We provide new insights into the autoregulation of nodulation that allow us to better understand how the legume Medicago truncatula restricts further nodulation once enough nodules have been formed.
Keywords: Autoregulation of nodulation (AON), CLAVATA signaling peptide, F-box protein, Nod factor perception (NFP), rhizobia, symbiotic nodulation
Abstract
The number of legume root nodules resulting from a symbiosis with rhizobia is tightly controlled by the plant. Certain members of the CLAVATA3/Embryo Surrounding Region (CLE) peptide family, specifically MtCLE12 and MtCLE13 in Medicago truncatula, act in the systemic autoregulation of nodulation (AON) pathway that negatively regulates the number of nodules. Little is known about the molecular pathways that operate downstream of the AON-related CLE peptides. Here, by means of a transcriptome analysis, we show that roots ectopically expressing MtCLE13 deregulate only a limited number of genes, including three down-regulated genes encoding lysin motif receptor-like kinases (LysM-RLKs), among which are the nodulation factor (NF) receptor NF Perception gene (NFP) and two up-regulated genes, MtTML1 and MtTML2, encoding Too Much Love (TML)-related Kelch-repeat containing F-box proteins. The observed deregulation was specific for the ectopic expression of nodulation-related MtCLE genes and depended on the Super Numeric Nodules (SUNN) AON RLK. Moreover, overexpression and silencing of these two MtTML genes demonstrated that they play a role in the negative regulation of nodule numbers. Hence, the identified MtTML genes are the functional counterpart of the Lotus japonicus TML gene shown to be central in the AON pathway. Additionally, we propose that the down-regulation of a subset of LysM-RLK-encoding genes, among which is NFP, might contribute to the restriction of further nodulation once the first nodules have been formed.
Introduction
Under nitrogen-limited conditions, legume plants interact with bacteria, rhizobia, to form symbiotic root nodules, in which the rhizobia fix nitrogen for the benefit of the plant (Oldroyd, 2013; Suzaki et al., 2015; Gamas et al., 2017). Nodulation is initiated through a chemical signal exchange between both symbiotic partners. After the host-derived flavonoids are perceived, the rhizobia secrete lipo-chitooligosaccharides, designated nodulation factors (NFs), which are sensed by the plant root epidermis to activate downstream symbiotic signaling (Fliegmann and Bono, 2015). In the model legume, Medicago truncatula (barrel medic), mutation of the lysin motif (LysM) receptor-like kinase (RLK) gene NF Perception (NFP) abolished nodulation from the earliest initiation stage onward and eliminated root hair curling and activation of (pre-)infection markers, such as Early NODulin 11 (ENOD11) (Ben Amor et al., 2003). Additionally, a related protein, LysM domain RLK 3 (LYK3), has been shown to be involved in NF perception, because its down-regulation by RNAi inhibited early nodulation (Limpens et al., 2003). Also in Lotus japonicus, two receptor proteins, NF receptor 5 (LjNFR5) and LjNFR1, that are related to MtNFP and MtLYK3, respectively, control the NF signaling (Madsen et al., 2003; Radutoiu et al., 2003). LYK3 and NFR1 are functional kinases that can form a complex with the NFR5/NFP non-functional kinases (Fliegmann and Bono, 2015). Moreover, knock-down of MtLYK4 also affects rhizobial infections, hinting at a functional redundancy between MtLYK3 and MtLYK4 for nodulation initiation (Limpens et al., 2003). In L. japonicus, direct binding of NFs on an NFR5/NFR1 heterodimeric complex was reported. An additional LysM-RLK gene, the high-affinity lipo-chitooligosaccharide-binding protein 3 gene (LYR3), was detected as a high-affinity NF-binding protein that interacts with MtLYK3, although the lyr3 mutants did not reveal any nodulation phenotype (Fliegmann et al., 2013, 2016; Fliegmann and Bono, 2015). Other LysM-RLKs have been connected with early nodulation: the ExoPolysaccharide Receptor 3 (LjEPR3) that is linked to the perception of another type of rhizobial signals, the exopolysaccharides, and that acts downstream of NFR1/NFR5 (Kawaharada et al., 2017) and the epidermal LysM receptor (NFRe) that has been proposed to maintain the response of epidermal cells to rhizobia along the expanding root system (Murakami et al., 2018). In M. truncatula as well as in other legumes, several LysM-RLK-encoding genes exist, the potential functions of which in relation to symbiotic nodulation remain to be established.
Nitrogen-fixing symbiotic nodulation is an adaptation of legume plants to nitrogen-starved conditions and is, therefore, tightly controlled by the host plant. One of the negative regulatory pathways that limits the nodule formation dependent on the metabolic status of the shoot (carbon) and root (nitrogen) is the long-distance (systemic) autoregulation of nodulation (AON) pathway (Suzaki et al., 2015). Early after rhizobial infection, the transcription of a subset of genes encoding CLAVATA3/Embryo Surrounding Region (CLE) peptides, the so-called MtCLE12 and MtCLE13 in M. truncatula, is triggered to activate the AON (Mortier et al., 2010). Similarly, in L. japonicus, the LjCLE-root signaling (RS)1, LjCLE-RS2, and LjCLE-RS3 peptide-encoding genes are induced by rhizobia and/or nitrogen (Okamoto et al., 2009; Nishida et al., 2016). Interestingly, these genes are orthologous to MtCLE12 and MtCLE13, as well as to the rhizobia-induced CLE 1 (RIC1) and RIC2 peptide-encoding genes in Glycine max (soybean) and Phaseolus vulgaris (common bean) (Reid et al., 2011; Lim et al., 2011; Hastwell et al., 2017). CLE peptides are 12–13 amino acids long and are secreted as signaling peptides derived from the C-terminal region of pre-proproteins that have been shown to act as non-cell-autonomous signals in various developmental contexts (Araya et al., 2016; Yamaguchi et al., 2016). In Arabidopsis thaliana (thale cress), their function is mostly associated with the regulation of cell proliferation and differentiation during plant development, notably in the shoot and root apical meristems and in the cambium meristem in relation to tracheary element differentiation. The MtCLE12, but not MtCLE13, peptides are tri-arabinosylated, possibly by an enzyme from the hydroxyproline O-arabinosyltransferase (HPAT) family encoded by the M. truncatula Root Determined Nodulation 1 (RDN1) gene, mutation of which leads to excessive nodulation (Schnabel et al., 2011; Kassaw et al., 2017). The negative effect of AON-related CLE peptides on the nodule number relies on a shoot-acting leucine-rich repeat (LRR) RLK, designated Super Numeric Nodules (SUNN) in M. truncatula, Hypernodulation Aberrant Root Formation (HAR1) in L. japonicus, Nodule Autoregulation Receptor Kinase (NARK) in soybean, and Symbiosis29 (SYM29) in Pisum sativum (pea) (Krusell et al., 2002; Searle et al., 2003; Schnabel et al., 2005; Suzaki et al., 2015). A transcript profiling analysis of inoculated and uninoculated nark soybean leaves revealed a differential expression of the jasmonic acid biosynthesis and response genes (Kinkema and Gresshoff, 2008), suggesting that a shoot-specific down-regulation of the jasmonic acid response genes by rhizobial inoculation might mediate the AON, at least in soybean. The AON may involve a SUNN-dependent regulation of the long-distance shoot-to-root polar auxin transport in M. trunculata (van Noorden et al., 2006) and the down-regulation of a specific mobile miRNA in L. japonicus (Tsikou et al., 2018).
Regarding the downstream targets of the MtCLE12/MtCLE13–MtSUNN pathway in roots, only very few fragmentary data are currently available (Schnabel et al., 2010; Suzaki et al., 2015). Therefore, there is a real need to expand knowledge of the downstream effectors of the AON pathway. In M. truncatula, the systemic AON pathway has been shown possibly to inhibit the NF signaling pathway, because the early nodulation marker MtENOD11 was no longer activated in roots ectopically expressing MtCLE13 after rhizobial inoculation (Mortier et al., 2010). In contrast, in L. japonicus, the AON might act on nodule organogenesis (Takahara et al., 2013). Genetic analyses in L. japonicus revealed that the too much love (tml) mutant is affected in a gene acting downstream of the LjCLE–RS/LjHAR1 pathway in roots (Magori et al., 2009; Takahara et al., 2013). The LjTML gene encodes a Kelch-repeat F-box protein that is probably involved in the targeted ubiquitin-dependent proteolysis of still unknown nuclear proteins that are expected to be critical for early nodulation. Recently, the TML transcript level has been shown to be controlled in roots by a shoot-derived systemic miRNA, the miR2111, the expression of which is up-regulated during nodulation in an LjHAR1-dependent manner (Tsikou et al., 2018), but whether the CLE RS1/RS2 expression affects the TML expression is still unknown.
To investigate the downstream molecular pathways activated by AON-related CLE peptides in M. truncatula, we analyzed the transcriptome of roots ectopically expressing MtCLE13. MtCLE13 was selected because its induction during nodulation occurs earlier than that of MtCLE12 (Mortier et al., 2010). We show that only a limited set of root genes were differentially regulated, including NFP and some LysM-RLK-related genes, as well as two TML orthologs. Together, these results suggest that in M. truncatula AON-related CLE peptides act through the root activity of TML F-box proteins and could inhibit nodule formation via the down-regulation of genes involved in NF perception, such as NFP and other related LysM-RLK genes.
Materials and methods
Biological material
Medicago truncatula Gaertn. cv Jemalong A17, the pNFP:GUS stable transgenic line (Arrighi et al., 2006), and the sunn-4 mutant (Sagan et al., 1995; Schnabel et al., 2005) were grown and inoculated as described (Mergaert et al., 2003). The Sinorhizobium meliloti Sm1021 strain and the Agrobacterium rhizogenes Arqua1 strain were grown at 28 °C in a yeast extract broth (YEB) medium supplemented with 50 mg l–1 streptomycin.
For the quantitative reverse transcription–PCR (qRT–PCR) analysis, plants were grown in vitro in square Petri dishes (12×12 cm) on a low nitrogen ‘i’ agar medium (0.125 mM KNO3; Blondon, 1964). For the nodulation kinetics, nodules were harvested 1–15 d after inoculation with S. meliloti from plants grown in vitro on nitrogen-poor ‘i’ medium. The symbiotic rhizobia-responsive zone, located above the root tip, was isolated from uninoculated roots and used as control. For the Agrobacterium rhizogenes-mediated transgenic root transformation, nitrogen-deprived Fahraeus medium was used (0.01 mM KNO3; Truchet et al., 1989).
Cloning procedures
The 35S:MtCLE4, 35S:MtCLE12, and 35S:MtCLE13 vectors were generated as described (Mortier et al., 2010). The ORFs of MtTML1 and MtTML2 were amplified from M. truncatula genomic DNA. Primers containing the attB sequences at their 5' end were used for amplification of Gateway cloning ready products (see Supplementary Table S1 at JXB online) into the pDONR207 vector by means of the Gateway BP recombinase (Invitrogen, Carlsbad, CA, USA). After verification by sequencing, constructs were transferred via an LR recombinase reaction into the pK7WG2D binary vector (Karimi et al., 2002), allowing the expression of the MtTML1 or MtTML2 genes under the control of the Cauliflower mosaic virus 35S promoter.
RNAi constructs were designed to target both the MtTML1 and MtTML2 genes within the region that is the most conserved at the nucleotide level between the two genes. The MtTML1 and MtTML2 RNAi constructs were amplified with primers specific for the MtTML1 or MtTML2 genes, respectively (Supplementary Table S1). These PCR products were cloned with the Gateway technology in the pENTR/D-TOPO vector (Thermo Scientific, Waltham, MA, USA) and then in the pFRN destination vector (Gonzalez-Rizzo et al., 2006). The final RNAi constructs were sequenced for validation. The β-glucuronidase (GUS) RNAi control vector has been published previously (Gonzalez-Rizzo et al., 2006).
RNA extraction, cDNA synthesis, and qRT–PCR analysis
Total RNA was isolated with the RNeasy Plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. After a DNase treatment, samples were purified by NH4Ac (5 M) precipitation, quality controlled, and quantified with a Nanodrop spectrophotometer (Isogen, Hackensack, NJ, USA). RNA (2 μg) was used for cDNA synthesis with the Superscript Reverse Transcriptase Kit (Invitrogen). The qRT–PCR experiments were done on a LightCycler 480 (Roche Diagnostics, Brussels, Belgium), and SYBR Green was used for detection. Cycle threshold values were obtained and analyzed by the 2-ΔΔCT method (Livak and Schmittgen, 2001). The values from at least three biological replicates and three technical replicates were normalized against genes encoding histone 3-like, actin 11, or ubiquitin, that had been selected as reference genes with the Genorm software (https://genorm.cmgg.be/). Values were calibrated with the control sample (control vector, untreated condition) to highlight fold changes. All primers used are indicated in Supplementary Table S1.
Agrobacterium rhizogenes-mediated transgenic root transformation
The protocol used has been described previously (Boisson-Dernier et al., 2001). Transgenic roots overexpressing either the MtCLE or MtTML genes were selected based on a green fluorescent protein marker present in the binary vector with an MZFLII stereomicroscope (Leica Microsystems,, Wetzlar, Germany) equipped with a blue light source and a Leica green fluorescent protein plus filter set. Transgenic roots expressing RNAi constructs were selected on 25 mg l–1 kanamycin (Sigma-Aldrich, St. Louis, MO, USA).
Transcriptomic analysis
Roots expressing either the 35S:GUS or the 35S:MtCLE13 constructs were obtained by means of an A. rhizogenes transformation, as described previously (Mortier et al., 2010), and grown in an aeroponic system on a nitrogen minimal medium ‘i’ (Blondon, 1964). At 40 days post-germination, transgenic roots of ~15 plants were harvested and pooled for RNA isolation as described (Mortier et al., 2010). Samples were hybridized to the Affymetrix Gene Chip M. truncatula genome array (http://www.affymetrix.com). RNA processing, probe hybridization, washing, and scanning of the arrays were carried out at the VIB MicroArray Facility (Leuven, Belgium).
Histochemical localization of GUS activity
The GUS activity of pNFP:GUS roots was analyzed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as substrate (Sigma-Aldrich) (Mortier et al., 2010). Roots and nodule primordia were vacuum infiltrated for 20 min and subsequently incubated at 37 °C for 4 h. After staining, roots and root nodule primordia were fixed and dehydrated in 70% ethanol as described (Mortier et al., 2010). Photographs were taken with an MZFLII stereomicroscope (Leica Microsystems).
Protein similarity tree analysis
To identify MtTML1-related proteins, the full amino acid sequence of MtTML1 was analyzed with the BLASTP algorithm on the NCBI database (https://www.ncbi.nlm.nih.gov/BLAST/) for M. truncatula and A. thaliana, and with the Lotus Genome Browser (https://lotus.au.dk/genome/) for L. japonicus. The sequences from the three proteins most closely related to MtTML1 were retrieved for each species, and the F-box protein Transport Inhibitor Response 1 (AtTIR1) from A. thaliana (At3g62980, TAIR database; https://www.arabidopsis.org/) was used to anchor the tree. For the generation of a similarity tree by means of the SeaView version 4 program (Gouy et al., 2010), the complete amino acid sequence was utilized for multiple alignments with MUSCLE (Edgar, 2004). Poorly aligned positions and divergent regions were eliminated with the G-blocks software (Castresana, 2000). A maximum-likelihood phylogenetic tree was obtained with the PhyML program (Guindon et al., 2010) as well as an approximate likelihood-ratio test for branch support.
Statistical analyses
For the microarray analysis, data were preprocessed with the ‘Robust Multiarray Averaging’ (RMA) algorithm (Irizarry et al., 2003) that involves three steps: background adjustment of RMA convolution, quantile normalization, and summarization with the median polish algorithm—in which the median values per probe set, adjusted for slide differences, are calculated. Based on an empirical Bayes moderated t-test (Smyth, 2004), as implemented in the Bioconductor package Linear Models for Microarray Data (LIMMA) (Ritchie et al., 2015), P-values were calculated to measure the differential expression between 35:GUS (control) and 35S:MtCLE13 transgenic roots. The P-values were then corrected for multiple testing problems to control the false discovery rate (FDR) (Benjamini and Hochberg, 1995). Threshold values for differential expression fold changes were set at 1.5 and at 0.05 for adjusted P-values (FDR).
For the frequency of GUS-positive roots, three biological repeats were analyzed and significant differences were identified with a χ2 test (P<0.01). For the nodulation phenotypes, a Kruskal–Wallis test was done (α<0.01) with the Xlstat software (https://www.xlstat.com/fr/). For qRT–PCR analyses, normalized values were examined with an ANOVA mixed model procedure with the SAS Enterprise 5.1 software (https://support.sas.com/en/support-home.html).
Results
Identification of downstream effectors of CLE peptide signaling during nodulation by means of a transcriptome analysis
Previously, MtCLE13 overexpression in M. truncatula roots had been shown to inhibit nodulation before the induction of MtENOD11 expression by S. meliloti (Mortier et al., 2010). We hypothesized that the inhibitory factor(s) could already be present in roots overexpressing the MtCLE13 gene prior to inoculation. Hence, the transcriptome of non-inoculated roots expressing either a 35S:GUS (control) or a 35S:MtCLE13 construct was compared. This analysis revealed that only a limited number of genes were differentially expressed. Considering a stringent threshold for the adjusted P-values (|FDR|<0.05), 17 differentially expressed Affymetrix microarray probes (corresponding to 16 genes) were identified between the control and 35S:MtCLE13 roots (Supplementary Dataset S1).
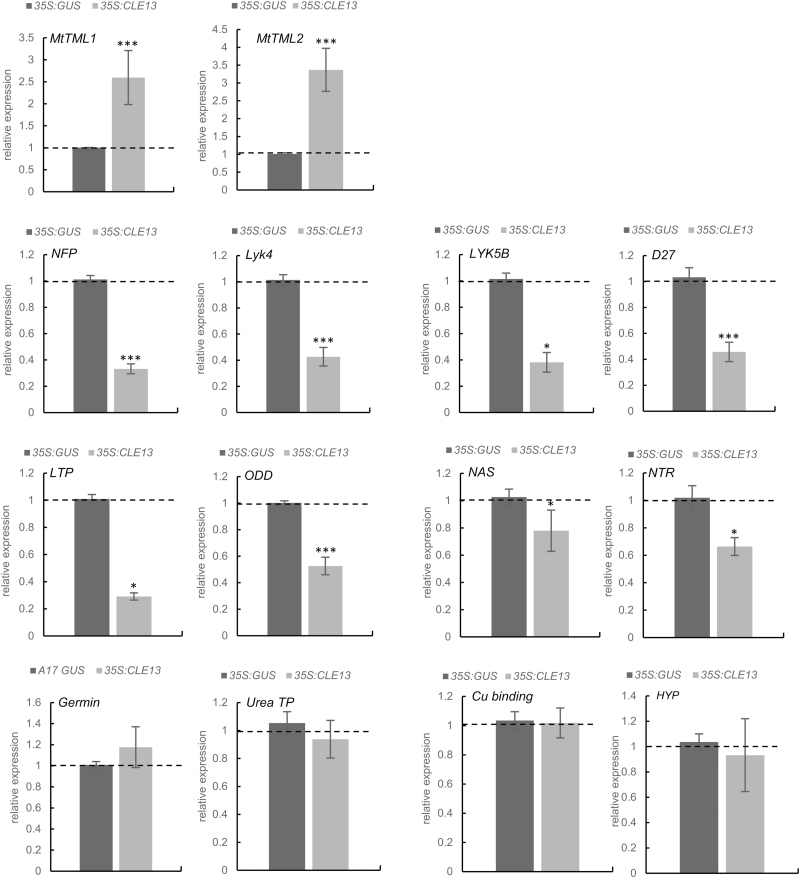
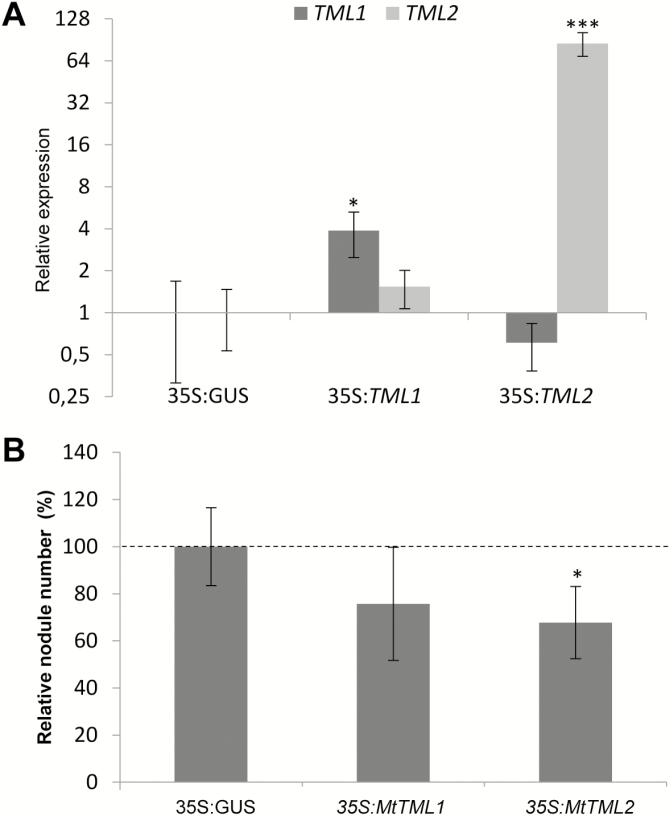
The most differentially expressed gene corresponded to MtCLE13, validating the efficiency of the overexpression strategy (Supplementary Dataset S1). Additionally, an up-regulation of two Kelch repeat-containing F-box protein-encoding genes in 35S:MtCLE13 roots was detected. Initially, three hits were discovered, but further sequence analyses uncovered that two Affymetrix probes (Mtr13058.1.S1_at and Mtr. 9802.1.S1_at) corresponded to the same gene. As the closest homolog in L. japonicus for these two genes was LjTML, we designated these genes MtTML1 (Medtr7g029290; MtGI v4.0) and MtTML2 (Medtr6g023805). Comparison of the proteins revealed that LjTML was 89% and 73% similar to MtTML1 and MtTML2, respectively. A similarity tree was generated with the three proteins from M. truncatula, L. japonicus, and A. thaliana most closely related to MtTML1 (Supplementary Fig S1). The analysis revealed that there was no equivalent duplication in L. japonicus compared with that identified for M. truncatula, suggesting that the nodulation-related function of TML proteins does not correlate with a specific duplication pattern. A qRT–PCR analysis validated the increased expression of TML1 and TML2 in independent 35S:CLE13 root samples (Fig. 1). Among the genes identified as down-regulated in the transcriptomic analysis of the 35S:MtCLE13 roots and validated by qRT–PCR on independent samples (Fig. 1), a striking enrichment for LysM-RLK-encoding genes was found, including NFP, LYK4, and a gene coding for a truncated LysM-RLK protein most closely related to LYK5 without a kinase domain, the so-called ‘LYK5B’ (Fig. 1) (Larrainzar et al., 2015; Bono and Cullimore, 2019).
Fig. 1.
qRT–PCR validation of genes differentially expressed in 35S:MtCLE13 roots. Expression analysis by qRT–PCR identified different genes as deregulated in 35S:MtCLE13 roots compared with 35S:GUS (control) roots with the microarray approach (see Suppementary Dataset S1). Gene expression was calibrated relative to the 35S:GUS control roots to highlight fold changes, as indicated by the dotted line. Error bars represent the SE of the mean of at least three biological repeats (n>15 plants per biological replicate). *P<0.05; **P<0.01; ***P<0.001 indicate significant differences between the two genotypes as measured with an ANOVA mixed model with a Tukey’s post-hoc comparison. The gene expression levels were calibrated against the expression found in the 35S:GUS control roots. TML1 and TML2, Too Much Love 1 and 2; NFP, Nod Factor perception; LYK4, LysM receptor-like kinase (RLK) 4; LYK5B, LysM-RLK 5B; D27, β-carotene isomerase; LTP, lipid transfer protein; ODD, oxoglutarate-dependent dioxygenase; NAS, nicotianamide synthase; NTR, MtNRT2.3 high-affinity nitrate transporter; Urea TP, urea transporter; Cu-binding, copper-binding protein; HyP, hypothetical protein.
In addition, genes coding for several enzymes were detected as down-regulated in both microarrays and the qRT–PCR analysis: a D27 β-carotene isomerase, a lipid transfer protein (LTP), a 2-oxoglutarate-dependent dioxygenase (ODD), and a nicotianamine synthase (NAS). Finally, the microarray analysis identified up-regulated genes in 35S:MtCLE13 roots encoding a high-affinity nitrate transporter (MtNRT2.3) (Pellizzaro et al., 2015), a sodium:solute symporter/urea-proton symporter family protein (Kojima et al., 2007), a copper-binding protein, a germin-like protein, and two hypothetical proteins. However, independent qRT–PCR validations could not confirm these expression patterns (Fig. 1) and no expression could be detected for one of the genes encoding a hypothetical protein. Overall, this transcriptomic analysis highlighted a limited number of genes, of which the expression is affected by the ectopic expression of MtCLE13 in roots, including genes encoding a subset of LysM-RLK proteins and TML-like proteins linked to the symbiotic NF perception and to the L. japonicus AON pathway, respectively.
MtCLE12 and MtCLE13 regulate the expression of NFP and TML in a SUNN-dependent manner
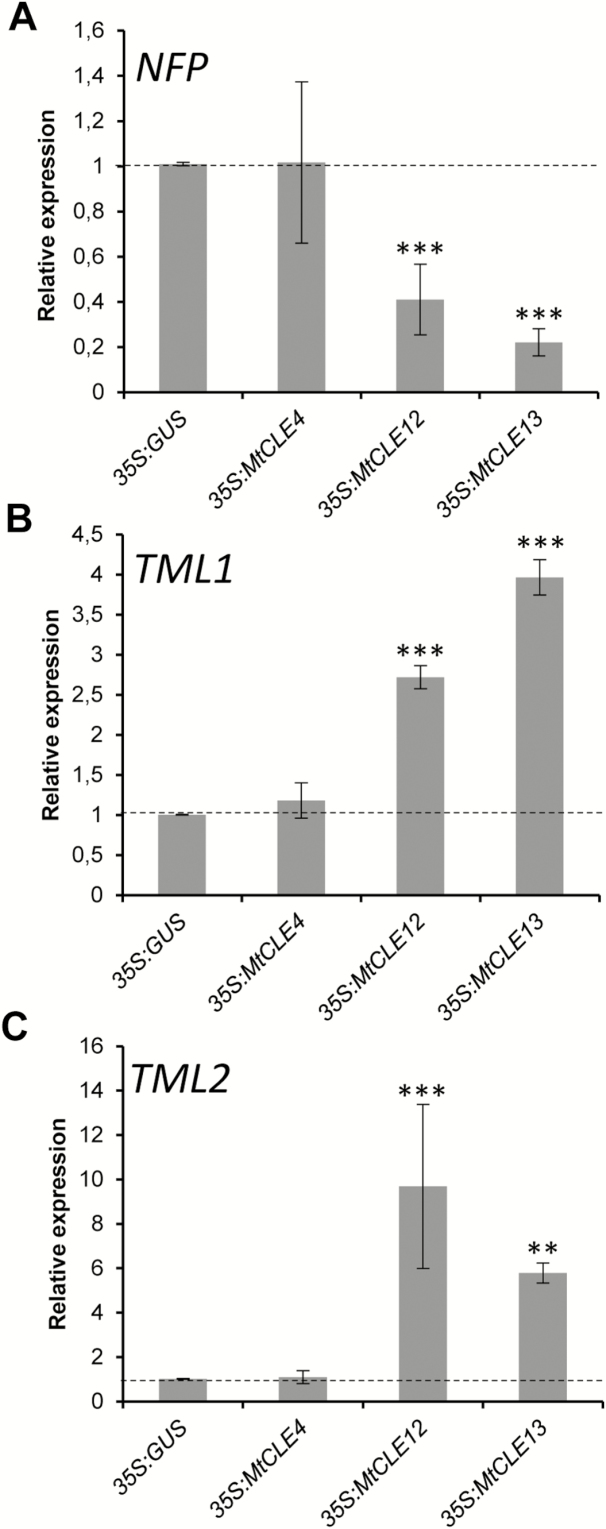
To determine whether the regulation of the previously nodulation-linked NFP and TML1/TML2 genes was specific for the ectopic MtCLE13 expression, we analyzed their expression in roots expressing MtCLE12 or the non-AON-related MtCLE4 gene (Mortier et al., 2010). First, the expression of the GUS, MtCLE4, MtCLE12, or MtCLE13 transgenes was controlled specifically in the corresponding overexpressing roots, validating the sampling procedure (Supplementary Fig S2). Interestingly, the ectopic expression of MtCLE12, like that of MtCLE13, also reduced NFP expression (Fig. 2A) and increased TML1 and TML2 expression (Fig. 2B, C). Importantly, the expression of NFP or TML1/TML2 was not altered by the ectopic MtCLE4 expression (Fig. 2).
Fig. 2.
Expression of the M. truncatula NFP, TML1, and TML2 genes in CLE peptide-overexpressing roots. (A–C) Expression analysis by qRT–CR of NFP (A), TML1 (B), and TML2 (C) in roots expressing a 35S:GUS (control), a 35S:MtCLE4 (a non-AON-related CLE control), a 35S:MtCLE12, or a 35S:MtCLE13 construct. The gene expression levels are shown relative to the expression found in the 35S:GUS control roots. To highlight the fold changes, these control levels are indicated by the dotted line. Error bars represent the SE of the mean of three biological repeats (n>15 plants per biological replicate). *P<0.05; **P<0.01; ***P<0.001, significant differences from the levels observed in the 35S:GUS genotype as found with an ANOVA mixed model with a Tukey’s post-hoc comparison.
As the nodulation inhibition provoked by the ectopic MtCLE13 expression depends on SUNN within the systemic AON pathway (Mortier et al., 2012), we wanted to determine whether the MtCLE13 regulation of NFP and TML1/TML2 expression was also SUNN dependent. The 35S:MtCLE13 or 35S:GUS constructs were introduced into wild-type and sunn-4 mutant roots, revealing that the down-regulation of NFP and the up-regulation of TML1/TML2 by the ectopic expression of MtCLE13 no longer occurred in sunn-4 mutants (Fig. 3). These data indicate that the MtCLE13-mediated regulation of NFP and TML1/TML2 expression relies on MtSUNN and support the hypothesis that down-regulation of NFP and up-regulation of TML1/TML2 are part of the AON mechanism.
Fig. 3.
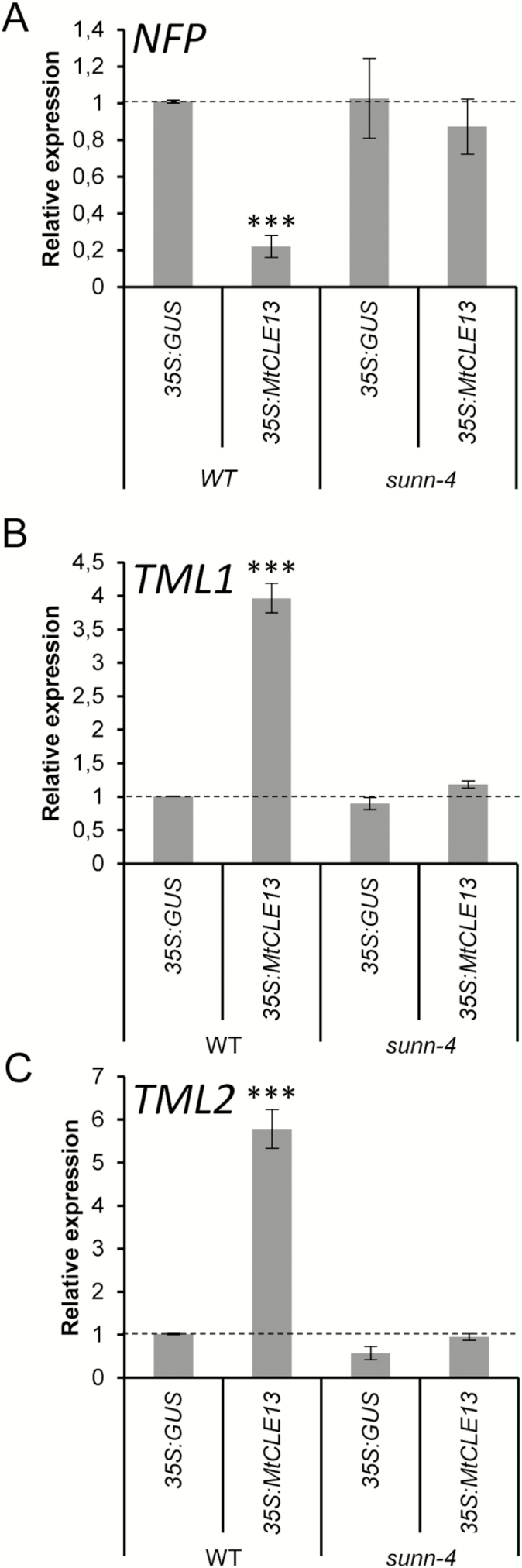
Expression of the M. truncatula NFP, TML1, and TML2 genes in the sunn mutant. (A–C) Expression analysis by qRT–PCR of NFP (A), TML1 (B), and TML2 (C) in wild-type (WT) or sunn-4 mutant roots, expressing a 35S:GUS (control) or a 35S:MtCLE13 construct. Gene expression is shown relative to the expression found in the 35S:GUS control roots and highlighted by the dotted line. Error bars represent the SE of the mean of three biological repeats (n>15 plants per replicate). The data of the WT plants are the same as those shown in Fig. 2. *P<0.05; **P<0.01; ***P<0.001, significant differences from the levels observed in the 35S:GUS genotype as found with an ANOVA mixed model with a Tukey’s post-hoc comparison.
The NFP promoter activity is down-regulated in 35S:MtCLE13 roots
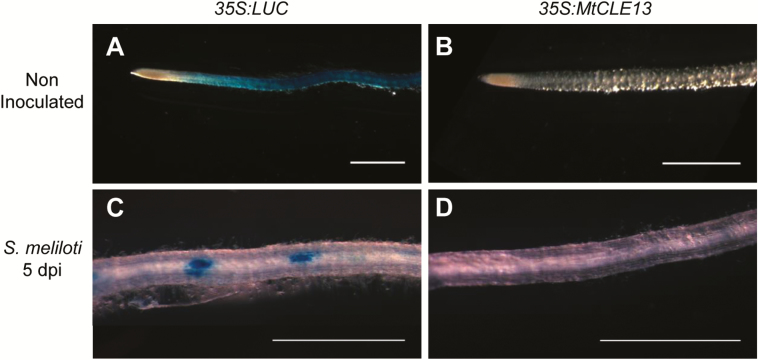
For independent corroboration of the transcriptomic and qRT–PCR datasets, roots of stable pNFP:GUS plants were transformed with the 35S:MtCLE13 construct. As a control, pNFP:GUS plants were generated with a 35S:LUCIFERASE (35S:LUC) construct. In 50% of the uninoculated control roots, a GUS activity was observed, whereas it occurred in only 18% of the 35S:MtCLE13 roots (Fig. 4A, B). Similarly, at an early stage after S. meliloti inoculation (5 dpi), the pNFP:GUS activity was observed in 61% of the 35S:LUC roots, but in only 4% of the 35S:MtCLE13 roots (Fig. 4C, D). Overall, these results support that the ectopic MtCLE13 expression leads to a down-regulated NFP expression already under non-inoculated conditions.
Fig. 4.
Down-regulated expression of a pNFP:GUS transcriptional fusion in roots overexpressing MtCLE13. Roots were stained with GUS before (A and B) and after (C and D) inoculation with S. meliloti (5 dpi). Representative roots shown express a 35S:LUC control vector (A and C) or a 35S:MtCLE13 construct (B and D) (n>44 independent roots analyzed for each construct and condition, from three different biological experiments). Scale bars=1 mm.
TML1 and TML2 negatively affect nodule number
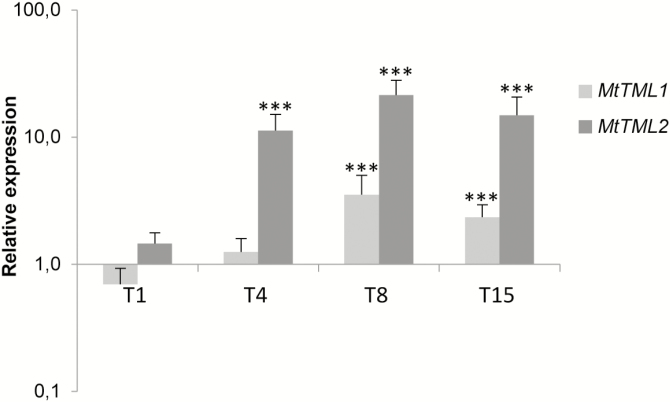
To determine whether the TML1 and TML2 genes could act during nodule development, we analyzed their expression by comparing the rhizobia-susceptible root regions before inoculation and at 1, 4, 8, and 15 dpi. The expression level of TML1 slightly increased at 4 dpi, but only significantly from 8 dpi onward (Fig. 5). At 4 dpi, the TML2 transcript level was already clearly higher than that in control roots and than that of TML1 (Fig. 5), hinting at a function for TML1 and TML2 in nodulation.
Fig. 5.
Expression analysis of the two M. truncatula genes, MtTML1 and MtTML2, during nodulation. Expression analysis by qRT–PCR of MtTML1 and MtTML2 at 1, 4, 8, and 15 dpi with S. meliloti. Gene expression was calibrated relative to non-inoculated control root regions susceptible to rhizobia to highlight fold changes. Error bars represent the SD of two biological repeats (n>20 plants per biological replicate). *P<0.05; **P<0.01; ***P<0.001, significant differences from the levels observed in the control sample as found with an ANOVA mixed model with a Tukey’s post-hoc comparison.
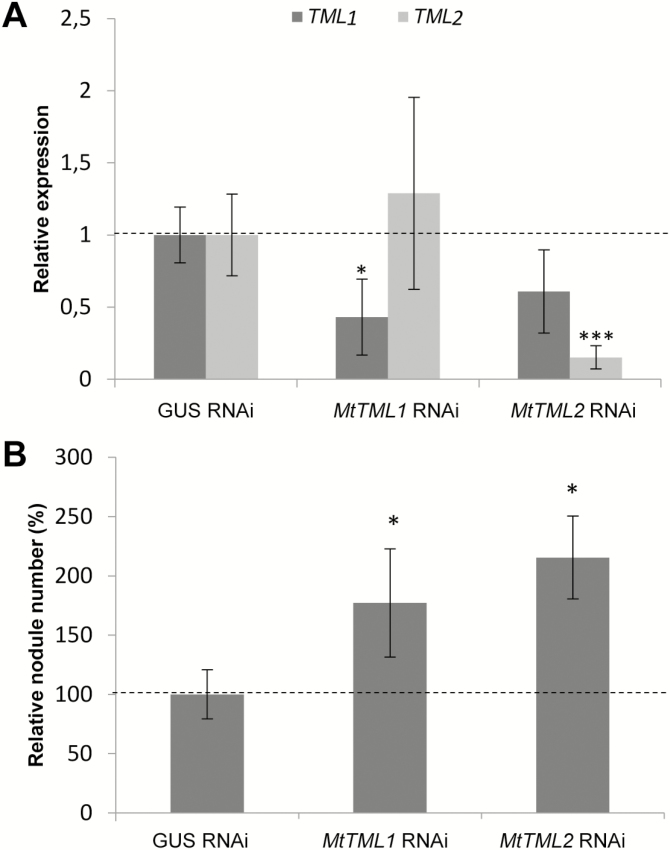
To evaluate such a potential symbiotic function, we applied an overexpression and an RNAi strategy. The expression of TML2 was induced at a much higher level in 35S:MtTML2 roots than that of TML1 in 35S:MtTML1 roots (Fig. 6A). Accordingly, the ectopic expression of TML2 significantly reduced the number of nodules formed (Fig. 6B). A non-significant reduction was observed in 35S:MtTML1 roots. RNAi constructs targeting preferentially either TML1 or TML2 were designed and validated (Fig. 7A), both leading to a significantly increased nodulation efficiency (Fig. 7B). Altogether, these observations indicate that TML1 and TML2 act in roots to negatively control the nodule number.
Fig. 6.
Reduced nodule formation of roots ectopically expressing MtTML1 and MtTML2. (A) Expression analysis of the MtTML1 and MtTML2 genes in 35S:MtTML1 and 35S:MtTML2 roots. Gene expression is shown relative to levels found in 35S:GUS control roots to highlight fold changes. Error bars represent the SD (n=6 per biological replicate). *P<0.05; **P<0.01; ***P<0.001, significant differences from the levels observed in the 35S:GUS genotype as found with an ANOVA mixed model with a post-hoc Tukey comparison. (B) Nodule number per plant in 35S:GUS (control), 35S:MtTML1, or 35S:MtTML2 roots at 14 dpi with S. meliloti. Data from two independent biological experiments were normalized, relative to the control. Error bars represent confidence intervals (α=0.05; n>16 per biological replicate). Significant statistical differences are indicated by asterisks based on a Kruskal–Wallis test (α<0.05).
Fig. 7.
Increased nodule number in M. truncatula roots silenced for MtTML1 and MtTML2. (A) Expression analysis of MtTML1 and MtTML2 genes in MtTML1 and MtTML2 RNAi roots. Gene expression was calibrated relative to GUS RNAi control roots to highlight fold changes, as indicated by the dotted line. Error bars represent the SD (n=4 per biological replicate). *P<0.05; **P<0.01; ***P<0.001, significant differences from the levels observed in the 35S:GUS RNAi genotype as found with an ANOVA mixed model with a post-hoc Tukey comparison. (B) Nodule number per plant in GUS RNAi (control), MtTML1 RNAi, or MtTML2 RNAi roots at 14 dpi with S. meliloti. Data from two independent biological experiments were normalized relative to the control. Error bars represent confidence intervals (α=0.05; n>13 plants per biological replicate). Significant statistical differences are indicated by asterisks based on a Kruskal–Wallis test (α<0.05).
Discussion
The two structurally and phylogenically related CLE peptides MtCLE12 and MtCLE13 that are up-regulated during early M. truncatula nodulation stages had previously been found to negatively control nodulation based either on ectopic expression or on simultaneous RNAi approaches (Mortier et al., 2010, 2012). A transcriptomic analysis in non-inoculated roots was carried out to analyze the downstream targets of MtCLE13, the nodulation-related CLE peptide that is encoded by the gene with the earliest symbiotic expression pattern. The ectopic expression of MtCLE13 resulted in the down-regulation of the NFP gene that encodes a presumptive NF receptor and of two homologous LysM-RLK-encoding genes, LYK4 and LYK5B, the latter of which is a LYK5 homolog with a truncated kinase domain, indicating that these genes might be putative AON targets. NFP mutation and LYK4 knock-down both reduce nodulation at the initiation stage (Ben Amor et al., 2003; Limpens et al., 2003), clearly implying their symbiotic function, whereas the truncated LYK5B had been shown previously to be rapidly controlled by S. meliloti inoculation and this in relation to the negative ethylene regulatory pathway (Larrainzar et al., 2015). Accordingly, the use of a pNFP:GUS fusion revealed that the ectopic MtCLE13 expression repressed the NFP promoter activity already before S. meliloti inoculation in the nodulation-susceptible root zone, possibly contributing to nodulation inhibition. This observation is consistent with the previously reported negative effect of these CLE peptides on nodulation at an early stage, because the S. meliloti-induced ENOD11 expression, an early epidermal infection marker, was reduced in roots ectopically expressing MtCLE13 (Mortier et al., 2010). In addition, we demonstrated the SUNN-dependent impact of the ectopic MtCLE13 expression on the NFP expression, further supporting the hypothesis that one of the downstream effects of the negative AON pathway might be a reduced NFP expression.
Besides MtCLE13, another CLE peptide-encoding gene, MtCLE12, was linked to the SUNN-dependent AON inhibition of nodulation (Mortier et al., 2010). Accordingly, roots overexpressing MtCLE12 also led to a down-regulated NFP expression. Importantly, ectopic MtCLE4 expression that does not lead to a decrease in nodule number (Mortier et al., 2010) also does not affect NFP expression, hinting at a regulation specificity toward nodulation-related CLE genes (Mortier et al., 2010).
The current AON model proposes that once nodulation is initiated, CLE peptides are produced in the roots and are systemically transported to the shoot, where they are perceived by the SUNN receptor, whereafter a feedback signal is delivered to the roots to inhibit nodulation further (Suzaki et al., 2015). Here, we show that the expression of a subset of presumptive NF receptors, including NFP and LYK4, is seemingly targeted by this shoot-to-root signal. Hence, we present a model in which down-regulation of NF perception would inhibit further nodulation events from the initiation stage. Because in L. japonicus, nodule primordia and rhizobial infections might both be targets of AON systemic signals (Suzaki et al., 2012; Tsikou et al., 2018), and because depending on the legume species, such as soybean and Medicago sativa (alfalfa), an AON effect on infections or on cortical cell divisions had been reported, respectively (Mathews et al., 1989; Caetano-Anollés and Gresshoff, 1991), the AON pathway might inhibit different nodulation steps, even depending on legume species.
In addition to genes encoding LysM-RLK receptors, two genes that code for Kelch repeat-containing F-box proteins most closely related to the L. japonicus TML genes were up-regulated by the ectopic expression of MtCLE13, as well as MtCLE12, depending on the SUNN receptor kinase, but not by the non-AON-related MtCLE4 peptide. Accordingly, the expression of both the MtTML1 and MtTML2 genes is up-regulated during nodulation. Both gain- and loss-of-function experiments collectively indicated that the MtTML1 and MtTML2 genes negatively regulate nodule numbers, as previously reported for L. japonicus (Magori et al., 2009). Surprisingly, very different transgene overexpression levels were consistently observed in 35S:MtTML1 or 35S:MtTML2 roots across different independent experiments, possibly hinting at a differential regulation of these two TML genes. Such a difference may rely on a post-transcriptional regulation exerted by a miR2111 systemic miRNA, homologous to the one described in L. japonicus (Tsikou et al., 2018). Given that the overexpression of AON-related CLE peptides causes TML transcript accumulation, it would also be interesting to determine whether the link between the AON-dependent down-regulation of the miR2111 accumulation is associated with known CLE-RS peptides in L. japonicus.
Thus, the M. truncatula genome contains two closely related genes, MtTML1 and MtTML2, which may be functionally redundant for the negative regulation of nodulation, whereas L. japonicus has only one single TML gene. This discrepancy might explain why no supernodulating mutant corresponding to a TML locus has been identified in M. truncatula based on forward genetic screens. Overall, these results suggest that the TML1 and TML2 genes are involved in the MtCLE/MtSUNN systemic AON pathway. These genes encode Kelch repeat-containing F-box proteins that are subunits of E3 ubiquitin ligase complexes that specify protein substrates for degradation by the 26S proteasome (Kelley, 2018). A remaining question to be addressed is which nodulation-related proteins interact with and are targeted for degradation by the TML1/TML2 F-box proteins.
Of the 10 other putative target genes of the MtCLE13 pathway identified, four were confirmed independently by qRT–PCR. One interesting gene that encodes a β-carotene isomerase D27 enzyme involved in strigolactone biosynthesis (Lin et al., 2009) was detected. Strigolactones have previously been linked to symbiotic nodulation in M. truncatula, notably based on applications of the synthetic strigolactone rac-GR24: low concentrations slightly increased the number of nodules, whereas high concentrations decreased nodulation (De Cuyper et al., 2017). In addition, MtD27 was rapidly up-regulated by a treatment with NFs in the root epidermis and in nodule primordia (van Zeijl et al., 2015). As the down-regulation of MtD27 by RNAi did not reveal any nodulation phenotype (van Zeijl et al., 2015), the observed change in MtD27 expression could be indirectly due to the down-regulation of the NFP expression. Hence, a potential link between strigolactones and the AON systemic pathway remains to be established based on alternative approaches. Another gene of which the expression is down-regulated by ectopic MtCLE13 expression codes for an enzyme from the ODD family that is involved in various aspects of plant metabolism, including the biosynthesis of gibberellins, flavonoids, and flavonoid derivatives (Prescott and John, 1996; Hedden and Phillips, 2000). These different signals are all tightly connected to early nodulation, notably in M. truncatula, in which they regulate the production of bacterial NF signals as well as plant NF signaling and nodule organogenesis (Ng et al., 2015; Fonouni-Farde et al. 2016a, b, 2017; Liu and Murray, 2016). Finally, genes coding for a lipid transfer protein and a nicotianamine synthase were additionally down-regulated by ectopic MtCLE13 expression. However, speculation about their involvement in the AON pathway is less obvious, and investigation would be needed to examine putative functions.
Altogether, this study provides molecular insights into how the AON–CLE systemic pathway might negatively impact on the nodulation in M. truncatula roots. The two functional LjTML homologs identified probably act downstream of the MtSUNN receptor and have a conserved function in the AON, similarly to the L. japonicus TML protein (Magori et al., 2009). The detection of the three related LysM-RLK genes, including NFP and LYK4 that are linked to early NF signaling, is very intriguing and suggests that the repression of the NF perception might be one of the early AON targets, explaining the negative regulation of the nodule number.
Supplementary data
Supplementary data are available at JXB online
Fig. S1. Protein similarity tree of MtTML1 and MtTML2 with the closely related proteins of M. truncatula, L. japonicus, and A. thaliana.
Fig. S2. Expression of MtCLE genes in the different MtCLE-overexpressing lines.
Table S1. List of primers used.
Dataset S1. List of genes differentially expressed between 35S:GUS and 35S:MtCLE13 roots.
Acknowledgments
We thank Clare Gough (Institut National de la Recherche Agronomique, Toulouse, France) and Julia Frugoli (Clemson University, Clemson, SC, USA) for stable transgenic lines and M. truncatula mutants, respectively, Christa Verplancke for skillful technical assistance, Wilson Ardiles for sequence analysis, Marnik Vuylsteke (GNOMIXX bvba, Melle, Belgium) for help with statistical analyses and transcriptomic data processing, Martine De Cock and Annick Bleys for help preparing the manuscript, and Julie Cullimore (Institut National de la Recherche Agronomique, Toulouse, France) for suggestions on the M. truncatula LysM-RLK family nomenclature. This work was supported by the Labex ‘Saclay Plant Science’ and the Lidex ‘Plant Phenotyping Pipeline’ (3P) to FF and the Research Foundation-Flanders (grant nos G.0350.04N and G.0066.07N) to SG. PG was the recipient of a Paris-Saclay University fellowship.
References
- Araya T, von Wirén N, Takahashi H. 2016. CLE peptide signaling and nitrogen interactions in plant root development. Plant Molecular Biology 91, 607–615. [DOI] [PubMed] [Google Scholar]
- Arrighi J-F, Barre A, Ben Amor B, et al. . 2006. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiology 142, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Shaw SL, Oldroyd GED, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C. 2003. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. The Plant Journal 34, 495–506. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B: Statistical Methodology 57, 289–300. [Google Scholar]
- Blondon F. 1964. Contribution à l’étude du développement de graminées fourragères: ray-grass et dactyle. Revue Générale de Botanique 71, 293–381. [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Molecular Plant-Microbe Interactions 14, 695–700. [DOI] [PubMed] [Google Scholar]
- Bono J-J, Cullimore J. 2019. Expression and function of the Medicago truncatula lysin motif receptor-like kinase (LysM-RLK) gene family in the legume–rhizobia symbiosis. In: de Bruijn, FJ, ed. The model legume Medicago truncatula. Hoboken, NJ: John Wiley & Sons Inc. (in press). [Google Scholar]
- Caetano-Anollés G, Gresshoff PM. 1991. Alfalfa controls nodulation during the onset of Rhizobium-induced cortical cell division. Plant Physiology 95, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552. [DOI] [PubMed] [Google Scholar]
- De Cuyper C, Struk S, Braem L, Gevaert K, De Jaeger G, Goormachtig S. 2017. Strigolactones, karrikins and beyond. Plant, Cell & Environment 40, 1691–1703. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J, Bono J-J. 2015. Lipo-chitooligosaccharidic nodulation factors and their perception by plant receptors. Glycoconjugate Journal 32, 455–464. [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Canova S, Lachaud C, et al. . 2013. Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chemical Biology 8, 1900–1906. [DOI] [PubMed] [Google Scholar]
- Fliegmann J, Jauneau A, Pichereaux C, Rosenberg C, Gasciolli V, Timmers ACJ, Burlet-Schiltz O, Cullimore J, Bono J-J. 2016. LYR3, a high-affinity LCO-binding protein of Medicago truncatula, interacts with LYK3, a key symbiotic receptor. FEBS Letters 590, 1477–1487. [DOI] [PubMed] [Google Scholar]
- Fonouni-Farde C, Diet A, Frugier F. 2016a Root development and endosymbioses: DELLAs lead the orchestra. Trends in Plant Science 21, 898–900. [DOI] [PubMed] [Google Scholar]
- Fonouni-Farde C, Kisiala A, Brault M, Emery RJN, Diet A, Frugier F. 2017. DELLA1-mediated gibberellin signaling regulates cytokinin-dependent symbiotic nodulation. Plant Physiology 175, 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonouni-Farde C, Tan S, Baudin M, Brault M, Wen J, Mysore KS, Niebel A, Frugier F, Diet A. 2016b DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nature Communications 7, 12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas P, Brault M, Jardinaud M-F, Frugier F. 2017. Cytokinins in symbiotic nodulation: when, where, what for? Trends in Plant Science 22, 792–802. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. The Plant Cell 18, 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27, 221–224. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hastwell AH, de Bang TC, Gresshoff PM, Ferguson BJ. 2017. CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Scientific Reports 7, 9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. 2000. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science 5, 523–530. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kassaw T, Nowak S, Schnabel E, Frugoli J. 2017. ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiology 174, 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, Nielsen MW, Kelly S, et al. . 2017. Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordia. Nature Communications 8, 14534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR. 2018. E3 ubiquitin ligases: key regulators of hormone signaling in plants. Molecular & Cellular Proteomics 17, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Gresshoff PM. 2008. Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Molecular Plant-Microbe Interactions 21, 1337–1348. [DOI] [PubMed] [Google Scholar]
- Kojima S, Bohner A, Gassert B, Yuan L, von Wirén N. 2007. AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. The Plant Journal 52, 30–40. [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, et al. . 2002. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420, 422–426. [DOI] [PubMed] [Google Scholar]
- Larrainzar E, Riely BK, Kim SC, et al. . 2015. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiology 169, 233–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CW, Lee YW, Hwang CH. 2011. Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant & Cell Physiology 52, 1613–1627. [DOI] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. [DOI] [PubMed] [Google Scholar]
- Lin H, Wang R, Qian Q, et al. . 2009. DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. The Plant Cell 21, 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-W, Murray JD. 2016. The role of flavonoids in nodulation host-range specificity: an update. Plants 5, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, et al. . 2003. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. [DOI] [PubMed] [Google Scholar]
- Magori S, Oka-Kira E, Shibata S, Umehara Y, Kouchi H, Hase Y, Tanaka A, Sato S, Tabata S, Kawaguchi M. 2009. TOO MUCH LOVE, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Molecular Plant-Microbe Interactions 22, 259–268. [DOI] [PubMed] [Google Scholar]
- Mathews A, Carroll BJ, Gresshoff PM. 1989. Development of Bradyrhizobium infections in supernodulating and non-nodulating mutants of soybean (Glycine max [L.] Merrill). Protoplasma 150, 40–47. [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E. 2003. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiology 132, 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S. 2010. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiology 153, 222–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S. 2012. Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. The Plant Journal 70, 367–376. [DOI] [PubMed] [Google Scholar]
- Murakami E, Cheng J, Gysel K, et al. . 2018. Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. eLife 7, e33506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng JLP, Hassan S, Truong TT, Hocart CH, Laffont C, Frugier F, Mathesius U. 2015. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. The Plant Cell 27, 2210–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H, Handa Y, Tanaka S, Suzaki T, Kawaguchi M. 2016. Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. Journal of Plant Research 129, 909–919. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ohnishi E, Sato S, Takahashi H, Nakazono M, Tabata S, Kawaguchi M. 2009. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant & Cell Physiology 50, 67–77. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews Microbiology 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Pellizzaro A, Clochard T, Planchet E, Limami AM, Morère-Le Paven M-C. 2015. Identification and molecular characterization of Medicago truncatula NRT2 and NAR2 families. Physiologia Plantarum 154, 256–269. [DOI] [PubMed] [Google Scholar]
- Prescott AG, John P. 1996. Dioxygenases: molecular structure and role in plant metabolism. Annual Review of Plant Physiology and Plant Molecular Biology 47, 245–271. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, et al. . 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. [DOI] [PubMed] [Google Scholar]
- Reid DE, Ferguson BJ, Gresshoff PM. 2011. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Molecular Plant-Microbe Interactions 24, 606–618. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan M, Morandi D, Tarenghi E, Duc G. 1995. Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Science 111, 63–71. [Google Scholar]
- Schnabel E, Journet E-P, de Carvalho-Niebel F, Duc G, Frugoli J. 2005. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Molecular Biology 58, 809–822. [DOI] [PubMed] [Google Scholar]
- Schnabel EL, Kassaw TK, Smith LS, Marsh JF, Oldroyd GE, Long SR, Frugoli JA. 2011. The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiology 157, 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli J. 2010. The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiology 154, 1390–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM. 2003. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299, 109–112. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3, Article 3. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. 2012. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139, 3997–4006. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yoro E, Kawaguchi M. 2015. Leguminous plants: inventors of root nodules to accommodate symbiotic bacteria. International Review of Cell and Molecular Biology 316, 111–158. [DOI] [PubMed] [Google Scholar]
- Takahara M, Magori S, Soyano T, et al. . 2013. TOO MUCH LOVE, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume–Rhizobium symbiosis. Plant & Cell Physiology 54, 433–447. [DOI] [PubMed] [Google Scholar]
- Truchet G, Barker DG, Camut S, de Billy F, Vasse J, Huguet T. 1989. Alfalfa nodulation in the absence of Rhizobium. Molecular and General Genetics 219, 65–68. [Google Scholar]
- Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, Madsen LH, Bhasin H, Sexauer M, Stougaard J, Markmann K. 2018. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science 362, 233–236. [DOI] [PubMed] [Google Scholar]
- van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. 2006. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiology 140, 1494–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A, Liu W, Xiao TT, Kohlen W, Yang WC, Bisseling T, Geurts R. 2015. The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biology 15, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S. 2016. CLE peptides and their signaling pathways in plant development. Journal of Experimental Botany 67, 4813–4826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.