A ribonucleotide reductase large subunit mutant isolated from Setaria italica had a Gly737 to Glu substitution in the C-terminus of the protein and exhibited growth retardation and striped leaves.
Keywords: Cell cycle progression, chloroplast biogenesis, DNA replication, growth retardation, ribonucleotide reductase, SiSTL1, striped leaf
Abstract
The activity of ribonucleotide reductase (RNR), which catalyses the transformation of four ribonucleoside diphosphates (NDPs) to their corresponding deoxyribonucleoside diphosphates (dNDPs), is the main determiner of the cellular concentration of dNTP pools and should be tightly coordinated with DNA synthesis and cell-cycle progression. Constitutively increased or decreased RNR activity interferes with DNA replication and leads to arrested cell cycle progression; however, the mechanisms underlying these disruptive effects in higher plants remain to be uncovered. In this study, we identified a RNR large subunit mutant, sistl1, in Setaria italica (foxtail millet), which exhibited growth retardation as well as striped leaf phenotype, i.e. irregularly reduced leaf vein distances and decreased chloroplast biogenesis. We determined that a Gly737 to Glu substitution occurring in the C-terminus of the SiSTL1 protein slightly affected its optimal function, leading in turn to the reduced expression of genes variously involved in the assembly and activation of the DNA pre-replicative complex, elongation of replication forks and S phase entry. Our study provides new insights into how SiSTL1 regulates plant growth, chloroplast biogenesis, and cell cycle progression in Poaceae crops.
Introduction
The cellular concentration of deoxyribonucleoside triphosphate (dNTP) pools fluctuates with cell cycle progression (Chabes and Stillman, 2007). Correct levels of dNTP pools are critical for the accomplishment and high fidelity of DNA replication (Poli et al., 2012). The level of dNTP pools is generally restricted during the G1 phase. Upon entry into the S phase, the concentration increases sharply—by approximately 3-fold in Saccharomyces cerevisiae—and then drops to the same levels as in G1 phase during the G2 and M phases (Chabes et al., 2003; Poli et al., 2012). dNTPs are generated by two pathways. In the de novo synthesis pathway, presumed to be the main biosynthetic pathway, dNTPs are synthesized from simple substances such as ribose phosphate, amino acids, and CO. In the second pathway, termed the salvage pathway, dNTPs are generated by simple transfer reactions involving deoxyribose phosphates derived from the de novo synthesis pathway. The activity of ribonucleotide reductase (RNR), which catalyses the transformation of NDPs to their corresponding dNDPs in the de novo synthesis pathway, is the main determiner of cellular dNTP-pool concentrations and is tightly coordinated with DNA replication, cell cycle progression, and DNA repair (Chabes et al., 2003; Poli et al., 2012).
Most eukaryotic RNRs are α2β2 heterotetramers comprising two large subunits (R1/RNRL) and two small subunits (R2/RNRS) (Nordlund and Reichard, 2006; Reichard, 2010; Sanvisens et al., 2013). The R1 subunit contains both catalytic and allosteric regulation domains, while R2 contains a non-heme dinuclear iron center. During each reduction reaction, a stable tyrosyl radical is created and transferred to the catalytic cysteine pair of R1 (Cys218 and Cys443 in S. cerevisiae) (Kolberg et al., 2004). The catalytic cysteine pair is then converted from the reduced form to the oxidized (disulfide-bonded) form. The disulfide-bond is subsequently reduced by thioredoxin and glutaredoxin to regenerate active R1 (reduced state) for the next catalytic cycle (Kolberg et al., 2004). A conserved cysteine pair at the R1 C-terminal end (designated the CX2C motif in eukaryotic R1s) is indispensable for the regeneration of the R1 catalytic cysteine pair, as it mediates the interaction of this cysteine pair and thioredoxin/glutaredoxin. In addition to the CX2C motif, the last ~100 amino acids located before the CX2C motif at the R1 C-terminus, designated the C-terminal insertion (CI) region, is also important for optimal R1 activity (Zhang et al., 2007).
Because the cellular dNTP pool is sufficient for only a small fraction of DNA replication, up-regulation of RNR activity is necessary when cells enter the S phase or experience DNA damage (Poli et al., 2012). Strategies used by cells for RNR up-regulation include the transcriptional induction of RNR genes, degradation of RNR inhibitors, and subcellular redistribution of RNR subunits (Nordlund and Reichard, 2006; Sanvisens et al., 2013). The E2F family of transcription factors, which is regulated in a cell cycle-dependent manner, plays a central role in controlling the expression of genes required for cell cycle progression, particularly DNA synthesis (Stevens and La Thangue, 2003). Up-regulation of both RNRL and RNRS during the S phase is mediated by E2F transcription factors. In human and mouse cells, R1 levels are almost constant and are present in excess during the cell cycle (Chabes et al., 2004; Shao et al., 2006). S-phase-specific RNR activity is determined by the E2F-dependent cell cycle regulation of R2 genes (Chabes et al., 2004). Other studies, however, have shown that R1 genes also exhibit S-phase-specific expression mode and are regulated by MBF/E2F transcription factors in S. cerevisiae and tobacco (Chabouté et al., 2002; Chabes et al., 2004; Lincker et al., 2004; Sanvisens et al., 2013). In addition to this transcriptional regulation, RNR activity is controlled by the Mec1/Rad53 protein kinase-dependent proteolysis of Sml1 (Zhao et al., 2000, 2001). The concentration of the Sml1 protein, a RNR large subunit inhibitor, also fluctuates during the cell cycle and is lowest during the S phase (Zhao et al., 2001). During G0 and G1 phases, Sml1 competitively combines with the catalytic site of the R1 subunit and thus blocks the reduction activity of R1 (Zhang et al., 2007). When cells enter the S phase or encounter DNA replication stress, Sml1 is phosphorylated and degraded in a Mec1/Rad53-dependent manner, thereby relieving RNR inhibition. To summarize, one conserved theme of RNR activity, albeit controlled by different mechanisms, is that it is cell cycle regulated, restricted during G0 and G1 phases and peaking at the S phase (Nordlund and Reichard, 2006; Guarino et al., 2014).
DNA synthesis begins with the assembly and activation of replication origins (Sheu et al., 2016). During this process, a double hexameric minichromosome maintenance (MCM) complex, composed of two Mcm2–Mcm7 hexamers, is loaded onto the replication origin to form a pre-replicative complex (pre-RC) with the help of an origin recognition complex (ORC) and the licensing factors Cdc6 and Cdt1 (Das et al., 2015; Sheu et al., 2016). Cdc45 is then recruited to activate the MCM complex with the assistance of S-phase cyclin-dependent kinases (CDKs) and Dbf4-dependent Cdc7 kinase (Rossbach et al., 2017). After these two steps, the replication origins are fully activated, which enables the recruitment of DNA polymerase and other replisome components to form the replication forks needed to start DNA elongation (Sheu et al., 2016).
Because RNR activity and DNA replication are interconnected, much research has been performed to explain how disruption of RNR activity impedes DNA replication and cell cycle progression (Chabes and Stillman, 2007; Poli et al., 2012; Giannattasio and Branzei, 2017). One proposed mechanism, conserved among budding yeast, fission yeast and human cells, is that disruption of RNR activity activates the S-phase checkpoint, which subsequently delays S-phase entry, increases dNTP synthesis and prevents late replication-origin firing (Giannattasio and Branzei, 2017). The S-phase checkpoint, known as the Mec1/Rad53 pathway in budding yeast and the ATR–CHK1 pathway in human cells, is composed of multiple serine/threonine kinases (Sun et al., 1995; Guo et al., 2000). In human cells, ATR–CHK1-mediated phosphorylation events inhibit the CDK activators Cdc25A, Cdc25B, and Cdc25C, thereby inhibiting the activities of CDK2–cyclin A/E and CDK1–cyclin B to delay S-phase entry (Krek et al., 1995; Giannattasio and Branzei, 2017). In addition, ATR can induce dNTP production by up-regulating E2F1. Late replication-origin firing is prevented by the phosphorylation events of ATR–CHK1 on proteins required for replication fork formation, such as MCM2, RPA2, ExoI, and BLM (Giannattasio and Branzei, 2017). Furthermore, disturbed RNR activity can impede DNA replication and cell cycle progression by mechanisms independent of the Mec1/Rad53 pathway. In S. cerevisiae, continuous induction of R1 alleles transiently arrests cell cycle progression in the late G1 phase by affecting the assembly of Cdc45 into the pre-RC and thus delays the activation of the pre-RC at the origins of DNA replication (Chabes and Stillman, 2007). Moreover, inhibition of RNR activity with hydroxyurea impedes DNA replication and cell cycle progression by inducing a slow DNA replication mode with a 25-fold reduction of the initiation rate and a 10-fold reduction of the elongation rate, thus extending the time required for S-phase completion by at least 8 h in budding yeast (Poli et al., 2012).
Although extensive effort has been invested in studying the effects of disrupted RNR activity on DNA replication and cell cycle progression in yeast and human cells, this phenomenon has been unclear in higher plants. Although rnr mutants of both large and small subunits have been described in Arabidopsis (large subunit, cls8; small subunits, tso2, rnr2a, and rnr2b) (Wang and Liu, 2006; Garton et al., 2007) and Oryza sativa (large subunit, v3; small subunit, st1) (Yoo et al., 2009), the cited studies were mainly concerned with their effects on chloroplast biogenesis. All rnr mutants characterized in higher plants have been found to exhibit decreased dNTP pools and reduced chloroplast biogenesis (Wang and Liu, 2006; Garton et al., 2007; Yoo et al., 2009), which suggests a strong correlation between cellular dNTP concentrations and chloroplast biogenesis. In Arabidopsis, the cls8 mutant and RNAi lines with a disrupted large subunit gene produce bleached leaves and siliques (Garton et al., 2007), while rice v3 and st1 mutants develop striped leaves in a growth stage-dependent manner (Yoo et al., 2009). In addition to producing bleached leaves and siliques, tso2-1 and tso2-1 rnr2a-1 exhibit obvious developmental defects, including callus-like floral organs, fasciated shoot apical meristems, and defects in cell cycle progression (Wang and Liu, 2006). However, although interesting, these results do not explain how reduced dNTP pools affect cell cycle progression.
In this study, we identified a RNR large subunit mutant, sistl1, which produced defective RNRL protein (SiSTL1) and exhibited growth retardation and striped leaf phenotype. Cross sections and microscopic observations of the striped leaves revealed that reduced chloroplast biogenesis and asymmetric leaf cell development occurred in sistl1. Yeast two-hybrid (Y2H) analysis revealed that Gly737 to Glu substitution of the SiSTL1 protein weakened its interaction with the RNR small subunit. RNA-seq analysis suggested that genes involved in DNA replication and cell cycle progression were repressed in sistl1.
Materials and methods
Plant materials and growth conditions
A sistl1 mutant was isolated from ethylmethane sulfonate (EMS)-treated S. italica ‘Yugu1’ (foxtail millet). After isolation, the mutant was backcrossed with Yugu1, and recessive derivatives from the backcrosses were used in subsequent experiments. All plants were grown in experimental fields in Beijing or Hainan, China, during the foxtail millet growth season.
Germination trials
For germination trials, seeds of Yugu1 and sistl1 were placed on two layers of wet filter paper. Root and shoot lengths and numbers of germinated seeds were determined every 24 h. In addition, germinated seeds of sistl1 and Yugu1 were photographed at 24, 36, and 48 h after placement on wet filter paper. Each germination trial involved 100 seeds per container, with three replicates. For root and shoot length measurements, 15 Yugu1 and sistl1 seedlings were used each (each seedling as a biological replicate).
Leaf structure and chloroplast ultrastructural observation
Fragments of fifth leaves of Yugu1 and sistl1 were observed under optical (DMLB, Leica, Wetzlar, Hessen, Germany) and confocal (LSM700, Zeiss, Oberkochen, Baden-Wurttemberg, Germany) microscopes. Leaf fragments were gradiently dehydrated with 75%, 85%, 95%, and 100% ethanol and then rendered transparent with 1:1 ethanol: xylene followed by 100% xylene. After washing with 100%, 85%, 65%, 30% ethanol and water, the fragments were stained with I2–KI solution. To generate resin-embedded sections, the leaf tissues were fixed with 2.5% glutaraldehyde, washed three times with 0.2 M phosphate buffer, fixed in 1% osmium tetroxide for 1 h, stained with uranyl acetate and subjected to dehydration using an ethanol gradient. After dehydration, the leaf tissues were embedded into resin. The resin blocks were sectioned with a glass blade, and the slices were observed under a transmission electron microscope (JEM 1230, JEOL, Tokyo, Japan). Density curves of bundle sheath cells (BSCs)/mesophyll cells (MCs) containing zero to six chloroplasts were constructed with resin sections of the fifth leaves of Yugu1 and sistl1. For each density curve, we counted the number of chloroplasts in four vascular bundles which containing >20 BSCs and >70 MCs.
Map-based cloning and whole-genome resequencing
For map-based cloning, we used 891 recessive individuals of an F2 population generated from a cross between sistl1 and S. italica ‘SSR41’. A total of 132 markers were used to localize the SiSTL1 gene to a 91-kb interval on chromosome 4. Details of simple sequence repeat (SSR) markers CAAS 4023, CAAS 4019, and CAAS 4033 are given in Zhang et al. (2014), and primer sequences of insertion–deletion (InDel) markers and cleaved amplified polymorphic sequence (CAPS) markers are described in Supplementary Table S1 at JXB online.
For the whole-genome resequencing, two DNA pools were constructed with 30 Yugu1 and sistl1 individuals each. Raw data were obtained using the Illumina HiSeq 2500 platform and uploaded with EMBL-EBI in the European Nucleotide Archive database under the accession number PRJEB27720. After quality control, clean data were generated as described in the ‘RNA-seq analysis’ section. Picard tools v1.41 (http://broadinstitute.github.io/picard/) and samtools v0.1.18 (http://www.htslib.org/) were used to sort, remove duplicated reads from and merge the BAM alignment results. For single nucleotide polymorphism (SNP) calling, reads of sistl1 were input into GATK2 software with S. italica v2.2 as the reference genome. Raw vcf files were filtered with the GATK standard filter method and other parameters (cluster Window Size: 10; MQ0≥4 and (MQ0/(1.0×DP))>0.1; QUAL<10; QUAL<30.0 or QD<5.0 or HRun>5), and only SNPs with distance >5 were retained. SNPs present within the 91-kb interval of sistl1 were then filtered by the same SNP calling steps with reads of Yugu1 and other Setaria mutants, SiDWARF3, and Loose Panicle1 (Fan et al., 2017; Xiang et al., 2017).
Knock-out of SiSTL1 homologous gene in rice
Sequences of RNRL proteins from Arabidopsis (AtRNRL, At2G21790), O. sativa (OsRNRL1, LOC_Os06g07210.1 and OsRNRL2, LOC_Os02g56100), Zea mays (GRMZM2G304362 and GRMZM2G340527), Sorghum bicolor (Sobic.010G054600 and Sobic.004G336100), and Setaria italica (SiSTL1, Seita.4G058800; SiSTL1-2, Seita.5G216600; and SiSTL1-3, Seita.1G356500) were downloaded from Phytozome v12. A phylogenetic analysis was carried out using MEGA 5 software.
For knock-out of the OsRNRL1 gene (LOC_Os06g07210) in rice, pYLCRISPR/Cas9-MH vectors were constructed as described by Ma et al. (2015) and subsequently transferred into rice cultivar Kitaki (japonica). To verify whether the transgenic plants contained the pYLCRISPR/Cas9-MH vector, PCR was performed with a primer pair specific for Cas9 gene amplification (crispr V3 F and R). To verify whether sequence variation occurred, OsRNRL1 was amplified and sequenced with primer pair OsCas9-C F and R. To verify what kinds of variation occurred in the striped T0 transgenic plants, OsCas9-C PCR products from OsC1-2, OsC1-8, OsC1-4, and OsC1-8 were cloned using the pEASYTM-Blunt Zero Cloning Kit (CB501-02, Transgen Biotech, Beijing, China) and sequenced with monoclone. To verify whether variations occurred in the descendants of the aforementioned transgenic plants, OsCas9-C PCR products of these transgenic descendants were sequenced and the results were analysed using DSDecodeM (http://skl.scau.edu.cn/dsdecode/). Sequence variation of OsC1-8 T1-1 was too complicated to be resolved by DSDecodeM, and thus its OsCas9-C PCR product was cloned using the pEASY-Blunt Zero Cloning Kit and sequenced with monoclone. Primers used for vector construction and transgenic plants verification are listed in Supplementary Tables S2 and S3.
Quantitative real-time RT-PCR and subcellular localization
To investigate the expression patterns of SiSTL1, SiSTL1-2, and SiSTL1-3 along leaf developmental gradients, four leaf fractions of striped fourth leaves of sistl1 and normal fourth leaves of Yugu1 were extracted as described by Li et al. (2010). After extraction of total mRNA with a Pure Link RNA mini kit (cat. no. 12183018; Invitrogen, Carlsbad, CA, USA), cDNAs were obtained using a PrimeScript first-strand cDNA synthesis kit (cat. no. 6210A; TakaRa, Otsu Shiga, Japan). Quantitative real-time RT-PCR (qRT-PCR) was performed using Fast Start Universal SYBR Green Master Mix (ROX) (cat. no. 04913914001, Roche, Mannheim, Germany) on an Applied Biosystems 7300 Analyser (Applied Biosystems, Foster City, CA, USA). Relative gene expression levels were calculated with the 2−ΔCt method. cullin (Seita.3G037700), described in Martins et al. (2016), was used as the reference gene. Relative expression levels of E2F1 and E2F2 in the leaf base of sistl1 and Yugu1 were also obtained in the same way as described above. For qRT-PCR to study the expression changes in sistl1 and Yugu1 of 11 genes considered to be involved in cell cycle progression, the relative expression of these genes were calculated with cullin as reference gene and the 2−ΔΔCt calculation method, as described by Winer et al. (1999). For the qRT-PCR conducted with rice transgenic plants, RNA was extracted from the basal region of the seventh leaf, and relative expression of the genes was determined in the same way as described above but using 2−ΔCt calculation method and Actin as reference gene, as described by Wang et al. (2017). Primers used for qRT-PCR are listed in Supplementary Table S4.
For determination of subcellular localization, SiSTL1 was fused to a p16318:GFP vector, which was then transferred into protoplasts isolated from fresh leaves of 7-day-old foxtail millet seedlings by a polyethylene glycol-mediated method (Kim et al., 2015b).
Y2H analysis
A Y2H assay was conducted using a Matchmaker Gold Yeast Two-Hybrid system (cat. no. 630489; Clontech, Mountain View, CA, USA). The wild-type SiSTL1 (Gly737) allele and the mutant SiSTL1 (Glu737) allele were separately fused to AD vectors, while the RNR small subunit gene (SiRNRS, Seita.4G114600) was fused to a BD vector. The fused AD and BD vectors were then co-transferred into Gold S. cerevisiae. The transformed yeast strains were tested for viability on SD/−Ade/−His/−Leu/−Trp/X-α-gal plates.
RNA-seq analysis
For RNA-seq libraries, we used mRNA of the basal region from the striped seventh leaves of sistl1, a leaf zone that was wrapped in the sixth leaf sheath and 1 cm above the leaf seven ligule, which was supposed to be undergoing active cell division. The corresponding leaf region of Yugu1 was used as a control. A total of six cDNA libraries (three of sistl1 and three of Yugu1) were sequenced with the Illumina NovaSeq 6000 system, and 150-bp paired-end reads were generated. Clean data were obtained by removing reads containing adapters, reads containing ploy-N and low-quality reads from the raw data. Q20, Q30, GC-content and the sequence duplication level of the clean data were calculated for quality control. High-quality clean reads were then mapped to the reference genome (Setaria italica v2.2) using Hisat2 tools (Kim et al., 2015a), and only reads with a perfect match or one mismatch were counted. Quantification of gene expression abundances was estimated by reads per kilobase of transcript per million fragments mapped (RPKM). For differential expression analysis, clean reads of sistl1 and Yugu1 were analysed with the DESeq R package (1.10.1) (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html). Genes with adjusted |log2RPKMsistl1/Yugu1|>0.5 and P value<0.01 found by DESeq were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages described by Young et al (2010). To validate the Illumina data, relative expression of 27 genes was investigated in sistl1 and Yugu1 by qRT-PCR. A high correlation (R2=0.95) was found between the RNA-seq and qRT-PCR data (see Supplementary Table S5). Raw data were uploaded with EMBL-EBI into the European Nucleotide Archive database under the accession numbers PRJEB25717 and PRJEB26878.
Flow cytometry
For flow cytometry, approximately 30 three-day-old first leaves were cut into pieces in nuclear extraction buffer as described by Lin et al. (2012). The extract was stained with 2.5 mg ml−1 4′,6-diamidino-2-phenylindole for 5–10 min and then analysed on a MoFlo XDP cytometer (Beckman Coulter, CA, USA). A total of 8000 nuclei were counted per trial, with three repeats.
Results
sistl1 exhibits delayed growth and a striped leaf phenotype
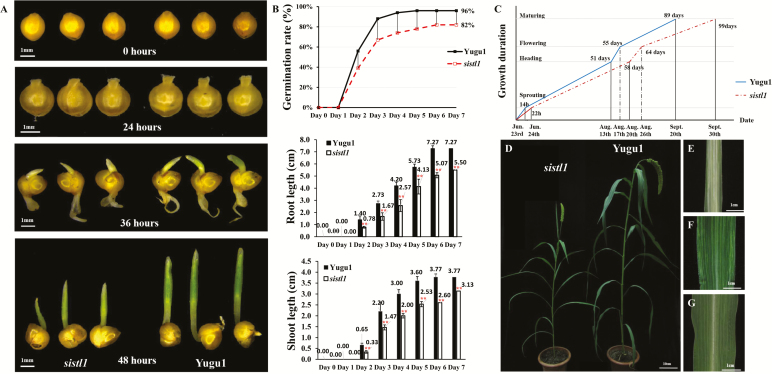
Compared with wild-type Yugu1, sistl1 displayed obvious developmental retardation. According to our germination trials, the germination rate of sistl1 (82%) was much lower than that of Yugu1 (96%), and root and shoot lengths of sistl1 were significantly shorter than those of Yugu1 at each observation time point (Fig. 1B). In addition, the germination time of sistl1 was delayed by 8 h. The radicles of 50% of wild-type Yugu1 seeds penetrated the episperm within 14 h after placement on wet filter paper, whereas most sistl1 radicles did not emerge for 22 h (Fig. 1A). Furthermore, a comparative examination of the overall growth period of sistl1 and Yugu1 revealed that developmental stages of sistl1 plants were delayed to varying degrees: sprouting time by 8 h, heading date by 7 days, and flowering and maturation dates by 9 and 10 days, respectively (Fig. 1C). These observations indicate that sistl1 experienced developmental retardation.
Fig. 1.
Striped-leaf phenotypes and delayed germination and growth of sistl1. (A) Germination status of sistl1 (left) and Yugu1 (right) at 0, 24, 36, and 48 h. (B) Germination rate, root length, and shoot length of sistl1 and Yugu1. Asterisks indicate a significant difference between root length and shoot length of sistl1 and Yugu1; error bars, ±SD (n=15 seedlings), Student’s t-test, P<0.01. (C) Growth duration of sistl1 and Yugu1 indicating dates of different developmental stages. (D) Morphology of sistl1 and Yugu1 grown in Beijing (40°N, 116°E; high temperatures and long days). (E) Fifth leaves of sistl1 grown in Hainan in the winter (19°N, 110°E; low temperatures and short days). (F, G) Fifth leaves of sistl1 (F) and Yugu1 (G) grown in Beijing in the summer.
Another characteristic of sistl1 was the production of striped leaves in a growth-stage- and environment-dependent manner. For example, in the summer in Beijing (40°N, 116°E) under high-temperature and long-day field conditions, sistl1 exhibited the normal green leaf phenotype up to the third-leaf stage, and then produced striped fourth and fifth leaves (Fig. 1F). In contrast, in the winter in tropical Hainan (19°N, 110°E) under low-temperature and short-day conditions, sistl1 produced striped second and third leaves, and the striped area was much larger (Fig. 1E). Leaves generated after the late shooting stage, such as ninth and later leaves, were much less prone to being striped. In favorable field conditions, striped leaves were sometimes nearly absent after the late shooting stage.
sistl1 had an abnormal leaf vein arrangement and reduced chloroplast biogenesis
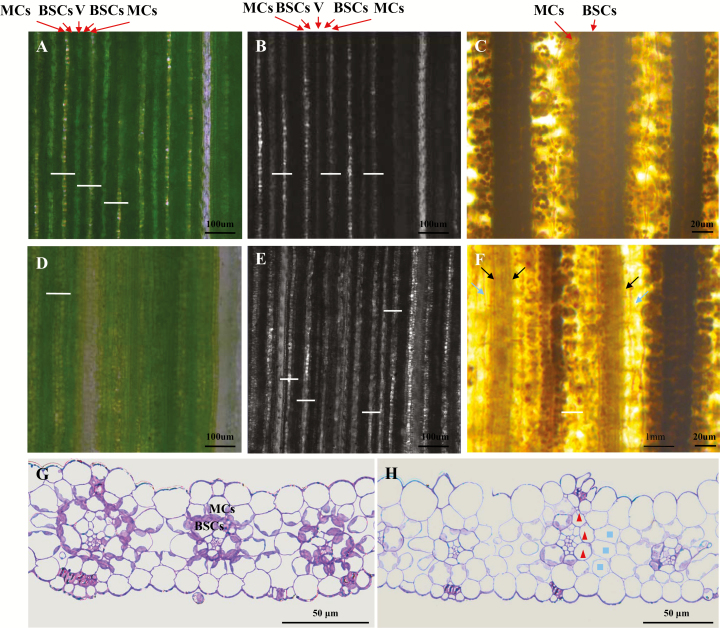
In C4 plants such as foxtail millet, vascular bundles, which are surrounded by a layer of bundle sheath cells (BSCs) plus another layer of mesophyll cells (MCs), are arranged in a MC–BSC–V–BSC–MC pattern, an organization referred to as a Kranz structure. Because BSCs are packed with large chloroplasts and tightly organized, they appear deep green under an optical microscope (Fig. 2A). Mesophyll cells have fewer, smaller chloroplasts and are loosely arranged. When MCs are observed under an optical microscope, light spots can be seen as light transmitted from MCs to the eyepiece (Fig. 2A). Vascular bundles are surrounded by BSCs and MCs and thus appear pale green. Similarly, BSCs are very dark under a confocal microscope, MCs are light-spotted and vascular bundles are gray because of their different transmittances (Fig. 2B).
Fig. 2.
Abnormal leaf vein arrangement and asymmetric cell development of sistl1. (A, B) Fifth-leaf fragments of Yugu1 observed by optical (A) and confocal (B) microscopy. Arrows indicate the locations of mesophyll cells (MCs) and bundle sheath cells (BSCs) in one vascular bundle. White bars show the distance between two adjacent veins. (D, E) Fifth-leaf fragments of sistl1 observed by optical (D) and confocal (E) microscopy. (C, F) I2–KI-stained fifth-leaf fragments of Yugu1 (C) and sistl1 (F). Dark and light arrows indicate unstained BSCs and MCs. (G, H) Resin-embedded sections of Yugu1 (G) and sistl1 (H) fifth-leaf fragments. Triangles and squares denote abnormal BSCs and MCs in one vascular bundle.
To characterize striped sistl1 leaves in more detail, fragments of the fifth leaves of Yugu1 and sistl1 were observed under optical and confocal microscopes (Fig. 2A, B, D, E). Yugu1 leaves were found to possess a well-organized MC–BSC–V–BSC–MC pattern (Fig. 2A, B), with a uniform distance between adjacent veins (white bars in Fig. 2A, B). In sistl1 leaf fragments, in contrast, the MC–BSC–V–BSC–MC pattern was disrupted, and the distance between adjacent veins decreased irregularly (Fig. 2D, E). After staining with I2–KI, many unstained MCs and BSCs were observed in sistl1 (Fig. 2F), which suggests that these cells lacked chloroplasts and thus could not accumulate starch. Observations of resin-embedded sections of sistl1 striped leaf fragments also identified some MCs and BSCs with no chloroplasts; consequently, they were not stained by toluidine blue and appeared as ‘empty’ cells (triangles and squares in Fig. 2H). In addition, some veins of sistl1 were only half normal in structure, indicating the possibility of asymmetric cell development along the vein axis.
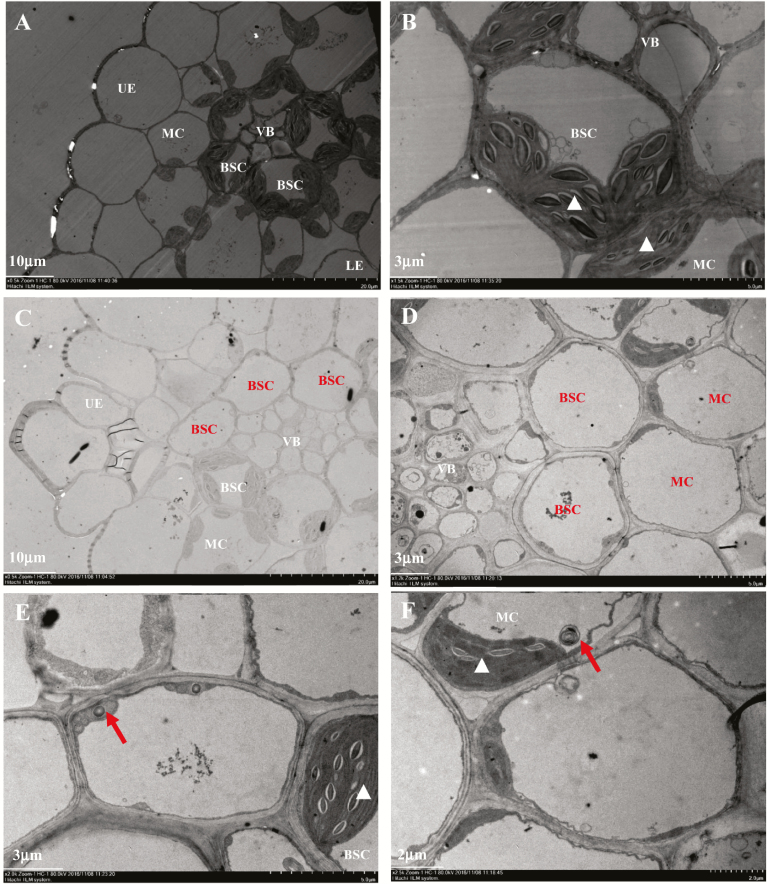
Ultrastructural observation revealed that the chloroplasts of sistl1 were indistinguishable from those of Yugu1 (white triangles in Fig. 3B, E, F), but the number of chloroplasts per BSC and MC was reduced dramatically (Figs 2G, H, 3A, C). According to our observation, most BSCs and MCs of Yugu1 contained two to four chloroplasts, whereas in sistl1, BSCs and MCs with no chloroplasts accounted for the largest proportion of cells (see Supplementary Fig. S1). Furthermore, we found that cells lacking chloroplasts produced many lysosome- and peroxisome-like organelles (Fig. 3E, F). All of these observations indicate that sistl1 had reduced chloroplast biogenesis and exhibited asymmetric leaf cell development.
Fig. 3.
Reduced chloroplast biogenesis in sistl1. (A, B) Ultrastructure of Yugu1 fifth-leaf fragments observed by transmission electron microscopy (TEM) under ×0.5K (A) and ×1.5K (B) magnification. (C–F) Ultrastructure of fifth-leaf sistl1 fragments observed by TEM under ×0.5K (C), ×1.2K (D), ×2.0K (E) and ×2.5K (F) magnification. Normal cells in Yugu1 and sistl1 are labeled with white letters, with white triangles indicating normally developed chloroplasts in BSCs and MCs. Unusual empty cells in sistl1 are labeled with dark letters. Dark arrows indicate lysosome- or peroxisome-like organelles.
Map-based cloning of the SiSTL1 gene
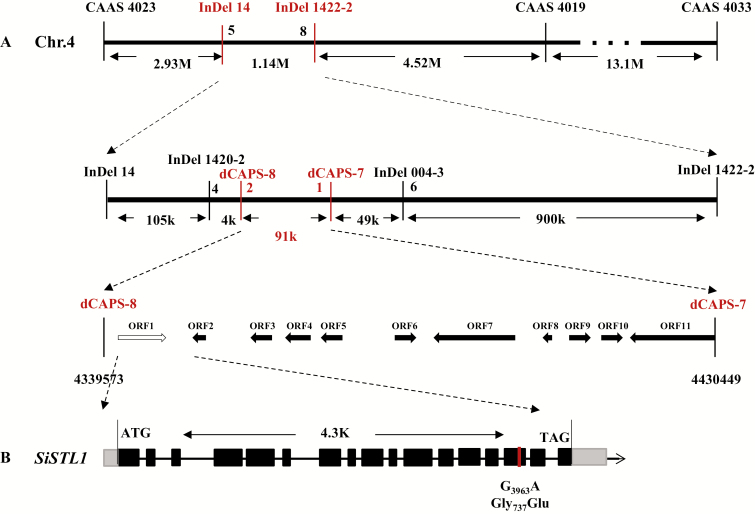
Genetic mapping of the SiSTL1 gene was performed using F2 individuals generated from a cross between the sistl1 mutant and foxtail millet cultivar SSR41. Using PCR-based markers, the SiSTL1 locus was initially mapped to an 8.6-Mb region between two SSR markers, CAAS 4023 and CAAS 4019, on chromosome 4 (Fig. 4). To generate a fine mapping, InDel and CAPS markers were developed by comparing the genomic sequences of Yugu1 and SSR41. The SiSTL1 locus was finally narrowed to a 91-kb interval between CAPS-8 (4339573 on chromosome 4) and CAPS-7 (4430449 on chromosome 4). In this region, three SNPs were identified by whole-genome resequencing of Yugu1 and sistl1 (see Supplementary Table S6) using Illumina next-generation sequencing technology. The first SNP, in the 15th exon of Seita.4G058800 (Chr4: 4347392), was a non-synonymous G3963A mutation leading to a missense mutation (Gly737 to Glu) in the encoded RNR large subunit protein (SiSTL1). The second SNP occurred in an intergenic region (Chr4: 4412036). According to the genome annotation of Setaria italica v2.2, the region 5000 bp upstream and downstream of this position does not contain a gene or ncRNA. The third SNP was located in the first exon of Seita.4G059700. It was a synonymous mutation also found in the Setaria mutants SiDWARF3 and Loose Panicle1 (Fan et al., 2017; Xiang et al., 2017). As described in Fan et al. (2017) and Xiang et al. (2017), SiDWARF3 and Loose Panicle1 both have dwarf and loose panicle phenotypes, but neither demonstrated the growth retardation or striped leaf phenotype that was seen for sistl1. The causal genes of the SiDWARF3 and Loose Panicle1 mutations were located in chromosomes 8 and 2, respectively. We therefore confirmed that the third SNP in the SiSTL1 locus was a background SNP. Notably, like sistl1, mutants of the RNR large subunit gene in Arabidopsis (cls8) and rice (v3) exhibit bleached and striped leaves (Garton et al., 2007; Yoo et al, 2009). We therefore confirmed that the non-synonymous SNP occurring in the 15th exon of Seita.4G058800 was responsible for the observed mutant phenotypes of sistl1.
Fig. 4.
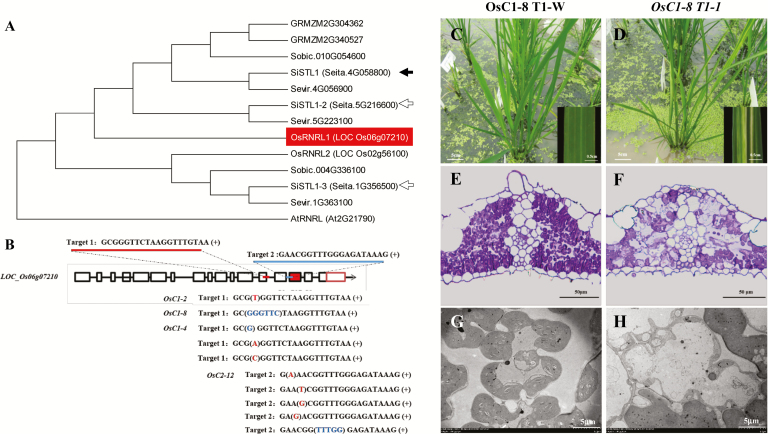
Map-based cloning of the SiSTL1 gene. (A) Map-based cloning of the SiSTL1 gene. Thick arrows indicate 11 candidate open reading frames (ORF) within the 91-kb interval. The white arrow indicates the mutant gene. (B) Schematic diagram of SiSTL1. Black boxes indicate the exons and black lines indicate introns of SiSTL1.
Functional verification of SiSTL1 in rice
Because a transfection system has not been fully established for Setaria, functional verification of Seita.4G058800 was carried out in rice. According to our phylogenetic analysis, LOC_Os06g07210 is orthologous to SiSTL1 in rice (Fig. 5A). We therefore constructed two pYLCRISPR/Cas9-MH vectors respectively targeting exons 13 and 15 (Fig. 5B) of LOC_Os06g07210. Of the subsequently generated positive transgenic T0 plants, one homozygous mutant (OsC1-2) and three heterozygous mutants (OsC1-4, OsC1-8, and OsC2-12) of LOC_Os06g07210 were obtained and exhibited striped leaves just like the sistl1 and rice v3 mutants (Yoo et al, 2009) (see Supplementary Fig. S2A). OsC1-2 was homozygous for a single T-nucleotide insertion in exon 13 (Fig. 5B) that was responsible for the loss of 168 amino acids from the C-terminus of the RNRL protein. We were unable to obtain T1 plants of this mutant, however, as all of them were albino and died at the early seedling stage, suggesting that the frame-shift mutation that occurred in this locus is lethal in homozygotes. We therefore observed the T1 lines of the heterozygous mutants. OsC1-8 had a 6-bp deletion in exon 13 (Fig. 5B), which resulted in the loss of two amino acids from the protein. It also produced albino T1 seedlings, and we only obtained one striped T1 plant (OsC1-8 T1-1) (Fig. 5D). Resin-section and ultrastructural assays of its leaf fragments showed that some of the mesophyll cells lacked chloroplasts (Fig. 5F, H). OsC1-4 had a G deletion in exon 13 (Fig. 5B), and three striped T1 plants were obtained among its T1 descendants (Supplementary Fig. S2B). A variation of OsC2-12 was an A insertion in exon 15 (Fig. 5B), and we also obtained three T1 plants that displayed pronounced stripe-leaf phenotype (Supplementary Fig. S2C). What is more, although in a small proportion, two striped descendants were also obtained among the descendants of OsC1-8 T1-1 (Supplementary Fig. S2D). We thus confirmed that the striped-leaf phenotype of all these transgenic plants was heritable. To study whether the striped leaf phenotype was tightly linked to the CRISPR-induced mutation, the targeted gene of the striped T1 and T2 descendants was sequenced. The result showed all of them contained variations in the targeted gene. OsC1-8 T1-1 was a chimeric mutant that contained the wild-type gene sequence, a 6-bp deletion in exon 13 that was inherited from OsC1-8, and a G deletion in exon 13 (Supplementary Fig. S3B). Genotypes of all three striped T1 descendants of OsC1-4 were the same as each other. As shown in Supplementary Fig. S3C, they were also chimeric mutants, containing the wild-type sequences, an A insertion in exon 13, and a T insertion in exon 13. Striped T1 descendants of OsC2-12 were heterozygous mutants. As showed in Supplementary Fig. S3D, variation of OsC2-12 T1-1 was the same as that of its parent OsC2-12; OsC2-12 T1-2 contained a C insertion in exon 15. OsC2-12 T1-3 had an A deletion in exon 15. As new variations were found in the striped T1 and T2 descendants, PCR was carried out with primers specific for Cas9 gene amplification, which showed that all the striped T1 and T2 descendants still contained the pYLCRISPR/Cas9-MH vector (Supplementary Fig. S4), suggesting that new variations occurring in these descendants might result from secondary editing by Cas9 in the plants. Taking all these results together, we tentatively concluded that mutations of LOC_Os06g07210 result in a striped-leaf phenotype and the striped leaf phenotype of sistl1 was caused by the G3963A base substitution in the SiSTL1 gene.
Fig. 5.
Knock-out of SiSTL1 homologous gene in rice. (A) Phylogenetic tree of RNRLs in Arabidopsis, Oryza sativa, Zea mays, Sorghum bicolor, and Setaria italica based on protein sequences. Black arrow indicate the causal gene for sistl1. White arrows indicate two other genes encoding SiSTL1 homologues in Setaria (SiSTL1-2 and SiSTL1-3). The homolog of SiSTL1 in rice (OsRNRL1, LOC Os06g07210) is highlighted by shading. (B) Position of two CRISPR targets in OsRNRL1 and sequence variations in the striped transgenic T0 plants, OsC1-2, OsC1-4, OsC1-8, OsC2-12, and three other chimeric plants. (C, D) T1 plants of OsC1-8 grown in a paddy field. OsC1-8 T1-W, normal green individual; OsC1-8 T1-1, striped-leaf individual. (E, F) Resin sections of OsC1-8 T1-W (E) and striped OsC1-8 T1-1 (F) leaf fragments. (G, H) Ultrastructures of OsC1-8 T1-W (G) and striped OsC1-8 T1-1 (H) leaf fragments.
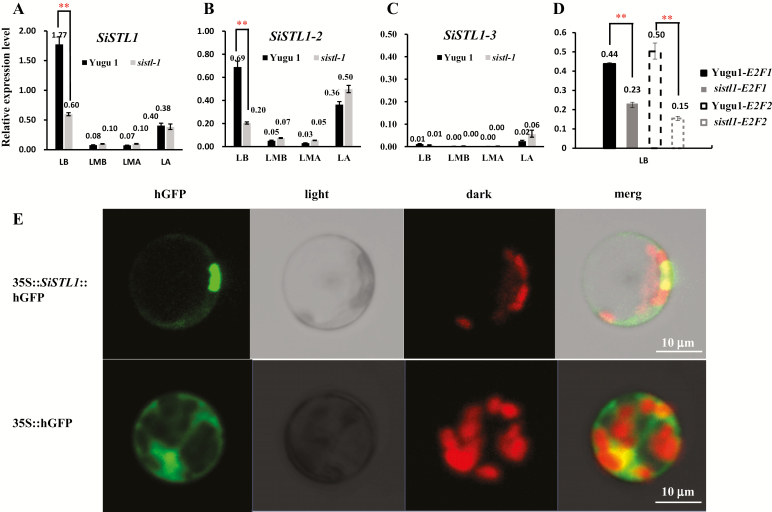
SiSTL1 is preferentially expressed in younger leaf tissues.
Phylogenetic analysis revealed the presence of three genes encoding RNRL in Setaria: Seita.4G058800 (SiSTL1), Seita.5G216600 (SiSTL1-2), and Seita.1G356500 (SiSTL1-3) (black and white arrows in Fig. 5A). To investigate the expression pattern of these genes along leaf developmental gradients, four leaf fractions of the striped fourth leaves of sistl1 and normal fourth leaves of Yugu1 were extracted as described in Li et al. (2010). The first fraction (LB) was taken from the 1 cm basal region above the fourth leaf ligule. This leaf zone, which was undergoing the most active cell division and was wrapped in the third leaf sheath, had not yet developed any chloroplasts and represented the earliest stage of leaf development (Li et al., 2010). The fourth fraction (LA) was obtained from the 1 cm region beneath the leaf tip. This leaf portion was totally expanded and contained well-developed chloroplasts (Li et al., 2010). The second and third fractions (LMB and LMA) corresponded respectively to the 1 cm regions beneath and above the third leaf ligule. These fractions represented transition stages for proplastid development into chloroplasts (Li et al., 2010).
According to our qRT-PCR analysis, SiSTL1 was preferentially expressed in LB, followed by LA (Fig. 6A). Fewer transcripts were detected in LMB and LMA fractions. These results suggest that active RNR activity is required during early leaf cell division. The expression pattern of SiSTL1-2 was the same as that of SiSTL1, but the abundance was much lower (Fig. 6B). Virtually no transcripts of SiSTL1-3 were detected in any of the leaf tissues (Fig. 6C), indicating that SiSTL1-3 may be a pseudogene or have functions in other tissues. Notably, compared with Yugu1, relative expression levels of SiSTL1 as well as its upstream regulators, E2F1 and E2F2, were greatly reduced in the LB fraction of sistl1 (Fig. 6D), indicating that a feedback mechanism involving the upstream genes E2F1, E2F2, and SiSTL1 may operate in sistl1.
Fig. 6.
Expression pattern of SiSTL1 and subcellular location of SiSTL1. (A–C) Expression patterns of SiSTL1 (A), SiSTL1-2 (B), and SiSTL1-3 (C) along leaf developmental gradients in Yugu1 and sistl1 fourth leaves. LB, basal 1 cm region above the fourth leaf sheath; LMB, 1 cm region beneath the third leaf sheath; LMA, 1 cm region above the third leaf sheath; LA, 1 cm region beneath the leaf tip. Asterisks indicate a significant difference between the relative expression level of genes in Yugu1 and sistl1 fourth leaf base; error bars, ±SD (n=3 replicates), Student’s t-test, P<0.01. (D) Expression patterns of E2F1 and E2F2 in Yugu1 and sistl1 fourth-leaf bases. (E) Subcellular localization of the SiSTL1 protein in Setaria protoplast.
In a SiSTL1 subcellular localization experiment, hGFP signals were detected in both the nucleus and cytoplasm (Fig. 6E). This observation is consistent with the results of Lincker et al. (2004), who reported that RNRL is primarily present in the cytoplasm and can be transferred to the nucleus when RNR activity is needed.
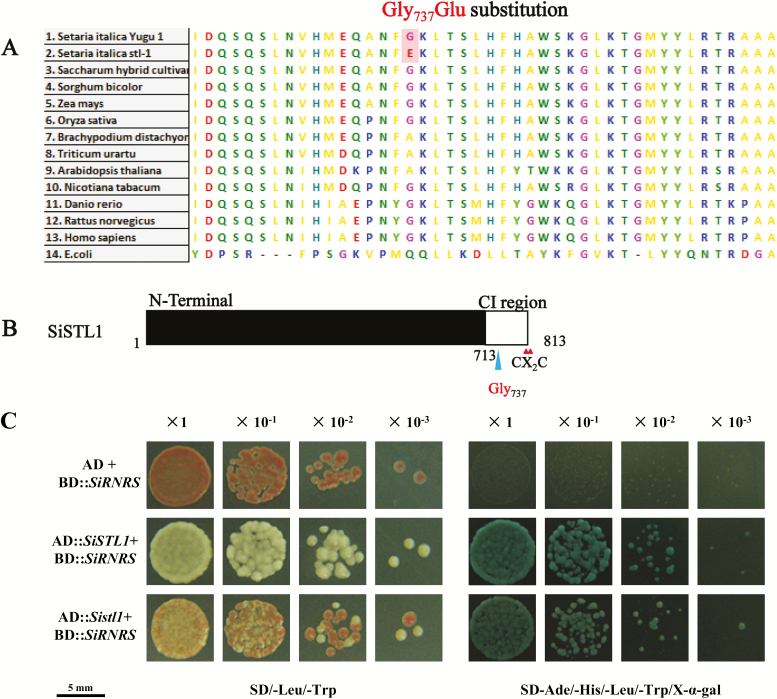
Gly to Glu substitution in the C-terminus of SiSTL1 weakens its interaction with the RNR small subunit
Sequence analysis indicated that Gly737 is not a highly conserved residue. Among 100 eukaryotic RNRL proteins, 78% have Gly in the homologous location (Fig. 7A). Ala was present in 9% of such proteins, with a Lys or Ser in the remainder. Both Gly and Ala are uncharged amino acids, with –H or –CH3 as the side chain, respectively. In contrast, Glu is an acidic amino acid that has a negatively charged –(CH2)2–COO− side chain. The Gly737Glu substitution thus changed the charge properties of this part of the protein.
Fig. 7.
Sequence analysis of SiSTL1 and results of a yeast two-hybrid assay of SiSTL1 and SiRNRS. (A) Comparison of aligned sequences of SiSTL1 and homologous proteins from other species. (B) Schematic diagram of SiSTL1. Small triangles indicate the positions of the CX2C motif of SiSTL1. The last ~100 amino acids before the CX2C motif are designated as the C-terminal insertion (CI) region. The large triangle indicates the position of Gly737 in the CI region of SiSTL1. (C) Yeast two-hybrid analysis of SiSTL1 and SiRNRS. Dilutions are shown at the top (×10−1, yeast diluted 10 times; ×10−2, yeast diluted 100 times; ×10−3, yeast diluted 1000 times).
The CI region, which comprises approximately the last 100 amino acids before the CX2C motif at the RNRL C-terminus, is important for optimal RNRL activity (Zhang et al., 2007). An rnr1 rnr3 double mutant of Saccharomyces cerevisiae transformed with rnr1 mutant alleles lacking the CI region grew more slowly than the wild-type (Zhang et al., 2007). We thus speculated that the Gly737Glu substitution in the SiSTL1 CI region (Fig. 7B) would affect the function of this protein. To test this hypothesis, Y2H tests were conducted. Wild-type SiSTL1 (Gly737) and mutant SiSTL1 (Glu737) alleles were fused to AD vectors, and the RNR small subunit (SiRNRS, Seita.4G114600) was fused to a BD vector. Y2H Gold yeast cells transformed with the Glu737 SiSTL1 allele were visible on SD/−Ade/−His/−Leu/−Trp/X-α-gal medium (Fig. 7C). However, compared with yeast transformed with the wild-type Gly737 SiSTL1 allele, they grew slowly, with blue substrate appearing approximately 6 h later. Similar results were obtained from Y2H tests using Gly737 and Glu737 SiSTL1 alleles respectively fused to BD vectors and SiRNRS fused to an AD vector. These consistent results indicate that the Gly737 to Glu substitution in the C-terminus of SiSTL1 does not block the interaction of this protein with the small subunit, but weakens its optimal functioning.
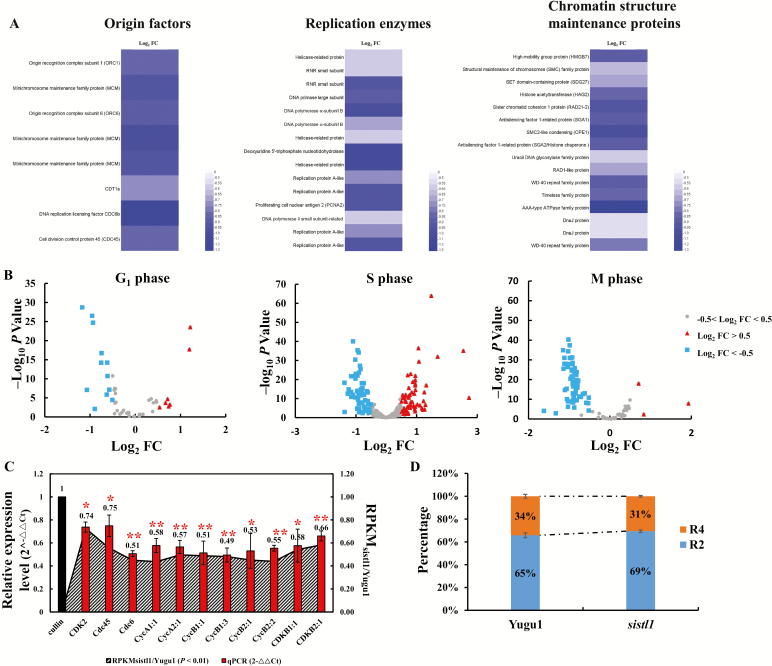
DNA replication activities are affected in sistl1
Because inhibition of RNR activity with hydroxyurea slows DNA replication in budding yeast (Poli et al., 2012), we presumed that DNA replication activity in sistl1 would also be affected. To verify this assumption, we analysed expression abundances of genes involved in replication activities by RNA-seq analysis using the basal zone of the striped seventh leaves of sistl1 and corresponding leaf region of Yugu1. Expression abundances of genes involved in initiation and activation of replication origins, including MCMs, ORC1, ORC6, Cdc6, Cdt1 and Cdc45, and genes encoding multiple replication enzymes and chromatin structure maintenance proteins, including DNA polymerase α- and δ-subunits, DNA primase large subunit, helicase-related proteins and structural maintenance of chromosomes family proteins, were significantly reduced in sistl1 (Fig. 8A; Supplementary Table S7). These observations strongly suggest that DNA replication activities are impeded in sistl1.
Fig. 8.
RNA-seq and flow cytometric analysis of Yugu1 and sistl1. (A) Heatmaps of genes encoding DNA replication origin factors, replication enzymes or chromosome structural maintenance proteins that significantly reduced gene expression abundances in sistl1 (log2FC<−0.5 and P<0.01, log2FC=log2RPKMsistl1/Yugu1. RPKM, reads per kilobase of transcript per million fragments mapped). (B) Expression changes of 49, 324 and 107 genes that were preferentially expressed during G1, S, and M phase, respectively, between sistl1 and Yugu1 within the RNA-seq analysis. The x-axis is log2FC, indicating log2RPKMsistl1/Yugu1 of the discussed genes, and the y-axis is −log10P. The triangles indicate genes where log2FC>0.5 and −log10P>2, which means that the expression level of these genes in sistl1 was >1.41 times higher than that in Yugu1. The squares indicate genes where log2FC<−0.5 and −log10P>2, which means that the expression level of these genes in sistl1 was >1.41 times down-regulated in Yugu1. The gray circles represent genes where −0.5<log2FC<0.5, which means that the expression level of these genes in sistl1 had no significant difference compared with that of in Yugu1. Detailed information, such as gene ID, log2FC, P-value and preferential expression phase of the genes shown in the figure is listed in Supplementary Table S8. (C) Relative expression levels of 11 representative cell cycle regulatory genes in sistl1. Left y-axis shows the 2−ΔΔCt values for these genes with a qRT-PCR assay (with Cullin as the reference gene). Right y-axis shows the RPKMsistl1/Yugu1 values for these genes extracted from RNA-seq. Asterisks indicate a significant difference between the relative expression level of genes in Yugu1 and sistl1; error bars, ±SD (n=3 replicates), Student’s t-test, **P<0.01; *P<0.05. (D) Results of flow cytometric analysis of 3-day-old first-leaf cells. R4 indicate 4C cells, and R2 indicate 2C cells.
Cell cycle progression is arrested in sistl1
To verify whether cell cycle progression is affected in sistl1, we further explored the RNA-seq results. Menges et al. (2003) identified a set of 1082 cell cycle-regulated genes, among which 129, 669, 20, and 198 had expression peak in the G1, S, G2, and M phase, respectively, by analysis of gene expression profiles during synchronous cell cycle progression with Arabidopsis cell suspension. We blasted these 1082 genes against the Setaria italica genome and found 486 Setaria homologs present in our RNA-seq list (Supplementary Table S8). And among these 486 genes, 214 genes exhibited significantly different expression patterns (P<0.01, |log2RPKMsistl1/Yugu1|>0.5) between sistl1 and Yugu1. Among the 214 genes, in the research of Menges et al. (2003), 20 had peak expression in the G1 phase, 120 in the S phase, 3 in G2 phase, and 71 in M phase. Interestingly, Setaria homologs of the G1- and S-phase-specific expressed genes in the research of Menges et al. (2003) were approximately half up-regulated (40% and 50.8%) and half down-regulated (60% and 49.2%) in sistl1, whereas Setaria homologs of the genes preferentially expressed during the M phase were largely down-regulated in sistl1 (3 (4.2%) up-regulated and 68 (95.8%) down-regulated; Fig. 8B). Because only a few genes were preferentially expressed during the G2 phase, they were not taken into account. We thus speculated that a decrease of cells in the M phase in sistl1 might be the cause of the reduced expression of genes believed to preferentially express during the M phase. To verify this, we conducted a flow cytometric analysis with 3-day-old first-leaf cells. The results indicated that the percentage of 4C cells in sistl1 was much lower than that of Yugu1 (Fig. 8D), consistent with the assumption that the number of G2/M-phase cells was significantly decreased in sistl1.
In addition, we performed GO term enrichment analysis for the 1082 cell cycle regulated genes described by Menges et al. (2003) and the 214 Setaria homologous genes that were differentially expressed between Yugu1 and sistl1 using the most current annotations (Tair 10 for Arabidopsis, and Setaria italica v2.2 for foxtail millet). As we expected, cell cycle-related GO terms were the most abundant terms. Supplementary Fig. S5A exhibits the top 20 most enriched non-redundant biological process GO terms for the 1082 Arabidopsis cell cycle regulated genes. The enriched GO term containing most abundant genes was ‘response to chemical stimulus’, which was consistent with the result described in Menges et al. (2003), where the data were generated with aphidicolin- and sugar starvation-treated MM2d cell suspensions. The remaining enriched GO terms were all cell cycle-related (see Supplementary Fig. S5A). The top 20 most enriched GO terms for the 214 DEGs in sistl1 were also all cell cycle-related (Supplementary Fig. S5B), with the three most significant terms being ‘regulation of mitotic cell cycle’, ‘regulation of cell cycle phase transition’, and ‘DNA replication initiation’. To further explore how these biological processes were affected in sistl1, we investigated how many genes were down-regulated and how many up-regulated in these biological processes. For 15 out 20 terms, the genes were all down-regulated in sistl1 (Supplementary Fig. S5C). The remaining five terms included only two up-regulated genes (three terms) or one up-regulated gene (two terms). These results are highly consistent with our finding that DNA replication and cell cycle progression were impeded in sistl1.
Moreover, qRT-PCR analysis of 11 genes considered to be related to cell cycle regulation, specifically, genes promoting S-phase entry (CDK2, Cdc45, Cdc6, CycA1:1, CycA2:1, CycB1:1, CycB1:3, CycB2:1, CycB2:2, CDKB1:1, and CDKB2:1) (Stevens and and La Thangue, 2003; Yang et al., 1999), revealed that they were significantly down-regulated in sistl1 (Fig. 8C). We also checked the RPKM values of all these genes in our RNA-seq list. The results were completely consistent. RPKMsistl1/Yugu1 values of all these genes were <1 (P-value<0.01), indicating that the relative expression level of all these genes was significantly reduced in sistl1. Taken together, our results suggest that cell cycle progression is arrested in the G1/S phase in sistl1.
Discussion
The phenotype of sistl1 is comparable to v3 and cls8 mutants
Mutants of RNRL have been characterized in both Arabidopsis (cls8) and rice (v3). v3 produces chlorotic leaves in a growth stage-dependent and temperature-conditional manner (Yoo et al., 2009). In favorable conditions, v3 generally exhibits a normal green phenotype up to the third-leaf stage, produces chlorotic leaves at the tillering stage, and then produces nearly normal green leaves after heading. If v3 is grown at a constant 20 °C, a temperature not optimal for growth, the bleached leaf phenotype is more severe, and striped leaves may appear beginning from the second leaf. The Arabidopsis mutant cls8-1 produces bleached, crinkled leaves because of reduced chloroplast numbers in leaf cells and asymmetric cell development along the vein axis (Garton et al., 2007).
In this study, we identified a RNRL mutant in Setaria (sistl1) that exhibits a phenotype comparable to v3 and cls8-1 (Fig. 1). As described above, sistl1 produces striped leaves in the same way as v3. Compared with fourth and fifth leaves generated during the shooting stage, the first three leaves of sistl1 and those produced after the late shooting stage are much less likely to exhibit the striped phenotype. When sistl1 is grown in poor conditions, the striped leaf phenotype is much more pronounced (Fig. 1E). We also observed an abnormal leaf vein arrangement and irregularly reduced leaf vein distances in sistl1 that may be due to the same phenomenon causing crinkled leaves and asymmetrical flowers in cls8-1, namely, asymmetrical development of cells along the vein axis (Fig. 2). Unlike cls8-1, however, sistl1 exhibited obvious growth retardation throughout the entire growth period (Fig. 1A–C), a behavior consistent with the reduced root growth of cls8-1 and the delayed plant growth of AtRNRL-disrupted RNAi lines (Garton et al., 2007).
SiSTL1 has two homologs in Setaria (Fig. 5A). The identity of protein sequences between SiSTL1 and SiSTL1-2 is 98% and the identity between SiSTL1 and SiSTL1-3 is 88%. In sistl1, a G3963A mutation in SiSTL1 caused a striped leaf phenotype regardless of the presence of the other two wild-type SiSTL1 homologs, SiSTL1-2 and SiSTL1-3, indicating that these proteins are not redundant. The reason might be as follows. First, in the qRT-PCR analysis to investigate the expression pattern of these genes along leaf developmental gradients, virtually no transcripts of SiSTL1-3 were detected in any leaf tissues (Fig. 6C; Supplementary Fig. S6). We thus think that SiSTL1-3 might be a pseudogene or functional in other tissues, and is not redundant with SiSTL1. Although the expression pattern of SiSTL1-2 was similar to that of SiSTL1, its abundance was much lower (Fig. 6B). Thus, it cannot fully complement the functional defects caused by the G3963A mutation of SiSTL1 in sistl1. In addition, relative expression levels of SiSTL1 as well as its upstream regulators E2F1 and E2F2 were greatly reduced in the LB fraction of sistl1 (Fig. 6A, D), indicating that a positive feedback other than negative feedback mechanism involving the upstream genes of E2F1, E2F2 and SiSTL1 may operate in sistl1. We thus presume that SiSTL1-2 might not be redundant with SiSTL1. However, it remains unclear why SiSTL1 and SiSTL1-2 are not functionally reductant although they share such high identity (98%). In the future, we need to undertake more experiments to draw stronger conclusions.
The function of RNRL is indispensable for plant growth and survival
No frame-shift mutations of RNRL have previously been identified in higher plants. In a study involving AtRNRL-disrupted RNAi lines, a quarter of the T2 seedlings had a pronounced cls8 phenotype, and most failed to develop beyond the four-leaf stage (Garton et al., 2007). Only heterozygotes were able to produce seeds, and homozygotes were unlikely to survive. In the current study, OsRNRL1 (LOC_Os06g07210) was targeted using the Cas9 protein. The T1 seedlings of OsC1-2, a homozygous frame-shift mutant, were unable to survive to the two-leaf stage. The T1 plants we obtained, which produced striped leaves, were descendants of heterozygous and chimeric T0 lines and were heterozygous and chimeric mutants (Supplementary Figs S2, S3). Taking all of this together we presume that activity of RNRL is indispensable for plant growth and survival, and transgenic descendants with either homozygous frame-shift mutations of the gene or dramatically decreased gene expression are unable to survive. In fact, heterozygous mutants OsC1-8, OsC1-4 and OsC2-12 also produced albino descendants. We presumed that these albino seedlings were homozygous frame-shift variations that derived from their heterozygous parents.
SiSTL1 functionally corresponds to OsRNRL1
In the paper describing the v3 mutant, Yoo et al. (2009) proposed that upon insufficient activity of RNR, plastid DNA synthesis is preferentially arrested to allow nuclear genome replication in developing leaves, enabling continuous plant growth. In other words, chloroplast biogenesis is vulnerable to insufficient activity of RNR. To verify the functionally corresponding relationship of SiSTL1 and OsRNRL1, we selected five genes (OsRNRL1 and four genes involved in cell cycle progression, corresponding to genes that were down-regulated in sistl1—OsE2F1, OsCDK2, OsCycA1:1 and OsCycB1:1) to perform qRT-PCR using the RNA of striped and normal T1 lines of OsC1-4, OsC2-12, and striped and normal T2 lines of OsC1-8. Compared with normal descendants, relative expression of all these genes was down-regulated in the striped descendants (see Supplementary Fig. S7). Thus, we confirmed that indeed SiSTL1 functionally corresponds to OsRNRL1. Taking all of these results together, we tentatively conclude that mutations of LOC_Os06g07210 result in a striped-leaf phenotype and the striped leaf phenotype of sistl1 was also caused by the G3963A base substitution in the SiSTL1 gene.
The CI region is important for the function of the RNR large subunit.
Higher plants with defective RNRL proteins feature reduced chloroplast biogenesis (Garton et al., 2007; Yoo et al., 2009). Notably, both cls8 and v3 are missense mutants with only a single amino acid alteration, namely, Gly718Ala in cls8-1 and Gly291Ser in v3. These residues are conserved in higher plants, but are not considered key residues in the catalytic site (Garton et al., 2007; Yoo et al., 2009). In this study, we identified another missense RNRL mutant, sistl1 in Setaria. The altered Gly737 is likewise not designated a key residue in allosteric regulation or any catalytic reaction. Indeed, in contrast to the above-mentioned mutations, Gly737 is not even highly conserved in higher plants (Fig. 7A). However the Gly737Glu substitution changed the charge of the protein and thus may disrupt the function of the CI region. sistl1 also exhibits a striped leaf phenotype and obvious growth retardation. In a study of Saccharomyces cerevisiae, deletion of the whole CI region has been found to affect the optimal function of the RNR large subunit (Zhang et al., 2007). In the present study, we observed that an amino acid substitution within the CI region also affects the function of SiSTL1. Our findings provide new evidence that the CI region is important to the function of the RNR large subunit.
Slight defects in SiSTL1 affect chloroplast DNA biosynthesis but not chloroplast development
Both v3 and cls8 exhibit reduced chloroplast biogenesis, consistent with our observations for sistl1. In this study, we conducted a qRT-PCR analysis of three SiSTL1 gene homologues during the transition to chloroplast from proplastids during leaf development. Relatively few gene transcripts were detected during this phase (Fig. 6A–C), suggesting that RNRL is not crucial for the development of chloroplasts from proplastids. Ultrastructural observation of sistl1 revealed that cells in striped areas lacked chloroplasts, whereas cells in green areas produced chloroplasts that were indistinguishable from those of Yugu1 (Fig. 3). No undifferentiated chloroplasts were observed in either leaf area. We thus propose that defects in SiSTL1 affect chloroplast DNA synthesis, but not chloroplast development.
Activation of the S-phase checkpoint may occur in sistl1
In yeast and human cells, inhibition of RNR activity leads to reduced cellular dNTP pools and subsequent DNA replication stress and activation of the S-phase checkpoint, thereby restricting the formation of later replication forks in the S phase, increasing dNTP production, and arresting the cell cycle transition (Giannattasio and Branzei, 2017; Pardo et al., 2017). In this study, many genes involved in DNA replication and cell cycle regulation were down-regulated in sistl1 (Fig. 8A, C). Given the close causal relationship between disrupted RNR activity and the S-phase checkpoint in yeast and human cells (Giannattasio and Branzei, 2017), we propose that the S-phase checkpoint is also activated by the defective SiSTL1 protein in sistl1 and plays a role in the reduced expression of genes involved in DNA replication and cell cycle regulation.
Notably, DNA replication stress and activation of the S-phase checkpoint always induced increased RNR activity in previous studies (Pardo et al., 2017). However, in our study, relative expression levels of two SiSTL1 genes and their regulators, E2F1 and E2F2, were all reduced in sistl1 (Fig. 6A, B, D). Expression abundance of genes encoding DP proteins, which combine with E2F to generate the functional E2F–DP complex, were also decreased in sistl1 (see Supplementary Table S7). Because ATR–CHK1 up-regulates E2F1 and thus increases RNR activity in human cells (Giannattasio and Branzei, 2017), we believe that other mechanisms exist in sistl1 to down-regulate the E2F transcription factors when cells encounter DNA replication stress caused by defective SiSTL1 protein.
Finally, the ability of the S-phase checkpoint to prevent late replication-origin firing is mediated by ATR–CHK1-dependent phosphorylation of replication origin proteins (Giannattasio and Branzei, 2017). This regulation, however, is at the protein level. In this study, we have provided new perspectives on how defective SiSTL1 protein impedes DNA replication at the transcriptional level.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Density curves of BSCs/MCs containing zero to six chloroplasts in Yugu1 and sistl1.
Fig. S2. Photographs showing four striped T0 lines, striped T1 lines of OsC1-4 and OsC2-12, and T2 lines of OsC1-8.
Fig. S3. Sequencing analysis of OsC1-8, OsC1-4, OsC2-12, and their descendants.
Fig. S4. Agarose gel electrophoresis for the PCR products amplified with Cas9 gene-specific primers from the T1 and T2 descendants of OsC1-4, OsC2-12, and OsC1-8.
Fig. S5. GO term enrichment analysis for the 1082 cell cycle-regulated genes described by Menges et al. (2003) in Arabidopsis and 214 Setaria homologs differentially expressed between Yugu1 and sistl1.
Fig. S6. Verifying the relative expression level of SiSTL1, SiSTL1-2, SiSTL1-3, E2F1, and E2F2 along leaf developmental gradients in Yugu1 and sistl1 fourth leaves with semi-quantitative RT-PCR.
Fig. S7. Relative expression levels of OsRNRL1, OsE2F1, OsCDK2, OsCycA1:1, and OsCycB1:1 in T1 descendants of OsC1-4 and OsC2-12, and T2 descendants of OsC1-8.
Table S1. Locus and primer sequences of InDel and CAPS markers.
Table S2. Primers used for vectors construction.
Table S3. Primers used for and transgenic verification.
Table S4. Primers used for qRT-PCR.
Table S5. Verification of RNA-seq result by qRT-PCR.
Table S6. SNPs identified within the candidate interval (91 kb).
Table S7. Differentially expressed genes involved in DNA replication in sistl1 and Yugu1.
Table S8. 486 cell cycle regulated genes expressed in sistl1 and Yugu1.
Table S9. Genes differentially expressed in sistl1 and Yugu1.
Acknowledgements
This work was supported by Fundamental Research Funds of CAAS (CAAS-XTCX2016002, Y2017JC15, S2018PY03), the National Natural Science Foundation of China (31501324, 31871692), the China Agricultural Research System (CARS06-13.5-A04), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences, and the Science and Technology Cooperation Program of Henan Province (172106000078). We thank Yaoguang Liu, PhD, from the Institute of Life Science, South China Agricultural University, for providing pYLCRISPR/Cas9-MH vectors and helpful instructions for their construction. We thank Chuanyin Wu, PhD, from the Institute of Crop Science, Chinese Academy of Agricultural Sciences, for providing a rice transgenic platform. We thank Jinrong Liu and Suying Wang, from Anyang Academy of Agriculture Sciences, for their contributions to foxtail millet mutant screening and identification. We offer our gratitude to Yanchao Jia for conducting field work. We thank Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Data deposition
Data collected from the germination trials, flow cytometic analysis and expression changes on sistl1 and Yugu1 are available at Dryad Digital Repository (https://doi.org/10.5061/dryad.t6v7t8s; Tang et al. 2018)
References
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112, 391–401. [DOI] [PubMed] [Google Scholar]
- Chabes A, Stillman B. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences, USA 104, 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes AL, Björklund S, Thelander L. 2004. S Phase-specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F-binding site and an upstream promoter activating region. The Journal of Biological Chemistry 279, 10796–10807. [DOI] [PubMed] [Google Scholar]
- Chabouté ME, Clément B, Philipps G. 2002. S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. The Journal of Biological Chemistry 277, 17845–17851. [DOI] [PubMed] [Google Scholar]
- Das SP, Borrman T, Liu VW, Yang SC, Bechhoefer J, Rhind N. 2015. Replication timing is regulated by the number of MCMs loaded at origins. Genome Research 25, 1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Tang S, Zhi H, He M, Ma W, Jia Y, Zhao B, Jia G, Diao X. 2017. Identification and fine mapping of SiDWARF3 (D3), a pleiotropic locus controlling environment independent dwarfism in foxtail millet. Crop Science 57, 2431–2442. [Google Scholar]
- Garton S, Knight H, Warren GJ, Knight MR, Thorlby GJ. 2007. crinkled leaves 8 – A mutation in the large subunit of ribonucleotide reductase–leads to defects in leaf development and chloroplast division in Arabidopsis thaliana. The Plant Journal 50, 118–127. [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Branzei D. 2017. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cellular and Molecular Life Sciences 74, 2361–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino E, Salguero I, Kearsey SE. 2014. Cellular regulation of ribonucleotide reductase in eukaryotes. Seminars in Cell & Developmental Biology 30, 97–103. [DOI] [PubMed] [Google Scholar]
- Guo Z, Kumagai A, Wang SX, Dunphy WG. 2000. Requirement for atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes & Development 14, 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015a. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Moon SJ, Min MK, Choi EH, Kim JA, Koh EY, Yoon I, Byun MO, Yoo SD, Kim BG. 2015b. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Frontiers in Plant Science 6, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolberg M, Strand KR, Graff P, Andersson KK. 2004. Structure, function, and mechanism of ribonucleotide reductases. Biochimica et Biophysica Acta 1699, 1–34. [DOI] [PubMed] [Google Scholar]
- Krek W, Xu G, Livingston DM. 1995. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell 83, 1149–1158. [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nature Genetics 42, 1060–1067. [DOI] [PubMed] [Google Scholar]
- Lin Q, Wang D, Dong H, et al. 2012. Rice APC/C(TE) controls tillering by mediating the degradation of MONOCULM 1. Nature Communications 3, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincker F, Philipps G, Chabouté ME. 2004. UV-C response of the ribonucleotide reductase large subunit involves both E2F-mediated gene transcriptional regulation and protein subcellular relocalization in tobacco cells. Nucleic Acids Research 32, 1430–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, et al. 2015. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant 8, 1274–1284. [DOI] [PubMed] [Google Scholar]
- Martins PK, Mafra V, De Souza WR, Ribeiro AP, Vinecky F, Basso MF, Cunha BADB, Kobayashi AK, Molinari HBC. 2016. Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Scientific Reports 6, 28348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. 2003. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Molecular Biology 53, 423–442. [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annual Review of Biochemistry 75, 681–706. [DOI] [PubMed] [Google Scholar]
- Pardo B, Crabbe L, Pasero P. 2017. Signaling pathways of replication stress in yeast. FEMS Yeast Research 17, 1–11. [DOI] [PubMed] [Google Scholar]
- Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. 2012. dNTP pools determine fork progression and origin usage under replication stress. The EMBO Journal 31, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. 2010. Ribonucleotide reductases: substrate specificity by allostery. Biochemical and Biophysical Research Communications 396, 19–23. [DOI] [PubMed] [Google Scholar]
- Rossbach D, Bryan DS, Hesselberth JR, Sclafani R. 2017. Localization of Cdc7 protein kinase during DNA replication in Saccharomyces cerevisiae. G3 7, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanvisens N, de Llanos R, Puig S. 2013. Function and regulation of yeast ribonucleotide reductase: cell cycle, genotoxic stress, and iron bioavailability. Biomedical Journal 36, 51–58. [DOI] [PubMed] [Google Scholar]
- Shao J, Zhou B, Chu B, Yen Y. 2006. Ribonucleotide reductase inhibitors and future drug design. Current Cancer Drug Targets 6, 409–431. [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Kinney JB, Stillman B. 2016. Concerted activities of Mcm4, Sld3, and Dbf4 in control of origin activation and DNA replication fork progression. Genome Research 26, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB. 2003. E2F and cell cycle control: a double-edged sword. Archives of Biochemistry and Biophysics 412, 157–169. [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF. 1995. Spkl/Rad53 is regulated by Mecl-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes & Development 10, 395–406. [DOI] [PubMed] [Google Scholar]
- Tang C, Tang S, Zhang S, Luo M, Jia G, Zhi H, Diao X. 2018. Data from: SiSTL1 encoding a large subunit of RNR, is crucial for plant growth, chloroplast biogenesis, and cell cycle progression in Setaria italica. Dryad Digital Repository. doi: 10.5061/dryad.t6v7t8s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z. 2006. Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. The Plant Cell 18, 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Khoshravesh R, Karki S, Tapia R, Balahadia CP, Bandyopadhyay A, Quick WP, Furbank R, Sage TL, Langdale JA. 2017. Re-creation of a key step in the evolutionary switch from C3 to C4 leaf anatomy. Current Biology 27, 3278–3287.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer J, Jung CK, Shackel I, Williams PM. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Analytical Biochemistry 270, 41–49. [DOI] [PubMed] [Google Scholar]
- Xiang J, Tang S, Zhi H, Jia G, Wang H, Diao X. 2017. Loose Panicle1 encoding a novel WRKY transcription factor, regulates panicle development, stem elongation, and seed size in foxtail millet [Setaria italica (L.) P. Beauv.]. PLoS One 12, e0178730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Müller C, Huynh V, Fung YK, Yee AS, Koeffler HP. 1999. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Molecular and Cellular Biology 19, 2400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC. 2009. Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiology 150, 388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Tang C, Zhao Q, et al. 2014. Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 15, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yang K, Chen CC, Feser J, Huang M. 2007. Role of the C terminus of the ribonucleotide reductase large subunit in enzyme regeneration and its inhibition by Sml1. Proceedings of the National Academy of Sciences, USA 104, 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. The EMBO Journal 20, 3544–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Georgieva B, Chabes A, Domkin V, Ippel JH, Schleucher J, Wijmenga S, Thelander L, Rothstein R. 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Molecular and Cellular Biology 20, 9076–9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.