Abstract
The field of epigenetics describes the relationship between genotype and phenotype, by regulating gene expression without changing the canonical base sequence of DNA. It deals with molecular genomic information that is encoded by a rich repertoire of chemical modifications and molecular interactions. This regulation involves DNA, RNA and proteins that are enzymatically tagged with small molecular groups that alter their physical and chemical properties. It is now clear that epigenetic alterations are involved in development and disease, and thus, are the focus of intensive research. The ability to record epigenetic changes and quantify them in rare medical samples is critical for next generation diagnostics. Optical detection offers the ultimate single-molecule sensitivity and the potential for spectral multiplexing. Here we review recent progress in ultrasensitive optical detection of DNA and histone modifications.
Introduction
Optical detection and quantification is widely used throughout all fields of biology and biotechnology. Specifically, it has been instrumental to the field of genetics, where the majority of sequencing instruments use optical contrast in order to read genetic information. With genetic DNA sequencing solved and established, epigenetic analysis is becoming accessible and is considered a new frontier for discovery. Even with identical genetic content, cells may differ in their function due to different epigenomic patterns that regulate gene expression. Two of the most-pronounced epigenetic regulation mechanisms involve DNA and histone tail modifications, with direct physical impact on the process of transcription. Chemical modifications such as methylation of DNA may lead to repelling or recruiting various functional binding proteins. Modifying histone tails affects their packing density and thus the compaction of chromatin and its accessibility. The stochastic and dynamic nature of epigenetic modifications yields a great degree of variability in the epigenetic profile of different cells. This in turn requires reading epigenetic information on the single-cell and single-molecule level.
Despite a plethora of molecular biology-based techniques for assessing epigenetic modifications [1–5], light-based detection holds two unique advantages for such analysis. The first is the extreme sensitivity offered by modern optical detection, allowing the detection and quantification of individual molecules. The second is the inherent multiplexing offered by the spectral properties of light. This may be accomplished either by use of labels of different colors to distinguish different modifications or by directly detecting the specific spectral signatures of different epigenetic moieties as they interact with light. High sensitivity and the capability to multiplex are especially advantageous for biomedical applications, where only minute amounts of sample are available, notably for personalized medicine and liquid biopsies. Ideally, the maximum amount of diagnostic information should be extracted from the minimum amount of patient sample. Light-based methods offer key properties for achieving this goal. This review seeks to present current advances in optics-based analytical epigenetics with a clear path towards clinical utility. As such, we will not cover a range of single-molecule biophysical work that were applied to epigenetic systems [6–9]. Discussion is focused on reports that provide real quantitative data on the epigenetic state of DNA and chromatin. Specifically, we survey work on the detection of histone-tail modifications and the DNA modifications 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC). We divide our review by the three labeling modalities for epigenetic information (Figure 1):
Figure 1.
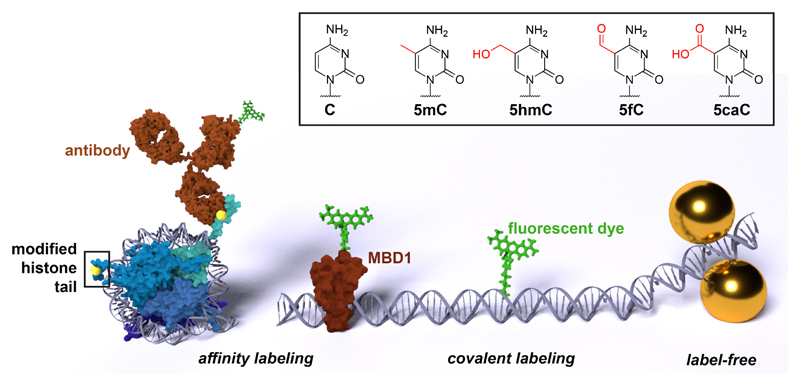
The detection schemes discussed in this review: affinity labeling via fluorescently-labeled antibodies or other binding proteins such as the methyl binding domain protein 1 (MBD1), direct covalent labeling with a fluorescent dye, and the label-free detection via surface-enhanced Raman scattering (left to right, sizes not to scale). The chemical structure of cytosine (C), 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC) is shown in the box.
The first is the attachment of a fluorescent reporter to the target mark by affinity binding. The second focuses on covalent attachment of fluorescent molecules to epigenetic targets, and finally, we discuss recent advances in epigenetic label-free detection offered by surface-enhanced Raman scattering (SERS).
Affinity labeling
The most-established mode of specific molecular labeling in biology and medicine is immunostaining, the use of antibodies to detect a specific protein or molecular species in a sample [10]. Fluorescent labeling is achieved by primary antibodies conjugated to fluorophores or by fluorescently-labeled secondary antibodies. The development of antibodies against epigenetic modifications opened new avenues for single-molecule detection of these marks. Recombinant epigenetic reader proteins with natural affinity to their native target marks are an alternative to antibodies. Methyl binding domain (MBD) proteins, for example, specifically bind methylated DNA [11] and were used for fluorescent labeling by conjugation with a fluorophore or fusion with a fluorescent protein [12]. Aptamers specific for histone modifications have also been reported and may function as non-peptide-based labeling agents [13].
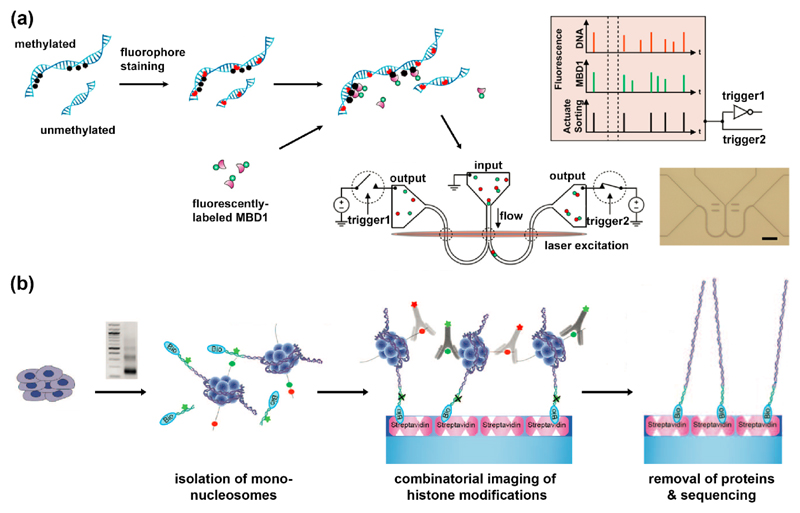
Only few affinity-based single-molecule detection schemes have provided evidence for clinical potential. A set of reports by Craighead, Soloway and Coworkers [6,14,15] describes a method for single-molecule sorting and analysis of chromatin fragments in nano-channel devices (Figure 2 (a)). Single-chromatin molecule analysis at the nanoscale (SCAN) is analogous to fluorescent activated cell sorting (FACS). While FACS sorts cells tagged with fluorescent antibodies, SCAN sorts chromatin fragments according to their fluorescent signatures. These are dictated by the colors of fluorescent tags on the DNA and the epigenetic marks it carries. The team used fluorescent antibodies against specific histone modifications or fluorescently-labeled MBD1 to mark methylation sites on DNA. Combinations of antibodies, each recognizing distinct epigenomic marks and carrying distinct fluorophores, might enable the parallel assessment of the levels of different epigenetic marks. Soloway et al. detected epigenetic differences between normal and immortalized genomes, implying that SCAN may be used for diagnosis and monitoring of cancer [16]. With appropriate calibration, the optical signal could convey the global amount of epigenetic marks, and with further improvement in throughput, each sorted fraction may be collected for further analysis such as DNA sequencing.
Figure 2.
Affinity-based methods for optical detection of epigenetic marks. (a) Single-molecule detection of DNA methylation sites. Methylation sites on stained DNA are labeled with fluorescent MBD1 protein. Two-color labeled DNA is then loaded on a nano-fluidic device, detected and sorted according to its fluorescent signature [15]. (b) Single-molecule detection of post translational modifications on nucleosomes. Nucleosomes are prepared by MNase digestion. DNA ends are ligated to fluorescent biotinylated adaptors, purified and captured on PEG-streptavidin-coated slides. Attached nucleosomes are incubated with fluorescently-labeled antibodies to histone modifications which can be then imaged. Finally, histones are removed, and their genomic position is determined by single-molecule sequencing-by-synthesis [18]. Reprinted with permission from AAAS.
The ability to not only quantify the global epigenetic status, but to characterize the state of specific genomic loci, provides additional information with high biomedical potential. Two reports from the Bernstein lab have been able to integrate antibody-based epigenetic characterization with optical genetic mapping techniques. Oren et. al. developed ChIP-String, for characterizing the chromatin state at several hundreds of positions in the genome [17]. Transcript identity is achieved by hybridizing specific fluorescent barcodes, offering several hundreds of unique patterns. The team used antibodies to pull-down chromatin fragments with specific histone modifications and then used the nCounter (NanoString Inc. Seattle, USA) to analyze the DNA released from the captured nucleosomes. Despite the high sensitivity, this method is limited by the number of available fluorescent barcodes and is not suitable for genome-wide analysis. Shema et al. [18] overcome this limitation by utilizing single-molecule sequencing-by-synthesis [19] (Figure 2 (b)). Chromatin fragments containing a single nucleosome are tethered to a microscope slide and the attachment and detachment of fluorescent antibodies against two different histone states are monitored. Once the combinatorial state at each nucleosome location is determined, nucleosomes are dissociated and the remaining tethered DNA is sequenced on the slide to determine the genomic positions of modified nucleosomes.
The single-molecule sensitivity of these techniques offers analysis of unamplified, native DNA, potentially suitable for limited medical samples such as liquid biopsies or circulating tumor cells. However, quantitative analysis at low concentrations is limited by the inherent unbinding kinetics of the affinity reagents.
Covalent labeling
Conjugation stability of covalently attached labels is not affected by the analyte concentration. Therefore, they are more suited for analyses in the low concentration regime and particularly at the single-molecule level. The small size of the covalently-attached probes presents another advantage over antibodies: Being bulky molecules, antibodies cannot accurately quantify closely-spaced epigenetic marks and therefore might underestimate their occurrence. With covalently attached molecules of similar size as nucleotides, an accurate quantification even at high densities of epigenetic modifications is possible. Whereas affinity labeling can be evolved towards any molecular residue, covalent labeling relies on the existence of specific chemical or chemoenzymatic coupling reactions for each epigenetic modification. In the following, labeling strategies for the most-important cytosine modifications will be discussed. Covalent labeling schemes for histone tails are still in early development and were not yet utilized for analytical work [20,21].
5-hydroxymethylcytosine (5hmC)
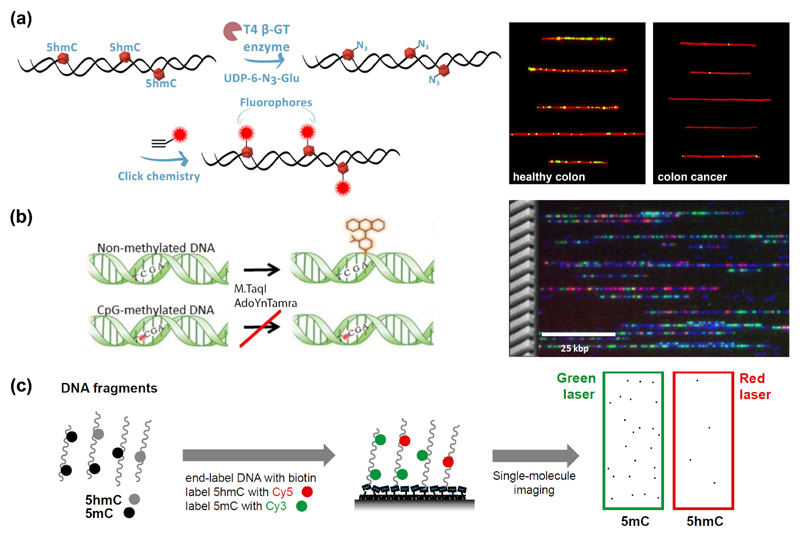
The nucleobase most-thoroughly studied by covalent epigenetic labeling is 5hmC, owing to the ability of T4 β-glucosyltransferase (βGT) to selectively place a glucose moiety onto the hydroxyl group of 5hmC. When the respective cofactor in this reaction, uridine diphosphoglucose (UDPG), is modified, new chemical functionalities can be introduced [22,23]. In particular, an azide-modified glucose offers a universal linkage point through click chemistry (Huisgen cycloaddition). When fluorophores are attached, the 5hmC content can be read out with optical means: either globally, with a spectrometer, or at the single-molecule level, by optical mapping. Both were demonstrated by Michaeli et al. [24]. For global monitoring of 5hmC levels, the absorbance of the dye was compared with that of DNA, in a simple UV/Vis spectrometer. If the fluorescence of the label is measured, the sensitivity improves. This allowed the detection of 5hmC levels as low as 0.002 % of the total nucleotides in mouse tissues at high throughput with a common plate reader [25]. Single-molecule optical mapping approaches offer the ultimate sensitivity. Typically, fluorescently-labeled DNA is deposited or stretched on a microscope slide and imaged by a fluorescence microscope [26,27]. This enables precise quantification by single-molecule counting, while requiring only minute amounts of sample (~50 ng DNA) [28]. Thus, blood and colon cancer cells could be identified by their decreased 5hmC content despite the naturally low levels in these tissues [28] (Figure 3 (a)). Additionally, the method enables multiplexing, as demonstrated by assessing 5hmC positions and fluorescently-labeled DNA damage sites on the same DNA molecules [29]. The first whole-genome single-molecule epigenetic profiles were recently reported for human blood by combining 5hmC labeling with optical genome mapping in nanochannels (commercialized by BioNano Genomics Inc.) [30]. Such information may provide locus-specific 5hmC patterns with future diagnostic value such as the epigenetic status of long variable genomic regions such as the human leukocyte antigen (HLA) genes.
Figure 3.
Covalent-based methods for optical detection of epigenetic marks. (a) Enzymatic glycosylation of 5hmC is performed, then a fluorophore is attached. Fluorescence microscopy images on the right compare 5hmC levels in single DNA molecules from healthy colon and colorectal cancer tissue [28]. (b) Fluorescent labeling of non-methylated CpGs. M.TaqI catalyzes the transfer of a fluorophore onto the adenine in its recognition site. If the CpG is methylated, the reaction is blocked. Genomic DNA (blue) from a human lymphocyte cell line was tagged with genetic (red) and methylation-sensitive labels (green), then imaged on a nanochannel array chip [38]. (c) Simultaneous detection of 5mC and 5hmC. 5hmC is labeled with Cy5, 5mC with Cy3 and DNA fragments are end-labeled with biotin for immobilization. Single DNA molecules are then imaged by TIRF microscopy [41].
5-methylcytosine (5mC)
Considering the multitude of possible epigenetic marks and, in part, their high abundance (up to 80 % of all CpG dinucleotides are methylated) [31], the interrogation of non-modified nucleobases becomes an important task in its own right. In the case of unmodified CpGs, a selective labeling with alkyne or amine groups can be achieved by ‘methyltransferase-directed transfer of activated groups’ (mTAG) [32–34]. The universal methylation cofactor S-adenosyl-methionine (SAM) can be chemically modified such that a methyltransferase enzyme is tricked into transferring a group of interest to the target methylation site on DNA [35–37]. This reaction is only successful if the site is unmethylated. After tagging with fluorophores, the unmethylated sites (or, the ‘unmethylome’) can be read out by genome-wide optical methylation mapping [38] (Figure 3 (b)), allowing the analysis of large structural aberrations such as pathogenic macrosatellite arrays. Future implementation of super-resolution mapping promises to locate the epigenetic modifications more precisely, possibly resolving the detailed methylation status of CpG islands [39]. The lack of direct covalent labeling strategies for 5mC can also be bypassed by transforming 5mC into 5hmC with Tet enzymes [40]. Selectivity between 5hmC and 5mC can be maintained if the native 5hmC is blocked by βGT-mediated glucose-transfer beforehand. This enabled a multiplexed study, where 5mC and 5hmC were tagged with different fluorophores and their relative distance was assessed by Förster resonance energy transfer (FRET) [41] (Figure 3 (c)).
5-formylcytosine (5fC)
The chemical reactivity of the aldehyde group in 5fC enables a plethora of covalent labeling strategies. A detailed account is given in a recent review by Dietzsch et al. [42]. A challenge for these approaches is that abasic sites and 5-formyluracil (5fU) also display respective aldehyde groups. Selectivity towards the aldehyde group of 5fC can be achieved through fine-tuning of reaction conditions. Still, considering the low abundance of 5fC in natural samples, even low levels of background noise are problematic [31]. An elegant strategy to increase selectivity and sensitivity is to form a fluorophore with the nucleobase of interest in situ. Liu et al. presented a bifunctional labeling agent that forms a fluorophore by reacting with both the aldehyde and the amine group of 5fC [43]. The reaction products with 5fU and abasic sites are not fluorescent. Another, intrinsic advantage is that no purification from the unreacted, non-fluorescent labeling agent is necessary. While not relying on common coupling chemistry (NHS, click reactions), this has potential to be a valuable, orthogonal labeling strategy, especially for multiplexing in optical mapping techniques.
Epigenetic modifications for which no specific covalent coupling reactions were reported can be marked indirectly by selective glycosylases: After excision of the nucleotide of interest, a modified derivative of the nucleotide can be incorporated at the position of the lesion [44]. Several reports have used this strategy to introduce fluorescent nucleotides instead of various DNA damage lesions for optical detection of DNA damage [29,45]. 5-Carboxycytosine (5caC), for example, can be selectively excised by thymine DNA glycosylase (TDG) [46] and could be replaced by a fluorescent nucleotide via in-vitro DNA repair.
Surface-enhanced Raman scattering
Fluorescence-based techniques for epigenetic analysis have very high sensitivity, but rely on labeling and provide only limited possibilities for multiplexing due to a limited number of fluorescent dyes that can be detected in parallel within the currently accessible spectral window. Surface-enhanced Raman scattering (SERS) is a promising technique in this respect because it can be sensitive enough to detect single molecules while providing a chemical fingerprint allowing for label-free analyte detection [47,48]. SERS is based on the strong Raman signal enhancement in close vicinity of gold or silver nanostructures upon excitation of their surface plasmon resonance.
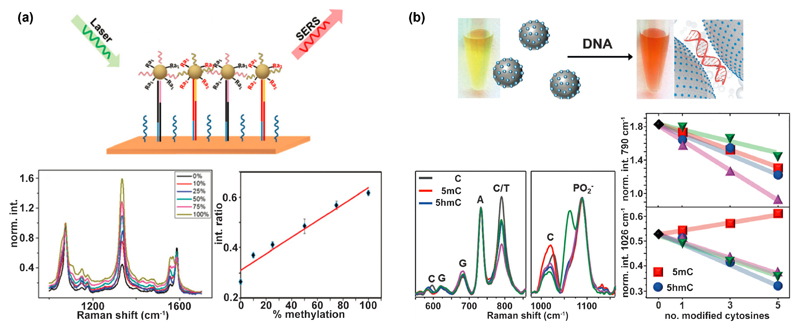
Gold or silver nanoparticles coated with dye molecules (‘SERS tags’) represent a labeling alternative to fluorescent tags. SERS tags could quantify methylated DNA in breast cancer cell lines and a serum-derived DNA sample [49] (Figure 4 (a)). These tags consisted of gold nanoparticles coated with: (1) DNA oligos that bind either T- or C-rich DNA after bisulfite treatment, and (2) dye molecules that act as a barcode by having a strong and characteristic Raman signature. In a complementary approach, one SERS tag served as an internal normalizing control, while the other one was the methylation indicator [50]. Since Raman spectra have very narrow bandwidths, SERS tags are suitable multiplexing labels.
Figure 4.
SERS-based methods for detection of epigenetic marks. (a) Nanoparticles functionalized with DNA and dye molecules allow to determine the degree of methylation. Adapted from Ref. [49] with permission from The Royal Society of Chemistry. (b) Direct label-free detection of various DNA modifications by silver nanoparticles that aggregate in the presence of DNA, leading to a shift of their surface plasmon resonance and consequently a color change of the nanoparticle solution. Two SERS bands show an intensity change for the modified nucleobases 5mC and 5hmC [53].
SERS can also serve as a tool for the direct detection of DNA modifications. Barhoumi et al. demonstrated the label-free detection of N6-methyladenine (6mA), 5mC, 5hmC and 8-oxo-guanine by SERS [51]. Guerrini et al. extended this approach and demonstrated the detection of single base mismatches and base methylations (5mC and 6mA) in DNA duplexes [52]. At the same time, hybridization events could be quantified. In another study, several C modifications (among them 5mC and 5hmC) were quantified in ssDNA and dsDNA [53] (Figure 4 (b)). The authors applied positively-charged spermine-coated silver nanoparticles, which aggregate upon addition of DNA due to electrostatic interactions. In this way, DNA is trapped in the plasmonic hot spot and detection of pg-amounts of DNA is possible, making pre-amplification of DNA samples obsolete. The method is also suitable to quantify the relative amount of DNA base modifications within a DNA strand. C modifications lead to an intensity decrease and band shifts in the SERS spectra. The initial proof-of-concept studies demonstrate the capabilities of SERS and its potential for label-free detection of DNA modifications. When data are analyzed with chemometric techniques, there is no fundamental limit for multiplexing. In the future, robust procedures for sample preparation and analysis need to be developed.
Summary and outlook
Epigenetic analysis is emerging as an important field for biotechnological development. Optical detection schemes adapted to quantify various epigenetic marks are becoming available and will potentially become accessible to the wider research and biomedical community. Clearly, the single-molecule sensitivity offered by fluorescence has enabled extremely precise measurements on small amounts of genetic material. Nevertheless, some of the advantages of optical detection have not been fully exploited. Most noticeable is the lack of multiplexing experiments, with a maximum of three-color detection reported [38]. Understanding the relations between the various epigenetic marks is one of the most exciting prospects in the field. With the right choice of optics and dyes, multiple modifications may be recorded simultaneously, may it be by global quantification of fluorescence or by optical mapping. In any case, the number of observables is limited by the orthogonality of the labeling reactions and the spectral width of the fluorophores. Alternative optical labels such as Raman tags, with their unique spectral fingerprint, promise to further expand this scope. Ultimately, label-free detection will provide analytical solutions that decrease to a minimum the processing time and effort of the analyzed sample. It is now to be seen if any of the concepts described here will establish itself as a gold standard for analytical epigenetics.
Acknowledgements
YE would like to thank financial support from the BeyondSeq consortium (EC program 63489), the European Research Council starter grant (grant no. 337830), the European Research Council Proof of Concept grant by the EU-Horizon2020 program (grant no. 767931) and the NIH-R21 grant (1R21ES028015-01). IB acknowledges an ERC Consolidator Grant (grant no. 772752) and the German Research Foundation (DFG; BA4026/5-2).
References
- 1.Soloway PD. Analysis of Combinatorial Epigenomic States. ACS Chem Biol. 2016;11:621–631. doi: 10.1021/acschembio.5b00833. [∎ Comprehensive review highlighting existing and future technologies for epigenetic analysis.] [DOI] [PubMed] [Google Scholar]
- 2.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 3.Zentner GE, Henikoff S. High-resolution digital profiling of the epigenome. Nat Rev Genet. 2014;15:814–827. doi: 10.1038/nrg3798. [DOI] [PubMed] [Google Scholar]
- 4.Lisanti S, Omar WAW, Tomaszewski B, De Prins S, Jacobs G, Koppen G, Mathers JC, Langie SAS. Comparison of Methods for Quantification of Global DNA Methylation in Human Cells and Tissues. PLoS One. 2013;8:e79044. doi: 10.1371/journal.pone.0079044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Önder Ö, Sidoli S, Carroll M, Garcia BA. Progress in epigenetic histone modification analysis by mass spectrometry for clinical investigations. Expert Rev Proteomics. 2015;12:499–517. doi: 10.1586/14789450.2015.1084231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyun BR, McElwee JL, Soloway PD. Single molecule and single cell epigenomics. Methods. 2015;72:41–50. doi: 10.1016/j.ymeth.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha T. Probing Nature’s Nanomachines One Molecule at a Time. Biophys J. 2016;110:1004–1007. doi: 10.1016/j.bpj.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boettiger AN, Bintu B, Moffitt JR, Wang S, Beliveau BJ, Fudenberg G, Imakaev M, Mirny LA, Wu CT, Zhuang X. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature. 2016;529:418–422. doi: 10.1038/nature16496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy-Sakin M, Grunwald A, Kim S, Gassman NR, Gottfried A, Antelman J, Kim Y, Ho SO, Samuel R, Michalet X, et al. Toward single-molecule optical mapping of the epigenome. ACS Nano. 2014;8:14–26. doi: 10.1021/nn4050694. [∎ Review describing fundamental single-molecule work in the context of epigenetic mapping.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coons AH, Creech HJ, Jones RN. Immunological Properties of an Antibody Containing a Fluorescent Group. Exp Biol Med. 1941;47:200–202. [Google Scholar]
- 11.Jørgensen HF, Adie K, Chaubert P, Bird AP. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 2006;34:e96. doi: 10.1093/nar/gkl527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Rausch C, Hastert FD, Boneva B, Filatova A, Patil SJ, Nuber UA, Gao Y, Zhao X, Cardoso MC. Methyl-CpG binding domain protein 1 regulates localization and activity of Tet1 in a CXXC3 domain-dependent manner. Nucleic Acids Res. 2017;45:7118–7136. doi: 10.1093/nar/gkx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, Zichi D. Aptamers and the RNA world, past and present. Cold Spring Harb Perspect Biol. 2012;4:a003582. doi: 10.1101/cshperspect.a003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cipriany BR, Zhao R, Murphy PJ, Levy SL, Tan CP, Craighead HG, Soloway PD. Single molecule epigenetic analysis in a nanofluidic channel. Anal Chem. 2010;82:2480–2487. doi: 10.1021/ac9028642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cipriany BR, Murphy PJ, Hagarman JA, Cerf A, Latulippe D, Levy SL, Benitez JJ, Tan CP, Topolancik J, Soloway PD, et al. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc Natl Acad Sci. 2012;109:8477–8482. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy PJ, Cipriany BR, Wallin CB, Ju CY, Szeto K, Hagarman JA, Benitez JJ, Craighead HG, Soloway PD. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci. 2013;110:7772–7777. doi: 10.1073/pnas.1218495110. [∎ Single chromatin molecule analysis at the nanoscale (SCAN) is used to study the epigenetic state of tumor cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shema E, Jones D, Shoresh N, Donohue L, Ram O, Bernstein BE. Single-molecule decoding of combinatorially modified nucleosomes. Science. 2016;352:717–721. doi: 10.1126/science.aad7701. [∎ ∎ Combined nucleosome and sequence analysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, DiMeo J, Efcavitch JW, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 20.Nadal S, Raj R, Mohammed S, Davis BG. Synthetic post-translational modification of histones. Curr Opin Chem Biol. 2018;45:35–47. doi: 10.1016/j.cbpa.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 21.David Y, Muir TW. Emerging Chemistry Strategies for Engineering Native Chromatin. J Am Chem Soc. 2017;139:9090–9096. doi: 10.1021/jacs.7b03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–75. doi: 10.1038/nbt.1732. [∎ First report on utilizing a sythetic cofactor as a chemical handle for labeling 5hmC.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qing Y, Tian Z, Bi Y, Wang Y, Long J, Song CX, Diao J. Quantitation and mapping of the epigenetic marker 5-hydroxymethylcytosine. BioEssays. 2017;39 doi: 10.1002/bies.201700010. 1700010. [DOI] [PubMed] [Google Scholar]
- 24.Michaeli Y, Shahal T, Torchinsky D, Grunwald A, Hoch R, Ebenstein Y. Optical detection of epigenetic marks: Sensitive quantification and direct imaging of individual hydroxymethylcytosine bases. Chem Commun. 2013;49:8599–8601. doi: 10.1039/c3cc42543f. [DOI] [PubMed] [Google Scholar]
- 25.Shahal T, Gilat N, Michaeli Y, Redy-Keisar O, Shabat D, Ebenstein Y. Spectroscopic quantification of 5-hydroxymethylcytosine in genomic DNA. Anal Chem. 2014;86:8231–8237. doi: 10.1021/ac501609d. [DOI] [PubMed] [Google Scholar]
- 26.Müller V, Westerlund F. Optical DNA mapping in nanofluidic devices: principles and applications. Lab Chip. 2017;17:579–590. doi: 10.1039/c6lc01439a. [DOI] [PubMed] [Google Scholar]
- 27.Levy-Sakin M, Ebenstein Y. Beyond sequencing: Optical mapping of DNA in the age of nanotechnology and nanoscopy. Curr Opin Biotechnol. 2013;24:690–698. doi: 10.1016/j.copbio.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Gilat N, Tabachnik T, Shwartz A, Shahal T, Torchinsky D, Michaeli Y, Nifker G, Zirkin S, Ebenstein Y. Single-molecule quantification of 5-hydroxymethylcytosine for diagnosis of blood and colon cancers. Clin Epigenetics. 2017;9:70. doi: 10.1186/s13148-017-0368-9. [∎ First diagnostic demonstration of single-molecule epigenetics.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zirkin S, Fishman S, Sharim H, Michaeli Y, Don J, Ebenstein Y. Lighting up individual DNA damage sites by in vitro repair synthesis. J Am Chem Soc. 2014;136:7771–7776. doi: 10.1021/ja503677n. [DOI] [PubMed] [Google Scholar]
- 30.Gabrieli T, Sharim H, Nifker G, Jeffet J, Shahal T, Arielly R, Hoch L, Arbib N, Michaeli Y, Ebenstein Y. Epigenetic Optical Mapping of 5-Hydroxymethylcytosine in Nanochannel Arrays. ACS Nano. 2018;12:7148–7158. doi: 10.1021/acsnano.8b03023. [∎ Genome-wide optical epigenetic profiling of 5hmC at the single-molecule level.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carell T, Kurz MQ, Müller M, Rossa M, Spada F. Non-canonical Bases in the Genome: The Regulatory Information Layer in DNA. Angew Chemie - Int Ed. 2018;57:4296–4312. doi: 10.1002/anie.201708228. [DOI] [PubMed] [Google Scholar]
- 32.Lukinavičius G, Lapiene V, Staševskij Z, Dalhoff C, Weinhold E, Klimašauskas S. Targeted labeling of DNA by methyltransferase-directed transfer of activated groups (mTAG) J Am Chem Soc. 2007;129:2758–2759. doi: 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- 33.Hanz GM, Jung B, Giesbertz A, Juhasz M, Weinhold E. Sequence-specific labeling of nucleic acids and proteins with methyltransferases and cofactor analogues. J Vis Exp. 2014;93:e52014. doi: 10.3791/52014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klimašauskas S, Weinhold E. A new tool for biotechnology: AdoMet-dependent methyltransferases. Trends Biotechnol. 2007;25:99–104. doi: 10.1016/j.tibtech.2007.01.006. [∎ Describes the synthetic manipulation of methyltransferase cofactors for DNA labeling.] [DOI] [PubMed] [Google Scholar]
- 35.Grunwald A, Dahan M, Giesbertz A, Nilsson A, Nyberg LK, Weinhold E, Ambjörnsson T, Westerlund F, Ebenstein Y. Bacteriophage strain typing by rapid single molecule analysis. Nucleic Acids Res. 2015;43:e117. doi: 10.1093/nar/gkv563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kriukiene E, Labrie V, Khare T, Urbanavičiute G, Lapinaite A, Koncevičius K, Li D, Wang T, Pai S, Ptak C, et al. DNA unmethylome profiling by covalent capture of CpG sites. Nat Commun. 2013;4 doi: 10.1038/ncomms3190. 2190. [∎ First demonstration of utilizing methyltransferases for epigenetic mapping.] [DOI] [PubMed] [Google Scholar]
- 37.Gilboa T, Torfstein C, Juhasz M, Grunwald A, Ebenstein Y, Weinhold E, Meller A. Single-Molecule DNA Methylation Quantification Using Electro-optical Sensing in Solid-State Nanopores. ACS Nano. 2016;10:8861–8870. doi: 10.1021/acsnano.6b04748. [DOI] [PubMed] [Google Scholar]
- 38.Grunwald A, Sharim H, Gabrieli T, Michaeli Y, Torchinsky D, Juhasz M, Wagner KR, Pevsner J, Reifenberger J, Hastie AR, et al. Reduced representation optical methylation mapping (R2OM2) bioRxiv. 2017 doi: 10.1101/113522. [∎ Genome-wide optical profiling of DNA methylation on the single-molecule level.] [DOI] [Google Scholar]
- 39.Jeffet J, Kobo A, Su T, Grunwald A, Green O, Nilsson AN, Eisenberg E, Ambjörnsson T, Westerlund F, Weinhold E, et al. Super-Resolution Genome Mapping in Silicon Nanochannels. ACS Nano. 2016;10:9823–9830. doi: 10.1021/acsnano.6b05398. [DOI] [PubMed] [Google Scholar]
- 40.Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Commun. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song C-X, Diao J, Brunger AT, Quake SR. Simultaneous single-molecule epigenetic imaging of DNA methylation and hydroxymethylation. Proc Natl Acad Sci. 2016;113:4338–4343. doi: 10.1073/pnas.1600223113. [∎ ∎ One of the few examples for multiplexing.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dietzsch J, Feineis D, Höbartner C. Chemoselective labeling and site-specific mapping of 5-formylcytosine as a cellular nucleic acid modification. FEBS Lett. 2018;592:2032–2047. doi: 10.1002/1873-3468.13058. [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Wang Y, Yang W, Wu F, Zeng W, Chen Z, Huang J, Zou G, Zhang X, Wang S, et al. Fluorogenic labeling and single-base resolution analysis of 5-formylcytosine in DNA. Chem Sci. 2017;8:7443–7447. doi: 10.1039/c7sc03685j. [∎ Selective labeling of 5fC without side reactions.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riedl J, Ding Y, Fleming AM, Burrows CJ. Identification of DNA lesions using a third base pair for amplification and nanopore sequencing. Nat Commun. 2015;6:8807. doi: 10.1038/ncomms9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J, Kim Y, Lim S, Jo K. Single-molecule visualization of ROS-induced DNA damage in large DNA molecules. Analyst. 2016;141:847–852. doi: 10.1039/c5an01875g. [DOI] [PubMed] [Google Scholar]
- 46.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zrimsek AB, Chiang N, Mattei M, Zaleski S, McAnally MO, Chapman CT, Henry AI, Schatz GC, Van Duyne RP. Single-Molecule Chemistry with Surface- and Tip-Enhanced Raman Spectroscopy. Chem Rev. 2017;117:7583–7613. doi: 10.1021/acs.chemrev.6b00552. [DOI] [PubMed] [Google Scholar]
- 48.Heck C, Kanehira Y, Kneipp J, Bald I. Placement of Single Proteins within the SERS Hot Spots of Self-Assembled Silver Nanolenses. Angew Chemie - Int Ed. 2018;57:7444–7447. doi: 10.1002/anie.201801748. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Wee EJH, Trau M. Highly sensitive DNA methylation analysis at CpG resolution by surface-enhanced Raman scattering via ligase chain reaction. Chem Commun. 2015;51:10953–10956. doi: 10.1039/c5cc03921e. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Wee EJH, Trau M. Accurate and sensitive total genomic DNA methylation analysis from sub-nanogram input with embedded SERS nanotags. Chem Commun. 2016;52:3560–3563. doi: 10.1039/c6cc00547k. [DOI] [PubMed] [Google Scholar]
- 51.Barhoumi A, Halas NJ. Detecting chemically modified DNA bases using surface-enhanced raman spectroscopy. J Phys Chem Lett. 2011;2:3118–3123. doi: 10.1021/jz201423b. [∎ Fundamental study showing SERS spectra of a variety of base modifications.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerrini L, Krpetić Ž, Van Lierop D, Alvarez-Puebla RA, Graham D. Direct surface-enhanced Raman scattering analysis of DNA duplexes. Angew Chemie - Int Ed. 2015;54:1144–1148. doi: 10.1002/anie.201408558. [DOI] [PubMed] [Google Scholar]
- 53.Morla-Folch J, Xie HN, Gisbert-Quilis P, De Pedro SG, Pazos-Perez N, Alvarez-Puebla RA, Guerrini L. Ultrasensitive direct quantification of nucleobase modifications in DNA by surface-enhanced Raman scattering: the case of cytosine. Angew Chemie - Int Ed. 2015;54:13650–13654. doi: 10.1002/anie.201507682. [∎ Label-free quantification of 5hmC and 5mC levels via SERS.] [DOI] [PubMed] [Google Scholar]