Abstract
Data concerning the human microbiota composition during Clostridioides (Clostridium) difficile infection (CDI) using next-generation sequencing are still limited. We aimed to confirm key features indicating tcdB positive patients and compare the microbiota composition between subgroups based on toxin gene load (tcdB gene) and presence of significant diarrhea. Ninety-nine fecal samples from 79 tcdB positive patients and 20 controls were analyzed using 16S rRNA gene sequencing. Chao1 index for alpha diversity were calculated and principal coordinate analysis was performed for beta diversity using Quantitative Insights into Microbial Ecology (QIIME) pipeline. The mean relative abundance in each group was compared at phylum, family, and genus levels. There were significant alterations in alpha and beta diversity in tcdB positive patients (both colonizer and CDI) compared with those in the control. The mean Chao1 index of tcdB positive patients was significantly lower than the control group (P<0.001), whereas there was no significant difference between tcdB groups and between colonizer and CDI. There were significant differences in microbiota compositions between tcdB positive patients and the control at phylum, family, and genus levels. Several genera such as Phascolarctobacterium, Lachnospira, Butyricimonas, Catenibacterium, Paraprevotella, Odoribacter, and Anaerostipes were not detected in most CDI cases. We identified several changes in the microbiota of CDI that could be further evaluated as predictive markers. Microbiota differences between clinical subgroups of CDI need to be further studied in larger controlled studies.
Introduction
Clostridioides (Clostridium) difficile is a common cause of antibiotic-associated colitis. CDI rates have plateaued in the United States since about 2010 although rates have declined remarkably in England and other parts of Europe since their peak before 2010 [1–4]. CDI has a broad spectrum of clinical features, ranging from mild diarrhea to severe diseases such as toxic megacolon. Although toxigenic C. difficile is detected in patient samples, many patients do not meet the criteria for significant diarrhea [5–7].
The most important risk factor for CDI is antibiotic use [8]. In susceptible hosts, microbiota-mediated colonization resistance is diminished partly by a reduction in the diversity of the gut microbiota caused by antibiotics [1, 8–11]. After antibiotic treatment for CDI, there is a phase of the restoration of normal microbiota, which itself averts recurrence of CDI [8, 12]. Thus, recently developed therapy, such as fecal microbiota transplantation (FMT), tries to restores gut microbiota diversity instead of the direct eradication of the pathogen [13, 14].
Decreased diversity and alteration of the gut microbiota composition in CDI has been shown in previous studies using various techniques from culture-based methods to high throughput sequencing [13, 15, 12, 15–17]. However, data for the human microbiota composition during CDI are still limited and comparison between low and high toxin gene load or between colonizer and overt CDI rarely performed.
The present study aimed to compare the composition of the gut microbiota in healthy controls and C. difficile toxin positive patients using sequencing of the 16S rRNA gene. We attempted to find key features indicating CDI and to compare the microbiota composition between subgroups based on the toxin gene load and clinical criteria in C. difficile toxin positive patients.
Materials and methods
Clinical samples
This study was approved by the Institutional Review Board of the Konkuk University Medical Center, Seoul, Korea. This study included 99 fecal samples from patients, which were submitted to our center for laboratory tests. These included 79 tcdB positive samples by real-time PCR (Xpert C. difficile system, Cepheid, Sunnyvale, CA, USA) from March 2017 to October 2017 and the other 20 fecal samples were obtained from healthy controls whose samples were submitted for occult blood test of general health examination (controls). This study required neither study-specific nor any other interventions and the data were analyzed anonymously. Therefore, written informed consent from the enrolled patients was waived by the ethics committee.
Clinical data collection
We collected clinical data through chart review, including demographic data and laboratory data (white cell count, serum creatinine, and albumin concentrations tested within 3 days of fecal sample collection). We obtained the baseline serum creatinine concentrations from tests performed more than 6 months before study entry. The clinical characteristics are described in Table 1.
Table 1. Characteristics of study population.
| Control | tcdB positive | P | low tcdB | high tcdB | P | |
|---|---|---|---|---|---|---|
| Number | 20 | 79 | 49 | 30 | ||
| Female (%) | 50 (50%) | 41 (51.9%) | 1 | 23 (46.9%) | 17 (58.6%) | 0.401 |
| Age (years, SD) | 62.2 (14.4) | 62.5 (19.9) | 0.8724 | 59.9 (19.8) | 64.0 (17.5) | 0.3538 |
| WBC (109/L, SD) | 8,932 (7,886) | - | 8,633 (8,804) | 9,448 (6,097) | 0.6297 | |
| 50% rise in creatinine (%) | 9 (11.4%) | - | 5 (10.0%) | 4 (13.8%) | 1 | |
| Albumin (g/dL, SD) | 3.3 (0.47) | - | 3.3 (0.42) | 3.3 (0.55) | 0.8095 | |
| CDI* (%) | 58 (73.4%) | - | 40 (80.0%) | 18 (62.1%) | 0.3904 |
* (≥ 3 unformed stools in 24 hours)
Abbreviation: CDI, C. difficile infection
The tcdB positive samples were categorized according to tcdB gene load (low tcdB, n = 49 and high tcdB, n = 30) based on the cycle threshold (Ct) values of tcdB real-time PCR suggested from previous study [18] and the presence of significant diarrhea; colonizer (< 3 unformed stools in 24 hours, n = 21) and CDI (≥ 3 unformed stools in 24 hours, n = 58)[2, 8].
Library preparation and sequencing
Bacterial DNA was extracted from 200 mg of stool sample using a QIAamp DNA stool mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Bacterial 16S rRNA genes were amplified by polymerase chain reaction (PCR) using an Ion 16SMetagenomics Kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The kit includes 2 primer tubes and each tube includes 3 primer sets that amplify the hypervariable regions of 16S rRNA (V2, 4, 8 and V3, 6–7, 9, respectively). PCR amplicons were purified using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA). Sequencing libraries were then prepared using an Ion Plus Fragment Library Kit and Ion Xpress Barcode Adapters (ThermoFisher Scientific) according to the manufacturer’s protocol. Prepared libraries were quantified using a High Sensitivity DNA kit on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Template preparation and sequencing were performed using the Ion Chef System and Ion S5 XL system with Ion 530 Chip Kit (ThermoFisher Scientific).
Data analysis
Sequencing data were analyzed using the Torrent Suite software 5.8.0 (ThermoFisher Scientific) to filter out low quality and polyclonal reads, as well as to trim any adaptor sequences at the 3′ end. After filtering, the sequencing data were demultiplexed and exported as FASTQ files. The FASTQ files were processed using the Quantitative Insights into Microbial Ecology (QIIME) pipeline 1.9.1.[19]. After quality filtering, 10,530,756 sequences were obtained, with a mean of 96,918 sequences per sample (min: 6,183, max: 374,959). Operational taxonomic units (OTUs) were clustered based on 97% sequence similarity with at least 10 identical sequences and assigned against the curated Greengenes v13.8 reference database at the QIIME web site (http://qiime.org/home_static/dataFiles.html). The reference database was modified by excluding both the IDs and sequences of OTUs that are not assigned to a taxonomy level below order. Alpha diversity was assessed by observed OTUs and Chao1 and also included unidentified OTU. Alpha and beta diversity measures were calculated by QIIME [20]. To compare the microbial diversity between samples, qualitative (unweighted UniFrac) and quantitative distances (weighted UniFrac) were calculated. Microbial diversity was visualized using Principal Coordinate Analysis (PCoA) calculated by QIIME. The mean relative abundance in each group was compared at the phylum, family, and genus levels.
Statistical analysis
The difference between the continuous variables was analyzed using Student’s t-test or the Mann–Whitney U test, and that between categorical variables was analyzed using the chi-squared test, Fisher’s exact test, or the McNemar test. The Kruskal–Wallis test and one-way analysis of variance (ANOVA), followed by the Games–Howel's posthoc test, were used to assess the differences between groups. Permutational multivariate analysis of variance (PERMANOVA) analysis between groups was performed using QIIME. Statistical analysis was performed using MedCalc Statistical Software (version 15.8, MedCalc Software, Mariakerke, Belgium) and IBM SPSS Statistics 22.0 (IBM Corporation, Armonk, NY, USA). P values less than 0.05 were considered statistically significant.
Results
Comparison of alpha diversity of the gut microbiota among the control and tcdB positive patients
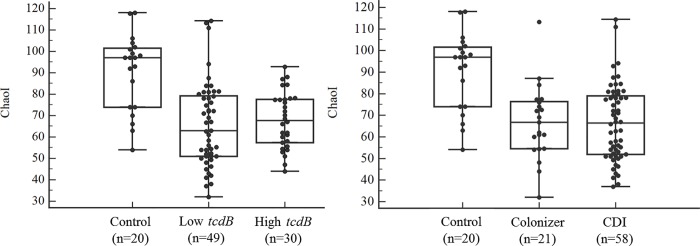
We evaluated the differences in intra-individual variability (alpha diversity) between the control and each category of tcdB positive patients. The distribution of the Chao1 indexes in each group is presented in Fig 1. The mean Chao1 index of the control group was significantly higher than that of the tcdB positive patients (P < 0.001). The mean Chao1 index between each category of tcdB positive patients was not significantly different (P = 0.808 and 0.999 between low and high tcdB groups and between colonizer and CDI, respectively).
Fig 1. Alpha diversity in control and each category of tcdB positive patients.
The distribution of ChaoI diversity index was presented between groups (A, control, low and high tcdB; B, control, colonizer and CDI, respectively). Black dot line, median value; gray horizontal line, interquartile value.
Comparison of beta diversity of the gut microbiota
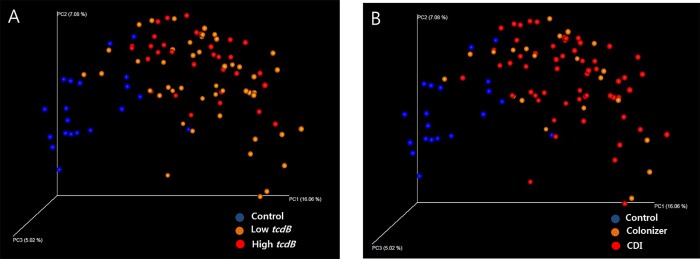
Principal Coordinate Analysis (PCoA) using weighted and unweighted UniFrac matrix was performed to evaluate the beta diversity among the samples in each group. In both analysis, the control and tcdB positive patients clustered separately (PERMANOVA P = 0.001), while the tcdB positive patients categorized by tcdB gene load (Fig 2A) or presence of significant diarrhea (colonizer vs. CDI)(Fig 2B) could not be separated (only analysis by unweighted UniFrac matrix was shown).
Fig 2.
Evaluation of beta-diversity in control (blue) and tcdB positive patients (A, low and high tcdB; B, colonizer and CDI, orange and red, respectively). Principal coordinate analysis (PCoA) was performed using unweighted UniFrac distances of 16S rRNA gene sequences. The each axis represents intersample variation.
The comparison of mean relative abundance in each group at the phylum level is shown in Table 2. The predominant phyla were Firmicutes and Bacteroidetes in the control group and Firmicutes, Bacteroidetes, and Proteobacteria in the tcdB positive patients. The mean proportion of Proteobacteria was significantly higher in the tcdB positive patients compared with that in the control group (32.44% vs. 21.44%, P = 0.008). The mean proportion of Firmicutes was significantly lower in the high tcdB group compared with that in the low tcdB group (27.67% vs. 37.90%, P = 0.038). The comparisons of mean relative abundance in each group at the family and at genus levels are shown in Tables 3 and 4. In all groups, the Bacteroidaceae family was predominant, followed by the Lachnospiraceae, Enterobacteriaceae, and Ruminococcaceae. The Lachnospiraceae, Ruminococcaceae, and Prevotellaceae showed a significantly lower mean proportion in the tcdB positive patients compared with that in the control (P = 0.003, 0.000, and 0.000, respectively). The Enterobacteriaceae, Porphyromonadaceae, and Enterococcaceae showed a significantly higher mean proportion in the tcdB positive patients compared with that in the control (P = 0.005, 0.000 and 0.000, respectively)(Table 3). Genera including Prevotella, Phascolarctobacterium, Haemophilus, Lachnospira, Coprococcus, Dialister, Butyricimonas, Catenibacterium, Faecalibacterium, Paraprevotella, Odoribacter, and Anaerostipes were present at a significantly lower proportion in the tcdB positive patients compared with that in the control (Table 4). The genera Parabacteroides, Enterococcus, Veillonella, Klebsiella, and Akkermansia were present at a significantly higher proportion in the tcdB positive patients compared with that in the control. The genera Klebsiella and Akkermansia were present at a significantly different proportion between high tcdB group and low tcdB, and Oscillospira was present at a significantly different proportion between colonizer and CDI.
Table 2. Comparison of the mean relative abundance (%) in each group at phylum level.
| Phylum | Control (n = 20) |
tcdB positive (n = 79) | P | tcdB positive | |||||
|---|---|---|---|---|---|---|---|---|---|
| low tcdB (n = 49) |
high tcdB (n = 30) |
P | colonizer (n = 21) |
CDI (n = 58) |
P | ||||
| Firmicutes | 38.73 | 34.02 | 0.196 | 37.90 | 27.67 | 0.038 | 36.30 | 33.19 | 0.595 |
| Bacteroidetes | 36.14 | 30.26 | 0.085 | 27.02 | 35.56 | 0.107 | 28.12 | 31.03 | 0.619 |
| Proteobacteria | 21.44 | 32.44 | 0.008 | 31.71 | 33.64 | 0.703 | 32.48 | 32.43 | 0.992 |
| Actinobacteria | 2.42 | 1.33 | 0.254 | 1.74 | 0.66 | 0.115 | 1.18 | 1.38 | 0.830 |
| Fusobacteria | 0.09 | 0.73 | 0.354 | 0.32 | 1.39 | 0.229 | 0.54 | 0.79 | 0.744 |
Abbreviation: CDI, C. difficile infection. Phyla with mean relative abundance >1.0 were described in Table 2.
Table 3. Comparison of the mean relative abundance (%) in each group at family level.
| Family | Control (n = 20) |
tcdB positive (n = 79) | P | tcdB positive | |||||
|---|---|---|---|---|---|---|---|---|---|
| low tcdB (n = 49) |
high tcdB (n = 30) |
P | colonizer (n = 21) |
CDI (n = 58) |
P | ||||
| Bacteroidaceae | 20.28 | 22.22 | 0.545 | 19.17 | 27.21 | 0.093 | 18.61 | 23.53 | 0.353 |
| Lachnospiraceae | 16.51 | 9.00 | 0.003 | 8.24 | 10.25 | 0.399 | 10.91 | 8.31 | 0.319 |
| Enterobacteriaceae | 13.75 | 28.09 | 0.005 | 27.55 | 28.98 | 0.769 | 27.82 | 28.19 | 0.945 |
| Ruminococcaceae | 11.39 | 4.22 | 0.000 | 3.52 | 5.36 | 0.176 | 6.86 | 3.27 | 0.063 |
| Prevotellaceae | 8.25 | 1.22 | 0.000 | 1.30 | 1.08 | 0.404 | 0.60 | 1.44 | 0.528 |
| Veillonellaceae | 4.40 | 3.34 | 0.435 | 2.43 | 4.84 | 0.093 | 2.77 | 3.55 | 0.593 |
| Alcaligenaceae | 2.91 | 2.00 | 0.396 | 1.39 | 2.99 | 0.154 | 2.60 | 1.78 | 0.476 |
| Pasteurellaceae | 2.23 | 0.23 | 0.034 | 0.17 | 0.32 | 0.586 | 0.01 | 0.31 | 0.323 |
| Bifidobacteriaceae | 2.13 | 0.53 | 0.124 | 0.60 | 0.43 | 0.677 | 0.37 | 0.59 | 0.602 |
| Porphyromonadaceae | 1.29 | 4.29 | 0.000 | 4.35 | 4.20 | 0.917 | 5.10 | 4.00 | 0.501 |
| Streptococcaceae | 1.19 | 0.68 | 0.253 | 0.84 | 0.40 | 0.178 | 0.51 | 0.74 | 0.610 |
| Erysipelotrichaceae | 1.13 | 1.10 | 0.938 | 1.15 | 1.01 | 0.755 | 0.67 | 1.25 | 0.233 |
| Rikenellaceae | 1.10 | 1.36 | 0.797 | 1.02 | 1.92 | 0.380 | 2.23 | 1.04 | 0.293 |
| Lactobacillaceae | 1.07 | 4.07 | 0.079 | 6.50 | 0.09 | 0.017 | 3.31 | 4.35 | 0.782 |
| Enterococcaceae | 0.56 | 8.70 | 0.000 | 12.87 | 1.89 | 0.001 | 8.86 | 8.65 | 0.964 |
Abbreviation: CDI, C. difficile infection. Familes with mean relative abundance >1.0 or with significant differences were described in Table 3.
Table 4. The mean relative abundance (%) of selected genera in each group.
| Genus | Control (n = 20) |
tcdB positive (n = 79) | P | tcdB positive | |||||
|---|---|---|---|---|---|---|---|---|---|
| low tcdB (n = 49) |
high tcdB (n = 30) |
P | colonizer (n = 21) |
CDI (n = 58) |
P | ||||
| Bacteroides | 20.25 | 22.22 | 0.539 | 19.17 | 27.21 | 0.093 | 18.61 | 23.53 | 0.353 |
| Prevotella | 8.22 | 1.22 | 0.001 | 1.29 | 1.01 | 0.844 | 0.60 | 1.44 | 0.483 |
| Sutterella | 2.91 | 2.00 | 0.396 | 1.39 | 2.99 | 0.154 | 2.60 | 1.78 | 0.447 |
| Bifidobacterium | 2.13 | 0.53 | 0.124 | 0.59 | 0.43 | 0.678 | 0.37 | 0.59 | 0.603 |
| Phascolarctobacterium | 1.97 | 0.55 | 0.033 | 0.51 | 0.63 | 0.759 | 0.54 | 0.56 | 0.962 |
| Haemophilus | 1.96 | 0.21 | 0.040 | 0.16 | 0.29 | 0.615 | 0.01 | 0.29 | 0.320 |
| Lachnospira | 1.59 | 0.37 | 0.004 | 0.39 | 0.34 | 0.900 | 0.74 | 0.24 | 0.361 |
| Coprococcus | 1.29 | 0.48 | 0.008 | 0.55 | 0.37 | 0.516 | 0.47 | 0.48 | 0.969 |
| Parabacteroides | 1.28 | 4.12 | 0.000 | 4.29 | 3.84 | 0.760 | 5.04 | 3.78 | 0.441 |
| Oscillospira | 1.22 | 2.04 | 0.061 | 1.39 | 3.11 | 0.070 | 3.99 | 1.34 | 0.032 |
| Streptococcus | 1.17 | 0.66 | 0.250 | 0.83 | 0.40 | 0.179 | 0.51 | 0.72 | 0.619 |
| Dialister | 0.88 | 0.18 | 0.031 | 0.07 | 0.36 | 0.092 | 0.22 | 0.17 | 0.743 |
| Butyricimonas | 0.64 | 0.18 | 0.009 | 0.15 | 0.21 | 0.688 | 0.53 | 0.05 | 0.067 |
| Enterococcus | 0.56 | 8.70 | 0.000 | 12.8 | 0.89 | 0.001 | 8.86 | 8.65 | 0.964 |
| Catenibacterium | 0.53 | 0.00 | 0.018 | 0.00 | 0.00 | - | 0.00 | 0.00 | - |
| Veillonella | 0.52 | 2.38 | 0.005 | 1.54 | 3.75 | 0.115 | 1.38 | 2.74 | 0.331 |
| Faecalibacterium | 0.48 | 0.10 | 0.000 | 0.10 | 0.10 | 0.981 | 0.09 | 0.10 | 0.826 |
| Paraprevotella | 0.48 | 0.08 | 0.019 | 0.05 | 0.13 | 0.361 | 0.04 | 0.10 | 0.567 |
| Odoribacter | 0.46 | 0.12 | 0.008 | 0.05 | 0.24 | 0.216 | 0.53 | 0.05 | 0.419 |
| Anaerostipes | 0.38 | 0.01 | 0.019 | 0.00 | 0.02 | 0.298 | 0.04 | 0.00 | 0.232 |
| Klebsiella | 0.05 | 0.75 | 0.000 | 0.68 | 0.89 | 0.596 | 0.46 | 0.85 | 0.302 |
| Akkermansia | 0.01 | 0.14 | 0.025 | 0.01 | 0.35 | 0.022 | 0.26 | 0.09 | 0.222 |
Abbreviation: CDI, C. difficile infection. Only genera with mean relative abundance >1.0 or >0.1 with significant differences were described in Table 4.
The proportion of patients within the control and CDI groups harbouring detectable levels of specific genera are shown in Table 5. Prevotella, Phascolarctobacterium, Haemophilus, Lachnospira, Coprococcus, Dialister, Butyricimonas, Catenibacterium, Faecalibacterium, Paraprevotella, Odoribacter, and Anaerostipes were not detected in a considerable proportion of the CDI group (26.6% to 100.0%); however, proportion of “no detection” were significantly lower in the control group (0.0% to 55.0%).
Table 5. The proportion of samples with no detection of several genera in control and CDI.
| Control (n = 20) |
CDI (n = 58) |
P | |
|---|---|---|---|
| No detection of | |||
| Prevotella | 3 (15.0%) | 27 (46.6%) | 0.0128 |
| Phascolarctobacterium | 4 (20.0%) | 42 (72.4%) | <0.0001 |
| Haemophilus | 4 (20.0%) | 35 (60.4%) | 0.0020 |
| Lachnospira | 1 (5.0%) | 45 (77.6%) | <0.0001 |
| Coprococcus | 0 (0.0%) | 21 (26.6%) | - |
| Dialister | 6 (30.0%) | 44 (75.9%) | 0.0002 |
| Butyricimonas | 3 (15.0%) | 50 (86.2%) | <0.0001 |
| Catenibacterium | 11 (55.0%) | 58 (100.0%) | - |
| Faecalibacterium | 1 (5.0%) | 35 (60.3%) | <0.0001 |
| Paraprevotella | 9 (45.0%) | 51 (87.9%) | 0.0001 |
| Odoribacter | 4 (20.0%) | 47 (81.0%) | <0.0001 |
| Anaerostipes | 4 (20.0%) | 57 (98.3%) | <0.0001 |
Discussion
The gut microbiota plays a key role in maintaining normal homeostasis by modulating the immune system [21]. An altered intestinal microbiota can result from various influences, including antibiotics, diet, lifestyle, and hygiene. The state of the gut microbiota is also related to certain disease states, especially chronic inflammation or metabolic dysfunction, such as obesity [21]. Disruption of the gut microbiota is a key mechanism of CDI, and a decrease in species abundance and diversity has been consistently observed in previous studies using various methods [8, 11, 15]. However, more data on microbial composition for human CDI is required and comparison between low and high toxin gene load or between colonizer and overt CDI rarely performed. In terms of diagnosis of CDI, qualitative tcdB gene positivity by PCR cannot distinguish asymptomatic colonization from symptomatic infection [5]. Recent many studies suggested that toxin gene load (low Ct) as a predictor for free toxin positivity [18, 22–26] although conflicting results also exist on correlation between toxin load and disease outcome [22, 24, 27]. In this study, we categorized tcdB positive patients by the tcdB gene load and the presence of significant diarrhea (colonizer and CDI), and compared the gut microbiota between them. Moreover, there are very few data on the gut microbiota profile of the Korean population, which might have different dietary habits, such as the consumption of kimchi.
As expected, the alpha diversity index, in this case Chao1, was significantly lower in the tcdB positive patients compared with that in the control group. Other studies also showed decreased alpha diversity in CDI or antibiotic exposure group compared with the control [11, 15]. However, the diversity between low and high tcdB gene load or between colonizer and CDI showed no significant difference (Fig 1). A study with small study population also showed similar alpha diversity between CDI and asymptomatic colonizers [15]. In this study, colonizer did not meet criteria of significant criteria but they could not be included as healthy population because they were hospitalized patients. Healthy toxin-producing C. difficile colonizers were not included in our study and could be evaluated in further study. Decreased species abundance and diversity might be features of CDI but could also occur in many hospitalized patients without CDI. The development of overt CDI or more severe disease can be affected by host factors, such as immunity, age, or hospital stay [8, 28]. In this study, alpha diversity analysis also included unidentified OTUs by 97% sequence identity because unidentified OTU should be counted for diversity. OTUs with less than 10 sequences were discarded due to the possibility of error like many previous studies [29, 30]. Chao1 values could be changed whether low-abundance read is included in analysis. We compared Chao1 values between groups and these rules were applied to each group in same conditions. Similarly, PCoA showed evident separation between the control and tcdB positive patients, but mixed patterns between low and high tcdB gene load or between colonizer and CDI. Data on the comparison between subgroups of tcdB positive is lacking and we need to confirm this finding in a further study.
The relative abundance of specific OTUs among the total OTUs showed different trends between the control and tcdB positive patients. At the phylum level, compared with the control, tcdB positive patients showed a significantly higher mean relative abundance of Proteobacteria (P = 0.003). Decreased Bacteroidetes and increased Proteobacteria in CDI have been observed in previous studies [15, 31, 32]. Our results were similar and seem to be recurrent findings in CDI but these features were also observed in colonizer in this study. In this study, the decrease in the abundance of Bacteroidetes in the tcdB positive patients was not statistically significant (P = 0.085), which might be resulted from low statistical power due to the low number of subjects or specific features in our population. Decreased Bacteroidetes and increased Proteobacteria have been also observed after vancomycin treatment in CDI [17]. Only the Firmicutes phylum demonstrated a significantly lower proportion in the high tcdB group compared with that in the low tcdB group (P = 0.038). It could be associated with other bacteria such as Enterococcaceae. In contrast to the low tcdB group, in the high tcdB group, we could assume that C. difficile replication is high and leads to high toxin production.
At the family level, Lachnospiraceae, Ruminococcaceae, and Prevotellaceae showed significantly lower proportions in tcdB positive patients and not significantly different between colonizer and CDI. Decreases in Lachnospiraceae and Ruminococcaceae have also been reported in other studies [33] and the presence of these families has been shown to correlate with protection against CDI [34]. Enterobacteriaceae, Porphyromonadaceae, and Enterococcaceae were present at a significantly higher proportion in tcdB positive patients, and the increased Enterobacteriaceae and Enterococcaceae agreed with the findings of previous studies [11, 35].
The significant decreases in Prevotella and Faecalibacterium, and in the genera of the Lachnospiraceae, such as Lachnospira, Odoribacter, Coprococcus, and Anaerostipes were also important findings in CDI [11, 15]. Faecalibacterium and Bifidobacterium have health-promoting activities and their low prevalence is associated with many intestinal disorders, such as inflammatory bowel diseases [36, 37]. We observed that many genera of the Lachnospiraceae, such as Lachnospira, Odoribacter, Coprococcus, and Anaerostipes, a butyrate-producing organism, were present at significantly lower proportions in tcdB positive patients. Butyric acid decreases intestinal permeability and improves defense against infection [16, 38]. The changes in the proportions of these genera observed in the colonizer and CDI and did not differ by tcdB gene load. This finding suggested that depletion of these health-promoting genera occurred not only in severe disease, but also in mild forms or in various other conditions in hospitalized patients. Several genera, including Parabacteroides, Enterococcus, Veillonella, Klebsiella, and Akkermansia were present at significantly higher proportions in tcdB positive patients. Increased Parabacteroides, Enterococcus, Klebsiella, and Akkermansia in CDI have been observed in other studies and reflect a blooming phenomenon resulting from reduced ecological niche competition [11, 15, 39, 40]. However, Akkermansia (A. muciniphila) is associated with a healthier metabolic status in different settings of recent studies [41, 42].
Importantly, many genera were not detected in our analysis platform in most CDI patients; however, “no detection” was rarely observed in the control group (Table 5), especially for Phascolarctobacterium, Lachnospira, Butyricimonas, Catenibacterium, Paraprevotella, Odoribacter, and Anaerostipes (P < 0.0001) (Table 5). These features could be signature changes of CDI. These changes also occurred in colonizer and could be further studied [34, 43].
This study had several limitations. First, this study could not assess the cause–effect relationship between specific alterations of the microbiota and clinical status. There are also many covariates that could affect the gut microbiota composition [44]. In this study, we simply tried to compare the microbiota composition between subgroups rather than exploring the cause or independent factors responsible for specific alterations of the microbiota. Second, many OTUs did not have a complete taxonomy label at the genus level; for example, many OTUs of the Lachnospiraceae family, which is a common feature of this kind of study. Moreover, the results could be different between algorithms or programs used for OTU analysis [44, 45].
In conclusion, there were significant alterations in the alpha and beta diversity in tcdB positive patients (both colonizer and overt CDI) compared with those in the control. We identified several changes in the microbiota of CDI that could be further evaluated as predictive markers. Microbiota differences between clinical subgroups of tcdB positive patients require further study in larger controlled studies.
Data Availability
Data are uploaded to a public repository (Dryad Digital Repository, doi:10.5061/dryad.30kk201).
Funding Statement
This research (HM) was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2017R1A2B4005402). SH and JK are employed by and receive salary from BioCore Co Ltd. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Martin JS, Monaghan TM, Wilcox MH. Clostridium difficile infection: epidemiology, diagnosis and understanding transmission. Nature reviews Gastroenterology & hepatology. 2016;13(4):206–16. Epub 2016/03/10. 10.1038/nrgastro.2016.25 . [DOI] [PubMed] [Google Scholar]
- 2.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infection control and hospital epidemiology. 2010;31(5):431–55. Epub 2010/03/24. 10.1086/651706 . [DOI] [PubMed] [Google Scholar]
- 3.Bartlett JG. Historical perspectives on studies of Clostridium difficile and C. difficile infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2008;46 Suppl 1:S4–11. Epub 2008/02/07. 10.1086/521865 . [DOI] [PubMed] [Google Scholar]
- 4.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2018;66(7):987–94. doi: 10.1093/cid/ciy149 . [DOI] [PubMed] [Google Scholar]
- 5.Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. The Lancet Infectious diseases. 2013;13(11):936–45. Epub 2013/09/07. 10.1016/S1473-3099(13)70200-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truong CY, Gombar S, Wilson R, Sundararajan G, Tekic N, Holubar M, et al. Real-Time Electronic Tracking of Diarrheal Episodes and Laxative Therapy Enables Verification of Clostridium difficile Clinical Testing Criteria and Reduction of Clostridium difficile Infection Rates. Journal of clinical microbiology. 2017;55(5):1276–84. doi: 10.1128/JCM.02319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2018;66(7):e1–e48. 10.1093/cid/cix1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagdasarian N, Rao K, Malani PN. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. Jama. 2015;313(4):398–408. Epub 2015/01/28. 10.1001/jama.2014.17103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, et al. Loss of Microbiota-Mediated Colonization Resistance to Clostridium difficile Infection With Oral Vancomycin Compared With Metronidazole. The Journal of infectious diseases. 2015;212(10):1656–65. Epub 2015/04/30. 10.1093/infdis/jiv256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seekatz AM, Young VB. Clostridium difficile and the microbiota. J Clin Invest. 2014;124(10):4182–9. 10.1172/JCI72336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, Mancabelli L, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016;6:25945 10.1038/srep25945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louie TJ, Byrne B, Emery J, Ward L, Krulicki W, Nguyen D, et al. Differences of the Fecal Microflora With Clostridium difficile Therapies. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2015;60 Suppl 2:S91–7. Epub 2015/04/30. doi: 10.1093/cid/civ252 . [DOI] [PubMed] [Google Scholar]
- 13.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2011;53(10):994–1002. 10.1093/cid/cir632 . [DOI] [PubMed] [Google Scholar]
- 14.Cammarota G, Ianiro G, Gasbarrini A. Fecal microbiota transplantation for the treatment of Clostridium difficile infection: a systematic review. Journal of clinical gastroenterology. 2014;48(8):693–702. 10.1097/MCG.0000000000000046 . [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Dong D, Jiang C, Li Z, Wang X, Peng Y. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe. 2015;34:1–7. 10.1016/j.anaerobe.2015.03.008 . [DOI] [PubMed] [Google Scholar]
- 16.Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH, et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. Journal of clinical microbiology. 2013;51(9):2884–92. 10.1128/JCM.00845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaac S, Scher JU, Djukovic A, Jimenez N, Littman DR, Abramson SB, et al. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. The Journal of antimicrobial chemotherapy. 2016. Epub 2016/10/07. 10.1093/jac/dkw383 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HN, Kim H, Moon HW, Hur M, Yun YM. Toxin positivity and tcdB gene load in broad-spectrum Clostridium difficile infection. Infection. 2018;46(1):113–7. 10.1007/s15010-017-1108-y . [DOI] [PubMed] [Google Scholar]
- 19.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71(12):8228–35. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer F, Backhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–38. 10.1038/nrmicro2974 . [DOI] [PubMed] [Google Scholar]
- 22.Schwenk HT, Bio LL, Kruger JF, Banaei N. Clinical Impact of Clostridium difficile PCR Cycle Threshold-Predicted Toxin Reporting in Pediatric Patients. J Pediatric Infect Dis Soc. 2018. 10.1093/jpids/piy117 . [DOI] [PubMed] [Google Scholar]
- 23.Kamboj M, Brite J, McMillen T, Robilotti E, Herrera A, Sepkowitz K, et al. Potential of real-time PCR threshold cycle (CT) to predict presence of free toxin and clinically relevant C. difficile infection (CDI) in patients with cancer. J Infect. 2018;76(4):369–75. 10.1016/j.jinf.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies KA, Planche T, Wilcox MH. The predictive value of quantitative nucleic acid amplification detection of Clostridium difficile toxin gene for faecal sample toxin status and patient outcome. PloS one. 2018;13(12):e0205941 10.1371/journal.pone.0205941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crobach MJT, Duszenko N, Terveer EM, Verduin CM, Kuijper EJ. Nucleic Acid Amplification Test Quantitation as Predictor of Toxin Presence in Clostridium difficile Infection. Journal of clinical microbiology. 2018;56(3). 10.1128/JCM.01316-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senchyna F, Gaur RL, Gombar S, Truong CY, Schroeder LF, Banaei N. Clostridium difficile PCR Cycle Threshold Predicts Free Toxin. Journal of clinical microbiology. 2017;55(9):2651–60. 10.1128/JCM.00563-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Origuen J, Orellana MA, Fernandez-Ruiz M, Corbella L, San Juan R, Ruiz-Ruigomez M, et al. Toxin B PCR amplification cycle threshold adds little to clinical variables to predict outcomes in Clostridium difficile infection: a retrospective cohort study. Journal of clinical microbiology. 2018. 10.1128/JCM.01125-18 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin J, Wilcox M. New and emerging therapies for Clostridium difficile infection. Curr Opin Infect Dis. 2016;29(6):546–54. 10.1097/QCO.0000000000000320 . [DOI] [PubMed] [Google Scholar]
- 29.Serrano-Silva N, Calderon-Ezquerro MC. Metagenomic survey of bacterial diversity in the atmosphere of Mexico City using different sampling methods. Environ Pollut. 2018;235:20–9. 10.1016/j.envpol.2017.12.035 . [DOI] [PubMed] [Google Scholar]
- 30.Li H, Li T, Beasley DE, Hedenec P, Xiao Z, Zhang S, et al. Diet Diversity Is Associated with Beta but not Alpha Diversity of Pika Gut Microbiota. Front Microbiol. 2016;7:1169 10.3389/fmicb.2016.01169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–8. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2(3):145–58. 10.4161/gmic.2.3.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, Young VB, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio. 2014;5(3):e01021–14. Epub 2014/05/08. 10.1128/mBio.01021-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YJ, Arguello ES, Jenq RR, Littmann E, Kim GJ, Miller LC, et al. Protective Factors in the Intestinal Microbiome Against Clostridium difficile Infection in Recipients of Allogeneic Hematopoietic Stem Cell Transplantation. The Journal of infectious diseases. 2017;215(7):1117–23. 10.1093/infdis/jix011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antharam VC, McEwen DC, Garrett TJ, Dossey AT, Li EC, Kozlov AN, et al. An Integrated Metabolomic and Microbiome Analysis Identified Specific Gut Microbiota Associated with Fecal Cholesterol and Coprostanol in Clostridium difficile Infection. PloS one. 2016;11(2):e0148824 10.1371/journal.pone.0148824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–61. 10.1016/j.mib.2013.06.003 . [DOI] [PubMed] [Google Scholar]
- 37.Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7(1):61–71. 10.1038/nrmicro2047 . [DOI] [PubMed] [Google Scholar]
- 38.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–43. . [DOI] [PubMed] [Google Scholar]
- 39.Theriot CM, Young VB. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annu Rev Microbiol. 2015;69:445–61. 10.1146/annurev-micro-091014-104115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stecher B, Maier L, Hardt WD. 'Blooming' in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11(4):277–84. 10.1038/nrmicro2989 . [DOI] [PubMed] [Google Scholar]
- 41.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–36. 10.1136/gutjnl-2014-308778 . [DOI] [PubMed] [Google Scholar]
- 42.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2017;106:171–81. 10.1016/j.micpath.2016.02.005 . [DOI] [PubMed] [Google Scholar]
- 43.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–7. 10.1126/science.aan3706 . [DOI] [PubMed] [Google Scholar]
- 44.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, et al. Conducting a microbiome study. Cell. 2014;158(2):250–62. 10.1016/j.cell.2014.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, et al. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J. 2012;6(1):94–103. 10.1038/ismej.2011.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are uploaded to a public repository (Dryad Digital Repository, doi:10.5061/dryad.30kk201).