GhNAC83 inhibits corm dormancy release in Gladiolus hybridus by promoting ABA signaling and inhibiting CK biosynthesis.
Keywords: ABA, corm, cytokinins, dormancy, gladiolus, NAC
Abstract
Corm dormancy is an important trait for breeding in many bulb flowers, including the most cultivated Gladiolus hybridus. Gladiolus corms are modified underground stems that function as storage organs and remain dormant to survive adverse environmental conditions. Unlike seed dormancy, not much is known about corm dormancy. Here, we characterize the mechanism of corm dormancy release (CDR) in Gladiolus. We identified an important ABA (abscisic acid) signaling regulator, GhPP2C1 (protein phosphatase 2C1), by transcriptome analysis of CDR. GhPP2C1 expression increased during CDR, and silencing of GhPP2C1 expression in dormant cormels delayed CDR. Furthermore, we show that GhPP2C1 expression is directly regulated by GhNAC83, which was identified by yeast one-hybrid library screening. In planta assays show that GhNAC83 is a negative regulator of GhPP2C1, and silencing of GhNAC83 promoted CDR. As expected, silencing of GhNAC83 decreased the ABA level, but also dramatically increased cytokinin (CK; zeatin) content in cormels. Binding assays demonstrate that GhNAC83 associates with the GhIPT (ISOPENTENYLTRANSFERASE) promoter and negatively regulates zeatin biosynthesis. Taken together, our results reveal that GhNAC83 promotes ABA signaling and synthesis, and inhibits CK biosynthesis pathways, thereby inhibiting CDR. These findings demonstrate that GhNAC83 regulates the ABA and CK pathways, and therefore controls corm dormancy.
Introduction
In most countries, summer-flowering Gladiolus cultivars are widely planted and are among the most important cut flowers. Summer-flowering Gladiolus shows great diversity in plant height, flower color, number of florets, and flower size. During the Gladiolus growing season, a new corm is produced over the mother corm. Afterwards, cormels are formed at the tips of branched stolons that develop from buds located at the base of the new corm (Le Nard, 1993). In autumn, the corms and cormels are lifted out of the ground and placed in a cold storage house to accelerate corm dormancy release (CDR; ~2–3 months) before the next planting (Wu et al., 2015). Understanding the mechanism of CDR to shorten the growth season is of great interest to the flower industry.
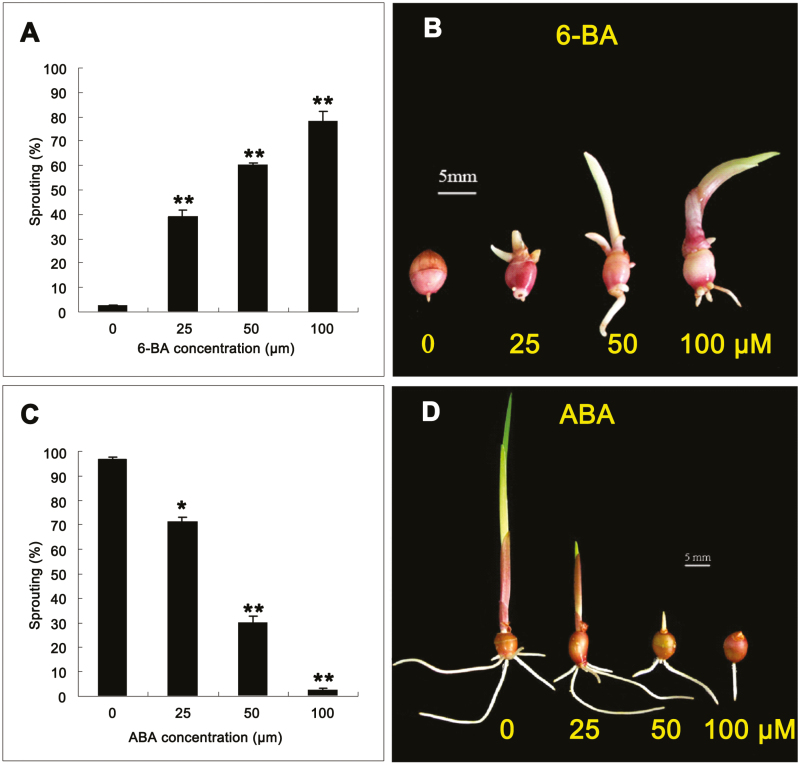
In Gladiolus, ABA (abscisic acid) is the key inhibitor of CDR, and GhABI5 (ABA INSENSITIVE 5) has been shown to delay CDR. GA (gibberellic acid) plays a minor role in this process (Ginzburg, 1973; Wu et al., 2015). Moreover, 6-BA [6-benzylaminopurine; an exogenous aromatic cytokinin (CK)] increases dark CO2 fixation rates in dormant Gladiolus cormels, indicating that 6-BA has a positive role in CDR (Ginzburg, 1981). However, the molecular mechanisms of ABA’s and CK’s antagonistic regulation of CDR are unknown.
In Arabidopsis, ABA controls seed dormancy by inhibiting the activities of clade A PP2Cs, a group of protein phosphatases (PPs) including ABI1/2 (ABA INSENSITIVE 1/2) and HAB1/2 (HYPERSENSITIVE TO ABA 1/2), which act as co-receptors with PYR1/PYL/RCAR (PYRABACTIN RESISTANT/PR1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR) in ABA signaling (Ma et al., 2009; X. Wang et al., 2018). These protein phosphatases play important roles in seed germination and abiotic stress responses (Gosti et al., 1999; Kong et al., 2015). When ABA levels increase, clade A PP2Cs lose the ability to inhibit the activity of SnRK2s (class II SNF1-related protein kinase 2) activating downstream ABA responses (Hubbard et al., 2010). In strawberries, silencing of FaABI1 promotes fruit ripening, indicating that ABI1 has an inhibitory role in fruit ripening (Jia et al., 2013). In recent years, upstream regulators of PP2Cs have been identified and shown to function in salt stress (MYB20), leaf senescence (AtNAP; NON-INTRINSIC ABC PROTEIN), drought response (AtHB7/12; HOMEOBOX 7/12), and water stress (ORA47; octadecanoid-responsive AP2/ERF-domain transcription factor 47) ( Valdes et al., 2012; Zhang and Gan, 2012; Cui et al., 2013; Chen et al., 2016).
CKs are involved in delaying leaf senescence, promoting differentiation of the shoot and root meristems, seed germination, and stress responses (Werner et al., 2003; Dong et al., 2008; Choi et al., 2010; Wang et al., 2011; Verslues, 2016). The relationship between ABA and CKs varies depending on the species and biological process (Bozhkov et al., 1992; Zubo et al., 2008; Wang et al., 2011). CKs have been shown to antagonize ABA’s role in seed dormancy by inhibiting ABI5 expression (Wang et al., 2011). Adenosine phosphate-isopentenyltransferase (IPT) and CYP735As (CYTOCHROME P450, FAMILY 735, SUBFAMILY As) catalyze important steps of CK biosynthesis to produce trans-zeatin (Takei et al., 2004; Sakakibara, 2006; Tarkowska and Strnad, 2018). Endogenous CKs activate the receptor gene Cytokinin Response 1 (CRE1) to initiate phosphorelay signaling (Inoue et al., 2001). In Arabidopsis, the core signaling pathway consists of His kinases (AHKs), His phosphotransfer proteins (AHPs), and response regulators (ARRs). ARR4/5/6 interact with, and negatively regulate, ABI5 expression during seed germination (Wang et al., 2011). The antagonistic roles of ABA and CKs/GA have also been shown in potato tuber sprouting and are possibly linked to altering SnRK1 (Sucrose non fermenting Related Kinase1)/T6P (TREHALOSE-6-PHOSPHATE) signaling (Subbaraj et al., 2010; Sonnewald and Sonnewald, 2014). However, the molecular mechanisms of CK–ABA interaction in dormancy release are still unclear.
NAC transcription factors (TFs) form one of the largest TF families in plants, and are classified into 24 groups (Jensen et al., 2010). The NACs recognize the consensus cis-acting elements CGT(G/A) and CACG (Cao et al., 2017). In Arabidopsis, NACs play roles in plant development (Ko et al., 2007), senescence (Yang et al., 2011), drought (Park et al., 2009; You et al., 2014), cold tolerance (Shan et al., 2014), and biotic stress (Seo et al., 2010). Some NACs (ATAF1) mediate signaling in response to both pathogen and abiotic stresses (Wu et al., 2009). NACs have been implicated in the regulation of an ABA biosynthesis gene (NCED; 9-CIS-EPOXYCAROTENOID DIOXYGENASE) and an ABA response gene (RD29; RESPONSIVE TO DESICCATION 29), further modulating drought stress (Wu et al., 2009; Jensen et al., 2013; Xu et al., 2013). In addition, a membrane-bound NAC (NTM1; NAC WITH TRANSMEMBRANE MOTIF1) has been reported to mediate CK signaling, specifically during cell division (Kim et al., 2006).
Currently, not much is known about how hormones control CDR, particularly the mechanisms behind the antagonistic role that ABA and CKs play in this process. In this study, transcriptome sequencing and functional analysis revealed that GhPP2C1 positively regulates the CDR. GhNAC83 was found to bind the GhPP2C1 promoter directly by yeast one-hybrid screening, and further evidence suggests that GhNAC83 is a negative regulator of GhPP2C1. We also show that GhNAC83 decreases zeatin content by inhibiting the expression of CK biosynthesis genes (GhCYP735A and GhIPT). Thus, GhNAC83 positively regulates ABA signaling through down-regulation of GhPP2C1 and inhibits CK biosynthesis through down-regulation of GhIPT, ultimately delaying CDR. Our findings uncover GhNAC83’s role in regulating ABA and CK pathways in the control of corm dormancy.
Materials and methods
Plant material and growth conditions
Gladiolus ‘Rose Supreme’ was planted and harvested as described previously (Wu et al., 2015). Harvested cormels were dried at 25 °C for 6 weeks, and then were kept in a cold storage room at 4 °C with 60–70% relative humidity.
For expression pattern analysis, tissues and organs were collected at the flowering stage with seven leaves. Cormels at different dormant stages were sampled after harvest (desiccation and cold storage) every 2 weeks.
Sprouting tests were used to determine dormancy release patterns under different hormone treatments. Dormant cormels used in 6-benzylaminopurine (6-BA) treatments measured 0.5 cm in diameter. These cormels were sterilized first and then were embedded in 0.6% (w/v) agar plates which contained different concentrations of 6-BA (0, 25, 50, and 100 μM) before being placed in a plant growth chamber at 25 °C with 12 h/12 h light/dark. The sprouting percentage was counted on the 20th day after plating. Sprouting was defined as a bud on the top of the cormel elongated >5 mm (Luo et al., 2012). Thirty cormels per sample were used for each sprouting test. Error bars in the figures represent the SD of three biological replicates. Non-dormant cormels were used for ABA treatments (0, 25, 50, and 100 μM), and the sprouting test was the same as explained above.
Transcriptome analysis
Samples for RNA sequencing (RNA-seq) were collected at deep dormancy (DD; 19 December 2012), weak dormancy (WD; 17 January 2013), and ecodormancy (ED; 9 May 2013) stages (Wu et al., 2015). Three biological samples were collected for each stage, frozen immediately in liquid nitrogen, and stored in a freezer at –80 °C until RNA extraction. The sprouting percentage was counted on the 20th day after planting on soil. Sprouting was defined as a bud on the top of the cormel elongated >5 mm (Luo et al., 2012). Thirty cormels per stage were used for each sprouting test. Error bars in the figures represent the SD of three biological repeats.
Total RNA from Gladiolus cormels was extracted using the Tiangen RNA extraction reagent kit (Tiangen, Beijing, China). RNA was quantified using a NanoDrop 2000 (Thermo Scientific, DE, USA) and its quality was determined by an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). High-quality RNA (RNA integrity number >9.0) was selected for cDNA library preparation. Strand-specific RNA libraries were constructed as previously described (Zhang et al., 2015).
The RNA-seq libraries were sequenced in a single lane of a Hiseq 2500 platform at the Novogene Company (Beijing, China) and 150 bp paired-end reads were generated (10-fold depth of RNA sequencing). The raw sequence reads were deposited in the NCBI Sequence Read Archive (SRA) database under the accession number PRJNA491310.
Raw data were filtered to remove low-quality reads, and adaptor sequences were trimmed using Trimmonmatic (Bolger et al., 2014). The resulting data were then aligned to the rRNA sequence databases (Quast et al., 2013) and the GenBank virus database using Burrows–Wheeler aligner (BWA) with default parameters (Li and Durbin, 2010). Mapped reads in these two databases were discarded. Only high-quality clean reads were used in the following analysis.
De novo transcriptome assembly was performed using the Trinity program (Grabherr et al., 2011). To delete the redundant Trinity-assembled contigs, the contigs were further assembled using iAssembler (Zheng et al., 2011). All assembled unigenes were subjected to the NCBI non-redudant protein (Nr) database, Swiss-prot database, Nucleotide database (Nt), Cluster of Orthologous Groups (COG) database, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) database with a typical cut-off E-value of 1E−5. Based on the annotation, BLAST2GO (Conesa et al., 2005) was assigned to obtain the GO annotation for describing the biological processes, cellular components, and molecular functions. The COG database was used to classify unigene functions (Tatusov et al., 2000). The KEGG pathway of unigenes was annotated by mapping the resulting sequences from BLAST2GO to the contents of the KEGG metabolism pathway database (Kanehisa and Goto, 2000).
Isolation of full-length GhPP2C1 and GhNAC83 cDNAs, and sequence analysis
The full-length GhPP2C1 sequence was cloned by RACE according to the manufacturer’s instructions (Clontech). The full-length GhNAC83 sequence was directly isolated from our transcriptome database by PCR (Supplementary Table S1 at JXB online).
Multiple amino acid alignments were performed using ClustalX1.8 and BioEdit7.0 (Chenna et al., 2003; Hall, 2005), and phylogenetic trees were constructed by the maximum likelihood method using the MEGA5.0 software (Tamura et al., 2011).
Quantitative real-time-PCR
Total RNA was extracted using the Tiangen RNA extraction reagent kit. A 1 μg aliquot of DNase-treated RNA was used to synthesize cDNA by M-MLV (Takara). About 400 ng of cDNA was used as the template for real-time PCRs (RT-PCRs) and was run by the Step One Plus real-time PCR system (Applied Biosystems) using the SYBR Premix Ex Taq kit (Takara). GhActin (accession no. JF831193) was used as the internal control. The PCR procedure was performed according the manufacturer’s instructions. Primers used are listed in Supplementary Table S1.
Virus-induced gene silencing
Silencing of GhPP2C1 or GhNAC83 in dormant cormels was conducted as previously described (Zhong et al., 2014; Wu et al., 2015), with some modifications. Freshly grown Agrobacterium tumefaciens GV3101 cells harboring pTRV1, pTRV2, pTRV2-GhPP2C1, or pTRV2-GhNAC83 vectors were collected and suspended in infiltration buffer (10 mM MgCl2, 200 mM acetosyringone, 10 mM MES, pH 5.6) to a final OD600 of 2.0. A mixture containing equal volumes of pTRV1 and pTRV2-GhPP2C1 or pTRV2-GhNAC83 cultures were used for the GhPP2C1-TRV2 and GhNAC83-TRV2 experiments, respectively. A mixture containing an equal volume of pTRV1 and pTRV2 cultures was used as the control (TRV2). The mixtures were stored at 25 °C for 3 h in darkness. Vacuum infiltration of dormant cormels and later growth stages was performed as previously described (Wu et al., 2015). Three independent experiments were conducted with 24 silenced cormels in each experiment. The silenced plantlets were imaged and analyzed after 10 d on soil.
Promoter analysis, cloning, and transient expression assay in Nicotiana benthamiana
The upstream regulatory sequence (URS) of GhPP2C1 was cloned using high-efficiency thermal asymmetric interlaced PCR (Hi-TAIL) (Liu and Chen, 2007). The cis-regulatory elements were annotated using PlantCARE (Lescot et al., 2002), and potential TF-binding sites were analyzed using PlantPan 2.0 (Chow et al., 2016).
The URS and truncated URSs were inserted into the pCAMBIA1391 binary vector. GhPP2C1:GUS was then introduced into GV3101 for N. benthamiana infiltration. Agrobacterium tumefaciens cells harboring the truncated promoter fragments were suspended in infiltration buffer (10 mM MgCl2, 200 mM acetosyringone, 10 mM MES, pH 5.6) to an OD600 of 0.8, then each suspension was infiltrated into different regions of the same N. benthamiana leaf. After 3 d, the infiltrated leaves were immersed in GUS (β-glucuronoidase) staining solution overnight and were decolorized using 70% ethanol (Chen et al., 2013). Three independent experiments were conducted with 12 leaves from six plants in each experiment.
Yeast one-hybrid screening
Yeast one-hybrid library screening was performed as previously described (Deplancke et al., 2006), with some modifications. The GhPP2C1 truncated promoter (base pairs –833 to –615) was recombined into the pDEST-HISi-2 vector by Gateway cloning. Then the linearized vector was transformed into yeast strain YM4271(a) using the PEG/LiAc method. Transformed yeast colonies were tested for background expression of the HIS3 reporter, and the appropriate 3-aminotriazole (3-AT) concentration was selected. An Arabidopsis thaliana TF library (Mitsuda et al., 2010) was transformed into yeast strain EGY48(α) by electroporation. Mutagenesis of the GhPP2C1 promoter was generated by PCR-driven overlap extension (Heckman and Pease, 2007). The same method of mutagenesis was used to generate the mutant GhIPT promoter used below. Primers are listed in Supplementary Table S1.
To test the interaction between GhNAC83 and the GhIPT promoter truncations, the GhIPT promoters (T1, T2, T3, and T2mut) and GhNAC83 were recombined into pDEST-HISi-2 and pDEST-GAD424, respectively, by Gateway technology. The recombined vectors were then transformed into yeast strain YM4271(a) (for pDEST-HISi-2) and EGY48(α) (for pDEST-GAD424). Transformed YM4271(a) containing the various truncated GhIPT promoter regions were first tested for the background HIS3 expression using increasing 3-AT concentrations (0, 5, 10, 20, and 40 mM). A single transformed YM4271(a) colony requiring the lowest 3-AT concentration (10 mM) from each transformed yeast (T1, T2, T3, and T2mut) was used for mating with EGY48(α) containing GhNAC83. Following mating on YPD plates for 16 h, the yeast cells were washed off with water and spread on yeast plates (SD-Ura-His-Leu). The plates were cultured at 28 °C for 3 d to select for diploids. Yeast cultures (OD600 diluted to 0.08) were spotted on selection plates (SD-Ura-His-Leu+10 mM 3-AT) and cultured at 28 °C for 3 d. The interaction was judged by the growth of yeast on selection media.
GUS/LUC assay in N. benthamiana
The transient GUS/luciferase (LUC) assay was performed as previously described (Zhao et al., 2016). The constructs (35S:GhNAC83/pSuper1300, pSuper1300, GhPP2C1:GUS/pCAMBIA1391, and 35S:LUC) were independently transformed into A. tumefaciens strain GV3101. Then, 35S:GhNAC83, GhPP2C1:GUS, and 35S:LUC (OD600=0.8 each; 1000:1000:5 v/v/v) were co-agroinfiltrated into N. benthamiana. After 3 d, GUS and LUC activities were measured using methyl umbelliferyl glucuronide (Sigma-Aldrich; 881005-91-0) and the Bio-Glo™ Luciferase Assay System kit (Promega; G7940), respectively. The LUC activity (35S:LUC) was used as an internal control and pSuper1300 was used as a negative control. The GUS/LUC ratio was used to reflect the promoter activity. Three biological replicates were conducted in this assay (n=5 leaves).
Subcellular localization assay
The GhNAC83 ORF was cloned into pCAMBIA1300-GFP (green fluorescent protein). Both the fusion construct (GhNAC83-GFP) and the control (GFP) were transformed into A. tumefaciens GV3101 and used to agroinfiltrate N. benthamiana leaves. GFP fluorescence was observed using confocal microscopy (Nikon Inc., Melville, NY, USA) at 3 d post-infiltration.
Transactivation domain analysis in yeast
For the transactivation assay in Saccharomyces cerevisiae strain AH109, different truncations of the GhNAC83 coding region were PCR amplified and inserted into the pGBKT7 vector (Clontech, Mountain View, CA, USA) with NdeI and XmaI sites. The different portions of GhNAC83 fused with the GAL4 DNA-binding domain are as follows: full length (FL; amino acids 1–219), C-terminal part (CP; amino acids 111–219), N-terminal part (NP; amino acids 1–110), and the C-terminus (CT; amino acids 161–219). The primers are listed in Supplementary Table S1. The positive control (pBD-AD; +) and the negative control (pBD; –) were also introduced into AH109 according to the manufacturer’s protocol (Stratagene). Transcriptional activation was tested as described in the yeast protocols handbook (PT3024-1; Clontech).
Extraction and quantification of phytohormones
The extraction of ABA from Gladiolus cormels was performed according to Wu et al. (2016). Gladiolus cormels (50 mg) were homogenized, and added to an extraction solvent (500 μl; isopropanol/H2O/concentrated HCl with a volume ratio of 2:1:2E-3) with 10 ng of internal standard (d6-ABA). Samples were inverted at 4 °C (100 rpm, 30 min), and then 1 ml of dichloromethane was added for a second round of inversion. After centrifugation (14000 rpm, 30 min), the lower phase of solvent was transferred to a new tube. The solvent was dried using a DNC-2000 concentrator (Beijing IDES Technology) and was re-dissolved in 100 μl of methanol. The extraction of CKs from cormels was based on the procedure described previously (Antoniadi et al., 2015) with some modifications. Samples (500 mg) were homogenized and extracted using a 5 ml mixture of methanol/water/methanoic acid (15:4:1, v/v/v) containing 20 mg l–1 sodium diethyldithiocarbamate. Deuterium-labeled CKs were added to serve as internal standards. Extractions were purified with a Sep-Pak Plus C18 cartridge and Oasis MCX column as described previously (Chen et al., 2010). Then, the column was washed with 1 M methanoic acid (5 ml), and pre-concentrated analytes were eluted by two-step elution using NH4OH (5 ml) and 5 ml of 0.35 M NH4OH in 60% methanol. The eluate was vacuum evaporated and kept at –80 °C until analysis. Quantitative analysis of ABA and CKs in crude extracts was determined by HPLC-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) (Pan et al., 2008; Farrow and Emery, 2012). At least three biological replicates were conducted.
Dual-luciferase reporter assay
The GhNAC coding sequence was cloned into pGreenII 62-SK. A promoter of the GhPP2C1p, GhPP2C1pMUT, GhIPTp, or GhIPTpMUT regions was cloned into pGreenII LUC vector (Wei et al., 2017). All constructs were transformed into A. tumefaciens strain GV3101 harboring the pSoup helper plasmid. The infiltration and LUC measurements were performed as previously described (Wei et al., 2017).
Results
GhPP2C1 promotes corm dormancy release in Gladiolus
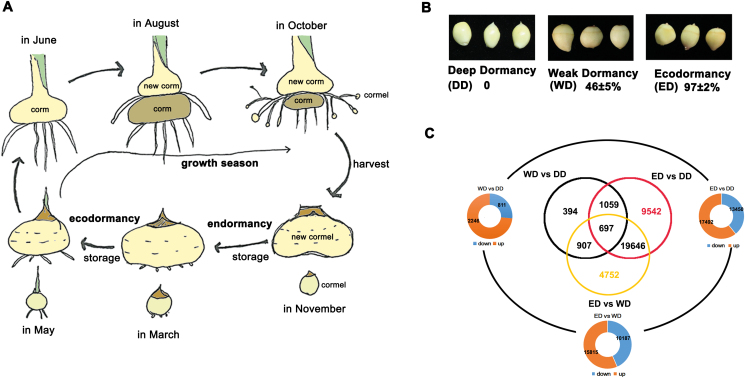
To investigate the molecular mechanism of Gladiolus CDR, we first tracked sprouting of cormels at different stages (Fig. 1A). We chose deep dormant (DD; unsprouting), weak dormant (WD; half-sprouting), and ecodormant (ED; all-sprouting) cormels for large-scale transcriptome sequencing on the Illumina Hiseq2500 platform using the paired-end protocol (Fig. 1B). To identify genes that are differentially regulated during CDR, differentially expressed genes (DEGs) were screened using a cut-off ratio of log2 < –1 or >1, and a q-value of <0.05, and 697 overlapping DEGs were found (Supplementary Table S2). The results in Fig. 1C indicate that the greatest change in gene expression occurred during the ED transition (ED versus WD; 26 002 unigenes) and not in the WD transition (WD versus DD; 3057 unigenes). During the WD transition, GO terms of phytohormone biosynthesis (zeatin and ABA) and plant hormone signal transduction were highly enriched (Supplementary Fig. S1), supporting the opposing roles of these hormones in CDR (Fig. 2).
Fig. 1.
Transcriptome analysis of Gladiolus corm dormancy release. (A) Life cycle of Gladiolus. Corms >1 cm in diameter are used for cut-flower production. Cormels are planted in the next growing season and develop into corms. (B) Different stages of corm dormancy. DD, deep dormancy; WD, weak dormancy; ED, ecodormancy. Sprouting rates were tested 20 d after planting on soil. Data are shown as means of three replicates ±SD (n=30). (C) Differentially expressed genes (DEGs) during Gladiolus dormancy release. Genes were considered to be DEGs when there was a cut-off ratio of log2 < –1 or >1 and a q-value <0.05. The 697 overlapping DEGs are listed in Supplementary Table S2. (This figure is available in color at JXB online.)
Fig. 2.
ABA and cytokinins are involved in corm dormancy release. (A) 6-BA promotes sprouting of dormant cormels. (B) The phenotype of dormant cormels exposed to 6-BA for 20 d. (C), ABA inhibits sprouting of non-dormant cormels. (D) The phenotype of non-dormant cormels exposed to ABA for 20 d (*P<0.05 and **P<0.01). Averages of three biological replicates with the SD are shown; n=30. (This figure is available in color at JXB online.)
With respect to phytohormones, ABA-related DEGs, including PP2C family genes, were the most abundant, showing strong up-regulation from DD to WD (Supplementary Table S3). Moreover, three PP2C unigenes (GlaUn030679, GlaUn052869, and GlaUn078852) maintained high transcriptional levels during CDR (Supplementary Table S3).
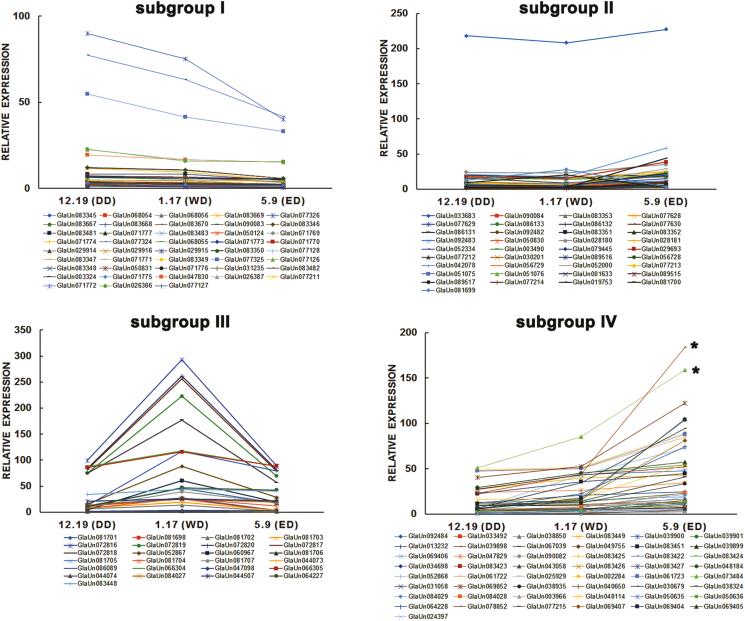
PP2Cs are a part of the core ABA signaling module and are involved in seed dormancy ( Seiler et al., 2011; Nee et al., 2017). In order to investigate PP2C’s function in CDR, 154 members were identified in the transcriptome and sorted into four subgroups by their expression pattern: subgroup I (43/154), subgroup II (37/154), subgroup III (25/154), and subgroup IV (49/154) (Fig. 3). When a threshold for change in expression level was set (fold <0.8 or >1.6 and relative expression value >20), only two members (GlaUn078852 and GlaUn073484) met the criteria. The full-length cDNAs of GlaUn078852 and GlaUn073484 were amplified from G. hybridus cv. ‘Rose Supreme’ cormels by RACE, and were found to be the same gene. Therefore, we selected this gene for further study.
Fig. 3.
Expression patterns of GhPP2C genes in Gladiolus CDR. An asterisk (*) represents the selected unigenes (GhPP2C1) from Gladiolus CDR transcriptome analysis. Expression of unigenes in the top left panel decreased during CDR (DD→WD→ED). Unigenes in the top right panel decreased in expression from DD to WD, but increased from WD to ED. Expression of unigenes in the bottom left panel increased from DD to WD, but decreased from WD to ED. Expression of unigenes in the bottom right panel increased during CDR (DD→WD→ED). The expression levels are based on a FPKM evaluation. DD, deep dormancy; WD, weak dormancy; ED, ecodormancy. (This figure is available in color at JXB online.)
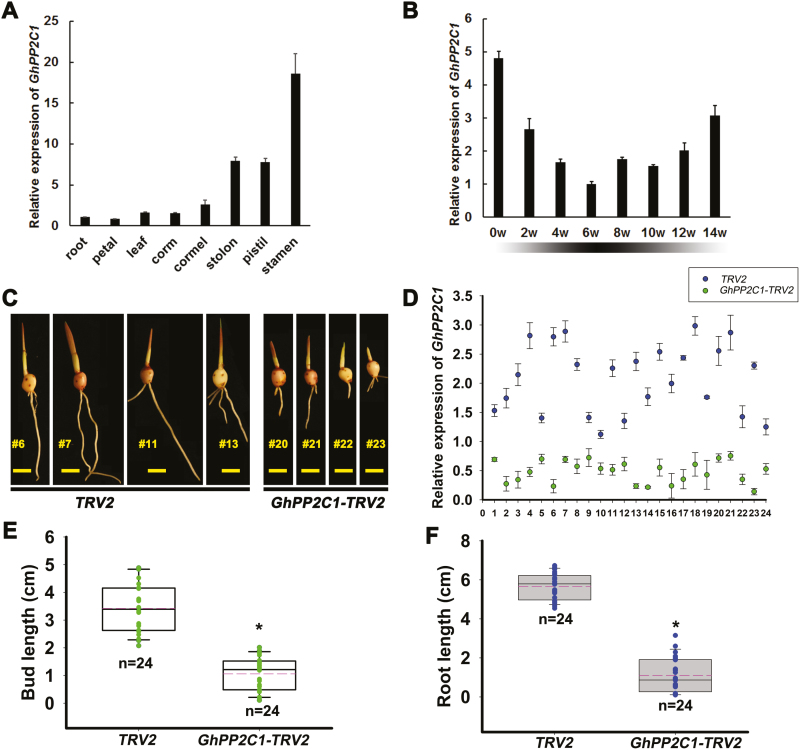
This PP2C member, which belongs to group A of the PP2C family and shares high sequence similarity with Arabidopsis HAB1 and HAB2 (Supplementary Fig. S2), was named GhPP2C1 (GenBank ID: KP710220). The expression of GhPP2C1 was evaluated in different organs of blooming plants. As shown in Fig. 4A, GhPP2C1 was expressed in all tested organs, including cormels and corms. GhPP2C1 was expressed throughout desiccation (weeks 0–6) and storage (weeks 6–14). The transcript levels began to decrease after harvest, and were lowest at the end of the desiccation period. However, the expression of GhPP2C1 gradually increased after cold storage for CDR (Fig. 4B). This result is in accordance with the transcriptome data and suggests that GhPP2C1 may regulate CDR.
Fig. 4.
GhPP2C1 is involved in corm dormancy release. (A) The expression of GhPP2C1 in different organs at blooming flower stage. (B) The expression pattern of GhPP2C1 during corm desiccation (weeks 0–6) and cold storage (weeks 6–14). Data in (A) and (B) are displayed as averages of three biological repeats with the SD. (C) Phenotype resulting from GhPP2C1 silencing 10 d after planting on soil. The scale bar represents 1 cm. (D) The expression of GhPP2C1 in 24 independent GhPP2C1-TRV2 lines. Data are shown as averages of three technical replicates with the SD. Bud length (E) and root length (F) in GhPP2C1-TRV2 and TRV2 lines; n=24 independent lines (*P<0.05; **P<0.01). (This figure is available in color at JXB online.)
Virus-induced gene silencing (VIGS) is widely used in functional analysis of horticultural plants, such as rose, apple, strawberry, and Gladiolus (Zhong et al., 2014; Wu et al., 2016; Ma et al., 2017; S. Wang et al., 2018). Therefore, we investigated the role of GhPP2C1 in CDR using a VIGS approach. We inserted a specific 3'-untranslated region (UTR) fragment of GhPP2C1 into the TRV2 vector for specific gene silencing in dormant cormels (Fig. 4C, D). After 10 d on soil, GhPP2C-silenced (GhPP2C-TRV2) cormels grew significantly more slowly than the control (empty TRV2 vector), and buds and roots were dramatically shorter than those of controls (Fig. 4C, E, F). These results indicate that down-regulation of GhPP2C1 in dormant cormels leads to delayed CDR, demonstrating that GhPP2C1 acts as a positive regulator of CDR.
GhNAC83 is a negative regulator of GhPP2C1
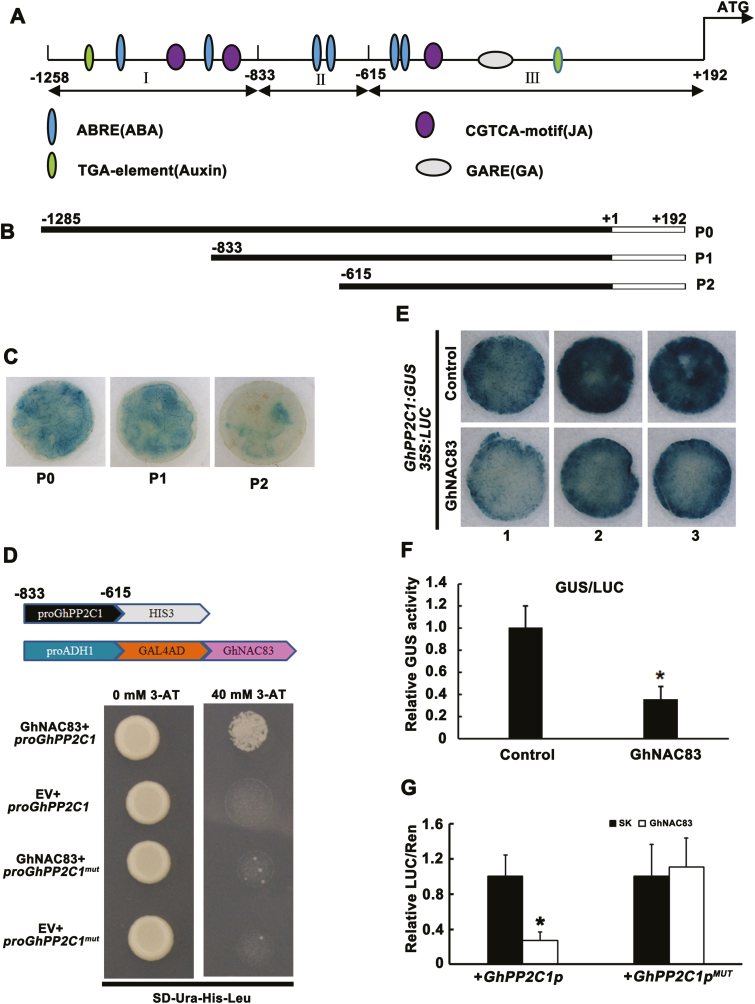
To explore the regulation of GhPP2C1 during CDR further, we isolated a 1.5 kb sequence of the GhPP2C1 regulatory region upstream of the translation start site (Fig. 5A) by Hi-TAIL PCR. Based on the distribution of cis-elements, we truncated the promoter (Fig. 5B) and performed transient expression assays in leaves of N. benthamiana. Our results show that the promoter activity is unaffected when region I is deleted (–1285 to –833; P1 construct); however, a deletion in region II (–833 to –615; P2 construct) led to a sharp decrease in promoter activity (Fig. 5C). Therefore, we focused our efforts on identifying regulators that bind region II of the GhPP2C1 promoter.
Fig. 5.
GhNAC83 binds the GhPP2C1 promoter and inhibits its transcription. (A) The distribution of cis-elements in the GhPP2C1 promoter. (B) Truncations of the GhPP2C1 promoter used in transient assays. (C) Deletion of base pairs –833 to –615 in the GhPP2C1 promoter (P2 construct shown in B) dramatically affects its activity in transient N. benthamiana assays. Three biological replicates were conducted and showed similar results. One biological replicate of leaf discs is shown. (D) The interaction of GhNAC83 and the GhPP2C1 promoter in yeast one-hybrid assays. The empty prey vector (pDEST-GAD424) was used as the control. The mutagenesis of NAC-binding sites (Supplementary Fig. S3) in the GhPP2C1 promoter (proGhPP2C1mut) was also tested. The interaction between the GhNAC83 protein and the GhPP2C1 promoter was determined by cell growth on synthetic dropout nutrient medium lacking Ura, His, and Leu, and containing 40mM 3-AT. (E) GhNAC83 represses GhPP2C1 promoter activity in transient expression assays in N. benthamiana leaves. 35S:GhNAC83 was used as an effector and GhPP2C1:GUS was used as a reporter. 35S:LUC was used as an internal control. GUS stains were performed on the third day post-infiltration. (F) The relative GUS activity (GUS/LUC) indicates that GhNAC83 represses GhPP2C1 transcription in planta. Three biological replicates were performed and are shown with the SD. (G) The interaction of GhNAC83 with the GhPP2C1 promoter using a dual-luciferase reporter assay in N. benthamiana leaves. A fragment of GhPP2C1’s promoter (base pairs +192 to –1285) was used in this assay. Mutated sites in GhPP2C1pMUT are shown in Supplementary Fig. S3. The empty vector (pGreenII 62-SK) was used as a control. Data are shown as the average of three biological replicates with the SD (n=5 leaves), *P<0.05. (This figure is available in color at JXB online.)
The 219 bp region II contains several conserved TF-binding sites (Supplementary Fig. S3A). To identify TFs that bind this region of the GhPP2C1 promoter, a yeast one-hybrid screen was performed using a TF library from Arabidopsis (Mitsuda et al., 2010). First, we selected yeast harboring the integrated 219 bp promoter that could not survive on selection medium containing 40 mM 3-AT. Then, we performed the yeast one-hybrid screen and isolated 12 TFs among 100000 cfu (Table 1). We then identified Gladiolus homologous genes using the Gladiolus transcriptome database, and five TFs were able to bind region II (Table 1). Taking into consideration the expression level during CDR and the number of clones identified from the yeast one-hybrid screen (Table 1), GhNAC83 (GlaUn057212) was selected for further study.
Table 1.
Potential upstream regulators of GhPP2C1 screened by yeast one-hybrid analysis
| Gene | Family | Repeatsa | Re-Y1Hb | DD | WD | ED |
|---|---|---|---|---|---|---|
| GhNAC83 | NAC | 42 | + | 60.55 | 28.08 | 7.32 |
| GhbZIP1 | bZIP | 28 | + | 22.98 | 58.48 | 74.44 |
| GhWRKY40 | WRKY | 16 | + | 331.05 | 347.76 | 93.83 |
| GhMYB1R1 | MYB | 12 | + | 26.29 | 33.28 | 49.16 |
| GhAPL-Like | MYB | 4 | + | 106.24 | 83.66 | 53.97 |
| GhDof1.8 | Dof | 18 | – | 6.56 | 4.28 | 7.12 |
| GhbZIP60 | bZIP | 14 | – | 23.12 | 36.86 | 59.18 |
| GhBPC1 | BPC | 12 | – | 1.19 | 1.09 | 3.40 |
| GhCIB4 | bHLH | 12 | – | – | – | – |
| GhWOX6 | HB | 11 | – | – | – | – |
| GhTCP4 | TCP | 10 | – | 17.69 | 17.12 | 12.51 |
| GhKNU | C2H2 | 1 | – | – | – | – |
a Number of colonies harboring the same gene isolated by yeast one-hybrid (Y1H) screens using an Arabidopsis TF library (Mitsuda et al., 2010).
b Positive or negative Y1H results when using Gladiolus clones.
DD, deep dormancy; WD, weak dormancy; ED, ecodormancy. The data correspond to the expression level (based on FPKM evaluation) in the Gladiolus transcriptome database. Values in bold indicate down-regulation and those in italics indicate up-regulation.
To test the binding ability of GhNAC83, we mutated the NAC-binding site in the promoter (Supplementary Fig. S3B). The result showed that GhNAC83 binds the native GhPP2C1 promoter, but cannot bind the mutant promoter (Fig. 5D). In addition, to test further the effect of GhNAC83 on GhPP2C1 transcription, we performed transient transactivation assays using the GhPP2C1 promoter driving GUS reporter expression. A GhNAC83 effector construct driven by the 35S promoter was co-agroinfiltrated with the reporter construct into leaves of N. benthamiana. The expression of GhPP2C1 was significantly inhibited by GhNAC83 (Fig. 5E, F). Furthermore, a dual-LUC reporter assay was performed to analyze the regulation of the GhPP2C1 promoter by GhNAC83 in planta. The results show that GhNAC83 decreases GhPP2C1 promoter activity; furthermore, when we mutated the binding sites of GhNAC83 in the GhPP2C1 promoter (GhPP2C1pMUT), GhPP2C1MUT promoter activity was unaffected (Fig. 5G). These results suggest that GhNAC83 directly binds the GhPP2C1 promoter and negatively regulates its expression in planta.
GhNAC83 is down-regulated during corm dormancy release
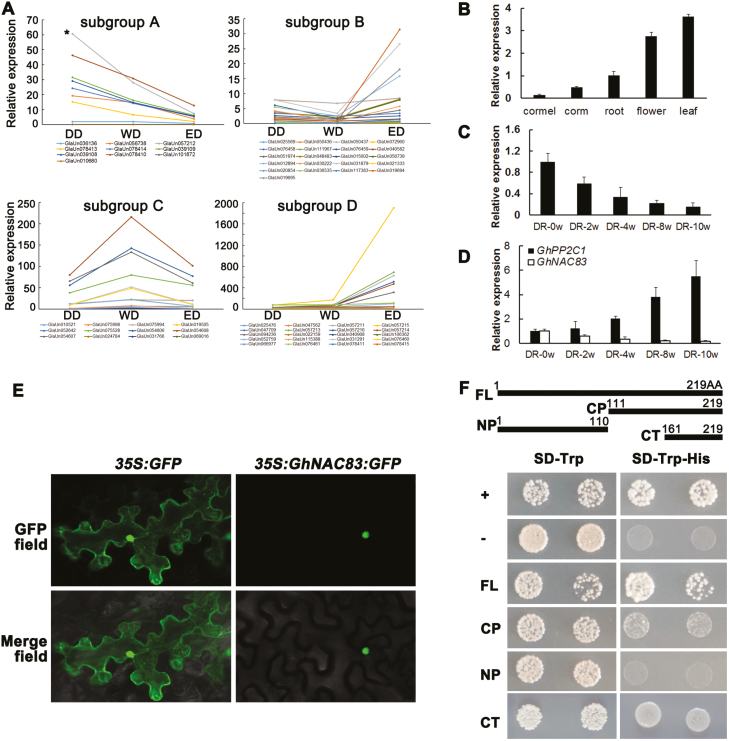
To better understand the function of GhNAC83, we analyzed its sequence and expression patterns. GhNAC83 encodes 219 amino acids with high similarity to NAC83-like in other species and VNI2 (VASCULAR-RELATED NAC-DOMAIN INTERACTING2) in Arabidopsis, and belongs to subgroup II VIII-3 cluster ( Ooka et al., 2003; Jensen et al., 2010) (Supplementary Fig. S4A), containing five conserved domains (A–E) (Supplementary Fig. S4B). In addition, GhNAC83 contains a transcription repressor motif ‘LVFY’ (Puranik et al., 2012; Wang et al., 2017). In our transcriptome database, GhNAC83 is one of 10 GhNACs down-regulated during CDR (Fig. 6A). GlaUn078410 (red line in subgroup A of Fig. 6A) showed a similar trend to GhNAC83, and had high sequence similarity to ATAF1 in Arabidopsis and OsNAC6 in rice. ATAF1 and OsNAC6 have been shown to participate in ABA signaling (Nakashima et al., 2007; Ton et al., 2009).
Fig. 6.
The expression pattern and protein characteristics of GhNAC83. (A) Expression pattern of GhNAC genes in the transcriptome database of Gladiolus CDR. The expression levels are based on a FPKM evaluation. The asterisk (*) represents GhNAC83. (B) The expression of GhNAC83 in different organs during the blooming stage. (C) The expression pattern of GhNAC83 during CDR. (D) The expression pattern of GhNAC83 shows a negative relationship with GhPP2C1 during CDR. Three biological replicates with the SD were performed. (E) Subcellular localization of GhNAC83 in the epidermal cells of N. benthamiana. (F) Transactivation activity of GhNAC83 in yeast. Yeast cells were grown on synthetic dropout nutrient medium lacking Trp (SD-Trp)and Trp/His (SD-Trp-His). DR, dormancy release; FL, full length; CP, C-terminal part; NP, N-terminal part; CT, C-terminus. (This figure is available in color at JXB online.)
At the blooming stage, GhNAC83 had high expression in leaves, flowers, and roots, and had low expression in cormels (Fig. 6B). In addition, the expression of GhNAC83 gradually decreased during the cold storage required for CDR (Fig. 6C). During dormancy release stages, the expression pattern of GhNAC83 was almost opposite to that of GhPP2C1 (Figs 4B, 6D). These results are in accordance with the results in planta which showed that GhNAC83 negatively regulates GhPP2C1 expression during CDR (Fig. 5E–G).
To provide evidence for potential roles of GhNAC83 in transcriptional regulation, we examined the subcellular localization of GhNAC83 in N. benthamiana epidermal cells. The results showed that the GhNAC83–GFP fusion protein localizes to the nucleus (Fig. 6E). Additionally, a transactivation activity assay was performed in yeast to examine the transactivation ability of GhNAC83. On selection medium, yeast colonies harboring pGAL4 (positive control), FL (full length), CP (C-terminal part), or CT (C-terminus) grew, whereas colonies harboring pBD (negative control) or NP (N-terminal part) did not grow (Fig. 6F). These data suggest that GhNAC83 contains a transactivation domain in its C-terminal region between amino acids 161 and 219. GhNAC83 contains a transcriptional repressor domain in the NP (LVFY; amino acids 105–108) in addition to a transactivation domain, suggesting that GhNAC83 is a bifunctional TF, similar to its homologous gene (VNI2) in Arabidopsis (Yang et al., 2011).
Silencing of GhNAC83 accelerates corm dormancy release
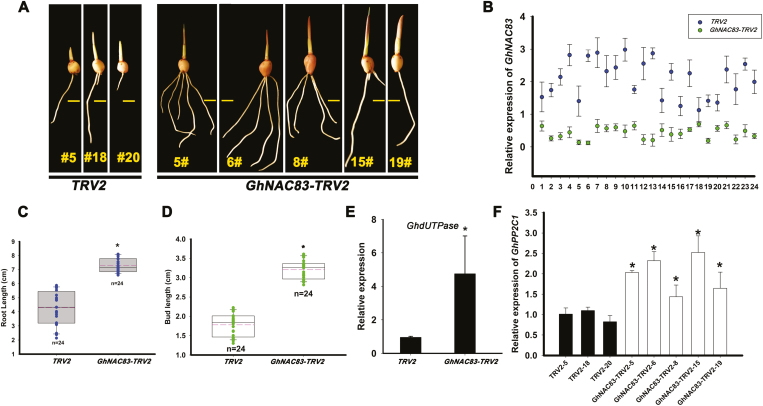
Since there is no report about NAC members participating in corm dormancy, we conducted VIGS in order to evaluate the potential role of GhNAC83 in Gladiolus CDR. Accelerated sprouting occurred when GhNAC83 was silenced in dormant cormels (Fig. 7A, B). Roots and buds of GhNAC83-TRV2 were also significantly longer than those of the TRV2 control (Fig. 7C, D). It has been shown that UTPase is a marker for tuber dormancy release in Solanum tuberosum (Senning et al., 2010; Hartmann et al., 2011). Here, the expression of a CDR marker (GhdUTPase) was dramatically higher in silenced lines than in the control, a result that confirmed the observed phenotype (Fig. 7E). To confirm that silencing of GhNAC83 can affect transcription of GhPP2C1, we determined the expression level of GhPP2C1 in GhNAC83 silenced lines. The results showed that the expression of GhPP2C1 was higher in GhNAC83 silenced lines (Fig. 7F). Together with the binding assay results in Fig. 5, our data suggest that GhNAC83 negatively regulates GhPP2C1 expression and inhibits CDR.
Fig. 7.
Silencing of GhNAC83 accelerates corm dormancy release. (A) The phenotype of GhNAC83-TRV2 independently silenced lines compared with TRV2 control lines 10 d after planting on soil. The scale bar represents 1 cm. (B) The expression of GhNAC83 in 24 independent GhNAC83-TRV2 lines. Data are shown as averages of three technical replicates with the SD. The root length (C) and bud length (D) in GhNAC83-TRV2 and TRV2 lines. Data are shown as averages of n=24 independent lines with the SD. (E) Expression of the dormancy release marker GhdUTPase in GhNAC83-TRV2 and TRV2 lines. (F) Expression of GhPP2C1 in five independent GhNAC8-silenced lines. Data are shown as averages of three technical replicates with the SD (*P<0.05; **P<0.01). (This figure is available in color at JXB online.)
GhNAC83 mediates CK biosynthesis by directly targeting the GhIPT promoter
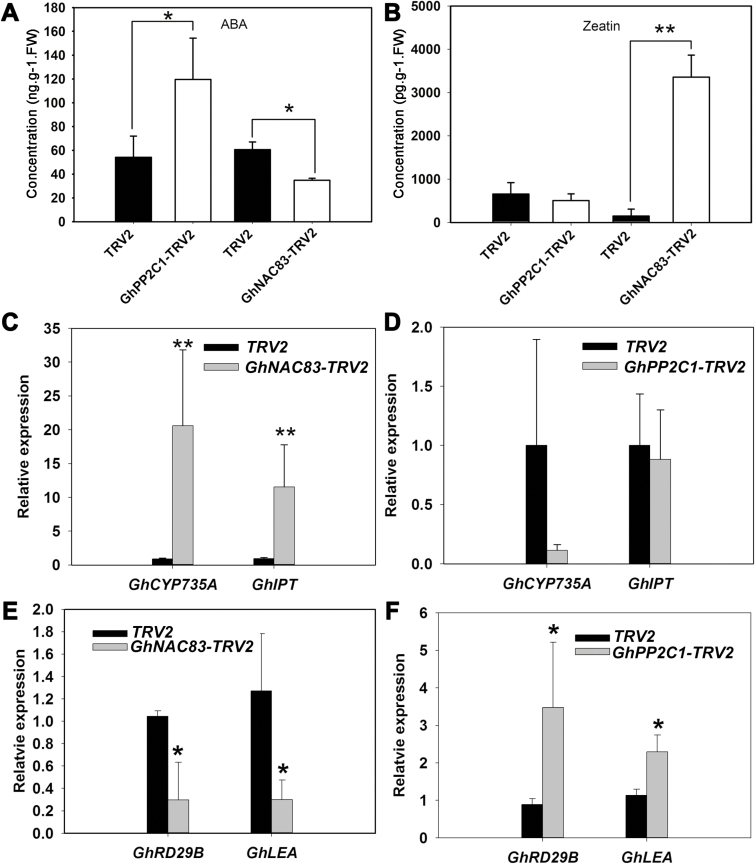
To investigate further how GhPP2C1 and GhNAC83 affect CDR, we measured endogenous phytohormone levels in GhNAC83-silenced cormels. GhNAC83-silenced cormels had half the ABA content and a dramatic increase in zeatin level compared with TRV2 cormels (Fig. 8A, B). This result suggests that GhNAC83 may be involved in the antagonism between ABA and CKs during CDR. In GhPP2C1-TRV2 cormels, zeatin was unaffected compared with TRV2 cormels (Fig. 8B). Moreover, the expression of CK biosynthesis gene homologs (GhCYP735A and GhIPT) was dramatically higher in GhNAC83-TRV2 lines and there was no difference in GhPP2C1-TRV2 lines when compared with TRV2 lines (Fig. 8C, D). Therefore, it is likely that GhNAC83 mediates zeatin biosynthesis during CDR independently of GhPP2C1. As for ABA, silencing of GhNAC83 decreased the expression of genes downstream of ABI5 (GhRD29B and GhLEA) and silencing of GhPP2C1 led to an opposite result (Fig. 8E, F). Overall, these results suggest that GhNAC83 can influence ABA signaling responses through GhPP2C1, and that GhNAC83 affects ABA and zeatin levels in an antagonistic manner during CDR.
Fig. 8.
GhNAC83 mediates CK biosynthesis during CDR in a GhPP2C1-independent manner. (A) ABA levels in GhNAC83- or GhPP2C1-silenced lines. (B) Zeatin levels in GhNAC83- or GhPP2C1-silenced lines. (C) The expression of important CK biosynthesis genes in GhNAC83-silenced lines. (D) The expression of important CK biosynthesis genes in GhPP2C1-silenced lines. (E) Expression of genes downstream from GhABI5 in GhNAC83-silenced lines. (F) Expression of genes downstream from GhABI5 in GhPP2C1-silenced lines. Averages of three biological replicates with the SD are shown (*P<0.05; **P<0.05). Five independent silenced lines and five independent control lines were used.
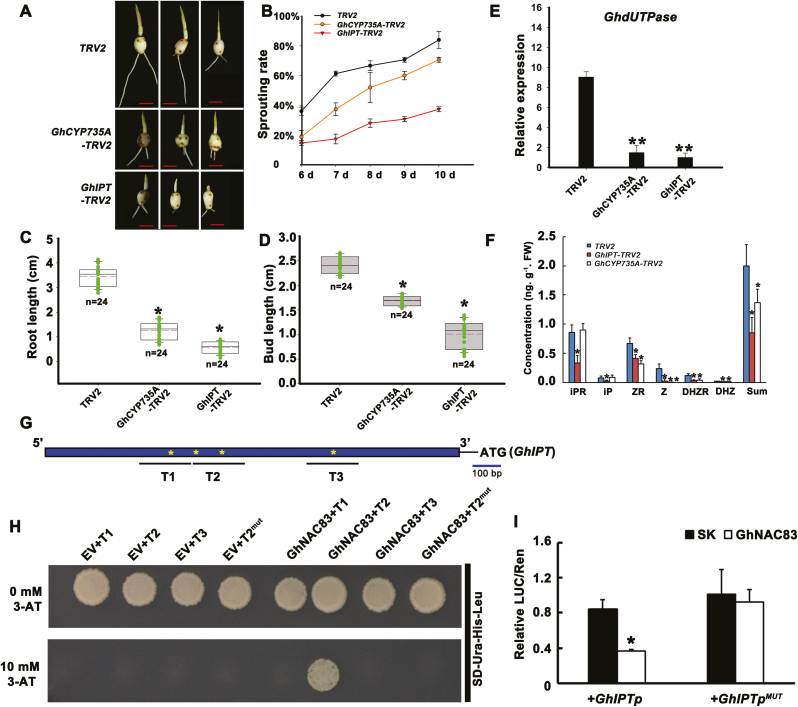
Presently, there is limited information concerning the relationship between CK biosynthesis and CDR. To test the role of CK biosynthesis genes GhCYP735A and GhIPT in CDR, VIGS was employed. The results showed that silencing GhCYP735A or GhIPT in dormant cormels led to delayed sprouting (Fig. 9A, B). The buds and roots of silenced cormels (GhCYP735A and GhIPT) were significantly shorter than those of the TRV2 control (Fig. 9C, D). Moreover, expression of the CDR marker, GhdUTPase, was dramatically lower than that of TRV2 (Fig. 9E). To test whether a decrease in CK levels was indeed responsible for CDR, we measured the levels of active forms of CKs in silenced cormels. We focused on active CKs as they function in CK signal transduction, and they can mainly reflect CK changes in plants (Wang et al., 1995; Yong et al., 2000; Werner et al., 2001; Aoyama and Oka, 2003; Yonekura-Sakakibara et al., 2004; Ashikari et al., 2005; Tao et al., 2010; Matsuo et al., 2012). Silencing of GhCYP735A decreased the levels of zeatin (free and riboside) and dihydrozeatin (free and riboside) in cormels (Fig. 9F). Furthermore, silencing of GhIPT also decreased the level of isopentenyladenosine and isopentenyladenine in cormels (Fig. 9F). These results indicate that silencing CK biosynthesis genes (GhCYP735A and GhIPT) reduces active CK contents in cormels and further inhibits CDR.
Fig. 9.
Silencing of cytokinin biosynthesis genes delays corm dormancy release. (A) Phenotypes associated with silencing of GhCYP735A and GhIPT in dormant cormels 10 d after planting on soil; scale bar=1 cm. (B) The sprouting rate of control (TRV2) and silenced cormels. Cormels were considered sprouted once bud length was longer than 0.5 cm. Averages of three biological replicates with the SD are shown (n=24 independent lines). Root length (C) and bud length (D) of the TRV2 control and GhCYP735A /GhIPT-TRV2 silenced cormels (n=24 independent lines). (E) Transcript levels of the dormancy release marker GhdUTPase in TRV2 and silenced cormels. Data are shown as averages of four independent lines with the SD. (F) Concentration of endogenous CKs in TRV2 and GhCYP735A/GhIPT-TRV2 cormels. iPR, isopentenyladenosine; iP, isopentenyladenine; Z, zeatin, ZR: zeatin riboside; DHZ, dihydrozeatin; DHZR, dihydrozeatin riboside; Sum, total amount of iPR, iP, Z, ZR, DHZ, and DHZR. Data are shown as averages of three independent lines with the SD. (G) Schematic representation of the GhIPT upstream regulatory region. Asterisks (*) correspond to putative NAC-binding sites and the lines indicate the fragments used in the yeast one-hybrid analysis shown in (H). T1, base pairs –1140 to –940; T2, base pairs –910 to –710; T3, base pairs –500 to –300 relative to the translation start site of GhIPT. (H) The interaction of GhNAC83 with the GhIPT promoter in yeast. EV, empty prey vector (pDEST-GAD424). The interaction between protein and promoter was determined by yeast growth on synthetic dropout nutrient medium lacking Ura, His, and Leu, and containing 10mM 3-AT. The mutagenesis of NAC-binding sites (Supplementary Fig. S5) in the GhIPT promoter (T2mut) was also tested. (I) The interaction of GhNAC83 with the GhIPT promoter using a dual-luciferase reporter assay in N. benthamiana leaves. A fragment of the GhIPT promoter (base pairs +56 to –1537) was used in this assay, and mutant sites in GhIPTpMUT are shown in Supplementary Fig. S5. The empty vector (pGreenII 62-SK) was used as a control. Data are shown as the average of three biological replicates with the SD (n=5 leaves) (*P<0.05 and **P<0.01). (This figure is available in color at JXB online.)
To determine whether GhNAC83 can directly regulate the expression of CK biosynthesis genes, we cloned a 1594 bp upstream sequence of GhIPT by Hi-TAIL PCR. There are four predicted NAC-binding sites in the GhIPT promoter (Fig. 9G). Using a yeast one-hybrid approach, we found that GhNAC83 binds the T2 fragment (base pairs –910 to –710) of the GhIPT promoter, while mutation of NAC-binding sites in the T2 fragment (Supplementary Fig. S5) resulted in no binding ability (Fig. 9H). In addition, we performed a LUC reporter assay to analyze the regulation of GhIPT expression by GhNAC83 in planta (Fig. 9I). Nicotiana benthamiana leaf cells co-transformed with 35S:GhNAC83 and GhIPT:LUC exhibited significantly lower LUC activity than cells transformed with empty vector and GhIPT:LUC. The activity of the GhIPT promoter with mutated NAC-binding sites in the T2 region (GhIPTpMUT) was no longer significantly different between treatments with empty vector and 35S:GhNAC83 (Fig. 9I). These data suggest that GhNAC83 negatively regulates GhIPT in planta.
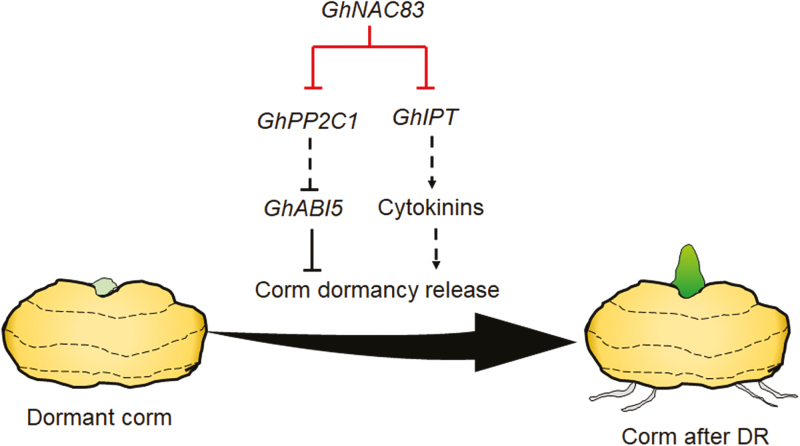
Together, our results demonstrate that GhNAC83 directly binds the GhIPT promoter and negatively regulates CK biosynthesis in a GhPP2C1-independent manner to control CDR in Gladiolus (Fig. 10).
Fig. 10.
Diagram of GhNAC83 in regulating corm dormancy release in Gladiolus. GhNAC83 directly binds GhPP2C1 and GhIPT promoters and represses their expression, modulating ABA signaling and CK biosynthesis during CDR. (This figure is available in color at JXB online.)
Discussion
In this study, we identified a novel NAC family member in Gladiolus, GhNAC83, and characterized its negative regulatory role in CDR. GhNAC83 expression decreases during CDR. GhNAC83 directly binds to and inhibits GhPP2C1 expression, thereby activating ABA downstream response, and additionally GhNAC83 inhibits CK biosynthesis through direct binding and down-regulation of GhIPT, thus delaying CDR (Fig. 10). Accordingly, silencing of GhNAC83 in dormant cormels leads to a higher zeatin content and lower ABA levels, thereby promoting CDR, while silencing of GhPP2C1 results in delayed CDR by enhancing ABA downstream response. Altogether, the data shown here indicate that GhNAC83 regulates ABA–CK crosstalk to inhibit CDR.
GhPP2C1 promotes CDR
Plant dormancy is a complex trait and is regulated by several phytohormones, with ABA being a central player (Finkelstein et al., 2008). In Arabidopsis, PP2Cs are sorted into 10 subgroups (A–J) and have been shown to play a role in diverse signaling pathways related to plant development and stress responses (Kerk et al., 2002). Members of subgroup A, including ABI1/2 and HAB1/2, are co-receptors of ABA signaling, and regulate seed germination and abiotic stress (Gosti et al., 1999; Kong et al., 2015).
Here, we isolated GhPP2C1, which belongs to subgroup A and shares high sequence similarities with HAB1 and HAB2 (Supplementary Fig. S2), in addition to conserved motifs, such as the PYL interaction site, PA (phosphatidic acid)-binding site, metal contact points, and a nuclear localization signal (NLS)-like motif at the C-terminus (Supplementary Fig. S6) (Moes et al., 2008; Zhang et al., 2014). GhPP2C1 was differentially expressed in a transcriptomic analysis of Gladiolus during CDR. The transcription of GhPP2C1 increases during CDR in Gladiolus, and further functional analysis showed that silencing of GhPP2C1 results in delayed CDR by enhancing ABA downstream response (Fig. 8F). Together with the transcriptome analysis data (Supplementary Table S3), our results present a role for the clade A PP2C, GhPP2C1, as a positive regulator of CDR.
GhNAC83 plays a role in ABA–CK crosstalk to inhibit CDR
Yeast one-hybrid screening is widely used for the identification of TFs that bind a specific cis-element in the promoter of a gene of interest. Also, employing this technique allows us to use a TF-specific library which is much more convenient than a traditional cDNA library given that it reduces false positives, enriches full-length TFs, and overall has higher efficiency (Mitsuda et al., 2010). Therefore, we performed yeast one-hybrid screening with an Arabidopsis TF library and identified homologs in Gladiolus from these results. We then confirmed the results by performing yeast one-hybrid analysis with the homologous TFs, proving the interaction with the GhPP2C1 promoter. One TF, GhNAC83, had the highest affinity for the GhPP2C1 promoter, and further analysis by transient reporter activation assays showed that GhNAC83 acts as a negative regulator of GhPP2C1 (Fig. 5E–G). These data are in accordance with the expression of GhPP2C1 in GhNAC83-silenced cormels (Fig. 7F).
NACs are a large TF family in plants and are associated with diverse processes. Despite their highly conserved DNA-binding domains, the remarkable diversification across plant species reflects their numerous functions (Puranik et al., 2012). Here, we identified a novel NAC member, GhNAC83, which has high similarity to NAC83 from Asparagus officinalis. Similar to its homologous protein (VNI2) in Arabidopsis, which functions in integrating ABA signaling into leaf senescence via the COLD-REGULATED (COR)/RD genes (Yang et al., 2011), GhNAC83 contains a transactivation domain in its C-terminal region and is localized to the nucleus (Fig. 6E, F). Further study of GhNAC83 showed that GhNAC83 is down-regulated during CDR and has an opposite expression pattern to GhPP2C1 (Fig. 6C, D). Silencing GhNAC83 results in earlier CDR with longer buds and roots (Fig. 7A, C, D), showing that NAC family members also contribute to the process of CDR in plants.
Previous studies have shown that NACs modulate drought stress, oxidative stress, and leaf senescence by regulating the expression of some PP2C family members (Zhang and Gan, 2012; You et al., 2014). In particular, in rice, SNAC1 (STRESS-RESPONSIVE NAC1) directly regulates OsPP18 (PROTEIN PHOSPHATASE18) and confers drought and oxidative stress tolerance by regulating ROS (reactive oxygen species) homeostasis through ABA-independent pathways. Although OsPP18 belongs to the PP2C family (subgroup F), ABA response genes and ABA sensitivity were not affected in the ospp18 mutant (You et al., 2014). In this study, we found that GhNAC83 negatively regulates the expression of GhPP2C1 (Fig. 7F) and up-regulates the expression of ABA-responsive genes (GhRD29B and GhLEA; Fig. 8E), indicating that GhNAC83 regulates CDR in an ABA-dependent pathway.
Previous research has shown that some NAC family members participate in ABA pathways, as explained above, and some NAC family members participate in CK pathways, such as NTM1, which is activated by proteolytic cleavage through regulated intramembrane proteolysis and tightly mediates CK signaling during cell division in Arabidopsis (Kim et al., 2006). In this study, we show that GhNAC83 is involved in both ABA (above) and CK pathways. GhNAC83 is a nuclear protein that negatively regulates GhIPT expression, inhibiting CK biosynthesis and resulting in partial repression of CDR. Given the large size of the NAC TF family, it will be interesting in the future to test if different NACs can integrate different environmental and endogenous signals to regulate growth rates in cormels and other organs by balancing ABA and CK levels and signaling.
Corm and seed dormancy release
Corm and seed dormancy release are two processes with similarities and differences. Seed dormancy release is regulated by two major hormones: ABA and GA (Finch-Savage and Leubner-Metzger, 2006). On the other hand, Gladiolus corm dormancy release is regulated by CKs and ABA. Moreover, previous research has shown that GA is not an essential hormone in promoting CDR in Gladiolus (Ginzburg, 1973). This research is in accordance with our transcriptome analysis, where we showed that GA-related DEGs are not in the top three of hormone metabolism-related DEG abundance (Supplementary Fig. S1C, D). Instead, ABA- and CK-related DEGs are enriched, suggesting that CKs may play a more prominent role than GA in Gladiolus CDR, and not GA, but the molecular mechanism is still largely unknown (Ginzburg, 1973; Wu et al., 2015). Another difference in corm and seed dormancy is that corms lack seed coats and an endosperm; therefore, due to these structural differences, corms do not undergo coat and endosperm dormancy as seeds do. Thus, factors related to coat or endosperm dormancy do not affect corm dormancy (Finch-Savage and Leubner-Metzger, 2006). Given that hormone crosstalk plays a major role in regulating seed dormancy, with most hormones contrasting the inhibitory role of ABA (Gazzarrini and Tsai, 2015; Shu et al., 2016), it will be interesting in the future to characterize the interaction between ABA, CK, and other hormones such as auxin in Gladiolus CDR.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primer sequences used in this study.
Table S2. Overlapping differentially expressed genes in deep, weak, and ecodormancy.
Table S3. List of differentially expressed cytokinin- and ABA-related genes.
Fig. S1. Gene Ontology (GO) enrichment analysis.
Fig. S2. Alignment of GhPP2C1 with the PP2C family in Arabidopsis.
Fig. S3 Predicted transcription factor-binding sites in the GhPP2C1 promoter (–833 to –615 bp).
Fig. S4. Phylogenetic analysis of GhNAC83.
Fig. S5. Mutagenesis of NAC-binding sites in the GhIPT promoter.
Fig. S6. Sequence alignment of GhPP2C1 with its homologs.
Acknowledgements
We thank Dr Nobutaka Mitsuda (Bioproduction Research Institute, Japan) for providing the transcription factor cDNA library and plasmids (pDEST-GAD424 and pDEST-HISi-2). Dr Jumi A. Shin (University of Toronto Mississauga, Canada) for kindly supporting yeast strain YM4271. Dr Junping Gao (China Agricultural University, China) for supporting the dual-luciferase plasmids (pGreenII 62-SK and pGreenII LUC), and undergraduate student Sebastian Dowhanik (University of Toronto Scarborough, Canada) for editing and revising this manuscript. This work was funded by National Natural Science Foundation projects (grants 31701952 to JW, 31300219 to JH, and 31171991 to MY) and the National Science and Engineering Research Council of Canada (NSERC) to SG. EV was supported by an NSERC-PGSD, JW was supported by China Scholarship Council and China Postdoctoral Council scholarships.
References
- Aoyama T, Oka A. 2003. Cytokinin signal transduction in plant cells. Journal of Plant Research 116, 221–231. [DOI] [PubMed] [Google Scholar]
- Antoniadi I, Plačková L, Simonovik B, Doležal K, Turnbull C, Ljung K, Novák O. 2015. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. The Plant Cell 27, 1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production. Science 309, 741–745. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozhkov PV, Lebedenko LA, Shiryaeva GA. 1992. A pronounced synergistic effect of abscisic acid and 6-benzyladenine on Norway spruce (Picea abies L. Karst) somatic embryo maturation. Plant Cell Reports 11, 386–389. [DOI] [PubMed] [Google Scholar]
- Cao L, Yu Y, Ding X, Zhu D, Yang F, Liu B, Sun X, Duan X, Yin K, Zhu Y. 2017. The Glycine soja NAC transcription factor GsNAC019 mediates the regulation of plant alkaline tolerance and ABA sensitivity. Plant Molecular Biology 95, 253–268. [DOI] [PubMed] [Google Scholar]
- Chen HY, Hsieh EJ, Cheng MC, Chen CY, Hwang SY, Lin TP. 2016. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytologist 211, 599–613. [DOI] [PubMed] [Google Scholar]
- Chen W, Gai Y, Liu S, Wang R, Jiang X. 2010. Quantitative analysis of cytokinins in plants by high performance liquid chromatography:electronspray ionization ion trap mass spectrometry. Journal of Integrative Plant Biology 52, 925–932. [DOI] [PubMed] [Google Scholar]
- Chen W, Yin X, Wang L, Tian J, Yang R, Liu D, Yu Z, Ma N, Gao J. 2013. Involvement of rose aquaporin RhPIP1;1 in ethylene-regulated petal expansion through interaction with RhPIP2;1. Plant Molecular Biology 83, 219–233. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. 2010. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Developmental Cell 19, 284–295. [DOI] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC. 2016. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Research 44, D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, Ok SH, Yoo SD, Shin JS. 2013. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Letters 587, 1773–1778. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Vermeirssen V, Arda HE, Martinez NJ, Walhout AJ. 2006. Gateway-compatible yeast one-hybrid screens. Cold Spring Harbor Protocols doi: 10.1101/pdb.prot4590. [DOI] [PubMed] [Google Scholar]
- Dong H, Niu Y, Li W, Zhang D. 2008. Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. Journal of Experimental Botany 59, 1295–1304. [DOI] [PubMed] [Google Scholar]
- Farrow SC, Emery RN. 2012. Concurrent profiling of indole-3-acetic acid, abscisic acid, and cytokinins and structurally related purines by high-performance-liquid-chromatography tandem electrospray mass spectrometry. Plant Methods 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsai AY. 2015. Hormone cross-talk during seed germination. Essays in Biochemistry 58, 151–164. [DOI] [PubMed] [Google Scholar]
- Ginzburg C. 1973. Hormonal regulation of cormel dormancy in Gladiolus grandiflorus. Journal of Experimental Botany 24, 558–566. [Google Scholar]
- Ginzburg C. 1981. Metabolic changes in gladiolus cormels during the break of dormancy: the role of dark CO2 fixation. Plant Physiology 68, 1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J. 1999. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, et al. . 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. 2005. BioEdit version 7.0.0.Raleigh, NC: Department of Microbiology, North Carolina State University. [Google Scholar]
- Hartmann A, Senning M, Hedden P, Sonnewald U, Sonnewald S. 2011. Reactivation of meristem activity and sprout growth in potato tubers require both cytokinin and gibberellin. Plant Physiology 155, 776–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. 2007. Gene splicing and mutagenesis by PCR-driven overlap extension. Nature Protocols 2, 924–932. [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. 2010. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes & Development 24, 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K. 2010. The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. The Biochemical Journal 426, 183–196. [DOI] [PubMed] [Google Scholar]
- Jensen MK, Lindemose S, de Masi F, et al. . 2013. ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3, 321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Lu D, Sun JH, Li CL, Xing Y, Qin L, Shen YY. 2013. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. Journal of Experimental Botany 64, 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. 2002. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiology 129, 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. 2006. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. The Plant Cell 18, 3132–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Park AH, Lerouxel O, Han KH. 2007. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. The Plant Journal 50, 1035–1048. [DOI] [PubMed] [Google Scholar]
- Kong L, Cheng J, Zhu Y, et al. . 2015. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nature Communications 6, 8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nard M. 1993. The physiology of flower bulbs: a comprehensive treatise on the physiology and utilization of ornamental flowering bulbous and tuberous plants. Amsterdam: Elsevier. [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Chen Y. 2007. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43, 649–650, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Luo X, Yi J, Zhong XH, et al. . 2012. Cloning, characterization and expression analysis of key genes involved in ABA metabolism in Gladiolus cormels during storage. Scientia Horticulturae 143, 115–121. [Google Scholar]
- Ma QJ, Sun MH, Lu J, Liu YJ, Hu DG, Hao YJ. 2017. Transcription factor AREB2 is involved in soluble sugar accumulation by activating sugar transporter and amylase genes. Plant Physiology 174, 2348–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M. 2010. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant & Cell Physiology 51, 2145–2151. [DOI] [PubMed] [Google Scholar]
- Matsuo S, Kikuchi K, Fukuda M, Honda I, Imanishi S. 2012. Roles and regulation of cytokinins in tomato fruit development. Journal of Experimental Botany 63, 5569–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moes D, Himmelbach A, Korte A, Haberer G, Grill E. 2008. Nuclear localization of the mutant protein phosphatase abi1 is required for insensitivity towards ABA responses in Arabidopsis. The Plant Journal 54, 806–819. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal 51, 617–630. [DOI] [PubMed] [Google Scholar]
- Née G, Kramer K, Nakabayashi K, Yuan B, Xiang Y, Miatton E, Finkemeier I, Soppe WJJ. 2017. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nature Communications 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, et al. . 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Research 10, 239–247. [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. 2008. Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography–electrospray tandem mass spectrometry. Phytochemistry 69, 1773–1781. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. . 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S, Sahu PP, Srivastava PS, Prasad M. 2012. NAC proteins: regulation and role in stress tolerance. Trends in Plant Science 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H. 2006. Cytokinins: activity, biosynthesis, and translocation. Annual Review of Plant Biology 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N. 2011. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. Journal of Experimental Botany 62, 2615–2632. [DOI] [PubMed] [Google Scholar]
- Senning M, Sonnewald U, Sonnewald S. 2010. Deoxyuridine triphosphatase expression defines the transition from dormant to sprouting potato tuber buds. Molecular Breeding 26, 525–531. [Google Scholar]
- Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, Kim J, Park CM. 2010. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. The Plant Journal 61, 661–671. [DOI] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Lu WJ, Chen JY. 2014. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant, Cell & Environment 37, 2116–2127. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH. 2016. Two faces of one seed: hormonal regulation of dormancy and germination. Molecular Plant 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Sonnewald S, Sonnewald U. 2014. Regulation of potato tuber sprouting. Planta 239, 27–38. [DOI] [PubMed] [Google Scholar]
- Subbaraj AK, Funnell KA, Woolley DJ. 2010. Dormancy and flowering are regulated by the reciprocal interaction between cytokinin and gibberellin in Zantedeschia. Journal of Plant Growth Regulation 29, 487–499. [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. 2004. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. Journal of Biological Chemistry 279, 41866–41872. [DOI] [PubMed] [Google Scholar]
- Tao G-Q, Letham DS, Yong JWH, Zhang K, John PCL, Schwartz O, Wong SC, Farquhar GD. 2010. Promotion of shoot development and tuberisation in potato by expression of a chimaeric cytokinin synthesis gene at normal and elevated CO2 levels. Functional Plant Biology 37, 43–54. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkowská D, Strnad M. 2018. Isoprenoid-derived plant signaling molecules: biosynthesis and biological importance. Planta 247, 1051–1066. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Research 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. 2009. The multifaceted role of ABA in disease resistance. Trends in Plant Science 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Valdés AE, Overnäs E, Johansson H, Rada-Iglesias A, Engström P. 2012. The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Molecular Biology 80, 405–418. [DOI] [PubMed] [Google Scholar]
- Verslues PE. 2016. ABA and cytokinins: challenge and opportunity for plant stress research. Plant Molecular Biology 91, 629–640. [DOI] [PubMed] [Google Scholar]
- Wang J, Letham D, Taverner E, Badenoch-Jones J, Hocart C. 1995. A procedure for quantification of cytokinins as free bases involving scintillation proximity immunoassay. Physiologia Plantarum 95, 91–98. [Google Scholar]
- Wang L, Li Z, Lu M, Wang Y. 2017. ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic tamarix and Arabidopsis. Frontiers in Plant Science 8, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Song M, Guo J, Huang Y, Zhang F, Xu C, Xiao Y, Zhang L. 2018. The potassium channel FaTPK1 plays a critical role in fruit quality formation in strawberry (Fragaria×ananassa). Plant Biotechnology Journal 16, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo C, Peng J, et al. . 2018. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytologist 221, 341–355. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ye T, Zhao S, Liu Z, Feng YQ, Wu Y. 2011. Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. The Plant Journal 68, 249–261. [DOI] [PubMed] [Google Scholar]
- Wei Q, Ma C, Xu Y, Wang T, Chen Y, Lü J, Zhang L, Jiang CZ, Hong B, Gao J. 2017. Control of chrysanthemum flowering through integration with an aging pathway. Nature Communications 8, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmülling T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell 15, 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Strnad M, Schmulling T. 2001. Regulation of plant growth by cytokinin. Proceedings of the National Academy of Sciences, USA 98, 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Seng S, Sui J, et al. . 2015. Gladiolus hybridus ABSCISIC ACID INSENSITIVE 5 (GhABI5) is an important transcription factor in ABA signaling that can enhance Gladiolus corm dormancy and Arabidopsis seed dormancy. Frontiers in Plant Science 6, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ma N, Jia Y, Zhang Y, Feng M, Jiang C-Z, Ma C, Gao J. 2016. An ethylene-induced regulatory module delays flower senescence by regulating cytokinin content. Plant Physiology 173, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng Z, Lai J, et al. . 2009. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Research 19, 1279–1290. [DOI] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon do Y, et al. . 2013. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. The Plant Cell 25, 4708–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SD, Seo PJ, Yoon HK, Park CM. 2011. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. The Plant Cell 23, 2155–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. 2004. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiology 134, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong JW, Wong SC, Letham DS, Hocart CH, Farquhar GD. 2000. Effects of elevated [CO2] and nitrogen nutrition on cytokinins in the xylem sap and leaves of cotton. Plant Physiology 124, 767–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zong W, Hu H, Li X, Xiao J, Xiong L. 2014. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiology 166, 2100–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Fu X, Lv Z, Shen Q, Yan T, Jiang W, Wang G, Sun X, Tang K. 2014. Type 2C phosphatase 1 of Artemisia annua L. is a negative regulator of ABA signaling. BioMed Research International 2014, 521794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Gan SS. 2012. An abscisic acid–AtNAP transcription factor–SAG113 protein phosphatase 2C regulatory chain for controlling dehydration in senescing Arabidopsis leaves. Plant Physiology 158, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Berkowitz O, Teixeira da Silva JA, Zhang M, Ma G, Whelan J, Duan J. 2015. RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album. Frontiers in Plant Science 6, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang ML, Ma TL, Wang Y. 2016. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. The Plant Cell 28, 3005–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhao L, Gao J, Fei Z. 2011. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Yuan X, Wu Z, et al. . 2014. Virus-induced gene silencing for comparative functional studies in Gladiolus hybridus. Plant Cell Reports 33, 301–312. [DOI] [PubMed] [Google Scholar]
- Zubo YO, Yamburenko MV, Selivankina SY, et al. . 2008. Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiology 148, 1082–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.