Abstract
Advances in sequencing technology have significantly expanded our understanding of the genetics of autism and neurodevelopmental disorders (NDDs). Continued technological improvements and cost reductions have now shifted the focus to investigations into the functional noncoding portions of the genome. There is a patient trend toward an excess of de novo and potentially disruptive mutation among conserved noncoding sequences implicated in the regulation of genes. The signals become stronger when restricting to genes already implicated in NDDs, but de novo mutation in such elements is estimated to account for <5% of patients. Larger sample sizes, improved variant detection, functional testing, and better approaches for classifying noncoding variation will be required to identify specific pathogenic variants underlying disease.
Keywords: autism, neurodevelopmental disorders, noncoding, regulatory mutations, genome sequencing, genetic architecture
GENETIC ARCHITECTURE AND GENOME TECHNOLOGY
Investigations into the genetic basis of autism and other neurodevelopmental disorders (NDDs) are limited by sample size and the scope and sensitivity of the genomic technology employed. Single-nucleotide polymorphism microarray data, for example, provided access to common variants under a genome-wide association study (GWAS) model as well as to large copy number variants (CNVs). Later, when whole-exome sequencing (WES) became commonplace the focus shifted to de novo and rare, inherited variants within the protein-encoding portions of our genome.
Each technological advance provided unique insights into the genetic architecture of autism and other NDDs. While GWAS has identified only a few consistent variants or loci associated with autism [1, 2], it did provide insights into its heritability, suggesting that common variants must contribute substantially to risk [3], or at a minimum, to sensitizing an individual to develop autism. There is an emerging paradigm of polygenic risk scores as playing an important role, and it is likely that with larger sample sizes more definitive autism risk loci will become apparent [4]. The discovery of an excess of large CNVs among 8%–15% patients with autism and NDD [5] was important because it suggested a genetic model where de novo and rare, inherited mutations created gene-expression dosage imbalances early in development leading to the development of disease. WES extended this model confirming the importance of de novo [6–10] and rare, inherited mutations [11] that disrupted gene function in an estimated 21% of autism [12] and 42% of NDD individuals [13]. Importantly, the nature of the mutations discovered by WES provided the specificity required to identify new genes and pathways underlying NDD, leading to the discovery of more than 124 genes reaching exome-wide significance, and 253 genes (5% FDR) with an excess of recurrent protein-damaging de novo mutations (DNMs) [14].
Combined rare/de novo coding variants and CNVs are now thought to contribute to about 20%–30% [8, 15] of individuals with autism. This means, of course, that for the majority of individuals with autism there is no obvious identified genetic cause. Given that the most recent heritability estimates for autism are now ~80% [16], other genetic risk variants, both rare and common, await discovery and characterization. Of note, it is possible that both rare and common variants might contribute to disease in the same patients. For example, large effect mutations such as CNVs might predispose to developmental delay, while the background common variants confer phenotypic specificity.
Whole-genome sequence (WGS) data provide, in principle, access to the complete spectrum of human genetic variation in an individual irrespective of the class or frequency of a variant. Among patients where no obvious genic or CNV cause has been identified, there has been a shift in focus to investigating the functional noncoding regions of the genome that are important in regulation of gene expression [15, 17–19]. This includes (but is not limited to) the untranslated regions (UTRs) of genes (3’ UTR, 5’ UTR), enhancers, promoters, noncoding RNAs (e.g., miRNA, piRNA), miRNA binding sites, and topologically associating domain (TAD) boundaries (Box 1, Figure I). Variants identified within noncoding DNA have long been known to cause Mendelian diseases [20, 21] and contribute to complex genetic traits [22–26]. For example, one of the most common causes of developmental delay and autism, Fragile X syndrome, is due to the hyperexpansion of a CGG repeat sequence located in the 5’ UTR of the X chromosome gene FMR1. The expansion of the CGG repeat promotes hypermethylation of the promoter region leading to silencing of FMR1 [27, 28]. Despite this simple model of functional effect, almost all cases of Fragile X syndrome are the result of CGG repeat expansion with relatively few examples of loss-of-function mutations in the protein-coding portion of FMR1 despite sequencing of tens of thousands of individuals with idiopathic autism and NDD (see denovo-db [29] version 1.6.1).
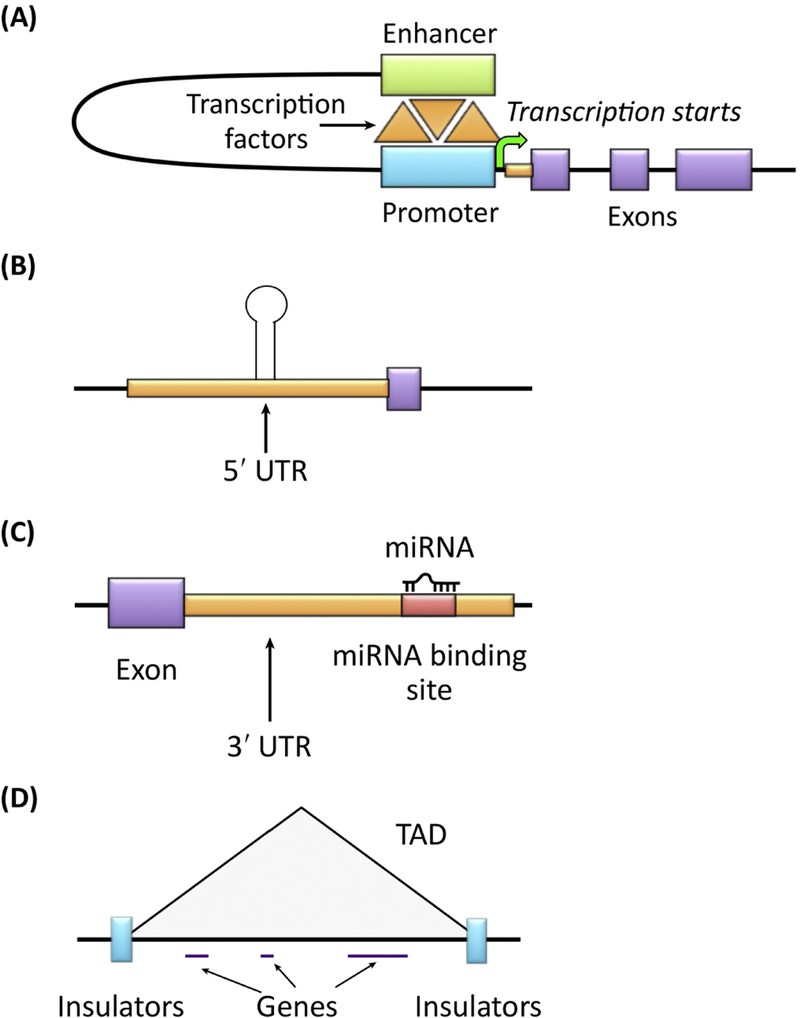
Box 1: Types of noncoding variants.
Some functional noncoding regions include promoters, enhancers, repressors, insulators, untranslated regions (UTRs), and noncoding RNA. Promoters (Figure I-A) are 5’ proximal to the transcription start site and are the location at which the core transcription machinery binds to the DNA. Enhancers (shown in Figure I-A bound by transcription factors and linked to the promoter) and repressors (not shown) are position-independent sequences involved in increasing and decreasing the transcriptional activity of genes, respectively. They can be close or far from the transcriptional start site. UTRs (Figure I-B) map to the 5’ and 3’ ends of genes and are part of the full-length transcript. The 5’ UTR contains the sequence for translation initiation and can also have other regulatory activity (exhibited as a hairpin here). The 3’ UTR (Figure I-C) contains the sequence for the termination of translation and frequently harbors miRNA binding sites important for repression of translation. At a higher level, the genome is organized into topologically associating domains (TADs) (Figure I-D) that comprise genes and regulatory elements. These TADs are flanked by insulator elements that can either block enhancer activity on a gene or maintain the boundaries for a set of genes or regions contained within the TADs. In addition to these elements, there are a variety of noncoding genes, such as transfer RNAs (tRNA), ribosomal RNA (rRNA), microRNA, siRNA, piRNA, snoRNA, lncRNA among many others, that regulate transcription, translation and splicing of genes or play a role in chromatin organization (e.g., XIST and X chromosome inactivation).
Figure I (for Box 1): Schematic of noncoding variants.
(A) Diagram of the promoter region of a gene where the promoter is bound by transcription factors and linked to an enhancer. The arrow indicates the location of the start of transcription. (B) Shown is an example 5’ UTR region with a hairpin initiating a site of regulatory activity. (C) An example 3’ UTR region is exhibited here with an miRNA binding site. An miRNA is also shown bound to this site to exemplify a location of regulatory activity. (D) Diagram of a TAD with genes inside of it and insulators shown at the boundaries of the TADs.
While there is no question that noncoding variation will play a role in human NDDs, a major challenge has been defining the functional elements and interpreting mutational effects. Large-scale efforts like the ENCODE [30] and Roadmap Epigenomics [31] projects have attempted to systematically catalog the noncoding regions of the genome in different cell types and tissues. Notwithstanding these valiant efforts, a definitive set of functional noncoding elements (NCEs) does not yet exist. In this Review, we consider the current evidence for the role of noncoding variants based on large-scale sequencing studies in autism and other NDDs and summarize the different approaches undertaken over the last few years to address this question. Our synthesis of the available data provides further support for specific NCE categories but also highlights potential pitfalls going forward. We lastly highlight some of the remaining questions in terms of classifying this variation, statistical testing, application of newer deep-learning-based approaches to further refine the NCEs, and the critical role of large-scale functional testing.
Large-scale next-generation sequencing studies
Simplistically, there have been two different sequencing-based approaches for assessment of noncoding variation: namely, WGS and targeted sequencing of affected individuals and/or their family members or other controls. Targeted sequencing includes WES studies because of its potential to recover regulatory variation within the UTR portions of genes but, of course, coding variation was the primary target of such efforts. Most published studies have focused on de novo variants assuming a dominant model of disease where de novo or disruptive mutation in regulatory DNA might interfere with normal expression of a gene. WGS is less biased because it puts no a priori knowledge on the initial experimental design when compared to targeted sequencing, which preselects putative functional regions for testing.
WGS studies of autism and NDD.
The first WGS studies were limited in scope to 20–85 parent–child trios [17, 32–34], were generated using a variety of sequencing platforms, and focused on establishing a framework for the discovery of new mutations and characterizing their patterns. The studies consistently reported increased detection sensitivity for DNM in protein-coding regions as well as smaller gene-disruptive CNVs increasing the diagnostic yield when compared to WES platforms [17, 33, 34]. Considerable genetic heterogeneity was noted even among families with multiple affected individuals where different autism- relevant mutations appear to be segregating (Figure 1) [34, 35]. In ~10% of families, WGS identified more than one potential risk variant suggesting a multifactorial rare variant model of disease for some individuals with autism (Figure 1), although the number of families was limited and still underpowered [17]. Turner and colleagues reported a nominal enrichment of de novo and private, disruptive mutations within putative regulatory regions of the fetal brain as defined by DNase I hypersensitivity when comparing probands to their unaffected siblings. The fetal brain had been strongly implicated previously based on gene expression and protein-protein interaction network analysis of autism risk genes identified from WES data [36–38] and reviewed in [39].
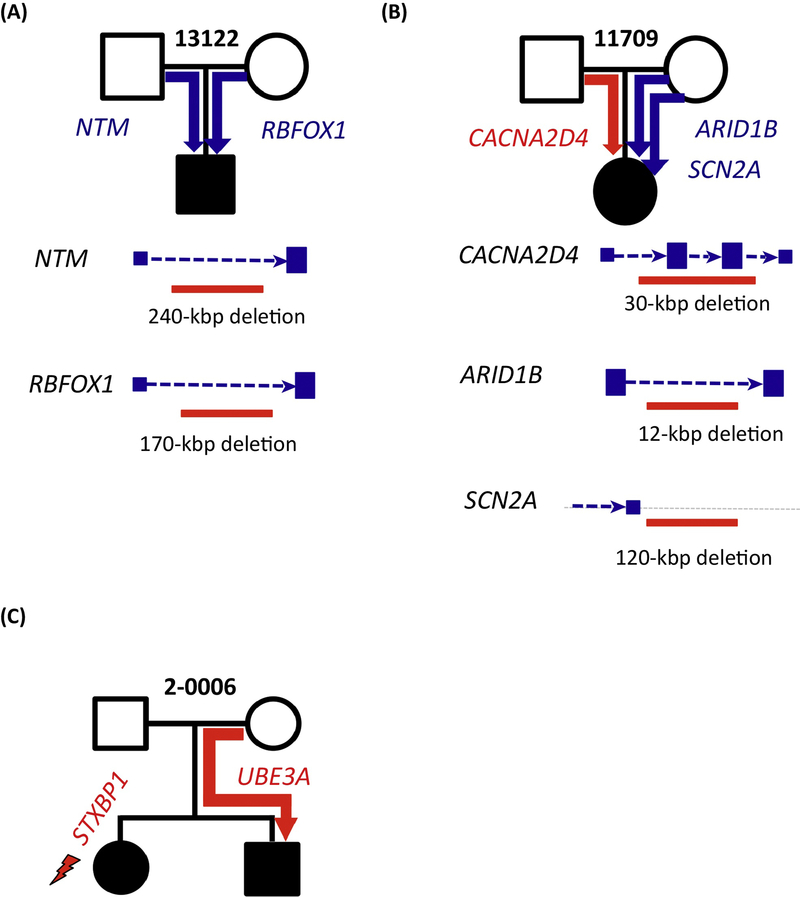
Figure 1: Genetic heterogeneity and multiple disruptive mutations in autism pedigrees.
In ~10% of patients with autism, genome sequencing reveals multiple de novo and disruptive mutations in different genes and their regulatory DNA. For example, (A) a child from SSC family 13122 inherits two large deletions affecting putative regulatory regions of autism risk genes NTM and RBFOX1. (B) Similarly, a child in SSC family 11709 inherits three different deletions with two affecting putative regulatory regions of the autism risk genes ARID1B and SCN2A. (C) Examination of families with multiple affected individuals frequently finds that a genetic risk variant segregates to only one of the two children or that different affected individuals each carry a different risk variant. For example, in MSSNG multiplex family 2–0006 [34], one autistic offspring carries a de novo loss-of- function event in STXBP1 while the other affected male sibling carries a maternally inherited loss-of-function event in UBE3A. Panels (A) and (B) are adapted from Turner et al. 2016 [17]. MSSNG family 2–0006 was studied in Yuen et al. 2015 [34].
Later WGS studies were significantly larger (200–500 families), focusing almost exclusively on autism [15, 18, 19, 40–43] with a subset emphasizing noncoding variation [15, 18, 19, 41–43] (Table 1). In one study, for example, de novo variants were assessed in 200 parent–child families from the MSSNG autism cohort [41] and compared to de novo variants in a published control cohort (258 families) called Genome of the Netherlands (GoNL) [44]. The study reported DNM enrichment for noncoding variants in the conserved part of UTRs, variants that caused exon skipping, and transcription factor binding sites located within DNase I hypersensitive sites (DHS) mapping near to genes. The experimental design was criticized, however, because the controls were sequenced at significantly lower sequence read-depth (13-fold vs. 32-fold) using a different sequencing platform as part of a different study, although steps were taken to minimize differences in sensitivity.
Table 1:
Autism and intellectual disability WGS cohorts
| Cohort | Family type(s) | Projected total number of families | Projected total number of individuals | Actual available samples as of July 2018 | Sequencer | Websites* | Publications |
|---|---|---|---|---|---|---|---|
| Simons Simplex Collection (SSC)& | simplex | 2,412# | 9,198# | 4675 | Illumina HiSeq X Ten |
https://www.sfari.org/ http://ccdg.rutgers.edu/ |
Turner et al. 2016@ [17], Brandler et al. 2016 [73], Turner et al. 2017@ [15], Brandler et al. 2018@ [19], Werling et al. 2018@ [18], Zhou et al. bioRxiv@ [42] |
| MSSNG | Simplex and multiplex | 2,756^ | 7,187^ | 7231 | Illumina HiSeq 2000 and Some Complete Genomics | https://www.mss.ng/ | Yuen et al. 2015 [34], Yuen et al. 2016@ [41], Yuen et al. 2017 [40] |
| Autism Genetic Resource Exchange (AGRE) | multiplex | 1,010# | 4,551# | 2308 | Illumina HiSeq X Ten | http://www.ihart.org/ | Ruzzo et al. bioRxiv@ [43] |
| NIMH | MZ twin pairs | 10 | 40 | 40 | Illumina HiSeq | https://ndar.nih.gov/study.html?id=322 | Michaelson et al. 2012 [32] |
| Intellectual disability patient cohort | trios | 50 | 150 | 150 | Complete Genomics | https://www.ebi.ac.uk/ega/studies/EGAS00001000769 | Gilissen et al. 2014 [33] |
| Total | 6238 | 21126 | 14404 | ||||
website link as of July 2018
from website as of July 2018
from readme file (mssngresearcherreadme_20171020.pdf) from website (July 2018)
indicates the study had a major focus on noncoding somewhere in the paper
families also being sequenced as part of the Centers for Common Disease Genomics (CCDG)
Phase I of the Simons Simplex Collection (SSC) consisted of ~520 families selected to be negative for “known” disease-causing mutation events. The study design had the advantage that an unaffected sibling was sequenced from each family using the same sequencing platform and same sequence depth. The genome sequence from the unaffected child served as a genetic control for the pattern of DNM when compared to the autism proband [45]. The WGS data were analyzed by three groups using different approaches and emphasizing different regions or classes of genetic variation [15, 18, 19]. In our own study, we applied multiple callers to identify de novo single-nucleotide variants (SNVs), indels, and SVs (structural variants). We reported an excess of smaller deletions that disrupted genes and a nominally significant genome-wide enrichment for de novo variants in UTRs and in putative, regulatory regions (transcription factor binding site [TFBS] in central nervous system [CNS] DHS) that are the most likely to function as enhancers and promoters [15]. Although the study was criticized for not considering all possible categories of noncoding functional DNA [18, 46], DNM signals became more significant if the analysis was restricted to autism- related genes. Interestingly, the study also found that patients are more likely to carry multiple coding and noncoding DNMs in different genes (Figure 2) and such genes with multiple hits are enriched for expression in striatal neurons. An excess of multiple DNMs in different noncoding DNA in patients would be consistent with this class of variation being pathogenic and/or support an oligogenic model of disease as has been suggested previously based on CNV studies [47, 48].
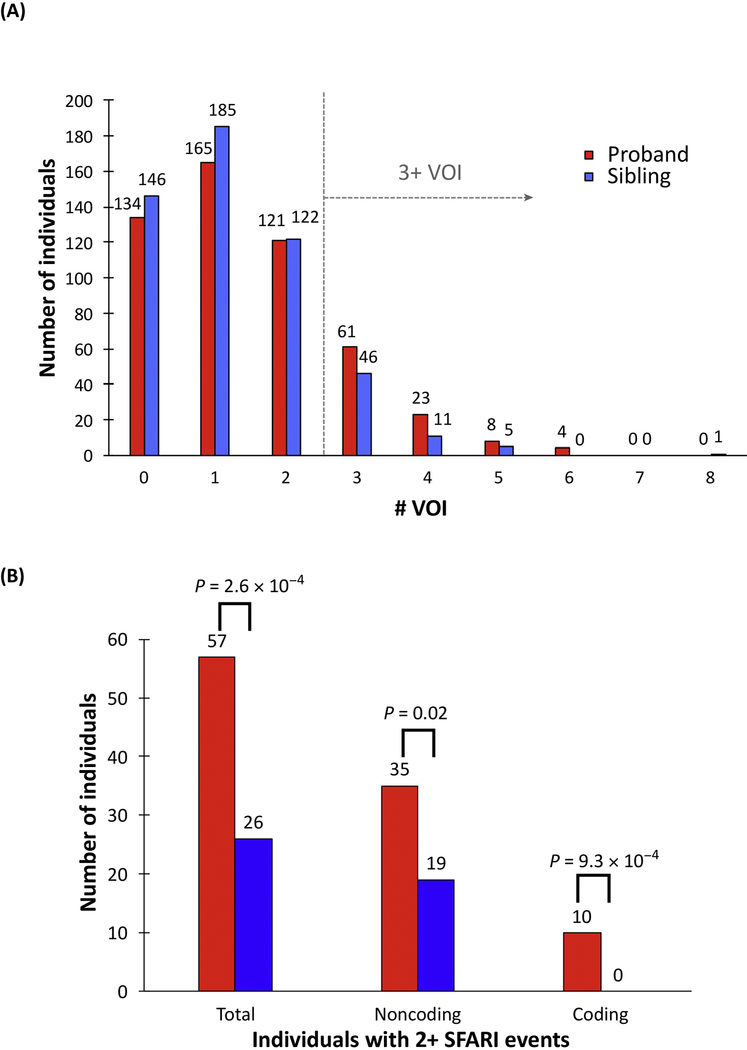
Figure 2: The pattern of multiple de novo mutations in autism genomes.
(A) The distribution compares the number of de novo variants of interest (VOIs) between affected individuals (red) and their siblings (blue) for 516 families from the SSC. VOIs are defined as severe DNMs that are likely to disrupt protein function and DNMs in putative regulatory DNA (promoter, 5’ UTR, 3’ UTR and DHS). Autism genomes tend to carry a greater number of such mutations creating a skewed distribution when compared to the genomes of their unaffected siblings. Multiple mutations would be expected if the individual mutations were pathogenic (increased probability of probands carrying multiple events based on a Poisson model) or if multiple mutations were necessary to reach a liability threshold of disease. (B) Restricting the analysis to known autism-risk genes (Simons Foundation Autism Research Initiative [SFARI]) shows a significant excess of two or more events in probands (red) compared to siblings (blue). The trend is observed when partitioning coding and noncoding portions of the genome, emphasizing the importance of WGS. Adapted from Turner et al. 2017 [15].
The second study focused exclusively on SVs [19] (>100 bp in size) because of their greater likelihood to disrupt gene function and expression when compared to SNVs. Although no difference was found for de novo SVs, the study did find a preferential transmission bias of cis-regulatory element SVs affecting promoters and the UTR of genes. They reported that these cis-regulatory element SVs were preferentially transmitted from fathers to their affected offspring in a study of 829 families (SSC and Relating Genes to Adolescent and Child Health [REACH]) that was subsequently replicated in a second study of 1,771 autism families (MSSNG and SSC cohort). These findings contrast previous reports that have observed a maternal transmission bias of private, putative-truncating mutations within protein-coding sequence from mothers to their affected sons [11, 49]. One possible explanation offered was that such paternal transmissions that affect regulatory mutations are less damaging than protein-coding mutations and that in the former cases multiple mutations (oligogenic or bilineal model) may be required to manifest in disease.
The final study to assess the SSC cohort [18] claimed a hypothesis-free approach where they assessed 51,801 annotation categories that reduced to 4,123 correlated ones for the purpose of multiple testing correction. Unlike other approaches that focused on the most plausible functional NCEs (e.g., promoters, UTRs, enhancers), they considered many more categories treating annotations such as long noncoding RNA (lncRNA) and pseudogenes as equivalent to promoters and enhancers. Dubbing their approach a category-wide association study (CWAS), they examined de novo and inherited SNVs and indels. They concluded that no category could achieve significance after multiple testing correction. There were some interesting trends noted, however, such as an enrichment among autists of de novo SNVs and indels for promoters and UTRs, especially of developmental delay genes (Figure 3), confirming earlier studies. Predictably, less functional categories (pseudogenes) showed enrichments among unaffected siblings. Importantly, the authors’ analytical framework established a statistical threshold on the order of 5 × 10−6 estimating that >8,000 families would be required to detect a genome-wide signal if all noncoding annotation categories are considered functionally equivalent.
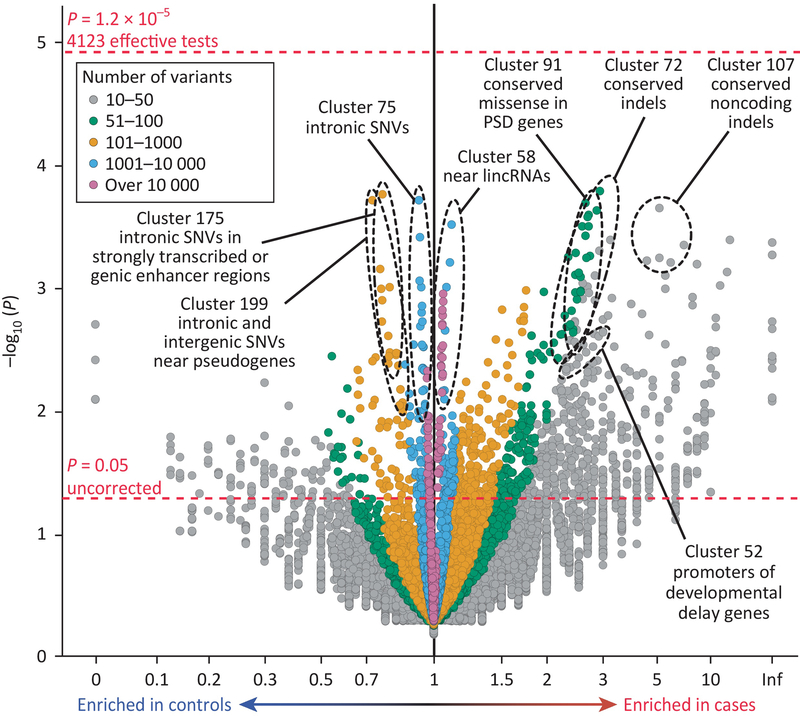
Figure 3: Category-wide association study for noncoding regulatory DNA.
The volcano plot depicts the burden of de novo SNVs and indels from a genome- wide analysis of 519 autism and 519 unaffected siblings. It considers 13,704 annotation categories (points) and computes both case–control enrichments and significance correcting for 4,123 effective independent tests. No individual category survives Bonferroni correction (top horizontal red line) under this analytical framework, although biologically plausible categories are highlighted. PSD = postsynaptic density. Adapted from Werling et al. 2018 [18].
Targeted sequencing of autism and other NDDs.
Comparatively, there have been relatively few targeted studies focused on mutational burden within noncoding regulatory DNA, although one study [50] did examine available WES data in a small number of individuals with autism (n = 48) and their parents, reporting an excess of putative inherited regulatory variants in autism-risk genes, fetal development genes, and microRNA genes. In two recent studies, experiments were designed to target and sequence specific noncoding portions with the hypothesis that these NCEs would be more likely to exhibit a functional effect [51, 52]. Doan et al. 2016 [51], for example, focused on human accelerated regions (HARs)—regions that have experienced a burst of mutation specifically in the human lineage and have been implicated in the regulation of genes important in human evolution, including neural genes. The authors found a 6.5-fold enrichment of rare, de novo CNVs within HARs among individuals with autism when compared to sibling-matched controls from the SSC cohort. Interestingly, within a consanguineous population, the authors reported a significant excess of rare, biallelic point mutations in these HARs suggesting compound heterozygotes could account ~5% of individuals with autism among consanguineous families. In a second study, Short et al. 2018 [52] focused on 6,139 putative regulatory elements corresponding to conserved noncoding elements as well as known enhancers from the VISTA browser and putative heart enhancers. Focusing on 6,239 children with developmental delay that were negative for obvious pathogenic events by exome sequence, they reported a nominal enrichment for DNMs in conserved NCEs. If they restrict to those that are active in the fetal brain, then the enrichment becomes significant (p = 8.1 × 10−4). They estimate that DNMs in this specific subset might contribute to 1.0% to 2.8% of “exome- negative” patients.
CONCLUDING REMARKS
The first genome-wide investigations into the role of noncoding regulatory mutation have highlighted both the potential and challenges of this class of variation in helping to explain neurodevelopmental disease. Notwithstanding the fact that most work is still underpowered due to limited sample size, some common findings have begun to emerge from some of the early-targeted sequencing and WGS studies. First, there is evidence for increased de novo or inherited disruptive mutation burden in putative regulatory regions in probands when compared to controls [15, 17, 19, 41, 52]. Although evidence for the role of inherited rare variation is still emerging and statistically underpowered, a recent study of transmission in multiplex families based on WGS data provides additional support [43]. Second, these signals often become more significant when restricting to autism and NDD risk genes or restricting to conserved regions active in the fetal brain (as determined by DNase I hypersensitivity) [15, 17]. Third, the type of mutation is an important consideration, with larger and more disruptive mutations (e.g., SVs and CNVs) showing potentially larger effects [15, 19, 51]. Finally, early estimates suggest such mutations account for a small fraction of patients (<5%) although these estimates are almost certainly a lower bound, in the absence of complete mutation ascertainment and a consideration of potential additive effects of different classes of mutation [15]. Collectively, most of the available data point to a model where single and multiple disruptions of regulatory DNA contribute to NDD risk by leading to downregulation and misexpression of genes important in fetal brain development. Although these findings are tantalizing, important challenges remain, as discussed next.
Improved variant discovery.
With respect to regulatory effects, not all mutations are created equal; deletions, in particular, have been shown experimentally to be more disruptive than SNVs [53, 54]. Most SNVs within unique regions of the genome are now readily detected using short-read sequencing platforms, but this is not the case for other forms of SV [55]. A recent comparison of genomes sequenced with both Illumina and long-read PacBio data showed that 50% of indels (10–49 bp in size), 51% of larger deletions (>50 bp), 83% of insertions (>50 bp in size) and nearly all inversions are not detected using short-read sequencing platforms [56]. Thus, there is the potential for large swaths of regulatory mutation to be missed even when genomes are sequenced deeply using short-read technologies. A major challenge will be to increase detection sensitivity for these understudied classes of mutation, especially variable number tandem repeat and short tandem repeat expansions (e.g., FMR1 CGG repeat) which already have a long-standing association with neurodevelopmental and neurodegenerative disorders. Notwithstanding these technological advances, it is likely that high- impact rare variants will only be diagnosed in a minority of cases. Another challenge will be developing appropriate methods to integrate both rare coding and noncoding variants with the pattern of common variation associated with polygenic risk scores and environmental exposure. Such models are critical for understanding both phenotypic variability and an individual’s true risk of disease.
Sample size and uniform variant calling.
Most researchers agree that much larger sample sizes will be required to prove and replicate these early genome- wide observations. No specific loci or associated genes are even close to significance, although it is interesting that recurrent DNMs and damaging, rare SVs have been identified among regulatory DNA of known autism risk genes [43]. We estimate that large-scale efforts such as the Centers for Common Disease Genomics (CCDG), MSSNG, and SSC along with some of the first multiplex cohorts from Autism Genetic Resource Exchange (AGRE) [43] will generate over 21,000 genomes from about 6,200 autism and 50 intellectual disability families (Table 1) by the end of 2018 (see Outstanding Questions regarding additional genome data resources [e.g., Gnomad [57] and Bravo]). Caution should be exercised, however, in naively combining datasets or making comparisons to control genomes where different sequencing platforms, variant callers, or thresholds of coverage can affect sensitivity. For example, simply combining published de novo variant lists from the MSSNG and the SSC (available in denovo-db 1.6.1; 2,204 autism probands and 521 unaffected siblings) would show, according to our estimates, a significant enrichment of DNMs in CNS DHS (10,526 proband variants vs. 2,637 unaffected variants, one-sided Fisher’s exact test p-value = 1.51 × 10−38, OR = 1.32). The enrichment increases if we restrict to variants within TFBS in these regions (CNS DHS TFBS) (739 in probands, 162 in unaffected siblings, one-sided Fisher’s exact test p-value = 8.51 × 10−7, OR = 1.50). Although each of these tests would pass the category-wide significance threshold of 5 × 10−6 proposed by Werling et al. 2018 [18], an examination of the data suggests that most of the signal originates from greater DNM variance and increased indel counts in the MSSNG dataset. By eliminating individuals sequenced by Complete Genomics technology, those with less than 50 DNMs, and by focusing on SNVs in regions of good mappability, the dataset drops to 1,623 autism probands and 519 unaffected siblings. This reduction significantly reduces observed levels of significance with CNS DHS having 7,024 proband variants and 2,367 unaffected variants (one-sided Fisher’s exact test p-value = 9.11 × 10−3, OR = 1.06) and CNS DHS TFBS having 491 proband variants and 152 unaffected variants (one-sided Fisher’s exact test p-value = 0.07, OR = 1.15). Ideally, cases and control WGS should be sequenced and processed identically to draw meaningful conclusions.
Outstanding Questions Box.
What is the relative contribution of noncoding variants to NDDs?
What are the most important noncoding regions to assess for NDDs?
What percentage of pathogenic variation is missed by short-read WGS?
Are the effects of noncoding mutations less severe than those in protein- coding regions?
Are the genes affected by coding variants the same as those affected by noncoding variants? How can one assess the rule of multiple rare variants (oligogenic) in contributing to disease outcome? What is the relative contribution of rare and common variants to autism risk?
What methods and assays could improve high-throughput functional testing of noncoding regulatory mutations?
How does the burden of mutation (both noncoding and coding) depend on sex of the affected individual?
Are the same genes and biological pathways implicated by noncoding variants as in protein-coding-region variants?
What is the best clinical approach to apply WGS information when no “known” coding event is identified?
What burden of proof is necessary to conclude pathogenicity for a noncoding variant? How will it fit into clinical standards (e.g., the American College of Medical Genetics and Genomics guidelines)?
Phenotypic data are often minimal or even lacking from population controls (e.g., the Genome Aggregation Database [http://gnomad.broadinstitute.org], the Trans-Omics for Precision Medicine Bravo Database [https://bravo.sph.umich.edu]). In light of this, what are the best strategies for their use and should additional investment be made to create valid disease-specific controls?
Refinement of functional noncoding regulatory DNA.
Although some have argued that a proper statistical framework of assessing the effect of NCE mutation should consider all possible annotation categories [18, 46], an alternative approach would be to refine the subset to those regions of largest biological effect. The ENCODE [30] and Roadmap Epigenomics [31] projects, in this regard, were an important first step that served to enrich for functional elements in specific cell types and tissues. Currently, the field continues to refine these maps to even greater resolution, driving down to the single-cell level [58] with methods such as ATAC-seq [58, 59] that will help define regulatory regions in specific cell types of the developing fetal brain [60]. A focus on defining fetal brain enhancers during cortical development using ATAC-seq/Hi-C methods [61], including those that have been gained in the human lineage [62], will be particularly powerful in refining the noncoding search space to the most functionally relevant portions for neurodevelopment. Others have applied deep machine learning algorithms (reviewed elsewhere, [63]) to better delineate and predict the potential effect of noncoding mutations. A recent preprint posted on bioRxiv [42], for example, applied such a deep-learning-based framework to 1,790 autism families and showed that ASD probands harbor transcriptional and post-transcriptional regulation-disrupting mutations of significantly higher functional impact than unaffected siblings. A third approach involves expanding the known list of autism- and NDD-risk genes [13, 14, 64], especially those associated with haploinsufficiency, and then systematically characterizing all long-range (by Hi-C) and short-range regulatory DNA associated with those high-impact targets.
High-throughput functional assays.
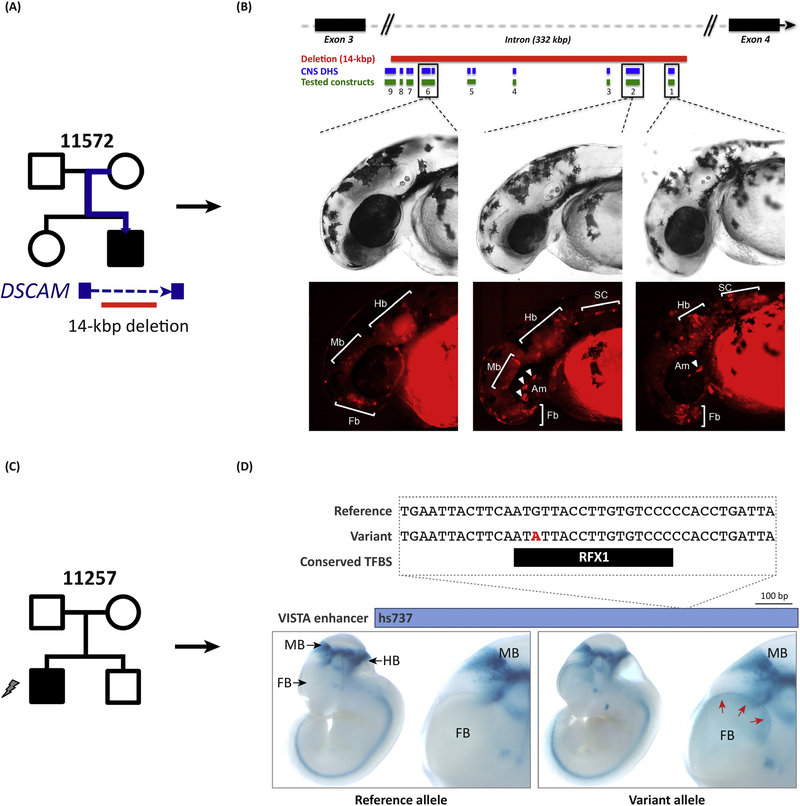
The ultimate litmus test for the relevance of specific NCE mutations to the etiology of neurodevelopmental disorders is demonstrating that they have biological consequences. Historically, researchers have utilized relatively low-throughput assays (e.g., luciferase or transgenic models) to assay function. One version of the transgenic assays utilizes a Tol- transposon in zebrafish and can relatively quickly provide a visual readout of the enhancer activity of a DNA sequence [65]. This has been utilized to test the spatial and temporal location of enhancer activity driven by NCE in an intron of DSCAM (Figure 4). The other major transgenic approach is a gold-standard experiment in mouse that assays the potential enhancer activity of a DNA sequence utilizing a lacZ reporter gene [66, 67]. Although these methods are powerful for providing insight into the effect of specific variants on the spatial and temporal dynamics of enhancers (Figure 4), the scale of discovery of thousands of NCE mutations demands technological advances in throughput.
Figure 4: Functional assessment of putative regulatory regions.
(A) A 14 kbp de novo deletion of the intron of the autism-risk gene DSCAM deletes multiple putative regulatory elements (DHS) in an autism patient (SSC family 11572). (B) Independent testing of different elements in a zebrafish enhancer- reporter assay shows that three elements mapping within the deleted region drive expression to different parts of the CNS of the developing embryo. (C) Discovery of a de novo variant in an autism proband (SSC family 11257) mapping to a functionally assessed enhancer (VISTA) conserved between mouse and human. The mutation maps to a TFBS and a fetal brain DHS. (D) A mouse lacZ reporter assay shows that the single-base-pair mutation causes a gain-of-function where novel expression is identified in the forebrain in addition to the expected expression in the midbrain and hindbrain. Adapted from Turner et al. 2016 [17] and Turner et al. 2017 [15].
Over the last five years, researchers have developed methods to rapidly assay noncoding variants using massively parallel reporter assays that leverage high- throughput sequencing (e.g., MPRA [68], STARR-seq [69], STAP-seq [70]) to look at variant effects on enhancers and promoters. While these assays are quantitative, such reporter construct assays suffer from relatively high false positive rates and are not by themselves definitive. Additional methods such as CRISPR-Cas genome-editing technologies that systematically introduce mutations into their native regulatory context and measure their effects on expression, or cellular/organismal phenotype, are being envisioned at a massive scale (reviewed in Montalbano et al. 2017 [71]). It is likely that various levels of high-throughput functional assay triage, followed by gold-standard transgenic assays, will need to be employed to systematically identify NCEs of the largest clinical (see Outstanding Questions; see also the ACMG guidelines [72]) and biological effect.
Highlights.
Recent sequencing advances have allowed for whole-genome sequencing (WGS) of large numbers of individuals with autism
WGS data are beginning to provide insight into the potential contribution of noncoding variants in neurodevelopmental disorders
A trend has been observed for an excess of de novo variants in conserved noncoding regions among autism patients with no obvious genic cause
The noncoding de novo mutation signature is stronger near genes already implicated in autism
Increased sample size is necessary to further our understanding of these noncoding signals and to refine the picture of their action at the gene level
Improved variant detection and functional classification of noncoding elements will increase our ability to detect pathogenic variants
Ultimately, functional testing will be critical to understanding the effect of mutations on gene expression and their relevance to disease.
ACKNOWLEDGEMENTS
We wish to thank Jay Shendure, Daniela Witten, Jessica Chong, and members of the Eichler laboratory for helpful discussions regarding the contents of this Review. We also thank Bradley Coe and Mark Chaisson for early access to data included in this Review. We also thank Tonia Brown for edits. The authors’ research is supported, in part, by grants from the US National Institutes of Health (NIH R01MH101221) to E.E.E. and the National Institute of Mental Health to T.N.T. (1K99MH117165). We are grateful to all of the families who participated in research studies to understand the genetic risk factors underlying autism and neurodevelopmental disorders. E.E.E. is an investigator of the Howard Hughes Medical Institute.
GLOSSARY
- CRISPR (clustered regularly interspaced short palindromic repeats) technology
molecular tool for editing the genome; can be used in a high-throughput manner.
- de novo variant:
genetic variant present in a child that is not present in either parent.
- DHS (DNase I hypersensitive site)
location in the genome where DNase I is able to cleave DNA.
- Enhancer
DNA sequences in the genome that raise the level of transcription of a gene.
- Genetic architecture
the complete understanding of the genetic factors underlying a phenotype.
- Indel (insertion/deletion)
small genetic variant (1 to 49 base pairs length) that either removes or adds bases to the genome.
- Machine learning
computational approach that uses artificial intelligence to train on input data and make future predictions without being explicitly programmed.
- Massively parallel reporter assay
high-throughput testing of thousands of DNA sequences for regulatory activity.
- Noncoding
the part of the genome that does not code for proteins.
- Oligogenic
intermediate between monogenic and polygenic models of disease where a few genes or loci of relatively large effect play a role in the resulting phenotype.
- Promoter
DNA sequences in the genome that are close to and encompass the start site of transcription of a gene.
- Regulatory
having an effect on transcription of a gene.
- SNV (single-nucleotide variant)
DNA variant that changes one base to another.
- STR (short tandem repeat)
DNA repeats with units that are typically 2 to 6 base pairs in length and vary polymorphically between individuals.
- SV (structural variant)
DNA variant that is greater than or equal to 50 base pairs in length and involves the deletion, duplication, inversion, or translocation of sequencing.
- TAD (topologically associating domains)
section of the genome that physically interacts only with itself and typically varies in size from thousands to millions of base pairs; noncoding, regulatory sequences in a TAD are thought to only regulate genes within the same TAD.
- TFBS (transcription factor binding site)
location in the genome where transcription factors bind to DNA.
- WES (whole-exome sequencing)
DNA sequencing approach that primarily targets and sequences the ~1.5% of the genome that is protein coding.
- WGS (whole-genome sequencing)
DNA sequencing approach that assesses “all” of the accessible portions of the genome; includes popular short-read sequencing approaches (e.g., Illumina) as well as longer read sequencing technologies (e.g., Oxford Nanopore, Pacific Biosciences).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST DISCLOSURE
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc.
REFERENCES
- 1.Wang K et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459 (7246), 528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss LA et al. (2009) A genome-wide linkage and association scan reveals novel loci for autism. Nature 461 (7265), 802–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaugler T et al. (2014) Most genetic risk for autism resides with common variation. Nat Genet 46 (8), 881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grove J et al. (2017) Common risk variants identified in autism spectrum disorder. bioRxiv
- 5.Cooper GM et al. (2011) A copy number variation morbidity map of developmental delay. Nat Genet 43 (9), 838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Roak BJ et al. (2011) Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 43 (6), 585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Roak BJ et al. (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485 (7397), 246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iossifov I et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature [DOI] [PMC free article] [PubMed]
- 9.Iossifov I et al. (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74 (2), 285–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders SJ et al. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485 (7397), 237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm N et al. (2015) Excess of rare, inherited truncating mutations in autism. Nat Genet 47 (6), 582–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iossifov I et al. (2014) The contribution of de novo coding mutations to autism spectrum disorder. Nature 515 (7526), 216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DDD (2017) Prevalence and architecture of de novo mutations in developmental disorders. Nature 542 (7642), 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coe BP et al. (in press) Neurodevelopmental disease genes implicated by de novo mutation and CNV morbidity. Nature Genetics [DOI] [PMC free article] [PubMed]
- 15.Turner TN et al. (2017) Genomic patterns of de novo mutation in simplex autism. Cell 171 (3), 710–722.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandin S et al. (2017) The heritability of autism spectrum disorder. JAMA 318 (12), 1182–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner TN et al. (2016) Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet 98 (1), 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werling DM et al. (2018) An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 50 (5), 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandler WM et al. (2018) Paternally inherited cis-regulatory structural variants are associated with autism. Science 360 (6386), 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lettice LA et al. (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12 (14), 1725–35. [DOI] [PubMed] [Google Scholar]
- 21.de Kok YJ et al. (1995) A duplication/paracentric inversion associated with familial X-linked deafness (DFN3) suggests the presence of a regulatory element more than 400 kb upstream of the POU3F4 gene. Hum Mol Genet 4 (11), 2145–50. [DOI] [PubMed] [Google Scholar]
- 22.Grant SF et al. (2006) Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38 (3), 320–3. [DOI] [PubMed] [Google Scholar]
- 23.Sladek R et al. (2007) A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445 (7130), 881–5. [DOI] [PubMed] [Google Scholar]
- 24.Scott LJ et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316 (5829), 1341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emison ES et al. (2010) Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet 87 (1), 60–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee S et al. (2016) Enhancer variants synergistically drive dysfunction of a gene regulatory network In Hirschsprung Disease. Cell 167 (2), 355–368.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu YH et al. (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67 (6), 1047–58. [DOI] [PubMed] [Google Scholar]
- 28.Pieretti M et al. (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66 (4), 817–22. [DOI] [PubMed] [Google Scholar]
- 29.Turner TN et al. (2016) denovo-db: a compendium of human de novo variants. Nucleic Acids Research [DOI] [PMC free article] [PubMed]
- 30.ENCODE (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306 (5696), 636–40. [DOI] [PubMed] [Google Scholar]
- 31.Kundaje A et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature 518 (7539), 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michaelson JJ et al. (2012) Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151 (7), 1431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilissen C et al. (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511 (7509), 344–7. [DOI] [PubMed] [Google Scholar]
- 34.Yuen RK et al. (2015) Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med 21 (2), 185–91. [DOI] [PubMed] [Google Scholar]
- 35.Guo H et al. (accepted) Genome sequencing identifies multiple deleterious variants in autism patients with more severe phenotypes. . Genetics in Medicine [DOI] [PMC free article] [PubMed]
- 36.Willsey AJ et al. (2013) Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155 (5), 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parikshak NN et al. (2013) Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155 (5), 1008–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hormozdiari F et al. (2015) The discovery of integrated gene networks for autism and related disorders. Genome Res 25 (1), 142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders SJ (2015) First glimpses of the neurobiology of autism spectrum disorder. Curr Opin Genet Dev 33, 80–92. [DOI] [PubMed] [Google Scholar]
- 40.Yuen RK et al. (2017) Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 20 (4), 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuen RK et al. (2016) Genome-wide characteristics of de novo mutations in autism. NPJ Genom Med 1, 160271–1602710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J et al. (2018) Whole-genome deep learning analysis reveals causal role of noncoding mutations in autism. bioRxiv
- 43.Ruzzo EK et al. (2018) Whole genome sequencing in multiplex families reveals novel inherited and de novo genetic risk in autism. bioRxiv
- 44.(2014) Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 46 (8), 818–25. [DOI] [PubMed] [Google Scholar]
- 45.Fischbach GD and Lord C (2010) The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68 (2), 192–5. [DOI] [PubMed] [Google Scholar]
- 46.Wray NR and Gratten J (2018) Sizing up whole-genome sequencing studies of common diseases. Nat Genet 50 (5), 635–637. [DOI] [PubMed] [Google Scholar]
- 47.Girirajan S et al. (2010) A recurrent 16p12.1 microdeletion supports a two- hit model for severe developmental delay. Nat Genet 42 (3), 203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duyzend MH et al. (2016) Maternal Modifiers and Parent-of-Origin Bias of the Autism-Associated 16p11.2 CNV. Am J Hum Genet 98 (1), 45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iossifov I et al. (2015) Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci U S A 112 (41), E5600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams SM et al. (2018) An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol Psychiatry [DOI] [PubMed]
- 51.Doan RN et al. (2016) Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 167 (2), 341–354.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Short PJ et al. (2018) De novo mutations in regulatory elements in neurodevelopmental disorders. Nature 555 (7698), 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osterwalder M et al. (2018) Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554 (7691), 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dickel DE et al. (2018) Ultraconserved Enhancers Are Required for Normal Development. Cell 172 (3), 491–499.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huddleston J and Eichler EE (2016) An Incomplete Understanding of Human Genetic Variation. Genetics 202 (4), 1251–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chaisson MJP et al. (2017) Multi-platform discovery of haplotype- resolved structural variation in human genomes. bioRxiv [DOI] [PMC free article] [PubMed]
- 57.Lek M et al. (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536 (7616), 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cusanovich DA et al. (2018) A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell [DOI] [PMC free article] [PubMed]
- 59.Cusanovich DA et al. (2015) Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348 (6237), 910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nowakowski TJ et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358 (6368), 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de la Torre-Ubieta L et al. (2018) The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172 (1–2), 289–304.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reilly SK et al. (2015) Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347 (6226), 1155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khurana E et al. (2016) Role of non-coding sequence variants in cancer. Nat Rev Genet 17 (2), 93–108. [DOI] [PubMed] [Google Scholar]
- 64.Stessman HA et al. (2017) Targeted sequencing identifies 91 neurodevelopmental-disorder risk genes with autism and developmental- disability biases. Nat Genet [DOI] [PMC free article] [PubMed]
- 65.Fisher S et al. (2006) Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1 (3), 1297–305. [DOI] [PubMed] [Google Scholar]
- 66.Pennacchio LA et al. (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444 (7118), 499–502. [DOI] [PubMed] [Google Scholar]
- 67.Visel A et al. (2007) VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35 (Database issue), D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melnikov A et al. (2012) Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30 (3), 271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arnold CD et al. (2013) Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339 (6123), 1074–7. [DOI] [PubMed] [Google Scholar]
- 70.Arnold CD et al. (2017) Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol 35 (2), 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montalbano A et al. (2017) High-throughput approaches to pinpoint function within the noncoding genome. Mol Cell 68 (1), 44–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richards S et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17 (5), 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandler WM et al. (2016) Frequency and Complexity of De Novo Structural Mutation in Autism. Am J Hum Genet 98 (4), 667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]