Abstract
Members of the Ski/Sno protein family are classified as proto-oncogenes and act as negative regulators of the TGF-ß/BMP-pathways in vertebrates and invertebrates. A newly identified member of this protein family is fussel (fuss), the Drosophila homologue of the human functional Smad suppressing elements (fussel-15 and fussel-18). We and others have shown that Fuss interacts with SMAD4 and that overexpression leads to a strong inhibition of Dpp signaling. However, to be able to characterize the endogenous Fuss function in Drosophila melanogaster, we have generated a number of state of the art tools including anti-Fuss antibodies, specific fuss-Gal4 lines and fuss mutant fly lines via the CRISPR/Cas9 system. Fuss is a predominantly nuclear, postmitotic protein, mainly expressed in interneurons and fuss mutants are fully viable without any obvious developmental phenotype. To identify potential target genes or cells affected in fuss mutants, we conducted targeted DamID experiments in adult flies, which revealed the function of fuss in bitter gustatory neurons. We fully characterized fuss expression in the adult proboscis and by using food choice assays we were able to show that fuss mutants display defects in detecting bitter compounds. This correlated with a reduction of gustatory receptor gene expression (Gr33a, Gr66a, Gr93a) providing a molecular link to the behavioral phenotype. In addition, Fuss interacts with Rpd3, and downregulation of rpd3 in gustatory neurons phenocopies the loss of Fuss expression. Surprisingly, there is no colocalization of Fuss with phosphorylated Mad in the larval central nervous system, excluding a direct involvement of Fuss in Dpp/BMP signaling. Here we provide a first and exciting link of Fuss function in gustatory bitter neurons. Although gustatory receptors have been well characterized, little is known regarding the differentiation and maturation of gustatory neurons. This work therefore reveals Fuss as a pivotal element for the proper differentiation of bitter gustatory neurons acting within a chromatin modifying complex.
Author summary
Ski/Sno proteins have been discovered as proto-oncogenes transforming chicken fibroblasts into cancer cells. They have been found to be ubiquitously expressed in embryonic and adult tissues and to interfere with TGF-ß/BMP signaling. More recently, a group of proteins has been discovered which belongs to the same protein family, the functional Smad suppressing elements (Fussel). They have a highly restricted, mainly neuronal expression pattern suggesting different functional importance compared to Ski/Sno. We have used Drosophila as a model organism to characterize the highly specific neuronal expression pattern and created knock-out mutations within the Drosophila fuss gene. Surprisingly, fuss mutants are fully viable, but they show defects in bitter taste perception, and indeed, we could prove that Fuss is expressed specifically in bitter sensing neurons, where it affects their terminal differentiation making these cells insensitive for bitter compounds. To understand the molecular process involved in Fuss function we started protein interaction studies and could show, that Fuss forms part of a chromatin modifying complex, which seems to be important for the proper differentiation of neurons in the adult nervous system, therefore, assigning Drosophila as an indispensable model to study the molecular function of the Fuss protein family.
Introduction
During development, the TGF-ß superfamily plays an important role in cell proliferation, differentiation, apoptosis, cell adhesion, wound healing, bone morphogenesis and cell motility [1]. Accordingly, there are multiple inhibitory factors taking care of proper regulation of TGF-ß pathways. Besides inhibitory Smads and Smurfs, which act by preventing the activation of TGF-ß receptors, another group of negative regulators of the TGF-ß pathway exists: The Ski/Sno protein family [2–4]. Although Ski/Sno proteins are classified as proto-oncogenes, their exact role in cancer progression is not fully understood. Various experimental approaches have identified pro- and anti-oncogenic features, where the tumor promoting function of Ski/Sno proteins seems to be mainly linked to their ability to counteract the anti-proliferative effects of TGF-ß signaling [5,6]. Physiologically, Ski and Sno have both been implicated in axonal morphogenesis, myogenesis and mammary gland alveogenesis [7–9]. Proteins of the Ski/Sno family are characterized by a Ski/Sno homology domain and a SMAD4 binding domain. These domains, although resembling DNA binding domains, mediate protein-protein interactions enabling binding of mSin3a, N-CoR, the histone deacetylase HDAC1, SMAD4 and different regulatory SMADs, thus leading to the recruitment of a repressive transcription complex, binding to target genes of the TGF-ß signaling pathway [10–12].
Whereas Ski and Sno are expressed mainly ubiquitously, two additional members of the Ski/Sno protein family, the functional smad suppressing elements (Fussel) 15 and 18 (Skor1 and Skor2 in mouse, respectively), are highly restricted to postmitotic neurons such as Purkinje cells [13–15]. Previous analysis showed that Skor1 interacts with Smad3 and acts as a transcriptional corepressor for LBX1, whereas Skor2 inhibits BMP signaling in overexpression assays and is required for the expression of Sonic Hedgehog in Purkinje cells. In addition, Skor2 is needed for proper differentiation of Purkinje cells and knockout mice die prematurely within 24 h after birth. However, there is no further insight into the functional mechanisms of Skor1 in mice [15–17].
In contrast to vertebrates, Drosophila melanogaster has only two Ski/Sno proteins: The Ski novel oncogene Snoo and the functional Smad suppressing element Fussel (Fuss). Snoo is the homologue of Ski and Sno and Fuss the homologue of Skor1 and Skor2. As in mice, Snoo is expressed broadly during development and adulthood, whereas Fuss expression is limited to a subset of cells in the nervous system during development [18]. Snoo has been found to be involved in eggshell patterning and in wing and tracheal development in Drosophila melanogaster [19–21]. Recent findings of Fuss function are highly controversial due to the lack of a reliable Drosophila fussel mutation. Loss of function experiments with a chromosomal deletion suggested, that Fuss acts as a cofactor for Smox signaling enabling ecdysone receptor (EcrB1) expression in developing mushroom bodies in the brain. In addition, a severe malformation of the adult mushroom bodies was detected [22]. Contrary to these results, in overexpression assays Fuss leads to an inhibition of BMP signaling via its interaction with Medea, the SMAD4 homologue in Drosophila melanogaster [18].
To clarify the molecular and physiological function of Fuss, we generated a complete loss of function allele via CRISPR/Cas9 editing. Interestingly, fuss mutants are fully viable, suggesting a modulatory function during development or/and adulthood, rather than an essential role for cell survival. To be able to better analyze Fuss expressing cells, we generated specific antibodies and reporter lines, which enabled us to further clarify the expression pattern of Fuss. During development, Fuss is expressed postmitotically in a highly restricted number of interneurons of the central nervous system (CNS). In our fuss mutant, we could show that Fuss, in contrast to previous studies, is neither acting as a negative regulator of BMP signaling endogenously, nor involved in mushroom body development. A targeted DamID (TaDa) experiment could not only confirm our findings molecularly, but also revealed, that Fuss is expressed in bitter sensory neurons. In consequence, fuss mutant flies lack the ability to sense bitter compounds and show reduced expression of bitter gustatory receptors. Furthermore, interaction studies show that Fuss can form a protein complex with Rpd3, a homologue of HDAC1, and indeed, downregulation of rpd3 in bitter gustatory neurons resembles loss of Fuss. Thus, we propose that the Fuss/Rpd3 complex is required for proper cell fate determination in gustatory neurons either by direct or indirect control of the expression of gustatory bitter sensing receptors.
Results
Generation and analysis of fuss mutants
The fuss gene is localized on the fourth chromosome and therefore, due to the limited genetic resources for this chromosome, fuss mutations have escaped discovery in genetic screens for developmental mutations and previous research on this gene focused either on overexpression studies or on a chromosomal deletion covering multiple genes [18, 22]. However, it was shown recently that genes of the fourth chromosome can be CRISPR/Cas9 edited via homology directed repair and thus, we decided to generate a fuss null allele using this system [23]. A prerequisite for proper mutant generation is a detailed analysis of the genomic organisation of the gene. The fuss gene locus is fairly complex as it overlaps N-terminally with the RNA gene sphinx and C-terminally with the RNA gene CR44030. In addition, the Pax6 homologue twin of eyeless (toy) lies downstream of fuss and is transcribed in the opposite direction. Although the two genes are over 10 kb away from each other, regulatory sites of toy are located in the vicinity of the transcription start site of fuss [24].
Three different transcriptional start sites of fuss are annotated, which lead to three transcripts fussB, fussC and fussD. FussB and fussD have an identical amino acid sequence in contrast to the fussC transcript, which differs in 25 amino acids N-terminally from fussB and fussD. FussC uses an artificial promoter sequence due to a transposon insertion, leading to the assumption that this transcript is rather insignificant as it is of very low abundance (see below). To reduce side effects by deleting a silencer/enhancer structure of sphinx or toy and to maximise the effect on fuss, we removed an 855bp fragment, which is shared by all three transcripts (Fig 1A). This fragment contains the conserved Ski/Sno/Dac homology and SMAD4 binding domains, which are important for protein interactions and function of the Ski/Sno proteins [25,26]. Simultaneously, an attP site was introduced in the open reading frame of all fuss transcripts, which additionally results in a premature stop codon. Successful deletion of the two domains and the presence of the attP site was confirmed by PCR and subsequent DNA sequencing (S1A Fig). We termed this deletion fussdelDS and it is a null allele. Due to the deletion and the premature stop codon no functional proteins can be made, which could be shown by anti-Fuss antibody stainings of heterozygous and homozygous fussdelDS embryos (S1B Fig and S1C Fig). As a second mutant allele, we used the MiMIC-line fussMi13731, which is a gene trap insertion leading to the expression of GFP under the fussB and fussD promotor and a premature transcriptional stop of the fussB and fussD transcripts (Fig 1A). qPCR revealed that transcript levels of fussB and fussD in homozygous fussMi13731 flies are reduced to ten percent in contrast to WTB flies (S1D Fig). With an anti-GFP and anti-Fuss antibody staining, we could confirm that heterozygous fussMi13731/+ flies express GFP in the correct Fuss expression pattern, whereas Fuss staining in homozygous fussMi13731 flies is reduced to background levels (S1E Fig and S1F Fig). We conclude that fussMi13731 is at least a strong hypomorph for fuss, which further suggests that fussC is not specifically expressed or only at very low levels. In addition to fussMi13731, which can be used as a GFP reporter line, we created a Gal4 line from fussMi13731 by recombination mediated cassette exchange [27]. This Gal4 line was named fussBD-Gal4 and it was edited with the CRISPR/Cas9 system following the same strategy as for the fussdelDS allele. This resulted in a line called fussdelDS-Gal4, which allows Gal4 expression in Fuss expressing cells in a mutant background enabling us to analyze the presence and integrity of these cells. At last, we generated a UAS-t::gRNA-fuss4x line, which allows cell specific gene disruption of fuss via the UAS-Gal4 system [28]. The gRNAs target four CRISPR target sites located in the DNA sequence of the Ski/Sno homology domain (S1G Fig). We could detect a strong loss of GFP signal in adult brains of flies overexpressing GFP tagged Fuss, Cas9 and t::gRNA-fuss4x by fussBD-Gal4 compared to adult brains only expressing GFP tagged Fuss and Cas9 by fussBD-Gal4 (S1H and S1I Fig).
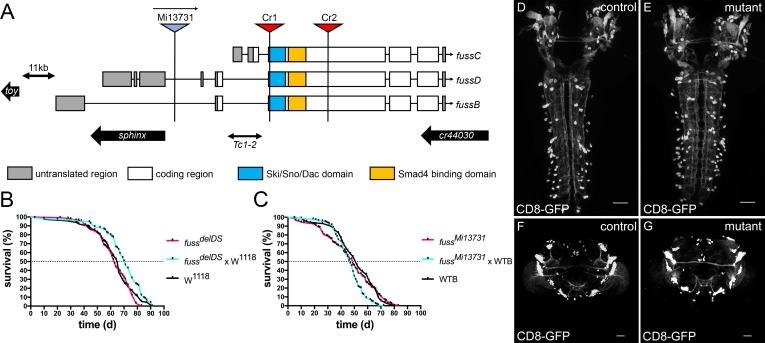
Fig 1. Generation of CRISPR/Cas9 induced fuss mutant.
(A) Generation of a fuss knockout allele, which lacks the Ski/Sno/Dac homology domain (blue box) and the SMAD4 binding domain (yellow box) by means of two CRISPR target sites Cr1 and Cr2. The location of the second mutant allele fussMi13731, a gene trap insertion in the FussB and FussD transcript is indicated. Approximate locations of toy, sphinx and cr44030 are depicted as black arrows. The location of the transposable element Tc1-2 is shown as a double-sided arrow. (B) Differences in the mean lifespan of homozygous fussdelDS flies (n = 133, purple) and the two controls W1118 (n = 116, black) and fussdelDS x W1118 flies (n = 109, blue) are not relevant. C) Longevity experiments show no differences between homozygous fussMi13731 (n = 164, purple), heterozygous fussMi13731 x WTB (n = 136, blue) and WTB flies (n = 85, black). (D) Projection pattern and cell bodies of larval brains visualized by expression of UAS-CD8-GFP by fussdelDS-Gal4/+ larvae (control). (E) Projection pattern and cell bodies of larval brains visualized by expression of UAS-CD8-GFP by fuss mutant fussdelDS-Gal4/fussdelDS larvae (mutant). (F) Projection pattern and cell bodies of adult brains visualized by expression of UAS-CD8-GFP by fussdelDS-Gal4/+ flies (control). (G) Projection pattern and cell bodies of adult brains visualized by expression of UAS-CD8-GFP by fuss mutant fussdelDS-Gal4/fussdelDS flies (mutant). Scale bars indicate 50 μm.
In a previous study by Takaesu et al. [22], a 40 kb spanning genomic deletion including the fuss gene (among several other genes) was used for functional studies of the fuss gene. They observed a strongly reduced survivability during development and a decreased lifespan, which was attributed to the loss of Fuss expression alone. In contrast to their results, we did not observe a reduced survivability during larval or pupal stages with our fuss mutant flies. Therefore, we conducted longevity experiments. Neither homozygous fussMi13731 nor fussdelDS–flies showed a significant reduction in lifespan compared to their controls (Fig 1B and Fig 1C).
Next, we compared the CD8-GFP expression pattern of heterozygous fussdelDS-Gal4/+ and mutant fussdelDS-Gal4/fussdelDS flies. We did neither observe an evident loss of GFP positive cells in the CNS of third instar larvae (Fig 1D and Fig 1E) nor in three to five-day old adult flies (Fig 1F and Fig 1G). Therefore, loss of fuss does neither lead to cell death, nor to a reduced survival during development or to a shortened lifespan.
Characterization of embryonic Fuss expressing cells reveals distinct neuronal identities
Due to the absence of any clear visible phenotype, we created specific polyclonal antibodies against a 16 kDa nonconserved fragment localized at the C-terminus of Fuss to characterize Fuss expressing cells and to draw conclusions about its function (S2A Fig). These anti-Fuss antibodies clearly detect a Fuss-GFP fusion protein on western blots and stainings mirror previously conducted RNA in situ hybridisations (S2B Fig) [18, 22].
In a first overview of Fuss staining during embryonic development, Fuss expression is mainly observed in the embryonic brain (Fig 2A, circles), the developing stomatogastric nervous system (Fig 2A, arrowhead), single cells lying anterior to the CNS (Fig 2A arrows), which will develop to inner gustatory neurons as shown later and the ventral nerve cord (VNC, Fig 2B). As Fuss is characterized by its conserved domains as a member of the Ski/Sno protein family, which are all considered to be transcription regulators, we observe Fuss protein, as expected, exclusively localized in the nucleus. During embryonic development, Fuss protein appears first at stage 13 and the number of Fuss positive cells increases continuously from early to late embryonic stages as previously observed (S2C Fig [22]). At embryonic stage 16, expression can be observed in two to five cells per hemineuromer with ascending numbers from posterior to anterior (Fig 2B and S2C Fig).
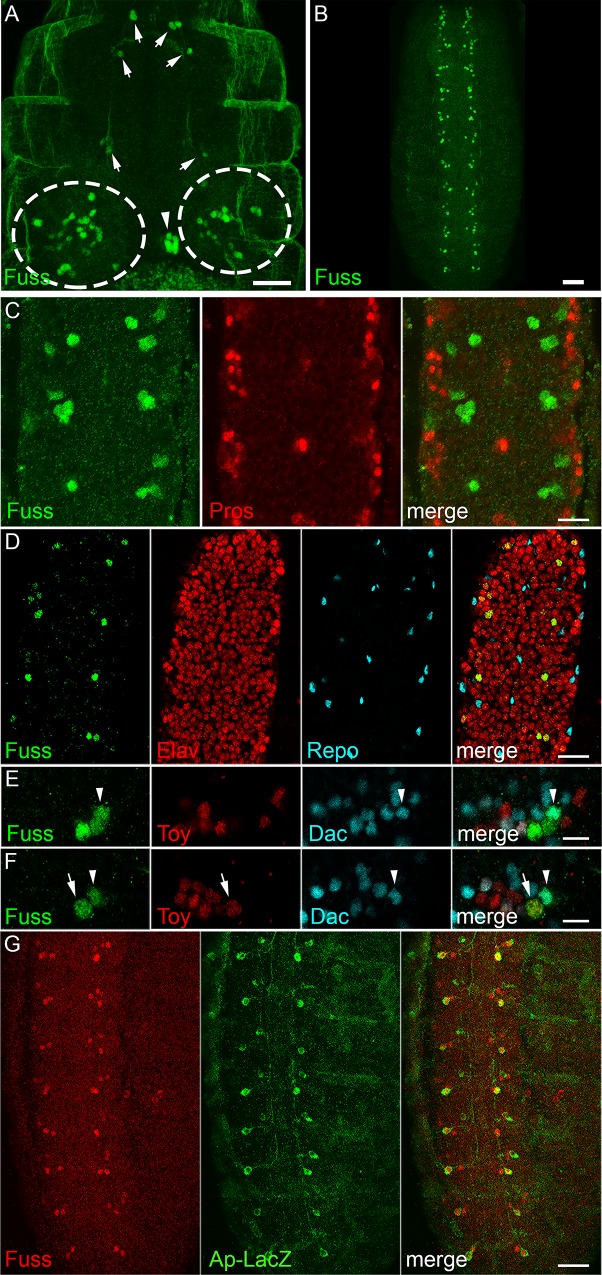
Fig 2. Fuss is expressed in postmitotic interneurons.
(A) In stage 16 embryos, anti-Fuss staining can be observed in a restricted number of cells in the CNS (dashed circles). Fuss is also found in individual cells of the stomatogastric nervous system (arrowhead) and anterior to the CNS (arrows). (B) In the ventral nerve cord (VNC), Fuss is expressed in two to five neurons per hemineuromer in ascending number from posterior to anterior. (C) Confocal microscopy images reveal that Fuss expressing cells in the VNC (green) do not overlap with ganglion mother cells stained with anti-Prospero (red). (D) Fuss is exclusively expressed in neurons (Elav, red) but not in glia (Repo, blue). (E, F) In one representative hemineuromer five Fuss (green) positive neurons are characterized regarding their Dac (blue, arrowhead) or Toy (red, arrow) expression. (G) In every hemineuromer one Fuss (green) neuron is positive for LacZ (red) expressed under the apterous promotor. Scale bars indicate 25 μm (A), 50 μm (B), 10 μm (C, E, F) and 20 μm (D, G).
The late appearance of the Fuss protein during development suggested, that Fuss might be expressed only postmitotically. We confirmed this hypothesis by visualizing ganglion mother cells in the embryo with anti-Prospero and Fuss cells with anti-Fuss antibodies and no overlapping stainings were detected (Fig 2C). As shown by colocalization studies with the glia marker Repo and the neuronal marker Elav, the staining pattern is exclusively neuronal (Fig 2D). To further identify neuronal subpopulations in hemineuromers of the VNC, prominent neuronal markers such as Engrailed (En), Even skipped (Eve), Apterous (Ap), Hb9, Dachshund (Dac) and Twin of eyeless (Toy) were utilised. No colocalization of Fuss with the interneuron marker En or with the motoneuron markers Eve or Hb9 was observed (S2D Fig, S2E Fig and S2F Fig). Because Eve and Hb9 label most of the embryonic motoneurons, Fuss is unlikely to be expressed in motoneurons [29,30]. We were especially interested if Dac and Fuss colocalize, because the interneuron marker Dac shares sequence similarity with Ski and Sno and consequently is a related protein to Fuss [31,32]. Interestingly, Dac and Fuss are partially coexpressed, which emphasizes that at least some Fuss neurons are interneurons (arrowhead, Fig 2E and Fig 2F). As the toy gene lies only 11 kb downstream of fuss and is transcribed in the opposite direction, it is reasonable that they partially share enhancer/silencer regions. Remarkably, we only found one Toy positive Fuss neuron per hemineuromer in the VNC (arrow, Fig 2E and Fig 2F) excluding extensive overlap of regulatory regions. Ap is expressed in three cells per abdominal hemineuromer. These cells are subdivided into one dorsal Ap and two ventral Ap interneurons [33]. Using the ap-tau-LacZ reporter, which only labels one ventral Ap cell and the ap-Gal4 driver line we showed, that both ventral AP interneurons are Fuss positive (Fig 2G and S2G Fig). Due to the location of the Toy positive Fuss neuron, we assume that it is one of the ventral Ap cells and therefore also an interneuron.
Taken together we could show that Fuss is expressed only postmitotically in interneurons in the developing CNS, which will be further confirmed later.
Transcriptional profiling of adult Fuss neurons in the head
Fuss is expressed in heterogenic neuronal populations, which are represented by differentially expressed markers and by their projection patterns. To develop new approaches to identify and study viable phenotypes in fuss mutants, it was of upmost importance to identify genes, which are regulated by Fuss. Therefore, we performed a targeted DamID (TaDa) experiment by expressing a Dam-PolII fusion protein with the fussdelDS-Gal4 driver line. RplI215, the large subunit of the RNA Polymerase II, is fused with the Dam methylase and thus this so called Dam-PolII fusion protein enabled us to detect the binding sites of the RNA Polymerase II similar to an RNA PolII ChIP and to detect transcribed genes in these neurons without cell sorting [34]. As a control, the unfused Dam protein was expressed with the fussdelDS-Gal4 driver line. Expression of UAS-Dam or UAS-Dam-PolII was inhibited by Gal80ts during development and expression of these proteins was allowed for 24 h at 29°C in one to three-day old flies. Next generation sequencing libraries were generated from three different biological replicates expressing Dam-PolII and from three replicates expressing Dam alone. Each experiment was compared to each control leading to nine individual datasets. Because the binding patterns of all nine files were highly similar, individual datasets were averaged to reduce the amount of false positive hits of expressed genes. Genes with a false discovery rate (FDR) lower than 0.01 were accounted as expressed resulting in 2932 genes (S1 Appendix). The TaDa data is represented as a log2 ratio of Dam-PolII/Dam. As expected, fuss was one of the genes with the lowest FDR and highest PolII coverage (Fig 3A). This clearly indicates that the approach was carried out successfully. Furthermore, genes already identified by antibody stainings such as elav, dac or toy, were also detected by the TaDa experiment. Toy was also expressed in some Fuss neurons in adult brains (Fig 3B). This again underlines, like already observed during embryonic development, that fuss and toy might share common silencer/enhancer elements with Fuss. To further verify the TaDa data, colocalization experiments were conducted. Two cell fate markers atonal (ato) and acj6 were enriched in our dataset and we could also detect the expression of these two proteins via immunofluorescense stainings in Fuss neurons (Fig 3B). Furthermore, we analyzed genes which show no or low PolII coverage e.g. pale (ple) and Insulin-like peptide 2 (Ilp2) via immunofluorescence and could not detect any staining in Fuss positive neurons (S3 Fig). In particular, the absence of Fuss in insulin producing cells is in disagreement with recent published results using enhancer/reporter constructs (S3B Fig, [35,36]). In summary, we can conclude, that using this strategy, we have successfully generated an adult Fuss neuron specific transcriptional profile.
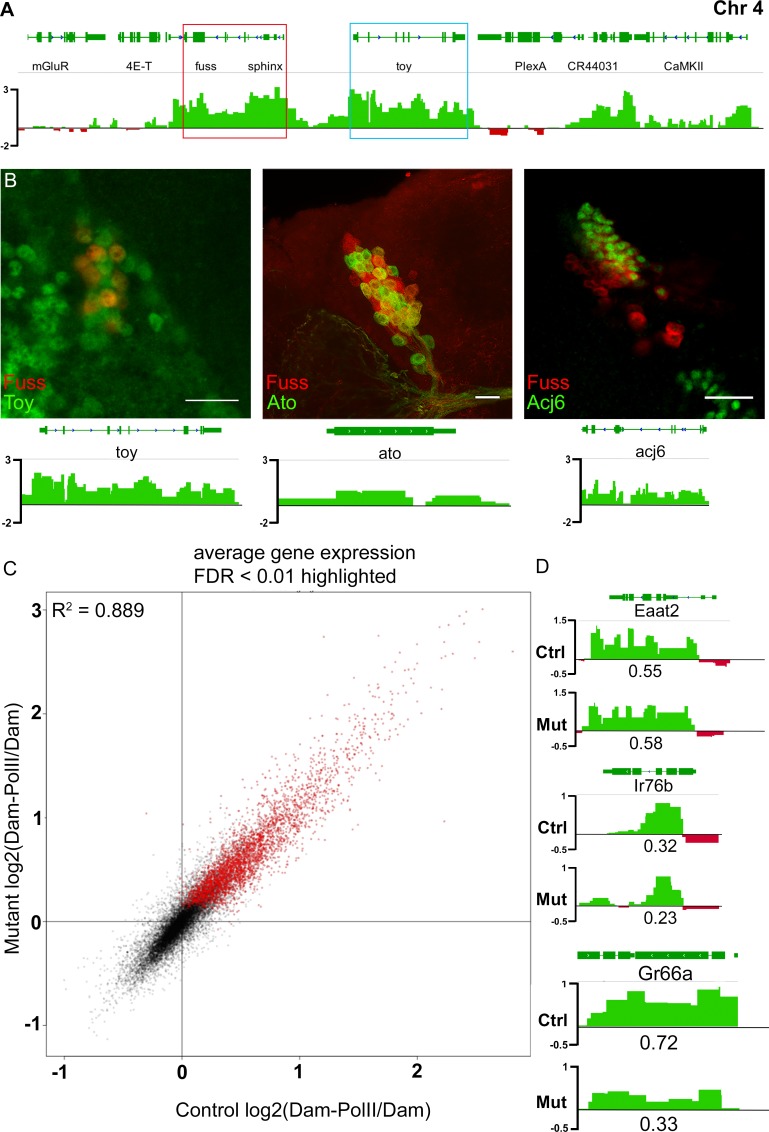
Fig 3. Targeted DamID of control and fuss mutants reveal sensory neuron markers as potential Fuss targets.
(A) A Dam-PolII/Dam binding pattern was generated from nine individual TaDa profiles and averaged to one single track. fuss (red box) and toy (blue box) are highly covered by Dam-PolII. Regions bound stronger by Dam-PolII than by Dam are depicted in green. Regions bound stronger by Dam than by Dam-PolII are depicted in red. (B) Verification of three TaDa positive genes, toy, atonal and acj6 by immunostaining. Toy was labelled by anti-Toy (green) and Fuss by anti-Fuss staining (red). Expression of LacZ by ato-Gal4 (green) and expression of GFP from the fussMi13731/+ reporter line (red). Labelling of Acj6 with anti-Acj6 antibody (green) and expression of CD8-GFP with fussBD-Gal4 (red). Scale bars indicate 10 μm. (C) log2(Dam-PolII/Dam) data from controls compared with log2(Dam-PolII/Dam) data from Fuss mutant neurons show only small deviations from each other. Coefficient of determination R2 = 0.889. (D) TaDa reveals sensory neuron marker expression of EAAT2, Ir76a, and Gr66a in both datasets (upper lane: control; lower lane: mutant) with a clear reduction in PolII coverage of Gr66a in the mutant dataset in contrast to the control dataset. PolII coverage is depicted under PolII binding pattern. Regions bound stronger by Dam-PolII than by Dam are depicted in green. Regions bound stronger by Dam than by Dam-PolII are depicted in red.
In the next step, we wanted to search for potential target genes of Fuss using the same strategy and conditions as above, but this time fussdelDS-Gal4 was kept over the fussdelDS allele to profile transcription of fuss mutant neurons. Again, individual datasets were averaged and genes with an FDR lower than 0.01 were accounted as expressed resulting in 3150 genes (S2 Appendix). The comparison of the log2(DamPolII/Dam) data of heterozygous fussdelDS/+ and homozygous fussdelDS flies showed, that there is not a strong deviation (coefficient of deviation R2 = 0.889) of the mutant transcriptional profile from the control (Fig 3C). Because Fuss is only expressed in a small number of CNS neurons, the acquired data can only be confirmed by antibody staining and not by semiquantitive qPCR or western blots from whole heads. There were three genes which attracted our attention: Eaat2, Ir76b and especially Gr66a as they provided a possible link to Fuss expression in gustatory sense neurons (Fig 3D). These genes could be found in both datasets, although only Eaat2 had an FDR lower than 0.01 in both datasets. The PolII coverage of Eaat2 and Ir76b was only slightly different between homozygous and heterozygous flies, whereas Gr66a, which is exclusively expressed in bitter gustatory sense neurons (GRNs), showed a significant reduction in mutant flies (S1 Appendix and S2 Appendix).
Fuss is expressed in a subset of gustatory neurons
It has been shown that the glutamate aspartate transporter Eaat2 is expressed in sensory neurons [37]. The ionotropic receptor Ir76b is expressed in gustatory neurons and the gustatory receptor Gr66a is specifically expressed in bitter GRNs, where Gr66a is a very important component for bitter taste sensation [38,39]. We already observed Fuss expression in cells outside of the larval CNS, therefore, to confirm the TaDa datasets, we analyzed gustatory neurons in larval and adult stages. In larvae, Fuss expression cannot be observed in the terminal or dorsal organ, but it can be found in the inner gustatory sense organs. We found Fuss expression in two pairs of neurons in the dorsal pharyngeal sensilia (DPS, Fig 4A), one neuron pair in the dorsal pharyngeal organ (DPO, Fig 4A) and two neuron pairs in the posterior pharyngeal sensilia (PPS, Fig 4A). None of the GRNs in the ventral pharyngeal sense organ (VPS) express Fuss. These cells have been already characterized by expression of different gustatory receptors and we found that larval Fuss expressing GRNs show a colocalization with a marker for bitter sensing neurons Gr33a [40]. In addition, one neuron pair in the DPS also shows an overlap with Gr93a which has been shown to be important for caffeine response in larvae (Fig 4B, [41,42]).
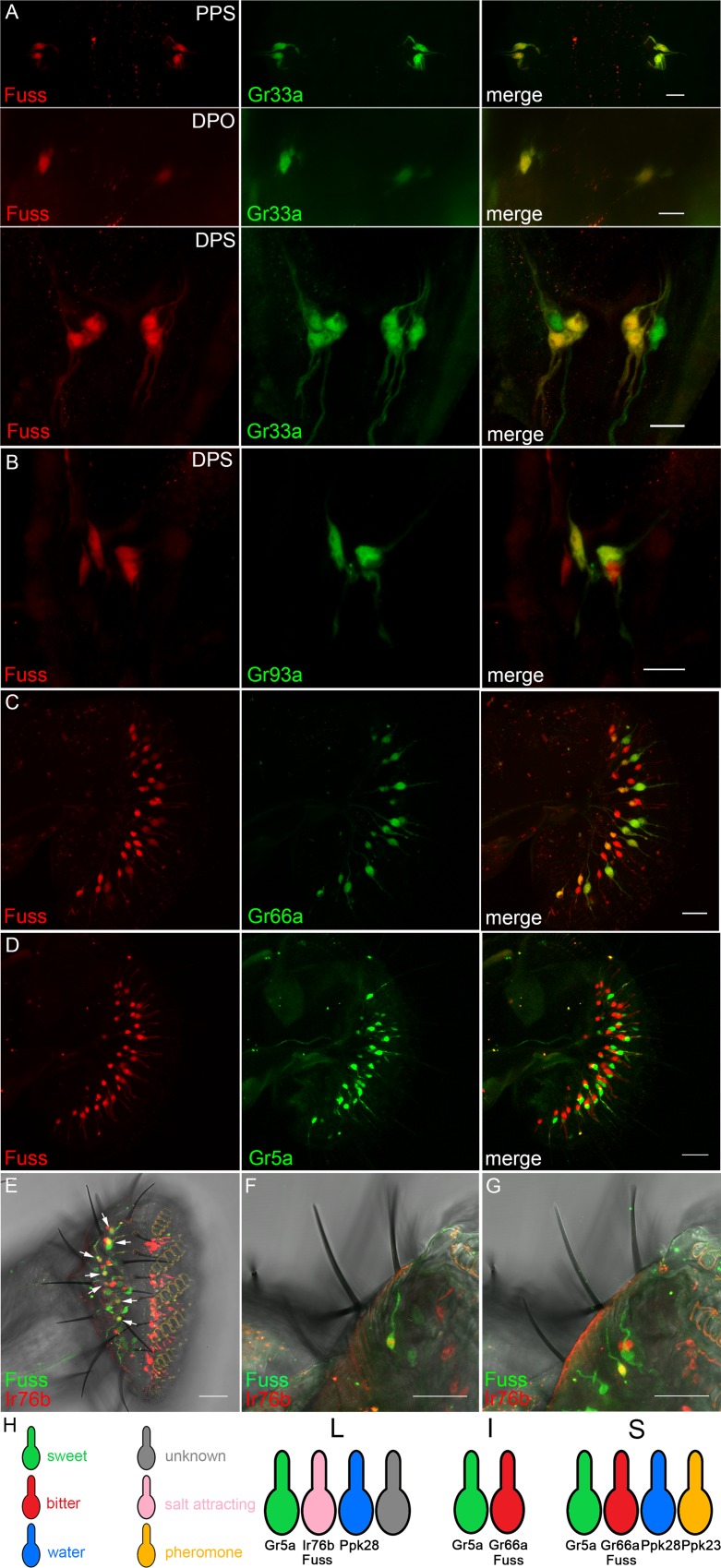
Fig 4. Fuss is expressed in bitter GRNs in inner gustatory organs of larvae and in bitter and salt attracting GRNs of the adult proboscis.
(A) GFP (red) expression from heterozygous fussMi13731/+ reporter line can be observed in bitter gustatory neurons marked with LacZ (green) expressed via Gr33a-Gal4 in the dorsal (DPS), posterior pharyngeal sensilia (PPS) and dorsal pharyngeal organ (DPO) identifying Fuss neurons as bitter gustatory neurons in these organs. (B) LacZ (green) expressed from a Gr93a-Gal4 line colocalizes with GFP (red) from a heterozygous fussMi13731/+ reporter line. Scale bars indicate 10 μm. (C) In adult flies, GFP (red) expression from the fussMi13731 gene trap line can be observed in the proboscis in one GRN per bristle. LacZ (green) driven by the bitter gustatory driver line Gr66a-Gal4 can be found in S- and I-type sensilla of GFP expressing GRNs. Scale bar indicates 20 μm. (D) No overlap between LacZ (green) driven by Gr5a-Gal4 and GFP (red) expressed from fussMi13731/+ can be observed. (E-G) LacZ (red) expressed by Ir76b-Gal4 and GFP (green) expressed from fussMi13731/+ overlap in L-type sensilla (arrows). Scale bars indicate 20 μm (C, D) and 25 μm (E-G), respectively. (H) Schematic representation of fuss expression in GRNs of L-, I- and S-type sensilla.
Later, in adulthood, Fuss expression continues in GRNs of the proboscis. In the adult labellum three different types of sensilla can be found divided into short (S-type), intermediate (I-type) and long sensilla (L-type). Intermediate sensilla are innervated by two GRNs and short and long sensilla by four GRNs [43]. Interestingly Fuss expression is observed in one GRN per gustatory sensilla and is consistently colocalized with the bitter GRN marker Gr66a in neurons innervating short and intermediate sensilia (Fig 4C [38]). Long sensilla do not contain a Gr66a positive GRN, therefore, all Gr66a neurons in the labellum are Fuss positive, but not vice versa. Another gustatory receptor which is broadly expressed and labels sweet GRNs is Gr5a, but no overlap with Fuss positive neurons was observed (Fig 4D). Besides Gr66a our TaDa dataset revealed that the ionotropic receptor Ir76b is expressed in Fuss neurons. Ir76b has been shown to be expressed by one GRN per L-type sensillum, which plays a role in attractive salt tasting [44]. We found that in L-type sensilla Fuss is coexpressed with Ir76b (Fig 4E–4G). Besides the expression in GRNs of the proboscis we found Fuss being expressed in two GRNs each in the last two tarsal segments in every leg (S4A Fig). In conclusion, we integrated Fuss expression into the GRN model from Freeman and Dahanukar (Fig 4H, [45]) and demonstrate that Fuss is expressed in bitter neurons in S- and I-type sensilla and in salt attracting neurons in L-type sensilla.
Loss of Fuss impairs bitter taste sensation
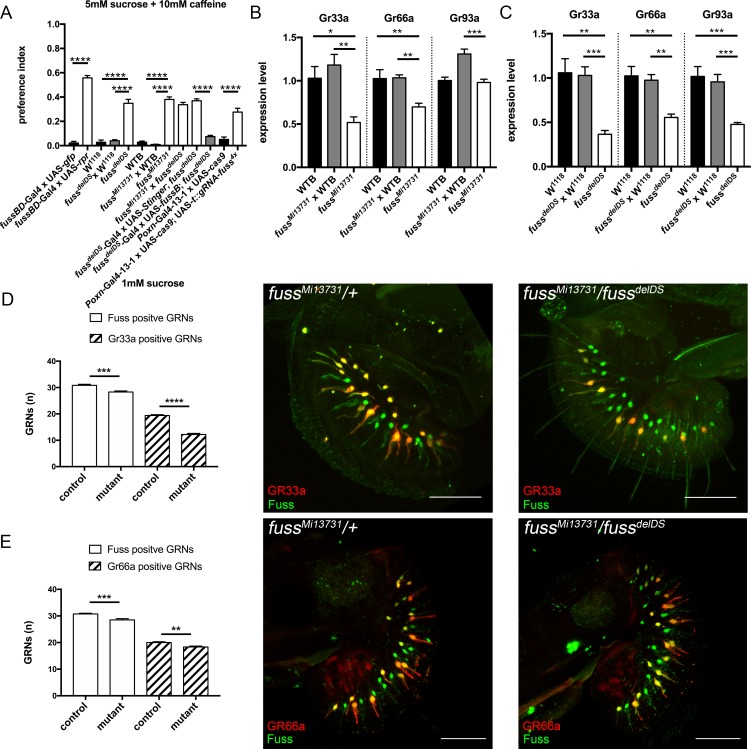
By its gustatory system Drosophila melanogaster can discriminate between valuable food sources for foraging or egg laying and toxic compounds which could harm the fly or its offspring [46]. To address if Fuss is required for the proper development of GRNs, we focused on the impact of Fuss mutation on differentiation of bitter GRNs, because Fuss is expressed in all bitter GRNs of the proboscis. To detect if fuss mutant flies display an impaired bitter taste sensation, we tested one to three-day old flies in a two-choice feeding assay. In our standard test, flies had to choose between 1mM sucrose or 5mM sucrose plus 10mM caffeine. We calculated a preference index ranging from zero to one, where zero indicates complete avoidance of the bitter compound and one a complete preference for it, due to the higher sugar concentration. First, Fuss expressing neurons were ablated by UAS-rpr expression with fussBD-Gal4 to show their importance in bitter sensing and indeed, these flies showed a strong impairment of bitter discrimination (Fig 5A). Furthermore, homozygous fussMi13731, fussdelDS and transheterozygous mutants (fussMi13731/fussdelDS) as well as their appropriate controls were tested. All mutant genotypes showed an increased preference for 5mM sucrose mixed with caffeine and by overexpression of Fuss in fuss mutant neurons we could revert preference to wildtype levels (Fig 5A). To show that the behavioural phenotype of fuss mutants is due to defects in GRNs and not derived from other higher order Fuss neurons in the CNS we specifically disrupted fuss in all GRNs with the Poxn-Gal4-13-1 driverline and our UAS-cas9; UAS-t::gRNA-fuss4x flies. Poxn-Gal4-13-1 expresses Gal4 early in development in all GRNs and in ellipsoid body neurons as well as interneurons of the antennal lobe of the brain (S4B Fig, [47]), therefore the only common neuronal populations between Fuss and Poxn-Gal4-13-1 are the GRNs and indeed, as shown in Fig 5A, these flies show the same bitter sensing deficits. We also tested different concentrations of caffeine as well as another bitter compound (denatonium benzoate) and fussMi13731 flies always displayed a higher preference towards the 5mM sucrose mixed with the bitter compound than controls except when the concentration of the bitter compound was too high (S4C and S4D Fig). Thus, not only detection of caffeine but more general bitter sensation is disturbed, because different GR multimers are needed for the detection of different aversive compounds, e.g. Gr93a which is expressed in a subset of S-type sensilla is needed for caffeine but not for denatonium benzoate sensation [48]. The gustatory receptor Gr66a showed a strong reduction in PolII coverage in mutant flies in contrast to control flies and is only expressed in a proportion of Fuss positive GRNs. The gustatory receptor GR33a has been found to be coexpressed with Gr66a in bitter GRNs and both are involved in bitter sensation, particularly together with Gr93a in caffeine sensation [40,48]. To validate GRN results from the TaDa experiment, we extracted RNA from adult proboscises and analysed the expression levels of those GRs via semiquantitative RT-PCR. In homozygous fussMi13731-flies Gr33a and Gr66a expression were strongly reduced as compared to WTB and heterozygous fussMi13731-flies. Gr93a expression levels of homozygous fussMi13731-flies were similar to WTB levels but reduced when compared to heterozygous fussMi13731-flies (Fig 5). The observed effects were enhanced in fussdelDS-flies. Gr33a, Gr66a and Gr93a expression levels were all reduced in fussdelDS-flies in contrast to both controls (Fig 5C). A similar downregulation of Gr33a, Gr66a and Gr93a expression levels was observed in transheterozygous fussMi13731/fussdelDS flies in contrast to WTB flies (S4E Fig). Next, we tested if the number of Gr33a and Gr66a positive GRNs is reduced in fuss mutant flies. We counted Fuss positive and Gr33a positive neurons in flies of the genotypes Gr33a-Gal4/UAS-LacZ; fussMi13731/+ and Gr33a-Gal4/UAS-LacZ; fussMi13731/fussdelDS. In this genetic combination, we counted 2.5 less Fuss positive cells and surprisingly 7.2 less Gr33a positive cells in transheterozygous mutants than in control flies (Fig 5D). Furthermore, we analysed the number of Fuss positive and Gr66a positive neurons in flies of the genotypes UAS-LacZ/+; Gr66a-Gal4/+; fussMi13731/+ and UAS-LacZ/+; Gr66a-Gal4/+; fussMi13731/fussdelDS. We found the same reduction in overall number of Fuss positive GRNs in transheterozygous mutants. But the number of Gr66a positive GRNs is decreased at the same level as the number of overall Fuss positive GRNs (Fig 5E). Thus, the total number of bitter GRNs is slighty reduced in fuss mutant flies, but interestingly Gr33a expression is completely abolished in some bitter GRNs, whereas the reduction of Gr66a expression found in qPCR experiments does not result in a reduced number of Gr66a positive GRNs. So, upon the loss of Fuss expression, bitter GRN differentiation is highly disturbed, which renders these flies inable to detect bitter compounds.
Fig 5. fuss mutant GRNs show impaired caffeine avoidance.
(A) Two-choice feeding assay reveals reduced caffeine sensation of homozygous fussMi13731 and fussdelDS flies in contrast to their appropriate controls. As a positive control, Fuss neurons were ablated by expression of rpr via fussBD-Gal4. Transheterozygous fussdelDS/fussMi13731 flies also have a reduced ability to sense bitter compounds comparable to levels of homozygous fussMi13731 and fussdelDS flies. Overexpression of fussB with fussdelDS-Gal4 reduces caffeine preference to wildtype levels. Flies with a GRN specific fuss gene disruption (Poxn-Gal4-13-1 x UAS-cas9; UAS-t::gRNA-fuss4x) show a reduced caffeine sensation compared to controls (Poxn-Gal4-13-1 x UAS-cas9) (n = 4–10 for each genotype). One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ****p<0.0001. Error bars indicate SEM. (B) Semiquantitative qPCR of bitter gustatory receptors Gr33a, Gr66a and Gr93a reveals a reduced expression of Gr33a and Gr66a in homozygous fussMi13731 flies in contrast to controls. Gr93a expression of homozygous fussMi13731 flies is only reduced if compared to heterozygous fussMi13731 x WTB but not WTB flies. n = 4–6 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. *p<0.05. **p<0.01. ***p<0.001. Error bars indicate SEM. (C) Analysis of Gr33a, Gr66a and Gr93a expression by semiquantitative qPCR reveals reduced expression of GRs in homozygous fussdelDS flies in contrast to heterozygous fussdelDS x W1118 and W1118 flies (n = 4–6 for each genotype). One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. *p<0.05 **p<0.01 ***p<0.001. Error bars indicate SEM. (D) Comparison of Fuss positive neurons and Gr33a positive neurons of the genotypes Gr33a-Gal4/UAS-LacZ;fussMi13731/+ (control) and Gr33a-Gal4/UAS-LacZ;fussMi13731/fussdelDS (mutant) shows a slight reduction in Fuss positive GRN numbers (30.8 vs 28.3) and a strong reduction in Gr33a positive GRN numbers (19.4 vs 12.2). Unpaired t-test was used to calculate p-values. n = 12–13 for each genotype. ***p<0.001. ****p<0.0001. Error bars indicate SEM. Adult proboscis of genotypes Gr33a-Gal4/UAS-LacZ;fussMi13731/+ (control, abbr: fussMi13731/+) and Gr33a-Gal4/UAS-LacZ;fussMi13731/fussdelDS (mutant, abbr: fussMi13731/fussdelDS). Scale bar indicates 50 μm. (E) Comparison of Fuss positive neurons and Gr66a positive neurons of the genotypes UAS-LacZ/+;Gr66a-Gal4/+;fussMi13731/+ (control) and UAS-LacZ/+;Gr66a-Gal4/+;fussMi13731/fussdelDS (mutant) shows a slight reduction in Fuss positive GRN numbers (30.75 vs 28.5) and an equal reduction in Gr66a positive GRN numbers (20 vs 18.3). n = 12 for each genotype. Unpaired t-test was used to calculate p-values. **p<0.01. ***p<0.001. Error bars indicate SEM. Adult proboscis of genotypes UAS-LacZ/+;Gr66a-Gal4/+;fussMi13731/+ (control, abbr: fussMi13731/+) and UAS-LacZ/+;Gr66a-Gal4/+; fussMi13731/fussdelDS (mutant, abbr: fussMi13731/fussdelDS). Scale bar indicates 50 μm.
Fuss interacts with the histone deacetylase Rpd3 to affect cell fate determination
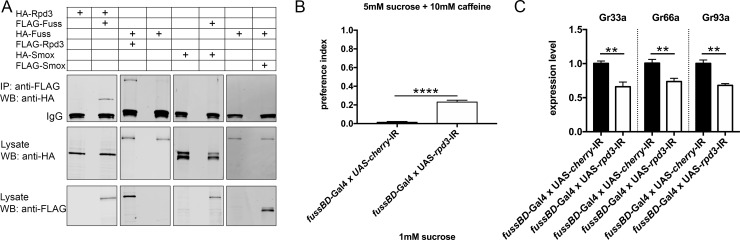
In mammals there are two homologues of Fuss, Skor1 and Skor2, which display a high sequence conservation within the Ski/Sno/Dac homology domain and the SMAD4 binding domain. In contrast, the conservation in the C-terminal region is very low, which shows a high degree of evolutionary divergence (S4F Fig). Although the I-loop of the SMAD4 binding domain, which has been implicated as an important structure for SMAD4 binding, is not very well conserved in Fuss and its homologues, we and others have detected an interaction between SMAD4 with Fuss and Skor2, respectively [11,14,18]. The repressive action of Ski/Sno proteins is generally exerted by the recruitment of a protein complex containing HDAC1 [10]. Skor1 and Skor2 also interact with HDAC1 and interestingly, it has been shown that the residues important for this interaction are localized in a segment reaching from amino acid 385–592 in mouse Skor2 [16,17]. Similar to the lack of the I-loop sequence, this segment is highly diverse between Fuss and Skor2 challenging if Fuss nevertheless is able to interact with Rpd3, the HDAC1 homologue in Drosophila melanogaster (S4F Fig). Therefore, we performed Co-Immunoprecipitations (CoIP) and transfected S2R+ cells with Fuss and Rpd3 tagged with FLAG or HA. Interaction between Fuss and Rpd3 could be shown independent of the type of the tags (Fig 6A). Skor1 and Skor2 have also been described to interact with Smad2 and Smad3, homologues of the Drosophila Smox, which executes the same function as Mad, but in the TGF-ß like signaling pathway [13,14,22]. Using the same methological approach as for the Fuss and Rpd3 interaction, we could not detect any interaction between Fuss and Smox, independent of the tags used (Fig 6A). Interestingly Smox is one of the genes specifically enriched in our TaDa datasets for Fuss neurons, so there would be a possibility for interaction in these cells.
Fig 6. Rpd3 interacts with Fuss and phenocopies fuss mutant phenotypes.
(A) Co-Immunoprecipitation experiments show that Fuss-HA binds to Rpd3-FLAG and Rpd3-HA interacts with Fuss-FLAG, respectively. No interaction between Smox and Fuss can be found regardless of the tags. (B) Knockdown of rpd3 results in an increased preference index towards 5mM sucrose mixed with 10mM caffeine compared to fussBD-GAL4 x UAS-cherry-IR flies. n = 4 for each genotype. Unpaired t-test was used to calculate p-values. ****p<0.001. Error bars indicate SEM. (C) Bitter gustatory receptors Gr33a, Gr66a and Gr93a are downregulated in fussBD-GAL4 x UAS-rpd3-IR flies compared to fussBD-GAL4 x UAS-cherry-IR flies. n = 4–5 for each genotype. Unpaired t-test was used to calculate p-values. **p<0.01. Error bars indicate SEM.
If Fuss is acting within a protein complex in concert with Rpd3, we should be able to mimic fuss mutant phenotypes with rpd3 depletion. Therefore, UAS-rpd3-IR was specifically expressed in Fuss neurons using the fussBD-Gal4 driver to reduce rpd3 expression throughout development. Adult flies were then tested again in a two-choice feeding assay for bitter sensing. Rpd3 knockdown flies showed a significant higher preference towards caffeine than control flies (fussBD-Gal4 x UAS-cherry-IR; Fig 6B). Because Rpd3 is involved in many different chromatin complexes, we analyzed again the expression levels of bitter gustatory receptors. Expression of all three tested GRs Gr33a, Gr66a and Gr93a was again diminished (Fig 6C) and therefore we conclude, that the Fuss/Rpd3 complex plays a key role in the final cell fate determination of gustatory neurons.
Fuss function in CNS neurons–a contentious issue
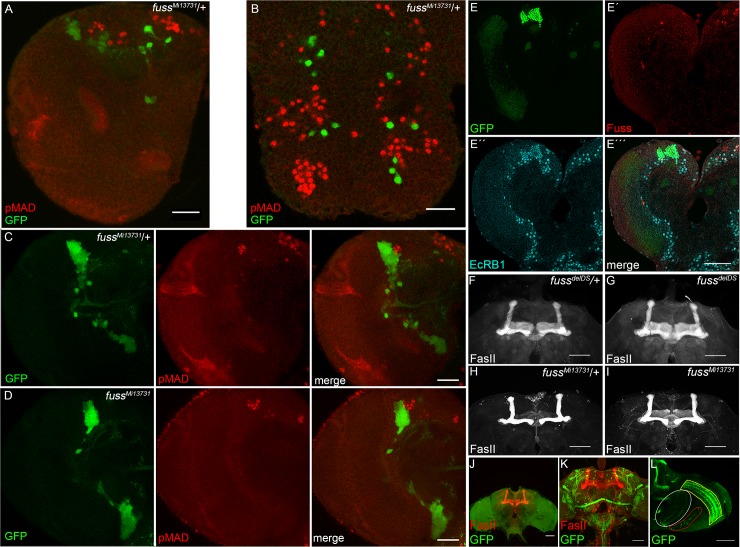
In overexpression experiments, Ski/Sno proteins have often been identified as negative regulators of TGF-ß or BMP-signaling [14,17]. In Drosophila, Dpp is the main homologue to vertebrate BMPs and it is involved in multiple developmental signaling events, in particular in the Drosophila wing [49]. We have previously shown, that an overexpression of Fuss during wing development indeed results in diminished expression of Dpp target genes and, concomitantly, induces a phenotype, which resembles loss of Dpp signaling, despite the fact, that we could only detect a physical interaction with the Co-Smad Medea but not with the R-Smad Mad [18]. In Dpp signaling, Mad gets phosphorylated by the type I receptors Saxophon and/or Thick veins and thus phosphorylated Mad is an excellent marker for active Dpp signaling and also for motoneurons or Tv neurons [50,51]. To analyse a possible role of Fuss in Dpp signaling, we used fussMi13731-flies, in which GFP is expressed under the fuss promotor to label Fuss expressing cells and we counterstained 3rd instar larval brains with an antibody against phosphorylated Mad (pMad) (Fig 7A and Fig 7B). These results clearly showed that Fuss expression is not overlapping with pMad in heterozygous fussMi1373/+ conditions. As there is a possibility that Fuss is acting upstream of Mad phosphorylation, we compared pMAD staining of heterozygous (Fig 7C) with homozygous fussMi13731-flies (Fig 7D). Again, there is no overlap of pMAD and GFP stainings in both genotypes, indicating that there is no increase of pMAD in fuss mutant neurons in the absence of Fuss. Importantly, this is in agreement with our overexpression studies, where Fuss had no influence on Mad phosphorylation [18]. Therefore, we conclude, that endogenously Fuss is not involved in Dpp signaling inhibition and it also emphasizes previous results, that Fuss is expressed in interneurons and not in motoneurons, which require pMad activity [51].
Fig 7. Fuss is neither a regulator of Dpp signalling nor involved in mushroom body formation.
(A) GFP (green) expressed from heterozygous fussMi13731/+ reporter line does not colocalize with pMAD (red) in larval brain. (B) GFP (green) expressed from heterozygous fussMi13731/+ reporter line does not colocalize with pMAD (red) in larval VNC. (C, D) GFP (green) expressed from heterozygous fussMi13731/+ reporter line and homozygous fussMi13731 marks Fuss neurons. Anti-pMAD (red) displays active Dpp signaling. No colocalization can be observed in any genotype indicating, that Fuss itself is not involved in Dpp signaling inhibition. All pictures depict slices of the larval brain or VNC and not full stacks to exclude false positive colocalization. Scale bars indicate 25 μm. (E) Representative picture of Kenyon cell nuclei of a third instar larval brain hemisphere. Nuclei of Kenyon cells are marked by colocalization of nuclear GFP driven by OK107-Gal4 (green, E) and EcRB1 (blue, E´´) in 3rd instar larval brains. Anti-Fuss (red, E´) staining cannot be observed in the Kenyon cell clusters. Fuss cells positive for EcRB1 expression, do not overlap with GFP expression from OK107-Gal4 driver (E´´´). (F-I) Mushroom bodies of an adult brain of heterozygous fussdelDS/+ flies (F), homozygous fussdelDS flies (G), heterozygous fussMi13731/+ flies (H) and homozygous fussMi13731 flies (I) visualized by anti-FasII staining. (J) Ablation of Fuss neurons removes all Fuss neurons but mushroom body (red) stained with anti-FasII is not affected in adult brains. (K) fussBD-Gal4 driven UAS-CD8-GFP visualises projection pattern of Fuss neurons in an adult brain. (L) fussBD-Gal4 driven UAS-CD8-GFP shows Fuss neurons strongly project to lobula (white), lobula plate (red) and medulla (yellow) in an adult brain. Scale bars indicate 50 μm.
Previously, the only loss of function data of fuss was generated using a genomic deletion of 40 kb including the fuss locus and additional genes [22]. This deletion lead to a reduced survivability during development, a shortened lifespan of the escapers and an impaired mushroom body development. All these phenotypes were attributed to the loss of Fuss expression. As we did not observe an impact on survivability or lifespan upon the loss of Fuss (see above), we wondered if Fuss is indeed involved in mushroom body development. Based on RNA in situ hybridisations Takaesu et al. assumed that Fuss is expressed in Kenyon cells during development and is required for the proper formation of the mushroom body [22]. Having now specific antibodies, gene trap constructs and fuss mutations in hand, we decided to carefully reevaluate this data on mushroom body expression and function during development. In a first step, we used OK107-Gal4 driven nuclear GFP as a marker for developing Kenyon cells and colabeled larval brains with EcRB1 and Fuss. We found that Fuss is not expressed in the developing mushroom body Kenyon cells, but it shows a partial overlap with EcRB1 expression outside of the Kenyon cell domain (Fig 7E–7E´´´). Next, we analysed adult mushroom body structures of fuss mutant flies using a FasII-antibody. As expected, due to the lack of Fuss expression in Kenyon cells, no deformation or loss of any of the lobes of the mushroom body was observed in homozygous fussMi13731 or fussdelDS-flies (Fig 7F–7I). In addition, the expression of rpr with the fussdelDS-Gal4 line lead to a complete ablation of Fuss neurons, but did not result in a malformation of adult mushroom bodies (Fig 7J). Furthermore, expression of CD8-GFP with fussBD-Gal4 in adult brains shows that Fuss neuron clusters are also localized distal to the mushroom body (Fig 7K). In fact, Fuss neuronal projections are localized outside of the mushroom body lobes in the adult brain and some Fuss neurons are targeting the optic lobe including different layers of the medulla, lobula and lobula plate but not the lamina (Fig 7L). From these results, we conclude that fuss has no impact on mushroom body development and that most of these neuronal populations such as the Fuss/Atonal positive neurons are higher order neurons of the visual system.
Discussion
The molecular and cellular functions of the fuss genes, which are members of the Ski/Sno protein family, are still poorly understood. The fact that Drosophila contains only one single fuss gene offers a great opportunity for a thorough analysis. However, this has been restrained due to its location on the 4th chromosome, where only limited genetic tools were available. As a consequence, previous reports have been focusing on the analysis of either overexpression studies or by using a multi-gene deficiency with contradictory results [18,22]. In the meantime, more recent methodological advances like the CRISPR/Cas9 genome editing [52] and the MiMIC gene trap technique [27] have expanded the Drosophila genetic toolbox and provided an appropriate genetic environment allowing a thorough and in-depth study of such genes. The availability of the fussMi13731 fly line, which is a gene trap of fuss, allowed us to study the expression pattern of Fuss. This line perfectly matches our Fuss-antibody stainings and was used to create a Gal4 line via RMCE as previously described [27]. A second independent mutant fuss allele, fussdelDS was created by CRISPR/Cas9 editing by deletion of the main functional protein domains. Although fussMi13731 and fussdelDS alleles are generated by different genetic approaches they share the same phenotypes, underlining that despite the complex genomic organization of fuss the observed phenotypes are due to the loss of fuss. Surprisingly, fuss mutant flies are fully viable and do neither show developmental lethality or reduced lifespans nor any other apparent phenotypes.
By means of our new tools, we could show that Fuss is expressed postmitotically in a small subset of neurons. All Fuss neurons in the CNS are interneurons, but they express different cell fate markers, suggesting that they represent a rather diverse group of neurons. These results were confirmed molecularly by a targeted DamID experiment, which, in addition, indicated a highly specific expression of gustatory receptor genes and indeed, Fuss is expressed in one GRN per sensillum. In S and I-type sensilla it is expressed in bitter GRNs and in L-type sensilla, which lack bitter GRNs, it is expressed in salt attracting GRNs. We investigated how the bitter GRNs react to the loss of Fuss and interestingly, this leads to an impairment of bitter sensation. Remarkably, this phenotype is correlated with a downregulation of bitter gustatory receptors Gr33a, Gr66a and Gr93a and in some bitter GRNs of fuss mutant flies no Gr33a expression can be observed anymore. The expression of Fuss in sensory neurons during development, and the adult phenotype, suggest that Fuss is needed for the proper maturation of these neurons and therefore is essential for bitter GRN differentiation. As there is a possibility, that the bitter sensation phenotype might be due to some higher order interneurons within the CNS, we generated a specific UAS-t::gRNA-fuss4x line to be able to perform cell type specific gene knockouts. Indeed, using an independent driver line (Poxn-Gal4-13-1) expressed in all GRNs, faithfully reproduced this phenotype indicating a direct association of bitter sensation and GRN defects. In fuss mutant flies morphology of bitter GRNs was not altered and cell number was just slightly changed compared to controls, while Gr33a expression was completely lost in 40% of all bitter GRNs and Gr66a expression was reduced in all GRNs, but was never completely absent from a bitter GRN. Therefore, in fuss mutant flies bitter GRNs are correctly specified but the terminal differentiation of this neurons is disturbed, which ultimately results in impaired bitter taste sensation. This is comparable to Fuss neurons in the larval and adult CNS, where loss of Fuss expression also did not have an impact on axonal projections or cell numbers and thus not on initial specification of these neurons. This supports the idea, that Fuss is required for fine tuning individual subgroups of neurons during development, a phenotype, which resembles loss of Skor2 in mice, where it is dispensable for initial Purkinje cell fate specification but is required for proper differentiation and maturation of Purkinje cells [15]. It is very likely that other genes will also be affected by the loss of Fuss, and the reduction of these gustatory receptors could lead to a cumulative effect, as it has been shown that they act in heteromultimers where a multimeric receptor consists of at least Gr66a, Gr33a and Gr93a, which are all required for caffeine sensation [53,54]. Whereas over the years many studies have dissected the function of single gustatory receptors, the complexes they establish, and genes which are involved in more common topics like sensory neuron formation, less is known about the differentiation and specification of subsets of GRNs [55–57]. To find further genes involved in differentiation of bitter GRNs and to clarify the molecular consequences of the fuss mutation in bitter GRNs we will conduct transcriptional profiling experiments specifically in Fuss positive GRNs.
Using the TaDa method, we were curious to see if this method is sensitive enough to pick up significant differences between fuss mutant and wildtype flies. This was indeed the case for GR66a. However, in general, the performed TaDa experiments showed only slight differences between mutant and control flies. This could be a consequence of Fuss being expressed in heterogenic neuronal clusters. We showed, that Fuss interacts with Rpd3, a histone deacetylase, and therefore, a chromatin modifier, which is preferentially associated with inhibitory gene regulating complexes [58]. This could be a common mechanism for Fuss in all Fuss expressing neurons. However, different neuronal populations have different open and closed chromatin and probably the Fuss/Rpd3 complex regulates different genes in different neuronal populations, which could lead to the masking of differential gene expression by individual neuronal cell groups. Additionally, although the TaDa technique functions very well to generate transcriptional profiles without cell isolation, data is nondirectional and at GATC fragment resolution, which decreases overall resolution. To overcome these limitations experiments are on the way to unravel the function of specific neuronal clusters as well as the function of fuss in these neuronal clusters, and to specifically profile transcription of these clusters and changes upon loss of fuss.
A careful analysis with our newly generated antibodies shows, that there is no expression of Fuss in larval or adult Kenyon cells as has been postulated recently [22]. To unequivocally show, that there is no requirement for Fuss in mushroom body development, neither autonomously nor non-autonomously, Fuss expressing neurons were ablated using a fuss-GAL4 line driving Reaper. Again, these flies, even without any fuss expressing cells, are fully viable and do not show mushroom body defects. Lastly, we also did not find any evidence of Fuss being expressed in insulin producing neurons by our antibody staining or DamID experiments as shown recently [36]. These discrepancies are most likely explained by the use of the specific knockout line fussdelDS, and the gene trap line fussMi13731 in our case, whereas a 40 kb genomic deletion Df(4)dCORL was used in Takaesu et al. [22] and Tran et al. [36]. This deletion covered the fuss locus as well as two more protein coding genes, 4E-T and mGluR, and three noncoding RNA genes, CR45201, CR44030 and sphinx. Any of these, or a combination of them, could be responsible for premature lethality or mushroom body defects. One additional possible explanation for their mushroom body defects in the deletion is an inappropriate fusion of a new transcriptional start site or enhancer region from the mGluR upstream to the toy gene creating a weak overexpression phenotype of toy in mushroom bodies, a phenotype, which has been described already [59]. Indeed, very recently Tran et al. [35] described a slight overexpression of Toy in their deficiency allele Df(4)dCORL.
We and others have shown that Ski/Sno proto-oncogenes have an inhibitory effect on TGF-ß or BMP signaling in overexpression assays [18,60]. This is often associated with the ability of Ski/Sno proteins to inhibit the antiproliferative effects of TGF-ß signaling in cancer and to promote their progression [61]. However, in an endogenous situation, Fuss is not expressed in cells, where the BMP/Dpp signaling pathway is active. This is displayed by the absence of the motoneuron marker pMad in Fuss neurons. Later in adulthood, Mad itself is also not specifically enriched in Fuss expressing neurons according to the TaDa dataset, clearly pointing against a function in BMP signalling. We also tested if Fuss is involved in the Activin signaling cascade, but we could not detect an interaction between Fuss and Smox in CoIP assays. However, we cannot rule out the possibility that the phosphorylated form of Smox is interacting with Fuss or the Fuss/Med complex. But since both, phosphorylated Smox and Fuss interact with Medea, we would potentially also get an artificial interaction [18,62]. At least according to the TaDa dataset, Smox is expressed in Fuss neurons. Unfortunately, there is currently no good marker available to test for an activated TGF-ß signaling pathway in Drosophila cells, like an antibody against phosphorylated Smox. What might be the main molecular mechanism for Fuss? Although the Ski/Sno/Dac homology domain and the SMAD4 binding domain in Ski have DNA binding character, they mainly have been shown to be involved in protein-protein interactions [11,63]. Furthermore, Ski/Sno proteins do not possess an intrinsic catalytic activity, they rather act as recruiting proteins [2]. In agreement, we could show that this is also the case for Fuss. Not only that Fuss binds to Medea, which is a DNA binding protein and therefore mediates the DNA binding, Fuss also interacts with Rpd3, a histone deacetylase. Thus, the Med/Fuss/Rpd3 complex is involved in chromatin silencing and plays a key role in terminal differentiation. Interestingly, the loss of bitter sensation and downregulation of bitter GRs could also be phenocopied by a knockdown of rpd3 in Fuss expressing gustatory neurons. One current hypothesis of Fuss/Rpd3 function in GRNs, which we propose, is, that this protein complex is inhibiting a repressor of GR genes and in the absence of either fuss or rpd3, the complex is inactivated and this repressor will inhibit bitter GR genes.
For Ski and Sno, the transcriptional repressor complexes have been reasonably well characterized [10,64], but for the Fuss-type proteins, very little is known about their complexes. It would be highly interesting if Fuss proteins act through repressor complexes identical to the complexes of Ski or Sno or a rather unique one. The most exciting question to solve regarding protein interaction will be, if the Fuss/Rpd3 complex plays a role in TGF-ß signalling, or if in contrast to its mammalian homologues, it is not only acting BMP independent, but also independent from the TGF-ß signalling cascade. Besides identifying further protein-protein interactions and investigating DNA-protein interactions more precisely, it will be very important to describe the exact function of the Fuss/Rpd3 complex. In mammals, Skor2 is thought to activate Sonic Hedgehog expression in Purkinje cells from direct binding to the Sonic Hedgehog promotor and this might be achieved by inhibition of the BMP pathway or by cooperation with the RORalpha pathway, a nuclear orphan receptor [15,17]. In contrast to that, Skor1 interacts with Lbx1, a homologue of the ladybird early or ladybird late in Drosophila, and acts as a transcriptional corepressor of Lbx1 target genes [16]. Our TaDa datasets strongly point towards another function for Fuss in Drosophila, as neither hedgehog nor the homologues of Lbx1, ladybird late and ladybird early, are enriched in Fuss expressing cells. Therefore, identifying target genes, interacting proteins, binding motifs of the Fuss complex and subsequent comparison with established models for other transcription factor complexes will elucidate the role of this complex in cell fate determination.
Material and methods
Drosophila genetics
Flies were kept under standard conditions (25°C, 12 h/12 h LD cycle). Flies from RNA interference crosses were kept at 29°C. Fly lines obtained from the Bloomington Stock Center were fussMi13731 (#60860), UAS-CD8-GFP (#5137), UAS-CD8-RFP (#32218), UAS-LacZ (#8529), tubulin-Gal80ts (#7108), UAS-Stinger (#65402), UAS-rpd3-IR (#33725), UAS-cherry-IR (#35785), ap-Gal4 (#3041), Gr33a-Gal4(#31425), Gr66a-Gal4 (#57670), Gr93a-Gal4 (#57679), Hb9-Gal4 (#32555), Gr5a-Gal4 (#57591), Ir76b-Gal4 (#51311), UAS-cas9 (#58985) and ato-Gal4 (#6480). UAS-Dam and UAS-Dam-PolII stocks were a gift from Andrea Brand. Poxn-Gal4-13-1 was a gift from Markus Noll. UAS-fussB, ap-tau-LacZ and UAS-rpr were from our stock collection. To generate the fussdelDS line two sgRNAs (GTAAGCTCCGTTTTGCTGTA and GGTGTTCCCTTTAACTTACA) were employed and cloned into pU6-BbsI-chiRNA. Homology arms were cloned into pHD-DsRed-attP and coinjected with pU6-BbsI-chiRNA as described in Gratz et al. [52]. The fussBD-Gal4 and the fussMi-cherry lines were created via RMCE with the vectors pBS-KS-attB1-2-GT-SA-GAL4-Hsp70pA and pBS-KS-attB1-2-GT-SA-mCherry-SV40, respectively [27]. To generate the mutant fussdelDS-Gal4 line, the fussBD-Gal4 line was additionally targeted with the same sgRNAs via CRISPR/Cas9, which were used for the fussdelDS line. Genomic DNA of CantonS and fussdelDS flies was extracted with QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). Successful indel mutation was confirmed by PCR with Cr1seqfw (CAAATCGACTGGGTAAATGGT) and Cr2seqrv (GTAGTCCACTACAAAGTTCCTG) oligonucleotides und subsequently sequenced (GATC Biotech, Konstanz, Germany). hs-fussB-GFP was generated by cloning the ORF of fussB-GFP into pCaSpeR. hs-fussB-GFP flies were generated via P-element integration of pCaSpeR-hs-fussB-GFP vector into w; +/Δ2–3, Ki and subsequent crossed to W1118 flies and transformants were balanced. For generation of UAS-t::gRNA-fuss4x flies we followed the protocol from Port et al. [28] and used primers, which allow the targeting of the CRISPR target sites GTAAGCTCCGTTTTGCTGTACGG, ATTGTATCCCTGCACATTGAAGG, CCAGTGAGTTCCCGACGATGTGG and TTGAAATTTGCGCCAAGCAAAGG. The pCFD6-t::gRNA-fuss4x was injected into y[1],M{vas-int.Dm}ZH-2A,w[*]; M{3xP3-RFP.attP}ZH-86Fb flies to generate UAS-t::gRNA-fuss4x flies. All Drosophila strains generated in this publication are available upon request.
Polyclonal anti-Fuss antibody generation
Fulllength fuss ORF was codon optimized at GeneArt, Regensburg, Germany. An appropriate fragment of the codon optimized fuss gene was cloned into pQE60 resulting in a 16 kDa 6xHis tagged Fuss fragment called Fuss16-6xHis (S1 Fig). Transformed Rosetta2 cells were grown to an OD 0.6 and protein expression was induced with 0.5 mM IPTG. Cells were incubated for 2.5 h at 37°C, harvested, resuspended in PBS supplemented with Protein Inhibitors (Roche, Switzerland) and lysed via sonication. Fuss16-6xHis was purified with an Äktapurifier10 (GE Healthcare, Life sciences) and was used for immunization of two rabbits at Davids Biotechnologie, Regensburg, Germany. The resulting antiserum was purified against Fuss16-6xHis to reduce nonspecific binding. Before using the anti-Fuss antibodies for immunostainings or western blots they were preabsorbed using 0–6 h embryos treated with 4% PFA in PBST 0.1% as follows: The antibody was diluted to 1:50 in 500ml PBST 0.1%, NGS 5% and incubated with 100 μl fixed embryos on a rotator at 4°C over night. Anti-Fuss antibody was further diluted to 1:200 in PBST 0.1%, NGS 5% for immunostainings and 1:1000 in TBST 0.1% for western blots.
Real time PCR
Sixty proboscises from each genotype (equal number of males and females) per biological replicate were dissected on ice and snap-frozen in liquid nitrogen. RNA was extracted by adding lysis buffer from the MicroSpin Total RNA Kit (VWR) and the tissue was extracted with a bead mill and it was proceeded according to the manufacturer’s protocol. cDNA was generated with the QuantiTect Reverse Transcription Kit (QIAGEN). For subsequent real time PCR ORA qPCR Green ROX L Mix (HighQu, Kralchtal, Germany) was employed. RP49 was used as a housekeeper control, with the primers RP49fw (CCAAGCACTTCATCCGCCACC) and RP49rv (GCGGGTGCGCTTGTTCGATCC). Primer sequences for Gr33afw (CCACCATCGCGGAAAATAC), Gr33arv (ACACACTGTGGTCCAAACTC), Gr66afw (ACAGGAATCAGTCTGCACAA), Gr66arv (AATGTTTCCATGTCCAGGGT), Gr93afw (CCACGTCACAAACTCATTCC), Gr93rv (GCCATCACAATGGACACAAA), fussBDfw (TGGCTTCTATATCTGTGGCTCA) and fussBDrv (CAAAGGCGCTCTTGACCTTC) were generated with PrimerBlast. For relative quantification, we applied the ΔΔCT method. Every experiment has been repeated at least four times.
Protein expression analysis
Developmental studies Hybridoma Bank (DSHB) antibodies were: Acj6 (1:50), Dac (Mabdac1-1, 1:20), EcRB1 (AD4.4, 1:50), LacZ (JIE7, 1:20), Pros (MR1A, 1:10), Elav (7E8A10, 1:50), Engrailed (4D9, 1:20), Even skipped (3C10, 1:20) Repo (8D12, 1:20), and Fas2 (1D4 1:10). Additional antibodies were: Pale (AB152, 1:500, Millipore), Ilp2 (1:400, gift from Pierre Leopold), Toy (1:200, gift from Uwe Walldorf), GFP (goat 1:100, Rockland; rabbit 1:1000, ThermoFisher), RFP (rabbit 1:20, ThermoFisher) and anti-phospho-SMAD1/5 (1:50, Cell signaling). Secondary antibodies were goat anti-mouse, anti-rabbit, anti-rat and anti-guinea pig Alexa Fluor 488, 555 and 594 (ThermoFisher). Samples were analysed with a Leica SP8 microscope. To confirm functionality of anti-Fuss antibodies hs-fussB-GFP third instar larvae were heatshocked for one hour at 37°C and were allowed to recover for another hour at room temperature. RIPA buffer was added to ten larvae and they were mechanically disrupted. Insoluble fragments were removed by centrifugation and supernatant was incubated at 95°C for five minutes. Supernatant was analysed via SDS-Page and Western blotting. As a housekeeper mouse anti-tubulin (B-5-1-2, MERCK) was utilised and secondary antibodies were goat anti-mouse 680nm and goat anti-rabbit 800nm (Li-Cor, Lincoln, USA). Signals were detected using an Odyssey infrared imaging system (Li-Cor, Lincoln, USA).
Co-Immunoprecipitation
S2R+ cells were cultured in Schneider’s Drosophila Medium (Pan Biotech, Aidenbach, Germany) supplemented with 10% Fetal Bovine Serum (Pan Biotech, Aidenbach, Germany). The coding regions of fussB, smox and rpd3 were inserted into pFSR11.58 3xHA and pFSR12.51 4xFlag (Frank Sprenger, Regensburg, Germany). Cells were transfected in 6 well plates at 70% confluency with 2 μg of pFSR11.58 Fuss-HA and pFSR12.51 Rpd3-Flag (or Smox-Flag), or pFSR11.58 RPD3-HA (or Smox-HA) and pFSR12.51 Fuss-Flag, respectively, using Lipofectamine 3000 (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s protocol and incubated for another 24 h. Transfected cells were harvested using a plastic scraper. For Rpd3 and Fuss interaction experiments nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Waltham, MA, USA) and only nuclear fraction was used. For Fuss and Smox interaction whole cell extracts were prepared with 400 μl lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl 150, 1% Triton X-100, 10% Glycerol, 1 mM EGTA, 10 mM NaF) supplemented with cOmplete Mini Protease Inhibitor Cocktail (Roche, Switzerland). After preclearing the extracts with 30 μl Protein A-Agarose beads (Santa Cruz, Dallas, TX, USA) and conjugating 1.5 μl Anti-Flag M2 antibody (Sigma, St. Luis, Mo, USA) to 30 μl Protein A/G Plus beads (Santa Cruz, Dallas, TX, USA), the volume of the nuclear extract was brought up to 400 μl using RIPA buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 1% (v/v) NP-40, 0.5% (w/v) Deoxycholat) supplemented with cOmplete Mini Protease Inhibitor Cocktail (Roche, Switzerland). 5% of the precleared extracts were saved for input analysis. Immunoprecipitation was conducted for 2 h at 4°C. Following three washing steps with RIPA buffer, the precipitated proteins, as well as the precleared nuclear extracts, were analyzed by SDS-PAGE and western blotting. As primary antibodies Anti-Flag M2 and Anti-HA.11 (Covance Inc., USA) were used. Secondary antibody was goat anti mouse 680 (Li-Cor). Signals were detected using an Odyssey infrared imaging system (Li-Cor, Lincoln, USA).
Targeted DamID and Bioinformatics
Targeted DamID to profile transcription in Fuss expressing neurons was performed as previously described [34,65,66]. UAS-Dam, UAS-DamPolII, UAS-Dam; fussdelDS or UAS-DamPolII; fussdelDS flies were crossed to tubulin-Gal80ts; fussdelDS-Gal4 flies. Three biological replicates of DamPolII expressing flies and three biological replicates of Dam expressing flies were conducted. Per replicate 100 one to three-day old flies (50 females and 50 males) were incubated for 24 h at 29°C and snap-frozen in liquid nitrogen. Heads were detached by vortexing and separated with sieves. Processing of genomic DNA from heads and data analysis were performed as described and NGS libraries libraries were prepared with NEBNext UltraII DNA Library Prep Kit for Illumina [34,65,66]. Sequencing was carried out by the Biomedical Sequencing Facility at CeMM. For aligning reads, dm6 release from UCSC was used. Data tracks from same genotype were averaged with the average_tracks script and 3150 genes were called with an FDR < 0.01 for mutant flies and 2932 genes for control flies. log2(Dam-PolII/Dam) ratio datasets were visualized with the Integrative Genomic Browser.
Life span
For life span determination, male flies were collected within 24 h after eclosion and were raised at 25°C under a 12 h∶12 h light/dark cycle. These flies were transferred to fresh food vials every two to three days.
Two choice feeding assay
Feeding behaviour was analysed as previously described at 25°C [38]. Fly age at time of testing ranged from one to three days and experiments were only accounted if at least 30% of all flies showed clear evaluable coloured abdomen. As bitter compounds caffeine and denatonium benzoate were utilised at the indicated concentrations. Because feeding behaviour was influenced by temperature, fussBD-Gal4 x UAS-cherry-IR and fussBD-Gal4 x UAS-rpd3-IR flies were shifted to 25°C two hours prior testing. Every experiment has been repeated at least four times.
Preparation of Figures
All figures were assembled with Adobe Photoshop CC (Adobe Systems) by importing microscopy images from Fiji and graphs from Prism.
Statistics
Survival data were analyzed using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. Significance was determined by two-tailed t-test or by One-way ANOVA with post hoc Tukey Multiple Comparison Test (****p<0.001; ***p<0.001; **p<0.01 and *p<0.05). Statistical analysis was carried out using Prism version 7.0a for MacOs, GraphPad Software, La Jolla, CA, USA.
Availability
Raw sequencing data are accessible via Gene Expression Omnibus: GEO Series GSE115347.
Supporting information
(A) Genotyping of CantonS and homozygous fussdelDS flies with fuss crispr1 seq fw and fuss crispr2 seq rv oligonucleotides via PCR of genomic DNA shows reduction of around 700bp in fussdelDS mutants as expected in contrast to control (B). Staining of heterozygous fussdelDS/+ embryos with anti-Fuss (red) and anti-Elav (green) antibodies. Scale bar indicates 25 μm. (C) Staining of homozygous fussdelDS embryos with anti-Fuss (red) and anti-Elav (green) antibodies. Scale bar indicates 25 μm. (D) Analysis of fussB and fussD transcript levels with fussBD fw and fussBD rv oligonucleotides via qPCR reveals a reduction of fussB and fussD transcript levels to 10% in homozygous fussMi13731 flies in contrast to WTB flies. n = 4 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ****p<0.0001. **p<0.01. Error bars indicate SEM. (E) Anti-Fuss staining colocalizes with GFP in larval brains of heterozygous fussMi13731/+ line. (F) No anti-Fuss staining in larval brains of homozygous fussMi13731 line can be detected. Arrowhead indicates magnified cell cluster. (G) Location of the four CRISPR target sites of the UAS-t::gRNA-fuss4x construct in the DNA sequence of the Ski/Sno homology domain. (H) Adult brains of UAS-cas9/UAS-fussB-GFP; fussBD-Gal4/+ flies show normal Fuss expression pattern. Scale bar indicates 50 μm. (I) In flies of the genotype UAS-cas9/UAS-fussB-GFP; UAS-t::gRNA-fuss4x; fussBD-Gal4/+ fussB-GFP is strongly reduced. Scale bar indicates 50 μm.
(TIF)
(A) Schematic representation of conserved domains and localization of the Fuss16 fragment used for immunization. Exact sequence of Fuss16-His fragment shown in red. (B) Detection of Fuss-GFP (green) from heatshock induced Fuss-GFP flies in western blots by anti-GFP and anti-Fuss antibodies. As a negative control CantonS is used and Tubulin as a housekeeper protein (red). Both antibodies recognize a predicted protein size of 112 kDa for the fusion protein. Endogenous levels of the Fuss protein cannot be detected on western blots due to the low abundance of the protein. (C) Comparison of VNC of stage 13 embryo with VNC of stage 16 embryo shows increase in number of Fuss (green) or Toy (red) cells, but only one cell per hemineuromer shows colocalization of both markers. (D) Comparison of expression pattern of interneuron marker Engrailed (red) and Fuss (green) visualized by antibody staining in embryonic VNC. (E) Fuss expression pattern as revealed by expression of GFP (green) by heterozygous fussMi13731/+ in larval brains does not colocalize with LacZ (red) driven by Hb9-GAL4 line. (F) Even skipped (red), a motor neuron marker, is not expressed in Fuss neurons (green) visualized by antibody staining in embryonic VNC. (G) Ventral Apterous cells marked by expression of CD8-RFP (red) with ap-Gal4 are positive for Fuss (green) expression in larval VNC. Scale bars indicate 30 μm (C, D, E, F) and 10 μm (G).
(TIF)
(A) pale (ple) is weakly bound by Dam-PolII as revealed by TaDa and no colocalization is observed between Ple positive cells (red) and GFP expressed by the heterozygous fussMi13731/+ reporter line (green) in whole adult brains. Overlap between signals arises from different optical slices and not from colocalization. (B) insulin like peptide 2 (ilp2) is weakly bound by Dam-PolII as revealed by TaDa. Confocal slices covering the pars intercerebralis and a part of the adult brain hemisphere show no colocalization between insulin producing cells labeled with anti-Ilp2 antibody (red) and Fuss neurons labeled with anti-Fuss antibody (green). In (A) and (B) regions bound stronger by Dam-PolII than by Dam are depicted in green, whereas regions bound stronger by Dam than by Dam-PolII are depicted in red. Scale bars indicate 50 μm.
(TIF)
(A) Expression of UAS-CD8-GFP with fussBD-Gal4 reveals four GRNs located in the two last tarsal segments of the prothoracic, mesothoracic and metatoracic leg. Scale bars indicate 50 μm. (B) GFP expression from Poxn-Gal4-13-1 is not overlapping with Cherry expression from fussMi-cherry reporter line in neurons of the adult CNS. Overlap can only be observed in GRN nerve fibers from proboscis. EBN = ellipsoid body neurons. ALI = Antennal lobe interneurons. Scale bar indicates 50 μm. (C) Homozygous fussMi13731 flies show reduced caffeine sensation also at lower concentrations compared to heterozygous fussMi13731 x WTB and WTB flies. n = 4–9 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. **p<0.01 ****p<0.0001. Error bars indicate SEM. (D) Homozygous fussMi13731 mutant flies show reduced sensation of denatonium benzoate compared to heterozygous fussMi13731 x WTB and WTB flies at a concentration of 100 μm. At 500 μm denatonium benzoate effect of homozygous fussMi13731 flies is reversed to control levels. n = 4–5 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ****p<0.0001. Error bars indicate SEM. (E) Transheterozygous fussMi13731/fussdelDS mutants show reduced transcript levels for Gr33a, Gr66a and Gr93a in contrast to W1118 control. n = 4 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ***p<0.001. **p<0.01. *p<0.05. Error bars indicate SEM. (F) Alignment of Drosophila Fuss with mouse Skor1 and Skor2. Ski/Sno/Dac homology domain, SMAD4 binding domain and proposed Rpd3 interaction fragment in Skor2 are displayed by colored lines as described.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Juan A. Navarro for critically reading the manuscript. We thank Renate Reng for technical assistance. We thank Lars Kullmann as well as Caroline Iglhaut for the generation of HS-FussB-GFP flies and Matthias Schramm for the generation of the appropriate vectors for the fussdelDS flies. We thank Frank Sprenger for providing pFSR vectors, Markus Noll for Poxn-Gal4-13-1 flies and Andrea H. Brand for UAS-Dam and UAS-Dam-PolII vectors and flies. We thank Michael Rehli and Owen J. Marshall for support regarding NGS library preparation and bioinformatical analysis. We would like to thank the Biomedical Sequencing Facility at CeMM for sequencing. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and antibodies from Developmental Studies Hybridoma Bank were used in this study.
Data Availability
Raw sequencing data are available via NCBI's Gene Expression Omnibus under accession number GSE115347. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (FI2133/1-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998; 67: 753–791. 10.1146/annurev.biochem.67.1.753 [DOI] [PubMed] [Google Scholar]
- 2.Deheuninck J, Luo K. Ski and SnoN, potent negative regulators of TGF-beta signaling. Cell Res. 2009; 19: 47–57. 10.1038/cr.2008.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001; 276: 12477–12480. 10.1074/jbc.C100008200 [DOI] [PubMed] [Google Scholar]
- 4.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y-Y, Grinnell BW, et al. The MAD-Related Protein Smad7 Associates with the TGFβ Receptor and Functions as an Antagonist of TGFβ Signaling. Cell. 1997; 89: 1165–1173. 10.1016/S0092-8674(00)80303-7 [DOI] [PubMed] [Google Scholar]
- 5.Boone B, Haspeslagh M, Brochez L. Clinical significance of the expression of c-Ski and SnoN, possible mediators in TGF-β resistance, in primary cutaneous melanoma. Journal of Dermatological Science; 53: 26–33. 10.1016/j.jdermsci.2008.07.010 [DOI] [PubMed] [Google Scholar]
- 6.Shinagawa T, Dong HD, Xu M, Maekawa T, Ishii S. The sno gene, which encodes a component of the histone deacetylase complex, acts as a tumor suppressor in mice. EMBO J. 2000; 19: 2280–2291. 10.1093/emboj/19.10.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahchan NS, Wang D, Bissell MJ, Luo K. SnoN regulates mammary gland alveologenesis and onset of lactation by promoting prolactin/Stat5 signaling. Development. 2012; 139: 3147–3156. 10.1242/dev.079616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berk M, Desai SY, Heyman HC, Colmenares C. Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial patterning, and skeletal muscle development. Genes & Development. 1997; 11: 2029–2039. 10.1101/gad.11.16.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stegmüller J, Konishi Y, Huynh MA, Yuan Z, DiBacco S, Bonni A. Cell-Intrinsic Regulation of Axonal Morphogenesis by the Cdh1-APC Target SnoN. Neuron. 2006; 50: 389–400. 10.1016/j.neuron.2006.03.034 [DOI] [PubMed] [Google Scholar]
- 10.Nomura T, Khan MM, Kaul SC, Dong H-D, Wadhwa R, Colmenares C, et al. Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor. Genes & Development. 1999; 13: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J-W, Krawitz AR, Chai J, Li W, Zhang F, Luo K, et al. Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-Mediated Repression of TGF-β Signaling. Cell. 2002; 111: 357–367. 10.1016/S0092-8674(02)01006-1 [DOI] [PubMed] [Google Scholar]
- 12.Wilson JJ, Malakhova M, Zhang R, Joachimiak A, Hegde RS. Crystal Structure of the Dachshund Homology Domain of Human SKI. Structure; 12: 785–792. 10.1016/j.str.2004.02.035 [DOI] [PubMed] [Google Scholar]
- 13.Arndt S, Poser I, Moser M, Bosserhoff A-K. Fussel-15, a novel Ski/Sno homolog protein, antagonizes BMP signaling. Molecular and Cellular Neuroscience. 2007; 34: 603–611. 10.1016/j.mcn.2007.01.002 [DOI] [PubMed] [Google Scholar]
- 14.Arndt S, Poser I, Schubert T, Moser M, Bosserhoff A-K. Cloning and functional characterization of a new Ski homolog, Fussel-18, specifically expressed in neuronal tissues. Lab Invest. 2005; 85: 1330–1341. 10.1038/labinvest.3700344 [DOI] [PubMed] [Google Scholar]
- 15.Nakatani T, Minaki Y, Kumai M, Nitta C, Ono Y. The c-Ski family member and transcriptional regulator Corl2/Skor2 promotes early differentiation of cerebellar Purkinje cells. Developmental Biology. 2014; 388: 68–80. 10.1016/j.ydbio.2014.01.016 [DOI] [PubMed] [Google Scholar]
- 16.Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y. Corl1, a novel neuronal lineage-specific transcriptional corepressor for the homeodomain transcription factor Lbx1. J Biol Chem. 2005; 280: 3645–3655. 10.1074/jbc.M411652200 [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Harrison W, Overbeek PA, Zheng H. Transposon mutagenesis with coat color genotyping identifies an essential role for Skor2 in sonic hedgehog signaling and cerebellum development. Development. 2011; 138: 4487–4497. 10.1242/dev.067264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer S, Bayersdorfer F, Harant E, Reng R, Arndt S, Bosserhoff A-K, et al. fussel (fuss)—A Negative Regulator of BMP Signaling in Drosophila melanogaster. PLOS ONE. 2012; 7: 1–12. 10.1371/journal.pone.0042349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrio R, López-Varea A, Casado M, de Celis JF. Characterization of dSnoN and its relationship to Decapentaplegic signaling in Drosophila. Developmental Biology. 2007; 306: 66–81. 10.1016/j.ydbio.2007.02.039 [DOI] [PubMed] [Google Scholar]
- 20.Djabrayan NJ-V, Casanova J. Snoo and Dpp Act as Spatial and Temporal Regulators Respectively of Adult Progenitor Cells in the Drosophila Trachea. PLOS Genetics. 2016; 12: 1–15. 10.1371/journal.pgen.1005909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shravage BV, Altmann G, Technau M, Roth S. The role of Dpp and its inhibitors during eggshell patterning in Drosophila. Development. 2007; 134: 2261–2271. 10.1242/dev.02856 [DOI] [PubMed] [Google Scholar]
- 22.Takaesu NT, Stinchfield MJ, Shimizu K, Arase M, Quijano JC, Watabe T, et al. Drosophila CORL is required for Smad2-mediated activation of Ecdysone Receptor expression in the mushroom body. Development. 2012; 139: 3392–3401. 10.1242/dev.079442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Koolhaas WH, Schnorrer F. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 (Bethesda, Md.). 2014; 4: 2409–2418. 10.1534/g3.114.013979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco J, Pauli T, Seimiya M, Udolph G, Gehring WJ. Genetic interactions of eyes absent, twin of eyeless and orthodenticle regulate sine oculis expression during ocellar development in Drosophila. Developmental Biology. 2010; 344: 1088–1099. 10.1016/j.ydbio.2010.05.494 [DOI] [PubMed] [Google Scholar]
- 25.Ueki N, Hayman MJ. Signal-dependent N-CoR requirement for repression by the Ski oncoprotein. J Biol Chem. 2003; 278: 24858–24864. 10.1074/jbc.M303447200 [DOI] [PubMed] [Google Scholar]
- 26.Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, et al. The Ski oncoprotein interacts with the Smad proteins to repress TGFβ signaling. Genes & Development. 1999; 13: 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nature methods. 2011; 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked Cas9 and Cpf1 sgRNAs. Nature methods. 2016; 13: 852–854. 10.1038/nmeth.3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broihier HT, Skeath JB. Drosophila Homeodomain Protein dHb9 Directs Neuronal Fate via Crossrepressive and Cell-Nonautonomous Mechanisms. Neuron; 35: 39–50. 10.1016/S0896-6273(02)00743-2 [DOI] [PubMed] [Google Scholar]
- 30.Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. even-skipped Determines the Dorsal Growth of Motor Axons in Drosophila. Neuron. 1999; 22: 43–52. 10.1016/S0896-6273(00)80677-7 [DOI] [PubMed] [Google Scholar]
- 31.Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mechanisms of Development. 1998; 74: 121–131. 10.1016/S0925-4773(98)00071-9 [DOI] [PubMed] [Google Scholar]
- 32.Miguel-Aliaga I, Allan DW, Thor S. Independent roles of the dachshund and eyes absent genes in BMP signaling, axon pathfinding and neuronal specification. Development. 2004; 131: 5837–5848. 10.1242/dev.01447 [DOI] [PubMed] [Google Scholar]
- 33.Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995; 121: 1769–1773. [DOI] [PubMed] [Google Scholar]
- 34.Southall TD, Gold KS, Egger B, Davidson CM, Caygill EE, Marshall OJ, et al. Cell-Type-Specific Profiling of Gene Expression and Chromatin Binding without Cell Isolation: Assaying RNA Pol II Occupancy in Neural Stem Cells. Developmental Cell. 2013; 26: 101–112. 10.1016/j.devcel.2013.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran NL, Takaesu NT, Cornell EF, Newfeld SJ. CORL Expression in the Drosophila Central Nervous System Is Regulated by Stage Specific Interactions of Intertwined Activators and Repressors. G3 (Bethesda, Md.). 2018. 10.1534/g3.118.200282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran NL, Goldsmith SL, Dimitriadou A, Takaesu NT, Consoulas C, Newfeld SJ. CORL Expression and Function in Insulin Producing Neurons Reversibly Influences Adult Longevity in Drosophila. G3 (Bethesda, Md.). 2018; 8: 2979–2990. 10.1534/g3.118.200572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Besson M.T., Sinakevitch I., Melon C., Iché‐Torres M., Birman S. Involvement of the drosophila taurine/aspartate transporter dEAAT2 in selective olfactory and gustatory perceptions. J. Comp. Neurol. 2011; 519: 2734–2757. 10.1002/cne.22649 [DOI] [PubMed] [Google Scholar]
- 38.Moon SJ, Köttgen M, Jiao Y, Xu H, Montell C. A Taste Receptor Required for the Caffeine Response In Vivo. Current Biology; 16: 1812–1817. 10.1016/j.cub.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 39.Ahn J-E, Chen Y, Amrein H. Molecular basis of fatty acid taste inDrosophila. Elife. 2017; 6 10.7554/eLife.30115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon SJ, Lee Y, Jiao Y, Montell C. A Drosophila Gustatory Receptor Essential for Aversive Taste and Inhibiting Male-to-Male Courtship. Current Biology; 19: 1623–1627. 10.1016/j.cub.2009.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apostolopoulou AA, Köhn S, Stehle B, Lutz M, Wüst A, Mazija L, et al. Caffeine Taste Signaling in Drosophila Larvae. Frontiers in Cellular Neuroscience. 2016; 10: 193 10.3389/fncel.2016.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi J, van Giesen L, Choi MS, Kang K, Sprecher SG, Kwon JY. A Pair of Pharyngeal Gustatory Receptor Neurons Regulates Caffeine-Dependent Ingestion in Drosophila Larvae. Frontiers in Cellular Neuroscience. 2016; 10: 1523 10.3389/fncel.2016.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amrein H, Thorne N. Gustatory Perception and Behavior in Drosophila melanogaster. Current Biology. 2005; 15: R673–R684. 10.1016/j.cub.2005.08.021 [DOI] [PubMed] [Google Scholar]
- 44.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013; 340: 1334–1338. 10.1126/science.1234133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freeman EG, Dahanukar A. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. 2015; 34: 140–148. 10.1016/j.conb.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebbs ML, Amrein H. Taste and pheromone perception in the fruit fly Drosophila melanogaster. Pflugers Arch. 2007; 454: 735–747. 10.1007/s00424-007-0246-y [DOI] [PubMed] [Google Scholar]
- 47.Boll W, Noll M. The Drosophila Pox neuro gene: control of male courtship behavior and fertility as revealed by a complete dissection of all enhancers. Development. 2002; 129: 5667–5681. 10.1242/dev.00157 [DOI] [PubMed] [Google Scholar]
- 48.Lee Y, Moon SJ, Montell C. Multiple gustatory receptors required for the caffeine response in Drosophila. Proceedings of the National Academy of Sciences. 2009; 106: 4495–4500. 10.1073/pnas.0811744106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bosch PS, Ziukaite R, Alexandre C, Basler K, Vincent J-P. Dpp controls growth and patterning in Drosophila wing precursors through distinct modes of action. Elife. 2017; 6: 375 10.7554/eLife.22546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allan DW, Pierre SES, Miguel-Aliaga I, Thor S. Specification of Neuropeptide Cell Identity by the Integration of Retrograde BMP Signaling and a Combinatorial Transcription Factor Code. Cell. 2003; 113: 73–86. 10.1016/S0092-8674(03)00204-6 [DOI] [PubMed] [Google Scholar]
- 51.Marqués G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, et al. The Drosophila BMP Type II Receptor Wishful Thinking Regulates Neuromuscular Synapse Morphology and Function. Neuron. 2002; 33: 529–543. 10.1016/S0896-6273(02)00595-0 [DOI] [PubMed] [Google Scholar]
- 52.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014; 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shim J, Lee Y, Jeong YT, Kim Y, Lee MG, Montell C, et al. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat Commun. 2015; 6 10.1038/ncomms9867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poudel S, Lee Y. Gustatory Receptors Required for Avoiding the Toxic Compound Coumarin in Drosophila melanogaster. Mol Cells. 2016; 39: 310–315. 10.14348/molcells.2016.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang M-L, Hsu C-H, Chien C-T. The Proneural Gene amos Promotes Multiple Dendritic Neuron Formation in the Drosophila Peripheral Nervous System. Neuron. 2000; 25: 57–67. 10.1016/S0896-6273(00)80871-5 [DOI] [PubMed] [Google Scholar]
- 56.Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993; 73: 1307–1321. 10.1016/0092-8674(93)90358-W. [DOI] [PubMed] [Google Scholar]
- 57.Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn Finger Transcription Factor, Is Necessary and Sufficient for Sensory Organ Development in Drosophila. Cell. 2000; 102: 349–362. 10.1016/S0092-8674(00)00040-4 [DOI] [PubMed] [Google Scholar]
- 58.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes & Development. 1999; 13: 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Furukubo-Tokunaga K, Adachi Y, Kurusu M, Walldorf U. Brain patterning defects caused by mutations of the twin of eyeless gene in Drosophila melanogaster. Fly. 2009; 3: 263–269. 10.4161/fly.10385 [DOI] [PubMed] [Google Scholar]
- 60.Akiyoshi S, Inoue H, Hanai J-i, Kusanagi K, Nemoto N, Miyazono K, et al. c-Ski Acts as a Transcriptional Co-repressor in Transforming Growth Factor-β Signaling through Interaction with Smads. J Biol Chem. 1999; 274: 35269–35277. 10.1074/jbc.274.49.35269 [DOI] [PubMed] [Google Scholar]
- 61.Sun Y, Liu X, Eaton EN, Lane WS, Lodish HF, Weinberg RA. Interaction of the Ski Oncoprotein with Smad3 Regulates TGF-β Signaling. Molecular Cell. 1999; 4: 499–509. 10.1016/S1097-2765(00)80201-4 [DOI] [PubMed] [Google Scholar]
- 62.Das P, Inoue H, Baker JC, Beppu H, Kawabata M, Harland RM, et al. Drosophila dSmad2 and Atr-I transmit activin/TGFbeta signals. Genes Cells. 1999; 4: 123–134. 10.1046/j.1365-2443.1999.00244.x [DOI] [PubMed] [Google Scholar]
- 63.Nyman T, Trésaugues L, Welin M, Lehtiö L, Flodin S, Persson C, et al. The Crystal Structure of the Dachshund Domain of Human SnoN Reveals Flexibility in the Putative Protein Interaction Surface. PLOS ONE. 2010; 5: e12907 10.1371/journal.pone.0012907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tecalco-Cruz AC, Ríos-López DG, Vázquez-Victorio G, Rosales-Alvarez RE, Macías-Silva M. Transcriptional cofactors Ski and SnoN are major regulators of the TGF-β/Smad signaling pathway in health and disease. Signal Transduction and Targeted Therapy. 2018; 3: 15 10.1038/s41392-018-0015-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marshall OJ, Brand AH. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015; 31: 3371–3373. 10.1093/bioinformatics/btv386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall OJ, Southall TD, Cheetham SW, Brand AH. Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat Protoc. 2016; 11: 1586–1598. 10.1038/nprot.2016.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Genotyping of CantonS and homozygous fussdelDS flies with fuss crispr1 seq fw and fuss crispr2 seq rv oligonucleotides via PCR of genomic DNA shows reduction of around 700bp in fussdelDS mutants as expected in contrast to control (B). Staining of heterozygous fussdelDS/+ embryos with anti-Fuss (red) and anti-Elav (green) antibodies. Scale bar indicates 25 μm. (C) Staining of homozygous fussdelDS embryos with anti-Fuss (red) and anti-Elav (green) antibodies. Scale bar indicates 25 μm. (D) Analysis of fussB and fussD transcript levels with fussBD fw and fussBD rv oligonucleotides via qPCR reveals a reduction of fussB and fussD transcript levels to 10% in homozygous fussMi13731 flies in contrast to WTB flies. n = 4 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ****p<0.0001. **p<0.01. Error bars indicate SEM. (E) Anti-Fuss staining colocalizes with GFP in larval brains of heterozygous fussMi13731/+ line. (F) No anti-Fuss staining in larval brains of homozygous fussMi13731 line can be detected. Arrowhead indicates magnified cell cluster. (G) Location of the four CRISPR target sites of the UAS-t::gRNA-fuss4x construct in the DNA sequence of the Ski/Sno homology domain. (H) Adult brains of UAS-cas9/UAS-fussB-GFP; fussBD-Gal4/+ flies show normal Fuss expression pattern. Scale bar indicates 50 μm. (I) In flies of the genotype UAS-cas9/UAS-fussB-GFP; UAS-t::gRNA-fuss4x; fussBD-Gal4/+ fussB-GFP is strongly reduced. Scale bar indicates 50 μm.
(TIF)
(A) Schematic representation of conserved domains and localization of the Fuss16 fragment used for immunization. Exact sequence of Fuss16-His fragment shown in red. (B) Detection of Fuss-GFP (green) from heatshock induced Fuss-GFP flies in western blots by anti-GFP and anti-Fuss antibodies. As a negative control CantonS is used and Tubulin as a housekeeper protein (red). Both antibodies recognize a predicted protein size of 112 kDa for the fusion protein. Endogenous levels of the Fuss protein cannot be detected on western blots due to the low abundance of the protein. (C) Comparison of VNC of stage 13 embryo with VNC of stage 16 embryo shows increase in number of Fuss (green) or Toy (red) cells, but only one cell per hemineuromer shows colocalization of both markers. (D) Comparison of expression pattern of interneuron marker Engrailed (red) and Fuss (green) visualized by antibody staining in embryonic VNC. (E) Fuss expression pattern as revealed by expression of GFP (green) by heterozygous fussMi13731/+ in larval brains does not colocalize with LacZ (red) driven by Hb9-GAL4 line. (F) Even skipped (red), a motor neuron marker, is not expressed in Fuss neurons (green) visualized by antibody staining in embryonic VNC. (G) Ventral Apterous cells marked by expression of CD8-RFP (red) with ap-Gal4 are positive for Fuss (green) expression in larval VNC. Scale bars indicate 30 μm (C, D, E, F) and 10 μm (G).
(TIF)
(A) pale (ple) is weakly bound by Dam-PolII as revealed by TaDa and no colocalization is observed between Ple positive cells (red) and GFP expressed by the heterozygous fussMi13731/+ reporter line (green) in whole adult brains. Overlap between signals arises from different optical slices and not from colocalization. (B) insulin like peptide 2 (ilp2) is weakly bound by Dam-PolII as revealed by TaDa. Confocal slices covering the pars intercerebralis and a part of the adult brain hemisphere show no colocalization between insulin producing cells labeled with anti-Ilp2 antibody (red) and Fuss neurons labeled with anti-Fuss antibody (green). In (A) and (B) regions bound stronger by Dam-PolII than by Dam are depicted in green, whereas regions bound stronger by Dam than by Dam-PolII are depicted in red. Scale bars indicate 50 μm.
(TIF)
(A) Expression of UAS-CD8-GFP with fussBD-Gal4 reveals four GRNs located in the two last tarsal segments of the prothoracic, mesothoracic and metatoracic leg. Scale bars indicate 50 μm. (B) GFP expression from Poxn-Gal4-13-1 is not overlapping with Cherry expression from fussMi-cherry reporter line in neurons of the adult CNS. Overlap can only be observed in GRN nerve fibers from proboscis. EBN = ellipsoid body neurons. ALI = Antennal lobe interneurons. Scale bar indicates 50 μm. (C) Homozygous fussMi13731 flies show reduced caffeine sensation also at lower concentrations compared to heterozygous fussMi13731 x WTB and WTB flies. n = 4–9 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. **p<0.01 ****p<0.0001. Error bars indicate SEM. (D) Homozygous fussMi13731 mutant flies show reduced sensation of denatonium benzoate compared to heterozygous fussMi13731 x WTB and WTB flies at a concentration of 100 μm. At 500 μm denatonium benzoate effect of homozygous fussMi13731 flies is reversed to control levels. n = 4–5 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ****p<0.0001. Error bars indicate SEM. (E) Transheterozygous fussMi13731/fussdelDS mutants show reduced transcript levels for Gr33a, Gr66a and Gr93a in contrast to W1118 control. n = 4 for each genotype. One-way ANOVA with post hoc Tukey´s test was used to calculate p-values. ***p<0.001. **p<0.01. *p<0.05. Error bars indicate SEM. (F) Alignment of Drosophila Fuss with mouse Skor1 and Skor2. Ski/Sno/Dac homology domain, SMAD4 binding domain and proposed Rpd3 interaction fragment in Skor2 are displayed by colored lines as described.
(TIF)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Raw sequencing data are available via NCBI's Gene Expression Omnibus under accession number GSE115347. All other relevant data are within the paper and its Supporting Information files.