Abstract
Background
Gestational diabetes mellitus (GDM) is one of the most common problems during pregnancy. Lack of international consistent diagnostic procedures has limit improvement of current therapeutic effectiveness. Here, we aimed to screen potential gene biomarkers that might play vital roles in GDM progression for assistance of its diagnostic and treatment.
Methods
Gene expression profiles in four GDM placentae at first trimester, four GDM placentae at second trimester, and four normal placentae were obtained from the publicly available Gene Expression Omnibus (GEO). Weighted gene coexpression network analysis (WGCNA) indicated two gene modules, that is, black and brown module, that was significantly positively and negatively correlated with GDM progression time points, respectively. Additionally, a significant positive correlation between module membership (MM) and degree in protein–protein interaction network of brown module genes was observed.
Results
KIF2C, CENPE, CCNA2, AURKB, MAD2L1, CCNB2, CDC20, PLK1, CCNB1, and CDK1 all have degree larger than 50 and MM larger than 0.9, so they might be valuable biomarkers in GDM. Gene set enrichment analysis inferred tight relations between carbohydrate metabolism or steroid biosynthesis‐related processes and GDM progression.
Conclusions
All in all, our study should provide several novel references for GDM diagnosis and therapeutic.
Keywords: GDM, gene module, GEO, GSEA, WGCNA
1. INTRODUCTION
Gestational diabetes mellitus (GDM) represents the most common pathoglycemia form in pregnant women that might also cause hypertension and cardiovascular disease (Oliveira et al., 2015). GDM's morbidity is about 2%–5% across worldwide (Ashwal & Hod, 2015), and which is deeply affected by pathological and environment factors. For example, dangerousness of GDM could be increased up to 3.8 times in obese pregnant women than those with normal body mass index (Pantham & Aye, 2015). Previous study even reported that social capital, such as neighborhood trust, emotional support, has obvious influences on prevalence of GDM (Mizuno et al., 2016). What is more, GDM could increase incidence of type 2 diabetes mellitus and obesity in not only women, but also their offsprings (Catalano, 2010; Coustan, 2013; Harreiter, Dovjak, & Kautzky‐Willer, 2014; Nikolic et al., 2016).
Gene expression profiling has been an important mean for exploring disease mechanism and identifying valuable diagnostic and treatment biomarkers particularly after the development of gene microarray and high‐throughput sequencing technologies (Janikova et al., 2011; Kanda et al., 2015; Vu et al., 2015). As a polygenic disease, GDM progression was generally considered to be promoted by aberrant expression of multiple genes in a gestational age‐dependent manner (Uuskula et al., 2012). Insulin has been a key agent with effective treatment results for GDM mainly through regulating cholesterol transport in human placenta (Dube & Ethier‐Chiasson, 2013). Differences exist in placenta surfaces that exposed to bloodstreams of mother and fetus which corresponding to trophoblasts and endothelial cells, respectively, and could yield large amount of insulin receptor. Through interacting with those receptors, insulin could strikingly control placental gene expression shifts from mother to fetus over the time course of pregnancy, which might shed new light on exploring therapeutic targets for GDM (Hiden et al., 2006). It was also previously reported that reduced trophoblast apoptosis along with elevated inflammation could significantly result in aberrant expression of associated placental transcripts or proteins followed by GDM‐related increased placenta and newborn weights (Magee et al., 2014).
Genetic variation including single nucleotide polymorphisms (SNPs) and structure variation makes up the most common variations across the human genome, and it has been applied for multiple disease diagnostics and treatments. Vitamin D is closely associated with β‐cell function and impaired glucose absorption in GDM through transportation to placenta by vitamin D‐binding protein, that is, vitamin D receptor, and SNPs in which have been previously reported to be correlated with GDM clinical parameters (Wang, Wang, et al., 2015). Several previous studies even developed SNP‐based risk score for GDM prediction. For example, through including type 2 diabetes‐related risk variants, Kawai et al. (2017) developed a risk score formula that could stably predict GDM risk; Chawla et al. (2014) proposed a model comprised of 48 SNPs along with several common clinicopathological features including ancestry, sex, gestational age, and so on, that could improve prediction of large‐for‐gestational‐age and newborn adiposity. Although the recent advancement of understanding about GDM mechanisms, the efficacy of conventional therapy is still poor and further studies for identifying valuable biomarkers are still needed.
In this study, we conducted weighted gene coexpression network analysis (WGCNA) for time series gene expression profiles in GDM samples at first trimester and second trimester as well as control samples. Compared with traditional methodologies that take every transcript in the microarray alone and only capture two few information than that the microarray could provide, WGCNA takes correlations among those transcripts into account and identified potential disease‐related gene coexpression modules (GCMs) by considering associations between GCMs and disease's traits as well as intramodular associations. We identified several gene modules that closely associated with GDM progression and screened potential biomarkers via combination with protein–protein interaction (PPI) network. Additionally, carbohydrate metabolism or steroid biosynthesis‐related pathways were obtained through gene set enrichment analysis (GSEA), which has been previously reported to associate with GDM development.
2. MATERIALS AND METHODS
2.1. Gene expression profile dataset
We searched the Gene Expression Omnibus (GEO) with the keywords of “(gestational diabetes mellitus)” AND “Homo sapiens[porgn:__txid9606]” and only retained gene expression datasets that profiled by using Affymetrix HG‐U133 Puls2.0 platform (ref no.: GPL570). As a result, we obtained one dataset that deposited by Mikheev et al. (2008) with the accession number of GSE9984, which consisted of placenta expression profiles of four GDM patients at first trimester (45–59 days), four GDM patients at second trimester and four control samples.
2.2. Gene expression preprocessing
R and Bioconductor packages were applied for preprocessing raw gene expression profiles. Background correction, normalization, and logarithm transformation were conducted by using the affy package (Gautier, Cope, Bolstad, & Irizarry, 2004). Probes were annotated as gene symbols based on the GPL570 annotation file, and average expression value was used for genes annotated by multiple probes.
2.3. Weighted gene coexpression network analysis
Gene coexpression analysis is a powerful mean for exploring correlations among genes at specific conditions, particularly in time series analysis. Weighted gene coexpression network analysis (WGCNA) assigns a connection weight to each gene pair in the coexpression network and uses soft thresholds that should be more biologically meaningful compares with traditional methods that use binary information (0 = unconnected, 1 = connected) (Zhang & Horvath, 2005). WGCNA package is a collection of R functions for soft power selection, GCM detection, identification of module eigengene (first principle component of the module), assessment of associations between modules and clinical traits, and so on (Langfelder, 2008). Here, we screened GCM based on gene expression profiles of the 12 samples and assessed associations between modules and GDM by using pregnancy age as clinical variable.
2.4. Protein–protein interaction analysis
Protein–protein interactions among genes were obtained from the STRING database (Szklarczyk et al., 2015), containing interactions that with reliability scores according to means by which they were presented, such as high‐throughput, bioinformatics prediction, or low‐throughput methods. Here, we used reliability score >0.9 as the threshold for screening of valuable gene pairs. Cytoscape version3.6.0 (Su, Morris, Demchak, & Bader, 2014) was applied for visualizing PPI network.
2.5. Gene set enrichment analysis
Gene set enrichment analysis was performed by using the GSEA software (Subramanian, Kuehn, Gould, Tamayo, & Mesirov, 2007). “C2.CP.V6.0.ENTREZ.gmt” was used as the gene set for the analysis. Permutation number was set to 1,000, and p value <0.05 was considered as statistically significant.
3. RESULTS
3.1. Gene coexpression modules
Coefficient of variation (CV) of every gene between GDM samples at first‐trimester or second‐trimester pregnancy age and control samples was calculated, and expression profiles of overlapping genes between the top 5,000 genes with largest CV in GDM samples at first and second trimester were used for identification of valuable GCMs. As a result, 3,731 genes were screened and sample clustering based on those genes’ Euclidean distance separated samples into two distinct clusters that contained control and GDM samples, respectively, as shown in Figure 1a.
Figure 1.
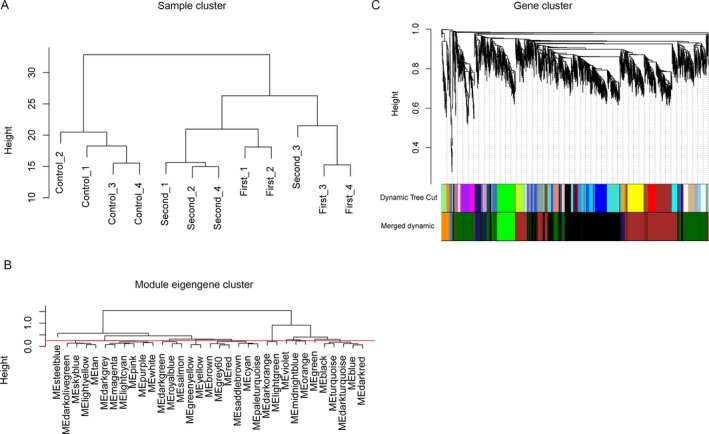
Sample and gene clustering analysis based on the 3,731 gene expression profiles. (a) Sample clustering identified two main clusters that containing gestational diabetes mellitus and control samples, respectively. (b) Gene coexpression modules (GCMs) obtained through weighted gene coexpression network analysis and GCM clusters based on their eigengenes. The red line represents the height of 0.2. (c) Visualization of GCMs before and after merging closer GCMs according to the height of 0.2. Colors indicate GCMs, and leaves represent genes
A total of 33 GCMs were identified based on the soft threshold of 11. Module clustering analysis was further performed based on correlations among eigengenes of the 33 GCMs, which produced seven merged GCMs according to height of 0.2 (red line in Figure 1b). Figure 1c illustrated the GCMs before and after merging closer GCMs.
3.2. Module‐trait correlation analysis
To screen candidate GCMs that might contribute GDM progression, we used GDM patients’ pregnancy age as clinical variable and estimated their correlations with GCMs’ eigengene. As a result, brown and black modules were closely correlated with pregnancy age of GDM patients by using correlation p value ≤0.01 as threshold (Figure 2a). Additionally, we calculated correlation between every gene's module membership (MM, correlation between a specific gene and module's eigengene) and gene significance (GS, correlation between a specific gene and clinical variable) in brown and black modules. As expected, significant positive correlations between gene's MM and GS were obtained in black as well as brown module as shown in Figure 2b,c, respectively.
Figure 2.
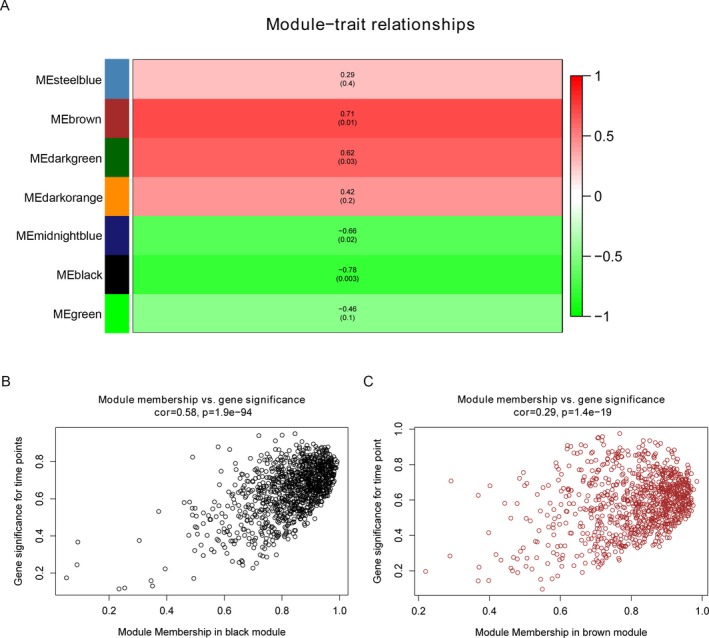
Gene coexpression module (GCM)‐gestational diabetes mellitus (GDM) progression correlation analysis. (a) A heatmap visualization of correlation between GCM and GDM patients’ pregnancy age. Numbers outside and inside brackets represent correlation coefficients and p value, respectively. (b,c) was the correlation plot of GS versus module membership for gene contained in black and brown module, respectively
3.3. Protein–protein interaction network
Identification of GCMs was purely based on mathematical correlations among genes in specific module, so we further explored potential biological associations among genes in brown and black modules. As a result, a total of 842 and 2,654 gene pairs were, respectively, obtained for genes contained in black and brown module. Figure 3a,b illustrate PPI network that comprised of pairs among genes in black and brown module, respectively. Additionally, degree of brown module genes (direct neighborhood in PPI network) was significantly positively correlated with their MM (Figure 3c), which might indicate closer relation between brown module and GDM progression than that of black module. Table 1 shows the genes that with PPI degree larger than 50 and their MM.
Figure 3.
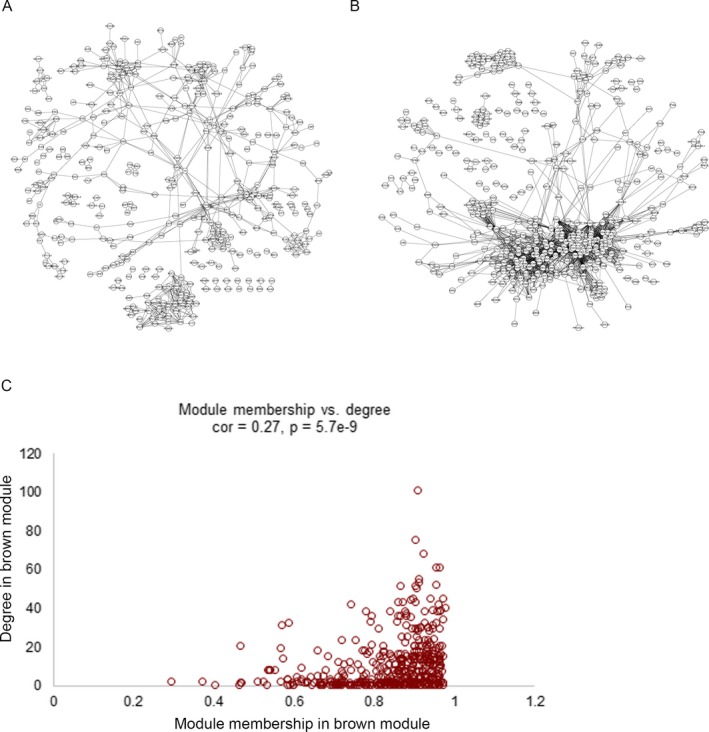
Protein–protein interaction (PPI) network analysis of genes contained in black and brown modules. (a) PPI network of genes contained in the black module. (b) PPI network of genes contained in the brown module. (c) Scatter plot indicated the positive correlation between network degree and module membership of genes contained in the brown module
Table 1.
Module membership (MM) and degree of genes that with degree larger than 10 in protein–protein interaction network
| Gene | MM | Degree | Gene | MM | Degree |
|---|---|---|---|---|---|
| CDK1 | 0.90 | 102 | AURKB | 0.91 | 54 |
| CCNB1 | 0.90 | 76 | CCNA2 | 0.95 | 53 |
| PLK1 | 0.92 | 69 | CENPE | 0.90 | 52 |
| CCNB2 | 0.96 | 62 | PAFAH1B1 | 0.86 | 52 |
| CDC20 | 0.95 | 62 | KIF2C | 0.90 | 51 |
| MAD2L1 | 0.91 | 56 |
3.4. Gene set enrichment analysis
We divided samples into first trimester and control group or second trimester and control group and subjected them to GSEA. Figure 4a,b illustrated the full list of KEGG pathways and top five KEGG pathways that significantly up‐regulated in first‐trimester and second‐trimester GDM samples compared with control samples, respectively. Table 2 is the full list of KEGG pathways significantly up‐regulated in second‐trimester GDM samples. Lysine degradation, carbon pool by folate, and steroid biosynthesis pathways were found to be significantly up‐regulated in both first‐trimester and second‐trimester GDM samples. Strikingly, cancer‐related pathways, such as colorectal cancer, cell cycle, were also significantly up‐regulated in second‐trimester GDM samples.
Figure 4.
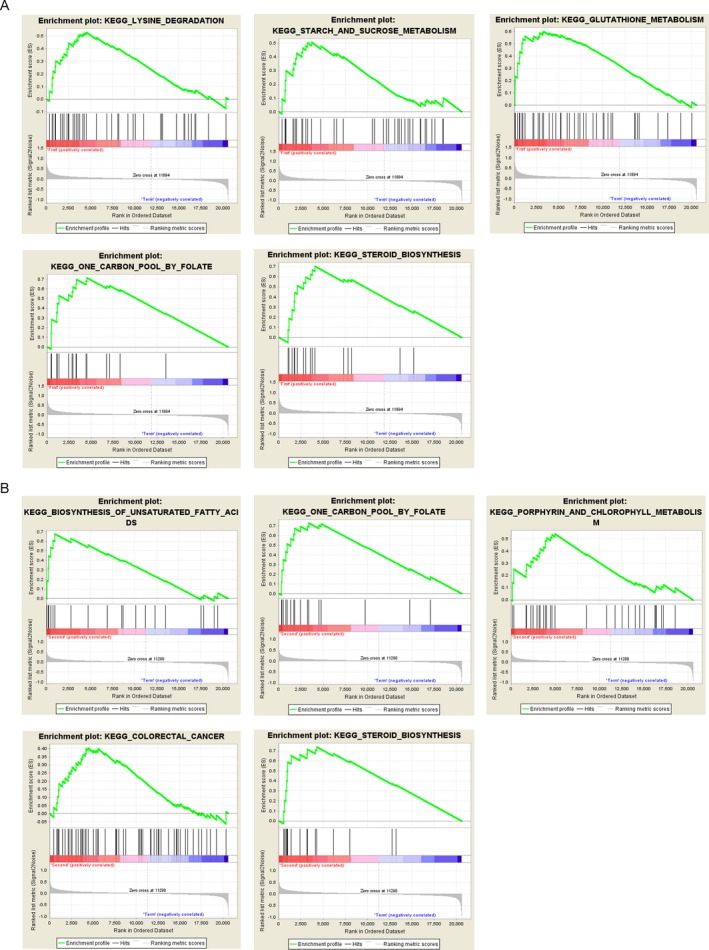
Gene set enrichment analysis. (a) The full list of significantly up‐regulated KEGG pathways in first‐trimester gestational diabetes mellitus (GDM) samples compared with normal samples. (b) The top five most significantly up‐regulated KEGG pathways in second‐trimester GDM samples compared with normal samples
Table 2.
Full list of KEGG pathways that significantly up‐regulated in second‐trimester gestational diabetes mellitus samples
| Pathway name | ES | Nom p value |
|---|---|---|
| KEGG_BIOSYNTHESIS_OF_UNSATURATED_FATTY_ACIDS | 0.677 | 0 |
| KEGG_ONE_CARBON_POOL_BY_FOLATE | 0.727 | 0 |
| KEGG_PORPHYRIN_AND_CHLOROPHYLL_METABOLISM | 0.539 | 0 |
| KEGG_COLORECTAL_CANCER | 0.404 | 0 |
| KEGG_STEROID_BIOSYNTHESIS | 0.738 | 0.0192 |
| KEGG_LYSINE_DEGRADATION | 0.556 | 0.0195 |
| KEGG_GLYCEROLIPID_METABOLISM | 0.417 | 0.0200 |
| KEGG_CELL_CYCLE | 0.577 | 0.0303 |
| KEGG_AMINOACYL_TRNA_BIOSYNTHESIS | 0.567 | 0.0306 |
| KEGG_VIBRIO_CHOLERAE_INFECTION | 0.412 | 0.0446 |
| KEGG_RNA_POLYMERASE | 0.460 | 0.0497 |
ES: enrichment score; Nom p value: normalized p value.
4. DISCUSSION
Gestational diabetes mellitus represents the most prevalence form of pathoglycemia in pregnancy that deeply affects the life of mothers as well as their offspring. In this study, we identified some potential GDM‐related genes by analyzing GDM time series gene expression profiles through WGCNA along with PPI network analysis. GSEA strikingly obtained some cancer‐related pathways in addition to several well‐known GDM‐associated pathways which were significantly up‐regulated in GDM samples compared with control samples. This study should shed some new light on the understanding of GDM mechanisms and its diagnosis or treatment.
Abnormal insulin secretion and metabolism contribute greatly to GDM initiation and progression. Here, lysine degradation, carbon pool by folate, and steroid biosynthesis pathways were significantly up‐regulated in first as well as second‐trimester GDM samples compared with control samples. Lysine represents a major component of histone, and modifications such as acetylation, methylation, and so on in it play vital roles in major cellular functions, for example, posttranscriptional proteins’ modification. In addition, lysine acetylation was also previously reported to affect both immunological and metabolic pathways, which could then induce type II diabetes and cardiovascular disease (Iyer & Fairlie, 2012; Kosanam et al., 2014). Nε‐(carboxymethyl) lysine‐conjugated bovine serum albumin is an essential component of advanced glycation end products which could damage mitochondrial functions and result in reducing insulin secretion followed by the incidence of diabetes (Lo et al., 2015). Diabetes could control many aspects of endocrine, including steroidogenesis, whose perturbation could in turn induce the initiation of diabetes (Hwang, 2014; Jangir, 2014). In addition to some well‐known pathways related to GDM, we strikingly identified the up‐regulation of several cancer‐related pathways in second‐trimester GDM samples, such as colorectal cancer, cell cycle. It has been widely reported that diabetes could result in elevated colorectal cancer risk (Jarvandi & Davidson, 2013; Wu et al., 2013), which was also confirmed in a rat model (Jia et al., 2014).
MM and degree are important for evaluating genes’ associations with specific trait in gene coexpression and PPI network, respectively. In this study, several genes have both high MM in brown module and high degree in PPI network, which might serve as important diagnosis or treatment biomarkers for GDM. CDK1 and CCNB1 are the top two genes with larger degree in PPI network, and CDK1 and CCNB1 interaction was previously proved to coordinates mitochondrial respiration and affect G2/M cell cycle progression (Wang et al., 2014). Cell cycle perturbation is closely associated with multi diseases’ progression including diabetes (Saavedra‐Avila et al., 2014; Wang, Fiaschi‐Taesch, et al., 2015). Besides, aberrant expression of CDK1 and CCNB1 in diabetes patients were also proved by previous studies (Page, Morris, Williams, Ruhland, & Malik, 1997; Su et al., 2015). CCNB2’s MM is the largest one compared with the other nine genes in Table 1. In Zhang's study (2017), CCNB2 was found to be significantly up‐regulated in diabetes mice compared with normal mice and was also the core gene in PPI network of DEGs. Although no previous study about direct association between some of those genes with GDM progression, lots of studies have proved associations between those genes with the well‐known GDM‐associated biological processes, such as CDC20 with metabolism (Martin, Mebarki, Paradis, Friguet, & Radman, 2014), CENPE with insulin absorption (Zhu, Ai, Wang, Xu, & Teng, 2012), and so on. So, those genes might be potential biomarkers for GDM.
5. CONCLUSIONS
In conclusion, we identified several potential biomarkers for GDM through WGCNA and PPI network analysis. GSEA identified some cancer‐related pathways in addition to several well‐known GDM‐related pathways, which might provide novel clues for GDM experimental research and clinical treatment.
CONFLICT OF INTEREST
I declare that there is no conflict between authors.
ACKNOWLEDGMENT
I would like to thank Wen Li for her help in data analysis.
Zhao X, Li W. Gene coexpression network analysis identified potential biomarkers in gestational diabetes mellitus progression. Mol Genet Genomic Med. 2019;7:e515 10.1002/mgg3.515
REFERENCES
- Ashwal, E. , & Hod, M. (2015). Gestational diabetes mellitus: Where are we now? Clinica Chimica Acta; International Journal of Clinical Chemistry, 451, 14–20. 10.1016/j.cca.2015.01.021 [DOI] [PubMed] [Google Scholar]
- Catalano, P. M. (2010). The impact of gestational diabetes and maternal obesity on the mother and her offspring. Journal of Developmental Origins of Health and Disease, 1, 208–215. 10.1017/S2040174410000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, R. , Badon, S. E. , Rangarajan, J. , Reisetter, A. C. , Armstrong, L. L. , Lowe, L. P. , … Lowe, W. L. Jr (2014). Genetic risk score for prediction of newborn adiposity and large‐for‐gestational‐age birth. The Journal of Clinical Endocrinology and Metabolism, 99, E2377–E2386. 10.1210/jc.2013-4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustan, D. R. (2013). Gestational diabetes mellitus. Clinical Chemistry, 59(9), 1310–1321. 10.1373/clinchem.2013.203331 [DOI] [PubMed] [Google Scholar]
- Dube, E. , & Ethier‐Chiasson, M. (2013). Lafond J Modulation of cholesterol transport by insulin‐treated gestational diabetes mellitus in human full‐term placenta. Biology of Reproduction, 88, 16 10.1095/biolreprod.112.105619 [DOI] [PubMed] [Google Scholar]
- Gautier, L. , Cope, L. , Bolstad, B. M. , & Irizarry, R. A. (2004). affy–analysis of affymetrix GeneChip data at the probe level. Bioinformatics, 20, 307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Harreiter, J. , Dovjak, G. , & Kautzky‐Willer, A. (2014). Gestational diabetes mellitus and cardiovascular risk after pregnancy. Women's Health, 10, 91–108. 10.2217/whe.13.69 [DOI] [PubMed] [Google Scholar]
- Hiden, U. , Maier, A. , Bilban, M. , Ghaffari‐Tabrizi, N. , Wadsack, C. , Lang, I. , … Desoye, G. (2006). Insulin control of placental gene expression shifts from mother to foetus over the course of pregnancy. Diabetologia, 49, 123–131. 10.1007/s00125-005-0054-x [DOI] [PubMed] [Google Scholar]
- Hwang, J. L. (2014). Weiss RE Steroid‐induced diabetes: A clinical and molecular approach to understanding and treatment. Diabetes/metabolism Research and Reviews, 30, 96–102. 10.1002/dmrr.2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, A. , & Fairlie, D. P. (2012). Brown L Lysine acetylation in obesity, diabetes and metabolic disease. Immunology and Cell Biology, 90, 39–46. 10.1038/icb.2011.99 [DOI] [PubMed] [Google Scholar]
- Jangir, R. N. (2014). Jain GC Diabetes mellitus induced impairment of male reproductive functions: A review. Current Diabetes Reviews, 10, 147–157. [DOI] [PubMed] [Google Scholar]
- Janikova, A. , Tichy, B. , Supikova, J. , Stano‐Kozubik, K. , Pospisilova, S. , Kren, L. , … Mayer, J. (2011). Gene expression profiling in follicular lymphoma and its implication for clinical practice. Leukemia and Lymphoma, 52, 59–68. 10.3109/10428194.2010.531412 [DOI] [PubMed] [Google Scholar]
- Jarvandi, S. , & Davidson, N. O. (2013). Schootman M Increased risk of colorectal cancer in type 2 diabetes is independent of diet quality. PLoS ONE, 8, e74616 10.1371/journal.pone.0074616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y. , Xu, G. , Zhou, W. , Wang, Z. , Meng, L. , Zhou, S. , … Tian, K. (2014). Diabetes promotes DMH‐induced colorectal cancer by increasing the activity of glycolytic enzymes in rats. PLoS ONE, 9, e110455 10.1371/journal.pone.0110455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda, M. , Oya, H. , Nomoto, S. , Takami, H. , Shimizu, D. , Hashimoto, R. , … Kodera, Y. (2015). Diversity of clinical implication of B‐cell translocation gene 1 expression by histopathologic and anatomic subtypes of gastric cancer. Digestive Diseases and Sciences, 60, 1256–1264. 10.1007/s10620-014-3477-8 [DOI] [PubMed] [Google Scholar]
- Kawai, V. K. , Levinson, R. T. , Adefurin, A. , Kurnik, D. , Collier, S. P. , Conway, D. , & Stein, C. M. (2017). A genetic risk score that includes common type 2 diabetes risk variants is associated with gestational diabetes. Clinical Endocrinology, 87, 149–155. 10.1111/cen.13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosanam, H. , Thai, K. , Zhang, Y. , Advani, A. , Connelly, K. A. , Diamandis, E. P. , & Gilbert, R. E. (2014). Diabetes induces lysine acetylation of intermediary metabolism enzymes in the kidney. Diabetes, 63, 2432–2439. 10.2337/db12-1770 [DOI] [PubMed] [Google Scholar]
- Langfelder, P. (2008). Horvath S WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, M. C. , Chen, M. H. , Lee, W. S. , Lu, C. I. , Chang, C. R. , Kao, S. H. , & Lee, H. M. (2015). Nepsilon‐(carboxymethyl) lysine‐induced mitochondrial fission and mitophagy cause decreased insulin secretion from beta‐cells. American Journal of Physiology Endocrinology and Metabolism, 309, E829–E839. 10.1152/ajpendo.00151.2015 [DOI] [PubMed] [Google Scholar]
- Magee, T. R. , Ross, M. G. , Wedekind, L. , Desai, M. , Kjos, S. , & Belkacemi, L. (2014). Gestational diabetes mellitus alters apoptotic and inflammatory gene expression of trophobasts from human term placenta. Journal of Diabetes and Its Complications, 28, 448–459. 10.1016/j.jdiacomp.2014.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F. A. , Mebarki, M. , Paradis, V. , Friguet, B. , & Radman, M. (2014). Hepatocellular carcinoma protein carbonylation in virus C and metabolic syndrome patients. Free Radical Biology and Medicine, 75(Suppl 1), S40 10.1016/j.freeradbiomed.2014.10.789 [DOI] [PubMed] [Google Scholar]
- Mikheev, A. M. , Nabekura, T. , Kaddoumi, A. , Bammler, T. K. , Govindarajan, R. , Hebert, M. F. , & Unadkat, J. D. (2008). Profiling gene expression in human placentae of different gestational ages: An OPRU Network and UW SCOR Study. Reproductive Sciences, 15, 866–877. 10.1177/1933719108322425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, S. , Nishigori, H. , Sugiyama, T. , Takahashi, F. , Iwama, N. , Watanabe, Z. , … Yaegashi, N. (2016). Association between social capital and the prevalence of gestational diabetes mellitus: An interim report of the Japan Environment and Children's Study. DiabetesResearch and Clinical Practice, 120, 132–141. 10.1016/j.diabres.2016.07.020 [DOI] [PubMed] [Google Scholar]
- Nikolic, D. , Al‐Rasadi, K. , Al Busaidi, N. , Al‐Waili, K. , Banerjee, Y. , Al‐Hashmi, K. , … Al‐Dughaishi, T. (2016). Incretins, pregnancy, and gestational diabetes. Current Pharmaceutical Biotechnology, 17, 597–602. [DOI] [PubMed] [Google Scholar]
- Oliveira, A. P. , Calderon, I. M. , Costa, R. A. , Roscani, M. G. , Magalhães, C. G. , & Borges, V. T. (2015). Assessment of structural cardiac abnormalities and diastolic function in women with gestational diabetes mellitus. Diabetes and Vascular Disease Research, 12, 175–180. 10.1177/1479164114563302 [DOI] [PubMed] [Google Scholar]
- Page, R. , Morris, C. , Williams, J. , von Ruhland, C. , & Malik, A. N. (1997). Isolation of diabetes‐associated kidney genes using differential display. Biochemical and Biophysical Research Communications, 232, 49–53. 10.1006/bbrc.1997.6224 [DOI] [PubMed] [Google Scholar]
- Pantham, P. , & Aye, I. L. (2015). Powell TL Inflammation in maternal obesity and gestational diabetes mellitus. Placenta, 36, 709–715. 10.1016/j.placenta.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra‐Avila, N. A. , Sengupta, U. , Sanchez, B. , Sala, E. , Haba, L. , Stratmann, T. , … Mora, C. (2014). Cyclin D3 promotes pancreatic beta‐cell fitness and viability in a cell cycle‐independent manner and is targeted in autoimmune diabetes. Proceedings of the National Academy of Sciences of the United States of America, 111, E3405–E3414. 10.1073/pnas.1323236111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, G. , Morris, J. H. , Demchak, B. , & Bader, G. D. (2014). Biological network exploration with Cytoscape 3. Current Protocols in Bioinformatics, 47, 8.13.1–8.13.24. 10.1002/0471250953.bi0813s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H. , Wan, Q. , Tian, X. J. , He, F. F. , Gao, P. , Tang, H. , … Zhang, C. (2015). MAD2B contributes to podocyte injury of diabetic nephropathy via inducing cyclin B1 and Skp2 accumulation. American Journal of Physiology Renal Physiology, 308, F728–F736. 10.1152/ajprenal.00409.2014 [DOI] [PubMed] [Google Scholar]
- Subramanian, A. , Kuehn, H. , Gould, J. , Tamayo, P. , & Mesirov, J. P. (2007). GSEA‐P: A desktop application for gene set enrichment analysis. Bioinformatics, 23, 3251–3253. 10.1093/bioinformatics/btm369 [DOI] [PubMed] [Google Scholar]
- Szklarczyk, D. , Franceschini, A. , Wyder, S. , Forslund, K. , Heller, D. , Huerta‐Cepas, J. , … von Mering, C. (2015). STRING v10: Protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43, D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uuskula, L. , Männik, J. , Rull, K. , Minajeva, A. , Kõks, S. , Vaas, P. , … Laan, M. (2012). Mid‐gestational gene expression profile in placenta and link to pregnancy complications. PLoS ONE, 7, e49248 10.1371/journal.pone.0049248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu, L. T. , Keschrumrus, V. , Zhang, X. , Zhong, J. F. , Su, Q. , Kabeer, M. H. , … Li, S. C. (2015). Tissue elasticity regulated tumor gene expression: Implication for diagnostic biomarkers of primitive neuroectodermal tumor. PLoS ONE, 10, e0120336 10.1371/journal.pone.0120336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Fiaschi‐Taesch, N. M. , Vasavada, R. C. , Scott, D. K. , García‐Ocaña, A. , & Stewart, A. F. (2015). Diabetes mellitus–advances and challenges in human beta‐cell proliferation. NatureReviews Endocrinology, 11, 201–212. 10.1038/nrendo.2015.9 [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, O. , Li, W. , Ma, L. , Ping, F. , Chen, L. , & Nie, M. (2015). Variants in vitamin D binding protein gene are associated with gestational diabetes mellitus. Medicine, 94, e1693 10.1097/MD.0000000000001693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Fan, M. , Candas, D. , Zhang, T. Q. , Qin, L. , Eldridge, A. , … Li, J. J. (2014). Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell‐cycle G2/M progression. Developmental Cell, 29, 217–232. 10.1016/j.devcel.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , Yu, C. , Jiang, H. , Tang, J. , Huang, H. L. , Gao, J. , & Zhang, X. (2013). Diabetes mellitus and the occurrence of colorectal cancer: An updated meta‐analysis of cohort studies. DiabetesTechnology and Therapeutics, 15, 419–427. 10.1089/dia.2012.0263 [DOI] [PubMed] [Google Scholar]
- Zhang, B. , & Horvath, S. (2005). A general framework for weighted gene co‐expression network analysis. Statistical Applications in Genetics and Molecular Biology, 4(1), Article17. 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhao, T. , Li, Z. , Yan, M. , Zhao, H. , Zhu, B. , & Li, P. (2017). Transcriptional profile of kidney from type 2 diabetic db/db mice. Journal of Diabetes Research, 2017, 1–12. 10.1155/2017/8391253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X. L. , Ai, Z. H. , Wang, J. , Xu, Y. L. , & Teng, Y. C. (2012). Weighted gene co‐expression network analysis in identification of endometrial cancer prognosis markers. Asian Pacific Journal of Cancer Prevention, 13, 4607–4611. 10.7314/APJCP.2012.13.9.4607 [DOI] [PubMed] [Google Scholar]


