Abstract
The response of the lysosomes, the waste clearance machinery of the cell, to different environmental stimuli is coordinated by a gene network with a master regulator Transcription factor EB (TFEB) at the core. Disruption of multiple facets of the lysosomal and autophagic network has been linked to various neurodegenerative and lysosomal storage disorders, making TFEB an attractive therapeutic target to rescue or augment lysosomal function under pathological scenario. In this study, we demonstrate that cinnamic acid, a naturally occurring plant-based product, induces lysosomal biogenesis in mouse primary brain cells via upregulation of TFEB. We delineate that cinnamic acid activates the nuclear hormone receptor PPARα to transcriptionally upregulate TFEB and stimulate lysosomal biogenesis. Moreover, using in-silico and biochemical approaches we established that cinnamic acid serves as a potent ligand for peroxisome proliferator-activated receptor α (PPARα). Finally, cinnamic acid treatment in male and female 5× Familial Alzheimer’s disease (5XFAD) mice remarkably reduced cerebral amyloid-beta plaque burden and improved memory via PPARα. Therefore, stimulation of lysosomal biogenesis by cinnamic acid may have therapeutic implications for treatment of Alzheimer’s disease and other lysosomal disorders originating from accumulation of toxic protein aggregates.
Keywords: Lysosomal biogenesis, Cinnamic acid, PPARα, TFEB, Plaque clearance, 5XFAD model
1. Introduction
Enhancement of cellular clearance pathways via induction of the lysosomal machinery has emerged as an attractive therapeutic target for diseases originating from accumulation of toxic aggregates. Lysosomes are the terminal degradative compartment for macromolecules received from the endocytic, autophagic, secretory and phagocytic pathways (Ballabio, 2016; De Duve, 1963; Settembre et al., 2013). Lysosomal functions under different physiological and pathological conditions are regulated by a central coordinator, TFEB, a basic helix-loop-helix transcription factor (Martini-Stoica et al., 2016; Sardiello et al., 2009), making it a promising candidate for treatment of Lysosomal storage disorders (LSDs) and neurodegenerative diseases. Indeed, mounting evidence has established the beneficial role of TFEB targeted lysosomal modulation in Alzheimer’s disease (Xiao et al., 2014; Xiao et al., 2015), Parkinson’s disease (Decressac et al., 2013; Dehay et al., 2010), Huntington’s disease (Sardiello et al., 2009; Tsunemi et al., 2012) and several LSDs (Medina et al., 2011; Napolitano and Ballabio, 2016; Rega et al., 2016; Song et al., 2013; Spampanato et al., 2013).
Alzheimer’s disease (AD), the most prevalent neurodegenerative disorder with a multifactorial pathophysiology, is diagnosed by progressive cognitive decline ultimately leading to dementia. Two major neurological hallmarks, deposition of senile plaques and neurofibrillary tangles (NFTs) underline the pathogenesis of AD (Huang and Mucke, 2012; Querfurth and Laferla, 2010). Extracellular senile plaques are comprised of toxic aggregates of the protein amyloid-beta (Aβ), generated by sequential proteolytic cleavage of the Amyloid precursor protein (APP) by beta and gamma secretases whereas abnormal hyperphosphorylation of microtubule-associated protein tau leads to intraneuronal accumulation of NFTs (Grundke-Iqbal et al., 1986; Querfurth and Laferla, 2010; Yoon and Kim, 2016). Genetic mutations in the neuronal membrane protein APP and the component of the gamma secretase complex presenilins (PSEN1 and PSEN2), underlie the early-onset familial forms of the disease which constitute only 1–5% of all AD occurrences (Bekris et al., 2010; Whyte et al., 2017). However, the etiology of the more prevalent late-onset sporadic form of AD remains largely unknown posing challenges to the development of AD therapeutics. Impaired clearance of Aβ has been implicated as a major causative agent in the widespread sporadic AD patients (Mawuenyega et al., 2010), stimulating a new avenue of research targeted at induction of effective cellular clearance of Aβ as a therapeutic strategy. Positive lysosomal modulation by Z-Phe-Ala-diazomethylketone (PADK) was shown to have protective effects by enhancing activity of cathepsin B, reducing Aβ accumulation and improving behavioral deficit in APP/ PS1 mice (Butler et al., 2011). Moreover, enhancement of astrocytic lysosomal biogenesis via virus-mediated TFEB overexpression has been demonstrated to facilitate Aβ uptake and its lysosomal degradation mitigating the amyloid pathogenesis (Xiao et al., 2014). However, pharmacological compounds promoting Aβ clearance in AD are less explored and demands further studies. Given the immense implications of lysosomal stimulation in countering the pathogenesis of AD, we examined whether cinnamic acid, a natural compound, can induce lysosomal biogenesis and thus have beneficial effects in an AD mouse model.
Cinnamic acid is an aromatic carboxylic acid found ubiquitously in plant-based products. The deamination of phenylalanine by the enzyme phenylalanine ammonia lyase (PAL) yields cinnamic acid, which undergoes further enzymatic modifications to produce numerous phytochemical compounds including coumarins, flavonoids, isoflavonoids, phenyl-propanoids, and lignins. Cinnamic acid and its bioactive metabolites are abundantly present in human diet including vegetables, fruits, honey and whole grains (Adisakwattana, 2017; Guzman, 2014; Vogt, 2010). Several studies have highlighted the role of cinnamic acid and its pharmacologically active derivatives as anti-microbial, antioxidant (Guzman, 2014; Natella et al., 1999; Sova, 2012), anti-cancer (De et al., 2011; Su et al., 2015), anti-atherogenic (Lapeyre et al., 2005), anti-tuberculosis (De et al., 2012), and anti-fungal agents (Tawata et al., 1996). Here, we delineate that cinnamic acid is a ligand of peroxisome proliferator-activated receptor α (PPARα) and that cinnamic acid induces lysosomal biogenesis via PPARα. We also demonstrate that cinnamic acid can attenuate the amyloid plaque pathology in 5XFAD mouse model in PPARα-dependent manner.
2. Materials and methods
2.1. Reagents
Trans-cinnamic acid was obtained from Sigma Aldrich (C80857). Materials for primary astrocyte culture (DMEM/F-12, Hank’s balanced salt solution, 0.05% trypsin, and antibiotic-antimycotic) were obtained from Mediatech (Washington, DC). For primary neuron culture, neurobasal media, B27 supplement, B27 supplement minus antioxidant were purchased from Life Technologies (Grand Island, NY). Fetal bovine serum (FBS) was purchased from Atlas Biologicals. Thioflavin-S and all other molecular biology-grade chemicals were purchased from Sigma Aldrich. All primary antibodies sources and dilutions used are listed in Table S1. Secondory antibodies (Alexa-fluor 488 and 647-conjugated) for immunostaining were purchased from Jackson ImmunoResearch. For immunoblotting, 10× Tris/Glycine/SDS running buffer was obtained from BioRad (1610772) and IR-dye-labeled secondary antibodies were obtained from Li-Cor Biosciences.
2.2. Isolation of primary mouse astroglia
Mouse primary astroglia were isolated from 7 to 9 day old pups as described earlier (Brahmachari et al., 2006; Ghosh et al., 2012; Khasnavis and Pahan, 2012). Following isolation, the mixed glia cultures were grown for 8 days with change of medium every 3 days. On day 9, the cells were subjected to shaking at 240 rpm for 2 h at 37 °C on a rotary shaker to eliminate microglia. On day 11, another round of shaking was performed at 190 rpm for 18 h to eliminate oligodendroglia and residual microglia. This ensured removal of all non-astro-glial cells from the culture. Next, the attached cells were washed and seeded onto new plates for subsequent studies.
2.3. Isolation of primary cortical neuron
Mouse primary neurons were isolated from fetus (E18 to E16) as described previously (Corbett et al., 2012; Ghosh et al., 2012). Briefly, whole brain was removed and the cortical part was dissected. The cells were washed by centrifugation (1000 rpm for 10 min, 3 times) and plated at 10% confluency on square coverslips pretreated with poly-D-lysine in 6-well plate. After 5 min, the cell suspension containing non-adherent cells was aspirated followed by addition of 500 ml complete neurobasal media with B27 supplement. The cells were allowed to grow for at least 5 days. The cells were treated in neurobasal media with B27 supplement minus antioxidants. MAP2 was used as a neuronal marker for immunocytochemistry.
2.4. LysoTracker staining
Cultured cells were treated under serum free conditions followed by live cell staining using LysoTracker Red DND99 (Life Technologies, Grand Island, NY). Briefly, following treatments, cells were incubated with 75 nM LysoTracker Red for 45 mins (Chandra et al., 2018b; Ghosh et al., 2015). Next, cells were washed thoroughly with filtered PBS and fixed using chilled methanol. Cells were incubated in DAPI for staining the nuclei. The coverslip containing the cells was mounted on a glass slide and observed under BX41 fluorescence microscope.
2.5. Electron microscopy
It was performed as described previously (Chandra et al., 2018a). Briefly, cells were plated on 6-well plates and after treatment cells were fixed using a mixture of paraformaldehyde (2%) and glutaraldehyde (2.5%). Following primary fixation, samples were processed in the Electron Microscopy core facility of the University of Illinois at Chicago Research Resources Center. Briefly, to stabilize cell components, samples were treated with 1% osmium tetroxide in phosphate buffer. Next, an increasing concentration of ethanol was used to dehydrate the samples. The samples were passed through propylene oxide, infiltrated and then embedded in a liquid resin. The resin block was then sectioned by ultramicrotomy (50–70 nm thickness) and collected on metal mesh ‘grids’. The grids were then stained with electron dense stains and observed in the TEM (JEOL JEM-1220). Images of 8000× magnification were analyzed for the presence of authophagic vesicles and lysosomes using ImageJ.
2.6. Real-time PCR analysis
Real-time PCR was performed using SYBR Green-based detection method in the ABI-Prism7700 sequence detection system (Applied Biosystems). It was performed as described in previous studies (Corbett et al., 2012; Ghosh et al., 2012; Khasnavis and Pahan, 2012). The mRNA expression of the target gene was normalized with GAPDH expression and expressed as fold change with respect to the untreated control. Data were processed using the ABI Sequence Detection System 1.6 software.
2.7. Immunoblotting
Western blotting was performed as described in previous studies (Corbett et al., 2015; Ghosh et al., 2012). Briefly, for in-vitro experiments, cells were scraped in PBS, collected in a microfuge tubes and spun down at into a pellet. Next, cells were lysed in RIPA buffer containing protease phosphatase inhibitors. The supernatant was used to measure the protein concentration using the Bradford method (Bio-Rad). For hippocampal tissue, the weight of the tissue was measured and it was dissolved in CHAPS buffer followed by a spin of 30mins at 15000 rpm. 15 to 30 μg of total protein was mixed with SDS sample buffer, boiled for 5 min. The samples were electrophoresed on custom-made SDS-Polyacrylamide gels (10% or 15%) using the Tris/Glycine/SDS or the MES running buffer followed by transfer onto nitrocellulose membrane (Bio-Rad) using semi-dry transfer apparatus (Thermo-Pierce). The membrane was blocked using Odyssey blocking buffer for 1 h and incubated with primary antibody overnight under shaking conditions at 4 °C. The following day, the membrane was thoroughly washed in PBST (PBS+ Tween 20) and incubated in IR-dye conjugated secondary antibody at room temperature for 1 h. Following three 10 min washes in PBSTT, membranes were visualized in the Odyssey® Infrared Imaging System (Li-COR, Lincoln, NE).
2.8. Densitometric analysis
Densitometric quantification of the immunoblots was performed using ImageJ (NIH, Bethesda, MD). Target bands were normalized using their respective β-actin loading controls. Data are representative of the average fold change relative to the untreated control for three independent experiments.
2.9. Immunofluorescence analysis
It was conducted as described in previous studies (Brahmachari et al., 2009; Ghosh et al., 2015; Khasnavis and Pahan, 2012). Briefly, cells were seeded onto square coverslips in 6-well plates. Following treatment, cells were fixed using chilled methanol and washed in PBS. Cells were blocked in 3% BSA (Bovine serum albumin) in PBSTT (PBS + TritonX-100 + Tween-20) for 30 mins followed by incubation in primary antibody dissolved in 1% BSA in PBSTT. Next, cells are washed three times in PBSTT followed by incubation in Alexa-fluor secondary antibody (dilution 1:10000) (Jackson ImmunoResearch). Followed by three washes in PBSTT, DAPI was used for staining the nuclei. The coverslips were mounted on glass slides and visualized under BX41 fluorescence microscope.
2.10. Lysosomal enzyme (TPP1, Cathepsin B,D) assays
It was performed as described earlier (Chandra et al., 2018b). Briefly, treated astrocytes or cortical tissue were homogenized and the supernatant was subjected to enzymatic assay. For TPP1 assay, following homogenization in a buffer containing .15 M NaCl and TritonX-100, the supernatant was incubated with the substrate Ala-Ala-Phe 7-amido-4-methylcoumarin (200uM) (Sigma) at pH 4. The reaction plate was measured in Victor X2 microplate reader (Parkin Elmer) at excitation/emission of 360 nm/460 nm. For cathepsinB and D assays, following lysis in homogenization buffer (pH 5.5; 2.5 mM EDTA, TritonX-100, 2.5 mM DTT), the supernatant was collected. For Cathepsin B assay, the supernatant was incubated with substrate Z-Arg-Arg-7-amido-4-methylcoumarin hydrochloride (100uM) at pH 6 followed by measurement at excitation/emission of 355/460 nm. For cathepsin D, the supernatant was incubated with the substrate 7-Methoxycoumarin-4-Acetyl-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-D-Arg-amide (10uM) (Enzo lifesciences) at pH 4 followed by measurement at excitation/emission of 320/420 nm. Data is representative of relative fluorescence unit (RFU) fold change of the treated samples with respect to the untreated control.
2.11. Construction of mouse Tfeb promoter-driven reporter construct
It was conducted as described earlier (Ghosh et al., 2015).
2.12. Cloning of Tfeb promoter and site-directed mutagenesis
It was conducted as described earlier (Ghosh et al., 2015).
2.13. Assay of Tfeb promoter-driven reporter activity
As described earlier (Chandra et al., 2018b; Ghosh et al., 2015), cells plated in 12-well plates (70–80% confluent) were transfected with pTFEB(WT)-Luc or pTFEB(Mu)-Luc (0.25 μg) using Lipofectamine Plus (Life Technologies, Grand Island, NY) for 24 h. Next, cells were treated under serum free conditions with different doses of cinnamic acid for 6 h. Next, the cell extracts were subjected to analysis for firefly luciferase activities using the Luciferase Assay System kit (Promega) in TD-20/20 Luminometer (Turner Designs).
2.14. Assay of transcriptional activities
Cells plated in 12-well plates (70–80% confluent) were co-transfected with 0.25 μg of PPRE-Luc (an PPAR-dependent reporter construct) and 12.5 ng of pRL-TK using Lipofectamine Plus as described earlier (Chandra et al., 2018b; Ghosh et al., 2015). Following 24 h of transfection, cells were treated with different doses of cinnamic acid for 4 h followed by measuring luciferase activities.
2.15. In silico structural analyses of PPARα complexed with cinnamic acid
The 3D structure of trans cinnamic acid was achieved from Zinc database with ID number 16051516. On the other hand, the 3D structure of mouse PPARα protein was derived with the help of homology modeling strategy considering human PPARα (PDB ID: 3VI8.pdb) as a template. Homology modeling was performed in Swiss-model online tool and the most stable structure was identified based on its nearest match with human PPARα. The protein-ligand interaction of PPARα and cinnamic acid was analyzed with Swiss-dock rigid body docking tool. Among 50 different predicted structures, the most stable structure was displayed at its 5A0 resolution. The most stable structure with Rank #1 was adjudged on the basis of solvation energy, Vander Waal energy and total free energy as shown in figure legend. Visualization was performed in Chimera software.
2.16. Thermal shift assay
Thermal shift assay was conducted using SYBR green real-time melting strategy using the thermal shift dye kit (Life Technologies) in Applied Biosystems 7500 standard real time thermal cycler. As described earlier (Roy et al., 2015; Roy et al., 2016), each reaction contained purified protein (0.5 μg to 1 μg), 18 μl thermal shift buffer and 1–2 μl of dye and was performed in dark. The 96-well PCR plate was loaded in the thermal cycler for the following two-stage program ([25 °C for 2 min] 1 cycle; [27 °C for 15 s, 26 °C for 1 min] 70 cycles; auto increment 1 °C for both stages). The filter was set at ROX with no quencher filter and no passive filter.
2.17. TR-FRET analysis
As described previously (Roy et al., 2015; Roy et al., 2016), TR-FRET was conducted using Lanthascreen TR-FRET PPAR-alpha coactivator assay kit. Trans-cinnamic acid was incubated with GST-tagged recombinant PPARα LBD, Terbium (Tb)-tagged anti GST antibody, and fluorescein (FL)-tagged PGC-1α according to the manufacturer’s protocol. The entire reaction was performed in corning 384-well plate by an automated robotic injector. The plate was centrifuged followed by incubation in dark for 30 min and analysis in molecular devices analyst equipped with dichroic mirror. The excitation and emission were fixed at 340 nm and 540 nm, respectively.
2.18. Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assay was performed as described before (Chandra et al., 2018b; Ghosh et al., 2015). Briefly, cultured primary astrocytes were treated under serum free conditions using 100 and 200uM of cinnamic acid for 2 h. Next, the cells were fixed with formaldehyde, pelleted down and lysed in IP buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, Nonidet P-40 (0.5% v/v), Triton X-100 (1.0% v/v)) and sonicated. The sheared chromatin was incubated with the PPARα, PPARβ, PPARγ, RNA polymerase, CBP, P300 and IgG antibodies overnight under shaking conditions at 4 °C. Next day, the samples were incubated with protein G-agarose for 2 h at 4 °C followed by wash with cold IP buffer, addition of Chelex (10 g/ 100 ml ofH2O) and boiled for 10 min. Next, the beads were incubated with Proteinase K and boiled and the supernatant was collected. This elute was used for conducting semi-quantitative and real-time PCR. The PPRE-containing fragment of the mouse Tfeb promoter was amplified using the following primers: sense: 5′-GAA CAT TCC AGG TGG AGG CA-3′, antisense: 5’-CCC CCA ACA CAT GCT TCT CT-3′. For real-time PCR, data were normalized with the input and the fold change with respect to the untreated control was calculated.
2.19. Animals and cinnamic acid treatment
Animal maintenance and experiments were conducted in accordance with National Institutes of Health guidelines and were approved by the Rush University Medical Center Institutional Animal Care and Use Committee. B6SJL-Tg(APPSwFlLon,PSEN1*M146 L*L286 V) 6799Vas/J transgenic (5×FAD) mice were obtained from Jackson Laboratories (Bar Harbor, ME). 5×FAD mice null for PPARα (5×/Ppara−/−) were generated and maintained (Corbett et al., 2015). Six months old male and female 5×FAD or 5×/Ppara−/− mice were orally administered with trans-cinnamic acid (100 mg/kg body weight/ day) or vehicle (0.5% Methylcellulose) via gavage for 30 days (for non-transgenic, 5×FAD transgenic (Tg), Tg + cinnamic acid and Tg + vehicle, n = 8 in two separate batches of experiments, for 5×/Ppara−/− + vehicle and 5X/Ppara−/− + cinnamic acid, n = 5). Non-transgenic mice from the same background were used as a control. Following treatment, behavioral tests were performed following which mice were sacrificed. Mice were injected with ketamine-xylazine injectables and anesthetized mice were perfused using PBS for 5 min. Hemi-brain was kept in 4% Paraformaldehyde and processed for immunohistochemistry whereas the hippocampus and cortex was dissected from the other half of the brain for biochemical analysis.
2.20. Immunohistochemistry
For immunohistochemistry, hemi brains were incubated with 4% paraformaldehyde (w/v) followed by incubation in 30% sucrose overnight at 4 °C. Next, brains were thoroughly washed in PBS and embedded in O.C.T (Tissue Tech) at −80 °C, and prepared for cryosectioning. Prior to staining, 40uM free floating hippocampal sections were washed thoroughly in PBS. The sections were blocked using 2% BSA in PBSTT (PBS + Triton X-100 + Tween-20) for 1 h. Next, the sections were incubated with primary antibody in 1% PBSTT at 4 °C overnight. The following day, sections were washed in PBSTT and incubated with 488 or 647- conjugated secondary antibody for 1 h at room temperature. Following washes in PBSTT, the sections were mounted on glass sides. The samples were visualized under Olympus BX41 fluorescence microscope.
2.21. Barnes maze
Barnes maze test was performed as described previously (Corbett et al., 2013; Modi et al., 2015; Roy et al., 2013) for assessing the spatial memory of the mice. Briefly, mice were trained for two days on a 20-hole Barnes maze where only one tunnel contained colored food bait. The mice were food deprived overnight prior to the training and test days. During training, mouse was placed in the middle of the maze in a cylindrical 10 cm high start chamber. After 10s, the cylinder was removed and the mouse was allowed to freely move and explore the maze and find the food baited tunnel. The maze was well lit with high wattage light to generate sufficient light and heat to motivate the mouse to find and escape into the tunnel. The tunnel was placed under the same hole and was stable within the spatial environment. Following two days training, mice were given rest for one day and tested on the maze on day 4. On the test day, food-deprived mouse was placed in the middle of the maze and their performance was recorded using the Noldus system. Memory of the mouse was analyzed by latency to the goal box (duration before all four paws were on the escape box floor) and errors (incorrect responses before all four paws were on the escape box floor). The maze was cleaned thoroughly after each trial.
2.22. T maze
T maze for assessing the memory of the mice was performed as described previously by us (Modi et al., 2014; Roy et al., 2013). Briefly, mice were trained and habituated on the T maze for two days and were food deprived prior to the training so that they reach for the food reward at least five times in a 10 min training period. During training, mice were placed at the start point for 30 s and allowed to make a forced entry into the right arm which was baited with colour food chips. Upon entering the right arm, mice were allowed to stay and habituate for 30 to 45 s. During training, the left arm of the maze was blocked. On day 3, the left arm was unblocked and food-deprived mouse was placed at the start point allowing it to either enter right arm (positive turn) which had the food bait or left arm (negative turn). The food reward side was always associated with a visual cue. The T maze was thoroughly cleaned after each trial.
2.23. Open field test
The open field test was conducted for analyzing the locomotor activity of mice. Mice were placed in the center of a square wooden open field arena (40×40cm, 30 cm high walls) and allowed to explore freely for 5 min. The movements of the mice were recorded using a camera linked to the Noldus system and EthoVisionXT software. Several parameters including velocity, total distance moved and movement cumulative duration were analyzed to assess the general locomotor activity of the mice. The open field was cleaned thoroughly after each trial.
2.24. Statistical analysis
Statistical analyses were performed in GraphPad Prism. Data sets were analyzed by one-way ANOVA followed by Tukey’s post hoc test or student’s t-test. Data represented as mean ± SD or mean ± SEM. A level of p < .05 was considered statistically significant. * denotes a p value < .05, ** denotes p < .01 and *** denotes p < .001.
3. Results
3.1. Cinnamic acid stimulates lysosomal biogenesis in mouse primary brain cells
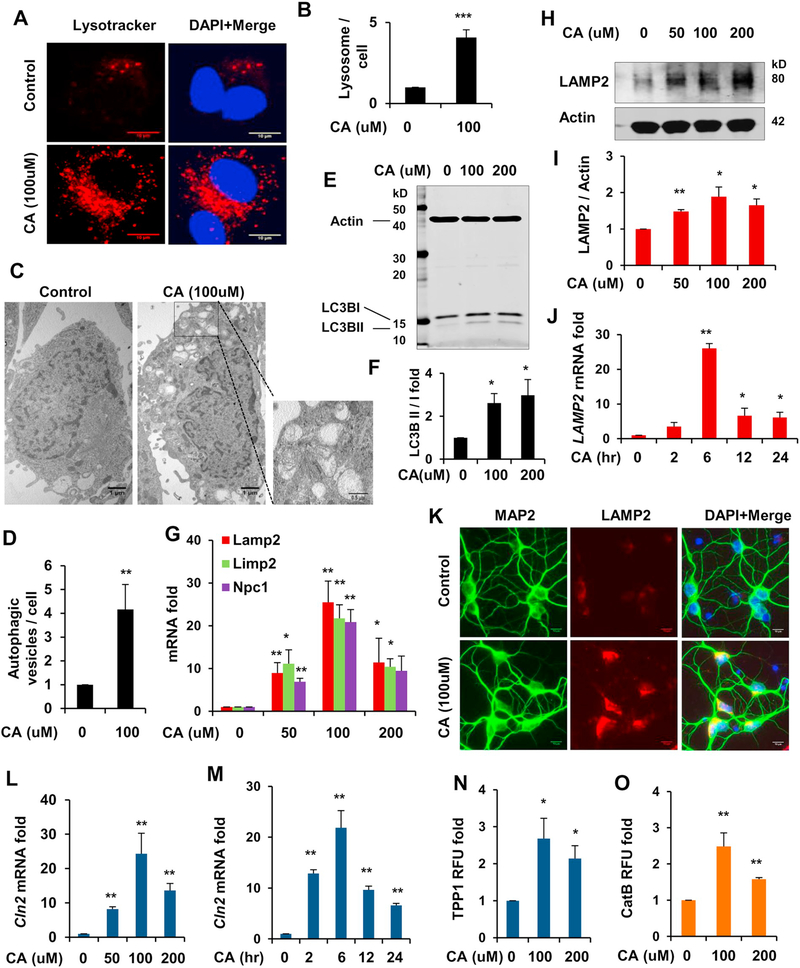
To begin with, we tested whether cinnamic acid could stimulate lysosomal biogenesis in mouse primary brain cells. First, we examined the total lysosome content of the cell by selectively labeling the acidic lysosomal organelles using Lysotracker Red. Cultured astrocytes were treated under serum free conditions with cinnamic acid for 24 h followed by staining with Lysotracker. Cinnamic acid treatment resulted in marked increase in the lysosomal abundance in astrocytes (Fig. 1A, B). We further explored the role of cinnamic acid in autophagy by electron microscopy and observed a profound upregulation in the number of autophagic vesicles of different stages (Fig. 1C, D). To confirm this, we analyzed the expression of autophagy marker LC3B and observed that cinnamic acid (100 and 200 μM) remarkably enhanced the autophagic flux (LC3BII/LC3BI) in primary astrocytes (Fig. 1E, F). Next, we treated the astrocytes with different doses of cinnamic acid (50, 100 and 200 μM) and checked the expression of various lysosomal markers by quantitative real time PCR. Data showed that cinnamic acid upregulated the mRNA levels of the genes encoding lysosomal membrane proteins (LAMP2, LIMP2, NPC1) in a dose dependent manner (Fig. 1G) with 100 μM having the highest effect. This was accompanied by a similar pattern of increase in the LAMP2 protein levels demonstrated by western blot analysis (Fig. 1H, I). We next performed a time point analysis using 100 μM cinnamic acid in cultured astrocytes and observed that cinnamic acid was able to upregulate the LAMP2 mRNA in a time dependent fashion (Fig. 1J). We further checked the expression of LAMP2 in cultured primary cortical neurons by immunohistochemistry. LAMP2 expression was markedly enhanced following cinnamic acid treatment in cortical neurons (Fig. 1K). We further explored the effect of cinnamic acid on expression and activity of lysosomal enzymes tripeptidyl-peptidase 1 (TPP1), a lysosomal protease that destabilizes β-sheet region of amyloid-β and cathepsin B, a cysteine protease that degrades amyloid-β and hence is beneficial for AD. Our data showed that cinnamic acid significantly upregulated the mRNA expression of Cln2, the gene encoding TPP1, in dose dependent and time course analysis (Fig. 1L, M). Induction of TPP1 by cinnamic acid was almost similar to that by gemfibrozil, a known activator of PPARα (Ghosh et al., 2012). Interestingly, the activity of both TPP1 and Cathepsin B were augmented with different doses of cinnamic acid treatment indicating enhanced lysosomal functionality (Fig. 1N, O). Together, our results reveal that cinnamic acid induces lysosomal biogenesis in mouse primary brain cells.
Fig. 1.
Cinnamic acid stimulates lysosomal biogenesis in mouse primary brain cells. (A-D) Cultured mouse primary astrocytes were treated under serum free condition with 100uM cinnamic acid for 24 h followed by (A) monitoring the lysosomal abundance by LysoTracker Red staining. DAPI was used for staining the nuclei. Scale bar 10um; (B) Quantification of lysotrcaker puncta per cell. (C) Transmission electron microscopy of primary astrocytes treated with 100uM cinnamic acid for 24 h for monitoring autophagy; (D) Quantification of the number of autophagic vesicles per cell; (E-F) Primary astrocytes were treated with 100 and 200uM of cinnamic acid for 24 h followed by (E) western blot analysis for autophagy marker LC3B and (F) analysis of the autophagic flux (LC3BII/LC3BI); (G-I) Primary astrocytes were treated with different doses (50, 100 and 200 uM) of cinnamic acid (G) for 8 h followed by mRNA expression analysis of lysosomal membrane genes (lamp2, limp2, npc1) by real time PCR; (H) for 24 h followed by checking the protein levels of LAMP2 by immunoblot and (I) densitometric analysis relative to β-actin. (J) Primary astrocytes were treated with 100uM cinnamic acid for different time points (2, 6, 12, 24 h) followed by monitoring the mRNA expression of lamp2; (K) Cultured primary cortical neurons were treated with 100uM cinnamic acid for 24 h and LAMP2 expression was monitored by immunocytochemistry. Scale bar 10um; (L-M) Primary astrocytes were treated with (L) for 8 h with 50, 100, 200uM cinnamic acid and (M) 100uM cinnamic acid for 2, 6, 12, 24 h followed by analyzing the mRNA expression of cln2 by real time PCR; (NeO) Primary astrocytes were treated with 100uM cinnamic acid for 24 h followed by monitoring the activity of the lysosomal enzymes (N) TPP1, coded by the gene cln2 and (O) Cathepsin B. All data represents fold change mean ± SD with respect to the untreated control. Statistical analysis was performed by student’s paired t-test. * p < .05; ** p < .01; *** p < .001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Cinnamic acid activates the nuclear receptor PPARα
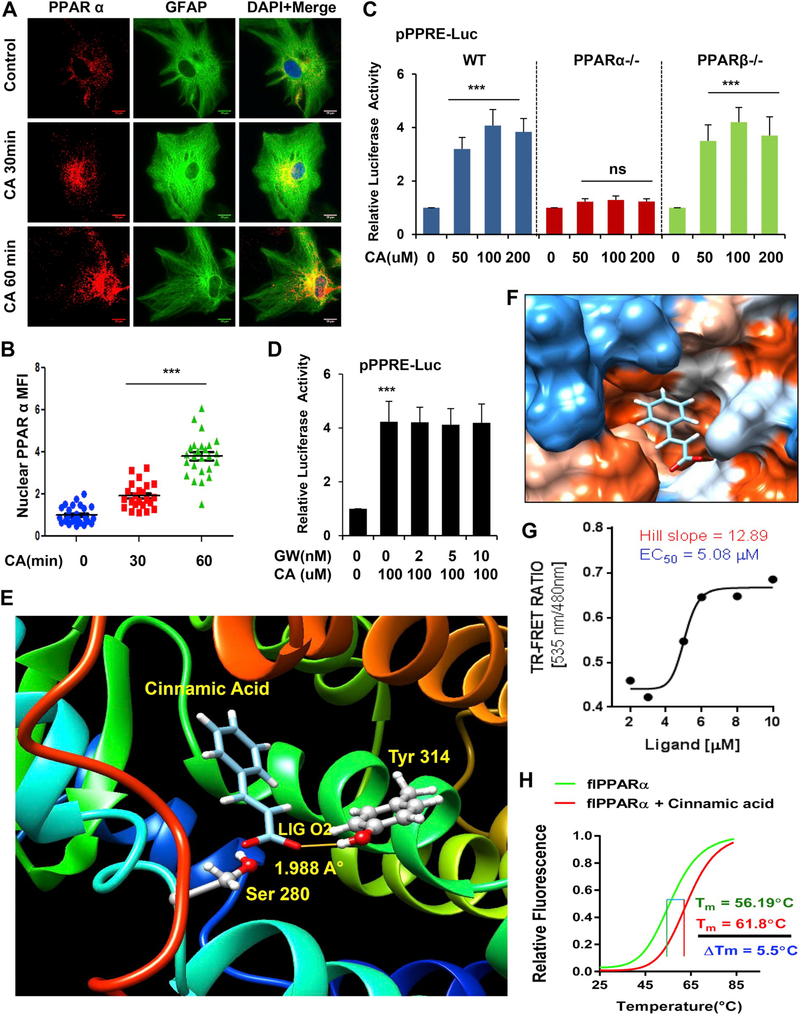
After establishing that cinnamic acid induces lysosomal biogenesis, we intended to explore the upstream mechanisms. TFEB is known as the central regulator of lysosomal biogenesis. Interestingly, the Tfeb promoter harbors a PPRE (peroxisomal proliferator-response element), binding site for the transcription factors PPAR (Peroxisome proliferator-activated receptor) 480 base pair upstream of transcription start site, which led us to investigate the involvement of the PPARs in the process. Importantly, previously activation of PPARα by its agonist gemfibrozil has been shown to transcriptionally upregulate Tfeb and induce lysosomal biogenesis (Ghosh et al., 2015). Hence, we tested our hypothesis that cinnamic acid activates the transcription factor PPARα to stimulate TFEB. To establish this, we first checked the effect of cinnamic acid on nuclear translocation of PPARα in WT primary astrocytes. Data showed that 30 mins and 60 mins treatment with 100 μM cinnamic acid markedly increases the nuclear and perinuclear localization of PPARα (Fig. 2A, B). We further analyzed the activation status of different PPARs following cinnamic acid treatment. Cultured primary astrocytes isolated from WT, PPARα−/− and PPARβ−/− mice were transfected with the pPPRE-luciferase construct for 24 h prior to cinnamic acid treatment (50, 100, 200 μM) for 6 h. Cinnamic acid markedly induced PPRE-driven luciferase activity in WT, PPARβ−/−, but not in PPARα−/− astrocytes, suggesting that cinnamic acid specifically activates PPARα and not PPARβ (Fig. 2C). To examine the role of PPARγ, we treated primary WT astrocytes with PPARγ antagonist GW9662 (2, 5, 10 nM) followed by treatment with 100uM cinnamic acid and observed that under PPARγ-inhibited conditions, cinnamic acid was still able to induce PPRE luciferase activity confirming that cinnamic acid does not require PPARγ (Fig. 2D). Together, our data indicate that cinnamic acid activates PPARα.
Fig. 2.
Cinnamic acid binds to and activates PPARα. (A-D) Cinnamic acid activates nuclear receptor PPARα. (A) Primary astrocytes were treated with 100uM of cinnamic acid for 30, 60mins and the nuclear translocation of PPARα was monitored by immunocytochemistry followed by (B) mean fluorescence intensity analysis of the nuclear level of PPARα. Scale bar 10um. (C) Primary astrocytes from WT, PPARα−/−, PPARβ−/− mice were transfected with pPPRE-luciferase construct for 24 h prior to treatment with different doses (50, 100, 200uM) of cinnamic acid for 6 h and were subjected to luciferase assay. (D) WT astrocytes were transfected with pPPRE-luciferase construct for 24 h, pretreated with PPARγ antagonist GW9662 (2, 5, 10 nM) followed by treatment with 100uM cinnamic acid for 8 h and monitoring luciferase activity. All data represents fold change mean ± SD with respect to the untreated control. Statistical analysis was performed by One way ANOVA followed by Tukey’s multiple comparison test. * p < .05; ** p < .01; *** p < .001. (E-H) Characterization of interaction of cinnamic acid with PPARα. (E) A rigidbody in silico docked pose of the PPARα LBD with cinnamic acid. The interaction was evaluated with free energy (ΔG) = −5.89 kcal/mol, desolvation energy (Esol) = −1832.9 kcal/mol, and total energy (Etot) = −1612.55 kcal/mol. (F) Electrostatic potential surface shows the distribution of charge of PPARα LBD around the backbone of cinnamic acid. Red = a negatively charged surface; blue = a positively charged surface; white = a neutral surface. (G) TRFRET analysis confirming the interaction between cinnamic acid and PPARα. The curve was plotted as 520-nm/490-nm ratio of response with increasing doses of ligand. Curve-fitting was done using GraphPad Prism software with the eq. Y = bottom + (XHill slope) × (top − bottom)/ (XHill slope + EC50Hill slope). (H) Thermal shift assay of flPPARα analyzed with 5 μM of cinnamic acid. The melting of PPARα was monitored using a SYBR Green real-time melting strategy. Results were analyzed and confirmed after three independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Cinnamic acid is a potent ligand of PPARα
Next, we wanted to study the mechanisms how cinnamic acid activates PPARα and whether cinnamic acid could serve as a ligand of PPARα. Crystal structure of mouse PPARα is unavailable. Therefore, first we constructed three-dimensional structure of mouse PPARα with the help of online homology modeling tool of Swiss-model server considering human PPARα (PDB ID: 1KKQ) as a template. After that, we applied Swissdock, a rigid body protein-ligand docking tool, to explore the interaction between cinnamic acid and ligand binding domain (LBD) of PPARα at a molecular level. Based on electrostatic (Etot) and desolvation (Esol) energies, fifty different structures were predicted. The most stable- docked structure was displayed with Chimera software. According to this analysis, we found that cinnamic acid docked in the interface of LBD of PPARα as shown in (Fig. 2E, F). The detailed view of that docking clearly indicated that cinnamic acid formed a strong hydrogen bond with Tyr314 residue of PPARα LBD at a distance of 1.988 A° (Fig. 2E). However, our in silico study needs to be supported with experimental evidences. Therefore, to validate the interaction between PPARα and cinnamic acid, we performed a time-resolved fluorescence resonance energy transfer or FRET assay as described earlier (Roy et al., 2015; Roy et al., 2016). Accordingly, our FRET analyses (Fig. 2G) confirmed that cinnamic acid indeed displayed a strong interaction with PPARα. The binding curve resulted EC50 value of 5.08 μM with Hill slope of 12.89. To further confirm, we performed thermal shift melting analyses of PPARα protein with 5 μM of cinnamic acid. Briefly, full length PPARα protein (flPPARα) was synthesized from HEK293FT cells transduced with lentiviral full-length PPARα. After that, its melting profile was monitored with the help of SYBR green reaction strategy at a range of 27 degree to 94 degree Celsius. The typical sigmoidal melting curve with melting temperature (Tm) of 56.19 °C clearly indicated that our in-house recombinant PPARα protein is conformationally stable. Our melting assay revealed that 5 μM of cinnamic acid strongly shifted the melting curve of PPARα to 61.8 °C with a difference of 5.95 °C (< 5 °C) (Fig. 2H), suggesting that cinnamic acid indeed serves as a ligand of PPARα.
3.4. Activation of PPARα by cinnamic acid transcriptionally regulates Tfeb
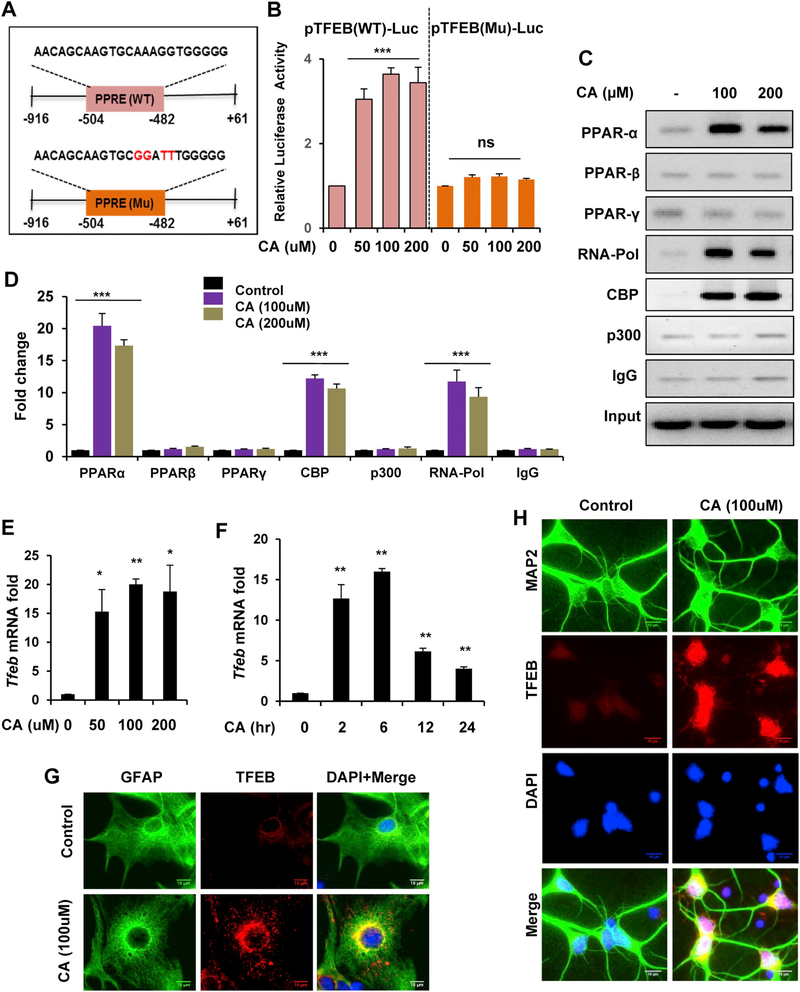
Next, we analyzed the effect of cinnamic acid-mediated PPARα activation on the transcription of Tfeb. We transfected WT astrocytes with either pTFEB(WT), a construct containing the PPRE site on the Tfeb promoter or pTFEB(Mu), a construct with mutated PPRE sequence (Fig. 3A). Different doses of cinnamic acid treatment (50, 100, 200 μM) showed robust induction of Wild type TFEB-driven luciferase activity, however a marked reduction was observed in the mutated TFEB-driven luciferase activity suggesting that cinnamic acid requires PPARα to transcriptionally upregulate Tfeb (Fig. 3B). When activated, PPARα is known to translocate into the nucleus and assemble a transcriptional complex that promotes transcription of the target gene. Therefore, we checked the recruitment of different PPARs and basal transcription apparatus to the Tfeb promoter following cinnamic acid treatment by performing Chromatin immunoprecipitation. Semi-quantitative PCR and real-time PCR data revealed that cinnamic acid treatment in the WT astrocytes resulted in specific recruitment of PPARα, RNA polymerase and coactivator CBP to the Tfeb promoter (Fig. 3C, D) confirming that PPARα forms a transcriptional complex to upregulate Tfeb expression. Together, our data demonstrate that cinnamic acid transcriptionally upregulates Tfeb expression via PPARα.
Fig. 3.
Upregulation of TFEB by cinnamic acid. (A-D) Cinnamic acid transcriptionally upregulate Tfeb. (A) Design of pTFEB(WT)-luciferase or pTFEB(Mutant)-luciferase construct. (B) WT astrocytes were transfected with pTFEB(WT)-luciferase or pTFEB(Mutant)-luciferase for 24 h followed by cinnamic acid treatment (50, 100, 200uM) for 6 h and subsequently subjected to luciferase assay. (C-D) WT astrocytes were treated with 100 and 200uM of cinnamic acid for 2 h followed by chromatin immunoprecipitation to analyze the recruitment of PPARα, β, γ, RNA polymerase, CBP and p300 to the Tfeb promoter by (C) semi-quantitative RT PCR and (D) real time PCR. (E-H) Cinnamic acid increases TFEB expression in primary brain cells. Mouse primary astrocytes were treated with (E) different doses (50, 100, 200uM) of cinnamic acid for 8 h, (F) 100uM cinnamic acid for 2, 6, 12, 24 h followed by Tfeb mRNA expression analysis by real time PCR; (G-H) TFEB expression level was monitored in (G) primary astrocyte and (H) cultured cortical neurons treated with 100uM cinnamic acid for 24 h. Scale bar 10um. Data represents fold change mean ± SD with relative to the untreated control. Student’s paired t-test was used for statistical analysis. * p < .05; ** p < .01; *** p < .001.
3.5. Cinnamic acid enhances TFEB expression in primary brain cells
We next explored whether cinnamic acid induces the expression of TFEB, which can transcriptionally upregulate various lysosomal genes and facilitate the lysosomal clearance machinery. We treated cultured astrocytes with different doses (50, 100, 200uM) of cinnamic acid and analyzed the mRNA expression by quantitative real time PCR. Cinnamic acid was able to significantly induce TFEB expression and it was found to be dose dependent (Fig. 3E). Time point analysis demonstrated that TFEB could be induced earlier than LAMP2, at 2 h, suggesting that TFEB is upstream of other lysosomal genes and cinnamic acid could induce lysosomal biogenesis via upregulation of TFEB (Fig. 3F). We next checked the expression of TFEB by immunocytochemistry in primary astrocytes and cortical neurons and observed that TFEB expression was markedly increased with cinnamic acid treatment (Fig. 3G, H). Collectively, these results suggest that cinnamic acid stimulates the master regulator TFEB to induce lysosomal biogenesis in primary brain cells.
3.6. Cinnamic acid stimulates lysosomal biogenesis via PPARα
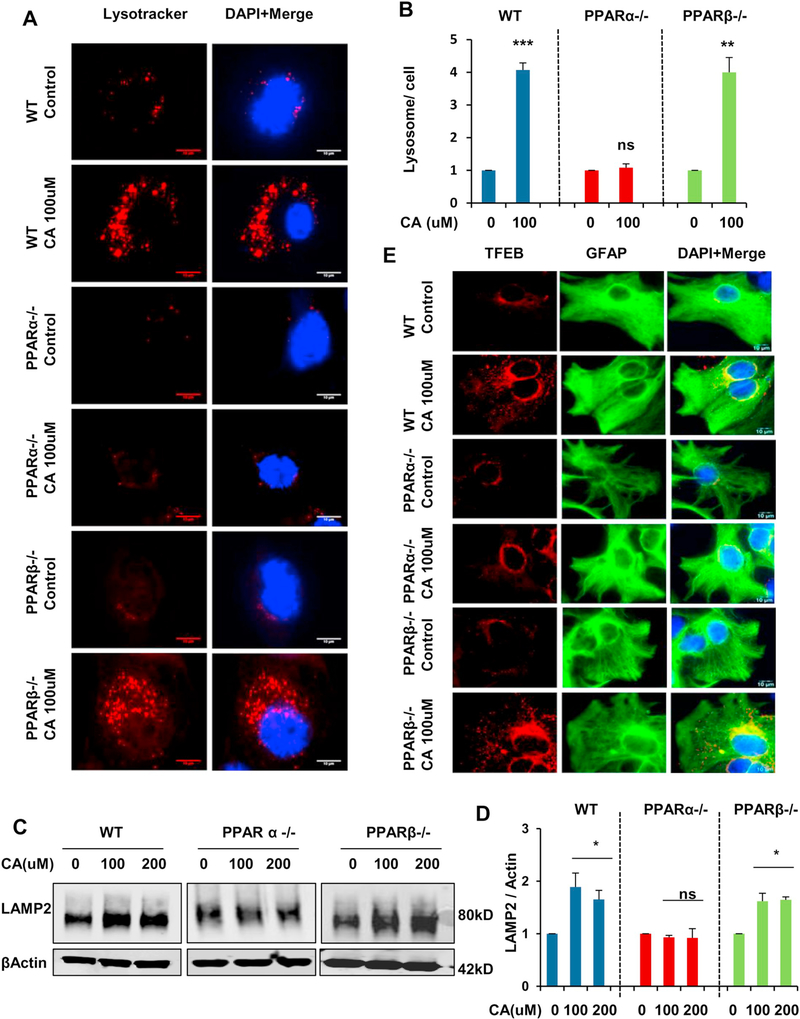
To further ascertain the role of PPARα in cinnamic acid mediated lysosomal biogenesis, we analyzed the lysosome content and different lysosomal markers under WT, Ppara-null and Pparb-null conditions. Primary astrocytes were isolated from WT, Ppara-null and Pparb-null mice and cultured astrocytes were treated under serum free conditions with cinnamic acid for 24 h. We first checked the total lysosome content by staining live cells with LysotrackerRed followed by quantification of the lysosome number per cell. Data showed that cinnamic acid markedly augmented the abundance of acidic lysosomes in WT and Pparbnull, but not in Ppara-null astrocytes (Fig. 4A, B) indicating cinnamic acid requires PPARα for inducing lysosomal biogenesis. This was further confirmed by a similar pattern of result in the expression of lysosomal membrane protein LAMP2. Immunoblot analysis demonstrated that cinnamic acid, at 100 and 200 μM doses, upregulated LAMP2 expression in WT and Pparb-null astrocytes but did not have any such effect in case of Ppara-null astrocytes (Fig. 4C, D). Finally, we analyzed the expression level of TFEB in these cells and observed an enhancement in TFEB expression in WT and Pparb-null astrocytes following treatment with cinnamic acid, however no such effect was observed in Ppara-null astrocytes. Taken together, our results establish a specific involvement of PPARα and indicate that cinnamic acid particularly requires PPARα for stimulating lysosomal biogenesis.
Fig. 4.
Cinnamic acid stimulates lysosomal biogenesis in WT, PPARβ−/− but not in PPARα−/− astrocytes. (A-C) Mouse primary astrocytes isolated from WT, PPARα−/− and PPARβ−/− mice were treated with cinnamic acid (100uM) for 24 h followed by (A) lysotracker staining and (B) quantification of the number of lysosomes per cell for monitoring lysosomal abundance. Scale bar 10um; (C) analysis of LAMP2 protein levels by immunoblot analysis and (D) densitometric analysis of LAMP2 expression; (E) double labeling TFEB and GFAP for monitoring TFEB expression. Scale bar 10um. All data represents fold change mean ± SD with respect to untreated control of the corresponding genotype. Statistical analysis was performed by student’s paired t-test. * p < .05; ** p < .01; *** p < .001.
3.7. Cinnamic acid increases lysosomal markers in vivo in 5XFAD mice
Following in vitro demonstration of stimulation of lysosomal biogenesis by cinnamic acid, we next intended to explore the effect of cinnamic acid on lysosomal markers in vivo in the 5XFAD model of AD. Following oral administration of cinnamic acid for one month, we examined the levels of TFEB, the central coordinator of the lysosomal system, in different regions of the hippocampus, the most affected brain region in AD, and the cortex. Recent studies have highlighted the importance of astrocyte specific lysosomal stimulation in facilitating the clearance leading to attenuation of pathology in AD model (Xiao et al., 2014). Indeed, astrocytes can uptake Aβ via cell surface receptors and target it for efficacious lysosomal degradation (Wyss-Coray et al., 2003). Hence, in order to analyze the astrocytic lysosomal biogenesis following cinnamic acid treatment, we performed co-labeling of free floating hippocampal sections with TFEB and GFAP, a marker of astrocytes. We observed that cinnamic acid treated mice had distinctly enhanced TFEB levels in the CA1 and Dentate gyrus (DG) regions of the hippocampus (Fig. S1 A-D) compared to the untreated transgenic (Tg) mice. Parallel to this, a marked upregulation in the TFEB expression in the cortex of the cinnamic acid treated mice was observed (Fig. S1 E, F). We also analyzed the activity of lysosomal enzyme cathepsin B, an Aβ-degrading cysteine protease, following cinnamic acid treatment. We observed that cinnamic acid treatment was able to significantly upregulate cathepsin B activity in the cortex of 5×FAD mice (Fig. S1 G). Next, we examined the activity of another lysosomal protease, TPP1, which destabilizes Aβ. We found that following cinnamic acid treatment, the activity of TPP1 was significantly upregulated in the cortex of these mice (Fig. S1 H). Together, these results establish that cinnamic acid induces the expression of TFEB and stimulates lysosomal proteolysis in 5×FAD mice.
3.8. Oral administration of cinnamic acid attenuates amyloid plaque load in 5×FAD mice
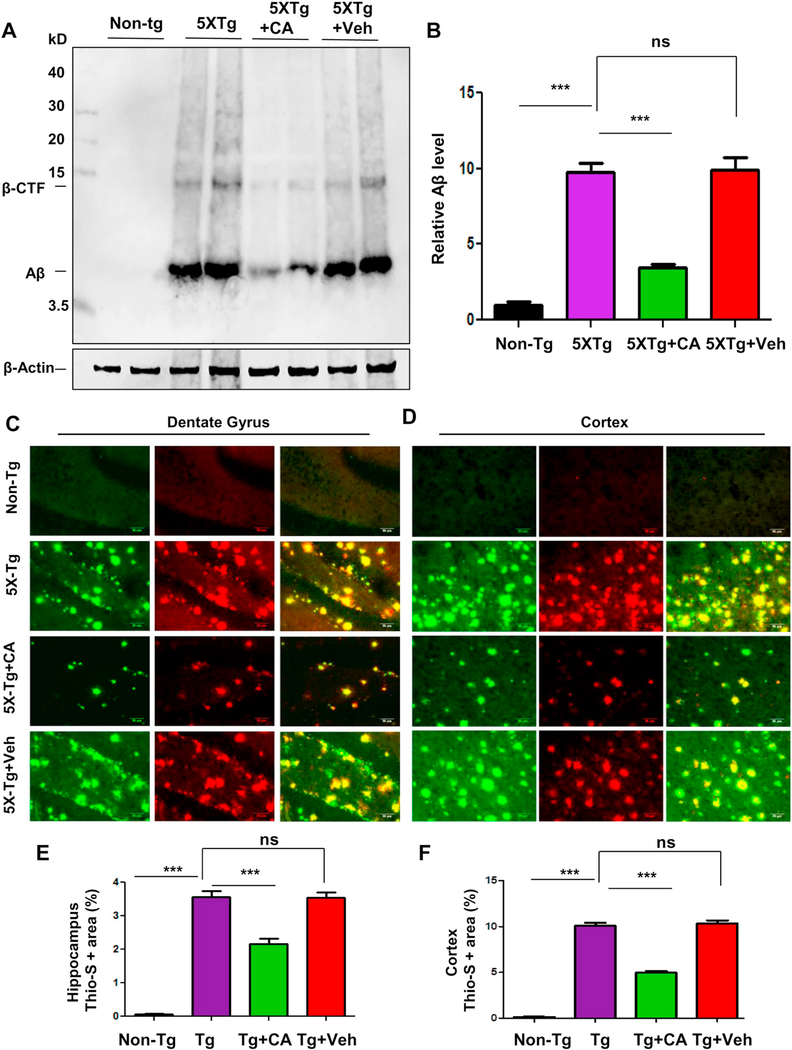
Next we evaluated the efficacy of cinnamic acid mediated lysosomal induction on the amyloid plaque pathology in the 5×FAD mice. The 5×FAD mice harbors five familial mutations linked to AD in APP and PS1 and develop amyloid deposition starting at two months of age (Oakley et al., 2006). Growing evidence have documented a beneficial role of exogenous TFEB induction in promoting lysosomal degradation of Aβ and APP leading to attenuation of pathology (Xiao et al., 2014; Xiao et al., 2015) suggesting TFEB stimulation is a potential therapeutic strategy in AD. We treated six month old 5×FAD mice with cinnamic acid (100 mg/kg/day) or vehicle (0.5% methylcellulose) followed by monitoring the amyloid beta levels in the hippocampus and cortex. As expected, immunoblot analysis of hippocampal homogenate using the 6E10 Aβ monoclonal antibody revealed a robust induction of Aβ and the β-CTF fragment in the transgenic (Tg) mice compared to the non-transgenic (Non-Tg). Interestingly, cinnamic acid was able to remarkably reduce the levels of Aβ along with a parallel decrease in βCTF, whereas vehicle treatment did not show any such effect (Fig. 5A, B). We further performed diaminobenzidine staining of hippocampal sections using 6E10 antibody and observed a marked decrease in the Aβ accumulation in the hippocampus of the cinnamic acid treated group compared to the Tg and vehicle groups (fig. S2A, B). Next, we analyzed the plaque deposition in these mice by colabeling hippocampal sections with thioflavin-S, a dye that detects the β-sheet region of amyloid plaques and Aβ 6E10 monoclonal antibody. Similar to our previous results, cinnamic acid treatment significantly reduced the thio-S positive Aβ plaques in CA1, CA3, dentate gyrus regions of the hippocampus (fig. S3A,B; Fig. 5C,E) as well as cortex (Fig. 5D, F). Collectively, our results demonstrate that cinnamic acid lowers the amyloid plaque burden in the brain of 5×FAD mice.
Fig. 5.
Administration of cinnamic acid reduces amyloid beta plaque deposition in the 5×FAD model of AD. Six months old 5×FAD mice (n = 8/group) were treated via oral gavage with cinnamic acid (100 mg/kg/day) or vehicle (0.5% methylcellulose) daily for one month following which they were sacrificed and (A-B) the cerebral amyloid-beta levels were analyzed by (A) immunoblot analysis of hippocampal homogenates using Aβ 6E10 monoclonal antibody and (B) densitometric analysis of relative Aβ (Aβ/Actin) levels with respect to non-transgenic; (C–F) Cerebral amyloid plaque deposition was monitored by colabeling free floating hippocampal sections with thioflavin-S and Aβ 6E10 antibody. Thio-S and Aβ positive plaques in (C) Dentate gyrus regions of the hippocampus and (D) Cortex are shown. Quantification of the thio-s area fraction (thio-S positive area represented as a percentage of total area) in the hippocampus (E) and cortex (F) was performed using ImageJ (analyze particle feature). All data represents mean ± SEM. All statistical analysis were performed by one way ANOVA followed by Tukey’s multiple comparison test; * p < .05; ** p < .01; *** p < .001.
3.9. Cinnamic acid lowers amyloid plaque burden through PPARα
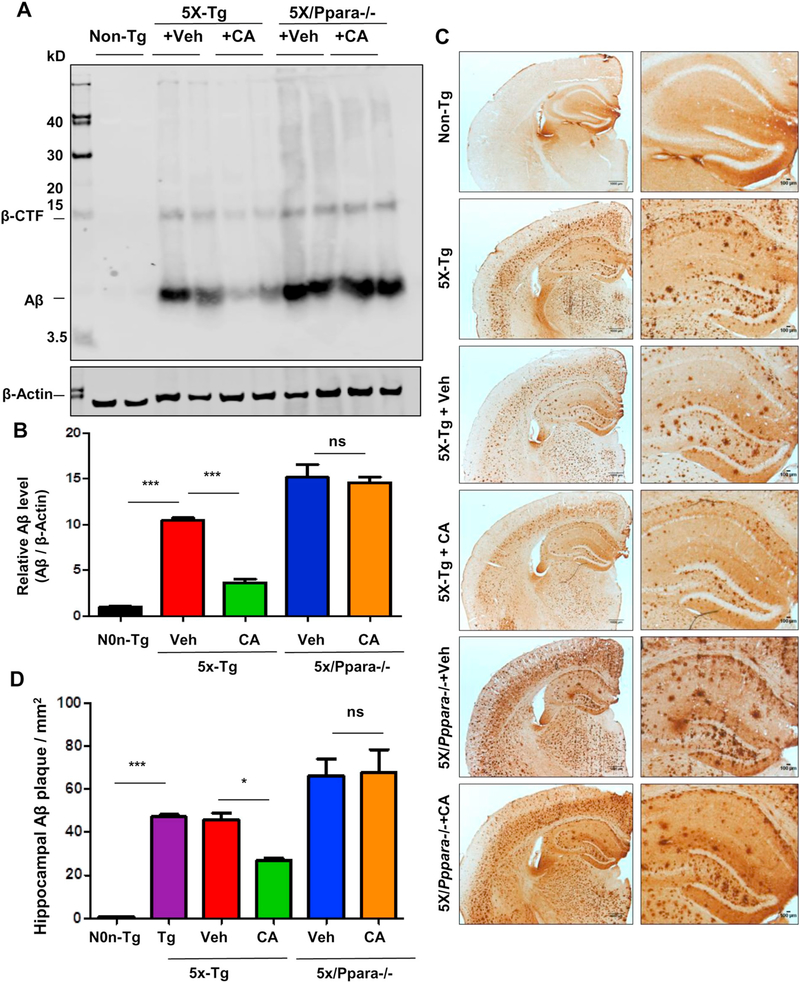
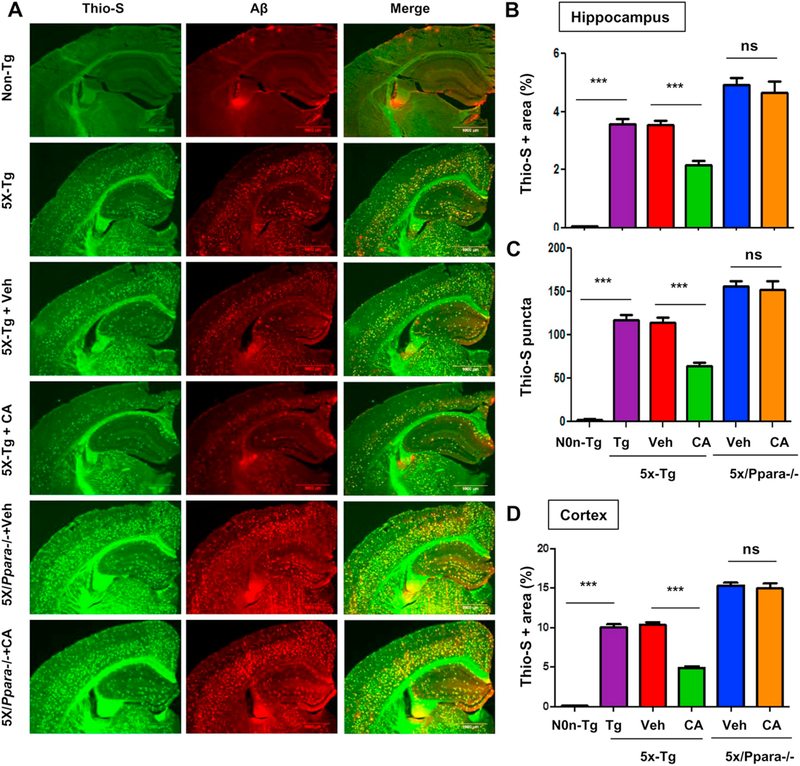
Having demonstrated that cinnamic acid activates PPARα, we examined whether it is the underlying mechanism of cinnamic acid’s amyloid attenuating effects. To explore the role of PPARα, we analyzed the effect of cinnamic acid on amyloid pathology of 5×FAD mice null for PPARα (5×/Ppara−/−). After one month oral administration of cinnamic acid, we first checked the Aβ levels in the hippocampal homogenates of the 5×FAD and 5×/Ppara−/− mice. As expected, cinnamic acid significantly reduced the Aβ levels in the 5×FAD mice compared to the vehicle group. However, cinnamic acid failed to decrease the Aβ levels in the 5×/Ppara−/− mice indicating that cinnamic acid requires PPARα to exhibit its amyloid lowering effects (Fig. 6A, B). It is noteworthy that the vehicle treated 5×/Ppara−/− mice presents significantly more Aβ levels compared to the 5×FAD mice, suggesting that ablation of PPARα contributes to the amyloid pathogenesis. Diaminobenzidine staining using Aβ 6E10 antibody also revealed a similar pattern of results with cinnamic acid markedly lowering the Aβ deposition in 5×FAD mice, but not having such effect in the 5×/Ppara−/− mice (Fig. 6C, D). Colabeling of hippocampal sections with thioflavin-S and Aβ showed that cinnamic acid reduced the thio-S positive amyloid plaques in the hippocampus and cortex of 5XFAD mice but was unable to do so in the 5X/Ppara−/− mice (Fig. 7A-D), further confirming that cinnamic acid requires PPARα for attenuating amyloid plaques in the 5XFAD mice. Taken together, these results suggest that cinnamic acid reduces amyloid plaque deposition in a PPARα dependent manner.
Fig. 6.
Cinnamic acid attenuates amyloid-beta burden via PPARα. Six months old 5×FAD mice (n = 8/group) and 5×FAD mice null for Ppara (5×/Ppara−/−) (n = 5/ group) were treated orally with cinnamic acid (7.5 mg/kg/day) or vehicle (.5% methylcellulose) daily for one month followed by which the amyloid-beta load in the hippocampus was monitored by (A) Immunoblotting of hippocampal homogenate with Aβ 6E10 monoclonal antibody and (B) densitometric analysis of relative Aβ levels (Aβ/Actin) and (C) Diaminobenzidine staining of hippocampal sections followed by (D) quantification of Aβ plaques per mm2 in the hippocampus using ImageJ. All data represents mean ± SEM. One way ANOVA followed by Tukey’s multiple comparison test was used for statistical analysis; * p < .05; ** p < .01; *** p < .001.
Fig. 7.
Cinnamic acid lowers plaque deposition in a PPARα-dependent manner. Six months old 5×FAD mice (n = 8/group) and 5XFAD mice null for Ppara (5X/Ppara−/−) (n = 5/group) were treated orally with cinnamic acid (100 mg/kg/day) or vehicle (0.5% methylcellulose) daily for one month after which (A) the cerebral amyloid plaque burden was monitored by colabeling hippocampal sections with thioflavin-S and Aβ monoclonal antibody 6E10. (B-D) Quantitative analysis of (B) hippocampal thio-S area fraction, (C) hippocampal thio-S positive puncta count and (D) thio-S positive area fraction in the cortex. Quantification was performed using the ‘analyze particle’ feature in ImageJ. All data represents mean ± SEM. One way ANOVA followed by Tukey’s multiple comparison test was used for statistical analysis; * p < .05; ** p < .01; *** p < .001.
3.10. Cinnamic acid improves behavioral deficit in 5XFAD mice via PPARα
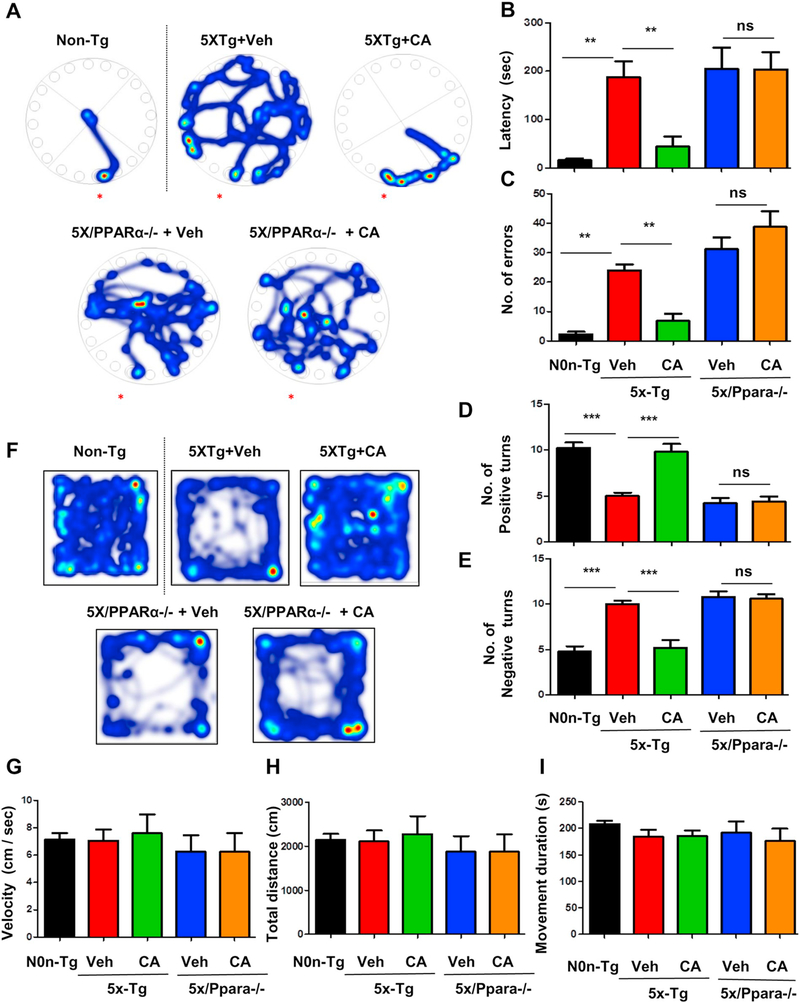
Finally, we analyzed the effect of cinnamic acid on the behavioral performance of the 5XFAD and 5X/Ppara−/− mice. The 5XFAD model successfully recapitulates memory deficit which is one of the main AD characteristics. We performed Barnes maze to test the spatial memory of the mice following cinnamic acid treatment. Our data showed that cinnamic acid treated 5XFAD mice performed better as they took significantly less time to reach the goal box (latency) and made fewer errors compared to the vehicle treated 5XFAD mice indicating that cinnamic acid improves the memory of 5XFAD mice (Fig. 8A-C). However, cinnamic acid treatment did not have any effect on memory deficit of the 5X/Ppara−/− mice suggesting that memory enhancing effect of cinnamic acid is dependent on PPARα (Fig. 8A-C). Next, we performed T-maze to further assess the memory of these mice. We observed that cinnamic acid-treated 5XFAD mice made significantly more number of positive turns and fewer negative turns compared to the vehicle treated 5XFAD mice (Fig. 8D, E) whereas no such improvement in memory was observed in 5X/Ppara−/− mice with cinnamic acid treatment (Fig. 8D, E) further confirming that cinnamic acid requires PPARα for behavioral improvement. We also examined the general locomotor activity of these mice in the open field test which showed no significant differences in the velocity, total distance traveled and cumulative movement duration of different cohorts of mice (Fig. 8G, H, I), indicating that cinnamic acid-mediated behavioral improvement is specifically attributed to the enhanced memory of these mice and is not due to any effect on locomotor function. Together, our results demonstrate that cinnamic acid improves memory of 5XFAD mice via PPARα.
Fig. 8.
Cinnamic acid improves behavioral deficit in 5XFAD mice via PPARα. After one month cinnamic acid treatment, behavioral tests (Barnes maze, T maze, Open field) were performed for assessing the memory of 5XFAD mice (n = 6–7/group) and 5XFAD mice null for Ppara (5X/Ppara−/−) (n = 5/group). (A-C) Barnes maze test showing (A) Representative heat maps, (B) latency to the goal box and (C) number of errors made; (D-E) T maze test showing (D) positive turns and (E) negative turns made by the mice on the test day; (FeH) Open field test demonstrating general locomotor activity (F) representative heat map, (G) velocity of mice, (H) total distance traveled (I) cumulative duration of movement in open filed. All data are represented as mean ± SEM. One way ANOVA followed by Tukey’s multiple comparison test was used for statistical analysis; * p < .05; ** p < .01; *** p < .001.
4. Discussion
At present there is no cure to halt or prevent the progression of Alzheimer’s disease and the available treatments only provide transient symptomatic relief. Although AD is a multifactorial disease with several features contributing to the pathophysiology, according to the widely recognized amyloid cascade hypothesis, accumulation of Aβ due to an imbalance between its production and clearance is the primary event that initiates and drives AD (Hardy and Selkoe, 2002; Hardy and Higgins, 1992; Heppner et al., 2015). Therefore, lowering the cerebral amyloid plaque burden has immense therapeutic potential for treatment of AD. In this study, we demonstrate that cinnamic acid, a naturally occurring plant-based product, can attenuate amyloid pathology in the brain of 5XFAD mice. We also delineate a mechanism, induction of lysosomal biogenesis and thus cellular clearance, by which cinnamic acid could exhibit its amyloid-lowering effects. Finally, cinnamic acid treatment also improves the memory and behavioral performance of 5XFAD mice. Hence, cinnamic acid might be beneficial in countering the toxic protein accumulation observed in AD as well as other neurodegenerative disorders and LSDs.
While exploring the mechanism underlying cinnamic acid-mediated lysosomal biogenesis, we observed that cinnamic acid specifically activates PPARα, a class of nuclear hormone receptors that act as lipid sensors (Kummer and Heneka, 2008). Previously, we have demonstrated a role of PPARα in stimulating lysosomal biogenesis (Ghosh et al., 2015). PPARs are ligand-inducible transcription factors with multiple functional domains: a highly conserved, zinc finger containing DNA binding domain, a hinge region, C-terminal ligand-binding domain, the E/F domain for dimerization and transactivation of the receptor and N-terminal domain for ligand-independent regulation. They form heterodimers with retinoid-X-receptors (RXRs) and bind to specific PPRE on the promoter of the target genes to regulate gene expression (Heneka and Landreth, 2007; Heneka et al., 2011; Kummer and Heneka, 2008). The α isoform of these receptors, PPARα is primary involved in regulation of energy homeostasis (Heneka and Landreth, 2007; Roy and Pahan, 2009). Multiple evidence in this study indicate that cinnamic acid activates PPARα to induce lysosomal biogenesis. Firstly, cinnamic acid treatment results in nuclear translocation of PPARα in primary astrocytes. Secondly, cinnamic acid stimulates PPRE luciferase activity in WT and PPARβ−/−, but not in PPARα−/−, astrocytes. Thirdly, cinnamic acid promotes the recruitment of specifically PPARα, but neither PPARβ nor PPARγ, to the Tfeb promoter. Finally, cinnamic acid stimulates lysosomal abundance in PPARαdependent manner.
Next, we examined how cinnamic acid could activate PPARα. Interestingly, using in-silico and biochemical approaches, we observed that cinnamic acid could bind to the ligand-binding domain of PPARα. Hence, here, for the first time, we established that cinnamic acid might serve as a ligand of PPARα. Our In-silico docking analysis revealed that cinnamic strongly interacted with a tyrosine residue (Y314) of PPARα. Consistently, TR-FRET analysis and Thermal shift assay further demonstrated that cinnamic acid indeed is a very strong agonist/ligand of PPARα. Therefore, our study establishes that cinnamic acid binds to and activates PPARα.
We further explored the downstream targets of cinnamic acid-mediated PPARα activation which could promote lysosomal biogenesis. Growing evidence has shed light on the mechanisms of lysosomal biogenesis. Lysosomal response to different environmental stimulus is coordinated by the central regulator, TFEB and the regulatory lysosomal gene network CLEAR (coordinated lysosomal expression and regulation) (Sardiello et al., 2009; Settembre et al., 2013). TFEB is a MiT family transcription factor that binds to the CLEAR sequence present on multiple lysosomal genes and thereby positively regulates gene expression, lysosomal abundance and degradation of lysosomal substrates (Napolitano and Ballabio, 2016; Settembre et al., 2013). Interestingly, the Tfeb promoter harbors a PPAR-response element (PPRE) and activation of PPARα by its agonist Gemfibrozil leads to transcriptional upregulation of TFEB and stimulates lysosomal biogenesis (Ghosh et al., 2015). In this study, we show that cinnamic acid transcriptionally enhances TFEB expression and induces lysosomal biogenesis via PPARα. Moreover, oral administration of cinnamic acid upregulates TFEB levels in the hippocampus of 5XFAD mice. Several studies have documented a beneficial role of TFEB induction in animal models of AD and tauopathy. Targeted TFEB expression in the astrocytes was shown to enhance lysosomal biogenesis and promote uptake and degradation of Aβ leading to attenuation of amyloid pathology (Xiao et al., 2014). In addition to that, activation of TFEB in the neurons facilitates lysosomal degradation of holo-APP and thereby reduces Aβ generation and amyloid plaque pathology in APP/PS1 mice (Xiao et al., 2015). In another study, inhibition of Glycogen kinase 3β (GSK3β) caused lysosomal biogenesis via nuclear translocation of TFEB and promoted lysosomal degradation of APP (Parr et al., 2012). Moreover, in an AD mouse model demonstrating tauopathy, TFEB was shown to effectively reduce NFT pathology along with improvement of behavioral deficit and neurodegeneration (Polito et al., 2014).
Besides AD, disruption of multiple facets of the lysosomal and autophagic network has identified aberrant lysosomal function as a common underlying mechanism of various neurodegenerative diseases. Therefore stimulation of TFEB to rescue or augment lysosomal function has been widely explored as a therapeutic strategy (Napolitano and Ballabio, 2016). Accumulated evidence has reported that in cellular and mouse models of Parkinson’s disease (PD), overexpression or pharmacological activation of TFEB improved lysosomal function and ameliorated α-synuclein aggregate mediated neurotoxicity (Decressac et al., 2013; Dehay et al., 2010; Kilpatrick et al., 2015). Overexpression of TFEB was shown to protect dopaminergic neurons by promoting α-synuclein clearance (Decressac et al., 2013). Moreover, genetic and chemical activation of TFEB facilitated autophagosome clearance and reduced cell death in a MPTP mouse model (Dehay et al., 2010). In addition, in cellular and different animal models of Huntington’s disease (HD), multiple studies have revealed the efficacy of augmenting the lysosomal function and autophagy in countering the disease pathogenesis. Overexpression of TFEB in cultured cells was observed to induce lysosomal biogenesis and increase degradation of pathogenic huntingtin proteins (Sardiello et al., 2009). Importantly, in a mouse model of HD, TFEB activation by PGC-1α facilitated huntingtin aggregate clearance (Tsunemi et al., 2012). Given the ability of TFEB to effectively coordinate lysosomal biogenesis with autophagy and lysosomal exocytosis, stimulation of TFEB has enormous therapeutic potential for various Lysosomal storage disorders (LSDs). TFEB has been established as a specific modulator of lysosomal proteostasis in Gaucher disease and Tay-Sachs disease, LSDs characterized by deficient activity of beta-glucocerebrosidase and hexosaminidase-A respectively (Song et al., 2013). Moreover, in case of Pompe disease, characterized by dysfunctional autophagy and lysosomal abnormality, overexpression of TFEB was shown to reduce lysosomal size and improve overall autophagy by inducing exocytosis of the autophagolysosomes in cellular as well as mouse model (Spampanato et al., 2013). Furthermore, in cystinosis, caused by accumulation of cystine in lysosomes, TFEB overexpression or stimulation of TFEB by genistein lowered the cystine levels and rescued the abnormalities of lysosomal compartments (Rega et al., 2016). Therefore, TFEB induction is an attractive therapeutic strategy to augment lysosomal function under pathological conditions and stimulation of TFEB and lysosomal biogenesis by cinnamic acid has therapeutic implications for various neurodegenerative diseases and LSDs.
In addition to TFEB, interestingly, in our study, we observed that cinnamic acid enhances the activity of lysosomal proteases Cathepsin B and TPP1 in cultured primary astrocytes as well as in the cortex of 5XFAD mice. Mounting evidence has highlighted the role of Cathepsin B and TPP1 have as therapeutic targets in AD models (Mueller-Steiner et al., 2006) (Sun et al., 2008). Enhancement of cathepsin B activity has been demonstrated to have anti-amyloidogenic and neuroprotective effects. In aged hAPP mice, induction of Cathepsin B reduces the amyloid plaque deposits including thioflavin-S plaques (Mueller-Steiner et al., 2006). Recent study has highlighted the role of another lysosomal protease TPP1 in destabilizing and degrading fibrillar Aβ by cleaving it within the β-sheet region (Sole-Domenech et al., 2018). Therefore, cinnamic acid mediated induction of lysosomal proteases might play roles in countering the amyloid pathology.
Currently, there is no cure or effective treatment for preventing or halting AD. The available therapies are designed to provide only symptomatic relief. Furthermore, these therapeutics often exhibit many side effects and inadequate outcomes. Enhancement of lysosomal function has been explored as a therapeutic strategy in several animal models of AD. In one study, positive lysosomal modulation by PADK demonstrated reduction of amyloid pathology and improvement of memory (Butler et al., 2011). Moreover, virus-mediated TFEB overexpression have been shown to attenuate amyloid burden by facilitating lysosomal function (Xiao et al., 2014; Xiao et al., 2015). Cinnamic acid has several benefits over available AD therapeutics. Firstly, it is a natural plant-based product and is relatively non-toxic. Cinnamic acid is present in huge amount in vegetables, fruits and grains which are regularly consumed by people all over the world. Secondly, cinnamic acid can be taken orally, the least painful route. Thirdly, cinnamic acid is readily available and much cheaper compared to available AD therapeutics. Importantly, cinnamic acid derivatives have been demonstrated to function as cholinesterase inhibitors and might have therapeutic beneficial in AD (Chen et al., 2018; Lan et al., 2017). In-vitro study by Zhang et al. has shown that one of the derivatives, compound 5I, inhibits choloinesterase activity, prevents aggregation of Aβ42 and is neuroprotective against amyloid-stimulated cell toxicity (Lan et al., 2017). These findings further suggest that cinnamic acid could be a potential therapeutic target in AD.
In conclusion, our study delineates a role of cinnamic acid in inducing lysosomal biogenesis via upregulation of the master regulator, TFEB. Furthermore, our study identifies cinnamic acid as a ligand for the nuclear hormone receptor PPARα and establishes activation of PPARα by cinnamic acid as the underlying mechanism through which it transcriptionally upregulate TFEB to promote lysosomal abundance. Finally, our study demonstrates that cinnamic acid treatment attenuates amyloid plaque pathology and improves memory in 5XFAD mice through PPARα. Therefore, cinnamic acid may have therapeutic implications for lowering the pathogenesis of AD and other lysosomal storage disorders.
Supplementary Material
Acknowledgements
This study was supported by a merit award from Veteran Affairs (1I01BX003033), the Zenith Fellows Award (ZEN-17-438829) from Alzheimer’s Association and a grant (AG050431) from NIH.
Abbreviations:
- TFEB
Transcription factor EB
- PPARα
Peroxisome proliferator-activated receptor α
- 5XFAD
5X Familial Alzheimer’s disease
- Aβ
amyloid-beta
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nbd.2018.12.007.
References
- Adisakwattana S, 2017. Cinnamic Acid and its Derivatives: Mechanisms for Prevention and Management of Diabetes and its Complications. Nutrients 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A, 2016. The awesome lysosome. EMBO Mol. Med 8, 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekris LM, et al. , 2010. Genetics of Alzheimer disease. J. Geriatr. Psychiatry Neurol 23, 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S, et al. , 2006. Induction of glial fibrillary acidic protein expression in astrocytes by nitric oxide. J. Neurosci 26, 4930–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S, et al. , 2009. Sodium benzoate, a metabolite of cinnamon and a food additive, reduces microglial and astroglial inflammatory responses. J. Immunol 183, 5917–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D, et al. , 2011. Protective effects of positive lysosomal modulation in Alzheimer’s disease transgenic mouse models. PLoS One 6, e20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra G, et al. , 2018a. Increase in Mitochondrial Biogenesis in Neuronal Cells by RNS60, a Physically-Modified Saline, via Phosphatidylinositol 3-Kinase-Mediated Upregulation of PGC1alpha. J. NeuroImmune Pharmacol 13, 143–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S, et al. , 2018b. Aspirin Induces Lysosomal Biogenesis and Attenuates Amyloid Plaque Pathology in a Mouse Model of Alzheimer’s Disease via PPARalpha. J. Neurosci 38, 6682–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. , 2018. Synthesis and bioevaluation of new tacrine-cinnamic acid hybrids as cholinesterase inhibitors against Alzheimer’s disease. J. Enzyme Inhib. Med. Chem 33, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, et al. , 2012. Gemfibrozil, a lipid-lowering drug, upregulates IL-1 receptor antagonist in mouse cortical neurons: implications for neuronal self-defense. J. Immunol 189, 1002–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, et al. , 2013. Sodium phenylbutyrate enhances astrocytic neurotrophin synthesis via protein kinase C (PKC)-mediated activation of cAMP-response elementbinding protein (CREB): implications for Alzheimer disease therapy. J. Biol. Chem 288, 8299–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett GT, et al. , 2015. Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc. Natl. Acad. Sci. U. S. A 112, 8445–8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve C, 1963. The lysosome. Sci. Am 208, 64–72. [DOI] [PubMed] [Google Scholar]
- De P, et al. , 2011. Cinnamic acid derivatives as anticancer agents-a review. Curr. Med. Chem 18, 1672–1703. [DOI] [PubMed] [Google Scholar]
- De P, et al. , 2012. Recent advances in the development of cinnamic-like derivatives as antituberculosis agents. Expert Opin Ther Pat. 22, 155–168. [DOI] [PubMed] [Google Scholar]
- Decressac M, et al. , 2013. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc. Natl. Acad. Sci. U. S. A 110, E1817–E1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay B, et al. , 2010. Pathogenic lysosomal depletion in Parkinson’s disease. J. Neurosci 30, 12535–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, et al. , 2012. Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor alpha: implications for late infantile Batten disease therapy. J. Biol. Chem 287, 38922–38935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, et al. , 2015. Activation of peroxisome proliferator-activated receptor alpha induces lysosomal biogenesis in brain cells: implications for lysosomal storage disorders. J. Biol. Chem 290, 10309–10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I, et al. , 1986. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U. S. A 83, 4913–4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman JD, 2014. Natural cinnamic acids, synthetic derivatives and hybrids with antimicrobial activity. Molecules 19, 19292–19349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy JA, Higgins GA, 1992. Alzheimer’s disease: the amyloid cascade hypothesis. Science 256, 184–185. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ, 2002. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Landreth GE, 2007. PPARs in the brain. Biochim. Biophys. Acta 1771, 1031–1045. [DOI] [PubMed] [Google Scholar]
- Heneka MT, et al. , 2011. Impact and Therapeutic potential of PPARs in Alzheimer’s Disease. Curr. Neuropharmacol 9, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner FL, et al. , 2015. Immune attack: the role of inflammation in Alzheimer disease. Nat. Rev. Neurosci 16, 358–372. [DOI] [PubMed] [Google Scholar]
- Huang Y, Mucke L, 2012. Alzheimer mechanisms and therapeutic strategies. Cell 148, 1204–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasnavis S, Pahan K, 2012. Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates neuroprotective Parkinson disease protein DJ-1 in astrocytes and neurons. J. NeuroImmune Pharmacol 7, 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick K, et al. , 2015. Genetic and chemical activation of TFEB mediates clearance of aggregated alpha-synuclein. PLoS One 10, e0120819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer MP, Heneka MT, 2008. PPARs in Alzheimer’s disease. PPAR Res. 2008, 403896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan JS, et al. , 2017. Design, synthesis and evaluation of novel cinnamic acid derivatives bearing N-benzyl pyridinium moiety as multifunctional cholinesterase inhibitors for Alzheimer’s disease. J. Enzyme Inhib. Med. Chem 32, 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapeyre C, et al. , 2005. Design, synthesis, and evaluation of pharmacological properties of cinnamic derivatives as antiatherogenic agents. J. Med. Chem 48, 8115–8124. [DOI] [PubMed] [Google Scholar]
- Martini-Stoica H, et al. , 2016. The autophagy-lysosomal pathway in neurodegeneration: a TFEB perspective. Trends Neurosci. 39, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega KG, et al. , 2010. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science 330, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina DL, et al. , 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi KK, et al. , 2014. A physically-modified saline suppresses neuronal apoptosis, attenuates tau phosphorylation and protects memory in an animal model of Alzheimer’s disease. PLoS One 9, e103606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi KK, et al. , 2015. Cinnamon and its Metabolite Sodium Benzoate Attenuate the Activation of p21rac and Protect memory and Learning in an Animal Model of Alzheimer’s Disease. PLoS One 10, e0130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Steiner S, et al. , 2006. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron 51, 703–714. [DOI] [PubMed] [Google Scholar]
- Napolitano G, Ballabio A, 2016. TFEB at a glance. J. Cell Sci 129, 2475–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natella F, et al. , 1999. Benzoic and cinnamic acid derivatives as antioxidants: structureactivity relation. J. Agric. Food Chem 47, 1453–1459. [DOI] [PubMed] [Google Scholar]
- Oakley H, et al. , 2006. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci 26, 10129–10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, et al. , 2012. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol. Cell. Biol 32, 4410–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polito VA, et al. , 2014. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med 6, 1142–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth HW, Laferla FM, 2010. Alzheimer’s disease. N. Engl. J. Med 362, 329–344. [DOI] [PubMed] [Google Scholar]
- Rega LR, et al. , 2016. Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int. 89, 862–873. [DOI] [PubMed] [Google Scholar]
- Roy A, Pahan K, 2009. Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol. Immunotoxicol 31, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, et al. , 2013. Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep. 4, 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, et al. , 2015. HMG-CoA reductase inhibitors bind to PPARalpha to upregulate neurotrophin expression in the brain and improve memory in mice. Cell Metab. 22, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, et al. , 2016. Identification and characterization of PPARalpha ligands in the hippocampus. Nat. Chem. Biol 12, 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, et al. , 2009. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Settembre C, et al. , 2013. Signals from the lysosome: a control Centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol 14, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole-Domenech S, et al. , 2018. Lysosomal enzyme tripeptidyl peptidase 1 destabilizes fibrillar Abeta by multiple endoproteolytic cleavages within the beta-sheet domain. Proc. Natl. Acad. Sci. U. S. A 115, 1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, et al. , 2013. TFEB regulates lysosomal proteostasis. Hum. Mol. Genet 22, [DOI] [PubMed] [Google Scholar]
- Sova M, 2012. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini Rev. Med. Chem 12, 749–767. [DOI] [PubMed] [Google Scholar]
- Spampanato C, et al. , 2013. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med 5, 691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P, et al. , 2015. Anticancer agents derived from natural cinnamic acids. Anti Cancer Agents Med. Chem 15, 980–987. [DOI] [PubMed] [Google Scholar]
- Sun B, et al. , 2008. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron 60, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawata S, et al. , 1996. Synthesis and antifungal activity of cinnamic acid esters. Biosci. Biotechnol. Biochem 60, 909–910. [DOI] [PubMed] [Google Scholar]
- Tsunemi T, et al. , 2012. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med 4, 142ra97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, 2010. Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20. [DOI] [PubMed] [Google Scholar]
- Whyte LS, et al. , 2017. Endo-lysosomal and autophagic dysfunction: a driving factor in Alzheimer’s disease? J. Neurochem 140, 703–717. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T, et al. , 2003. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat. Med 9, 453–457. [DOI] [PubMed] [Google Scholar]
- Xiao Q, et al. , 2014. Enhancing astrocytic lysosome biogenesis facilitates Abeta clearance and attenuates amyloid plaque pathogenesis. J. Neurosci 34, 9607–9620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q, et al. , 2015. Neuronal-Targeted TFEB Accelerates Lysosomal Degradation of APP, reducing Abeta Generation and Amyloid Plaque Pathogenesis. J. Neurosci 35, 12137–12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SY, Kim DH, 2016. Alzheimer’s disease genes and autophagy. Brain Res. 1649, 201–209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.