antibiotic therapy is an independent determinant of postinfection ICU and hospital lengths of stay in patients with sepsis. Crit Care Med 2015; 43: 2133–40. [DOI] [PubMed] [Google Scholar]
- 13. Accelerate Diagnostics, Inc. Accelerate PhenoTest BC™ Kit: Instructions for Use. Tucson, AZ, 2018. https://www.online-ifu.com/ADX000005/4311/EN-US. [Google Scholar]
- 14. de Cardenas JB, Su Y, Rodriguez A. et al. Evaluation of rapid phenotypic identification and antimicrobial susceptibility testing in a pediatric oncology center. Diagn Microbiol Infect Dis 2017; 89: 52–7. [DOI] [PubMed] [Google Scholar]
- 15. Charnot-Katsikas A, Tesic V, Love N. et al. Use of the Accelerate Pheno system for identification and antimicrobial susceptibility testing of pathogens in positive blood cultures and impact on time to results and workflow. J Clin Microbiol 2018; 56: e01166-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lutgring JD, Bittencourt C, McElvania TeKippe E. et al. Evaluation of the Accelerate Pheno system: results from two academic medical centers. J Clin Microbiol 2018; 56: e01672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pantel A, Monier J, Lavigne JP.. Performance of the Accelerate Pheno system for identification and antimicrobial susceptibility testing of a panel of multidrug-resistant Gram-negative bacilli directly from positive blood cultures. J Antimicrob Chemother 2018; 73: 1546–52. [DOI] [PubMed] [Google Scholar]
- 18. Pancholi P, Carroll KC, Buchan BW. et al. Multicenter evaluation of the Accelerate PhenoTest BC kit for rapid identification and phenotypic antimicrobial susceptibility testing using morphokinetic cellular analysis. J Clin Microbiol 2018; 56: e01329-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marschal M, Bachmaier J, Autenrieth I. et al. Evaluation of the Accelerate Pheno system for fast identification and antimicrobial susceptibility testing from positive blood cultures in bloodstream infections caused by Gram-negative pathogens. J Clin Microbiol 2017; 55: 2116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sofjan AK, Casey BO, Xu BA. et al. Accelerate PhenoTest(TM) BC kit versus conventional methods for identification and antimicrobial susceptibility testing of Gram-positive bloodstream isolates: potential implications for antimicrobial stewardship. Ann Pharmacother 2018; 52: 754–62. [DOI] [PubMed] [Google Scholar]
- 21. Descours G, Desmurs L, Hoang TLT. et al. Evaluation of the Accelerate PhenoTM system for rapid identification and antimicrobial susceptibility testing of Gram-negative bacteria in bloodstream infections. Eur J Clin Microbiol Infect Dis 2018; 37: 1573–83. [DOI] [PubMed] [Google Scholar]
- 22. Giordano C, Piccoli E, Brucculeri V. et al. A prospective evaluation of two rapid phenotypical antimicrobial susceptibility technologies for the diagnostic stewardship of sepsis. BioMed Res Int 2018; 2018: 6976923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bard JD, Lee F.. Why can’t we just use PCR? The role of genotypic versus phenotypic testing for antimicrobial resistance testing. Clin Microbiol Newsl 2018; 40: 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 26th Edition. M100-S26. CLSI, Wayne, PA, USA, 2016. [Google Scholar]
- 25. Jones E, Oliphant E, Peterson P. et al. SciPy: Open Source Scientific Tools for Python 2001. http://www.scipy.org/.
- 26. Travis E, Oliphant E.. Python for scientific computing. Comput Sci Eng 2007; 9: 10–20. [Google Scholar]
- 27. Mann HB, Whitney DR.. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Statist 1947; 18: 50–60. [Google Scholar]
- 28. Raman K, Nailor MD, Nicolau DP. et al. Early antibiotic discontinuation in patients with clinically suspected ventilator-associated pneumonia and negative quantitative bronchoscopy cultures. Crit Care Med 2013; 41: 1656–63. [DOI] [PubMed] [Google Scholar]
- 29. Cremers AJ, Sprong T, Schouten JA. et al. Effect of antibiotic streamlining on patient outcome in pneumococcal bacteraemia. J Antimicrob Chemother 2014; 69: 2258–64. [DOI] [PubMed] [Google Scholar]
- 30. Retamar P, Portillo MM, Lopez-Prieto MD. et al. Impact of inadequate empirical therapy on the mortality of patients with bloodstream infections: a propensity score-based analysis. Antimicrob Agents Chemother 2012; 56: 472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamma PD, Avdic E, Li DX. et al. Association of adverse events with antibiotic use in hospitalized patients. JAMA Intern Med 2017; 177: 1308–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bouza E, Sousa D, Munoz P. et al. Bloodstream infections: a trial of the impact of different methods of reporting positive blood culture results. Clin Infect Dis 2004; 39: 1161–9. [DOI] [PubMed] [Google Scholar]
- 33. Battle SE, Bookstaver PB, Justo JA. et al. Association between inappropriate empirical antimicrobial therapy and hospital length of stay in Gram-negative bloodstream infections: stratification by prognosis. J Antimicrob Chemother 2017; 72: 299–304. [DOI] [PubMed] [Google Scholar]
- 34. Baggs J, Jernigan JA, Halpin AL. et al. Risk of subsequent sepsis within 90 days after a hospital stay by type of antibiotic exposure. Clin Infect Dis 2018; 66: 1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Donner LM, Campbell WS, Lyden E. et al. Assessment of rapid-blood-culture-identification result interpretation and antibiotic prescribing practices. J Clin Microbiol 2017; 55: 1496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walker T, Dumadag S, Lee CJ. et al. Clinical impact of laboratory implementation of Verigene BC-GN microarray-based assay for detection of Gram-negative bacteria in positive blood cultures. J Clin Microbiol 2016; 54: 1789–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Blaschke AJ, Hersh AL, Beekmann SE. et al. Unmet diagnostic needs in infectious disease. Diagn Microbiol Infect Dis 2015; 81: 57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.