Abstract
We developed a computational method based on polyploid phasing of long sequence reads to resolve collapsed regions of segmental duplications within genome assemblies. The approach, Segmental Duplication Assembler (SDA), constructs graphs where paralogous sequence variants define the nodes and long-read sequences provide attraction and repulsion edges allowing us to partition and assemble long reads corresponding to distinct paralogs. We apply it to single-molecule, real-time sequence data from three human genomes and recover 33–79 Mbp of duplications where approximately half of the loci are diverged (<99.8%) when compared to the reference genome. We show that the corresponding sequence is highly accurate (>99.9%) and that the diverged sequence corresponds to copy number variable paralogs that are absent from the human reference. Our method can be applied to other complex genomes to resolve the last gene-rich gaps, improve duplicate gene annotation, and better understand copy number variant genetic diversity at the base-pair level.
Keywords: gene duplication, segmental duplication, long-read, single-molecule, real-time (SMRT) sequence, PacBio sequencing, Oxford Nanopore Technologies (ONT)
INTRODUCTION
Advances in sequencing technologies and the development of novel computational assembly algorithms are central to the complete characterization of the content of complex genomes. Recent developments in long-read sequencing technology have dramatically improved the contiguity and speed at which de novo assemblies of complex genomes can be generated1–8. Individual labs, for example, can now accurately assemble >90% of the euchromatin in less than 1,000 contigs within a few months5,6,9,10. Despite these recent advances, significant portions of the genome remain unresolved. This is especially true for larger, highly identical repetitive regions, including heterochromatin and gene-rich regions associated with segmental duplications (SDs), which are larger than majority of long reads11–17.
SDs in most mammalian genomes are organized into complex regions typically >100 kbp in length and, by definition, are present at multiple locations. They contribute to dosage imbalance associated with disease18,19 and are ten times more likely to contribute to normal copy number variation20. They are also a reservoir for gene innovations associated with species adaptations21–23. The size, copy number, and sequence identity of SDs means that they are usually the last regions of the genome to be sequenced and assembled often using large-insert BAC (bacterial artificial chromosomes)24,25. More than half the gaps that remain in FALCON-based genome assemblies of single-molecule, real-time (SMRT) sequence data correspond to regions of SD. We estimate that the architecture of only 29.2% of SD bases are resolved in an assembly of CHM1 (Figure S1, Table S1, Methods) with most disease-associated regions unresolved (Table S2)18,26. Similarly, an assembly of NA12878 using longer Oxford Nanopore Technologies (ONT) ultra-long reads27 shows moderate improvement (32.9% resolved) but leaves most SDs unresolved (Figure S1, Table S1).
Here, we develop and apply the Segmental Duplication Assembler (SDA) method that takes advantage of paralogous sequence variants (PSVs) and correlation clustering28 to uniquely assemble different paralogs of SDs that were previously collapsed in long-read human genome assemblies. We apply it to actual SMRT and ONT long-read datasets to resolve SDs in recent assemblies and generate >30 Mbp of highly accurate, novel human genome sequence data. This method is computationally tractable and a generalizable solution for resolving collapsed repeat content in de novo assemblies of other mammalian genomes.
RESULTS
The problem: Unresolved SDs
While ONT and PacBio sequencing platforms generate long sequence reads, they also typically suffer from high error rates between 10–15%16,17. The predominant long-read assembly methods for whole-genome shotgun sequence assembly (WGSA) are based on read correction and overlapping corrected reads to construct larger sequence contigs, e.g., Canu and FALCON7,8. The high error rate of long-read sequencing platforms is particularly problematic for distinguishing paralogous and allelic sequence because the duplications are highly identical (>95%) and well within the range of error from long-read sequencing. This leads to sequence reads being recruited and merged from both paralogs and alleles during the assembly process creating collapses (Figure 1) where the assembled sequence and corrected sequence contig are in error. To quantify the effect of collapse and misassembly, we compared several recent assemblies generated using both ONT and SMRT sequence data (Figure S1, Supplementary Note). Requiring a 50 kbp extension into unique sequence, we estimate that only 49.0–51.3 Mbp of SDs are fully resolved (Figure S2) leaving 71% (~125/175 Mbp) of SDs associated with gaps. We note that even without requiring an extension into unique sequence, 59.5–69.8% of SDs remain unresolved (Figure S1). We estimate that ~50 Mbp of the duplications correspond to regions where the assembly algorithm has collapsed highly identical duplications into the same contig. Analysis of an ONT assembly generated with ultra-long reads (2.5-fold coverage of reads over 100 kbp)27 showed a modest 8% improvement in SD assembly; however, most of the SDs still remained unresolved (Figure S1). As expected, the largest (>10 kbp) and most identical duplications (>95% identity) are particularly enriched in unresolved SDs (Figure S2a) and frequently correspond to annotated human genes (Figure S2b).
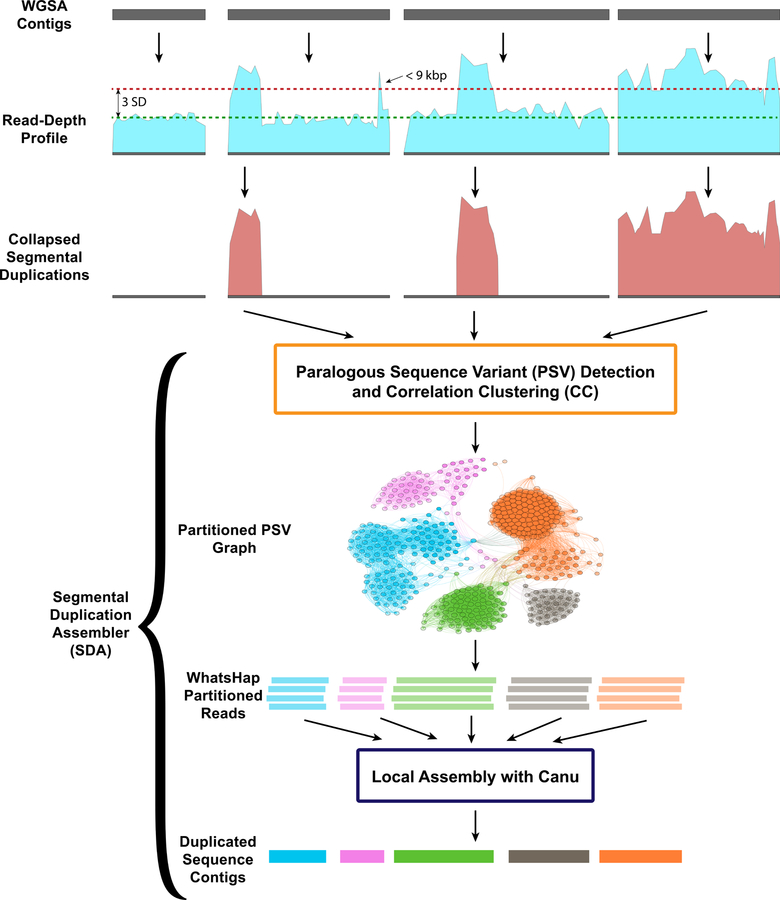
Figure 1. Flowchart of Segmental Duplication Assembler (SDA) method.
Regions of collapsed SDs are defined by assessing whole-genome shotgun (WGS) sequence read-depth profiles using BLASR across sequence contigs generated from a de novo WGSA. Regions (>9 kbp in length) with elevated sequence coverage (three standard deviations plus the mean) and not entirely composed of common repeats are considered collapsed SDs. Sequence reads corresponding to the collapsed SDs are recovered and examined for variants at each position along the collapse. Single-base-pair substitutions that appear at the same threshold as unique sequencing depth are identified and flagged as paralog-specific variants (PSVs) effectively partitioning reads into PSV clusters (WhatsHap). Sequence reads assigned to each PSV cluster are independently assembled using Canu and error-corrected using Quiver.
The approach: Segmental Duplication Assembler (SDA)
Previously, we presented a computational algorithm28 that could, in principle, assemble multi-copy duplications de novo using polyploid phasing29–33 and demonstrated its efficacy based on simulated datasets. Here, we develop SDA and apply it to WGSA collapsed duplications generated within existing human genome datasets. We specifically develop SDA to deal with different long-read datasets (Supplementary Note) and the generation of high-quality sequence contigs. Our method (Figure 1) identifies high-confidence PSVs ab initio and groups them using correlation clustering with defined attraction and repulsion edges into PSV graphs. We then assemble the partitioned reads independently, distinguishing the paralogous copies. Empirically we observe that we are able to assemble large duplications with less than 0.5% sequence divergence (Supplementary Note). As a measure of reproducibility, we apply this method to four human genomes and validate the results and accuracy based on targeted BAC sequencing and analyses of specific duplicated loci.
We begin by identifying all collapsed duplications within each assembly based on an excess of sequencing read depth11,34 (Methods). Within the CHM1 assembly9, for example, we identify 283 regions of collapse averaging 43 kbp in length (Table 1). When the 12.2 Mbp of collapsed CHM1 duplications are mapped back to the reference, they span 52.3 Mbp of sequence—93% (48.6 Mbp) are annotated as SDs and 88% of which (45.9 Mbp) overlap with regions of unresolved SDs in CHM1. Next, we define PSVs corresponding to each collapsed segment. We define candidate PSVs by classifying the second most frequent base at every position within the collapsed alignment and requiring sequence coverages consistent with a single-copy locus in order to distinguish PSVs from allelic variants (Methods). We next apply correlation clustering to filter false positive variants arising from sequencing error and uniquely assign each remaining PSV to the paralog from which it originates. For each collapsed region, we construct a graph where the PSVs define the nodes and the sequence reads define the edges. Attraction edges are formed when a read contains two or more PSVs connecting two or more nodes. Similarly, repulsion edges are formed when PSVs are mutually exclusive across all the sequence reads.
Table 1. SDA assembly statistics.
| De novo Assembly | Segmental Duplication Assembler (SDA) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Assembly Accession | Contig N50 (Mbp) | Sequence Coverage | Read N50 (kbp) | Unresolved SDs (Mbp) |
Collapses (count / Mbp) | Matched (count / Mbp) | Diverged (count / Mbp) | Multiple Assemblies (count / Mbp) | Failed |
| CHM19 | GCA_001297185.1 | 26.9 | 61 | 20.5 | 124.1 | 283 / 52.3 | 286 / 17.98 | 304 / 15.51 | 19 / 1 | 59 |
| CHM139 | GCA_002884485.1 | 29.3 | 67 | 18.2 | 126.5 | 527 / 86.6 | 685 / 39.1 | 755 / 35.0 | 69 / 3.1 | 339 |
| NA1924042 | GCA_001524155.4 | 29.1 | 61 | 17.5 | 124.1 | 489 / 82.4 | 789 / 38.8 | 983 / 40.9 | 107 / 5.8 | 257 |
| NA1287827 | GCA_900232925.1 | 7.7 | 35 | 12.5 | 117.7 | 365 / 52.5 | 38 / 0.066 | 792 / 22.1 | 8 / 0.21 | 1062 |
Genome summary statistics for four human genomes sequenced (SMRT/ONT) and assembled (FALCON/Canu) with long-read data.
Collapses from the assemblies were subjected to SDA and the number and Mbp of “matched” and “diverged” contig assemblies to the human reference genome (GRCh38) are shown.
With this formulation of the problem, it is possible to address the correlation clustering objective, which is to minimize the number of repulsion edges within clusters and minimize the number of attraction edges between clusters. Correlation clustering offers a distinct advantage over many other clustering algorithms because it does not require the number of clusters as a starting input. It is therefore ab initio and defined entirely by the underlying sequence data. However, correlation clustering is an NP (nondeterministic polynomial) complete problem; thus, we developed a heuristic to approximate the solution modeling after previous work35. The heuristic randomly assigns PSVs to clusters and then iteratively increases the size of the cluster by following positive edges that decrease the score of the entire graph (Methods).
Resolving SDs using SDA
We applied correlation clustering to each of the 283 collapsed regions in the CHM1 WGSA and generated a total of 668 distinct groupings. We created separate assemblies corresponding to each PSV graph partition using Canu followed by Quiver error correction. We successfully generated 590 assemblies where a single contig was produced corresponding to 33.1 Mbp of assembled sequence (Table 1, Figure 2) with an average sequence contig length of 60.7 kbp. The median assembly length was 53.0 kbp (mean 60.7 kbp), and the maximum sized assembly was 255.5 kbp. In general, the length of the assembly correlates (r = 0.67, Pearson’s correlation) with the size of the collapse (Figure S3). Of the 668 PSV graphs, 59 failed to generate an assembly and 19 assembled into multiple contigs. An inspection of those clusters that failed to assemble showed that the majority did so due to an insufficient number of reads while clusters with multiple contigs were the result of either incomplete PSV separation among multiple contigs or variable sequence coverage.
Figure 2. SDA results of the CHM1 human genome assembly.
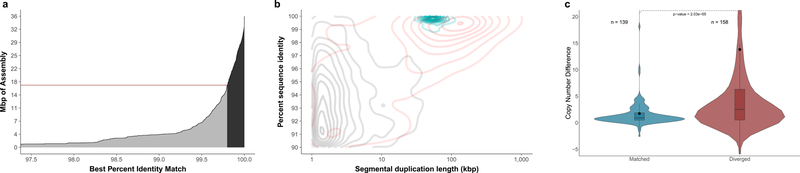
a) A cumulative distribution of the SDA assemblies and their percent identity to their best match in the reference. There is 16.4 Mbp of diverged assembly (<99.8% identity, gray) and 18.8 Mbp that map to the reference at high identity (>99.8% identity, black). The number of assembly Mbp is calculated independently of a mapping to the reference, unlike in Table 1. b) Density plot of SDs plotted by length and percent identity. Black represents duplications resolved in the CHM1 assembly, red shows unresolved duplications in the CHM1 assembly, and blue represents paralogs assembled using SDA. Resolved SDA sequences are “content” resolved and not ordered within the genome, whereas SDs in the assembly must extend into unique sequence on both sides to be considered resolved. c) Copy number difference (CND) between CHM1 and the reference genome (CHM1 copy number – reference genome copy number) comparing n=139 SD regions that match (>99.8%) versus n=158 diverged SD regions (<99.8% identity). The mean CND of the matched sequence is 1.75, and the mean CND of the diverged sequence is 13.82 (black dot) indicating that the diverged sequences are much more likely to represent additional duplicate copies that are unrepresented reference genome (GRCh38) (two-sided Mann-Whitney test; p=2.03*10–5). The boxes indicate the range between the first and third quartiles, with the bold line specifying the median. The whiskers show the minimum and maximum within 1.5 times the interquartile range extending from the first and third quartiles. Copy number was estimated in CHM1 examining k-mer frequency found in Illumina WGS reads; methods are described in Sudmant et al. 2015. A similar approach was used for estimating copy number in the reference except we generated simulated reads using the reference and then estimated copy number in the same fashion using the simulated reads.
In order to assess the accuracy and contiguity of the assembled SDs, we mapped each sequence contig back to the human reference genome (GRCh38). Of these assemblies, 48.5% (286/590) mapped to the human reference with at least 99.8% sequence identity over >90% of the contig length and accounted for ~18 Mbp of sequence. Interestingly, a similar fraction of assembled contigs (51.5% (304/590) (corresponding to 15.5 Mbp) showed greater sequence divergence ranging from 96% to 99.8% sequence identity (Figure 2a). We consider the contigs that “match” at high identity to GRCh38 to be correctly assembled and classify those with lower sequence identity than expected based on allelic variation (<99.8%)36 to be “diverged.” Since >0.2% divergence lies outside the typical range of human allelic variation, such diverged sequence may represent different copies of the duplication not yet represented in the human genome. We examined, in detail, a few human-specific gene families (e.g., SRGAP2 and NOTCH2NL) associated with neuroadaptation22,37–41 that have been the target of detailed BAC-based sequence assemblies (Table S3, Figures 3 and S4). Our analysis shows that we have successfully resolved the collapsed assemblies recreating the sequence and gene models present in the reference. This includes the identification and characterization of paralog-specific structural variation with most sequence assemblies matching ~99.8%−99.9% to their respective paralogs. Among these gene families, we estimate that 91%−93% of all PSVs have been correctly assigned.
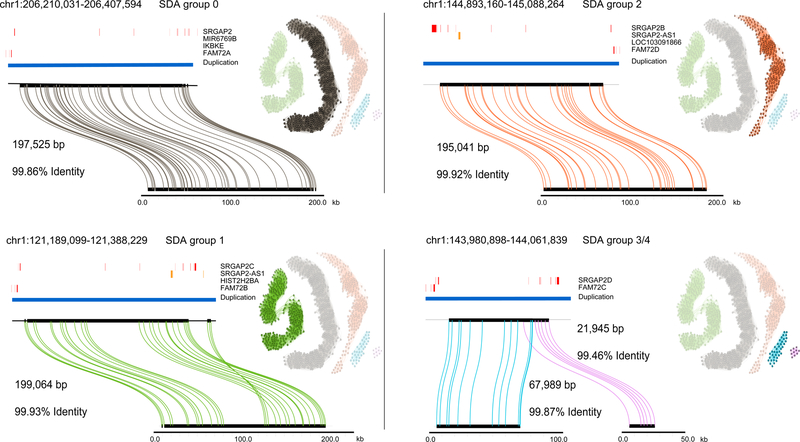
Figure 3. Sequence and assembly of SRGAP2 loci in the CHM13 human genome.
SDA sequence contigs from CHM13 aligned to the GRCh38 loci for SRGAP2(A/B/C/D) using Miropeats49. The length and percent identity of each alignment is shown. Similarly, in CHM1 we found that, on average, our sequence is 99.91% identical over all four loci and >99.999% identical if only mismatched bases are counted as errors as opposed to including indels. Adjacent to each alignment is the PSV graph with the relevant PSVs highlighted. Each node represents a PSV and loci are colored and numbered to reflect the grouping determined by correlation clustering. An edge is added between two nodes (PSVs) when a sequencing read contains both PSVs. The opacity of each node scales from 25% to 100% to reflect the position of the PSV along the collapse: 25% opacity reflects the first position along the collapse and 100% reflects the final position. For a more detailed view of the opacity of the nodes, see Figure S12. Clusters 3 and 4 in the PSV graph represent the fourth paralog (SRGAP2D), which carries a large deletion in the middle relative to the other paralogs.
We repeated this analysis for three additional long-read human genome assemblies, including a second haploid genome (CHM13)9, a diploid genome of African descent (YRI19240)42, and a diploid genome assembled with ONT (NA12878)27 (Table 1, Supplementary Note, Figures S5-S7). The proportion of matched and diverged sequence assemblies as well as resolved SD regions was very similar among the PacBio genomes. For example, 83% (1,772/2,136) of clusters resolved into single contig assemblies for the African diploid genome assembly. In contrast, an analysis of a human genome assembly (NA12878) generated with ultra-long ONT reads showed more failed SD assemblies, although we note that the coverage of this genome was significantly less than that of the PacBio genome assemblies (Figure S7). Combining both the “matched” and “diverged” sequences, we estimate that the SDA method adds an additional 72.6 and 78.6 Mbp of sequence corresponding to duplicated regions of the CHM13 and NA19240 human genomes, respectively.
Characterization of diverged duplications
We focused on the diverged duplications and considered two possibilities: the sequence could represent misassembled sequence or, alternatively, may represent additional copies not yet present in the human reference genome. The latter may be expected given that SD regions are 10-fold more likely to be copy number polymorphic20 than unique regions of the genome. If diverged sequences resulted from the sequence and assembly of additional copies, we would expect a significant increase in the copy number difference for diverged sequences when compared to duplicated sequences that matched the human reference genome (>99.8% sequence identity). Indeed, a comparison of the copy number difference for these two categories clearly showed that diverged copies were more likely (p = 2.0 × 10–5) to have a higher copy number in CHM1 (Figure 2c) than duplicated sequences that matched the reference genome assembly.
As a more direct test, we sequenced and assembled 1,253 large-insert BAC clones (Table S4) corresponding to regions of SD from a genomic library (CHORI-17) derived from CHM143,44 (Methods). Restricting our analysis to the 304 diverged sequences assembled by SDA from CHM1, we identify 105 diverged duplications that match the CHORI-17 clones. Each of these 105 sequences aligned to a clone over at least 90% of its length and at >99.8% sequence identity (mean sequence identity of 99.97%) (Figure 4, Table S5). If we assume that our method targeted all SDs evenly across the whole genome, then we would expect approximately 37.4% of the bases across our diverged sequences to validate. We observe that 105 of our diverged sequences, or 36.3% of the bases, validate and show significantly better alignment to the CHM1 clone inserts when compared to GRCh38. We estimated the sequence accuracy for our assembled duplications as 99.989 (quality value (QV) = 38.4) considering only single-base-pair mismatches and 99.857% (QV = 28.4) if indels and mismatches are counted. We note that many of the 105 validated assemblies contain sequences associated with gene families and, thus, have the potential to recover missing genic sequence not yet annotated. For example, we assembled a paralog of NBPF1 that is 1.2% diverged from the human reference but maps with >99.99% sequence identity to a CHM1 clone (Figure 4, Table S6). Similarly, Sudmant and colleagues45 identified an additional duplication in 16p12.1 that exists in most individuals but was absent from the reference. Using SDA, we recovered the proposed duplication46 (Figure S8) with only one mismatched base pair across a 95 kbp alignment to the BAC-generated contig.
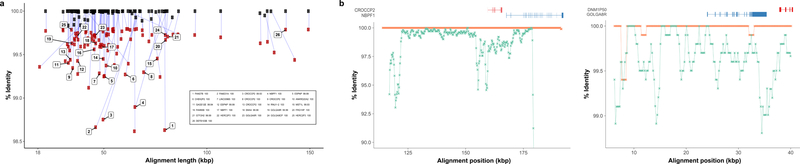
Figure 4. Correspondence between SDA sequence-diverged contigs and BACs.
a) The figure depicts the alignment length and percent identity sequence match for n=105 diverged SDA contigs compared to BAC clones (black) sequenced from the same source individual (CHM1) and the human reference genome (GRCh38) (red). The 15 most diverged sequences with respect to the reference and those containing duplicons as described by Jiang et al., 2007 and a more recent analysis of the human genome (n = 26) are shown. (See Tables S5 and S6 for more details.) b) Two examples of genes corresponding to diverged duplications are shown where the SDA sequence is aligned to both the reference genome (blue) and the CHM1 BACs (orange). BLASR alignments are computed in 1000 bp windows sliding 500 bp (steps).
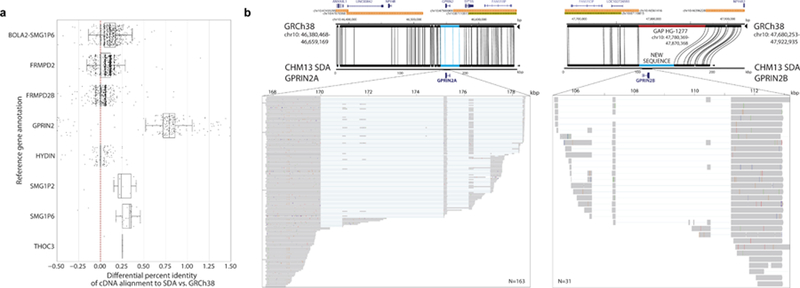
We analyzed more systematically the utility of these orphan SDA contigs to generate more accurate gene models for 37 human-specific segmental duplication (HSD) gene families. We selected 213,450 bulk single-molecule sequencing RNA reads (Iso-Seq) from fetal and adult human brain enriched for HSDs47. We aligned Iso-Seq data and compared their mapping between SDA contigs versus previous collapsed contigs in the CHM13 assembly. Transcripts showed improved mapping to the SDA contigs for 11 gene families to varying degrees (Figure S9). We identified six gene families (Figure 5a) where transcripts mapped better to the SDA assemblies than the human reference genome. A subset of transcripts from the GPRIN2 (G-coupled protein inducer of neurite outgrowth) gene family are most striking with a 1.5% improvement. We aligned the second SDA GPRIN2 contig that appears to be missing from the reference and found that it spans a gap in GRCh38 flanked by SDs (Figure 5b). Moreover, a previous analysis of Illumina whole-genome shotgun (WGS) sequence shows that GPRIN2 is polymorphic with copy number ranges from 3–7 copies with most humans carrying four in contrast to other apes which carry only one (diploid copy number = 2). Our analysis shows that both copies, GPRIN2A and GPRIN2B, are transcribed and encode similar open reading frames, although GPRIN2B has a 3-amino-acid insertion as well as several amino acid differences when compared to the ancestral GPRIN2A (Figure S10). Interestingly, these PSVs have been erroneously classified as single-nucleotide variants (SNVs; with near 50% “allele” frequency in dbSNP) because the reference is missing this second copy (Table S7). Thus, the SDA contig not only improves gene annotation but also improves interpretation of human genetic variation.
Figure 5. Gene discovery.
a) The percent identity differential of the mapping of full-length Iso-Seq transcripts (n=4,718) from human-specific duplications (HSDs) to both GRCh38 and SDA results on CHM13. The red dotted line represents equal mapping between the two; whereas points to the right represent an improved mapping with the SDA contigs. Six HSD gene families showed significantly (p < 0.05, two-sided Wilcoxon signed-rank test) improved mapping to the SDA-resolved contigs with the biggest difference occurring for GPRIN2. The boxes indicate the range between the first and third quartiles, with the bold line specifying the median. The whiskers show the minimum and maximum within 1.5 times the interquartile range extending from the first and third quartiles. b) GPRIN2 SDA contigs compared (Miropeats) to the human reference assembly (GRCh38) with gene and SD annotation. The SDA contigs close a gap (red) in GRCh38, which contains a duplicate copy of GPRIN2A denoted here as GPRIN2B. Mapping of individual Iso-Seq transcripts (inset) from the brain show that both loci are transcribed but that GPRIN2B has several coding differences, including a 3-amino-acid insertion at position 239 in GPRIN2B compared to GPRIN2A, the ancestral copy (Figure S10, Table S7).
DISCUSSION
In this study, we develop a method to accurately assemble high-identity SDs from long-read WGS sequence. There are three strengths to SDA. First, our approach does not require PSVs to be predefined and, as such, can be applied to any genome assembly where long-read data of sufficient depth has been generated. A similar concept was recently applied to partition viral quasispecies48. Second, our validation results suggest that the paralog-specific assemblies are highly accurate (99.86%−99.99%). Importantly, the approach allows missing paralogs to be sequenced especially within regions of extensive copy number variation. This is particularly exciting because it allows previously uncharacterized forms of human genetic variation to be sequence-resolved for the first time. Finally, our analysis of the human genome suggests that the majority of collapsed duplications are at least partially resolved (Figure 2). Since unassembled SDs typically represent ~70–90 Mbp of sequence per genome, recovery of 33–79 Mbp is the equivalent of recovering an entire chromosome’s worth of DNA for which accurate gene models can be constructed (Tables 1 and S8). The method we have developed can be effectively applied to any genome for which long-read WGSA data exist providing access to the duplicated regions and the genes therein.
Notwithstanding these advances, limitations remain. The majority of the sequence contigs we generated with SDA are small (~54 kbp) and are not yet commensurate with the average contig lengths generated by long-read sequence and assembly of unique regions of the genome. Only a small fraction (22%) of SDA contigs transition into unique sequence such that overlaps can be unambiguously assigned into the main genome assembly (Figure S11). Our new duplicated sequence contigs are not yet fully integrated into the genome and many of the resolved duplications remain “orphan” contigs in the absence of additional long-range mapping data. Directly integrating our SDA tool into popular long-read assemblers, to create long-range linkage information, may not be advisable even if it were possible. Optimizing parameters for SD assembly would likely come with costs for the remaining 95% of the genome. There are distinct advantages to performing bulk WGSA followed by a second-tier analysis to focus on the collapsed regions of the assembly. This is because overlap stringency should differ for high-identity duplications, and because PSVs provide important information for determining overlaps in these more difficult-to-assemble regions.
While we have shifted the accessible portions of SDs to larger (>50 kbp) and more identical regions (~99%), not all regions can be resolved using this approach. Duplications that are virtually identical cannot be distinguished and will require even longer read data, such as the ultra-long reads (>100 kbp) possible using ONT27. While we have developed and benchmarked SDA primarily with PacBio sequence data, we have also applied it to long-read sequence data from other platforms such as ONT (Supplementary Note). Our initial analysis of the ultra-long-read genome assembly of NA1287827, for example, showed a slight improvement of 8% in SD assembly (Figure S1). However, most of the high-identity SDs remained unresolved with a similar number of collapsed duplications (n = 365) when compared to PacBio genome assemblies. Application of SDA to the ONT dataset resulted in far fewer resolved assemblies (Figure S7) with an overall lower accuracy of the assembled sequence contigs. An important difference, however, is sequence coverage. The NA19240 PacBio assembly was sequenced at 73-fold sequence coverage versus the 35-fold ONT genome assembly. We note that while ultra-long ONT sequence reads were less successful in resolving SDs, they were useful as orthogonal data to validate PacBio SDA contigs (Supplementary Note). If long reads in excess of 200 kbp can be routinely generated with sufficient coverage to correct sequence error, it is possible that most SDs could be resolved by WGSA. The rapid advance of long-read sequencing technology may make the routine generation of ultra-long reads from low quantities of DNA a reality in the near future. Such advances would open up the possibility that other highly repetitive regions, such as centromeres and acrocentric DNA, could be routinely sequenced and assembled for the first time.
ONLINE METHODS
Human genome assemblies.
We analyzed three human genome assemblies derived from haploid (CHM1 and CHM13)9 and diploid source material (NA1924042) of African descent. FALCON genome assemblies were previously generated from at least 61-fold SMRT sequence using P6C4 chemistry generated on the PacBio RS II sequencing platform. We also analyzed one recent human genome assembly (NA12878) generated with ultra-long ONT sequence reads27.
SD characterization.
We mapped each human de novo assembly to the human reference genome GRCh38 using MashMap 2.0 (default settings)50 and defined SD regions based on intersection with annotated SDs in GRCh38. Sequence contigs overlapping SDs were defined as resolved if the contig completely contained the SD sequence and extended at least 50 kbp on either side into unique sequence. We compared the number of resolved and unresolved contigs (Figure 1a) for each assembly as a function of SD block length and maximum percent identity. Scripts are available at https://github.com/mvollger/segDupPlots, as well as a more detailed description of the analysis in the README.
Assembly collapse and PSV definition.
Within each assembly, we identified collapsed SDs by mapping SMRT or ONT sequencing reads back to each genome using BLASR51 (version rc46) or minimap252 (version 2.11) for ONT. Using unique regions, we computed the read coverage and standard deviation across 100 bp windows using the following BLASR settings (blasr $READS $ASM -sa $ASMSA \ -sdpTupleSize 13 -sdpMaxAnchorsPerPosition 10 -maxMatch 25 \ -minMapQV 30 -bestn 2 -advanceExactMatches 15 \ -clipping subread –sam). We excluded regions with >75% common repeat elements (RepeatMasker version 2004/03/06 –e wublast) and regions in the bottom or top two percentiles. We defined collapsed regions as those with a mean sequence coverage >3 standard deviations beyond the mean coverage and that were at least 9,000 bp in length (as smaller regions were routinely sequence and assembled). We examined all regions of collapse for the presence of SNVs and cataloged the second most common base at each position within the collapsed region using a more sensitive BLASR settings (blasr {input.basreads} {input.ref} \ -sam -preserveReadTitle -clipping subread \ -bestn 1 \ -mismatch 3 -insertion 9 -deletion 9 -minAlignLength 500). We defined these SNVs as potential PSVs if the sequence coverage was consistent with the read depth of unique regions. Three thresholds were applied to determine if an SNV was also a PSV. First, the total depth at the given position had to be at least the mean coverage plus three standard deviations. Second, the frequency of the second most frequent base had to be less than the mean coverage. Finally, the frequency of the second most frequent base had to be greater than the mean coverage minus three standard deviations or half the mean coverage, whichever was greater. This process favors the selection of PSVs over allelic variants (Figure S4). We developed a Snakemake pipeline for this analysis ProcessCollapsedAssembly.py, which can be found at https://github.com/mvollger/SDA.
PSV graph construction.
We constructed graphs for collapsed regions where each PSV corresponds to a node and sequence reads represent edges. Attraction edges are created when two PSV nodes have a substantial number of sequencing reads that contain both PSVs. Among reads containing both PSVs, we test whether each PSV is more likely to be real or a sequencing error using the ratio of two binomial tests. If at each PSV the log base 10 ratio of the two binomial tests was at least 1.5 (i.e., ~31 times more likely to be real than error), then an attraction edge was formed. Repulsion edges were created between any PSVs where less than 10% of the mean coverage of sequencing reads carried both PSVs.
Correlation clustering.
We initially added all nodes to an unclustered set from which a node was randomly selected and then expanded upon by iteratively searching neighbors of this node that reduce the overall score of the PSV graph (i.e., minimize the objective function). As nodes that meet this criterion are added to the cluster, they are removed from the unclustered set. This process was repeated until there were no unclustered nodes as previously described28. Next, all pairwise clusters are examined to see if they would improve the score of the graph if combined into a single cluster. Clusters are combined starting with the pairwise cluster that most improves the score of the correlation clustering objective. Clusters of three or fewer nodes are removed. The correlation clustering heuristic is run independently 15 times each with different random initializations and the clustering that best minimizes the correlation clustering objective is used to construct the final PSV clusters. It can be the case that in the construction of the PSV graph the PSVs are already clustered appropriately as unconnected components in the graph. In this case the application of correlation clustering is unnecessary to phase PSVs.
PSV read partition and assembly.
In order to partition SMRT or ONT sequencing reads according to the PSV clusters defined by correlation clustering, we apply WhatsHap53 (version 0.16) using the following parameters (whatshap haplotag $INPUT_VCF $INPUT_BAM -o $OUTPUT_BAM). Phasing was run on the entire set of reads for each PSV cluster, i.e., if there were five PSV clusters, WhatsHap was run five times to create five partitions of reads. After partitioning the reads into different paralogs, we independently assemble each correlation cluster with Canu version 1.5, followed by error correction (Quiver v 1.1.0) using the same set of reads. Specialized parameters are applied such that Canu can execute on such short contigs (https://github.com/mvollger/SDA/blob/master/SDA.2.snakemake.py).
BAC clone insert sequencing.
BAC clones from CHORI-17 (CH17) clone libraries (http://bacpac.chori.org) were hybridized with probes targeting complex or highly duplicated regions of the human genome reference (GRCh38) (n = 727) or based on previously sequenced clones (n = 526)43,44. DNA from positive clones was isolated by a modified alkaline lysis miniprep procedure as follows: cell pellet was resuspended in 200 μL Qiagen buffer P1 with RNase and lysed with 200 μL of 0.2M NaOH/1%SDS solution for five minutes. Lysis was neutralized with 280 μL 3M NaOAc, pH 4.8. Neutralized lysate was incubated on ice for up to 20 minutes, collected by centrifugation for 30 min at 4000 rpm, concentrated by standard isopropanol and then ethanol precipitation, and resuspended in 25 μL 10 mM Tris-Cl pH 8.5. We prepared barcoded libraries from clone DNA using Illumina-compatible Nextera DNA sample prep kits (Epicentre, Cat. No. GA09115) as described previously54 and paired-end sequenced (125 bp reads) on an Illumina HiSeq 2500. Reads were then mapped to the reference genome (GRCh38) to identify singly unique nucleotide k-mers (SUNKs), defined as 30-mers that identify a region of the genome and can be used in conjunction with short-read sequencing data to genotype highly identical paralogs55. This SUNK mapping was used to select a subset of positive clones for PacBio sequencing. BAC DNA from selected clones was isolated using a High Pure Plasmid Isolation Kit from Roche Applied Science per manufacturer instructions using 6 mL LB media with Chloramphenicol selective marker. We pooled non-overlapping BACs at equal molar amounts before library preparation. Approximately 1 µg of DNA per BAC was pooled and sheared using a Covaris® g-TUBE®. Libraries were processed using the PacBio SMRTbell Template Prep kit following the protocol ‘Procedure and Checklist −20 kb Template Preparation Using BluePippin™ Size-Selection System’. Libraries were size-selected on the Sage PippinHT with a start value of 10,000–12,000 and an end value of 50000. DNA/Polymerase Binding Kit (P6-C4 chemistry) was used to bind DNA template to DNA polymerase and the MagBead kit was used to capture DNA polymerase/template complexes for loading. Libraries were sequenced on the PacBio RS II platform. We performed de novo assembly of pooled BAC inserts using Canu v1.57. Reads were masked for vector sequence (pBACGK1.1) and assembled with Canu followed by consensus sequence calling with Quiver. Canu is specifically designed for assembly with long error-prone reads, while Quiver is a multi-read consensus algorithm that uses the raw pulse and base call information generated during SMRT sequencing for error correction. PacBio assemblies were reviewed for misassembly by visualizing read depth of PacBio reads in Parasight (http://eichlerlab.gs.washington.edu/jeff/parasight/index.html) using coverage summaries generated during the resequencing protocol.
Statistical information.
Statistical information for analysis of copy number differences is provided in Figure 2. The statistical analysis used to link PSVs with long-read data is described above in the section “PSV graph construction.”
Reporting summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
DATA AVAILABILITY
SMRT WGS for CHM1, CHM13, and NA12940 from this study are available at the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP044331 for CHM1; SRX818607, SRX825542, and SRX825575-SRX825579 for CHM13; and SRX1093000, SRX1093555, SRX1093654, SRX1094289, SRX1094374, SRX1094388, and SRX1096798 for NA19240. ONT WGS data are available at https://github.com/nanopore-wgs-consortium/NA12878/blob/master/Genome.md. De novo assemblies of CHM1, CHM13, NA12940, and NA12878 from this study are available at the NCBI Assembly database (Assembly; https://www.ncbi.nlm.nih.gov/assembly/) under accession numbers GCA_001297185.1, GCA_000983455.2, GCA_001524155.4, and GCA_900232925.1, respectively. Assembled CHORI-17 BACs are available at the NCBI Clone DB (Clone; https://www.ncbi.nlm.nih.gov/clone/) under the accession numbers listed in Table S4. Information about length, PSVs, and mapping location in GRCh38 can be found for all the SDA contigs generated in Table S8. Additional data that support the findings of this study are available from the corresponding author upon request.
CODE AVAILABILITY
Code for analyzing the resolved and unresolved SDs in a de novo assembly can be found at https://github.com/mvollger/segDupPlots. Code for processing de novo assemblies to find collapses and running SDA can be found at https://github.com/mvollger/SDA.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Cantsilieris and D. Gordon for technical assistance, J. Underwood for recommendations regarding the analysis of HSDs and Iso-Seq data, and T. Brown for help in editing this manuscript. This work was supported, in part, by grants from the U.S. National Institutes of Health (NIH grants HG002385 to E.E.E.; HG007635 to R.K.W. and E.E.E.; and HG003079 to R.K.W.). M.R.V. was supported by a National Library of Medicine (NLM) Big Data Training Grant for Genomics and Neuroscience (5T32LM012419–04). P.C.D. was supported by a National Human Genome Research Institute (NHGRI) Training Grant (5T32HG000035–23). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
CONFLICTS OF INTEREST
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc.
REFERENCES
- 1.Alkan C, Coe BP & Eichler EE Genome structural variation discovery and genotyping. Nat. Rev. Genet 12, 363–376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkan C, Sajjadian S & Eichler EE Limitations of next-generation genome sequence assembly. Nat. Methods 8, 61–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seo JS et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Shi L et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickhart DM et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet 49, 643–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D et al. Long-read sequence assembly of the gorilla genome. Science 352, aae0344–aae0344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koren S et al. Canu: scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res 1–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin C-S et al. Phased diploid genome assembly with Single Molecule Real-Time Sequencing. Nat. Methods 13, 056887 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huddleston J et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kronenberg ZN et al. High-resolution comparative analysis of great ape genomes. Science 6343, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley DR & Salzberg SL Detection and correction of false segmental duplications caused by genome mis-assembly. Genome Biol 11, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pop M Shotgun Sequence Assembly. Adv. Comput 60, 193–248 (2004). [Google Scholar]
- 13.Pevzner PA, Tang H & Waterman MS An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci 98, 9748–9753 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pevzner PA, Tang H & Tesler G De novo repeat classification and fragment assembly. Genome Res 14, 1786–1796 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers EW The fragment assembly string graph. Bioinformatics 21, 79–85 (2005). [DOI] [PubMed] [Google Scholar]
- 16.PacBio Available at: https://www.pacb.com/. (Accessed: 3rd March 2018)
- 17.Oxford Nanopore Technologies Available at: https://nanoporetech.com/. (Accessed: 3rd March 2018)
- 18.Stankiewicz P & Lupski JR Genome architecture, rearrangements and genomic disorders. Trends Genet 18, 74–82 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Sharp AJ et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet 38, 1038–1042 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Sudmant PH et al. Global diversity, population stratification, and selection of human copy-number variation. Science 349, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J et al. Bovine NK-lysin: Copy number variation and functional diversification. Proc. Natl. Acad. Sci. U. S. A 7223–7229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis MY & Eichler EE Human adaptation and evolution by segmental duplication. Current Opinion in Genetics and Development 41, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abegglen LM et al. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. Jama 314, 1850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Church DM et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander ES et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Emanuel BS & Shaikh TH Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet 2, 791–800 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Jain M et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol 36, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaisson MJ, Mukherjee S, Kannan S & Eichler EE Resolving multicopy duplications de novo using polyploid phasing. RECOMB 3, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das S & Vikalo H SDhaP: Haplotype assembly for diploids and polyploids via semi-definite programming. BMC Genomics 16, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aguiar D & Istrail S Haplotype assembly in polyploid genomes and identical by descent shared tracts. Bioinformatics 29, 352–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger E, Yorukoglu D, Peng J & Berger B HapTree: A novel bayesian framework for single individual polyplotyping using NGS data. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 8394 LNBI, 18–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puljiz Z & Vikalo H Decoding genetic variations: communications-inspired haplotype assembly. IEEE/ACM Trans. Comput. Biol. Bioinforma 13, 518–530 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Bonizzoni P et al. On the minimum error correction problem for haplotype assembly in diploid and polyploid genomes. J. Comput. Biol 23, 718–736 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Bailey JA Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Ailon N, Charikar M & Newman A Aggregating inconsistent information. J. ACM 55, 1–27 (2008). [Google Scholar]
- 36.Auton A et al. A global reference for human genetic variation. Nature 526, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiddes IT et al. Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florio M et al. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife 7, e32332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis MY et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuttle X et al. Rapid and accurate large-scale genotyping of duplicated genes and discovery of interlocus gene conversions. Nat. Methods 10, 903–909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis MY et al. The evolution and population diversity of human-specific segmental duplications. Nat. Ecol. Evol 1, 0069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinberg KM et al. High-quality assembly of an individual of yoruban descent. bioRxiv 067447 (2016). doi: 10.1101/067447 [DOI] [Google Scholar]
- 43.Chaisson MJP et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The CHORI-17 BAC Library from a hydatidiform (haploid) mole Available at: https://www.ncbi.nlm.nih.gov/clone/library/genomic/76/.
- 45.Sudmant PH et al. An integrated map of structural variation in 2,504 human genomes. Nature 526, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuttle X et al. Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature 536, 205–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dougherty ML, Underwood JG, Nelson BJ, Tseng E & Katherine M Transcriptional fates of human-specific segmental duplications in brain. Genome Res (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Artyomenko A et al. Long Single-Molecule Reads Can Resolve the Complexity of the Influenza Virus Composed of Rare, Closely Related Mutant Variants. J. Comput. Biol 24, 558–570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parsons JD Miropeats: graphical DNA sequence comparisons. Comput. Applic. Biosci 615–619 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Jain C, Koren S, Dilthey A, Phillippy AM & Aluru S A fast adaptive algorithm for computing whole-genome homology maps. Bioinformatics 259986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chaisson MJ & Tesler G Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H Minimap2: pairwise alignment for nucleotide sequences 34, 3094–3100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patterson M et al. WhatsHap: Weighted haplotype assembly for future-generation sequencing reads. J. Comput. Biol 22, 498–509 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Steinberg KM et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet 44, 872–880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudmant PH et al. Diversity of human copy number. Science 11184, 2–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SMRT WGS for CHM1, CHM13, and NA12940 from this study are available at the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP044331 for CHM1; SRX818607, SRX825542, and SRX825575-SRX825579 for CHM13; and SRX1093000, SRX1093555, SRX1093654, SRX1094289, SRX1094374, SRX1094388, and SRX1096798 for NA19240. ONT WGS data are available at https://github.com/nanopore-wgs-consortium/NA12878/blob/master/Genome.md. De novo assemblies of CHM1, CHM13, NA12940, and NA12878 from this study are available at the NCBI Assembly database (Assembly; https://www.ncbi.nlm.nih.gov/assembly/) under accession numbers GCA_001297185.1, GCA_000983455.2, GCA_001524155.4, and GCA_900232925.1, respectively. Assembled CHORI-17 BACs are available at the NCBI Clone DB (Clone; https://www.ncbi.nlm.nih.gov/clone/) under the accession numbers listed in Table S4. Information about length, PSVs, and mapping location in GRCh38 can be found for all the SDA contigs generated in Table S8. Additional data that support the findings of this study are available from the corresponding author upon request.