Supplemental Digital Content is available in the text.
Key Words: probiotics, celiac disease, gluten-free diet, gut microbiota
Abstract
Goals:
The goals of this study were to evaluate the efficacy and safety of a probiotic mixture in patients with celiac disease (CD) with irritable bowel syndrome (IBS)-type symptoms despite a strict gluten-free diet (GFD).
Background:
About 30% of patients with CD adherent to a GFD suffer from IBS-type symptoms; a possible cause resides in the imbalances of the intestinal microbiota in CD. Probiotics may represent a potential treatment.
Study:
CD patients with IBS-type symptoms entered a prospective, double-blind, randomized placebo-controlled study. A 6-week treatment period was preceded by a 2-week run-in and followed by a 6-week follow-up phase. Clinical data were monitored throughout the study by validated questionnaires: IBS Severity Scoring System (IBS-SSS); Gastrointestinal Symptom Rating Scale (GSRS); Bristol Stool Form Scale (BSFS); and IBS Quality of Life Questionnaire (IBS-QOL). The fecal microbiota were assayed using plate counts and 16S rRNA gene-based analysis.
Results:
In total, 109 patients were randomized to probiotics (n=54) or placebo (n=55). IBS-SSS and GSRS decreased significantly in probiotics, as compared with placebo [(−15.9%±14.8% vs. 8.2%±25.9%; P<0.001) and (−19.8%±16.6% vs. 12.9%±31.6%; P<0.001)], respectively. Treatment success was significantly higher in patients receiving probiotics, as compared with placebo (15.3% vs. 3.8%; P<0.04). Presumptive lactic acid bacteria, Staphylococcus and Bifidobacterium, increased in patients receiving probiotic treatment. No adverse events were reported.
Conclusions:
A 6-week probiotic treatment is effective in improving the severity of IBS-type symptoms, in CD patients on strict GFD, and is associated with a modification of gut microbiota, characterized by an increase of bifidobacteria.
Celiac disease (CD) is an autoimmune disorder that occurs in genetically predisposed individuals who develop an immune reaction to gluten, characterized by gluten-dependent clinical manifestations, specific antibodies, HLA-DQ2 and/or DQ8 haplotypes, and enteropathy.1 Although the adherence to a strict gluten-free diet (GFD) usually leads to the remission of major clinical symptoms, irritable bowel syndrome (IBS)-type symptoms occur frequently in patients with CD despite a strict GFD.2
The association of CD and IBS-type symptoms is biologically plausible, with several mechanisms being involved, such as motor dysfunction, low-grade gut inflammation, small intestinal disease, visceral hypersensitivity,3 and, recently, altered gut microbiota.
Gut microbiota play a key role in maintaining intestinal homeostasis and promoting health. Several recent studies report imbalances in the intestinal microbiota of patients with CD,4 mainly characterized by an increase of gram-negative bacteria (Bacteroides and enterobacteria) and a decrease of gram-positive bacteria, such as bifidobacteria.5,6 It is still debated whether such modifications enter in the pathogenesis of the disease or are just a consequence; however, the intestinal dysbiosis persists irrespective of the adherence to a GFD and in part is related to this particular diet. Indeed, GFD influences gut microbiota composition mainly because of a reduction in polysaccharide intake,7 which constitutes one of the main energy sources for commensal components of the gut microbiota.8
Wacklin et al9 have shown that dysbiosis of gut microbiota is associated with persistent gastrointestinal (GI) symptoms in treated CD patients, opening new possibilities to treat this subgroup of patients. On the basis of these observations, we hypothesized that probiotics might exert a beneficial effect in the treatment of IBS-type symptoms in patients with CD. The aim of the present study was to evaluate the efficacy and safety of a new probiotic mixture in a randomized, double-blind, placebo-controlled trial in CD patients with IBS-type symptoms despite a strict GFD.
MATERIALS AND METHODS
This was a prospective, double-blind, randomized placebo-controlled parallel group study. The patient recruitment was carried out between 2013 and 2015 at the Gastroenterology Units of University of Bari, Castellana Grotte (BA), Foggia and Taranto, by inviting volunteer adult (age 18 y and above) CD patients, who had been treated for a long term (GFD≥2 y), to participate to the study.
Inclusion criteria for patients were as follows: (a) they complained of persistent IBS-type symptoms according to the ROME III criteria with no clinical evidence of other medical conditions to explain the symptoms,10 (b) were strictly adherent to a GFD, and (c) had the diagnosis confirmed by chart review, showing elevated serum tissue transglutaminase immunoglobulin-A (tTG-IgA), in the presence of histologic evidence of villous atrophy with crypt hyperplasia, and an increase in intraepithelial lymphocytes on a gluten-containing diet.11
Exclusion criteria were as follows: clinically significant cardiovascular, respiratory, endocrine, renal, hematologic, hepatic, neurological or psychiatric disease; previous GI malignancy and/or surgery; pregnancy or lactation; alcohol abuse or drug addiction; current use of medications including corticosteroids or anti-inflammatory drugs, proton-pump inhibitors, antibiotic treatment, and participation in another clinical trial within 6 months before enrolment.
All participants underwent a thorough clinical examination, dietary measurements, CD serology, and basic laboratory parameters; an experienced dietician assessed the strict adherence to a GFD. Only subjects with negative celiac antibodies and on a strict GFD entered the trial.
Study Design and Study Product
A randomization list was computer generated in blocks of 8 patients (ie, 4 receiving placebo and 4 active treatments in random order within the block, software by randomization.com). Labeling of study products were performed by an outside partner not taking part in the study. The active study product consisted of a mixture of 5 strains of lactic acid bacteria and bifidobacteria: Lactobacillus casei LMG 101/37 P-17504 (5×109 CFU/sachet), Lactobacillus plantarum CECT 4528 (5×109 CFU/sachet), Bifidobacterium animalis subsp. lactis Bi1 LMG P-17502 (10×109 CFU/sachet), Bifidobacterium breve Bbr8 LMG P-17501 (10×109 CFU/sachet), B. breve Bl10 LMG P-17500 (10×109 CFU/sachet). The probiotic was given as a sachet once per day. The active study product and the placebo had identical appearance and taste with the placebo only lacking the viable bacteria. All study products were provided free of charge by Probioresearch (Rome, Italy), which monitored the stability of the probiotic formulation throughout the study. Group assignment was concealed from participants and investigators. The randomization list defining the given treatment was kept in a sealed envelope that was not opened unless there was a serious adverse event that was deemed by the principle investigators to be directly associated with the study product. Furthermore, the envelopes were kept sealed until all study-related data had been digitalized and the resulting computer file locked. An independent medical staff member assigned subjects to the 2 schedules. Each patient received the next pack of study product stored in the center following ascending order of labels.
Study Plan and Assessment of Patients
The 6-week treatment period (week 3 to 8) was preceded by a 2-week run-in (week 1 to 2) and followed by a 6-week follow-up phase (week 9 to 14) (Fig. 1). According to the recommendation of the Trials for Functional Gastrointestinal Disorders,12 only patients who had persistence of symptoms during the run-in period [irritable bowel syndrome severity scoring system (IBS-SSS)>75] proceeded to participate in the study. To assess compliance to study treatment and to record adverse events, patients were interviewed on a regular basis by medical personnel blinded to the regimen of each patient. Compliance was calculated as the percentage of ingested study product, and a rate >80% was set as the minimum.
FIGURE 1.
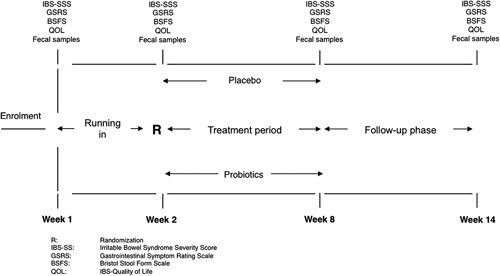
Study design. BSFS indicates Bristol Stool Form Scale; GSRS, Gastrointestinal Symptom Rating Scale; IBS-SSS, Irritable Bowel Syndrome Severity Scoring System; QOL, Quality of Life; R, randomization.
From week 1 to 14, patients filled-in the following questionnaires at 2-week intervals:
IBS-SSS consists of 5 questions that generate a maximum score of 100, each using a visual analogue scale and leading to a total possible score of 500. Mild, moderate, and severe cases were indicated by scores of 75 to 175, 175 to 300, and >300, respectively; controls scored below 75, and patients scoring in this range can be considered to be in remission.13
The 15-item Gastrointestinal Symptom Rating Scale (GSRS), a structured and validated questionnaire widely used in the research of GI diseases, was used to assess the severity of current GI symptoms.14
Bristol Stool Form Scale (BSFS): stool frequency and type of stool passed were recorded with the BSFS being the reference, which was provided to the parents.15
Irritable Bowel Syndrome Quality of Life Questionnaire (IBS-QOL): this is a well-established 34-item measure assessing the degree to which IBS interferes with patient quality of life. Each item is rated on a 5-point Likert scale, yielding a total score that ranges from 34 to 170, with higher scores indicating worse QOL.16
Outcome Measures, Adverse Events, and Disallowed Medication
The primary outcome was to determine whether probiotics, as compared with placebo, were able to improve GI symptoms, as assessed by IBS-SSS; this test was preferred to GSRS, because most of the patients mention IBS-type symptoms, and one third of the score of GSRS refers to the upper GI tract.
Secondary outcomes were to investigate whether probiotics, as compared with placebo, were able to (1) decrease at least 50% the symptom scores, as assessed by IBS-SSS (treatment success); (2) improve GRSR; (3) modify stool parameters, as assessed by BSFS; (4) improve IBS-QOL; and (5) modify gut microbiota and metabolomic fecal profile.
Adverse events were monitored throughout the study. Patients were not allowed to consume any probiotic, other than those provided, or prebiotics, and they were instructed to continue their eating and physical exercise habits. Concomitant use of medications affecting GI motility and/or pain perception was allowed, provided that the patients registered the intake.
Before and during the trial, staff members instructed patients to guarantee a well-balanced diet, with proper gluten avoidance, without modification to the dietetic habits.
Fecal Microbiota
Fecal samples were collected at the end of week 2, 8, and 14. Fecal samples (5 g) were mixed with 45 mL sterilized physiological solution and homogenized. Counts of viable bacterial cells were carried out as described by De Angelis et al.17
Fecal samples were also used for DNA and RNA extractions.17 The sample was subjected to mechanical disruption with the FastPrep instrument (BIO 101), and total DNA was extracted with the FastDNA Spin Kit for Soil (MP Biomedicals, Illkrich, France), according to the manufacturer’s instructions. An aliquot of ca. 200 mg of fecal sample was used for RNA extraction with the stool total RNA purification kit (Norgen Biotek Corp., ON, Canada). An aliquot of 1 μg of total RNA extracted was transcribed to cDNA, using random examers and the tetro cDNA synthesis kit from Bioline (Bioline USA Inc., Tanunton, MA), according to the manufacturer’s instructions.18
For each subject, the 3 DNA samples, were pooled and used for 16S-based tag-encoded FLX amplicon pyrosequencing analysis. 16S-based tag-encoded FLX amplicon pyrosequencing was performed by Research and Testing Laboratories (Lubbock, TX), according to standard laboratory procedures, and using the 454 FLX Sequencer (454 Life Sciences, Branford, CT).17,19 Raw sequence data were screened, trimmed, and filtered with default settings, using the QIIME pipeline version 1.4.0 (http://qiime.sourceforge.net). Chimeras were excluded by using the B2C2 (www.researchandtesting.com/B2C2.html).20 Sequences <250 bp were removed. Sequences are available at www.researchandtesting.com/docs. FASTA sequences for each sample, without chimeras, were evaluated using BLASTn against a database derived from GenBank (http://ncbi.nlm.nih.gov).21 DNA was also amplified using primer pair Probio_Uni and/Probio_Rev, which targets the V3 region of the 16S rRNA gene sequence.22 DNA was amplified under the polymerase chain reaction conditions described previously.22 Sequencing analyses were carried out according to the protocol of the Ion Torrent PGM system and using the Ion Sequencing 200 kit. The sequences were first clustered into operational taxonomic unit clusters with 97% identity (3% divergence), using USEARCH.23 To determine the identities of bacteria, sequences were first queried, using a distributed BLASTn.NET algorithm24 against 16S bacterial sequences, which were derived from NCBI. Database sequences were characterized as high quality on the basis of criteria that were originally described by Ribosomal Database Project (RDP, version 10.28).25 Alpha diversity (rarefaction, Good’s coverage, Chao1 richness, and Shannon diversity indices) and beta diversity measures were calculated and plotted using QIIME.
The study adhered to the Declaration of Helsinki and was approved by the institutional ethical committee. The full trial protocol can be accessed at www.ClinicalTrials.gov (identifier NCT01699191). All study participants provided written informed consent.
Statistical Analysis
All data are expressed as median±SD with 95% confidence intervals unless differently specified. Given the exploratory nature of the study, the sample size calculation was based on the intent to detect a 25% difference in the proportion of responders between treatment and placebo groups; 50 patients for each arm were required on the basis of a 0.90 power and a 2-sided type 1 error rate of 5% while compensating for just over a 10% drop out rate. The χ2 test or the Fisher exact test was used, as appropriate, to compare percentages and nominal variables. For continuous variables, differences between patients in the 2 treatment arms were compared using analysis of variance, and the Wilcoxon test was used for comparison of the mean values. All statistical tests were 2 tailed and performed at the 5% level of significance. All analyses were performed on the intention-to-treat basis and per protocol analysis. The statistical analyses were performed using the JMP SAS Institute program version 9 (in Cary, NC).
RESULTS
The flow of patients involved in the trial from assessment for eligibility through follow-up is shown in Figure 2. Of the 279 patients who participated in the first visit, 161 were excluded, because they did not meet the inclusion/exclusion criteria. Of the remaining 118 patients, 5 (4.2%) refused to participate, and 113 entered the running-in phase without protocol deviations; 3 patients referred a significant improvement of symptoms (IBS-SSS<75) and exited the study, whereas 1 withdrew consent. Therefore, 109 were available for randomization: 54 were assigned to probiotics and 55 to placebo. At the final assessment, complete data were available for 105 of the 113 participants (93%): 3 patients were noncompliant (2 placebo and 1 probiotics), and 1 was lost to follow-up (probiotics). These 4 patients were excluded from further analysis. The baseline characteristics of the participants in the 2 groups are reported in Table 1. By chance, patients in the probiotic group had significantly worse clinical scores, as compared with those receiving placebo. No differences in the composition of the diet and in the amount of FODMAP consumption were detected during the trial. No adverse events related to the study product were reported.
FIGURE 2.
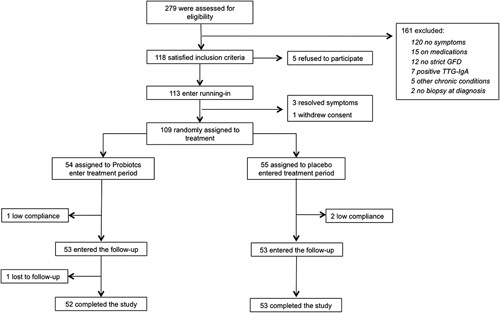
Flow diagram of patients in the trial from eligibility to the end of follow-up. GFD indicates gluten-free diet; TTG-IgA, tissue transglutaminase immunoglobulin-A.
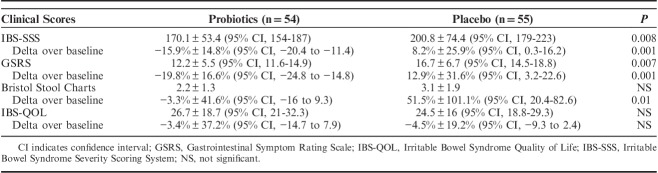
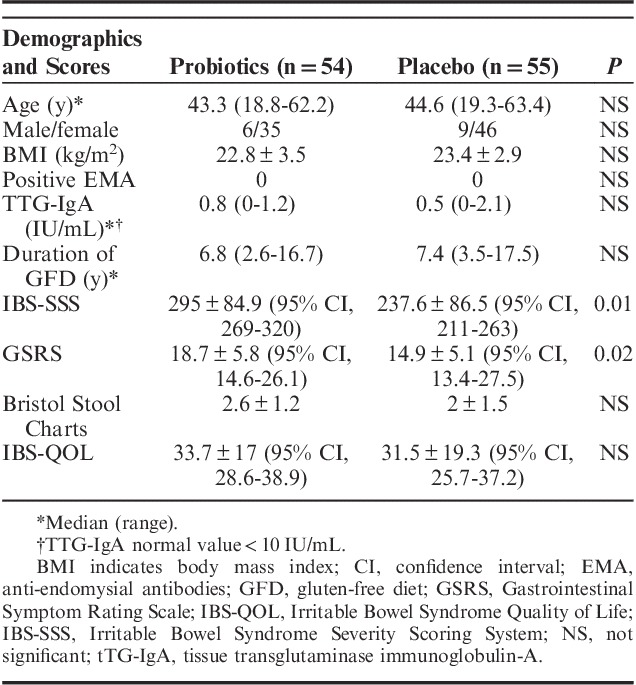
TABLE 1.
Baseline Characteristics of the Patients in the Full Analysis Set
IBS-SSS
The IBS-SSS was at baseline 295±84.9 in the probiotic and 237.6±86.5 in the placebo group (P<0.01); at the end of treatment, it decreased to 170.7±53.4 and 200.8±74.4, respectively (P<0.008). When expressed as variation over the pretreatment value, IBS-SSS in probiotics was significantly decreased, as compared with placebo (−21.4%±15.5% vs. −6.8%±21.7%; P<0.001). At the end of follow-up, IBS-SSS was 176.7±45.1 in the probiotic and 173.6±45.5 in the placebo group, respectively (P=NS).
GSRS, BSFS, and IBS-QOL
The GSRS was at baseline 18.7±5.8 in the probiotic and 14.9±5.1 in the placebo group (P<0.02); at the end of treatment, GSRS changed to 12.2±5.5 and 16.7±6.7, respectively (P<0.007) (Table 2). When expressed as variation over the pretreatment value, GSRS in probiotics was significantly decreased, as compared with placebo (−19.8%±16.6% vs. 12.9%±31.6%; P<0.001). At the end of follow-up, GSRS was 10.1±4.1 in the probiotic and 9.6±4.2 in the placebo group (P=NS).
TABLE 2.
Clinical Scores at the End of Treatment and Delta Values Expressed as Variation Over the Pretreatment Value (in Percentage)
After treatment, BSFS scores did not differ in patients receiving probiotics or placebo as absolute values (2.2±1.3 vs. 3.1±1.9, respectively; P=NS). When expressed as variation over the pretreatment value, BSFS score in probiotics was significantly decreased, as compared with placebo (−3.3%±41.6% vs. 51.5%±101.1%; P<0.01). At the end of follow-up, BSFS scores were 2.3±1.5 in the probiotic and 2.9±1.6 in the placebo group, respectively (P=NS).
After treatment, IBS-QOL scores did not differ in patients receiving probiotics or placebo, either as absolute values (26.7±18.7 and 24.5±16, respectively; P=NS) or when expressed as variation over the pretreatment values (−3.4%±37.2% vs. −4.5%±19.2%; P=NS). At the end of follow-up, QOL was 23±14.6 in the probiotic and 24±15.7 in the placebo group, respectively (P=NS).
Treatment Success
Treatment success was significantly higher in patients receiving probiotics, as compared with placebo, at both intention-to-treat (14.8% vs. 3.6%; P<0.04) and per protocol (15.3% vs. 3.8%; P<0.04) analysis; according to our data, 9 patients need to be treated to reach treatment success in 1 (number needed to treat: 9).
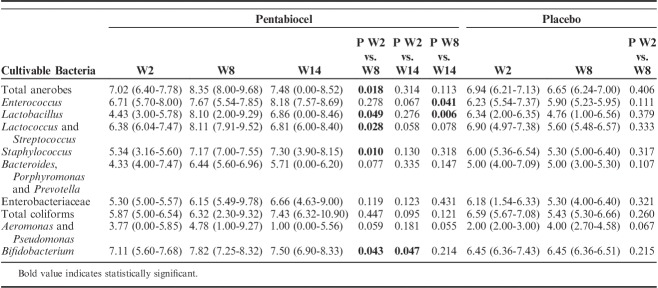
Enumeration of Fecal Cultivable Bacteria and Microbiome
Compared with baseline value (W2), total anerobes increased from 7.02 to 8.35 log CFU/g (median values, P=0.018) after 6 weeks of treatment with probiotics. The treatment with probiotics drives also the increase of presumptive lactic acid bacteria (Lactobacillus, Lactococcus, and Streptococcus), Staphylococcus and Bifidobacterium. Compared with W2, higher level of presumptive Bifidobacterium was also found after 6 weeks (W8) of treatment with probiotics. On the contrary, no statistical difference (P>0.05) was found between the cultivable microbes detected at W2, W8, and W14 for the placebo group (Table 3).
TABLE 3.
Median Values and Range of Cultivable Bacterial Cells (log CFU/g) of the Main Microbial Groups in the Fecal Samples of Celiac Disease With Irritable Bowel Syndrome Patients at Baseline (W2), After 6 Weeks (W8) of Treatment With Probiotics (Pentabiocel) or Placebo, and at the End of Follow-up (W14)
The total bacterial community richness was analyzed by rarefaction curves (Fig. S1, Supplemental Digital Content 1, http://links.lww.com/JCG/A401), richness estimator (Chao1) (Fig. S2A, Supplemental Digital Content 2, http://links.lww.com/JCG/A402), and diversity index (Shannon) (Fig. S2B, Supplemental Digital Content 2, http://links.lww.com/JCG/A402). No statistically significant (P>0.05; false discovery rate>0.05) differences were found between probiotic and placebo groups. A similar trend was also found for metabolically active fecal microbiome (data not shown). According to alpha diversity, the 3 phylogeny-based β-diversity measures did not show clear separation between the microbiome composition of placebo and probiotic groups in UniFrac distance principal coordinates analysis plots (Fig. S3, Supplemental Digital Content 3, http://links.lww.com/JCG/A403).
No significant (P>0.05; false discovery rate>0.05) differences were found for the total bacterial phyla (data not shown) and relative abundances of genera between placebo and probiotic groups (Fig. S4, Supplemental Digital Content 4, http://links.lww.com/JCG/A404). Similar results were found for metabolically active bacteria. The only exception was the phylum Actinobacteria and the related genus Bifidobacterium, which was higher in the probiotic compared with the placebo group (Fig. 3). Compared with W2, higher relative amount of metabolically active Bifidobacterium was found in the probiotic group during the washout period (W8) (Fig. 4).
FIGURE 3.
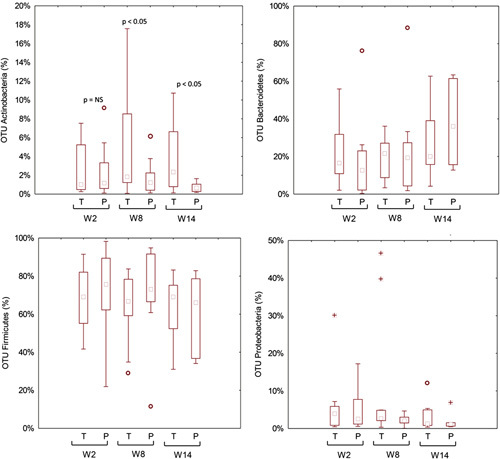
Relative abundance of the most relevant metabolically active bacterial phyla found in feces of the fecal samples of celiac disease patients with irritable bowel syndrome at baseline (W2), after 6 weeks (W8) of treatment with probiotics or placebo, and at the end of follow-up (W14). NS indicates not significant; OTU, operational taxonomic unit; P, placebo; T, treated.
FIGURE 4.
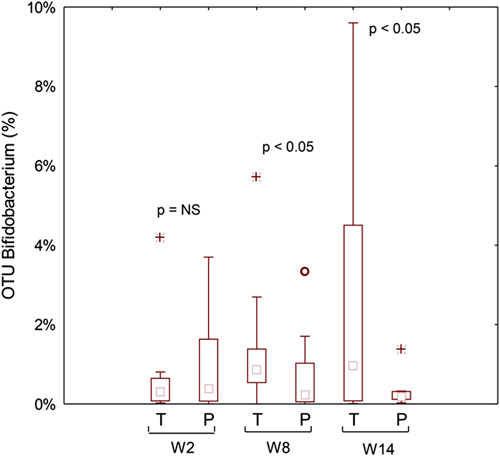
Metabolically active Bifidobacterium genus found in feces of the fecal samples of celiac disease patients with irritable bowel syndrome at baseline (W2), after 6 weeks (W8) of treatment with probiotics or placebo, and at the end of follow-up (W14). NS indicates not significant; OTU, operational taxonomic unit; P, placebo; T, treated.
DISCUSSION
This large prospective, randomized, study showed that the probiotic mix under study is effective in improving the severity of IBS-type symptoms in patients with CD adherent to a strict GFD. After 6 weeks treatment, we found a significantly higher proportion of treatment success, a decreased perception of pain according to different clinical scales, and a modification of gut microbiota, characterized by an increase of bifidobacteria that is still detectable 6 weeks after discontinuation of probiotics.
The persistence of GI symptoms despite a strict adherence to a GFD is not unusual and is mainly characterized by symptoms compatible with IBS.26,27 In a recent meta-analysis of 7 studies with 3383 participants, Sainsbury et al2 showed that, in patients who were adherent to a GFD, the pooled odds ratios for IBS-type symptoms, compared with controls without CD, was 4.3 with a prevalence of IBS symptoms of 38%: a strict adherence to a GFD was associated with a reduction but not a resolution of symptoms. Why CD and IBS might coexist in some patients is still debated; however, it is possible that the low-grade CD inflammation,28 similar to that seen in IBS,29 does not subside completely after implementation of GFD, causing motor dysfunction and/or visceral hypersensitivity.
We30,31 and others4–7,9 have reported aberrations in intestinal microbiota in celiac patients compared with healthy controls, irrespective of the adherence to GFD. Overall, although the studies available might differ in terms of microbiological methods, sample sizes, and patient’s characteristics, there is a substantial agreement on the presence of an unbalance between proinflammatory and anti-inflammatory species, with a prevalence of the first. Recently, Sanz32 elegantly reported the strength of existing evidence of the association between CD and intestinal dysbiosis, summarized as follows: increased levels of Bacteroides spp (stools and biopsies), low levels of bifidobacteria (stools), and increased levels of proteobacteria and Staphylococcus (stools and biopsies), regardless of the state of the activity of disease. The GFD is able to restore in part the gut microbiota; indeed, while increased numbers of enterobacteria or staphylococci are restored, other alterations such as decreased bifidobacteria and lactobacilli and increased Bacteroides and virulent E. coli still tend to persist and, therefore, could play a more prominent role in the maintenance of symptoms.7,9,30,31 The demonstration that a GFD itself determines marked reduction in intake of naturally occurring fructans, which have prebiotic action, might in part explain the reduction in beneficial gut bacteria populations, and may constitute a variable to be considered in treated CD patients, for its possible effects on gut health.33
The presence of a sustained intestinal dysbiosis might be associated with inadequate clinical response and persistence of GI symptoms. To address this hypothesis, Wacklin et al9 analyzed the gut microbiota of CD patients on a strict GFD with persistent symptoms, and found a higher abundance of proteobacteria and a lower abundance of bacteroidetes and firmicutes, as compared with patients without symptoms, suggesting that intestinal dysbiosis might play a role.
Whether or not these alterations are a cause or a consequence of CD, the similarity of microbiota modification among patients with CD and IBS34 might in part explain the presence of an overlap of clinical manifestation of these 2 conditions.35 Interestingly, in IBS, it has been shown that a state of mucosal immune activation is responsible for increased release of mediators acting on epithelial, neuronal, and muscle cells, leading to intestinal dysfunction and symptom development.28 According to some authors, this immune activation may be the consequence of a perturbation of gut microbiota,36 which might also be operating in CD. On the basis of the dysbiosis hypothesis of GI symptoms in celiac patients, we pursued the idea that a mixture of probiotic strains formulated according to the deficiencies revealed by microbiological data in our celiac population30,31,37 might be of help in this particular setting of patients. Therefore, according to our data on the fecal and duodenal microbiota of CD patients, we selected a pool of 3 bifidobacteria and 2 lactobacilli, to restore the alteration of the microbiota. When using a probiotic mixture, it is fundamental to show its efficacy, as different strains can act as antagonists, both inhibiting probiotic activity of other microorganisms and slowing the microbial growth rate.38
Few human clinical trials have been published to study the effect of a probiotic supplementation in CD with controversial results. Smecuol et al39 randomized 22 adult patients to receive either Bifidobacterium infantis or placebo before the initiation of a GFD, and, although they found no effect on celiac serology and intestinal permeability, patients on B. infantis experienced a significant improvement in GI symptoms, suggesting a role for probiotics in alleviating symptoms in untreated CD. Olivares et al40 conducted a double-blind, randomized, placebo-controlled trial in 33 children receiving either Bifidobacterium longum CECT 7347 or placebo while on GFD. After 3 months, the authors were able to show, in the probiotic-treated group, a greater height percentile increase, a reduced serum tumor necrosis factor (TNF)-α concentration, and a decrease of the Bacteroides fragilis group bacteria and fecal secretory IgA, suggesting a beneficial effect in celiac children. Klemenak et al41 investigated the effect of 2 B. breve strains (BR03 and B632) on serum interleukin-10 and TNF-α in 49 children with CD on a GFD and found that the probiotic intervention was able to decrease the production of proinflammatory cytokine. Finally, Harnett et al42 randomized 45 celiac patients, reporting only partial symptom improvement despite a strict GFD, to receive either 5 g of VSL#3 or placebo for 12 weeks, and found no changes in the fecal microbiota counts, blood safety parameters, and clinical improvement in symptoms over the course of the study.
We herein report that the probiotic combination, given daily for 6 consecutive weeks, is superior to placebo in decreasing the severity of IBS-type symptoms in patients with CD despite a GFD. We believe that part of this effect might be secondary to a positive modification of gut microbiota shown by the steady and persistent increase of the bifidobacteria count in fecal samples of CD patients. The evidence that both IBS-SSS and GSRS scores significantly decrease in the probiotic group, despite the fact that the values of both tests at entry were higher in those who received the probiotic, as compared with the placebo, further supports the effect of this probiotic combination. No clinically significant improvement on stool appearance was observed between treatment groups.
We believe that our study has some strength. To the best of our knowledge, this is the first clinical trial investigating the effect of probiotics in celiac patients suffering from IBS-type symptoms despite a strict GFD through both a clinical and microbiological study. Our results provide evidence that a long-term intervention on gut microbiota might determine a healthy clinical outcome in patients with symptoms not responsive to a GFD.
The limitations of the present study are that we have been unable to identify an improvement on QOL despite an improvement of symptoms after probiotic consumption; this might be explained by the complexity of the disease, supporting the hypothesis that patients’ perceived health status is multidimensional, and depends not only on symptoms but also on how diet restriction interacts with various factors, in particular, social life.43 Finally, our study is mainly based on subjective evaluations, and we did not perform intestinal biopsies to investigate the link between persistence of symptoms and markers of mucosal inflammation. We do not have data to support the hypothesis that the modification of gut microbiota might persist beyond the period of observation.
In recent years, an increasing amount of data have appeared with regard to the role of gut microbiota in CD patients, suggesting that dysbiosis could result in a modification of the mucosal homeostasis, causing persistent immune activation and clinical symptoms. If we consider the alterations of gut microbiota as an environmental factor involved in CD expression, probiotic administration may have a primary role in the overall manifestation of the disease.
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.jcge.com.
ACKNOWLEDGMENTS
The authors thank Dr Noemi Cavallo and Silvia Cutillo (Department of Soil, Plant and Food Science, University of Bari) for technical and scientific assistance.
Footnotes
R.F., F.I., M.D.A.: designed the research study and wrote the paper. M.P., L.P., F.S., M.B.: collected and analyzed the data. A.F. and M.G.: critical review of the article for important intellectual content. F.C. and S.C.: contributed to the design of the study.
R.F., F.I., M.G., and M.D.A. are the inventors of the patent N 0001425900, released on November 17, 2016 (Italy). The remaining authors declare that they have nothing to disclose.
REFERENCES
- 1.Lebwohl B, Sanders DS, Green PHR. Lancet. Coeliac disease. 2017;S0140-6736:31796–31798. [Google Scholar]
- 2.Sainsbury A, Sanders DS, Ford AC. Prevalence of irritable bowel syndrome-type symptoms in patients with celiac disease: a meta-analysis. Clin Gastroenterol Hepatol. 2013;11:359–365. [DOI] [PubMed] [Google Scholar]
- 3.Aziz I, Sanders DS. The irritable bowel syndrome-celiac disease connection. Gastrointest Endosc Clin N Am. 2012;22:623–637. [DOI] [PubMed] [Google Scholar]
- 4.Pagliari D, Urgesi R, Frosali S, et al. The interaction among microbiota, immunity, and genetic and dietary factors is the condicio sine qua non celiac disease can develop. J Immunol Res. 2015;2015:123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado MC, Donat E, Ribes-Koninckx C, et al. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. Austin J Clin Pathol. 2009;62:264–269. [DOI] [PubMed] [Google Scholar]
- 6.Nadal I, Donat E, Ribes-Koninckx C, et al. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56:1669–1674. [DOI] [PubMed] [Google Scholar]
- 7.De Palma G, Nadal I, Collado MC, et al. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br J Nutr. 2009;102:1154–1160. [DOI] [PubMed] [Google Scholar]
- 8.De Graaf AA, Venema K. Gaining insight into microbial physiology in the large intestine: a special role for stable isotopes. Adv Microb Physiol. 2008;53:73–168. [DOI] [PubMed] [Google Scholar]
- 9.Wacklin P, Laurikka P, Lindfors K, et al. Altered duodenal microbiota composition in celiac disease patients suffering from persistent symptoms on a long-term gluten-free diet. Am J Gastroenterol. 2014;109:1933–1941. [DOI] [PubMed] [Google Scholar]
- 10.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. [DOI] [PubMed] [Google Scholar]
- 11.Husby S, Koletzko S, Korponay-Szabó IR, et al. ESPGHAN Working Group on Coeliac Disease Diagnosis; ESPGHAN Gastroenterology Committee; European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr. 2012;54:136–160. [DOI] [PubMed] [Google Scholar]
- 12.Irvine EJ, Whitehead WE, Chey WD, et al. Design of treatment trials for functional gastrointestinal disorders. Gastroenterology. 2006;130:1538–1551. [DOI] [PubMed] [Google Scholar]
- 13.Francis CY, Morris J, Whorwell PJ. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Ther. 1997;11:395–402. [DOI] [PubMed] [Google Scholar]
- 14.Svedlund J, Sjödin I, Dotevall G. GSRS—a clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig Dis Sci. 1988;33:129–134. [DOI] [PubMed] [Google Scholar]
- 15.Heaton KW, Lewis SJ. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–924. [DOI] [PubMed] [Google Scholar]
- 16.Drossman DA, Patrick DL, Whitehead WE, et al. Further validation of the IBS-QOL: a disease-specific quality-of-life questionnaire. Am J Gastroenterol. 2000;95:999–1007. [DOI] [PubMed] [Google Scholar]
- 17.De Angelis M, Piccolo M, Vannini L, et al. Faecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowen CM, Fong SS. Genome-scale metabolic model integrated with RNAseq data to identify metabolic states of Clostridium thermocellum. Biotechnol J. 2010;7:759–767. [DOI] [PubMed] [Google Scholar]
- 19.Suchodolski JS, Markel ME, Garcia-Mazcorro JF, et al. The faecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLoS One. 2012;7:e51907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gontcharova V, Youn E, Wolcott RD, et al. Black box chimera check (B2C2): a windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol J. 2010;4:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowd SE, Zaragoza J, Rodriguez JR, et al. Windows.NET network distributed basic local alignment search toolkit (W.ND-BLAST). BMC Bioinformatics. 2005;6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milani C, Hevia A, Foroni E, et al. Assessing the faecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One. 2013;8:68739–68739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scannapieco FA. The oral microbiome: its role in health and in oral and systemic infections. Clin Microbiol Newsl. 2013;35:163–169. [Google Scholar]
- 24.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Cardenas E, et al. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dewar DH, Donnelly SC, McLaughlin SD, et al. Celiac disease: management of persistent symptoms in patients on a gluten-free diet. World J Gastroenterol. 2012;18:1348–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Midhagen G, Hallert C. High rate of gastrointestinal symptoms in celiac patients living on a gluten-free diet: controlled study. Am J Gastroenterol. 2003;98:2023–2026. [DOI] [PubMed] [Google Scholar]
- 28.O’Leary C, Wieneke P, Buckley S, et al. Celiac disease and irritable bowel-type symptoms. Am J Gastroenterol. 2002;97:1463–1467. [DOI] [PubMed] [Google Scholar]
- 29.El-Salhy M. Irritable bowel syndrome: diagnosis and pathogenesis. World J Gastroenterol. 2012;18:5151–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Cagno R, Rizello CG, Gagliardi F, et al. Different faecal microbiotas and volatile organic compounds in treated and untreated children with celiac disease. Appl Environ Microbiol. 2009;75:3963–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Cagno R, De Angelis M, De Pasquale I, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 2011;11:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanz Y. Microbiome and gluten. Ann Nutr Metab. 2015;2:28–41. [DOI] [PubMed] [Google Scholar]
- 33.Jackson FW. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects-comment by Jackson. Br J Nutr. 2010;104:773. [DOI] [PubMed] [Google Scholar]
- 34.Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in faecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. [DOI] [PubMed] [Google Scholar]
- 35.Verdu EF, Armstrong D, Murray JA. Between celiac disease and irritable bowel syndrome: the “no man’s land” of gluten sensitivity. Am J Gastroenterol. 2009;104:1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol. 2017;312:G52–G62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Francavilla R, Ercolini D, Piccolo M, et al. Salivary microbiota and metabolome associated with celiac disease. Appl Environ Microbiol. 2014;80:3416–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toscano M, De Grandi R, Pastorelli L, et al. A consumer’s guide for probiotics: 10 golden rules for a correct use. Dig Liver Dis. 2017;49:1177–1184. [DOI] [PubMed] [Google Scholar]
- 39.Smecuol E, Hwang HJ, Sugai E, et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacterium infantis natren life start strain super strain in active celiac disease. J Clin Gastroenterol. 2013;47:139–147. [DOI] [PubMed] [Google Scholar]
- 40.Olivares M, Castillejo G, Varea V, et al. Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum cect 7347 in children with newly diagnosed coeliac disease. Br J Nutr. 2014;112:30–40. [DOI] [PubMed] [Google Scholar]
- 41.Klemenak M, Dolinšek J, Langerholc T, et al. Administration of Bifidobacterium breve decreases the production of TNF-α in children with celiac disease. Dig Dis Sci. 2015;60:3386–3392. [DOI] [PubMed] [Google Scholar]
- 42.Harnett J, Myers SP, Rolfe M. Probiotics and the microbiome in celiac disease: a randomised controlled trial. Evid Based Complement Alternat Med. 2016;2016:9048574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurppa K, Collin P, Mäki M, et al. Celiac disease and health-related quality of life. Expert Rev Gastroenterol Hepatol. 2011;5:83–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's website, www.jcge.com.