Abstract
Here, we discuss the expected hallmark(s) of the cancer cell of origin and how this may be related to a new tumor cell phenotype, namely “energetic” cancer stem cells (e-CSCs). e-CSCs show many features that would be characteristic of the cancer cell of origin, including the over-expression of p21-WAF (CDKN1A), a key marker of senescence. It is tempting to speculate that the cancer cell of origin and e-CSCs are closely related entities. e-CSCs possess a hybrid phenotype, sharing key hallmarks of senescence, “stemness” and cancer. e-CSCs are hyper-proliferative and have elevated mitochondrial metabolism, with an NRF2-mediated anti-oxidant response signature, including glutaredoxin (GLRX) and ALDH3A1 over-expression, possibly related to their escape from senescence. Finally, in e-CSCs, BCAS1 (Breast carcinoma-amplified sequence-1) protein expression was up-regulated by >100-fold. BCAS1 is a candidate oncogene associated with “stemness” and aggressive oncogenic behavior, such as Tamoxifen resistance.
Keywords: cancer cell of origin, cancer stem cells (CSCs), senescence, metabolism, anti-oxidant response, tamoxifen resistance
Hallmarks: cancer cell of origin vs. e-CSCs
Cancer cells are believed to originate from senescent cells that have undergone stress-induced cell cycle arrest [1–3]. During chronological aging, genetic mutations are thought to accumulate in the stem cell compartment, resulting in i) oncogene activation, ii) tumor suppressor inactivation, as well as iii) a variety of genetic chromosomal rearrangements (deletions, translocations and duplications) and other epigenetic modifications [4–6].
Presumably, cancer cells may arise from senescent cells through a process of re-activation, leading from cell cycle arrest to hyper-proliferation. It has been proposed that telomerase activity (hTERT) catalyzes this transition [3]. However, this process of re-activation can also be achieved through cellular metabolism [7,8]. For example, David Sinclair and his colleagues at Harvard Medical School have shown that senescent cells suffer from a deficiency of NADH (nicotinamide adenine dinucleotide), a key anti-oxidant, and that senescent cells can be revived simply by the addition of an NADH precursor metabolite (namely, nicotinamide riboside) to the cell culture media [7,8]. These results directly imply that the anti-oxidant response may also trigger the revival of senescent cells (Figure 1). Similarly, cancer stem cells (CSCs) are strictly dependent on NADH, for their propagation as 3D-spheroids [9]. Also, CSCs have been shown to over-express classical “embryonic” markers of stemness, such as Oct4, c-Myc and Nanog, among others.
Figure 1.
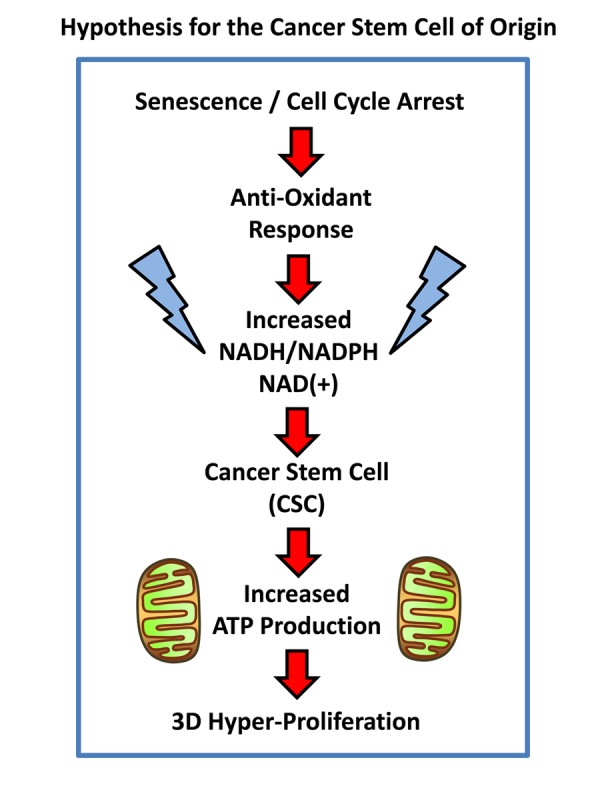
Hypothesis for how senescent cells can mechanistically become cancer stem cells. Senescent cells undergoing cell cycle arrest mount an anti-oxidant defense, to increase their levels of NADH. In turn, increased NADH levels are known to be sufficient to rescue senescent cells from cell cycle arrest, allowing new cell proliferation, by “re-activating” or “resuscitating” senescent cells. Increased mitochondrial power would then drive elevated ATP production and 3D anchorage-independent growth, fostering the generation and propagation of the cancer cell of origin.
Therefore, the cancer stem cell of origin [10,11] would be predicted to retain certain properties of senescent stem cells, while undergoing a gain-of-function process, thereby obtaining new properties of a cancer cell, resulting in a chimeric or hybrid phenotype (Figure 2). These properties would be expected to include: biological markers of senescence; a hyper-proliferative phenotype; a very active metabolic program to support anabolic growth and proliferation; an anti-oxidant response, for driving the revival program, to overcome senescence-induced cell cycle arrest [7,8,12]; and key stem cell features (See Table 1, Left).
Figure 2.
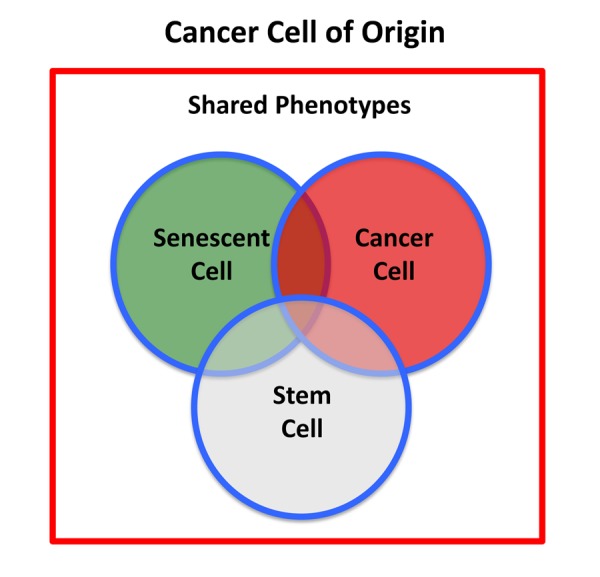
Cancer stem cell of origin. The cancer stem cell of origin would be predicted to have a chimeric- or hybrid-phenotype, retaining elements of i) senescent cells, ii) cancer cells, and iii) stem cells, as we observe in e-CSCs.
Table 1. Expected hallmarks of the cancer cell of origin: Comparison with e-CSCs.
| Cell Type: | Cancer Cell of Origin | e-CSCs |
| Properties: | Expected Features | Observed Features |
| 1) Cell Cycle Arrest: | Senescence Markers | Elevated p21-WAF (~17-fold) |
| 2) Propagation: | Hyper-Proliferative | G0/G1: ~35-37% S-phase: ~10-18% |
| G2/M: ~32-33% Polyploid: ~12-17% | ||
| 3) Metabolism: | Metabolically Active | Increased Mitochondrial Mass (~4-fold); High OXPHOS & Glycolysis |
| 4) REDOX: | Anti-Oxidant Response | ALDH Functional Activity (~9-fold Increased) |
| [Produces NADH] | Glutaredoxin-1 (GLRX) (~11-fold Increased) | |
| ALDH3A1 (~10-fold Increased) | ||
| QPRT (~4-fold Increased) | ||
| RRM2, GCLC, NQO2 (Each ~2-fold Increased) | ||
| 5) “Stemness”: | Stem Cell Markers | High Flavin-based Auto-fluorescence (FAD/FMN); Large Cell Size; Aldefluor (+); Anchorage-Independence; |
| BCAS1 (+) (>100-fold Increased) | ||
| 6) Inhibitors: | Unknown | Mitochondrial OXPHOS Inhibitors and CDK4/6 Inhibitors |
Recently, our laboratory may have fortuitously isolated a new tumor cell with a cancer cell of origin phenotype, by using flavin-derived auto-fluorescence as a selection marker, via flow-cytometry [13]. To functionally describe these cells, we coined the term “energetic” cancer stem cells (e-CSCs) [13]. Briefly, e-CSCs retain high expression of the senescence marker p21-WAF (CDKN1A), while paradoxically manifesting a hyper-proliferative phenotype (Table 1, Right). Based on Ingenuity Pathway Analysis (IPA) of e-CSC proteomics data, other upstream regulators of cell cycle arrest and senescence were activated, including p53, TGFB1, and p38 MAPK signaling [13].
Moreover, e-CSCs show a hyper-metabolic phenotype, with increased mitochondrial mass, elevated oxidative mitochondrial metabolism, as well as enhanced glycolytic function. e-CSCs mount a strong anti-oxidant response, characterized by increased levels of glutaredoxin-1 (GLRX) expression and ALDH isoform activity (Table 1, Right). Interestingly, e-CSCs also show a number of stem-like features, including large cell size and anchorage-independent growth, that is highly sensitive to inhibitors of both mitochondrial OXPHOS and CDK4/6 cell cycle progression (Figure 3). In addition, large cell size is a key feature commonly shared by CSCs, e-CSCs and senescent cells alike [13–16].
Figure 3.
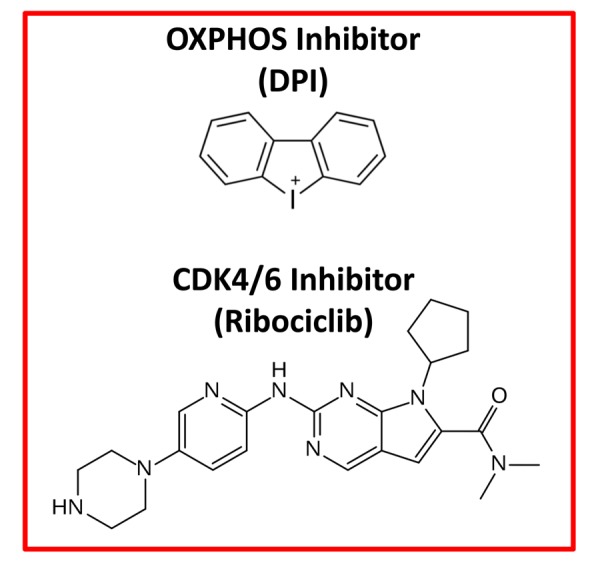
Therapeutic targeting of e-CSCs. Experimentally, both an OXPHOS inhibitor (Diphenyleneiodonium; DPI) and a CDK4/6 (Ribociclib) inhibitor were effective in abrogating the 3D-propagation of e-CSCs.
Lastly, in e-CSCs, BCAS1 (Breast carcinoma-amplified sequence-1) protein expression was up-regulated, by nearly 120-fold [13]. BCAS1 over-expression [17] is functionally associated with “stemness” [18], a more aggressive cancer cell phenotype [19] and Tamoxifen-resistance [20,21]. MCF7-TAMR cells, which were chronically selected for Tamoxifen-resistance, by including Tamoxifen in their tissue culture media, show a >50-fold increase in BCAS1 protein expression, promoting increased ATP production [21].
Conclusions
In summary, e-CSCs possess many of the predicted hallmarks that would be expected of the cancer cell of origin. This assertion could have broad implications for mechanistically understanding and more effectively targeting the cancer stem cell of origin, to prevent tumor recurrence and metastatic dissemination, throughout the body, to significantly improve clinical outcomes in cancer therapy.
ACKNOWLEDGEMENTS
We are tremendously grateful to Rumana Rafiq, for her kind and dedicated assistance, in keeping the Translational Medicine Laboratory at the University of Salford running very smoothly. We would like to thank the Foxpoint Foundation (Canada) and the Healthy Life Foundation (UK) for their philanthropic donations towards new equipment and infrastructure, in the Translational Medicine Laboratory at the University of Salford.
“Energetic” is defined as “showing great activity or vitality; full of energy”.
Synonyms are: “active, dynamic, vital, vibrant, tireless, indefatigable, full of vim and vigour, vigorous, high-powered, driving, powerful, potent”.
CONFLICTS OF INTEREST: MPL and FS hold a minority interest in Lunella Biotech, Inc.
FUNDING: This work was supported by research grant funding, provided by Lunella Biotech, Inc (to FS and MPL).
REFERENCES
- 1.Karimi-Busheri F, Rasouli-Nia A, Mackey JR, Weinfeld M, and Karimi-Busheri F1. Rasouli-Nia A, Mackey JR, Weinfeld M. Senescence evasion by MCF-7 human breast tumor-initiating cells. Breast Cancer Res. 2010; 12:R31 10.1186/bcr2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milanovic M, Fan DN, Belenki D, Däbritz JH, Zhao Z, Yu Y, Dörr JR, Dimitrova L, Lenze D, Monteiro Barbosa IA, Mendoza-Parra MA, Kanashova T, Metzner M, et al. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018; 553:96–100. 10.1038/nature25167 [DOI] [PubMed] [Google Scholar]
- 3.Patel PL, Suram A, Mirani N, Bischof O, Herbig U. Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence. Proc Natl Acad Sci USA. 2016; 113:E5024–33. 10.1073/pnas.1602379113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976; 194:23–28. 10.1126/science.959840 [DOI] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012; 481:306–13. 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013; 501:328–37. 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013; 155:1624–38. 10.1016/j.cell.2013.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schöndorf DC, Ivanyuk D, Baden P, Sanchez-Martinez A, De Cicco S, Yu C, Giunta I, Schwarz LK, Di Napoli G, Panagiotakopoulou V, Nestel S, Keatinge M, Pruszak J, et al. The NAD+ Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC and Fly Models of Parkinson’s Disease. Cell Reports. 2018; 23:2976–88. 10.1016/j.celrep.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 9.Bonuccelli G, De Francesco EM, de Boer R, Tanowitz HB, Lisanti MP. NADH autofluorescence, a new metabolic biomarker for cancer stem cells: identification of Vitamin C and CAPE as natural products targeting “stemness”. Oncotarget. 2017; 8:20667–78. 10.18632/oncotarget.15400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Visvader JE. Cells of origin in cancer. Nature. 2011; 469:314–22. 10.1038/nature09781 [DOI] [PubMed] [Google Scholar]
- 11.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017; 16:41. 10.1186/s12943-017-0600-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang JH, Lee SH, Hong D, Lee JS, Ahn HS, Ahn JH, Seong TW, Lee CH, Jang H, Hong KM, Lee C, Lee JH, Kim SY. Aldehyde dehydrogenase is used by cancer cells for energy metabolism. Exp Mol Med. 2016; 48:e272. 10.1038/emm.2016.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiorillo M, Sotgia F, Lisanti MP. “Energetic” Cancer Stem Cells (e-CSCs): A New Hyper-Metabolic and Proliferative Tumor Cell Phenotype, Driven by Mitochondrial Energy. Front Oncol. 2019; 8:677 10.3389/fonc.2018.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machado HL, Kittrell FS, Edwards D, White AN, Atkinson RL, Rosen JM, Medina D, Lewis MT. Separation by cell size enriches for mammary stem cell repopulation activity. Stem Cells Transl Med. 2013; 2:199–203. 10.5966/sctm.2012-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb R, Ozsvari B, Bonuccelli G, Smith DL, Pestell RG, Martinez-Outschoorn UE, Clarke RB, Sotgia F, Lisanti MP. Dissecting tumor metabolic heterogeneity: telomerase and large cell size metabolically define a sub-population of stem-like, mitochondrial-rich, cancer cells. Oncotarget. 2015; 6:21892–905. 10.18632/oncotarget.5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farnie G, Sotgia F, Lisanti MP. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget. 2015; 6:30472–86. 10.18632/oncotarget.5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins C, Rommens JM, Kowbel D, Godfrey T, Tanner M, Hwang SI, Polikoff D, Nonet G, Cochran J, Myambo K, Jay KE, Froula J, Cloutier T, et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci USA. 1998; 95:8703–08. 10.1073/pnas.95.15.8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fard MK, van der Meer F, Sánchez P, Cantuti-Castelvetri L, Mandad S, Jäkel S, Fornasiero EF, Schmitt S, Ehrlich M, Starost L, Kuhlmann T, Sergiou C, Schultz V, et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci Transl Med. 2017; 9:eaam7816. 10.1126/scitranslmed.aam7816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McFarlane M, Brettschneider J, Gelsthorpe A, James S, Snead D, Gopalakrishnan K, Mehenna H, Jankowski J, Arasaradnam R, Nwokolo C. An assessment of candidate genes to assist prognosis in gastric cancer. J Gastrointest Oncol. 2018; 9:303–10. 10.21037/jgo.2018.01.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker M, Sommer A, Krätzschmar JR, Seidel H, Pohlenz HD, Fichtner I. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther. 2005; 4:151–68. [PubMed] [Google Scholar]
- 21.Fiorillo M, Sotgia F, Sisci D, Cappello AR, Lisanti MP. Mitochondrial “power” drives tamoxifen resistance: NQO1 and GCLC are new therapeutic targets in breast cancer. Oncotarget. 2017; 8:20309–27. 10.18632/oncotarget.15852 [DOI] [PMC free article] [PubMed] [Google Scholar]


