Abstract
Coxsackievirus B is a significant human pathogen and is a leading cause of myocarditis. We and others have observed that certain enteroviruses including coxsackievirus B cause infected cells to shed extracellular vesicles containing infectious virus. Recent reports have shown that vesicle-bound virus can infect more efficiently than free virus. Though microRNAs are differentially regulated in cells following infection, few have been associated with the vesicles shed from infected cells. Here we report exclusive trafficking of specific microRNAs into viral vesicles compared to vesicles from non-infected cells. We found that the most highly-expressed unique microRNA in viral vesicles was miR-590–5p, which facilitates prolonged viral replication by blocking apoptotic factors. Cells over-expressing this miR were significantly more susceptible to infection. This may be a mechanism by which coxsackievirus B boosts subsequent rounds of infection by co-packaging virus and a select set of pro-viral microRNAs in extracellular vesicles.
Keywords: Coxsackievirus, microRNA, vesicles, apoptosis
Introduction
Coxsackievirus is an RNA virus of the Picornaviridae family and Enteroviridae genus. Whereas coxsackievirus type A infects mucous membranes and is known to cause hand, foot, and mouth disease, coxsackievirus B (CVB) tends to infect internal organs, causing systemic inflammation. Though both viruses typically cause mild, self-limiting symptoms such as fever, rash and upper respiratory illness, severe CVB infections can cause more life-threatening diseases. CVB is a leading cause of myocarditis [1–3], an inflammation of the myocardium which is typically marked by chest pain, listlessness, and arrhythmias. If left untreated, myocarditis can sometimes progress to congestive heart failure with death in about a third of such patients [4, 5]. The only effective treatment for this is heart transplantation. Myocarditis is also implicated in about 20% of sudden death cases in young adults [6]. Though CVB can cause disease in both young and old, infants are particularly susceptible to CVB3-elicited fulminant myocarditis, which is often lethal, while mortality declines in older children [7].
In addition to acute myocarditis, CVB infections have been linked to chronic dilated cardiomyopathy [8, 9]. Previously, our group tested whether low-grade neonatal CVB infection in mice could elicit chronic disease during adulthood [10]. Following infection, we saw that infected mice showed no sign of cardiac inflammation or other illness, gaining weight and thriving similarly to mock controls all the way through adulthood. However, when neonatally-infected adult mice underwent cardiac challenge, either via exercise or beta-adrenergic stimulation, they rapidly progressed to dilated cardiomyopathy. This was contrary to mock-infected controls which were tolerant of cardiac stress. Furthermore, in CVB-infected mice, we discovered rarefaction of myocardial capillary networks, which we believe resulted in inadequate blood supply during increased cardiac workload. This subtle alteration of the heart may mirror clinical cases of sudden cardiac death, wherein seemingly healthy young individuals succumb to spontaneous heart failure during intense physical activity [11–13].
CVB, like other viruses, subverts and hijacks host cell processes to enhance infection. For example, viruses require host ribosomes to synthesize viral proteins. This is accomplished by first shutting down translation of mRNAs with 5’ caps. Not only does this block production of host protein, but it also creates bias towards the translation of uncapped viral RNA while freeing up tRNAs for viral genome coding [14]. CVB itself possesses a capless positive-sense single stranded RNA genome which contains internal ribosomal entry sites allowing for direct recruitment of ribosomes despite the absence of a 5’ cap. Another element that several viruses hijack are intracellular membranes. To establish replication sites, many viruses rearrange host membranes to serve as a scaffold for viral production [15, 16]. Work investigating enteroviruses such as coxsackievirus, poliovirus, and enterovirus-71 showed that these viruses activate autophagy and hijack autophagosomal membranes to enhance viral replication [17–19]. Many of these viruses are thought to assemble replication factories onto autophagosomal membranes to enhance replication [20]. CVB itself prevents autophagic flux, causing virus-laden autophagosomes to accumulate and fuse into “megaphagosomes” [21]. Additionally, autophagy was shown to allow for non-lytic egress of enteroviruses from the host cell via released autophagosomes [22, 23].
Cells are equipped with innate antiviral immunity pathways which are triggered in response to the presence of viral material, namely double stranded RNA intermediates generated during viral replication. One mode of cellular antiviral defense is through the mitochondrial antiviral signaling (MAVS) protein [24]. In the presence of double-stranded RNA, MAVS induces an interferon-dependent cascade which leads to the initiation of apoptosis. By selectively compartmentalizing the virus within apoptotic bodies, the virus can then be cleared by macrophages, sparing the rest of the tissue from further viral spread. However, because host cell death limits the length of viral replication, viruses have evolved to circumvent antiviral apoptosis. The 3C protease of coxsackievirus has been shown to cleave MAVS, thereby prolonging cell viability during infection and ultimately resulting in increased viral replication [24]. Interestingly, cancer cells and other highly proliferative cells have been reported to be highly susceptible to coxsackieviral infection, and this may be attributable to their inherent resistance to apoptosis [25–28].
We and others have observed that a number of non-enveloped viruses including CVB can enclose themselves in hijacked membranes and disseminate from the cell in extracellular vesicles (EVs) which display autophagosomal markers [29–31]. Not only are these virus-laden vesicles infectious, but they have also been shown to be more effective at propagating infection than isolated virions. In the study described here, we find that EVs from CVB-infected cells (vEVs) contain a set of miRs which are absent in EVs (presumably classical exosomes) shed from control cells. A recurring characteristic amongst the miRs in the vEV milieu is that they commonly target pathways related to cell death. Therefore, the net effect of these miRs is enhanced cell survival, which would facilitate prolonged viral replication and increased susceptibility to infection. We hypothesize that CVB packages these pro-viral miRs into vEVs to prime downstream host cells to better support viral replication.
Results
CVB infection causes differential trafficking of miRs into EVs
We had previously reported that cells infected with CVB release EVs containing viral particles [29, 32]. Because these vesicles are capable of causing subsequent waves of infection, this suggests that shed viral EVs are indeed taken up by recipient cells. Other types of extracellular vesicles such as exosomes and apoptotic bodies are dense with miRs which allow for cell-to-cell crosstalk [33–35]. With this in mind, we investigated if the miR composition in viral EVs differs from that of EVs from mock-infected cells. This would allow us to determine if CVB infection can influence the types of miRs that are released from cells, and to interrogate their roles. We first infected HL-1 atrial cardiomyocytes with eGFP-CVB at MOI 10, while giving equivalent volumes of DMEM to mock-infected controls. We used eGFP-CVB to better visualize infection in real time. An MOI of 10 was used to ensure brisk infection within a short amount of time, thereby resulting in high yields of vEVs. Eight hours postinfection (8 h PI), we indeed observed robust expression of viral eGFP, yet cell death was minimal at this early point in infection (not shown). We isolated EVs and analyzed miR content via Taqman OpenArray MicroRNA Rodent Panels to assess the abundance of a wide-range of miRs involved in a broad spectrum of cellular functions. We found that out of over 750 miRs analyzed, 7 miRs could only be detected in viral EVs and were absent in mock EVs. These miRs were miR-125b*, miR-335, miR-582, miR-455, miR-99a*, miR-590–5p and miR-135b (Fig 1A). Among these, miR-590–5p was expressed at a considerably higher level than the rest of viral EV-associated miRs (~13 fold higher than the next most abundant miR). Interestingly, intracellular levels of miR-590–5p were not detectable in mock-infected cells and detected only in trace amounts in CVB-infected cells (Ct-values close to lowest threshold of detection), which may illustrate very low basal levels of miR-590–5p, and after the miR is synthesized following infection, it is rapidly trafficked into vEVs. Infection with non-eGFP-expressing CVB yielded similar results, which rules out the possibility that eGFP itself could contribute to this differential miR production (miR-590–5p Ct-values: Mock EV- 35.21, eGFP-CVB- 27.80, non-eGFP-CVB-27.06). This screen revealed that while most EV-bound miRs have conserved expression following CVB infection, there is a specific set which is exclusive to CVB EVs, and this served as the basis for downstream network analyses.
Figure 1. Viral EV-bound miRs promote cell survival.
(A) List of miRs expressed exclusively in EVs shed from CVB infected cells, and absent in EVs shed from mock infected cells. Black box indicates miR-590–5p, which was the most abundant miR in the list. (B) Ingenuity pathways analysis (IPA) of miRs expressed exclusively in EVs shed from CVB-infected HL-1 cardiomyocytes. Gray bars represent percentage of target mRNAs that overlap with IPA Knowledge Base for specific biological attributes. Numbers indicate total amount of molecules in IPA dataset that represent corresponding attribute. Yellow line corresponds to the padj (log(B-H p-value)) of attributes.
Viral EV-bound miRs promote cell survival
To investigate the effect of CVB-exclusive miRs in vEVs, we performed functional analyses on their potential mRNA targets. Experimentally observed targets previously described were identified by Ingenuity Pathway Analysis (IPA) using microRNA Target Filter analysis. In total, CVB-only miRs showed 55 validated targets. A core analysis was performed to associate our target dataset with the IPA Knowledge Base. Several targets were associated with apoptotic processes (Fig 1B). Because miRs function by inhibiting their mRNA targets, the net effect of these vEV-bound miRs is an overall preservation of cell survival. This is consistent with previous literature reporting that CVB suppresses antiviral apoptosis to prolong replication [24].
miR-590–5p plays roles in cell survival and viral infection
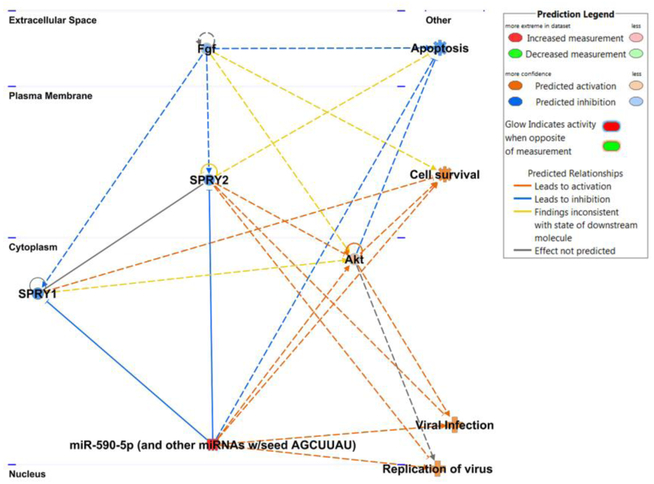
Having established that vEV-associated miRs have an influence on cell survival, we more closely examined the most abundant miR, 590–5p, and identified key proteins that miR-590–5p is predicted to inhibit (Fig 2). This miR shares the same seed region as miR-21 and is clustered in the same family, sharing the same targets. Using TargetScan, we found that sprouty-1 (Spry1) was among the top proteins inhibited by miR-590–5p. This tumor suppressor protein inhibits cell proliferation and increases apoptosis by suppressing fibroblast growth factor signaling, thus Spry1 could play a previously uncharacterized antiviral role [36–38]. Interestingly, our network also revealed indirect agonistic relationships between miR-590–5p with pathways relating to viral infection and replication. Additionally, the miR positively regulates Akt signaling, which allows for the expression of pro-survival genes. Thus, this analysis suggests that this highly abundant viral EV-bound miR may facilitate later rounds of infection by enhancing cell survival and prolonging viral replication.
Figure 2. miR-590–5p plays roles in cell survival and viral infection.
Network analysis of miR-590–5p and cellular targets. Blue lines indicate inhibited interactions while orange lines indicate activated interactions. General cellular pathways predicted to be affected by miR-590–5p are shown at the right of the network.
miR-590–5p enhances CVB infection
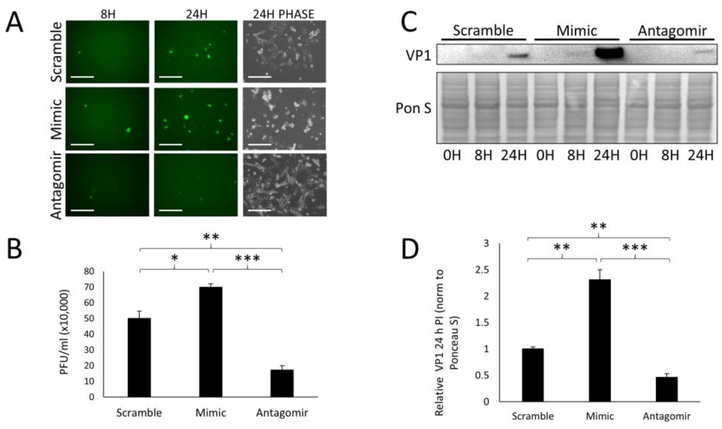
Because miR-590–5p was the highest-expressed miR exclusive to viral EVs, we investigated what effect it had on CVB infection. We transfected HL-1s with either a miR-590–5p mimic, a miR-590–5p-targeting antagomir, or scrambled miR and infected these cells with eGFP-CVB at MOI 1. This lower viral dose was used for longer infection timecourses in order for us to examine later timepoints wherein many rounds of infection could take place, without rampant cell death occurring. Prior to infection, the transfected cell groups were not observably different from each other in terms of cell viability and density (Supplemental Fig S1A). We found that at 24 h PI, eGFP+ cells were markedly more abundant in mimic-treated cells compared to scrambled-miR controls (Fig 3A). As mentioned previously, we were unable to detect basal levels of miR-590–5p in non-infected cells; however, in cells treated with the antagomir, very few eGFP+ cells could be observed (Fig 3A), which again may highlight a low but functional concentration of miR-590–5p in HL-1s under normal conditions. Plaque assays on culture media revealed a significant increase of extracellular infectious virus from mimic-treated cells compared to the controls, whereas the antagomir reduced these levels (Fig 3B). Western blots on whole cell lysates revealed a dramatic increase in VP1 viral capsid protein in mimic-treated cells, and a reduction in the antagomir-treated group 24 h PI (Figs 3C and D). These data illustrate that miR-590–5p increases CVB infection. Because this miR is enriched in vEVs, it may contribute to amplified infectivity of EV-bound virus.
Figure 3. miR-590–5p enhances CVB infection.
HL-1s were transfected with either scrambled miR, miR-590–5p mimic, or miR-590–5p antagomir and subsequently infected with eGFP-CVB at MOI 1. (A) Fluorescence microscopy of infected HL-1s at 8 h and 24 h postinfection (PI). Phase contrast images show cell number at 24 h PI. Scale bars represent 200 μm. (B) Plaque assays on culture media of infected HL-1s 24 h PI (*p<0.05, **p<0.01, ***p<0.001; Student’s t-test; n=3). (C) Western blots detecting VP1 in infected cells at 0 h, 8 h, and 24 h PI. (D) Densitometric quantification of VP1 levels in western blots of cells 24 h PI (**p<0.01, ***p<0.001; Student’s t-test; n=3–4).
CVB vEVs suppress Spry1
We next investigated whether the expression levels of one of the predicted targets of our vEVs, Spry1, would be altered upon infection. To test this, we first infected HL-1s at MOI 10 with multiply freeze-thawed viral stocks (naked virus). To confirm that these stocks lacked EVs, we attempted to isolate EVs from 107 PFU virus (a concentration of virus which exceeded our highest infection doses used in this study), and performed western blots on any material that precipitated. Western blots showed a lack of exosomal marker CD63 and Ponceau S staining showed very little protein in these isolates when compared to isolated vEVs (Supplemental Fig S2A). Surprisingly, we observed a gradual increase in Spry1 levels over the course of 10 h (Fig. 4A), which may illustrate a host response to limit infection. Next, we isolated vEVs from infected HL-1s, and immediately treated subsequent dishes of cells with the freshly collected vesicles. We observed that treatment with vEVs reduced Spry1 by 4 h following treatment, eventually recovering by 8 h. These data suggest that pro-viral cofactors may exist in CVB vEVs, and one mode of enhancing infection is by reducing Spry1, which we hypothesize functions as an antiviral protein.
Figure 4. miR-590–5p suppresses sprouty-1 which is an antiviral factor.
(A) Western blot detecting Spry1 in HL-1s infected with eGFP-CVB at MOI 10. (B) Western blot detecting Spry1 in HL-1s treated with EVs isolated from cells infected with eGFP-CVB. (C) Densitometric quantification of western blot in B (Student’s t-test; n=3). (D) Western blot detecting Spry1 in HL-1s transfected with scrambled miR or miR-590–5p mimic. (E) Densitometric quantification of western blot in D (****p<0.0001; Student’s t-test; n=3). (F) Fluorescence microscopy images on HL-1s transfected with either scrambled RNA (siSCRAMBLE) or siRNA targeting Spry1 (siSPRY1) infected with eGFP-CVB at MOI 1. Images were taken 24 h PI. Scale bars represent 200 μm. (G) Plaque assays on culture media of cells in F (*p<0.05; Student’s t-test; n=3). (H) Western blot detecting VP1 in cells in F. (I) Densitometric quantification of western blot in H (*p<0.05; Student’s t-test; n=3).
miR-590–5p suppresses sprouty-1 which is an antiviral factor
Earlier, we mentioned that a direct target of miR-590–5p is Spry1 which is a tumor suppressor and pro-apoptotic protein [39]. Because CVB preferentially infects highly proliferative cells, and also benefits from blockades in apoptosis [24], it is logical that miR-590–5p confers pro-viral effects by suppressing Spry1. To investigate this, we examined Spry1 expression in miR-590–5p mimic transfected cells and observed a significant reduction in Spry1 protein levels as measured by western blot (Figs 4B and C). Surprisingly, transfecting cells with lower concentrations of the miR-590–5p mimic yielded similar reductions in Spry1 in a dose-independent manner, suggesting even small concentrations of miR-590–5p can profoundly reduce Spry1 (Supplemental Fig S3). To confirm if Spry1 limits infection, we transfected cells with a SPRY1-targeting siRNA (siSPRY1) prior to infecting with eGFP-CVB at MOI 1. Western blotting confirmed a significant reduction in Spry1 following silencing (Supplemental Fig S4A and B). Similar to our miR-590–5p mimic and antagomir transfections, we observed no differences in cell viability and density with SPRY1 silencing compared to control cells transfected with scrambled RNA (siSCRAMBLE) (Supplemental Fig S1B). It should be noted however that due to differences in reagents and transfection duration when transfecting siRNAs versus transfecting miR mimics and antagomirs, cell densities for siRNA-treated cells were generally much higher than cells treated with the miR-related molecules. At 24 h PI, as expected, eGFP+ cells were more abundant in siSPRY1-transfected cells (Fig 4D). Plaque assays on culture media indicated that released infectious virus was significantly, albeit modestly increased with siSPRY1 treatment (Fig 4E), and western blots revealed that VP1 levels were greatly increased (Figs 4F and G). These data are consistent with our observations when we over-expressed miR-590–5p, thus our findings suggest that following CVB infection, miR-590–5p is packaged into released EVs to prime recipient cells to better support later rounds of infection. This is at least in part due to the miR suppressing Spry1, which we suspect plays a role in antiviral immunity. Disrupting vEVs with repeated freezing and thawing noticeably reduced the infection capability of these particles, suggesting that when vEV-bound cofactors such as miR-590–5p are lost, infection is attenuated (Supplemental Fig S2B).
Discussion
A number of viruses subvert cell death pathways in various ways in order to increase infection. Because cells can initiate apoptosis cascades upon detection of viral material, certain viruses have adapted to circumvent this to prolong viral replication. For example, the M11L protein of myxoma poxvirus interacts with the peripheral benzodiazepine receptor of mitochondria, inhibits membrane potential loss, and inhibits apoptosis, thereby prolonging cell survival and viral replication [40]. Hepatitis C virus encodes a serine protease NS3/4A which inhibits intracellular viral immunity by cleaving the mitochondria antiviral signaling (MAVS) protein and suppressing downstream IFN production [41]. Other viruses such as measles and HBV have been reported to induce autophagic elimination of mitochondria (mitophagy) to enhance replication, circumventing antiviral apoptosis [42, 43].
As can be seen, viruses have evolved to encode many proteins to subvert host cell function and bolster viral synthesis. In addition, EV-based viral dissemination has been suggested to intrinsically enhance later rounds of infection as well. In a recent study investigating egress of various enteroviruses, the authors described how these viruses can bundle multiple virions in phosphatidylserine-enriched EVs as a mode of cellular escape [30]. Additionally, they observed that these virus-laden vesicles infected cells more efficiently than free virus. The authors attributed this increased virulence to the fact that viral EVs can transfer large numbers of viral quasispecies allowing for even attenuated virions to continue to infect and pass on viral progeny through complementation.
In our current study, we present an additional layer of adaptation by which CVB (and likely other viruses) can enhance replication efficiency via vEV-bound miRs. We have found that 8 h following initial infection with “free” viral particles, EVs released from infected cell cultures were not only infectious, but they contained multiple miRs that were strongly upregulated. Functional analyses show that these miRs have predicted and previously validated targets pertaining to apoptosis and growth arrest, having a net effect of prolonging cell survival. When constructing functional networks pertaining to the function of miR-590–5p, the most enriched miR in viral EVs, we found that it is predicted to target apoptotic signaling molecules such as Spry1, and activate cell survival via Akt. The interaction between miR-590–5p and Akt signaling was previously implicated in gastric cancer as the miR was associated with increased tumor size, and metastasis [44]. By over-expressing miR-590–5p via miR mimic, we observed significantly increased CVB infection and consistent with this, Spry1 silencing conferred the same effect. Delivering a miR-590–5p-directed antagomir to cells blunted infection, and therefore may have implications in the development of clinical antiviral strategies. It should be noted that we cannot fully rule-out that free miRs or miR-protein complexes containing miR-590–5p may be present in our EV prep as well; however these types of molecules could presumably still confer similar proviral signaling.
Interestingly, sprouty proteins had previously been suggested to promote encephalomyocarditis virus infection in vitro, as the authors showed that sprouty 1,2, and 4 suppressed interferon signaling [45]. Simultaneously knocking these proteins out in mouse embryonic fibroblasts and pre-treating with IFNɑ caused the cells to exhibit decreased cytopathic effect following infection with the virus. Viral content was not measured in this study, and because the cells were primed with exogenous IFNɑ prior to infection, it is not entirely clear if viral propagation was altered, or if the cells exhibited differential sensitivity to cytotoxic factors. Additionally, because interferons limit infection partially by inducing apoptosis, gauging protection against infection based on cell death parameters can be somewhat confounding.
Though an increasing number of studies have shown that naked viruses such as coxsackievirus disseminate from the cell via EVs, it is still likely that the traditional cytolytic mode of viral egress is occurring as well. We hypothesize that during early stages of infection, EV-based viral dissemination is predominant. Because viral proteases disrupt antiviral apoptosis, this prolongs cell survival and sustains vEV release. If the cell were to have initially been infected with a vEV rather than a free virus, then prosurvival miRs like miR-590–5p will amplify this effect, as the cell would be further resistant to apoptosis. After the cell has succumbed to a high degree of viral burden, cytolysis may occur at this late-stage of infection, wherein free virus would be released.
Recently, another study had investigated differential extracellular miR trafficking following viral infection. Yogev’s group had shown that in lymphatic endothelial cells latently-infected with Kaposi’s sarcoma-associated herpesvirus, exosomes are released containing miRs that shift neighboring cells to a pro-angiogenic, glycolytic phenotype. Despite recipient cells not directly becoming infected, and in fact, becoming more resistant to infection, these cells release high energy metabolites that feedback to infected cells and further support viral replication [46]. This study provides additional insight into the unique ways viruses have evolved to manipulate extracellular miR trafficking in order to enhance infection.
The role of miR-590–5p in promotion of cell differentiation, proliferation and apoptosis through different mechanisms has been previously described [47–51]. However, to our knowledge, no reports have shown miR-590–5p to be secreted from virally infected cells, and its role in amplifying downstream infection has not been documented. Further studies are required to truly dissect how EV-bound miRs can alter the intracellular milieu to enhance infection. We showed that miRs 125b*, 335*, 582, 455, 99a*, 590–5p, and 135b were exclusively found in EVs released from infected cells, being absent in EVs from mock controls. This implicates a highly efficient and selective packaging mechanism to deliver these miRs into EVs during viral infection. These miRs represent important targets for future antagomir studies to determine their effects on infection. It will be important to determine whether interfering with one or more of these miRs could attenuate viral infection, first in vitro and subsequently in vivo to limit CVB-induced organ damage. Because CVB is a leading cause of myocarditis, antiviral inhibitors could help to limit infection, reduce cardiac damage, and preserve function.
Materials and Methods
Cell culture and virus.
HeLa RW cervical cancer cells (Dr. Rainer Wessely, UC San Diego) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, 11995–073) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, 16010–159) and antibiotic/antimycotic (Life Technologies, 15240–062). HL-1 mouse atrial cardiomyocytes (Dr. William Claycomb, Louisiana State University) were maintained in Claycomb medium (Sigma-Aldrich, 51800C) supplemented with 10% FBS, 0.1 nM norepinephrine (Sigma-Aldrich, A0937), 2 mM L-glutamine (Thermo Fisher Scientific, 25030–081), and antibiotic/antimycotic. Recombinant CVB (pMKS1) expressing enhanced green fluorescent protein (eGFP-CVB) was produced as previously described [29]. Briefly, eGFP was amplified from expression plasmids using sequence-specific primers which add flanking SfiI sequences. This was inserted into an infectious CVB clone (pH3) engineered to containing a unique SfiI restriction site (Dr. Kirk Knowlton, UC San Diego)[52]. Constructs were transfected into HeLa RW cells. Once cells exhibited ~50% cytopathic effect, cells were scraped, freeze-thawed three times, and centrifuged at 2000 rpm. Supernatants were collected and considered “passage 1”. Viral stocks were then expanded by infecting new HeLa RW cells with passage 1 virus. Once cells exhibited ~50% cytopathic effect, “passage 2” viral stocks were harvested as described earlier. Passage 2 stocks were used for all subsequent experiments.
EV Isolation.
EVs were isolated using ExoQuick-TC (Systems Biosciences, EXOTCxxA-1) as described previously [29]. Briefly, media was removed from infected HL-1 cells and centrifuged at 3000 × g for 15 min to sediment cells and debris. Supernatant was transferred to a new tube and ExoQuick-TC was added at 1:6 dilution. After refrigerating overnight, mixture was centrifuged at 1500 × g for 30 min to pellet EVs. Pellet was then washed in phosphate-buffered saline (PBS). EVs pellets were either processed directly for miR analysis, or resuspended in 100 μl PBS and added to culture medium for functional analysis. EV disruption was performed with 3 rapid freeze-thaw cycles using dry-ice cooled ethanol paths.[30]
Total RNA isolation and quality control.
Total RNA was isolated from EVs using miRNeasy Micro Kit (Qiagen, 217084), according to manufacturer’s protocol. RNA was eluted in 16 μl RNase-free water and quantified using Qubit Fluorometer (Thermo Fisher Scientific). The peak corresponding to the presence of miRNAs in the samples was assessed using an RNA 6000 Nano Kit (Agilent, 5067–1511) and analyzed with a 2100 Bioanalyzer (Agilent).
Reverse transcription and real-time PCR.
Ten nanograms of each RNA sample were reverse transcribed using Megaplex RT Primer Pools A and B (Applied Biosystems) and cDNA was pre-amplified using Megaplex PreAmp Primers (Applied Biosystems), according to manufacturer’s protocol. The pre-amplification reaction was set as 16 cycles. Samples were diluted, and real-time PCR was performed using TaqMan OpenArray MicroRNA Rodent Panels A and B and QuantStudio 12K Flex system (Applied Biosystems). The Ct value of each miR in each sample was normalized to average expression of all the miRs with valid Ct values in all samples (global normalization) [53]. Only Ct values < 28 showing amplification score higher than 1.0 and Ct confidence higher than 0.8 were accepted. For eGFP versus non-eGFP qPCR comparison analysis, miR was isolated from vEVs and as described. Ten nanograms of material was used for cDNA synthesis using TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems). Standard qPCR reactions were performed without pre-amplification using TaqMan Universal Master Mix II (444043, Applied Biosystems) and TaqMan MicroRNA Assay (001984, Applied Biosystems) according to manufacturer’s protocol. Ct values 35 or higher were considered undetectable.
miRNA-mRNA interaction analysis.
Predicted targets were identified by TargetScan release 7.1 and miR-mRNA interaction networks and canonical pathways were analyzed by Ingenuity Pathway Analysis (IPA) (Qiagen). Experimentally observed targets identified by IPA were submitted to the Core Analysis and the Benjamini-Hochberg (B-H) False Discovery Rate (FDR) was calculated to identify most important pathways related to these targets.
miR transfections.
HL-1s seeded in 60 mm culture dishes were transfected with miR-590–5p mimic (Thermo Fisher Scientific, MC11386), antagomir (Thermo Fisher Scientific, MH11386), or scrambled miR (Thermo Fisher Scientific, 4464058) using Lipofectamine 2000 (Thermo Fisher Scientific, 12566014) as per manufacturer’s suggestions. Two mixtures were first prepared. Mixture 1 was made by adding 17 μl Lipofectamine 2000 to 500 μl Opti-MEM medium and incubating at room temperature for 5 min. Mixture 2 was made by adding 166 pmol mimic, antagomir, or scrambled miR to 500 μl Opti-MEM. Mixtures 1 and 2 were then combined and mixed with repeat pipetting. Combined mixtures were then incubated for 20 min at room temperature. Following incubation, 1 ml of mixture was added to cells containing 5 ml fresh antibiotic-free Claycomb medium. After 48 h, media was refreshed with Claycomb medium, and subsequent experiments were performed.
siRNA transfections.
HL-1s seeded in 60 mm culture dishes were transfected with SPRY1 siRNA (Mouse; Santa Cruz Biotechnology, SC-41036) using Effectene Transfection Reagent (Qiagen, B00118) following manufacturer’s guidelines for reagent volumes. 72 h following transfection, media was refreshed with Claycomb medium, and subsequent experiments were performed.
Western blots.
Whole cell lysates were obtained by applying RIPA buffer directly to adherent cells and scraping. Detached cells were pelleted from culture media and combined with the rest of the corresponding lysate. Proteins were quantified using bicinchoninic acid solution (Sigma-Aldrich, B9643). Equal amounts of protein were run in 4–20% Tris-Glycine SDS PAGE gels (Life Technologies, EC6025) and transferred to nitrocellulose membranes. Membranes were blocked in 5% nonfat dry milk in tris-buffered saline with Tween-20 (TBS-T) for one hour at room temperature and then incubated in primary antibody diluted in 5% nonfat dry milk overnight at 4°C. Primary antibodies used were as follows: VP1 (1:142, Vector Laboratories, VP-E603), Spry1 (1:1000, Cell Signaling, 13013). Densitometry was performed using NIH Image J software. Quantifications were performed by measuring intensity of bands and subtracting adjacent white space in order to normalize for background signal. Values were then normalized to Ponceau S staining.
Supplementary Material
Supplemental Figure S1. Images of transfected HL-1s prior to infection. (A) Phase contrast images of HL-1s transfected with scrambled miR, miR-590–5p mimic, or miR-590–5p antagomir. (B) Phase contrast images of HL-1s transfected with scrambled RNA (siSCRAMBLE) or siRNA targeting Spry1 (siSPRY1) Scale bars represent 200 μm.
Supplemental Figure S2. Disrupted vEVs exhibit lowered infectivity. (A) Western blot detecting CD63 in ExoQuick isolates from 107 PFU free eGFP-CVB viral stock or media from eGFP-CVB-infected HL-1s at MOI 10. (B) Western blots detecting VP1 in cells treated with 3x freeze-thawed free virus at MOI 10 or vEVs.
Supplemental Figure S3. Sprouty-1 levels are reduced by miR-590–5p mimic in a dose-independent manner. Western blot detecting Spry1 in cells transfected with either scrambled miR or miR-590–5p mimic at 166 pmol, 83 pmol, 41.5 pmol, or 20.75 pmol.
Supplemental Figure S4. Sprouty-1 levels following transfection with scrambled RNA or siRNA targeting SPRY1. HL-1s were transfected with either scrambled RNA, or siRNA targeting SPRY1 (siSPRY1). (A) Western blot detecting Spry1 in transfected cells. (B) Quantification of Spry1 in western blot in A (*p<0.05; Student’s t-test; n=3).
Highlights:
Coxsackievirus B infection alters extracellular microRNA release
Extracellular vesicles from infected cells contain a unique population of microRNA
miR-590–5p is the most abundant microRNA exclusive to viral extracellular vesicles
miR-590–5p targets sprouty-1 and enhances infection
Acknowledgments
The CVB3 clone (pH3) was generously provided by Dr. Kirk Knowlton (University of California San Diego).
Funding
This work was supported by the National Institutes of Health [grant numbers P01 HL112730, T32 HL116273, KL2 TR001882]; and the Myocarditis Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cooper LT Jr. Myocarditis. N Engl J Med. 2009;360(15):1526–38. doi: 10.1056/NEJMra0800028. PubMed PMID: 19357408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu PP, Mason JW. Advances in the understanding of myocarditis. Circulation. 2001;104(9):1076–82. [DOI] [PubMed] [Google Scholar]
- 3.Henke A, Huber S, Stelzner A, Whitton JL. The role of CD8+ T lymphocytes in coxsackievirus B3-induced myocarditis. J Virol. 1995;69(11):6720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvy ML, Heerdink ER, Leufkens HG, Hoes AW. Predicting mortality in patients with heart failure: a pragmatic approach. Heart. 2003;89(6):605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pons F, Lupon J, Urrutia A, Gonzalez B, Crespo E, Diez C, et al. Mortality and cause of death in patients with heart failure: findings at a specialist multidisciplinary heart failure unit. Rev Esp Cardiol. 2010;63(3):303–14. [DOI] [PubMed] [Google Scholar]
- 6.Eckart RE, Scoville SL, Campbell CL, Shry EA, Stajduhar KC, Potter RN, et al. Sudden death in young adults: a 25-year review of autopsies in military recruits. Ann Intern Med. 2004;141(11):829–34. [DOI] [PubMed] [Google Scholar]
- 7.Dancea AB. Myocarditis in infants and children: A review for the paediatrician. Paediatr Child Health. 2001;6(8):543–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearney MT, Cotton JM, Richardson PJ, Shah AM. Viral myocarditis and dilated cardiomyopathy: mechanisms, manifestations, and management. Postgrad Med J. 2001;77(903):4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archard LC, Richardson PJ, Olsen EG, Dubowitz V, Sewry C, Bowles NE. The role of Coxsackie B viruses in the pathogenesis of myocarditis, dilated cardiomyopathy and inflammatory muscle disease. Biochem Soc Symp. 1987;53:51–62. [PubMed] [Google Scholar]
- 10.Sin J, Puccini JM, Huang C, Konstandin MH, Gilbert PE, Sussman MA, et al. The impact of juvenile coxsackievirus infection on cardiac progenitor cells and postnatal heart development. PLoS Pathog. 2014;10(7):e1004249. doi: 10.1371/journal.ppat.1004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Werf C, van Langen IM, Wilde AA. Sudden death in the young: what do we know about it and how to prevent? Circ Arrhythm Electrophysiol. 2010;3(1):96–104. doi: 10.1161/CIRCEP.109.877142. [DOI] [PubMed] [Google Scholar]
- 12.Fishbein MC. Cardiac disease and risk of sudden death in the young: the burden of the phenomenon. Cardiovasc Pathol. 2010;19(6):326–8. doi: 10.1016/j.carpath.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Solberg EE, Gjertsen F, Haugstad E, Kolsrud L. Sudden death in sports among young adults in Norway. Eur J Cardiovasc Prev Rehabil. 2010;17(3):337–41. doi: 10.1097/HJR.0b013e328332f8f7. [DOI] [PubMed] [Google Scholar]
- 14.Toribio R, Ventoso I. Inhibition of host translation by virus infection in vivo. Proc Natl Acad Sci U S A. 2010;107(21):9837–42. doi: 10.1073/pnas.1004110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva RA, Rouille Y, Dubuisson J. Interactions between virus proteins and host cell membranes during the viral life cycle. Int Rev Cytol. 2005;245:171–244. doi: 10.1016/S0074-7696(05)45006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasvari Z, Nagy PD. Making of viral replication organelles by remodeling interior membranes. Viruses. 2010;2(11):2436–42. doi: 10.3390/v2112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SC, Chang CL, Wang PS, Tsai Y, Liu HS. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J Med Virol. 2009;81(7):1241–52. doi: 10.1002/jmv.21502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson WT, Giddings TH Jr., Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, et al. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82(18):9143–53. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wileman T Aggresomes and autophagy generate sites for virus replication. Science. 2006;312(5775):875–8. doi: 10.1126/science.1126766. [DOI] [PubMed] [Google Scholar]
- 21.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, et al. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010;84(23):12110–24. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4(3):286–9. [DOI] [PubMed] [Google Scholar]
- 23.Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A. 2014;111(36):13081–6. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, et al. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7(3):e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto S, Inoue H, Nakamura T, Yamada M, Sakamoto C, Urata Y, et al. Coxsackievirus B3 is an oncolytic virus with immunostimulatory properties that is active against lung adenocarcinoma. Cancer Res. 2012;72(10):2609–21. doi: 10.1158/0008-5472.CAN-11-3185. [DOI] [PubMed] [Google Scholar]
- 26.Bradley S, Jakes AD, Harrington K, Pandha H, Melcher A, Errington-Mais F. Applications of coxsackievirus A21 in oncology. Oncolytic Virother. 2014;3:47–55. doi: 10.2147/OV.S56322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol. 2002;76(9):4430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry LJ, Au GG, Barry RD, Shafren DR. Potent oncolytic activity of human enteroviruses against human prostate cancer. Prostate. 2008;68(6):577–87. doi: 10.1002/pros.20741. [DOI] [PubMed] [Google Scholar]
- 29.Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10(4):e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160(4):619–30. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496(7445):367–71. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sin J, McIntyre L, Stotland A, Feuer R, Gottlieb RA. Coxsackievirus B Escapes the Infected Cell in Ejected Mitophagosomes. J Virol. 2017;91(24). doi: 10.1128/JVI.01347-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moldovan L, Batte K, Wang Y, Wisler J, Piper M. Analyzing the circulating microRNAs in exosomes/extracellular vesicles from serum or plasma by qRT-PCR. Methods Mol Biol. 2013;1024:129–45. doi: 10.1007/978-1-62703-453-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boon RA, Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. 2013;33(2):186–92. doi: 10.1161/ATVBAHA.112.300139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masoumi-Moghaddam S, Amini A, Wei AQ, Robertson G, Morris DL. Sprouty 1 predicts prognosis in human epithelial ovarian cancer. Am J Cancer Res. 2015;5(4):1531–41. [PMC free article] [PubMed] [Google Scholar]
- 37.Mekkawy AH, Pourgholami MH, Morris DL. Human Sprouty1 suppresses growth, migration, and invasion in human breast cancer cells. Tumour Biol. 2014;35(5):5037–48. doi: 10.1007/s13277-014-1665-y. [DOI] [PubMed] [Google Scholar]
- 38.Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y. Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development. 1999;126(11):2515–25. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Gong Y, Friesel R. Spry1 is expressed in hemangioblasts and negatively regulates primitive hematopoiesis and endothelial cell function. PLoS One. 2011;6(4):e18374. doi: 10.1371/journal.pone.0018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Everett H, Barry M, Lee SF, Sun X, Graham K, Stone J, et al. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J Exp Med. 2000;191(9):1487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102(49):17717–22. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SJ, Khan M, Quan J, Till A, Subramani S, Siddiqui A. Hepatitis B virus disrupts mitochondrial dynamics: induces fission and mitophagy to attenuate apoptosis. PLoS Pathog. 2013;9(12):e1003722. doi: 10.1371/journal.ppat.1003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia M, Gonzalez P, Li C, Meng G, Jiang A, Wang H, et al. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J Virol. 2014;88(9):5152–64. doi: 10.1128/JVI.03851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen B, Yu S, Zhang Y, Yuan Y, Li X, Zhong J, et al. miR-590–5p regulates gastric cancer cell growth and chemosensitivity through RECK and the AKT/ERK pathway. Onco Targets Ther. 2016;9:6009–19. doi: 10.2147/OTT.S110923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma B, Joshi S, Sassano A, Majchrzak B, Kaur S, Aggarwal P, et al. Sprouty proteins are negative regulators of interferon (IFN) signaling and IFN-inducible biological responses. J Biol Chem. 2012;287(50):42352–60. doi: 10.1074/jbc.M112.400721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yogev O, Henderson S, Hayes MJ, Marelli SS, Ofir-Birin Y, Regev-Rudzki N, et al. Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs. PLoS Pathog. 2017;13(8):e1006524. doi: 10.1371/journal.ppat.1006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q, Gao Q, Zhang Y, Li Z, Mei X. MicroRNA-590 promotes pathogenic Th17 cell differentiation through targeting Tob1 and is associated with multiple sclerosis. Biochem Biophys Res Commun. 2017;493(2):901–8. doi: 10.1016/j.bbrc.2017.09.123. [DOI] [PubMed] [Google Scholar]
- 48.Vishal M, Vimalraj S, Ajeetha R, Gokulnath M, Keerthana R, He Z, et al. MicroRNA-590–5p Stabilizes Runx2 by Targeting Smad7 During Osteoblast Differentiation. J Cell Physiol. 2017;232(2):371–80. doi: 10.1002/jcp.25434. [DOI] [PubMed] [Google Scholar]
- 49.Tao L, Bei Y, Zhou Y, Xiao J, Li X. Non-coding RNAs in cardiac regeneration. Oncotarget. 2015;6(40):42613–22. doi: 10.18632/oncotarget.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–81. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 51.Bao MH, Li JM, Zhou QL, Li GY, Zeng J, Zhao J, et al. Effects of miR‑590 on oxLDL‑induced endothelial cell apoptosis: Roles of p53 and NF-κB. Mol Med Rep. 2016;13(1):867–73. doi: 10.3892/mmr.2015.4606. [DOI] [PubMed] [Google Scholar]
- 52.Slifka MK, Pagarigan R, Mena I, Feuer R, Whitton JL. Using recombinant coxsackievirus B3 to evaluate the induction and protective efficacy of CD8+ T cells during picornavirus infection. J Virol. 2001;75(5):2377–87. doi: 10.1128/JVI.75.5.2377-2387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64. doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Images of transfected HL-1s prior to infection. (A) Phase contrast images of HL-1s transfected with scrambled miR, miR-590–5p mimic, or miR-590–5p antagomir. (B) Phase contrast images of HL-1s transfected with scrambled RNA (siSCRAMBLE) or siRNA targeting Spry1 (siSPRY1) Scale bars represent 200 μm.
Supplemental Figure S2. Disrupted vEVs exhibit lowered infectivity. (A) Western blot detecting CD63 in ExoQuick isolates from 107 PFU free eGFP-CVB viral stock or media from eGFP-CVB-infected HL-1s at MOI 10. (B) Western blots detecting VP1 in cells treated with 3x freeze-thawed free virus at MOI 10 or vEVs.
Supplemental Figure S3. Sprouty-1 levels are reduced by miR-590–5p mimic in a dose-independent manner. Western blot detecting Spry1 in cells transfected with either scrambled miR or miR-590–5p mimic at 166 pmol, 83 pmol, 41.5 pmol, or 20.75 pmol.
Supplemental Figure S4. Sprouty-1 levels following transfection with scrambled RNA or siRNA targeting SPRY1. HL-1s were transfected with either scrambled RNA, or siRNA targeting SPRY1 (siSPRY1). (A) Western blot detecting Spry1 in transfected cells. (B) Quantification of Spry1 in western blot in A (*p<0.05; Student’s t-test; n=3).