The pea auxin receptors PsTIR1a, PsTIR1b, and PsAFB2 are involved in auxin perception, and the TIR1s are implicated in the differential growth response to 4-Cl-IAA and IAA in pea fruit.
Keywords: Auxin, auxin receptors, 4-chloroindole-3-acetic acid, fruit development, hormonal interaction, indole-3-acetic acid, Pisum sativum, TIR1/AFB genes
Abstract
The auxins indole-3-acetic acid (IAA) and 4-chloroindole-3-acetic acid (4-Cl-IAA) occur naturally in pea (Pisum sativum); however, only 4-Cl-IAA mimics the presence of seeds in stimulating pericarp growth. To examine if this differential auxin effect is mediated through TIR1/AFB auxin receptors, pea TIR1 and AFB2 homologs were functionally characterized in Arabidopsis, and receptor expression, and auxin distribution and action were profiled in developing pea fruits. PsTIR1a, PsTIR1b, and PsAFB2 restored the auxin-sensitive root growth response to the mutant Arabidopsis seedlings Attir1-10 and/or Attir1-10 afb2-3. Expression of PsTIR1 or AtTIR1 in Attir1-10 afb2-3 mutants also restored the greater root inhibitory response of 4-Cl-IAA compared to that of IAA, implicating TIR1 receptors in this response. The ability of 4-Cl-IAA to stimulate a stronger DR5::GUS auxin response than IAA at the same concentration in pea pericarps was associated with its ability to enrich the auxin-receptor transcript pool with PsTIR1a and PsAFB2 by decreasing the transcript abundance of PsTIR1b (mimicking results in pericarps with developing seeds). Therefore, the markedly different effect of IAA and 4-Cl-IAA on pea fruit growth may at least partially involve TIR1/AFB receptors and the differential modulation of their population, resulting in specific Aux/IAA protein degradation that leads to an auxin-specific tissue response.
Introduction
Auxins are major regulators of plant reproductive and vegetative development. Auxins occurring naturally in plants include the most ubiquitous auxin indole-3-acetic acid (IAA), indole-3-butyric acid, phenylacetic acid (PAA, a weak auxin), and the chlorinated auxin 4-chloroindole-3-acetic acid (4-Cl-IAA; Reinecke et al., 1999). 4-Cl-IAA is a potent auxin tested in many different bioassays and reported to be 1.3–50 times more active than IAA (Reinecke, 1999). To date, species known to synthesize 4-Cl-IAA are restricted to the phylogenetic clades of the Fabeae and Trifoleae in the Fabaceae family, which includes Pisum sativum (pea), and it is found in high concentrations in the seeds of those plants (Reinecke, 1999; Lam et al., 2015). How different forms of natural auxins contribute to the overall auxin function in plants remains unclear. However, 4-Cl-IAA has been recognized as a regulator of fruit (Ozga and Reinecke, 2003; Ozga et al., 2009) and seed development in pea (McAdam et al., 2017).
The presence of developing seeds within the fruit is a requirement for normal ovary development in most flowering plants, including pea. The lack of ovule fertilization or seed removal results in ovary (pericarp) senescence and subsequent pea fruit abscission (Ozga et al., 1992; Ozga and Reinecke, 1999). Application of 4-Cl-IAA to the deseeded pericarp stimulates growth; in contrast, application of the other naturally occurring auxin IAA does not (Reinecke et al., 1995). A major determinant of 4-Cl-IAA-specific pericarp growth promotion is its ability to stimulate gibberellin (GA) biosynthesis (van Huizen et al., 1995, 1997; Ozga et al., 2003, 2009). In other species, such as tomato (Solanum lycopersicum; Serrani et al., 2008) and Arabidopsis (Dorcey et al., 2009), IAA stimulates fruit growth via GAs. Furthermore, in pea 4-Cl-IAA, but not IAA, also inhibits the ethylene response in deseeded pericarps, potentially via the inhibition of ethylene signaling (Johnstone et al., 2005; Jayasinghege et al., 2017), enhancing the promotion of pericarp growth. These differential interactions of IAA and 4-Cl-IAA with the GA and ethylene biosynthesis and signaling pathways likely play a primary role in determining the effects of the two auxins on pea pericarp development. However, the underlying signaling mechanisms that lead to these differential auxin effects on the hormonal pathways within the fruit are not known.
Repeated auxin application and dose–response experiments have demonstrated that the comparatively lower activity of IAA, or its lower chemical stability compared to 4-Cl-IAA, is not likely to be the primary reason for its inability to stimulate pea pericarp growth (Reinecke et al., 1995; Reinecke, 1999). Auxin analogs with varying substitutions at the fourth position of the indole ring, or analogs with chlorine or fluorine substituents at different positions of the indole ring, have been evaluated for their ability to induce pericarp development. These studies have shown that the position of the substituent on the indole ring, its size, and its lipophilicity are important factors in determining the activity of the analog, with the fourth position on the ring and a substituent size of approximately that of a chlorine atom being optimal for biological activity with respect to stimulating pea pericarp growth (Reinecke et al., 1995, 1999). Based on these observations, it has been speculated that the differential effects of IAA and 4-Cl-IAA on pea pericarp development may rest at the auxin-receptor level (Reinecke et al., 1999).
The TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX (TIR1/AFB) and AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) protein families act as co-receptors in auxin signaling. The interaction of the co-receptors in the presence of auxin initiates the proteasome-mediated degradation of Aux/IAAs and subsequent activation of the auxin response (Calderón Villalobos et al., 2012). The auxin-dependent interactions of the TIR1/AFB and Aux/IAA proteins appear to depend on which TIR1/AFB and Aux/IAA proteins combine to make the co-receptor, which type of auxin is present, and on the auxin concentration (Calderón Villalobos et al., 2012; Havens et al., 2012). For instance, the 50% effective concentration (EC50) of auxin required for the degradation of Arabidopsis Aux/IAAs differs depending on the Aux/IAA protein and F-box protein (TIR1 or AFB2) complex. The EC50 values are also higher with the weak auxin PAA compared to that of IAA or 4-Cl-IAA within a specific Aux/IAA protein and F-box protein complex (Shimizu-Mitao and Kakimoto, 2014).
The TIR1/AFB auxin-receptor protein family is conserved across land plants and phylogenetic analysis divides the family into four clades: TIR1, AFB2, AFB4, and AFB6. Arabidopsis contains three TIR1/AFB clades with two alleles per clade (AtTIR1 and AtAFB1; AtAFB2 and AtAFB3; AtAFB4 and AtAFB5; Parry et al., 2009). Pea contains TIR1/AFB gene members from four clades (TIR1a and TIR1b; AFB2; AFB4; AFB6; Ozga et al., 2010; Ligerot et al., 2017). Much of our current understanding of auxin-receptor function comes from the auxin response in Arabidopsis root growth assays. In these assays, the AtTIR1 and AtAFB1 proteins do not contribute equally to auxin signaling that affects growth, with a functional AtTIR1 being sufficient for normal auxin response in the absence of AtAFB1 (Parry et al., 2009). Although the absence of a functional AtTIR1 (Attir1-1 mutant) shows the highest auxin insensitivity in the Arabidopsis root elongation assays, the Attir1-1 Atafb2-3 double-mutant further enhances auxin insensitivity, demonstrating that AFB2 also has a role in root growth (Parry et al., 2009). While TIR1 followed by AFB2 are likely the most prominent auxin receptors regulating Arabidopsis root elongation, much less is known about the role of these receptors in Arabidopsis fruit development. The Arabidopsis tir1 and tir1afb2 mutants generally produce fruits similar to that of the wild-type (Dharmasiri et al., 2005); however, treatment with the auxin transport inhibitor N-1-naphthylphthalamic acid induces a greater frequency of reduced-valve and valveless silique phenotypes in these mutants compared with the wild-type (Ståldal et al., 2008; Zúñiga-Mayo et al., 2014), supporting a role of these auxin receptors in auxin-related fruit developmental processes. Little is known about the distinct functions of auxin receptors in species other than Arabidopsis due to the lack of available auxin-receptor mutants. Overexpression of auxin-receptor genes in tomato, including SlTIR1 (Ren et al., 2011) and CsTIR1 or CsAFB2 from cucumber (Cucumis sativus) (Xu et al., 2017), is reported to induce parthenocarpic fruit formation, showing that increasing ovary auxin sensitivity by elevating auxin-receptor abundance can modify tomato fruit development. To date, the only auxin-receptor mutant characterized in pea is an AFB4 mutant (also named PsAFB4/5 or ramosus2), which has been shown to be involved in the regulation of shoot branching (Ligerot et al., 2017).
In this study, in order to understand auxin perception in pea, the pea TIR1/AFB2 homologs were functionally characterized in Arabidopsis auxin-receptor mutant backgrounds to evaluate their role in the perception of 2,4-D. The TIR1 homologs were further tested for their perception of IAA and 4-Cl-IAA to determine whether they contribute to the differential action of IAA and 4-Cl-IAA in pea fruit development. The regulation of expression of pea pericarp auxin receptor genes by seeds and/or auxin was assessed to determine whether this could be a mechanism to modulate pericarp auxin responsiveness during fruit set. The expression of an auxin-responsive reporter gene (DR5::GUS) was also evaluated to determine whether modulation of auxin-receptor gene expression and/or auxin signaling are possible mechanisms involved in the differential action of IAA and 4-Cl-IAA during fruit development.
Materials and methods
Plant materials
The pea cultivar Pisum sativum L. cv. I3 (Alaska-type) was used for all pea studies. Plants were grown in a growth chamber with a 16/8 h light/dark photoperiod at 19/17 °C as described by Jayasinghege et al. (2017).
For the fruit developmental study, pistils (pericarp plus stigma and style; hereafter referred to as fruits) were used at the given developmental stage, located at flowering nodes 1–6 and within a specific pericarp length range (1 d after anthesis, DAA: 8–12 mm; 2 DAA: 15–20 mm; 3 DAA: 26–33 mm) and/or mean seed weight range (5 DAA: 1.2–2.5 mg; 6 DAA: 5–7 mg; 7 DAA: 14–16 mg; 8 DAA: 20–28 mg; 10 DAA: 70–100 mg). For non-pollinated fruits, floral buds at –2 DAA were emasculated, and tissues were collected at –2, 0, 1, 2, and 3 DAA. Fruits were collected onto ice and immediately dissected into seed/ovule, pericarp wall, pericarp dorsal vascular suture, and pericarp ventral vascular suture tissues (see Supplementary Fig. S1A at JXB online), except for those at –2 DAA, where the ovules were removed and the fruits were harvested.
For hormonal treatments, fruits at 2 DAA measuring 15–20 mm in length were split, deseeded, and treated with IAA or 4-Cl-IAA (50 μM in 0.1% aqueous Tween 80), ethephon (ethylene-releasing agent; 1000 mg l–1 in 0.1% aqueous Tween 80), or silver thiosulfate (STS, inhibitor of ethylene action; 1 mM in 0.1% aqueous Tween 80) either alone or in combination. Auxin or auxin–ethephon combinations were IAA plus 4-Cl-IAA, IAA plus ethephon, 4-Cl-IAA plus ethephon, all in 0.1% aqueous Tween 80. The split (split pericarps with seeds, SP) and split and deseeded (split pericarp, no seeds, SPNS) controls were treated with 0.1% aqueous Tween 80. All hormonal and control treatments were applied 12 h after pericarp splitting and seed removal, with one exception: STS was applied to the pericarp immediately after splitting and deseeding, with subsequent hormonal application occurring 12 h after STS application. Solutions (30 μl) were applied to the inside surface of the pericarp wall (endocarp), and the pericarps were attached to the plant throughout the experiment. Samples were collected into liquid nitrogen at 0, 2, 8, and 12 h after solution application (12, 14, 20, and 24 h after pericarp splitting, splitting and deseeding, or deseeding and STS treatment) and stored at –80 °C.
The DR5::GUS construct in the pRD400 vector (DeMason and Polowick, 2009) was transformed into pea as described by Reinecke et al. (2013), and T3 generation homozygous plants were studied. DR5-driven expression of the GUS (β-glucuronidase) marker gene was monitored over the course of development in pre-pollinated (–2 DAA), and pollinated fruits at 0, 3, 5, 8, and 10 DAA, and in 2 DAA deseeded pericarps treated with auxin.
For hormone quantification, non-pollinated fruits (ovules removed) at 3 DAA, pericarps from pollinated fruits (seeds removed) at 0, 3, 5, and 8 DAA, pericarp tissues from pollinated fruits (central wall, ventral vascular suture, dorsal vascular suture) at 5 DAA and 8 DAA (funiculus removed), and seeds at 8 DAA were harvested, immediately frozen in liquid nitrogen, and stored at –80 °C.
For gene expression analysis in different vegetative organs, tissues of 12-d-old pea seedlings were used. The plants had six or seven nodes below the shoot apex, and the cotyledonary node was designated as node 1. Leaves attached to the fourth node were collected as mature leaves (mean length 31±2 mm) and leaves attached to the sixth or seventh nodes were collected as immature leaves (mean length 8±2 mm). Internodes between nodes 3 and 4 were collected as mature tissues (mean length 25±5 mm) and the most apical internodes (between nodes 5 and 6, or 6 and 7) were collected as immature internodes (mean length 4±2 mm). The shoot apices were also collected from all the seedlings. For the auxin treatment of seedlings, seeds were germinated and grown in Magenta GA-7 vessels (Sigma-Aldrich) at room temperature (~22 °C) in continuous darkness. At 2 d after imbibition (DAI), water-grown seedlings were transferred to either water, IAA (1 µM), or 4-Cl-IAA (1 µM) and grown for an additional 2 d prior to harvesting of the plumule, epicotyl, and root-tip (6–7 mm) at 4 DAI. All seedling tissue samples were immediately frozen in liquid nitrogen and stored at –80 °C until RNA extraction.
Identification of pea TIR1/AFB family members
Five putative TIR1/AFB family members in the pea genome were identified by screening a small-scale Roche 454 Titanium sequencing database derived from the seed coats of the following pea cultivars at 10 DAA: I3 (Alaska-type), Courier, Canstar, Solido, and LAN 3017 (Ferraro, 2014). Complete coding sequences (CDSs) of all the genes were amplified from total RNA (primer sequences listed in Supplementary Table S1), cloned into the pCR8 vectors (Invitrogen), and verified by Sanger sequencing. Sequence comparisons and construction of a phylogenetic tree by the neighbor-joining method were conducted using the PRALINE sequence alignment program and the MEGA X program, respectively (Supplementary Protocol S1).
Gene expression analysis
RNA was isolated using a modified TRIzol (Life Technologies) method (Ayele et al., 2006). qRT-PCR primers, probes, and their reaction efficiencies are listed in Supplementary Table S2. All the probes were double-quenched and contained Iowa Black FQ (IBFQ) quencher at the 3′-end and 6-FAM fluorescent dye at the 5′-end (Integrated DNA Technologies). The only exception was the 18S rRNA control, which contained the fluorescent reporter VIC and the quencher TAMRA (Ozga et al., 2003). The CDSs of PsTIR1a and PsTIR1b share 82% identity; therefore, the target specificity of the qRT-PCR primers was confirmed by sequencing the PCR products. A TaqMan One-Step RT-PCR Master-Mix Reagents Kit or RNA-to-Ct 1-Step Kit (Applied Biosystems) was used for qRT-PCR analysis. The reactions were performed in a StepOnePlus Real-Time PCR system as described by Jayasinghege et al. (2017; Supplementary Protocol S2).
Transgenic Arabidopsis plants and root growth analyses
All Arabidopsis lines were from the Columbia (Col-0) ecotype. Wild-type Col-0 (CS 70000) and the T-DNA insertion mutants Attir1-10 (SALK_090445C) and Atafb2-3 (SALK_137151) were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). Backgrounds of the mutants were cleaned by backcrossing once with Col-0. Background-cleaned Atafb2-3 and Attir1-10 lines were crossed to obtain double-mutants. Transformation vectors were created by placing the PsTIR1a, PsTIR1b, PsAFB2 or AtTIR1 CDSs following the AtTIR1 3-kb promoter region (–1 bp to –3008 bp region from the start codon) into a modified version of pCAMBIA1300 named pCm1300-polyA (provided by Dr Enrico Scarpella, University of Alberta). Arabidopsis floral dip transformation was performed as described by Zhang et al. (2006).
Root growth assays were adapted from Parry et al. (2009). Transgenic lines homozygous for the transgene insert that expressed the transgene at reliable levels were selected for the assays (Supplementary Fig. S1). Seedlings were grown vertically under continuous light and aseptic conditions. Uniformly-sized seedlings at 4 d old were transferred aseptically to media with or without auxin (Arabidopsis lines expressing PsTIR1a, PsTIR1b, and AtTIR1 at 50 nM and 70 nM 2,4-D, or 400 nM and 800 nM IAA or 4-Cl-IAA; PsAFB2 at 70 nM and 90 nM 2,4-D) for an additional 3 d. The root elongation of each genotype in auxin medium is expressed as a percentage of the same genotype in the medium without auxin (Supplementary Protocol S3).
DR5::GUS expression analysis
For GUS staining, cross-sections of fresh tissue (<1 mm thick) of the mid-pericarp region from DR5::GUS expressing lines were made by hand using a scalpel. The staining procedure was adapted from Hull and Devic (1995). The sections were submerged in GUS staining solution (Supplementary Protocol S4) and placed in a vacuum desiccator for 30 min and then in an incubator at 37 °C for 12 h in darkness. After washing with 70% ethanol, the tissue sections were observed under a stereo microscope. To verify that the staining observed in the transgenic plants was due to the expression of DR5::GUS, SPNS pericarps treated with IAA, 4-Cl-IAA, or 0.1% aqueous Tween 80 were also examined in wild-type plants and no GUS staining was detected in the tissue 8 h after the hormone treatment (Supplementary Fig. S2).
For quantification of GUS enzyme activity, four pericarp wall discs per fruit (two from each side of the pericarp wall mid-region) were taken using a cork borer (6 mm diameter), immediately frozen in liquid nitrogen and stored at –80 °C. Tissues from two fruits were pooled as a biological replicate, and three biological replicates were assessed per treatment. GUS enzyme activity was determined using a fluorescent assay protocol employing the GUS substrate 4-methylumbelliferyl glucuronide (MUG) adapted from Jefferson (1987; Supplementary Protocol S4).
Hormonal analysis
Frozen tissues were lyophilized and ground to a fine powder. Each biological replicate consisted of tissues from a minimum of 10 fruits. Hormonal analysis was done by HPLC-tandem mass spectrometry at the National Research Council, Saskatoon, Canada (see Supplementary Protocol S5). We found that this method for extraction and quantification of auxins was sensitive for low levels of IAA and IAA-conjugates; however, it was not sensitive enough for quantification of low levels of 4-Cl-IAA (which occur in pea pericarp tissues; Magnus et al., 1997).
Statistical analyses
The data means in all experiments are the average of biological replicates (independent samples), and the number of biological replicates is given for each experiment within the specific data set. For the Arabidopsis root elongation and gene expression experiments, statistical significance was determined by performing a one-way ANOVA followed by a Holm–Sidak post hoc mean separation test (SigmaPlot 13; Systat Software Inc.). Two-tailed Student’s t-tests assuming unequal variance (Analysis ToolPak, Microsoft Excel 2013) were used for the pairwise comparisons of auxin-receptor gene expression in pollinated versus non-pollinated fruit tissues.
Results and discussion
TIR1/AFB family of auxin receptors in pea
Five genes belonging to the TIR1/AFB family of auxin receptors were identified in pea by screening a small-scale pea transcriptomic database. Two of these genes (PsAFB2 and PsAFB6, GenBank accession numbers KY829120 and KY829119, respectively) have already been sequenced and reported in a US patent (Ozga et al., 2010), one (PsAFB4, GenBank KX954126) was previously reported and characterized by Ligerot et al. (2017), and two were cloned and the sequences verified for the first time in this work (PsTIR1a and PsTIRb, GenBank KX954124 and KX954125, respectively). These genes represent four phylogenetic clades: TIR1, AFB2, AFB4, and AFB6 (Supplementary Fig. S3). Based on a transcriptome database screening, Ligerot et al. (2017) also reported five TIR1/AFB family members in pea. We further screened pea transcriptome sequences derived from diverse types of vegetative and reproductive tissues at different developmental stages (Franssen et al., 2011; Kaur et al., 2012; Duarte et al., 2014; Sudheesh et al., 2015) and found no additional putative pea auxin-receptor homologs, suggesting that the pea TIR1/AFB family is limited to only five members.
The leucine-rich repeat domain in the Arabidopsis TIR1 F-box protein contains a pocket for auxin binding. The bottom of this pocket contains an inositol hexakisphosphate (InsP6) molecule that probably acts as a structural co-factor. When auxin is bound in the TIR1 pocket, an Aux/IAA co-receptor protein fits into the pocket on top of the auxin molecule (Tan et al., 2007). Comparison of Arabidopsis and pea auxin-receptor proteins shows that all the amino acids important for auxin binding and interaction with InsP6 in AtTIR1 (Tan et al., 2007) are also conserved in the pea PsTIR1a, PsTIR1b, and PsAFB2 proteins, except for a single amino acid substitution, namely the V463 position in AtTIR1 represented by I457 in PsAFB2. A similar substitution was also seen in AtAFB2 (see amino acid alignment position 516 in Supplementary Fig. S4). The majority of AtTIR1 amino acids involved in the interactions with the Aux/IAA co-receptor were also conserved in the pea PsTIR1a, PsTIR1b, and PsAFB2 proteins (Supplementary Fig. S4).
Additionally, for the TIR1/AFB1 clade, a glycine (E) to lysine (K) change in the F-box domain of AtAFB1 compared to AtTIR1 reduces the ability of AtAFB1 to form a SKP1–CULLIN–F-box (SCF) ubiquitin ligase complex, which stabilizes the F-box protein and causes auxin resistance and associated growth defects, probably by protecting Aux/IAAs from degradation (Yu et al., 2015). The E-to-K substitution in the F-box domain is shared by AFB1 homologs from members of the Brassicaceae and Cleomaceae, but not in homologs from the outgroup species papaya (Carica papaya) and cacao tree (Theobroma cacao; Yu et al., 2015). Given this evolutionary grouping, the authors speculated that AFB1 and its orthologues retained a unique function after Brassicaceae diverged from the Capparaceae ~70 million years ago. Both PsTIR1 alleles possessed an E at this F-box domain position that is important for tethering TIR1 to SCF (Supplementary Fig. S5). However, the key E-to-K substitution was found in the F-box domain of the TIR1b allele of the Trifolium species in the subgenera Trifolium (Trifolium pratense and Trichocephalum (T. subterraneum), that as part of the Trifolieae tribe diverged from the Fabeae tribe in the Meso-Papilionoideae clade of the Fabaceae family ~16–23 million years ago (Ellison et al., 2006; Schaefer et al., 2012). Future detailed genetic studies in these or related legume species are required to determine whether the E-to-K change in the TIR1b orthologues in Trifolium imparts a unique function after the Trifolieae and Fabeae tribes diverged from their crown clade of Vicioid.
Pea auxin receptors functionally interact with 2,4-D, IAA, and 4-Cl-IAA for auxin response
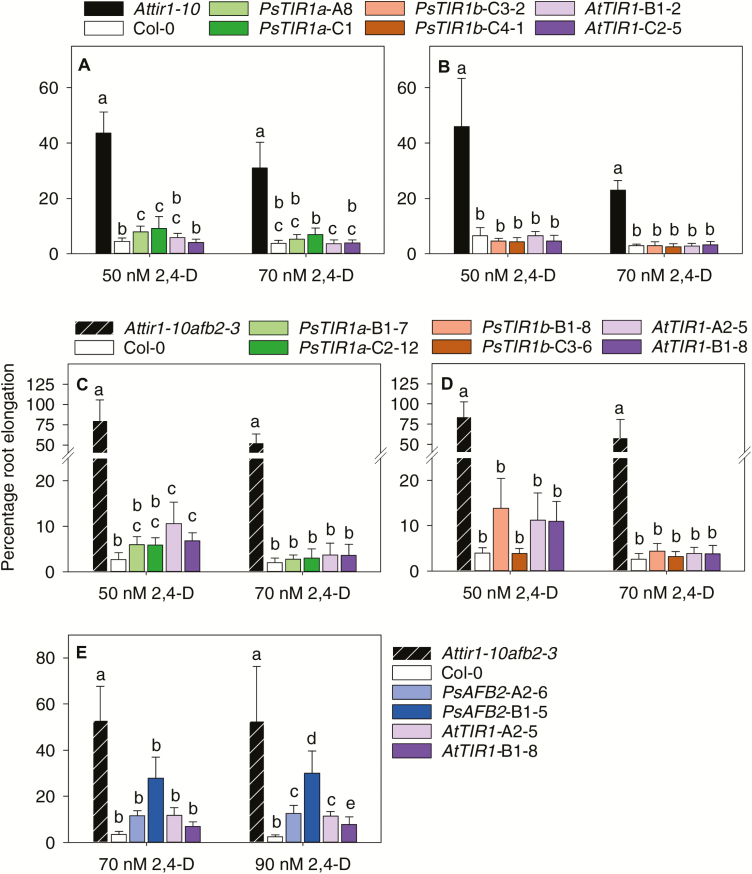
To determine whether PsTIR1a and PsTIR1b can complement AtTIR1 in root elongation assays, they were separately introduced into the Arabidopsis tir1-10 and Attir1-10 afb2-3 mutants under the regulation of the Arabidopsis TIR1 promoter. Attir1-10 has a T-DNA insertion near the 5′-end region of the CDS and therefore is probably a null mutation (Parry et al., 2009). In root growth assays, Attir1-10 shows 2,4-D-insensitive phenotypes similar to that of Attir1-1. In Atafb2-3, a T-DNA insertion in the promoter region reduces AtAFB2 expression (Parry et al., 2009). As a transgenic control, pAtTIR1::AtTIR1 was introduced into Attir1-10 and Attir1-10 afb2-3.
In root growth assays, Arabidopsis tir1-10 showed reduced auxin sensitivity (greater root elongation) in the presence of 2,4-D (50 nM and 70 nM) compared to that of the wild-type Col-0 seedlings (Supplementary Fig. S6). Compared to Attir1-10, the double-mutant Attir1-10 afb2-3 showed a further reduction in 2,4-D sensitivity, which was similar to that shown for Attir1-1 afb2-3 compared to Attir1-1 (Parry et al., 2009).
Transgenic Arabidopsis plants expressing PsTIR1a or PsTIR1b restored the 2,4-D inhibition of root growth in the Arabidopsis tir1-10 and tir1-10afb2-3 mutants to levels similar to those of mutants expressing the AtTIR1 transgene, and this was consistent with the auxin root-inhibiting activity exhibited by the wild-type seedlings (Fig. 1A–D). The restoration of auxin-sensitive root growth in the Arabidopsis auxin-resistant mutants indicates that both PsTIR1a and PsTIR1b code for functional auxin receptors.
Fig. 1.
Functional characterization of pea auxin receptors with 2,4-D in Arabidopsis. Seedlings at 4 d old were transferred to media containing 2,4-D for 3 d. The seedlings assessed were Col-0, Attir1-10, Attir1-10 afb2-3, or those expressing (A, B) pAtTIR1::cPsTIR1a (PsTIR1a-A8 and C1), pAtTIR1::cPsTIR1b (PsTIR1b-C3-2 and C4-1) or pAtTIR1::cAtTIR1 (AtTIR1-B1-2 and C2-5) in Attir1-10, or (C–E) pAtTIR1::cPsTIR1a (PsTIR1a-B1-7 and C2-12), pAtTIR1::cPsTIR1b (PsTIR1b-B1-8 and C3-6), pAtTIR1::cPsAFB2 (PsAFB2-A2-6 and B1-5) or pAtTIR1::cAtTIR1 (AtTIR1-A2-5 and B1-8) in Attir1-10 afb2-3. Root elongation is expressed as a percentage of the same genotype in medium without auxin. Data are means (±SD), n=8. Different letters indicate significant differences within the same auxin concentration as determined by one-way ANOVA and Holm–Sidak post hoc tests (P<0.05).
PsAFB2 under the regulation of the AtTIR1 promoter was introduced into Arabidopsis Attir1-10 afb2-3, as the AFB2 contribution to auxin response in the root was clearly shown in Attir1-10 afb2-3, but only a minor auxin sensitivity difference was observed in Atafb2-3 (Parry et al., 2009). The Attir1-10 afb2-3 seedlings expressing PsAFB2 exhibited reduced root length at 70 nM and 90 nM 2,4-D, indicating that PsAFB2 also codes for a functional auxin receptor (Fig. 1E).
In Arabidopsis tir1-1, transgenic expression of AtTIR1 under the AtTIR1 promoter rescues the mutant’s root growth response to 2,4-D to the level of wild-type seedlings; however, AtAFB1 driven by the AtTIR1 promoter does not (Parry et al., 2009). In contrast, both PsTIR1a and PsTIR1b were able to rescue the tir1-10 mutant root growth phenotype when regulated by the AtTIR1 promoter, and they both displayed a wild-type level of 2,4-D response at 70 nM (compare Col-0 to PsTIR1a and PsTIR1b transgenics; Fig. 1A, B). This difference in auxin response between AtTIR1/AFB1 and PsTIR1a/TIR1b may at least partially reside in an E-to-K substitution found in the F-box domain of AtAFB1 (reducing TIR1 tethering to SCF), but is not present in either PsTIR1a or PsTIR1b, as described above.
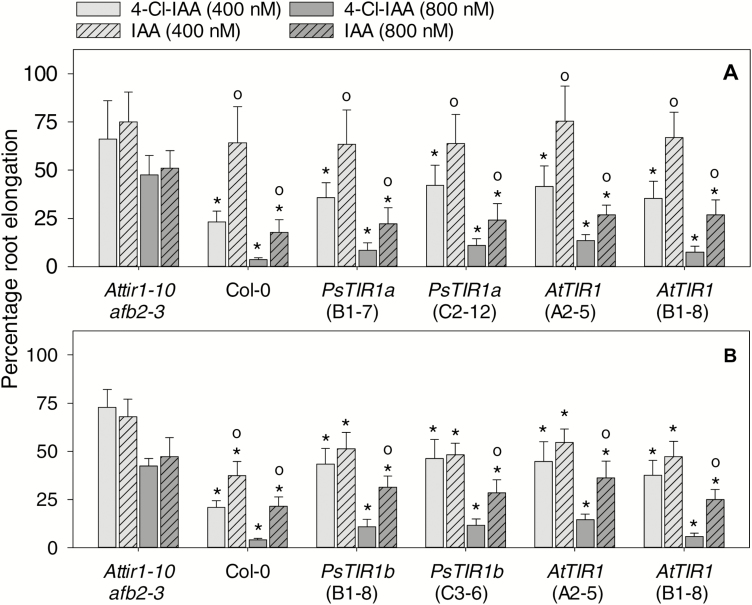
Both 4-Cl-IAA and IAA were also effective in inhibiting Arabidopsis root elongation, but in the wild-type seedlings inhibition was greater with 4-Cl-IAA than IAA at the same auxin concentration (Fig. 2). In contrast, IAA inhibited root elongation to the same extent as 4-Cl-IAA in the Attir1-10 afb2-3 mutant (Fig. 2). When PsTIR1a, PsTIR1b, or AtTIR1 were expressed in Attir1-10 afb2-3, the transgenes restored the greater root inhibitory effect of 4-Cl-IAA, particularly at 800 nM, compared to that of IAA (Fig. 2).
Fig. 2.
Functional characterization of pea auxin receptors with IAA and 4-Cl-IAA in Arabidopsis. Seedlings at 4 d old were transferred to media containing 0, 400, or 800 nM IAA or 4-Cl-IAA and grown for 3 d. Seedlings assessed were Col-0, Attir1-10 afb2-3 or those expressing (A) pAtTIR1::cPsTIR1a (PsTIR1a-B1-7 and C2-12), (B) pAtTIR1::cPsTIR1b (PsTIR1b-B1-8 and C3-6), or (A, B) pAtTIR1::cAtTIR1 (AtTIR1-A2-5 and B1-8). Root elongation in the auxin-containing medium is expressed as a percentage compared to the same genotype grown in medium without auxin. Data are means (±SD), n=8. Significant differences were determined by one-way ANOVA and Holm–Sidak post hoc tests (P<0.05): * means are significantly different from that of Attir1-10 afb2-3 within the same auxin concentration; ° means from media containing IAA are significantly different from those containing 4-Cl-IAA within the same concentration and genotype.
It is possible that the chlorine molecule at the 4-position of the indole ring of 4-Cl-IAA (compared to the hydrogen moiety in IAA) modifies the hydrophobic interactions and van der Waals contacts in the auxin-binding pocket of TIR1 (as described by Tan et al., 2007), leading to differences in auxin affinity to TIR1, in the binding of specific Aux/IAAs to the complex, and/or in the dissociation rate of the complexes. This is consistent with studies showing that the dissociation rate of the IAA7 degron motif DII and the TIR1 proteins differ depending on the type of auxin present (IAA, 1-NAA, picloram, or 2,4-D; Calderón Villalobos et al., 2012). However, we cannot exclude the possibility that part of the differential response of IAA and 4-Cl-IAA is due to differences in conjugation, metabolism, or transport of each auxin. Overall, the absence of a differential auxin response to IAA and 4-Cl-IAA in the tir1-10 afb2-3 mutant and the restoration of differential response in the mutants expressing AtTIR1, PsTIR1a, and PsTIR1b suggest that the stronger auxin response elicited by 4-Cl-IAA compared to IAA (at the same concentration) is at least partially mediated through TIR1.
Developmental regulation of the expression of auxin-receptor genes in pea
Seedling tissues
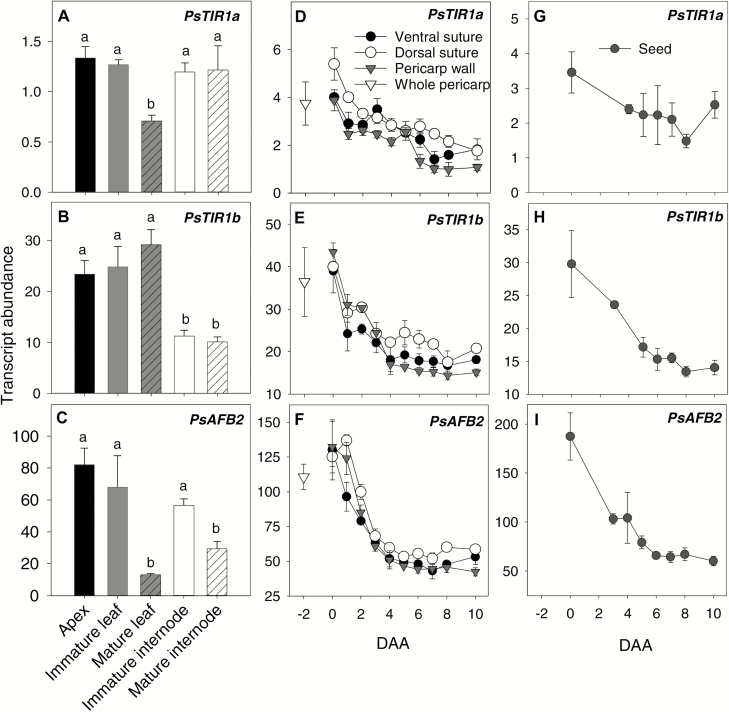
The TIR1a, TIR1b, and PsAFB2 genes were expressed widely in the tissues but with varying transcript abundance patterns depending on the tissue type and developmental stage. In pea seedlings at 12 d after imbibition (DAI), PsTIR1a transcript abundance was similar across the vegetative tissues (shoot apexes, leaves, and internodes), except that levels decreased in leaves upon maturity (Fig. 3A). The pattern of tissue-specific transcript abundance of PsTIR1b differed from that of PsTIR1a, with transcript levels being lower in the internodes than in the shoot apex and leaf tissues, but no variation was observed between young and older tissues within a tissue type (Fig. 3B). The pattern of PsAFB2 transcript abundance varied from that of PsTIR1a and PsTIR1b, with higher levels being observed in immature leaf and internode tissues than in the respective mature tissues (Fig. 3C).
Fig. 3.
Developmental regulation of PsTIR1a, PsTIR1b, and PsAFB2 transcript abundance in pea tissues. (A–C) Relative transcript abundance of auxin receptors in different tissues of 12-d-old seedlings. Data are means (±SD), n=3. Different letters indicate significant differences as determined by one-way ANOVA and Holm–Sidak post hoc tests (P<0.05). Relative transcript abundance of auxin receptors in (D–F) pericarps from pre-pollinated fruit at –2 d after anthesis (DAA) and in pericarp tissues (wall, dorsal vascular suture, and ventral vascular suture) from 0–10 DAA, and in (G–I) seeds from pollinated fruits from 0–10 DAA. Data are means (±SD), n=3 with the exception of dorsal suture at 0–3 DAA and seeds at 3 DAA and 4 DAA, where n=2. Each sample contained tissues from a minimum of three pericarps or from five seeds.
Early pea fruit development
In pea, flowers self-pollinate approximately 24 h before flower opening (–1 DAA), and fertilization takes place by 0 DAA (Cooper, 1938). The developmental regulation of PsTIR1a, PsTIR1b, and PsAFB2 transcript levels was analysed in –2 DAA pre-pollinated fruit, and in the pericarp wall, the dorsal and ventral pericarp vascular suture tissues, and the seeds of pollinated fruits from 0–10 DAA. In all pericarp tissues, PsTIR1a, PsTIR1b, and PsAFB2 transcript levels were highest immediately after fertilization (0 DAA) and declined gradually with fruit development (Fig. 3D–F). In the developing seeds, the transcript abundance of PsTIR1a did not substantially change from 0–10 DAA. However, both PsTIR1b and PsAFB2 gradually declined with development (by 2-fold and 5-fold from 0–10 DAA, respectively; Fig. 3G–I). Our data also suggested that PsTIR1b was more highly expressed (~10-fold) in pericarp tissues than PsTIR1a (Fig. 3D, E).
The presence of higher auxin-receptor transcript levels during early ovary growth followed by a gradual reduction with development appears to be a common trend in a number of species (CsTIR1 and CsAFB2 in cucumber, Cui et al., 2014; PslTIR1, PslAFB2, and PslAFB5 in plum, Prunus salicina, El-Sharkawy et al., 2014; SlTIR1 in tomato, Ren et al., 2011). The higher abundance of auxin-receptor transcripts that is observed closely following fertilization events, in precocious fruit set, and in parthenocarpic fruit phenotypes in TIR1 overexpression lines of tomato (Ren et al., 2011) supports the idea that an increased auxin sensitivity of the ovaries facilitates early ovary growth and development.
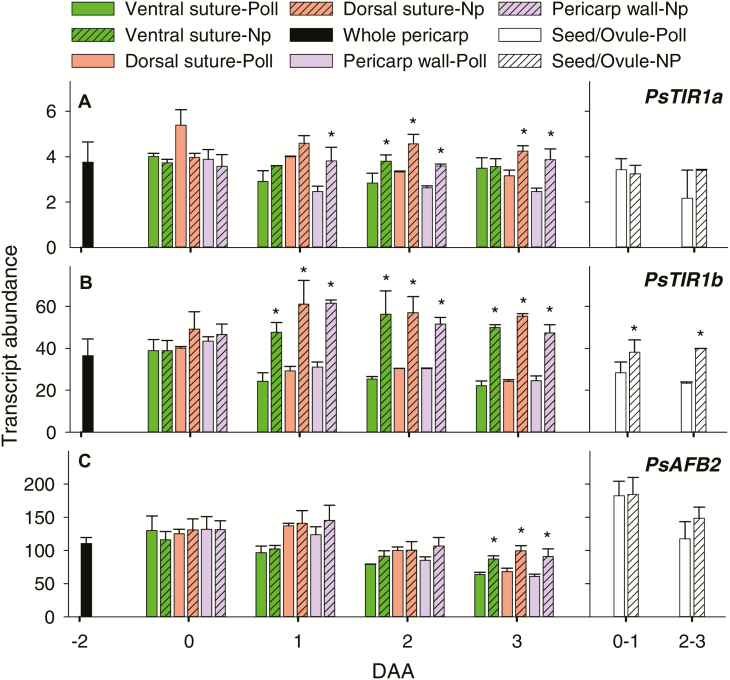
In pea, pericarp growth arrests by 1 DAA in the absence of pollination but pericarps remain green and turgid through to 3 DAA (Jayasinghege et al., 2017). In non-pollinated pericarps, PsTIR1b transcript abundance was elevated ~2-fold by 1 DAA and remained elevated through to 3 DAA in all pericarp tissues compared to pollinated fruits (Fig. 4). PsTIR1b transcript abundance was also higher in non-fertilized ovules compared to seeds. PsTIR1a and PsAFB2 expression in fruit/seed tissues was less tightly or not controlled by pollination events compared to that of PsTIR1b; however, by 3 DAA the transcript abundance of all three auxin receptors was higher in pericarp wall and dorsal suture tissues from non-pollinated compared to pollinated fruits (Fig. 4). Similarly, in the non-parthenocarpic cucumber cultivar 8419s-1, the transcript levels of CsTIR1 and CsAFB2 have been shown to increase in non-pollinated fruits but to decline in pollinated fruits during early fruit development (Cui et al., 2014).
Fig. 4.
Relative transcript abundance of (A) PsTIR1a, (B) PsTIR1b, and (C) PsAFB2 in different tissues of pollinated (Poll) and non-pollinated (Np) fruits of pea. Pre-pollinated pericarps are from fruits at –2 d after anthesis (DAA). Pericarp tissues (ventral vascular suture, dorsal vascular suture, and pericarp wall) are from pollinated or non-pollinated fruits at 0–3 DAA. Seeds (fertilized ovules from pollinated fruits) and non-fertilized ovules (from non-pollinated fruits) are from fruits at 0–1 DAA and 2–3 DAA. Data are means (±SD), n=3 for pre-pollinated pericarps and pericarp wall tissue; n=3 for pericarp ventral and dorsal sutures, except for a few samples where n=2 due to the limited size of tissue that was available. At 3 DAA, non-pollinated ovaries were still green and turgid. Pairwise mean comparisons were made using a two-tailed Student’s t-test (P<0.05): * means for non-pollinated fruits are significantly different from those for pollinated fruits within a tissue, gene, and DAA (P<0.05).
It is possible that increased expression of specific auxin-receptor genes is a response to low auxin levels or to signaling events within the pericarp tissue (due to the absence of developing seeds) that act to modify the auxin response in non-pollinated fruits. When Arabidopsis auxin receptors were studied using a synthetic system in yeast, the rates of IAA-induced Aux/IAA degradation were markedly different with TIR1 and AFB2 (Havens et al., 2012). These data support the hypothesis that the pool of specific TIR1/AFB receptors in a cell or tissues at any one time may dictate specific auxin signaling outputs, leading to specific auxin responses. Changing the gene expression of specific TIR1/AFBs within a tissue or cell is one possible mechanism to change the pool of specific TIR1/AFB receptors. The prominent up-regulation of PsTIR1b in non-pollinated pea pericarps has the potential to modify the pool of TIR1/AFB receptors and hence the auxin response in this tissue.
Developmental regulation of endogenous auxin levels and auxin activity in pea fruit
Due to the auxin-dependent activation of the DR5 promoter (Ulmasov et al., 1995, 1997), the localized GUS enzyme activity of DR5::GUS plants is commonly used as a marker of auxin maxima in plants (in pea; DeMason and Polowick, 2009). However, DR5::GUS expression relies on the transduction of the auxin signal to the DR5 promoter, which may be affected by changes in the auxin signaling pathway, including the type and abundance of auxin receptors (TIR1/AFBs and Aux/IAAs) and transcription factors (auxin response factors) present in the tissue. Therefore, DR5-driven GUS enzyme activity may not necessarily represent the localized auxin concentrations, but it is an indication of localized auxin response (Vernoux et al., 2011; Brunoud et al., 2012). Quantification of auxin levels in conjunction with DR5::GUS expression assays can provide a more comprehensive picture of how changes in auxin concentration are related to auxin action within the ovary.
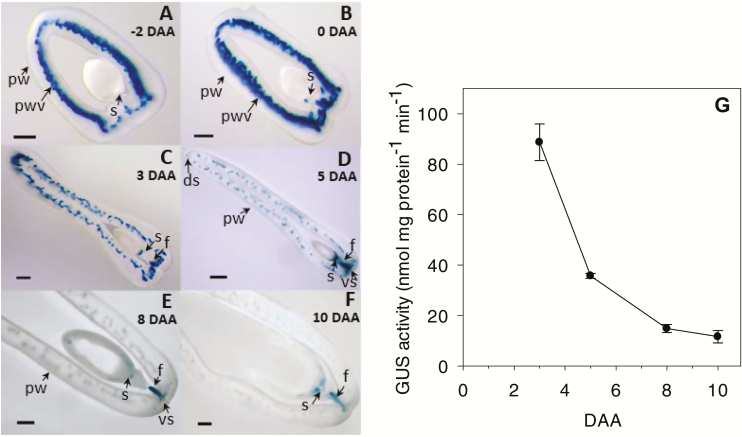
The IAA concentration in the pericarp of pollinated fruits was greatest at 0 DAA and decreased by ~2.5-fold by 3 DAA (on a fresh-weight basis, Table 1). In parallel with the decline in IAA, the concentration of the amide-conjugates IAA-aspartate (IAA-Asp) and IAA-glutamate (IAA-Glu) also declined from 0–3 DAA (Table 1). IAA-Asp and IAA-Glu are considered as irreversible IAA amide-conjugates destined for degradation (Ludwig-Müller, 2011), and IAA-Asp is the most abundant form of IAA amide-conjugate in pea (Sudi, 1964; Nordström and Eliasson, 1991). The reduction in the level of free IAA together with that of amide-conjugates suggests a reduction in pericarp IAA biosynthesis or a reduced IAA supply to the pericarp. Consistent with the reduction of IAA levels, the intense DR5-GUS staining observed in the pericarp vasculature of –2 DAA pre-pollinated and 0 DAA pollinated fruits also decreased by 3 DAA (Fig. 5A–C).
Table 1.
IAA, IAA-aspartate (IAA-Asp), and IAA-glutamate (IAA-Glu) levels in pollinated ovaries from 0–8 d after anthesis (DAA), in non-pollinated ovaries (NP) at 3 DAA, and in seeds at 8 DAA of pea (cv I3, Alaska type; DR5::GUS plants) determined using HPLC-MS/MS and heavy-labelled internal standards
| (ng g –1 DW) | (ng g –1 FW) | |||||
| IAA | IAA-Asp | IAA-Glu | IAA | IAA-Asp | IAA-Glu | |
| Whole pericarp | ||||||
| 0 DAA | 292±5 | 293±14 | 37±1 | 61±1 | 62±3 | 8±0 |
| 3 DAA | 168±10 | 43±16 | 19±3 | 25±1 | 6±2 | 3±0 |
| 3 DAA-NP | 38±13 | <5 | 23±6 | 6±2 | <1 | 4±1 |
| 5 DAA | 136±1 | 209±62 | 13±4 | 18±0 | 27±8 | 2±1 |
| 8 DAA | 135±11 | 124±47 | 8±0 | 23±2 | 21±8 | 1±0 |
| Pericarp wall | ||||||
| 5 DAA | 73±12 | 27±10 | 5±1 | 8±1 | 3±1 | 1±0 |
| 8 DAA | 145±24 | 34±11 | 4±0 | 24±4 | 6±2 | 1±0 |
| Ventral vascular suture | ||||||
| 5 DAA | 774±10 | 1266±262 | 40±2 | 123±2 | 200±41 | 6±0 |
| 8 DAA | 267±30 | 648±9 | 22±4 | 59±7 | 142±2 | 5±1 |
| Dorsal vascular suture | ||||||
| 5 DAA | 366 | 359 | 23 | 43 | 43 | 3 |
| 8 DAA | 148±7 | 70±65 | 20±2 | 31±1 | 15±14 | 4±0 |
| Seed | ||||||
| 8 DAA | 32 262±700 | 84 497±6280 | 2704±52 | 5148±112 | 13 484±1002 | 431 ± 8 |
Data are means (±SD), n=2 with the exception of dorsal vascular suture at 5 DAA where only a single sample was analysed. Each replicate contained more than 50 pericarps at early developmental stages to a minimum of 10 pericarps at later developmental stages. Seeds were without the funiculus. IAA-alanine (IAA-Ala), IAA-leucine (IAA-Leu) and indole-3-butyric acid (IBA) were also analysed, but the levels were minute to non-detectable, with the exception of IAA-Ala in the seeds of 8 DAA fruits where the mean was 47.7±0.4 ng g–1 DW.
Fig. 5.
Developmental regulation of auxin activity in pea fruit. (A–F) Representative micrographs of GUS-stained fruit cross-sections and (G) GUS enzyme activity in the pericarp wall as detected by MUG assays in plants expressing the GUS gene under the regulation of the auxin-responsive DR5 promoter (DR5::GUS). The micrographs show DR5::GUS expression in (A) pre-pollinated fruit at –2 d after anthesis (DAA) and (B–F) in pollinated fruit at 0, 3, 5, 8, and 10 DAA. ds, dorsal vascular suture; f, funiculus; pw, pericarp wall, pwv, pericarp wall vasculature; s, proximal end of the seed; vs, ventral vascular suture. Scale bars: (A, B) 500 µm and (C–F) 1000 µm. GUS enzyme activity was quantified in the central pericarp wall of pollinated fruits at 3–10 DAA. Data are means (±SD), n=3.
A further decrease in IAA levels (1.4-fold) in pericarps of pollinated fruits was observed from 3–5 DAA (Table 1). Similarly, Magnus et al. (1997) reported a 1.7-fold reduction in IAA levels from 3–6 DAA in pollinated pea pericarps (from 29 ng g–1 FW to 17 ng g–1 FW), whilst levels of 4-Cl-IAA in pollinated pericarps were lower than those of IAA and did not change from 3–6 DAA (~5 ng g–1 FW). The overall pericarp DR5-GUS staining intensity and enzyme activity also decreased from 3–5 DAA in the pericarps of pollinated fruits (Fig. 5C, D, G).
Young developing pea seeds contain high levels of auxin at 3 DAA and 6 DAA (ng g–1 FW, cv. Alaska I3; 3 DAA, IAA=393, 4-Cl-IAA=145; 6 DAA, IAA=1494, 4-Cl-IAA=231; Magnus et al., 1997), and we also found high levels at 8 DAA (IAA=5148; Table 1). Localized GUS-staining auxin activity was observed towards the proximal end of the seed adjacent to the attachment to the funiculus (intense DR5-GUS staining at 3 DAA and 5 DAA; Fig. 5C, D), and in the funiculus (3–10 DAA; Fig. 5C–F) during early fruit development. Higher IAA and IAA amide-conjugate levels (Table 1) and GUS staining intensity (Fig. 5D) were also observed in the ventral vascular suture (where the seeds are attached to the pericarp through the funiculus) compared to the dorsal vascular suture (IAA 3-fold and IAA-Asp 5-fold higher on a fresh-weight basis), or to the pericarp wall (IAA 15-fold and IAA-Asp 67-fold higher on a fresh-weight basis) at 5 DAA.
Auxin-sensitive promoter-gene marker assays in Arabidopsis support the concept that fertilization-stimulated endosperm initiation is coupled to the production of auxin in the central cell, followed by accumulation of auxin in the seed coat (Figueiredo et al., 2015, 2016) and the funiculus tissues (Robert et al., 2018). Furthermore, in the absence of developing seeds within the pea ovary, IAA levels were ~4-fold lower and IAA-Asp was minimally detectable in the pericarps (compare 3 DAA pericarps from pollinated and non-pollinated fruits in Table 1). These data support the hypothesis that following fertilization, developing seeds produce auxin that can be transported to the pericarp, as suggested by the auxin concentration and DR5::GUS staining gradients (high to low, from the seeds and adjacent vascular tissues to the other pericarp tissues). Within the pea pericarp, auxin stimulates GA biosynthesis and other auxin-regulated processes to promote pericarp growth (Magnus et al., 1997; Ozga et al., 2009). The higher transcript levels of PsTIR1b (and to a lesser extent PsTIRa and PsAFB2) in pericarps from non-pollinated compared to pollinated fruits and in non-fertilized ovules compared to seeds (Fig. 4) indicate that the expression of auxin receptors (particularly PsTIR1b) may be regulated to modify the auxin response under low auxin conditions in these tissues.
From 5 DAA to 8 DAA, the free IAA and IAA-Asp levels decreased ~1.5-fold in the pericarp dorsal vascular suture and 2-fold in the ventral vascular suture (Table 1). In contrast to the pericarp vascular sutures, levels of free IAA and IAA-Asp increased from 5 DAA to 8 DAA in the pericarp wall tissues (by ~3-fold and ~2-fold, respectively). By 8 DAA, the pericarp has reached most of its maximal length and width, but the diameter rapidly increases from 6–12 DAA to accommodate the developing seeds (Ozga and Reinecke, 2003). The reduction in IAA level in the vascular suture tissues and the increased level in the pericarp wall from 5–8 DAA may indicate that auxin (IAA and 4-Cl-IAA) is transported from the seed through the vascular suture to the pericarp wall, and the higher auxin concentration in the wall facilitates rapid expansion of the diameter of the pericarp. Taken together, these data support the hypothesis that auxin concentration and response (through the regulation of auxin-receptor type and abundance) are developmentally and seed-regulated to coordinate seed and ovary growth.
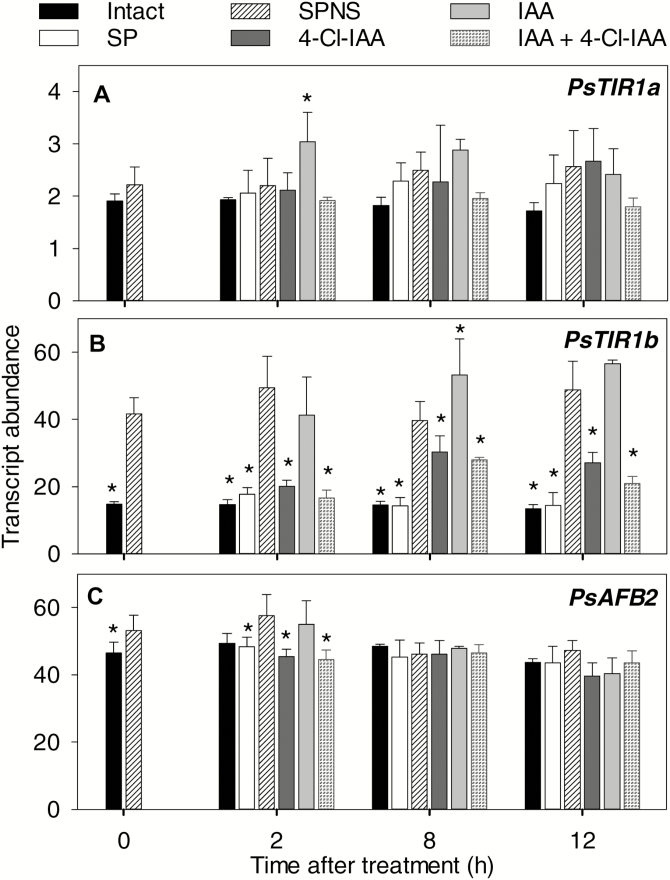
4-Cl-IAA and IAA differentially modulate PsTIR1 transcript abundance and auxin activity in pea fruit
When seeds were present (intact and split pericarps, SP), transcript levels of PsTIR1a, PsTIR1b, and PsAFB2 in the pericarps at 2 DAA remained relatively constant over the course of a 12-h assay (Fig. 6, Supplementary Fig. S7A–C). Seed removal (split pericarp no seeds, SPNS) increased the transcript abundance of PsTIR1b in the pericarp by ~3-fold but had a minimal or no effect on PsTIR1a and PsAFB2. Increased PsTIR1b transcript abundance in deseeded pericarps together with increased abundance in non-pollinated pericarps is consistent with the hypothesis that PsTIR1b expression is regulated in order to modify auxin signaling and response under low auxin conditions, and potentially in response to other seed-derived signals.
Fig. 6.
Effects of removal of pea seeds and auxin treatment on the relative transcript abundance of the pericarp auxin-receptor genes (A) PsTIR1a, (B) PsTIR1b, and (C) PsAFB2. At 2 d after anthesis, pollinated fruits were either left intact, split (split pericarps with seeds, SP), or split and deseeded (split pericarp no seeds, SPNS). Twelve hours after these treatments, the pericarps were treated with 4-Cl-IAA, IAA, or 4-Cl-IAA plus IAA (labelled as 0 h after treatment on the graphs). Data are means (±SD), n=3–8. * Means significantly different from that of the SPNS controls within time after treatment as determined by one-way ANOVA and Holm–Sidak post hoc tests (P≤0.05).
To assess auxins as a signal(s) regulating pericarp auxin-receptor expression, receptor abundance was determined in deseeded pericarps treated with IAA (which does not stimulate deseeded pericarp growth), 4-Cl-IAA (stimulates deseeded pericarp growth), or IAA plus 4-Cl-IAA. PsTIR1a and PsAFB2 transcript abundance remained relatively constant in pericarps treated with IAA, 4-Cl-IAA, or both auxins (Fig. 6). In contrast, 4-Cl-IAA reduced the PsTIR1b transcript abundance (4-Cl-IAA or IAA plus 4-Cl-IAA compared to SPNS controls), but IAA had either a minimal or no effect. The increased abundance of PsTIR1b transcripts under low pericarp growth conditions (in the absence of developing seeds) and the suppression of transcript accumulation only in response to the growth-active auxin 4-Cl-IAA indicate that 4-Cl-IAA is a potential seed signal that regulates PsTIR1b expression.
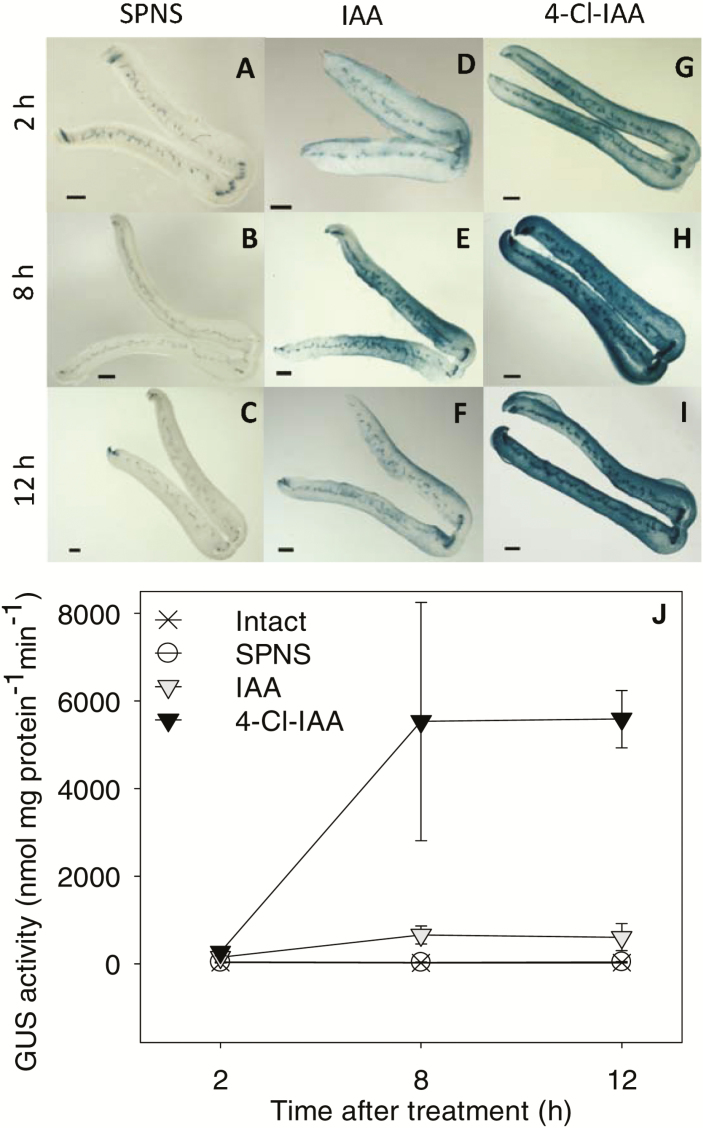
Auxin application to the pericarp increased the mesocarp GUS staining intensity and GUS enzyme activity by 2 h after treatment (Fig. 7). The increased GUS responses confirm that both IAA and 4-Cl-IAA diffuse from the endocarp application site into the mesocarp tissue and initiate auxin responses within 2 h. By 12 h after auxin treatment, GUS enzyme activity was 9-fold higher in pericarps treated with 4-Cl-IAA than in those treated with IAA (Fig. 7J). The ability of 4-Cl-IAA to stimulate a stronger auxin response (GUS enzyme activity) than IAA at the same concentration in pea pericarps was associated with its ability to enrich the auxin-receptor transcript pool with PsTIR1a and PsAFB2 transcripts (by decreasing PsTIR1b transcript abundance). It is possible that PsTIR1a and PsTIR1b have different binding affinities for 4-Cl-IAA and/or specific Aux/IAAs that vary the auxin response. Indeed, PsTIR1a and PsTIR1b have different amino acids at (respective) alignment positions 401 and 460 in their protein sequences (Supplementary Fig. S4), which have been identified as Aux/IAA (IAA7-)contacting amino acid residues of AtTIR1 (Tan et al., 2007). The differential effects of 4-Cl-IAA and IAA on DR5::GUS expression and GUS enzyme activity may also be due in part to other factors including differential catabolism of these auxins in the pericarp tissue.
Fig. 7.
Effect of IAA and 4-Cl-IAA on DR5::GUS activity in pea fruits. (A–I) Representative micrographs of GUS-stained fruit cross-sections and (J) GUS enzyme activity as detected by MUG assays. Fruits at 2 d after anthesis from plants expressing DR5::GUS were split and deseeded (split pericarp no seeds, SPNS). Twelve h after this treatment, pericarps were treated with (A–C) aqueous 0.1% Tween 80 (SPNS, controls), (D–F) IAA, or (G–I) 4-Cl-IAA. Samples were harvested at 2 h, 8 h, or 12 h after hormone application. Scale bars are 500 µm. GUS enzyme activities at the 2-h time point were as follows: intact, 26.6±3.4; SPNS, 40.1±6.4; IAA, 153.0±21.8; 4-Cl-IAA, 267.2±89.6. Data are means (±SD), n=3.
In the absence of pollination (or due to seed removal), ethylene evolution increases in the pericarp (Orzáez et al., 1999; Johnstone et al., 2005; Savada et al., 2017). Application of 4-Cl-IAA but not IAA to deseeded pericarps inhibits the action of ethylene (Johnstone et al., 2005). Therefore, to determine whether the regulation of auxin-receptor transcript abundance by IAA and 4-Cl-IAA was modulated through ethylene, deseeded pericarps were treated with either ethephon or STS alone or in the presence of IAA or 4-Cl-IAA. Neither the ethephon nor STS treatments had a clear effect on the transcript levels of PsTIR1a, PsTIR1b, or PsAFB2, indicating that their expression was not directly regulated by ethylene in this tissue (Supplementary Fig. S7 D–F).
Finally, to determine whether the differential regulation of auxin-receptor abundance by the two auxins was tissue dependent, we studied their transcript abundance in seedlings at 4 DAI exposed to IAA and 4-Cl-IAA. In contrast to pea pericarps where 4-Cl-IAA is stimulatory to growth and IAA is not (Reinecke et al., 1995), both auxins inhibited seedling root elongation (Supplementary Fig. S8A). However, neither IAA nor 4-Cl-IAA clearly affected the transcript abundance of PsTIR1a, PsTIR1b, or PsAFB2 in the plumule, epicotyl, or root-tip tissues of the seedlings (Supplementary Fig.S8B–D), suggesting that the modulation of auxin-receptor abundance by the two auxins is tissue specific.
Conclusions
In summary, the ability of the PsTIR1a, PsTIR1b, and PsAFB2 genes to complement the Attir1-10 afb2-3 mutant and to restore auxin-sensitive root growth in Arabidopsis indicates that these genes code for functional auxin receptors. The auxin-receptor proteins coded by PsTIR1a and PsTIR1b elicited an auxin response in the presence of IAA or 4-Cl-IAA in Arabidopsis root growth assays, with 4-Cl-IAA having a stronger auxin response than IAA at the same concentration (800 nM). Furthermore, the loss of a stronger auxin response elicited by 4-Cl-IAA compared to that of IAA in the absence of TIR1 (in Attir1-10 afb2-3) and the regaining of this effect upon reintroduction of TIR1 (AtTIR1, PsTIR1a, or PsTIR1b) into Attir1-10 afb2-3 suggest that at least part of this differential auxin response in Arabidopsis root growth assays is mediated through TIR1.
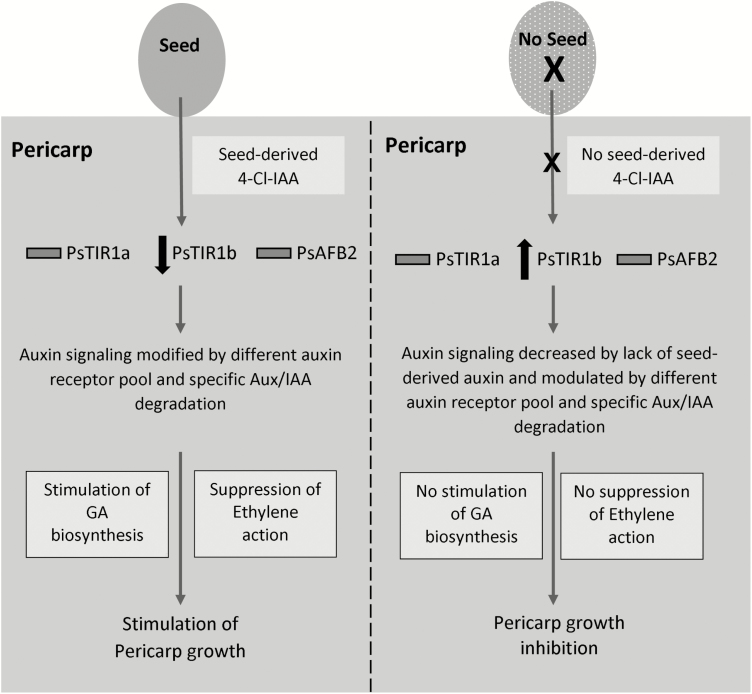
Our data suggest that the markedly different effects of IAA and 4-Cl-IAA on pea fruit growth may at least partially involve differential modulation of the pericarp auxin-receptor population, and the substantial reduction of PsTIR1b transcript abundance induced by 4-Cl-IAA in the pericarp could suggest its importance in this tissue. We propose the following working hypothesis that seed and auxin (4-Cl-IAA) regulation of TIR1/AFBs (at least partially through the modification PsTIR1b transcript abundance) in the pea pericarp tissue affects the composition of the TIR1/AFB-Aux/IAA co-receptor complexes and their stabilities. This can lead to IAA- and 4-Cl-IAA-specific degradation of Aux/IAA proteins, thus initiating an auxin type-specific response in the pericarp (Fig. 8). The 4-Cl-IAA-specific response includes the stimulation of GA biosynthesis and inhibition of ethylene action (Ozga et al., 2003, 2009; Johnstone et al., 2005; Jayasinghege et al., 2017) that facilitate pericarp growth. The lack of these 4-Cl-IAA-mediated pericarp responses (due to the absence of seeds) leads to pericarp senescence. The 4-Cl-IAA-dependent regulation of PsTIR1b transcript abundance in pea pericarps but not in seedling tissues supports a tissue-dependent modulation of auxin-receptor composition. The expression profiles of the Aux/IAA genes in different plant species also indicate tissue- and developmental stage-dependent modulation of Aux/IAA protein compositions (Kalluri et al., 2007; Song et al., 2009; Audran-Delalande et al., 2012; Singh and Jain, 2015). Future analysis of the expression profiles of Aux/IAAs and the remaining TIR1/AFBs (PsAFB4 and PsAFB6) and evaluation of the IAA- and 4-Cl-IAA-dependent interactions of different co-receptor combinations will help to determine the specific co-receptor combinations that mediate auxin-dependent pea pericarp growth.
Fig. 8.
A working model for seed and auxin (4-Cl-IAA) stimulation of pea fruit growth. 4-Cl-IAA in the seed is transported to the pericarp, which leads to modulation of the pericarp auxin-receptor pool (through a decrease in PsTIR1b transcript abundance) that targets the degradation of specific Aux/IAA proteins, resulting in auxin-related changes in gene expression that facilitate pericarp growth. These seed/4-Cl-IAA-induced changes include the stimulation of GA biosynthesis and the suppression of ethylene action in the pericarp. In the absence of developing seeds, the lack of seed-derived auxin (4-Cl-IAA) modifies the make-up of the pericarp auxin-receptor pool (as noted by an increase in PsTIR1b transcript abundance) and reduces auxin signaling and activity, thus inhibiting pericarp growth.
Supplementary Data
Supplementary data are available at JXB online.
Fig. S1. Diagram of pea fruit tissues and confirmation of transgenic gene expression in Arabidopsis.
Fig. S2. Comparison of GUS staining in the pericarps from non-transgenic and DR5::GUS-expressing plants.
Fig. S3. A phylogenetic tree of predicted P. sativum auxin-receptor proteins.
Fig. S4. Sequence alignment of pea and Arabidopsis auxin-receptor proteins.
Fig. S5. Conservation of the TIR1 clade F-box protein domain in the Fabaceae subfamily Papilionoideae.
Fig. S6. Root elongation of Arabidopsis tir1-10 and tir1-10 afb2-3 mutant seedlings grown in the presence of 2,4-D.
Fig. S7. Effect of seed removal and ethylene on the relative transcript abundance of pea pericarp auxin receptors.
Fig. S8. Effect of IAA and 4-Cl-IAA on pea seedling growth and auxin-receptor abundance at 4 DAI.
Table S1. PCR primers used for gene cloning and verification.
Table S2. Primers and probes used for qRT-PCR.
Protocol S1. Pea TIR1/AFB family sequence analysis and construction of the phylogenetic tree.
Protocol S2. qRT-PCR assays.
Protocol S3. Creation of transgenic Arabidopsis plants and root growth analysis.
Protocol S4. GUS staining and quantification of GUS enzyme activity.
Protocol S5. Hormonal analysis.
Authors contributions
CJ modified experimental designs for implementation and performed all experiments and data analysis except for those completed by HK and CN, and provided the first draft of the manuscript; JO conceived the original research, designed the experiments, interpreted the results, and was the primary editor of the manuscript; CN identified and sequenced PsAFB2 and performed the first characterization of PsAFB2 gene expression in pea fruit; HK performed qRT-PCR to verify the expression of the pea auxin receptors in transgenic Arabidopsis plants and edited the final manuscript; DR conceived the original research, created the DR5::GUS pea plants, identifying the pea PsTIR1a and PsAFB4 homologs, provided technical assistance to CJ, and complemented the writing and editing of the manuscript.
Acknowledgements
We would like to acknowledge Drs Mark Estelle and Michael Prigge for providing initial help in developing the CODEHOP primers used for cloning PsAFB2 and PsAFB6. We would also like to thank Dr Enrico Scarpella for providing the pCm1300-polyA vector, Dr Patricia Polowick for providing the DR5::GUS construct, and Dilini Adihetty for technical assistance. This research was supported by the Natural Sciences and Engineering Research Council of Canada through Discovery Grant no. 138166 to JO, and an S.M. Blair Graduate Student Scholarship to CJ.
References
- Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M. 2012. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant & Cell Physiology 53, 659–672. [DOI] [PubMed] [Google Scholar]
- Ayele BT, Ozga JA, Kurepin LV, Reinecke DM. 2006. Developmental and embryo axis regulation of gibberellin biosynthesis during germination and young seedling growth of pea. Plant Physiology 142, 1267–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunoud G, Wells DM, Oliva M, et al. 2012. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nature Chemical Biology 8, 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DC. 1938. Embryology of Pisum sativum. Botanical Gazette 100, 123–132. [Google Scholar]
- Cui L, Zhang T, Li J, Lou Q, Chen J. 2014. Cloning and expression analysis of Cs-TIR1/AFB2: the fruit development-related genes of cucumber (Cucumis sativus L.). Acta Physiologiae Plantarum 36, 139–149. [Google Scholar]
- DeMason DA, Polowick PL. 2009. Patterns of DR5::GUS expression in organs of pea (Pisum sativum). International Journal of Plant Sciences 170, 1–11. [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. 2005. Plant development is regulated by a family of auxin receptor F box proteins. Developmental Cell 9, 109–119. [DOI] [PubMed] [Google Scholar]
- Dorcey E, Urbez C, Blázquez MA, Carbonell J, Perez-Amador MA. 2009. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. The Plant Journal 58, 318–332. [DOI] [PubMed] [Google Scholar]
- Duarte J, Rivière N, Baranger A, et al. 2014. Transcriptome sequencing for high throughput SNP development and genetic mapping in Pea. BMC Genomics 15, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL. 2006. Molecular phylogenetics of the clover genus (Trifolium–Leguminosae). Molecular Phylogenetics and Evolution 39, 688–705. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif SM, Jones B, Mila I, Kumar PP, Bouzayen M, Jayasankar S. 2014. TIR1-like auxin-receptors are involved in the regulation of plum fruit development. Journal of Experimental Botany 65, 5205–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro KS. 2014. Comparative transcriptomics and proanthocyanidin metabolism in pea (Pisum sativum) seed coat. PhD thesis, University of Calgary, Alberta, Canada. [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C. 2016. Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5, e20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C. 2015. Auxin production couples endosperm development to fertilization. Nature Plants 1, 15184. [DOI] [PubMed] [Google Scholar]
- Franssen SU, Shrestha RP, Bräutigam A, Bornberg-Bauer E, Weber AP. 2011. Comprehensive transcriptome analysis of the highly complex Pisum sativum genome using next generation sequencing. BMC Genomics 12, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. 2012. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiology 160, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull GA, Devic M. 1995. The β-glucuronidase (gus) reporter gene system gene fusions. Spectrophotometric, fluorometric, and histochemical detection. In: Jones H, ed. Plant gene transfer and expression protocols. Methods in Molecular Biology, vol. 49. Totowa, NJ: Springer, 125–141. [DOI] [PubMed] [Google Scholar]
- Jayasinghege CPA, Ozga JA, Waduthanthri KD, Reinecke DM. 2017. Regulation of ethylene-related gene expression by indole-3-acetic acid and 4-chloroindole-3-acetic acid in relation to pea fruit and seed development. Journal of Experimental Botany 68, 4137–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Molecular Biology Reporter 5, 387–405. [Google Scholar]
- Johnstone MG, Reinecke D, Ozga J. 2005. The auxins IAA and 4-Cl-IAA differentially modify gibberellin action via ethylene response in developing pea fruit. Journal of Plant Growth Regulation 24, 214–225. [Google Scholar]
- Kalluri UC, Difazio SP, Brunner AM, Tuskan GA. 2007. Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biology 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Pembleton LW, Cogan NO, Savin KW, Leonforte T, Paull J, Materne M, Forster JW. 2012. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HK, McAdam SA, McAdam EL, Ross JJ. 2015. Evidence that chlorinated auxin is restricted to the Fabaceae but not to the Fabeae. Plant Physiology 168, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligerot Y, de Saint Germain A, Waldie T, et al. 2017. The pea branching RMS2 gene encodes the PsAFB4/5 auxin receptor and is involved in an auxin-strigolactone regulation loop. PLoS Genetics 13, e1007089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J. 2011. Auxin conjugates: their role for plant development and in the evolution of land plants. Journal of Experimental Botany 62, 1757–1773. [DOI] [PubMed] [Google Scholar]
- Magnus V, Ozga JA, Reinecke DM, Pierson GL, Larue TA, Cohen JD, Brenner ML. 1997. 4-chloroindole-3-acetic and indole-3-acetic acids in Pisum sativum. Phytochemistry 46, 675–681. [Google Scholar]
- McAdam EL, Meitzel T, Quittenden LJ, et al. 2017. Evidence that auxin is required for normal seed size and starch synthesis in pea. New Phytologist 216, 193–204. [DOI] [PubMed] [Google Scholar]
- Nordström A-C, Eliasson L. 1991. Levels of endogenous indole-3-acetic acid and indole-3-acetylaspartic acid during adventitious root formation in pea cuttings. Physiologia Plantarum 82, 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzáez D, Blay R, Granell A. 1999. Programme of senescence in petals and carpels of Pisum sativum L. flowers and its control by ethylene. Planta 208, 220–226. [DOI] [PubMed] [Google Scholar]
- Ozga JA, Brenner ML, Reinecke DM. 1992. Seed effects on gibberellin metabolism in pea pericarp. Plant Physiology 100, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga J, Reinecke D. 1999. Interaction of 4-chloroindole-3-acetic acid and gibberellins in early pea fruit development. Plant Growth Regulation 27, 33–38. [Google Scholar]
- Ozga J, Reinecke D. 2003. Hormonal interactions in fruit development. Journal of Plant Growth Regulation 22, 73–81. [Google Scholar]
- Ozga JA, Reinecke DM, Ayele BT, Ngo P, Nadeau C, Wickramarathna AD. 2009. Developmental and hormonal regulation of gibberellin biosynthesis and catabolism in pea fruit. Plant Physiology 150, 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga JA, Reinecke DM, Nadeau CD. 2010. Auxin receptors. US Patent No. US9441237B2. Patent application 2010; granted September 13, 2016.
- Ozga JA, Yu J, Reinecke DM. 2003. Pollination-, development-, and auxin-specific regulation of gibberellin 3β-hydroxylase gene expression in pea fruit and seeds. Plant Physiology 131, 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. 2009. Complex regulation of the TIR1/AFB family of auxin receptors. Proceedings of the National Academy of Sciences, USA 106, 22540–22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke D. 1999. 4-Chloroindole-3-acetic acid and plant growth. Plant Growth Regulation 27, 3–13. [Google Scholar]
- Reinecke DM, Ozga JA, Ilić N, Magnus V, Kojić-Prodić B. 1999. Molecular properties of 4-substituted indole-3-acetic acids affecting pea pericarp elongation. Plant Growth Regulation 27, 39–48. [Google Scholar]
- Reinecke DM, Ozga JA, Magnus V. 1995. Effect of halogen substitution of indole-3-acetic acid on biological activity in pea fruit. Phytochemistry 40, 1361–1366. [Google Scholar]
- Reinecke DM, Wickramarathna AD, Ozga JA, Kurepin LV, Jin AL, Good AG, Pharis RP. 2013. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiology 163, 929–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Li Z, Miao Q, Yang Y, Deng W, Hao Y. 2011. The auxin receptor homologue in Solanum lycopersicum stimulates tomato fruit set and leaf morphogenesis. Journal of Experimental Botany 62, 2815–2826. [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, et al. 2018. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nature Plants 4, 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savada RP, Ozga JA, Jayasinghege CPA, Waduthanthri KD, Reinecke DM. 2017. Heat stress differentially modifies ethylene biosynthesis and signaling in pea floral and fruit tissues. Plant Molecular Biology 95, 313–331. [DOI] [PubMed] [Google Scholar]
- Schaefer H, Hechenleitner P, Santos-Guerra A, Menezes de Sequeira M, Pennington RT, Kenicer G, Carine MA. 2012. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evolutionary Biology 12, 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrani JC, Ruiz-Rivero O, Fos M, García-Martínez JL. 2008. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. The Plant Journal 56, 922–934. [DOI] [PubMed] [Google Scholar]
- Shimizu-Mitao Y, Kakimoto T. 2014. Auxin sensitivities of all Arabidopsis Aux/IAAs for degradation in the presence of every TIR1/AFB. Plant & Cell Physiology 55, 1450–1459. [DOI] [PubMed] [Google Scholar]
- Singh VK, Jain M. 2015. Genome-wide survey and comprehensive expression profiling of Aux/IAA gene family in chickpea and soybean. Frontiers in Plant Science 6, 918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wang L, Xiong L. 2009. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229, 577–591. [DOI] [PubMed] [Google Scholar]
- Ståldal V, Sohlberg JJ, Eklund DM, Ljung K, Sundberg E. 2008. Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytologist 180, 798–808. [DOI] [PubMed] [Google Scholar]
- Sudheesh S, Sawbridge TI, Cogan NO, Kennedy P, Forster JW, Kaur S. 2015. De novo assembly and characterisation of the field pea transcriptome using RNA-Seq. BMC Genomics 16, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudi J. 1964. Induction of the formation of complexes between aspartic acid and indolyl-3-acetic acid or I-naphthalene acetic acid by other carboxylic acids. Nature 201, 1009–1010. [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. 1995. Composite structure of auxin response elements. The Plant Cell 7, 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM. 1997. Seed and hormonal regulation of gibberellin 20-oxidase expression in pea pericarp. Plant Physiology 115, 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Huizen R, Ozga JA, Reinecke DM, Twitchin B, Mander LN. 1995. Seed and 4-chloroindole-3-acetic acid regulation of gibberellin metabolism in pea pericarp. Plant Physiology 109, 1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Molecular Systems Biology 7, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li J, Cui L, et al. 2017. New insights into the roles of cucumber TIR1 homologs and miR393 in regulating fruit/seed set development and leaf morphogenesis. BMC Plant Biology 17, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhang Y, Moss BL, Bargmann BOR, Wang R, Prigge M, Nemhauser JL, Estelle M. 2015. Untethering the TIR1 auxin receptor from the SCF complex increases its stability and inhibits auxin response. Nature Plants 1, 14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. 2006. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zúñiga-Mayo VM, Reyes-Olalde JI, Marsch-Martinez N, de Folter S. 2014. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Frontiers in Plant Science 5, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.