Abstract
Thymine deprivation in thyA mutant E. coli causes thymineless death (TLD) and is the mode of action of popular antibacterial and anticancer drugs, yet the mechanisms of TLD are still unclear. TLD comprises three defined phases: resistance, rapid exponential death (RED) and survival, with the nature of the resistance phase and of the transition to the RED phase holding key to TLD pathology. We propose that a limited source of endogenous thymine maintains replication forks through the resistance phase. When this source ends, forks undergo futile break-repair cycle during the RED phase, eventually rendering the chromosome non-functional. Two obvious sources of the endogenous thymine are degradation of broken chromosomal DNA and recruitment of thymine from stable RNA. However, mutants that cannot degrade broken chromosomal DNA or lack ribo-thymine, instead of shortening the resistance phase, deepen the RED phase, meaning that only a small fraction of T-starved cells tap into these sources. Interestingly, the substantial chromosomal DNA accumulation during the resistance phase is negated during the RED phase, suggesting futile cycle of incorporation and excision of wrong nucleotides. We tested incorporation of dU or rU, finding some evidence for both, but DNA-dU incorporation accelerates TLD only when intracellular [dUTP] is increased by the dut mutation. In the dut ung mutant, with increased DNA-dU incorporation and no DNA-dU excision, replication is in fact rescued even without dT, but TLD still occurs, suggesting different mechanisms. Finally, we found that continuous DNA synthesis during thymine starvation makes chromosomal DNA increasingly single-stranded, and even the dut ung defect does not completely block this ss-gap accumulation. We propose that instability of single-strand gaps underlies the pathology of thymine starvation.
Keywords: thymineless death, chromosomal fragmentation, chromosomal replication, DNA-dU incorporation, base excision repair, ribonucleotide excision repair
Introduction
Death in the absence of dTTP synthesis in a thyA mutant of E. coli was first described in 1954 by Barner and Cohen and was called thymineless death (TLD) (Barner and Cohen 1954, Cohen and Barner 1954). The thyA mutants can still be propagated in media containing either thymine or thymidine (collectively referred to as “T”), whereas changing cells to media lacking thymine/thymidine induces T-starvation, a conditions that develops similarly in other thy mutant prokaryotes and eukaryotes that always ends with TLD (Ahmad, Kirk et al. 1998). TLD is a unique phenomenon, as starvation in E. coli typically causes cessation of metabolism and stasis, rather than loss of viability, be it for an essential metabolite, like an amino acid or an RNA base (Barner and Cohen 1954, Breitman, Finkleman et al. 1971), or general starvation for carbon or nitrogen source (McCann, Kidwell et al. 1991, Kabir, Sagara et al. 2004). In contrast, T-starvation is characterized by a severe inhibition of DNA synthesis in combination with with normal rates of RNA and protein synthesis — in fact, such an “unbalanced growth” was the first general explanation for the viability loss during TLD (Cohen and Barner 1954, Cohen 1971).
Synthesis of dTTP inside the cell requires additional steps compared to the other three DNA precursors. Ribonucleotide diphosphate reductase (the product of a generic nrd locus) converts ribonucleotide diphosphates (ADP, GDP, CDP and UDP) into deoxyribonucleotide diphosphates, an essential step in biosynthesis of DNA precursors (Fig. 1A) (Neuhard and Nygaard 1987). The deoxyribonucleotide diphosphates (dADP, dGDP, dCDP, dUDP) are then phosphorylated by the nucleotide diphosphate kinase (ndk) to become deoxyribonucleotide triphosphates, yielding three out of the four DNA precursors. Since DNA contains thymine instead of uracil, dUTP generated either by the action of Ndk on dUDP or by deamination of dCTP by deoxycytidine deaminase (dcd) is first hydrolyzed to dUMP by dUTPase (dut), while dUMP is then methylated to yield dTMP by thymidylate synthase (thyA) (Fig. 1A, the green pathway), the only pathway in E. coli for de novo production of dTTP (Neuhard and Nygaard 1987).
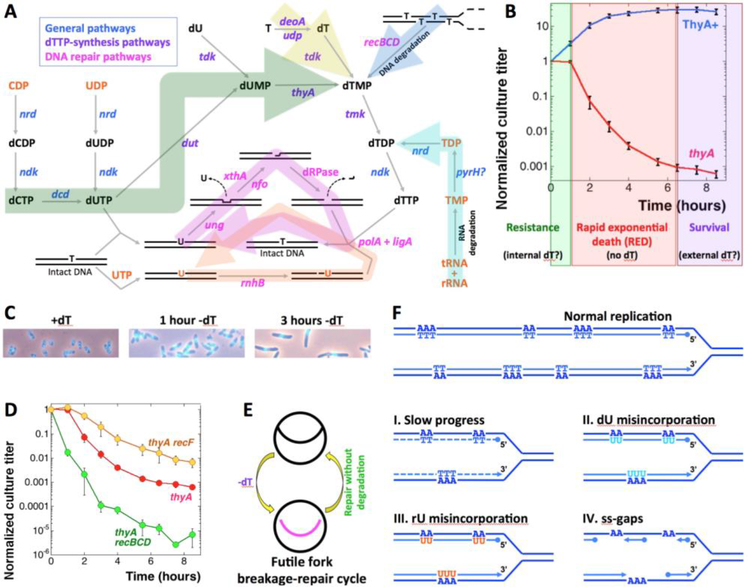
Fig. 1. The metabolism of dTTP production, the phenomenon of TLD, the futile cycles, and the expected changes in nucleotide metabolism mutants and kinetics of chromosomal fragmentation.
A. The metabolic pathways. Compounds of DNA metabolism are in black, compounds of RNA metabolism are in orange. Genes are colored according to functions: general (blue), dTTP-synthesis (purple) or DNA repair (magenta). The big arrow shades are: green, general biosynthesis; yellow, salvage; blue, DNA degradation; cyan, RNA degradation; magenta, futile DNA-dU misincorporation/excision cycle; orange, futile DNA-rU misincorporation/excision cycle. B. Extended time course of the culture titer during thymine starvation in our standard conditions, to highlight the survival phase. The strains are: ThyA+, KKW59; thyA, KKW58. The three phases of TLD are shown in color: green for the resistance phase, red for the RED phase, purple for the survival phase. The values in this and subsequent figures are means of 4–40 independent measurements ± SEM. C. DAPI-staining of cells grown in the presence of dT, as well as the same cells T-starved for 1 hour or for 3 hours. D. The thyA recBCD mutants lack the resistance phase, while the thyA recF mutants die slowly. The strains are: thyA, KKW58; thyA recBCD, KJK63; thyA recF, RA31. Here and henceforth: if error bars are not visible, they are masked by the symbols. E. A scheme of the futile fork-break-and-repair cycle. F. Various models of replisomes traversing A-runs in the template DNA during T-starvation that we tested in this work.
In contrast to thyA, the dut mutants (point mutations with no detectable dUTPase activity) are still viable (Hochhauser and Weiss 1978, Kouzminova and Kuzminov 2004), due to the dUMP production from dU via thymidine(deoxyuridine) kinase (tdk) (Neuhard and Nygaard 1987) (Fig. 1A), while the dut tdk combination is lethal, apparently due to its inability to produce dUMP (Ting, Kouzminova et al. 2008). Curiously, the dut tdk block cannot be circumvented by dT supplementation (Ting, Kouzminova et al. 2008), as thymidine kinase is also required to phosphorylate dT into dTMP (Neuhard and Nygaard 1987). In contrast to the dut tdk double mutants, thyA mutant grows normally on a medium supplemented with thymidine or thymine (Fig. 1A, the yellow pathway) (Barner and Cohen 1954, Cohen and Barner 1954). Thymine is converted to thymidine by deoxyribosyl transferases (deoA or udp), while thymidine is phosphorylated to dTMP by thymidine kinase (tdk) (Neuhard and Nygaard 1987). Then, dTMP is phosphorylated to dTDP by thymidine monophosphate kinase (tmk), while dTDP to dTTP phosphorylation is done by the non-specific nucleotide diphosphate kinase (ndk) (Fig. 1A) (Neuhard and Nygaard 1987).
Since the absolute bulk of thymine in the cell is in DNA (Neuhard and Nygaard 1987), T-starvation was always thought to kill via disbalancing DNA replication due to unavailability of dTTP (Cohen 1971). Starvation for all 4 DNA precursor by HU addition is only bacteriostatic (Kuong and Kuzminov 2009), emphasizing that the key to lethality is the absence of only one of the DNA precursors, rather than slow replication per se. Interestingly, starvation for another DNA precursor, dGTP, was recently characterized as a phenomenon that shares major features with TLD (Itsko and Schaaper 2014, Itsko and Schaaper 2016), further strengthening the idea of replication disbalance as the cause of death during T-starvation. Indeed, while actively replicating cells are susceptible to T-starvation, cells that cannot initiate a new replication round become immune to it (Maaloe and Hanawalt 1961). Further linking T-starvation with replication fork problems were reports of branched chromosomal structures (stalled replication intermediates?) (Nakayama, Kusano et al. 1994), massive degradation of the overinitiated chromosomal macrodomain around oriC (Fonville, Bates et al. 2010, Sangurdekar, Hamann et al. 2010, Kuong and Kuzminov 2012), induction of SOS response (Huisman and D’Ari 1981, Kuong and Kuzminov 2010) and correlation of severity of TLD with the number of chromosomal replication rounds before starvation (Martín and Guzmán 2011).
T-starvation in thyA mutant of E. coli begins with a resistance phase, that lasts an equivalent of two generations in cultures grown in the presence of thymine, during which the number of colony forming units remains stable (Kuong and Kuzminov 2010, Kuong and Kuzminov 2012). At 37°C, used throughout this work, the resistance phase lasts 1 hour, or about two divisions during uninhibited growth (Fig. 1B). Although the ongoing chromosomal replication in the E. coli cells exhausts the internal dTTP pools within several minutes into starvation (Neuhard and Munch-Petersen 1966, Ohkawa 1975), the cells continue to maintain their titer during the resistance phase, cell body widening and elongating around the brighter and compact nucleoid mass (Fig. 1C).
If thymidine is not added at the end of the resistance phase, the culture titer plunges over the period of the next several hours by several orders of magnitude during the so-called rapid exponential death (RED) phase (Kuong and Kuzminov 2010, Kuong and Kuzminov 2012) (Fig. 1B). During the RED phase, the cells elongate further, but nothing appears to happen with the centrally-located compact nucleoids (Fig. 1C). It is thus conceivable that the resistance phase of thymine starvation reflects cell’s limited ability to recruit thymine from minor sources to maintain a steady minimal rate of DNA synthesis producing normal nascent DNA, while the sudden onset of the RED phase may reflect a switch of T-starved cells to incorporation of thymine analogs, that will have to be excised from DNA, initiating a futile incorporation-excision cycle that eventually inactivates DNA (Khodursky, Guzmán et al. 2015).
In fact, the T-starvation pathology has been repeatedly explained by misincorporation of deoxy-uridine (dU) in place of thymidine (Breitman, Maury et al. 1972, Goulian, Bleile et al. 1980, Goulian, Bleile et al. 1980, Ahmad, Kirk et al. 1998, Khodursky, Guzmán et al. 2015) (Fig. 1A, the magenta pathway). Once incorporated, DNA-dU is rapidly removed by uracil-DNA-glycosylase (UDG), generating an abasic site, which is incised by exonuclease III (xthA) or endonuclease IV (nfo), generating a substrate for repair synthesis and ligation (Friedberg, Walker et al. 2006) (Fig. 1A). In the absence of dTTP, subsequent repair synthesis must again use dU in place of dT, initiating a futile DNA-dU insertion-excision cycle (Fig. 1A, the magenta cycle). Such futile insertion-excision cycle is proposed to kill either by itself (Ladner 2001), or via formation of double-strand gaps (Kunz and Haynes 1982), or via improper recombinational repair (Nakayama, Kusano et al. 1994).
The overall consideration of the DNA precursor biosynthesis, incorporation and excision pathways (Fig. 1A) naturally leads to an expectation that mutants affecting intracellular pools of thymidine or deoxyuridine should have significant effects on the TLD kinetics. The TLD kinetics curve under standard conditions is highly reproducible (note the tiny error bars in Fig. 1B), facilitating detection of either exacerbation or amelioration of TLD. Surprisingly, instead of defects in nucleotide metabolism, the only known defects strongly affecting TLD kinetics are those in recombinational repair (Nakayama, Nakayama et al. 1982). In particular, the recBCD mutants in double-strand break repair lack the resistance phase (Fig. 1D) (Nakayama, Nakayama et al. 1982, Kuong and Kuzminov 2010), suggesting formation of double-strand breaks in chromosomal DNA early on during T-starvation, which was indeed confirmed (Guarino, Salguero et al. 2007, Kuong and Kuzminov 2010, Kuong and Kuzminov 2012). At the same time, the recF, recO and recJ mutants in blocked ss-gap repair slow down the RED phase (Fig. 1D), suggesting formation of single-strand gaps (ss-gaps) in chromosomal DNA later on during T-starvation (Nakayama, Nakayama et al. 1982, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010). Since recombinational repair in bacteria is mostly used to address problems at replication forks (Kuzminov 1999), the strong TLD phenotypes of rec mutants support the view of thymineless death as “stalemate of starved replication forks”. Indeed, even if an inhibited and broken replication fork is successfully repaired, this cellular effort remains non-productive during T-starvation, because the restored forks are still inhibited and will break again, in another futile cycle, this one of fork breakage-and-repair (Fig. 1E). Persistent replication fork problems during T-starvation induce massive SOS-response (Kuong and Kuzminov 2010) (a transcription response of bacterial cells to replication problems (Kuzminov 1999)), which is proposed to be one of the killing mechanisms during TLD (Fonville, Bates et al. 2010).
If cells employ strategies to avoid or delay the futile fork breakage-repair cycle at the beginning of T-starvation, for example by manipulating nucleotide pools, the resistance phase and transition to the RED phase hold keys to understanding these strategies. Therefore, we sought to identify mutants affecting the (deoxy)thymidine and (deoxy)uridine pools in E. coli that would either accelerate or slow down TLD, similar to the recBCD or recF mutants (compare Fig. 1D with Fig. S1). We anticipated that functions disabled in TLD-accelerating mutants normally provide a major (non-biosynthetic) source of endogenous thymine/thymidine, while functions disabled in TLD-slowing mutants normally suppress such a source, including base analogs, or excise misincorporated base analogs from DNA (Fig. S1). In our search for such mutants, we were guided by the four possible explanations of the resistance phase (Fig. 1F): I) alternative internal sources of dT, maintaining slow fork progress; II) dU misincorporation; III) rU misincorporation; IV) discontinuous DNA synthesis, leading to accumulation of single-strand gaps (ss-gaps) in nascent DNA. Our findings reveal not only new survival strategies in response to T-starvation, but also a potential mechanistic cause of TLD pathology.
Materials and Methods
Bacterial strains.
E. coli strains (all K-12) and plasmids used in this study are described in Table S1. Strains were created by P1 transduction (Miller 1972), followed by transformation with pCP20 to remove antibiotic resistance when needed (Datsenko and Wanner 2000). Precise deletion-replacement mutations were obtained from E. coli genetic stock center and confirmed by PCR. The ΔthyA defect was additionally confirmed by inability to form colonies in the absence of thymidine. The recA, recB and recF mutant strains were verified by characteristic UV-sensitivities. The dut-1 mutation was confirmed by sensitivity to 10 mM uracil on plates (Kouzminova and Kuzminov 2004).
Growth conditions.
Overnight cultures were grown in M9 minimal medium supplemented with 0.2% casamino acids, 0.2% glucose (M9CAA) and 10 μg/ml thymidine (dT). The growth temperature was 37°C unless indicated. While screening for mutations linked to antibiotic resistance genes or cells carrying plasmids, the growth medium was supplemented with the following concentrations of antibiotics: 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml tetracycline or 10 μg/ml chloramphenicol.
Viability Assays.
Overnight cultures were diluted 100-fold in M9CAA + dT and grown to OD600 0.1– 0.15. Cells were collected on 0.2 μm nitrocellulose membrane by filtration and washed thrice with 5 ml of 1% NaCl to completely remove dT. Washed cells were suspended back into the same volume of M9CAA, but without dT. In some protocols, dT was removed by harvesting the cultures by centrifugation and resuspending the cell pellet and re-pelleting thrice with 1 ml of 1% NaCl in Eppendorf tubes before suspension in M9CAA. The resuspended cultures were vigorously shaken at 37°C. Culture aliquots taken at the indicated times were serially diluted in 1% NaCl and spotted on LB plates. Colony forming units were counted under a stereomicroscope after overnight growth at room temperature.
Direct cell counts in liquid cultures were obtained using Petroff-Hauser counting chamber and light microscope with phase contrast at magnification x400.
Fluorescent microscopy of DAPI-stained cells
was performed exactly as described (Kouzminova, Kadyrov et al. 2017).
Genomic DNA accumulation assay.
Cultures were grown to OD ~ 0.15 in M9CAA +dT from saturated overnight cultures, before filtration to start T-starvation. At indicated times, 1.5 ml of culture growing in the absence of dT was harvested to isolate DNA in plugs. The cells were suspended in 60 μl of TE and mixed with 2.5 μg of Proteinase K and 65 μl of lysis agarose (1.2% agarose in 20% lysis buffer) and poured into molds to set. Plugs were incubated overnight in at 65°C in lysis buffer (1% laurylsarcosine, 25 mM EDTA, 50 mM Tris-HCl), washed with 1 ml of TE thrice for 20 minutes, treated with 0.25 N HCl for 20 minutes, 0.5 N NaOH for 20 minutes and 1 M Tris-HCl pH = 8.0 for 20 minutes. The DNA from the plugs was vacuum transferred to nylon membrane, crosslinked and incubated in the hybridization buffer (5% SDS, 0.5 M sodium phosphate pH = 7.4) for an hour and probed with the whole genome probe in the same buffer overnight at 65°C. The probe was based on genomic DNA of stationary (aligned) cells of AB1157. The membranes were washed with 0.01x hybridization buffer thrice for 20 minutes each and exposed to phosphorimaging screen. The screens were developed in PhosphorImager FujiFilm FLA-3000.
Measurement of origin and terminus.
3 ml of culture were used to isolate DNA in duplicate agarose plugs as described above. After overnight lysis of cells in plugs and transfer of DNA to a nylon membrane, one set of plugs was hybridized to the ori probe, while the duplicate set of plugs was hybridized to the dif probe. This was followed by washing of the unhybridized probe, exposing to a phosphorimaging screen and analysis with a phosphorimager as above.
Single strand gap assay.
For cultures treated with AZT, the indicated concentrations of the drug were added once the strains reached OD600 ~ 0.15. For both AZT-treated and T-starved cultures, 1.5 ml aliquots were removed at the indicated times to isolate DNA in agarose plugs as described for genomic DNA isolation. After overnight lysis of plugs, they were washed with TE six times 10 minutes each, incubated with 100 μl of 2x NEB buffer 2.1 at 4°C for 1 hour, followed by restriction with 20 units of HaeIII and 4–32 μg of RnaseA in 1x NEB buffer 2.1 in 130 μl reaction overnight at 37°C. The next day the plugs were cut in half, and one set of them was denatured by treating with 0.25 N HCl for 20 minutes, 0.5 N NaOH for 20 minutes and 1 M Tris-HCl pH = 8.0 for 20 minutes. After this the DNA from both the native set and the denatured set of plugs was transferred onto nylon membrane by electric transfer at 80V for 1.5–3.0 hours, crosslinked and probed with the whole genomic probe. The membrane was exposed to an imaging screen and analyzed by a phosphorimager.
Measuring Stability of RNA.
The strain KKW58 was subcultured in MOPS-minimal phosphate medium (Bochner and Ames 1982) supplemented with casamino acids (CAA), 10 μg/ml dT and 2 μCi/ml 32P-orthophosphoric acid and grown to OD600 = 0.07 at 37°C. The label was removed by centrifugation, and the strain was grown in the same medium for another 30 minutes, until the 32P-redistribution in the cell mostly subsided. dT was removed from half of the culture by centrifugation, cells were washed by resuspension in 1 ml of 1% NaCl thrice, and the final pellet was suspended back into MOPS-CAA. 1 ml of thymidine-starved culture and gradually decreasing volumes of cultures growing in the presence of thymidine were harvested to isolate total RNA. RNA was isolated by RNAsnap protocol (Stead, Agrawal et al. 2012). One-fifth of the isolated RNA was run on 1.2% agarose gel in 0.5x TBE buffer at 40 V for 80 minutes. The gel was placed on a nylon membrane, dried at 50°C and exposed to an imaging screen to be analyzed by a phosphorimager.
Chromosomal Fragmentation assay.
Saturated overnight culture of KKW58 was diluted 100-fold in the presence of dT, 2 μCi/ml 32P-orthophosphoric acid in MOPS-CAA, and grown to OD600 ~ 0.15 at 37°C, when the culture was harvested by centrifugation to remove dT and the radioactive label. The cells were washed by resuspension in 1% NaCl thrice and suspended back in MOPS-CAA. At indicated times, 1.5 ml of culture aliquots were taken, and cells were collected and lysed as described for genomic DNA isolation. The conditions for running pulsed-field gel electrophoresis and quantification of chromosomal fragmentation were exactly as before (Kouzminova, Rotman et al. 2004).
Measurement of density of deoxyuridine and ribonucleotides in DNA.
Deoxyuridine (DNA-dU) densities were measured in pMTL20 plasmid, isolated from thyA, thyA dut and thyA ung strains. The strains were grown in 30 ml M9CAA +dT supplemented with 100 μg/ml ampicillin till early exponential phase (OD600 ~ 0.1 – 0.15), after which dT was removed from 20 ml of the culture by filtration and three washes with 5 ml of 1% NaCl each. The cells from the filter were suspended in 20 ml M9CAA (no dT), and after 5.5 hours plasmid was isolated by the standard alkaline lysis protocol (Birnboim 1983). 10 ml of strains growing in the presence of dT were grown to OD600 ~ 0.6 or for 5.5 hours before plasmid isolation. For DNA-dU density determination, 250 ng of the isolated pMTL20 was treated either with 1 unit of ExoIII alone, or with 1 unit each of UDG and ExoIII together, at 37°C for 30 minutes. The plasmid intermediates were separated in 1.2 % agarose gel in TAE buffer (Maniatis, Fritsch et al. 1982). The gels were gently rocked with 0.25 M HCl for 40 minutes, 0.5 M NaOH for 40 minutes and 1 M Tris-Cl for 40 minutes before capillary transfer of DNA onto nylon membrane (Hybond-N+) for hybridization with a pMTL20-specific 32P random-primer-labeling probe. The calculations to determine DNA-dU density, taking the signal of the supercoiled plasmid as the zero class of the Poisson distribution, were as before (Kouzminova and Kuzminov 2006).
DNA-dU density in the thyA dut ung mutant was measured using a small fragment released from pBR322 plasmid. The plasmid was isolated from this strain after growth in the absence of dT for 5.5 hours. 250 ng of plasmid was treated with either 1 unit of ExoIII, or 1 unit each of UDG and ExoIII together at 37°C for 30 minutes. This was followed by digestion of the plasmid with BglI and denaturation of the restricted DNA with 200 mM NaOH and 20 mM EDTA for 20 minutes on ice. The reaction products were run on 3% agarose gel, and DNA was transferred onto nylon membrane and probed against the 234 bp BglI fragment. The intact single stranded 234 bp fragment was used as the zero class of the Poisson distribution.
Ribonucleotide (DNA-rN) densities were measured using pEAK86-transformed strains. Strains were grown either in the presence of dT till OD600=0.6, or in the absence of dT for 5.5 hours before plasmid extraction by alkaline lysis protocol, which was done on ice after addition of S2 to inhibit DNA-rN hydrolysis (Kouzminova, Kadyrov et al. 2017, Cronan, Kouzminova et al. 2019). 250 ng of pEAK86 was treated with 1 unit of NEB RNase HII in 1X NEB thermopol buffer for 30 minutes at 37°C, loaded on 1.2% agarose gel, run in TAE buffer and transferred to nylon membrane to be probed with pEAK86-specific 32P-labeled probe. DNA-rN densities were determined using the Poisson distribution as described above and before (Kouzminova, Kadyrov et al. 2017).
Chromosomal DNA degradation.
The protocol was adapted from (Zahradka, Buljubasić et al. 2009) with some changes. Briefly, 10 ml of culture was chronically labelled with tritiated thymidine, grown to OD600 ~0.35 (if recA+) or OD600 ~ 0.6 (if recA-) and harvested by centrifugation. Cell pellets were washed with 10 ml of 67 mM phosphate buffer (pH=7.4) and suspended in 5 ml of the same phosphate buffer containing 0.01% Triton X-100. 2.5 ml of this suspension was mixed with 2.5 of 2x LB (unirradiated control). The other half was exposed to 50 J/m2 of UV radiation after spreading (in two portions of 1.25 ml each) as a thin layer in a sterile 100 mm x 150 mm Petri plate, and was mixed with 2.5 ml of 2x LB after UV and shaken at 37°C (Khan and Kuzminov 2012). At the indicated times, 500 μl aliquots were taken and mixed with 5 ml of ice-cold 10% trichloroacetic acid (TCA) and incubated on ice for 1 hour. The samples were filtered through Fisherbrand G6 glass fiber filters, washed with 5 ml of 10% TCA and 5 ml of EtOH. The filters were neutralized by spotting 100 μl of 1 M KOH, dried, submerged in scintillation fluid (Bio-Safe II, RPI) and counted for tritium on Beckman-Coulter LS 6500.
Results
The spike of chromosomal fragmentation could drive intrachromosomal DNA redistribution
If T-starvation is indeed the “stalemate of starved replication forks”, the cell could resolve it by reducing the number of active replication forks. Two major physical processes were reported during the resistance phase of T-starvation (Kuong and Kuzminov 2010, Kuong and Kuzminov 2012): 1) significant yet rapidly diminishing DNA synthesis; 2) chromosomal fragmentation, interpreted to reflect replication fork disintegration. However, the occurrence of chromosomal fragmentation during the resistance phase of TLD was dependent on conditions (Yoshinaga 1973, Nakayama, Kusano et al. 1994, Guarino, Salguero et al. 2007, Kuong and Kuzminov 2012), while its nature was unclear, so we decided to determine its kinetics.
The time course of chromosomal fragmentation in the repair proficient thyA mutants during thymine starvation showed an early peak with subsequent disappearance (Fig. 2AB), as reported before (Kuong and Kuzminov 2010). In continued T-starvation, chromosomal fragmentation was reported to reappear at 28°C (Kuong and Kuzminov 2010), but at 37°C we did not detect any significant increase at later times (Fig. 2AB). Thus, the peak of fragmentation at 37°C coincides with the resistance phase, while the disappearance occurs during the RED phase, — suggesting that either irreparable chromosomal fragmentation is not involved in TLD pathology, or that subsequent breaks in the replicating chromosome fail to generate linear subchromosomal fragments (detectable by pulsed-filed gels).
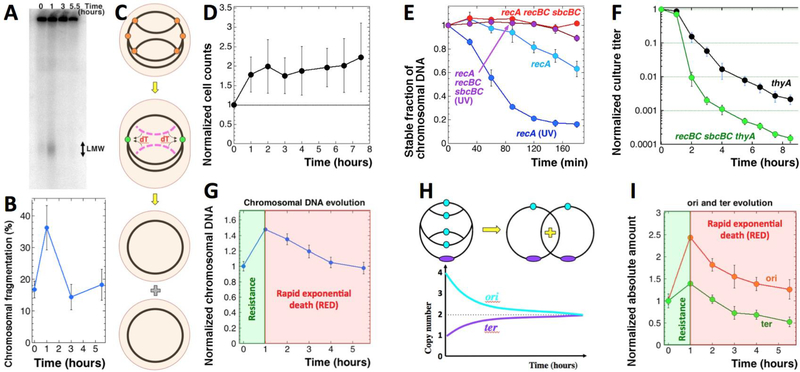
Fig. 2. Testing the intrachromosomal DNA redistribution idea.
A. A pulsed-field gel showing kinetics of chromosome fragmentation induced by thymine starvation. LMW, low molecular weight species, ~50–200 kbp in size. The strain is KKW58. B. Quantitative kinetics of chromosomal fragmentation, from several gels like in “A”. The data are means (n = 4 or 5) ± SEM. C. The scheme of how cannibalizing linear DNA (magenta arcs) generated by disintegration of some of the stalled forks (orange circles) could allow cells to keep the remaining forks active (green circles), explaining the resistance phase. D. Cell counts in T-starved cultures of the thyA mutant (KKW58) normalized to time of dT removal. E. Stabilization of prelabeled chromosomal DNA (TCA-precipitable counts) in UV-irradiated recA mutant cells by recBC sbcBC defect. The strains are: recA, RA46; recA recBCD sbcBC, RA47. The UV-dose was 50 J/m2. F. Time course of TLD in the recBCD sbcBC thyA mutant. The strains are: thyA, KKW58; recBCD sbcBC thyA, RA45. G. Change in the total amount of chromosomal DNA over time (“chromosomal DNA evolution”) in the thyA mutant (KKW58) during T-starvation. The first two phases of T-starvation are shown in the same colors as in Fig. 1A. H. Expectations about the origin and terminus copy number evolution during T-starvation according to the intrachromosomal DNA redistribution idea. I. Evolution of the amount of origin and terminus under the same conditions as in “G”.
Since previous measurements of the total chromosomal DNA during thymine starvation would uniformly report minimal changes (Barner and Cohen 1958, Gallant and Suskind 1961, Wachsman, Kemp et al. 1964, Breitman, Maury et al. 1972, Kuong and Kuzminov 2012), we reasoned that the coincidence of the residual DNA synthesis, double-strand breaks and the overall stability of DNA amount could reflect an intracellular zero-sum redistribution of the scarce thymidine. Specifically, the broken DNA, if degraded rather than repaired, could provide thymidine to support the still surviving replication forks during the resistance phase (Fig. 2C) (Fonville, Vaksman et al. 2011). This strategy of intrachromosomal DNA redistribution (“intrachromosomal cannibalism”) resolves the stalemate of starved replication forks by a combination of antagonistic methods to eliminate replication forks as the “problematic chromosomal points”. Indeed, the failed forks could be denied repair, targeting the resulting linear chromosomal branches for degradation, so that the released thymidine would allow the surviving forks to terminate the current replication round, — eventually eliminating all replication forks in the chromosomes (Fig. 2C). Termination of chromosomal replication should trigger cell division — and indeed direct counting of cells revealed cell doubling in the T-starved cultures within the resistance phase (Fig. 2D).
Reducing linear DNA degradation has no effect on the resistance phase
If the resistance phase of TLD is due to such intrachromosomal DNA redistribution, then minimizing linear DNA degradation, while keeping recombinational repair of disintegrated replication forks functional, should exacerbate the futile fork breakage-repair cycle (Fig. 1E). Indeed, even though the unstable replication forks are efficiently repaired, such repair becomes counterproductive without linear DNA degradation to generate any endogenous thymidine from extra chromosomal parts. If true, the “degradation-reduced” conditions should lead to the disappearance of the resistance phase during T-starvation and to deepening of the RED phase.
We tested this prediction in two ways. The recBC sbcBC mutant lacks the major linear duplex DNA degradase (RecBCD helicase/nuclease), as well as two major ssDNA degradases, Exo I and SbcCD nucleases (reviewed in (Kuzminov 1999)). The resulting mutant is grossly deficient in the linear DNA degradation (Rinken, Thoms et al. 1992, Zahradka, Buljubasić et al. 2009), as revealed by its inability to degrade chromosomal DNA after UV irradiation, if RecA is not available (Fig. 2E and S2A). At the same time, the mutant is fully proficient in recombinational repair, via the RecFOR pathway (Kuzminov 1999). Contrary to our expectations, TLD kinetics in the recBC sbcBC thyA mutant featured exactly the same 1-hour resistance phase as in the linear DNA degradation-proficient thyA mutant (Fig. 2F). At the same time, the recBC sbcBC thyA mutant does develop a faster RED phase and decreased survival, showing that cannibalizing the failed chromosomal branches indeed helps a small fraction (~1 out of 100) of starved cells to delay TLD.
To achieve a reduced linear DNA degradation with normal recombinational repair differently, we expressed (by arabinose induction off pKD46 plasmid (Datsenko and Wanner 2000)) phage lambda genes alpha, beta and gamma coding, correspondingly, for lambda exonuclease, ssDNA annealing protein and the inhibitor of the RecBCD enzyme (reviewed in (Kuzminov 1999)). This setup again shows no change in the resistance phase, yet a faster RED phase and decreased survival, repeating the effects of the recBC sbcBC mutant (Fig. S2B). Since the resistance phase was independent of the linear DNA degradation capacity of thyA mutant, counter to the prediction of the intrachromosomal DNA redistribution idea, we tested the idea with physical assays.
Genomic DNA increase during the resistance phase
A strong prediction of the intrachromosomal cannibalism idea is that T-starvation should not change the total DNA amount, at least not during the resistance phase. As was mentioned above, previous multiple studies reported no significant chromosomal DNA change during T-starvation (Barner and Cohen 1958, Gallant and Suskind 1961, Wachsman, Kemp et al. 1964, Breitman, Maury et al. 1972, Kuong and Kuzminov 2012), consistent with this idea. At the same time, during the resistance phase the average rate of residual replication is still 10% of the normal (+dT) rate (Kuong and Kuzminov 2012). Since the resistance phase was not due to linear DNA degradation, this significant DNA synthesis rate could translate into a measurable genomic DNA accumulation, apparently missed by the previous studies, perhaps because the standard DNA purification techniques become inadequate during T-starvation.
To resolve the issue, we used our plug-blotting technique for total cellular DNA content (Kouzminova and Kuzminov 2012), to minimize sample handling and DNA loss due to extraction methods. In contrast to the previous studies, we found that in a thyA mutant during T-starvation, the amount of DNA increases by 1.5 times in the first hour, but then steadily decreases to the original amount by 5.5 hours (Fig. 2G). In other words, the resistance phase of T-starvation is associated with a significant chromosomal DNA accumulation, whereas the RED phase coincides with disappearance of a similar amount of DNA (perhaps mostly affecting the new DNA synthesized during T-starvation?). This later DNA disappearance is not reduced (in fact appears stronger) in the thyA recBCD mutant (Fig. S2C), so its nature remains unclear at the moment. Important for our argument, though, the initial 1.5x increase is inconsistent with the zero-sum game of intrachromosomal DNA redistribution, suggesting instead non-chromosomal sources of dT.
Evolution of the copy number of origin versus terminus
Two parameters describe chromosomal replication: the rate of initiation from the replication origins and the rate of elongation of the resulting replication forks. In the bacterial chromosome, quantification of both parameters is facilitated by the unique replication origin and terminus. Indeed, initiation can be monitored by increase in the origin copy number, while elongation can be similarly monitored as completion of the chromosomal replication, by increase in the terminus copy number.
Perhaps the strongest prediction of the intrachromosomal DNA redistribution idea is the decrease of the cannibalized origin-containing parts of the chromosome, matched by a similar increase in the replicated terminus-containing parts of the chromosome (presented schematically in Fig. 2H). When we measured the evolution of the chromosomal origin copy number versus the terminus copy number during thymine starvation, in contrast to these predictions we found a sharp increase in the copy number of the origin during the resistance phase (Fig. 2I), suggesting adequate levels of dTTP for initiation of at least one round of chromosomal replication. At the same time, the subsequent gradual disappearance of the origin during the RED phase is consistent with the suspected instability of the DNA synthesized during the resistance phase. Thus, the observed evolution of the origin amounts (Fig. 2I) was at odds with predictions of the DNA redistribution model (Fig. 2H), but generally consistent with prior measurements at 28°C (Kuong and Kuzminov 2012).
The evolution of the terminus DNA copy number during thymine starvation was generally like the evolution of the overall DNA (Fig. 2G vs. I) (Kuong and Kuzminov 2012). Specifically, terminus accumulates to 1.5X of the original value during the resistance phase, reflecting significant progress of replication forks and again suggesting adequate availability of dTTP. During the RED phase, terminus experiences a gradual drop to ~60% of the original value, which translates into almost three-fold loss from the peak value at 1 hour (Fig. 2I).
Overall, we found that the chromosomal replication during the resistance phase of T-starvation affects not only the terminus half of the chromosome, but also its origin half, apparently making the whole chromosome unstable during the subsequent RED phase (Fig. S2D). In summary, our studies of the chromosomal DNA dynamics during thymine starvation suggest: 1) limited but adequate supplies of dTTP or its analog during the resistance phase; 2) RED-phase instability of the DNA synthesized during the resistance phase, suggesting that the nascent DNA is defective; 3) unclear nature of this RED-phase chromosomal DNA instability. Most importantly for our model (Fig. 2C), the initial increase followed by a similar decrease in both origin and terminus during T-starvation argue against the intrachromosomal DNA redistribution idea, but imply the existence of an intracellular source of dT or its analog (incorporation of which would make DNA unstable).
Endogenous thymine from stable RNA
An apparent endogenous source of thymine is the ribo-thymine from tRNAs and rRNAs, if the two stable RNA species are somehow destabilized during T-starvation (Fig. 1A, the cyan pathway). Rapidly-growing E. coli cells have ~50,000 ribosomes and 500,000 tRNA molecules per cell (Bremer and Dennis 1996). Every tRNA has a single thymine residue (T54), while every 23S rRNA has two thymines (Sergeeva, Bogdanov et al. 2015) (Fig. 3A). If all this thymine is somehow extracted from stable RNA, the 600,000 thymine residues could potentially support synthesis of four times as much DNA, ~2.4×106 nt, or about quarter of the genome equivalent, roughly half of the overall chromosomal DNA increase we detect during the resistance phase (Fig. 2G). As a quick check of the logic, we artificially destabilized tRNA and rRNA with a short sub-lethal heat-shock, — and found that such heat-shock does enhance survival during subsequent thymine starvation (Fig. 3B), — although other interpretations are possible with such a pleotropic treatment.
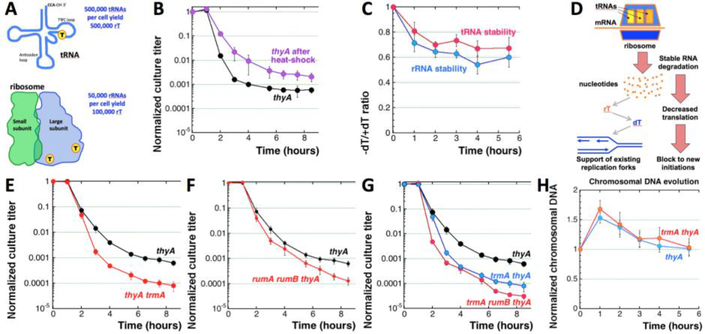
Fig. 3. Testing the contribution of ribo-thymine from stable RNA.
A. A scheme of stable RNAs (tRNA has one rT, 23S rRNA has two) and how many thymine residues they can yield per average rapidly-growing cell. B. Time course of TLD after 15’ @ 54°C heat shock. The strain is KKW58. In this case, the experiment-specific thyA TLD curve was used. C. Stability of tRNA and rRNA during thymine starvation. The strain is KKW58. Stability is expressed as the ratio of the corresponding RNA species in the cells incubated without dT to cells from the same culture incubated in the presence of dT. The actual measurements, normalized to time 0, are shown in Figures S3B and S3C. D. Recruitment of rT by degrading stable RNA kills two birds with one stone: it yields dT to support stalling replication forks and at the same time it inhibits translation, blocking new initiations and slowing down general metabolism. Both changes contribute to metabolism rebalancing. E. Time course of TLD in the thyA trmA mutant (RA9). F. Time course of TLD in the thyA rumA rumB mutant (RA13). G. Time course of TLD in the trmA thyA and trmA rumB thyA mutants. The strains are: thyA, KKW58; trmA thyA, RA9 (from “E”); trmA rumB thyA, RA14. H. Evolution of the chromosomal DNA absolute amount in the thyA and trmA thyA mutants (strains like in “E”) during T-starvation.
Then we checked stability of the two stable RNA species by growing thyA mutant cells in the medium +dT supplemented with 32P-orthophosphoric acid, changing the medium to the one without both the label and dT, and measuring the amount of the remaining 32P label in the rRNA and tRNA, separated on an agarose gel, at various time points during the ensuing T-starvation (Fig. S3A). We found that ~20% of both rRNA and tRNA are lost during the first hour of T-starvation, and ~40% is lost by 4 hours (Fig. 3C, Fig. S3BC). Thus, ribo-thymine does become available in T-starved cells, while any turnover of stable RNA, even though much slower than that of mRNA, should further contribute to this thymine source. Remarkably, this “mining” of ribo-thymine from stable RNA not only could support existing replication forks, but should also block further initiations from the origin by inhibiting proteins synthesis (Fig. 3D). In this respect, it could be an ingenious strategy to re-balance metabolism, thrown out of balance by T-starvation.
Specific RNA-uracil methylase TrmA methylates U54 in tRNA, while RumB and RumA methylases methylate, correspondingly, U747 and U1939 in 23S rRNA (Madsen, Mengel-Jørgensen et al. 2003). We deleted the trmA, rumB and rumA genes in the thyA mutant background, expecting that, if ribo-thymine is indeed used for DNA synthesis during thymine starvation, the mutants would show a shorter resistance phase, but likely no increase in the depth of the RED phase. The results again turned out to be the opposite of our expectation, as the thyA trmA strain shows no changes in the resistance phase, but a faster and deeper RED phase than in the thyA strain (Fig. 3E). The rumA and rumB defects made the TLD curve only slightly deeper, suggesting a truly minor thymine contribution from 23S rRNA (Fig. 3F, Fig. S3D). Combining the rum and the trmA defects together resulted in a detectable additivity (Fig. 3G), showing overall that ribo-thymine from stable RNA is indeed a dTTP source in a small sub-population (~1 in 103) of thyA cells, that makes these cells immune to TLD.
The lack of the ribo-thymine influence on the bulk population is supported by the physical measurements, as the trmA defect changes neither the overall pattern of chromosomal DNA evolution during TLD (Fig. 3H), nor the origin and terminus copy number initial increase and subsequent decrease (Fig. S3EF). Altogether, we conclude that thymine derived from stable RNA does not explain the resistance phase, but the way removal of this source of thymine accelerates the RED phase and decreases survival (Fig. 3G) indicates a small sub-population of cells that does recruit enough thymine from stable RNA, mostly from tRNA, to survive thymineless death.
Abasic site nuclease deficiency exacerbates TLD
Having found no internal source of dT to support the resistance phase, we turned to dT analogs. Uracil in the form of dUTP is incorporated into DNA in place of thymine during normal replication (Kouzminova and Kuzminov 2006). Much less frequently, cytosine in DNA spontaneously deaminates to uracil (Frederico, Kunkel et al. 1990). Although uracil-DNA incorporation is 105 times more frequent than cytosine deamination (Kouzminova and Kuzminov 2008), the former is not associated with any mutagenesis, in contrast to the latter scenario, where the next round of DNA replication yields transitions (G→A, C→T). To prevent this, the cell has a dedicated machinery to excise uracils from DNA (Friedberg, Walker et al. 2006) (Fig. 1A, the magenta pathway).
Inhibition of thymidylate synthase leads to a great expansion of dUTP pools in mammalian cells (Goulian, Bleile et al. 1980); in the ΔthyA mutants of E. col without dT supplementation, the intracellular dUTP pools must be also elevated, as judged by the increased DNA-dU incorporation (Warner, Duncan et al. 1981). 75% of dUTP in the E. coli cells is generated by deoxycytidine deaminase (Dcd) acting on dCTP (Fig. 1A, the green pathway) (Neuhard and Thomassen 1971). dUTP is converted to dTMP by the consecutive action of Dut and ThyA (Fig. 1A). When a sufficient pool of dTMP is generated, it inhibits the Dcd enzyme (Beck, Eisenhardt et al. 1975). In the absence of enough dTMP, Dcd remains active, elevating the pool size of dUTP,— which causes problems in the dut mutants, so that, for example, the dut thyA double mutants grow poorly even if supplemented with exogenous dT (Fig. S4A).
As mentioned in the introduction, the most popular explanation of TLD envisions that dUTP pool expansion in thyA mutants leads to repeated incorporation of dU into DNA in place of dT, followed by uracil excision that, via the futile repair cycle, somehow leads to TLD (Fig. 1A and 4A) (Breitman, Maury et al. 1972, Goulian, Bleile et al. 1980, Goulian, Bleile et al. 1980, Ahmad, Kirk et al. 1998, Khodursky, Guzmán et al. 2015). If during the resistance phase DNA-dU excision could be transiently suppressed (by unknown mechanisms), this would explain both the observed DNA synthesis and cell survival. Moreover, if suppression of this DNA-dU excision was then lifted during the RED phase, the resulting futile DNA-dU incorporation-excision cycle would provide an explanation for the RED phase.
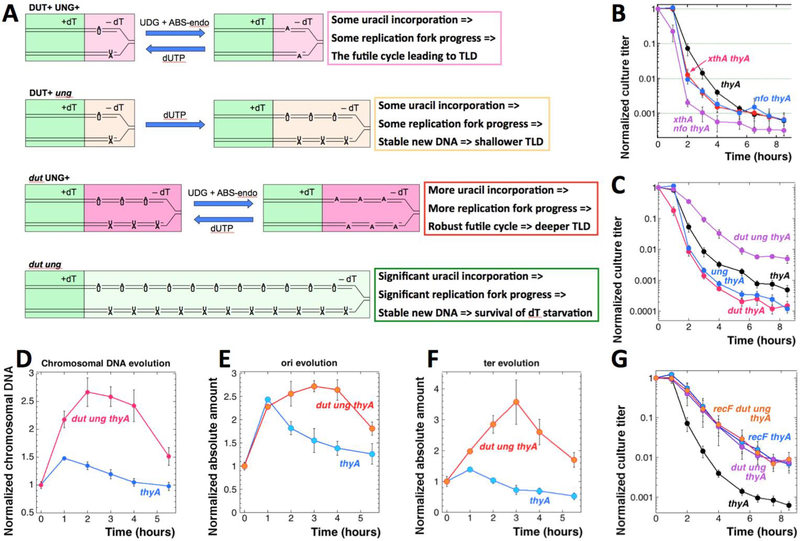
Fig. 4. The role of deoxy-uridine in TLD.
A. A scheme of DNA-dU misincorporation and its consequence in thyA, ung thyA, dut thyA and dut ung thyA mutants undergoing thymine starvation. Green fill, growth with dT. Light pink fill, attempted replication without dT using little dU (Dut+) with active DNA-uracil removal (Ung+). Yellow fill, replication without dT using little dU (Dut+), with no DNA-uracil removal (ung). Pink fill, attempted replication without dT but with plenty of dU (dut) with active DNA-uracil removal (Ung+). Light green fill, normal replication without dT but with plenty of dU (dut) and without DNA-uracil excision (ung). B. Time course of TLD in ABS-endo mutants. Strains are: thyA, KKW58; xthA thyA, RA22; nfo thyA, RA23; xthA nfo thyA, RA24. C. Time course of TLD in the dut thyA and ung thyA mutants. The +dT/-dT medium switch in this case was by centrifugation. Strains are: thyA, KKW58; dut thyA, RA16; ung thyA, KJK78; dut ung thyA, RA18. D. Evolution of the chromosomal DNA amount in the thyA (KKW58) and dut ung thyA (RA18) mutants during thymine starvation. E. Evolution of the replication origin copy number in the thyA and dut ung thyA mutants (strains like in “D”) during thymine starvation. F. Evolution of the chromosomal terminus copy number in the thyA and dut ung thyA mutants (strains like in “D”) during thymine starvation. G. TLD kinetics of the recF dut ung thyA mutant. The strains are thyA,KKW58; thyA recF, RA31; thyA dut ung, RA18; thyA dut ung recF, RA32.
The idea implies that, without dU incorporation into DNA of thymine-starved cells, there could be no thymineless death, only stasis. This reasoning predicts that dcd inactivation should ameliorate TLD (since 3/4 of all dUTP is generated via this route (Neuhard and Thomassen 1971)), but we found the TLD kinetic unaffected by the dcd defect (Fig. S4B). We have also inactivated Exo III and Endo IV, the two abasic site endonucleases of E. coli that are critical for completion of uracil excision repair (Friedberg, Walker et al. 2006) (Fig. 1A, the magenta pathway), alone or together. The RED phase was somewhat accelerated in the single mutants and dramatically accelerated in the double xthA nfo mutant, ending with a reduced resistance phase and a lower survival (Fig. 4B), — which indicates significant base-excision repair efforts during the RED phase. It should be noted, though, that base excision repair removes all kinds of DNA modifications, so incorporation of modified nucleotides other than dU, or processing of other types of DNA lesions during T-starvation, cannot be excluded.
Supposed replacement of dT with stable DNA-dU does not prevent TLD
To test the validity and limitations of the futile DNA-dU incorporation-excision cycle for TLD, we employed mutants deficient in either uracil excision (ung) or dUTPase activity (dut), or both (dut ung), in the thyA background. TLD was supposed to be slightly relieved in the ung thyA mutant, deficient in uracil-DNA-glycosylase, because incorporated DNA-dU becomes stable in this mutant (Fig. 4A). In contrast, we expected a deeper death in the dut thyA mutant because of the further elevated dUTP pool stimulating an even more frequent uracil-DNA incorporation and thus exacerbating the futile cycle. The situation should again change in the dut ung thyA triple mutant, where, because of the increased DNA-dU incorporation without excision, replication forks should be stabilized, preventing TLD (Fig. 4A).
Since the dut thyA mutant grows extremely poorly even in the presence of thymidine (Fig. S4A), we alleviated its growth problems by blocking the major route of dUTP biosynthesis with the dcd mutation, which by itself has no influence on the TLD kinetics (Fig. S4B). We found almost no difference in the TLD kinetics between the dut dcd thyA and ung dcd thyA triple mutants and their dcd thyA double mutant parent (Fig. S4C), whereas the dut ung dcd thyA quadruple mutant showed a two-times longer resistance phase and the slightly slower RED phase with higher survival (Fig. S4C). Since the unexpectedly modest effects of the dut and ung mutations could be due to the nucleotide pool changes in the dcd background, we decided to return to the Dcd+ thyA mutant set, while better managing its growth and handling.
The major handling problem of the slow-growing dut thyA mutant was the attachment of cells to filters during changing the medium from +dT to –dT, so in this set of strains we collected cells by centrifugation instead. The TLD curve of the thyA mutant was almost the same in the new protocol (Fig. 4C), but there was a noticeable aggravation of the RED phase by the ung defect (Fig. 4C) — as if uracil excision does not contribute to the pathology of thymine starvation, but does contribute a small alternative source of thymine. At the same time, as expected, we found that the dut defect shortens the resistance phase and lowers the survival, but without making the RED phase faster (Fig. 4C). Finally, the dut ung thyA mutant, that stably incorporates dU into its DNA in place of dT, after a “WT” resistance phase of one hour shows significant, though incomplete, relief of TLD, slowing the RED phase and elevating survival (Fig. 4C). The deeper TLD in the dut thyA and ung thyA mutants, and its 100-fold alleviation in the corresponding dut ung thyA combination, offer proof of principle for the toxicity of futile DNA-dU insertion-excision cycle. At the same time, even the dut ung defect does not eliminate TLD completely, — perhaps because at a high density DNA-dU incorporation becomes poisonous by itself (El-Hajj, Zhang et al. 1988, El-Hajj, Wang et al. 1992)?
Stable DNA-dU incorporation supports two rounds of chromosomal replication
The improved TLD survival of the dut ung thyA mutant implied that the disappearing thymidine was at least partially replaced in the nascent DNA with dU residues (not only numerous, but also stable in the chromosomal DNA in the ung background), — and therefore predicted more chromosomal DNA accumulation in the T-starved cells. Indeed, we found accumulation of up to 2.5-times of the original DNA in the dut ung thyA cells during 2 hours of thymine starvation (Fig. 4D). However, this DNA became eventually unstable and started rapidly disappearing after 4 hours of T-starvation (Fig. 4D). The origin copy number kinetics in the dut ung thyA mutant showed one round of initiation by 1 hour into thymine starvation, with the origin copy number essentially plateauing for three hours before going down sharply (Fig. 4E). The terminus copy number in the dut ung thyA mutant climbs steadily for three hours to the value almost 4x of the initial one, confirming uninterrupted and steady progress of replication forks (Fig. 4F). Thus, the dut ung thyA mutant completes not only the replication round that was ongoing during the removal of dT, but also the new round initiated during starvation, — however, its chromosome succumbs to degradation anyway (Fig. 4DEF). In other words, DNA-dU incorporation without excision supports what looks like stable and complete chromosomal replication — but after the resistance phase the dut ung thyA mutant still suffers an unknown problem, resulting in a rapid loss of the chromosomal DNA later on (Fig. S4D).
TLD in the dut ung mutant is not affected by the nfi or recF mutations
One possible reason for the eventual degradation of DNA containing dUs in the dut ung thyA mutant is that the uracil DNA glycosylase-deficient cells still have endonuclease V, the alternative excision repair enzyme specific against DNA-inosines (Yao, Hatahet et al. 1994), that has also a minor activity against DNA-dU in vitro (Yao and Kow 1997). However, inactivation of this minor uracil excision activity in the nfi mutant (Guo and Weiss 1998) failed to further improve the already shallow TLD kinetics of the dut ung thyA mutant (Fig. S4E). Also, the quadruple dut ung nfi thyA mutant showed essentially the same DNA accumulation during T-starvation compared to its progenitor dut ung thyA triple mutant, which was followed by a similarly dramatic loss of DNA (Fig. S4F). Thus, Endo V is neither the cause of the residual TLD in the dut ung thyA triple mutant, nor the reason for the eventual decline in the amount of genomic DNA in this mutant, — which is consistent with reports that Endo V does not attack DNA-dU in vivo (Guo and Weiss 1998).
The dut ung thyA mutants could still succumb to TLD due to the action of the RecFOR pathway for repair of persistent single-strand gaps. Because recF, recO and recR mutations alleviate TLD, persistent single-strand gaps were proposed to accumulate during T-starvation, triggering detrimental recombination (Nakayama, Nakayama et al. 1982, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010). Curiously, the TLD curve of the recF thyA mutant looks very similar to the TLD curve of the dut ung thyA mutant (Fig. 4G). If the remaining TLD in the dut ung thyA mutant is due to the action of persistent ss-gap repair pathway, the dut ung recF thyA quadruple mutant should become virtually resistant to TLD. However, we found that the quadruple mutant has the same TLD kinetics as the recF thyA or dut ung thyA mutants (Fig. 4G), suggesting that stable DNA-dU incorporation is epistatic over the recFOR pathway inactivation, probably because formation of persistent single strand gaps in the chromosomal DNA is prevented by filling them in with dU.
We conclude that stable DNA-dU incorporation during T-starvation significantly relieves TLD, most likely by reducing formation of persistent single-strand gaps. Nevertheless, 99% of the T-starved dut ung thyA cells still die, perhaps from remaining single-strand gaps, but also possibly from another, unknown reason.
The density of DNA-dUs is increased during T-starvation
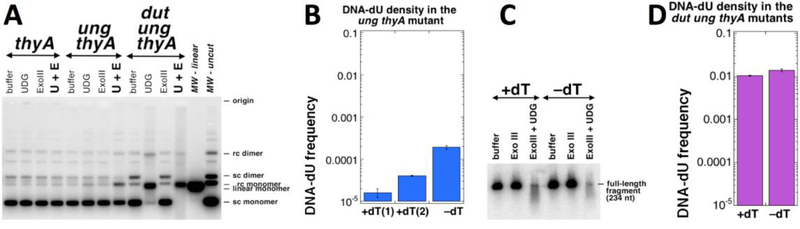
We quantified the density of DNA-dU residues by plasmid-relaxation in vitro (Kouzminova and Kuzminov 2006), treating supercoiled plasmid DNA, isolated from cultures under various conditions, with either Exo III alone (the background, no nicking expected) or UDG + Exo III (DNA backbone is nicked at the removed uracils, relaxing the plasmid) (Fig. 5A). The density of uracil is estimated by taking the supercoiled plasmid remaining after the UDG + Exo III treatment for the zero class of the Poisson distribution (Kouzminova and Kuzminov 2006).
Fig. 5. Density of DNA-dU in the ung thyA and dut ung thyA mutants, either starved or not for dTTP.
+dT, growth in the presence of thymidine; –dT, starvation without thymidine. Strains are: thyA, KKW58; ung thyA, KJK78; dut ung thyA, RA18. A. A representative gel (1.1% agarose) of DNA-dU density determination in plasmid DNA (pMTL20). UDG, treatment with uracil-DNA glycosylase; Exo III, treatment with exonuclease III; U + E, treatment with both UDG and Exo III. B. Quantification of the DNA-dU density (presented as frequency = 1/density) in the ung thyA mutant grown in the presence of absence of dT, from several gels like in “A”. The “–dT” culture is processed 5 hours after dT removal by filtration. There are two different conditions of growth in the presence of dT, though: “+dT #1” is also processed 5 hours after the filtration (but dT was re-added in this case), so the culture becomes stationary. In contrast, “+dT #2” is processed when the culture reaches OD=0.6. C. A representative gel (3% alkaline agarose) of DNA-dU density determination in a 234 nt long fragment of pBR322. D. Quantification of the DNA-dU density (presented as frequency, = 1/density, and in the same scale as in “B”, for direct comparison) in the dut ung thyA mutant grown in the presence of absence of dT for 5 hours, from several gels like in “C”.
Using this sensitive procedure, we could not detect any uracil in the plasmid DNA isolated from growing cultures of the thyA or the thyA dut mutants (Fig. 5A), confirming previous results (Kouzminova and Kuzminov 2006) and demonstrating the efficiency of uracil-DNA glycosylase in the DNA-dU removal. We did detect a modest density of DNA-dU in the plasmid DNA from ung thyA double mutant grown in the presence of thymidine (1 uracil in ~75,000 nucleotides in stationary cells, 1 uracil in ~25,000 nucleotides in growing cells) (Fig. 5AB), which is significantly increased to 1 uracil in ~6,000 nucleotides after five hours of T-starvation (Fig. 5B). However, this DNA-dU incorporation frequency in the Dut+ cells is clearly inadequate to sustain T-starved replication forks, explaining the lack of positive effect of the ung inactivation on TLD kinetics (Fig. 4C and S4C).
As expected, the high density of DNA-uracil in the dut ung thyA strain leads to the complete disappearance of the supercoiled species after the combined UDG and Exo III treatment (Fig. 5A), thus falling beyond the range of detection of the plasmid relaxation technique. We tried to release a much smaller supercoiled circle by employing Cre-loxP site-specific recombination (Hoess, Wierzbicki et al. 1985), but the efficiency of this in vitro reaction turned out to be inadequate. Eventually, we determined the DNA-dU density in the 234 bp fragment of a plasmid DNA after its denaturation and separation in 3% alkaline agarose, taking for the zero class of the Poisson distribution the remaining full-length ssDNA (Fig. 5C). As expected, the frequency of DNA-dU in the thyA dut ung mutant was extremely high (Fig. 5D), ~1,000 times higher than in the thyA ung mutant (cf. Fig. 5B and D) and within the ranges reported for this mutant before (Warner, Duncan et al. 1981). Unexpectedly, in the dut ung thyA mutant, T-starvation caused only ~25% further increase in DNA-dU density (from 1/95 to 1/72 dU/dN) (Fig. 5D), perhaps reflecting insufficient sensitivity of our detection system. We conclude that DNA-dUs are indeed stable in the ung mutants and massively incorporated in the dut mutants, but the unexpectedly small further increase in their density during T-starvation suggests that: 1) there is still some residual amount of dTTP around; 2) replicative polymerases discriminate against dUTP in favor of dTTP in vivo.
Overall, we conclude that significant dT replacement by dU is only possible in the dut mutants, which have elevated dUTP pools. From this perspective, the relief of TLD observed in the dut ung thyA mutant not only proves that dU can effectively substitute for dT in DNA synthesis during T-starvation, but also provides a strong evidence against contribution of DNA-dU misincorporation/excision to either the resistance or the RED phases in the Dut+ thyA mutant cells.
The role of translesion polymerases and rU incorporation
T-starvation strongly induces the SOS response (Huisman and D’Ari 1981, Kuong and Kuzminov 2010) — the increase in transcription of some 30 genes in E. coli in response to inhibition of DNA replication associated with accumulation of single-stranded DNA (Kuzminov 1999). Among other enzymes, the SOS response elevates expression of three translesion DNA polymerases of E. coli, Pol II, Pol IV (gp dinB) and Pol V (gp umuCD) (Napolitano, Janel-Bintz et al. 2000). Both Pol IV and Pol V have spacious active sites that allow them to accommodate template with bulky DNA lesions (Jarosz, Beuning et al. 2007). In addition, Pol V incorporates ribonucleotides during translesion synthesis in vitro (Vaisman, Kuban et al. 2012). It was therefore possible that during T-starvation, ribo-uridine (rU) could be misincorporated into DNA by Pol V across template adenine (Fig. 6A), which could exacerbate the RED phase of TLD via futile misincorporation/excision of DNA-rU (Fig. 1A, the orange pathway). However, we found no big difference in the TLD kinetics, whether the TLS polymerases are removed individually or together (with the possible exception of a slightly higher survival after 4 hours in the dinB thyA double mutant) (Fig. 6B).
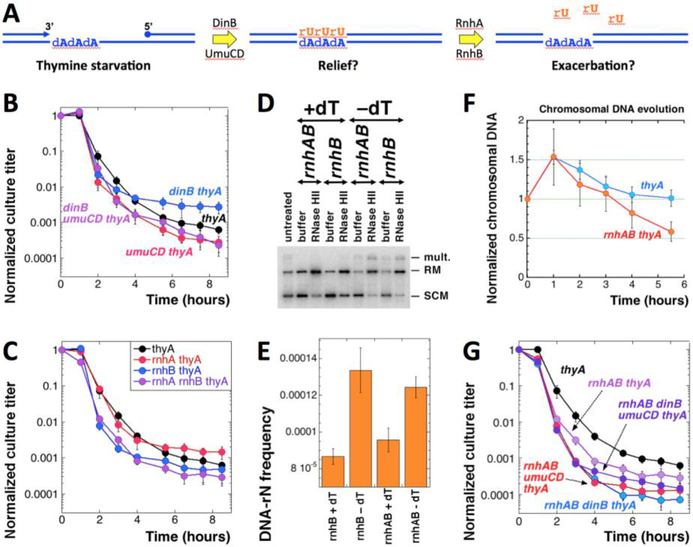
Fig. 6. The effect of rN incorporation and excision on TLD.
A. A scheme of the possible futile DNA-rU misincorporation/excision cycle. B. TLD kinetics of the dinB thyA and umuCD thyA mutants. The strains are: thyA, KKW58; dinB thyA, KJK90; umuCD thyA, KJK87; dinB umuCD thyA, RA36. C. TLD kinetics of the rnhA thyA and rnhB thyA mutants. The strains are: thyA, KKW58; rnhA thyA, RA33; rnhB thyA, RA34; rnhAB thyA, RA35. D. A representative gel (1.1% agarose) of DNA-rN density determination in plasmid DNA (plasmid is pEAK86) isolated from rnhB mutant cells. +dT, growth in the presence of thymidine; –dT, starvation without thymidine. RNase HII, in vitro treatment with RNase HII. Strains are: rnhB, RA34; rnhAB, RA35. E. DNA-rN density (presented as frequency = 1/density) in the rnhB and rnhAB mutants grown in the presence of absence of dT for 5 hours, from several gels like in “E”. F. Evolution of the chromosomal DNA amount in the thyA (KKW58) versus rnhAB thyA (RA35) mutants during dTTP starvation. G. TLD kinetics of the dinB rnhAB thyA and umuCD rnhAB thyA mutants. The strains are: rnhAB thyA, RA35; umuCD rnhAB thyA, RA40; dinB rnhAB thyA, RA39; dinB umuCD rnhAB thyA, RA41.
If rU residues are indeed incorporated in place of dT during T-starvation, they should be subsequently excised by the RNase H enzymes (Fig. 6A). There are two RNases H in E. coli, I and II, encoded by the rnhA and rnhB genes, respectively (Schroeder, Randall et al. 2015). The two RNase H enzymes remove all kinds of rNs from DNA: RNA primers for Okazaki fragments, R-loops, misincorporated single RNA nucleotides. If DNA-rU incorporation starts another futile misincorporation-excision cycle during T-starvation, inactivation of RNase H enzymes should alleviate TLD (Fig. 1A, the orange pathway, and Fig. 6A). However, we found that the rnhA thyA and rnhB thyA mutants do not have a significantly different TLD kinetics compared with the thyA single mutant (Fig. 6C). If anything, and in contrast to our expectations, the rnhB defect confers a faster RED phase and lowers the final survival (Fig. 6C), as if rN excision from DNA is beneficial for T-starved cells.
To verify the rnhB inactivation, which leads to accumulation of single rNs in the DNA (DNA-rNs are undetectable in the RnhB+ cells) (Kouzminova, Kadyrov et al. 2017), we isolated plasmid DNA from rnhB thyA and rnhAB thyA mutants, grown in the presence or absence of thymidine, and treated it with RNase HII in vitro, to relax rN-containing supercoiled plasmid molecules (Fig. 6D) (Kouzminova, Kadyrov et al. 2017). We confirmed a high density of DNA-rNs characteristic of the rnhB and rnhAB mutants (Kouzminova, Kadyrov et al. 2017, Cronan, Kouzminova et al. 2019) — 1 DNA-rN in 11,603 nt and in 10,644, correspondingly (Fig. 6E). We also found a modest increase in the density of DNA-rNs during T-starvation - 1 DNA-rN in 7,621 or in 8,106 nt, correspondingly (Fig. 6E). At the same time, this more frequent rU-DNA incorporation does not lead to more genomic DNA accumulation in the rnhAB thyA strain, where such DNA-rNs are stable, as compared to the thyA strain, but may lead to more DNA loss (Fig. 6F). Apparently, DNA-rU stability is mildly poisonous for the T-starved cells.
Since the effect of rU-incorporation by TLS polymerases could be masked by the subsequent efficient DNA-rU excision by RNase H enzymes, we also tested the effect of dinB or umuCD inactivation on TLD in the rnhAB thyA mutant background (Fig. 6G). None of the combinations has affected the resistance phase. We observed a slight detrimental effect of the UmuCD inactivation during the RED phase in the rnhAB thyA mutant, but this time dinB inactivation was also slightly detrimental (cf. Fig. 6B and G). Perhaps the most dramatic effect among all these mutants in TLS polymerases and RNase H enzymes is the two orders of magnitude lower survival of the thyA dinB rnhAB mutant compared to its thyA dinB progenitor (Fig. S5). We conclude that TLS polymerases might help tolerate T-starvation, but not by rU incorporation into DNA. In fact, survival of T-starved cells (most clearly seen with dinB thyA mutants) strongly depends on the functional RNase H enzymes (Fig. S5).
Accumulation of ssDNA during dTTP starvation
Chromosomal replication during T-starvation was expected to generate persistent ss-gaps in DNA (Ahmad, Kirk et al. 1998, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010, Sangurdekar, Hamann et al. 2010). The RecFOR pathway initiates recombinational repair of persistent single strand gaps in DNA by licensing RecA polymerization onto them (Kuzminov 1999, Kuzminov 2011). A major paradox of thymine starvation was that inactivation of the RecFOR DNA repair pathway leads to amelioration of TLD, rather than to its aggravation (Nakayama, Nakayama et al. 1982, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010) (Fig. 1D), supporting the concept of “poisonous repair”. Later epistatic analysis suggested that the major detrimental effect of the RecFOR pathway in TLD is via strong SOS induction (Fonville, Bates et al. 2010). Here we show that the recF mutants accumulate significantly more chromosomal DNA during the resistance phase of thymine starvation, even though they display a similar degree of the chromosomal DNA instability during the subsequent RED phase (Fig. 7A). We interpreted this accumulation of more genomic DNA in the absence of the system to repair persistent ss-gaps to mean that the chromosomal DNA synthesis during thymine starvation is associated with formation of ss-gaps, while the RecFOR system curbs this DNA accumulation by trying to repair the ss-gaps. Formation of ss-gaps in the chromosomal DNA of thymine-starved cells was suggested from electron microscopy observations, but prior efforts to quantify this ssDNA accumulation have been unsuccessful (Nakayama, Kusano et al. 1994).
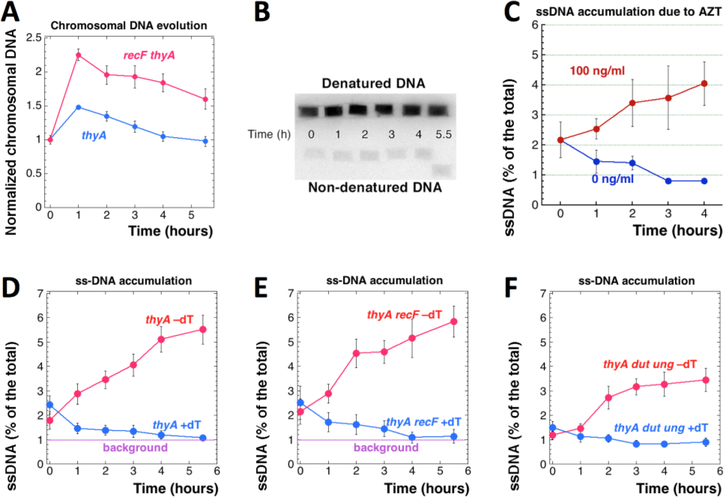
Fig. 7. Accumulation of ss-gaps during T-starvation.
A. Evolution of the chromosomal DNA amount in the thyA (KKW58) versus recF thyA (RA31) mutants during T-starvation. B. An example of plug-blot procedure to quantify ssDNA accumulation during T-starvation. After electric transfer to positively-charged nylon membrane, the genomic DNA was hybridized to the total genomic probe. C. Accumulation of ssDNA in percentage to the total DNA signal in the WT cell (AB1157) cultures grown in the presence of the indicated concentrations of AZT. D. The level of ssDNA in thyA mutant (KKW58) cultures grown in the presence (+dT) or absence (–dT) of thymidine. E. The level of ssDNA in the thyA recF (RA31) mutant. F. The level of ssDNA in the thyA dut ung (RA18) mutant.
To quantify ss-gaps in DNA during TLD, we have isolated DNA in duplicate agarose plugs and then transferred it, either without prior denaturation, or after standard denaturation, to nylon membrane for hybridization with a whole genome probe (Fig. 7B). The probe can hybridize only to ssDNA, so in the non-denatured samples, hybridization is only possible with ss-gaps 20 nt or longer (due to our 65°C hybridization temperature, using E. coli chromosomal DNA, which is 50% GC-rich). The ratio of the signal in the non-denatured DNA plug to the signal in the corresponding denatured DNA plug reveals the fraction of ssDNA in this DNA preparation.
As a positive control for our detection and quantification of ss-gaps in genomic DNA, we treated growing cultures of WT cells with 100 ng/ml azidothymidine (AZT), a chain-terminator nucleotide causing ss-gaps in DNA (Cooper and Lovett 2011). Without treatment, the cultures start with ~2% ssDNA, apparently reflecting active replication status of the cells, but several hours of normal growth steadily reduce ssDNA to the level just below 1% of the total (therefore, we took 1% as the background of the procedure), apparently due to the decline of DNA replication activity in stationary cells (Fig. 7C). In contrast, the AZT-treated cultures showed a steady increase in the amounts of single-stranded chromosomal DNA (Fig. 7C). Treatment with AZT leads to even more dramatic increase in ssDNA in the chromosomal DNA preparations of xthA and recA mutants (Fig. S6), as expected from their increased AZT sensitivity (Cooper and Lovett 2011).
Using this new method of ssDNA detection, we found accumulation of ss-gaps in the chromosomal DNA of thyA mutant during T-starvation, but not in the same cells growing with dT (Fig. 7D). Some ssDNA already forms during the resistance phase, but the main increase happens once the RED phase begins. As expected, proportion of ss-gaps in DNA of the same thyA mutant cells growing with dT approaches background over time, apparently reflecting cessation of the chromosomal replication as the culture saturates (Fig. 7D). The thyA recF mutant shows similar accumulation of ss-gaps during T-starvation (Fig. 7E). Interestingly, the thyA dut ung mutant accumulates less ss-gap DNA during T-starvation (Fig. 7F), suggesting that some of the ss-gaps are indeed prevented with the help of DNA-dU incorporation. We conclude that there is a significant ss-gap accumulation in the chromosomal DNA of thymine-starved cells.
If ss-gaps were linked to T-starvation toxicity, then AZT treatment of T-starved cells, by inducing even more ss-gaps in their DNA, should exacerbate TLD. We found it to be the case, as treatment with 100 ng/ml AZT, the dose that is mostly static for a Thy+ sibling in the same medium, reduces the resistance phase in half and deepens the RED phase for the thyA mutant (Fig. 8A). Remarkably, AZT treatment during T-starvation also rapidly kills the thyA recF mutant, that survives T-starvation without AZT much better than the thyA single mutant and is not extra-sensitive to AZT when Thy+ (Fig. 8B). We conclude that ss-gap accumulation contributes to T-starvation toxicity and may even be its major driver.
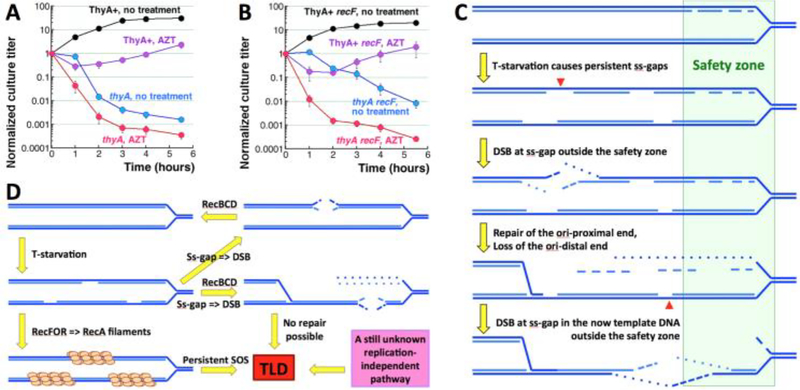
Fig. 8. Persistent single-strand gaps kill.
A. TLD kinetics of the thyA mutant (KKW58) in the presence of AZT. After growth in the medium +dT, cells were washed and resuspended in the same volume of the same medium, but without dT, supplemented or not with 100 ng/ml AZT. The ThyA+ strain (KKW59) is also shown, to demonstrate the toxicity of this AZT dose in this medium (without dT). B. TLD kinetics of the thyA recF mutant (RA31) in the presence of 100 ng/ml AZT (done like in “A”). The ThyA+ recF strain (RA48) is also shown, to illustrate AZT toxicity. C. A model to explain loss of the replication forks due to ss-gaps accumulating outside the “safety zone” (green rectangle) around the replication points, within which DNA with ss-interruptions is safe (note the Okazaki fragments on the lagging strand). Red arrowheads mark the position of double-strand breaks. D. The known and the suspected unknown aspects of TLD. The two known aspects, instability of ss-gaps and SOS-induction, are linked with replication forks. However TLD in the thyA dut ung mutant suggests an unknown, replication-independent pathway.
Discussion
To get insights into the mechanisms of TLD we sought mutants in the metabolism of nucleic acids and their precursors that would affect the resistance phase and transition to the RED phase the way the recBCD and recF mutants in recombinational repair do (Fig. 1D vs Fig. S1). To understand the chromosomal aspects of the expected defects, we precisely quantified the total chromosomal DNA and determined its ssDNA fraction. We started by determining the kinetics of chromosomal fragmentation caused by T-starvation, finding to our surprise that after peaking during the resistance phase, fragmentation disappears at the beginning of the RED phase, weakening the previously proposed link of double-strand breaks to TLD (Kuong and Kuzminov 2010). We report a considerable accumulation of the chromosomal DNA during the resistance phase (coincident with fragmentation), followed by return to the original DNA amount during the RED phase (coincident with the disappearance of fragmentation) (cf. Fig. 2AB vs G). The nature of this chromosomal DNA disappearance is unclear, as the recBCD mutant deficient in the main linear DNA degradation activity of E. coli cells still shows it (Fig. S2C). In general, double-strand breaks, even though numerous early on during T-starvation, do not appear to directly contribute to thymineless death. However, we revealed that the chromosomal DNA in T-starved cells becomes increasingly single-stranded (Fig. 7), — providing a potential mechanistic insight into the pathology of thymineless death.
In our experiments with specific mutants, we were guided by two questions: 1) what is the nature of the resistance phase? 2) what is the nature of the abrupt transition from the resistance to the RED phase? — and by two models (Fig. 1F): 1) the slow progress during the resistance phase due to alternative sources of dT (I); 2) dT analog dU incorporation and excision during the RED phase (II). For the resistance phase, we explored two potential sources of dT: 1) dTMP generated by linear chromosomal DNA degradation; 2) reduction of ribo-thymine derived from stable RNA degradation. For the transition to the RED phase, we tested increased dU and rU incorporation into DNA with subsequent excision (Fig. 1F, II and III). We found that, even though changes in the levels of all four sources of thymine or its uracil replacements usually make the RED phase faster and deeper, none of them have impact on the resistance phase. Thus, the significant accumulation of chromosomal DNA during the resistance phase must be supported by another source, whose identity is yet to be revealed, while the reason for the RED phase could be subsequent exhaustion of this mystery source marking the start of synthesis of defective DNA. In support of this idea, we have found that T-starved cells accumulate high levels of single-strand gaps in the chromosomal DNA (Fig. 1F, IV). We propose that accumulation of these ss-gaps, due to the suspected ss-gap instability outside the safety zone behind the replication fork (Kouzminova and Kuzminov 2012), leads to destruction of one of the replication forks in a T-starved cell (Fig. 8C). The destruction becomes possible due to ~10-times stronger degradation of the origin-distal end, compared with the origin-proximal end, during double-strand break repair in E. coli (Sivaramakrishnan, Sepúlveda et al. 2017). Remarkably, although the resulting formation of an irreparable double-strand gap in the template DNA ruins the chromosome (Fig. 8C), no chromosomal fragmentation is expected, as long as only one replication point was affected, and the chromosomal DNA stays one-piece and branched, — so it should not enter pulsed-field gels.
Instability of the chromosomal DNA during T-starvation
The peculiar chromosomal DNA stability during T-starvation was always one of the major mysteries of TLD. Traditionally, measurements of the total chromosomal DNA purified from the same volume of T-starved cells reported either no change or slightly increasing amounts during the time course of TLD ((Barner and Cohen 1958, Gallant and Suskind 1961, Wachsman, Kemp et al. 1964, Kuong and Kuzminov 2012), also works cited in (Breitman, Maury et al. 1972)). Moreover, pre-labeled chromosomal DNA shows surprising overall stability during T-starvation (Yoshinaga 1973, Kuong and Kuzminov 2012). This DNA amount constancy (or slight increase), in combination with numerous reports of accumulation of single-strand DNA breaks in T-starved cells (Ahmad, Kirk et al. 1998) and recent reports of complex patterns of double-strand DNA breaks (Guarino, Salguero et al. 2007, Kuong and Kuzminov 2010, Kuong and Kuzminov 2012) naturally leads to the idea of the intrachromosomal DNA redistribution (cannibalism) (Fig. 2C). According to this idea, progress of replication forks during T-starvation is supported by thymidine from the degraded chromosomal parts that have extra copies, and whose degradation therefore will not inactivate the chromosome. In effect, the above-average-copy-number chromosomal parts are cannibalized to support replication of the below-average-copy-number chromosomal parts (Fig. 2H). In support of the chromosomal DNA instability during T-starvation, during a phage infection of T-starved cells, there is appreciable incorporation of the chromosomal DNA material in the phage genome, which is not observed in the non-starved cells (Thakur and Poddar 1977). The potential for intrachromosomal cannibalism is further bolstered by the appearance in response to T-starvation of short-lived relatively small subchromosomal fragments (Fig. 2AB) (Kuong and Kuzminov 2010).
In fact, intrachromosomal DNA redistribution seems like a winning solution for the problem of starved replication forks, resulting in replication fork elimination by “fork disproportionation” and leading to equal copy number of all chromosomal parts. However, a strong prediction of the intracellular chromosomal cannibalism idea, — that the origin copy number should decrease, while the terminus copy number should increase, — was not confirmed. Instead, we found that the origin and terminus behavior was similar during T-starvation, first increasing during the resistance phase, and then both decreasing during the RED phase (Fig. 2I) (Kuong and Kuzminov 2012).
Moreover, employing the robust plug-blot procedure (Kouzminova and Kuzminov 2012), we for the first time accurately measured evolution of the total chromosomal DNA amount during T-starvation. In contrast to previously reported stable amounts (see above), we now show that the chromosomal DNA amounts significantly expand during the resistance phase, only to gradually shrink to the original level during the RED phase (Fig. 2G). A similar bi-modal DNA evolution curve was once reported before, when DNA-deoxyribose was followed (Breitman, Maury et al. 1972). The initial significant accumulation of the total chromosomal DNA (1.5x the original amount) and its subsequent disappearance are both incompatible with the intrachromosomal cannibalism idea, which (being a zero-sum game) predicts constancy of the DNA amount. Thus, it was not surprising that our genetic inactivation of linear DNA degradation did not affect the resistance phase of TLD (Fig. 2F) (but it was surprising that it failed to stop the DNA disappearance either (Fig. S2C)).
The short-lived chromosomal fragmentation in response to T-starvation and the peculiar small size of the chromosomal fragments that we found in this work offer explanation for the previous conflicting results in this area. Whereas single-strand breaks in chromosomal DNA of T-starved cells are not disputed ((Ahmad, Kirk et al. 1998) and also see below), there were reports of no double-strand breaks (Nakayama, Kusano et al. 1994), significant early (Guarino, Salguero et al. 2007) or late (Yoshinaga 1973) chromosomal fragmentation, as well as complex kinetics of chromosomal fragmentation in T-starved cells (the initial spike with subsequent slow increase (Kuong and Kuzminov 2010) or redistribution from LMW to HMW (Kuong and Kuzminov 2012)). Our current results (Fig. 2AB) explain how chromosomal fragmentation can be easily missed, or found to be significant, depending on the time points during T-starvation or on the size of the monitored DNA fragments.
Deoxy-uridine misincorporation
The most popular idea explaining both the resistance phase and its sudden change to the RED phase is dU misincorporation in place of dT, with subsequent excision initiated by UDG, which is followed by inevitable DNA synthesis that has to use dU again (Fig. 4A) (Breitman, Maury et al. 1972, Goulian, Bleile et al. 1980, Goulian, Bleile et al. 1980, Ahmad, Kirk et al. 1998, Khodursky, Guzmán et al. 2015). This so-called futile DNA-dU misincorporation/excision cycle (Goulian, Bleile et al. 1980) is proposed to degrade the chromosomal DNA beyond repair (several sub-scenarios reviewed in (Kuong and Kuzminov 2010)). This concept remains popular in the TLD field (Khodursky, Guzmán et al. 2015) not only because it elegantly explains the major features of TLD, but also because DNA-dU misincorporation is known to happen in thymine-requiring mutant of Bacillus subtilis (Tamanoi and Okazaki 1978), and because inactivation of various uracil-DNA-targeting glycosylases does alleviate toxicity of 5-fluorouracil (FU) in eukaryotic cells, offering proof of principle for the futile DNA-dU misincorporation/excision cycle (Seiple, Jaruga et al. 2006, An, Robins et al. 2007, Kunz, Focke et al. 2009).
However, the assumed mechanistic relationship between FU toxicity and TLD remains to be experimentally tested. One reason to suspect that TLD is different from FU toxicity is that TLD kinetics in E. coli is not improved by inactivation of uracil-DNA glycosylase ((Kuong and Kuzminov 2010), Figs. 4C and S4C). The apparent reason for this, found here, is the lack of significant increase in DNA-dU misincorporation during T-starvation, even when dUTP concentrations are artificially elevated by the dut defect (Fig. 5). This suggests existence of mechanisms limiting DNA-dU misincorporation, at least in E. coli. In fact, TLD survival is barely improved by inactivation of UDG in B. subtilis, where DNA-dU misincorporation during T-starvation is considerable (Makino and Munakata 1978).
To test the idea that DNA-dU misincorporation influences TLD parameters, we inactivated dcd (the major route of dUTP production in E. coli), but found no effect on TLD (Fig. S4B). At the same time, during T-starvation, genetic inactivation of the abasic site incision stage of dU removal in the xthA nfo mutant accelerates the RED phase and decreases survival (Fig. 4B), indicating active BER in T-starved cells. In yeast, loss of AP-endonuclease activity makes the organism profoundly sensitive to aminopterin – a drug that induces thymine starvation (Dornfeld and Johnson 2005). For TLD in E. coli, loss of abasic site repair pathway should have an even stronger effect in the dut thyA strain, due to the inability to complete excision of DNA-dU, but this could not be tested because of the co-lethality of the dut xthA combination (Taylor and Weiss 1982).
As a proof-of-concept that the futile DNA-dU misincorporation/excision cycle could be set up in E. coli, we increased the dUTP concentration in T-starved cells by inactivating the dut dUTPase (Hochhauser and Weiss 1978, Warner, Duncan et al. 1981), — and indeed observed disappearance of the resistance phase (Fig. 4C). In other words, DNA-dU incorporation, instead of supporting the initial fork progress in the absence of dTTP, was made predictably poisonous by active DNA-dU excision repair (Ung+). This means that one of the survival tactics during T-starvation could be further limitation of the DNA-dU misincorporation.
To test the idea that stabilization of DNA-dU misincorporation could save T-starved cells, we blocked DNA-dU excision in the dut mutants by ung mutation. The resulting dut ung mutants are known to accumulate significant amounts of dU in their DNA during normal growth, replacing up to 15% of DNA-dT with DNA-dU (Warner, Duncan et al. 1981). The expected effect during T-starvation was a complete suppression of TLD. Compared with the TLD kinetics of the thyA dut mutant, the TLD kinetics of the thyA dut ung mutant does have a prolonged resistance phase, a slower RED phase and 100-times higher survival (Fig. 4C). In fact, the “unlimited” DNA-dU incorporation without subsequent excision allows T-starved cells not only to finish the ongoing replication round, but also to initiate and finish one more replication round (Fig. 4DEF). Thus, stabilized DNA-dU misincorporation is indeed capable of ameliorating chromosomal problems of T-starved cells. Remarkably, even fully-replicated chromosomes cannot save 99% of the thyA dut ung mutant cells from TLD, suggesting an unrelated (non-chromosomal?) nature of its pathology, at least in this case.
It should be kept in mind that the surprising TLD of thyA dut ung mutant may still have chromosomal nature. For example, subsequent initiations of chromosomal replication appear blocked in the T-starved thyA dut ung cells, — is it because, in the presence of dU in DNA, oriC cannot initiate? Uncharacterized metabolic or regulatory problems were reported in cells with chromosomal DNA that contains a lot of dU (Warner, Duncan et al. 1981). The peculiar inviability of mutants in which dT in chromosomal DNA was mostly replaced with dU was systematically explored before, in conjunction with inability to obtain deletions of the dut gene (El-Hajj, Zhang et al. 1988, El-Hajj, Wang et al. 1992).
Ribo-uridine incorporation?
Until fairly recently, ribo-uridine (rU) was not considered to incorporate into DNA even in T-starved cells because of the generally efficient discrimination of DNA polymerases against rNTPs. Characterization of RNase H mutants, deficient in excision of single rNs from DNA (Ohtani, Haruki et al. 1999, Cerritelli and Crouch 2009) suggested that this discrimination power of DNA polymerases is overrated. Indeed, studies with the three eukaryotic replicative polymerases in vitro showed incorporation from two to 16 rNs every 10,000 nt synthesized (Nick McElhinny, Watts et al. 2010); the replicative polymerase of E. coli in vitro incorporates ~4 rNs per 10,000 nt (Yao, Schroeder et al. 2013). We recently determined the in vivo density of rNs in the DNA of the E. coli rnhB mutant, unable to remove single DNA-rNs, as 1 rN per 14,000 nt (Kouzminova, Kadyrov et al. 2017) or 1 rN per ~9,000 nt (Cronan, Kouzminova et al. 2019). Giving all this information, it was not impossible that, during T-starvation, rU incorporation in place of missing dT would be significantly elevated, supporting chromosomal DNA replication.
However, we found only modest (~25%) increase in DNA-rN density in T-starved cells (Fig. 6E). Since this increase is, presumably, all specific to rU, which, assuming otherwise equal incorporation of the four rNs, represents 1/4 of the total, the specific rU density increase becomes ~2-fold, which is still modest. Moreover, the rnhAB mutant deficient in removal of any DNA-rN has a slightly lower TLD survival, meaning that DNA-rNs exacerbate, rather than alleviate TLD. Interestingly, inactivation of TLS polymerases tend to aggravate TLD in the rnhAB mutants further, suggesting that TLS polymerases help to synthesize across DNA-rNs. In summary, the modest increase in incorporation of rN into DNA of T-starved cells (Fig. 8, III) aggravates TLD, especially in the absence of TLS polymerases, which means that increasing the discrimination power of replicative DNA polymerases against DNA-rN incorporation could be another tactic to survive T-starvation.
Accumulation of ssDNA during T-starvation
Long single-strand gaps in chromosomal DNA were postulated to form during T-starvation (Buick and Harris 1975, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010, Kuong and Kuzminov 2012), due to the fact that, with little dTTP around, any A-run of three or longer in the template DNA becomes in essence a non-coding lesion (Kuong and Kuzminov 2012), to which the replisomes are known to react by reinitiating downstream and thus forming a “daughter-strand gap” (Rupp and Howard-Flanders 1968) (Fig. 1F, IV). Indeed, T-starved cells have problems maturating their Okazaki fragments, even though their DNA ligase is functional (Freifelder and Katz 1971), indicating formation of ss-gaps. However, quantification of long ss-gaps in chromosomal DNA of T-starved cells received surprisingly little experimental attention.
While numerous studies report accumulation of single-strand breaks in the chromosomal DNA during T-starvation (Freifelder 1969, Walker 1970, Bhattacharjee and Das 1973, Hill and Fangman 1973, Bousque and Sicard 1976), only three studies explicitly addressed the possibility of formation of long single-strand gaps. First, Nakayama and colleagues mentioned a significant single-stranded character of their non-migrating DNA without presenting experimental data (Nakayama, Kusano et al. 1994). Second, Khodursky and colleagues presented S1 nuclease treatment of the chromosomal DNA from T-starved cells, which is more consistent with ss-breaks, rather than gaps, as S1 treatment massively fragments this DNA only after ExoIII action (which converts nicks into ss-gaps) (Sangurdekar, Hamann et al. 2010). Finally, a recent report describes accumulation of SSB-YFP foci in T-starved cells; unfortunately, no quantification was attempted, and normalization controls were missing (Hong, Li et al. 2017). It looks like we are the first to quantify accumulation of significant single-strandedness in the chromosomal DNA of T-starved cells; we also propose how some of these ss-gaps could turn into irreparable double-strand gaps in the template DNA of T-starved cells (Fig. 8C).
Persistent single-strand gaps in the chromosomal DNA of E. coli initiate the RecFOR pathway of recombinational repair, that licenses RecA polymerization on such gaps, to catalyze their filling-in using the corresponding strand of the intact sister duplex (Kuzminov 1999). Since the recF defect alleviates TLD ((Nakayama, Nakayama et al. 1982, Fonville, Bates et al. 2010, Kuong and Kuzminov 2010), this work), the persistent ss-gap repair by recombination is apparently detrimental, rather than helpful, somehow leading to formation of dysfunctional chromosomes. However, modern schemes of such poisonous recombination (Nakayama, Kusano et al. 1994, Kuong and Kuzminov 2010, Fonville, Vaksman et al. 2011) still lack specific details to become experimentally testable. Alternatively, the main RecFOR contribution to TLD could well be via the induction of massive and persistent SOS response (Fonville, Bates et al. 2010, Kuong and Kuzminov 2010) (Fig. 8D).
Survival strategies and tactics in response to T-starvation
Our original focus was on finding mutants with a reduced resistance phase or with a slower RED phase (Fig. S1). We found two mutants without the resistance phase, dut (enhancing the futile cycle of DNA-dU misincorporation/excision) and the xthA nfo (blocked base-excision repair), as well as one mutant with a slower RED phase, dut ung (stabilized DNA-dU misincorporation). At the same time, several mutants we tested exhibited an unexpected third phenotype, — a decreased survival (a deeper RED phase). Before this finding, we assumed that the level of survival is a characteristic of particular growth conditions, being mostly determined by the level of persisters, — dormant cells with a minimal metabolic activity resistant to TLD because they do not replicate their chromosome (Helaine and Kugelberg 2014). Our finding of mutants with significantly reduced survival in the same growth conditions revealed existence of an active response to T-starvation, with defects leading to reduced survival identifying various strategies and tactics of this response.
By trying to find a source of dT for the resistance phase, we instead identified two possible strategies to increase survival during T-starvation: 1) degradation of linear DNA with a possible intra-chromosomal DNA redistribution of the degraded material (Fig. 2C), allowing cells to liquidate extra replication forks and at the same time to derive dTMP for termination of the remaining forks — thus getting rid of all replication forks, which are the points of chromosomal vulnerability during T-starvation; 2) degradation of stable RNA, allowing cells to both inhibit translation and at the same time to mine a significant amount of ribo-thymine to convert to dT in support of DNA replication (Fig. 3D) — thus, ameliorating the metabolic disbalance of T-starved cells. Both tactics represent an attempt of the cell to derive thymine from dispensable molecules (whether DNA or RNA), so as to redistribute it to the critical molecules in dire need of it (surviving replication forks), while inhibiting the processes that contribute to the disbalance, whether in the chromosome or in the general metabolism. The benefit for the T-starved cells of the degradation of extra chromosomal parts to generate dT to support the surviving replication forks was considered before (Fonville, Vaksman et al. 2011).
We have also noticed certain DNA damage avoidance and repair tactics that affect the survival of T-starved cells: 1) ABS-endonuclease activities reduce accumulation of abasic sites in the chromosomal DNA; 2) dUTP hydrolase Dut controls the dUTP pool to minimize DNA-dU misincorporation and to avoid the futile misincorporation/excision cycle, while uracil-DNA-glycosylase Ung ensures timely removal of uracils from DNA; 3) the RNase H enzymes control accumulation of rNs in DNA. These tactics all work to decrease chances of replication fork encounters with non-coding DNA lesions that would otherwise lead to even more ss-gaps in chromosomal DNA. However, a possible alternative explanation for the decreased survival of all these excision repair mutants (xthA nfo, ung, rnhB) is that DNA repair causes dT release from DNA, which is then used for some other, still unknown, critical cellular process. From this perspective, all these strategies and tactics become ways of releasing thymine, either from DNA or from stable RNA.
Conclusion
The overall picture of TLD pathology keeps evolving as a result of the latest round of investigations (Fonville, Bates et al. 2010, Kuong and Kuzminov 2010, Sangurdekar, Hamann et al. 2010, Fonville, Vaksman et al. 2011, Martín and Guzmán 2011, Kuong and Kuzminov 2012) and now begins with accumulation of persistent ss-gaps that are either converted into double-strand breaks, to be repaired by the RecBCD pathway, or are recognized by the RecFOR + RecQJ pathway as substrates for RecA polymerization (Fig. 8D). Both pathways become potentially lethal as T-starvation progresses. The RecBCD-promoted degradation of the origin-distal end of the double-strand break may lead to the destruction of the intact template for repair of a similar double-strand break in the other sister chromatid, ruining the replication point (Fig. 8C). At the same time, robust RecA filament formation at ss-gaps, in conditions continuously generating new ss-gaps, makes it a positive-feedback loop that could kill the cell via runaway SOS-induction. Finally, death of 99% of cells of the thyA dut ung mutants, that successfully complete two chromosomal replication rounds and accumulate two times less DNA in ss-gaps, suggest an additional, still unknown, TLD mechanism, perhaps independent of DNA replication altogether (Fig. 8D).
Our finding of a significant accumulation of ss-gaps in the chromosomal DNA of T-starved cells raises immediate questions about the nature of this accumulation. Our original assumption all along this project was that it is important for the cell to maintain certain minimal rates of the replication fork progress for the fork stability. In other words, we automatically assumed that, when there was nothing to incorporate opposite “A” in template DNA, ss-gaps would accumulate behind slowly-progressing replication forks. In addition, RecQ and RecJ, due to their enzymatic activities, could lengthen these gaps (Fonville, Bates et al. 2010, Sangurdekar, Hamann et al. 2010). In the future, we will be testing the assumed replication-dependence of this ss-gap accumulation, its chromosomal position relative to the replication forks, as well as its genetics. Also, the origin and terminus instability during TLD (Fonville, Bates et al. 2010, Sangurdekar, Hamann et al. 2010, Kuong and Kuzminov 2012) should be related to this ssDNA accumulation. The apparent insensitivity of the replication forks to incompletely-replicated nascent DNA, reflected in the accumulation of ssDNA in T-starved cells, is another paradox that requires experimental attention. Finally, the source of thymidine that supports significant chromosomal replication during the resistance phase awaits identification.
Supplementary Material
Highlights.
Mutants lacking resistance phase of TLD should identify sources of endogenous dT
Neither chromosomal DNA, nor stable RNA degradation, are the sources of endogenous dT
The futile cycle of DNA-dU incorporation/excision operates only in the dut mutants
The dut ung thyA cells undergo two normal rounds of DNA replication, but die anyway
T-starved cells accumulate long ss-gaps in their chromosome, explaining TLD pathology
Acknowledgements
We would like to thank Susan Rosenberg (Baylor) for critical reading of the manuscript and many helpful suggestions. This work was supported by grant # GM 073115 from the National Institutes of Health. The authors have no conflict of interest to declare.
Abbreviations:
- TLD
thymineless death
- T-starvation
growth of thyA mutants without exogenous thymine/thymidine
- dT
deoxythymidine
- dU
deoxyuridine
- rU
ribouridine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad SI, Kirk SH and Eisenstark A (1998). “Thymine metabolism and thymineless death in prokaryotes and eukaryotes.” Annu. Rev. Microbiol 52: 591–625. [DOI] [PubMed] [Google Scholar]
- An Q, Robins P, Lindahl T and Barnes DE (2007). “5-Fluorouracil incorporated into DNA is excised by the Smug1 DNA glycosylase to reduce drug cytotoxicity.” Cancer Res. 67: 940–945. [DOI] [PubMed] [Google Scholar]
- Barner HD and Cohen SS (1954). “The induction of thymine synthesis by T2 infection of a thymine requiring mutant of Escherichia coli.” J. Bacteriol 68: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barner HD and Cohen SS (1958). “Protein synthesis and RNA turnover in a pyrimidine-deficient bacterium.” Biochim. Biophys. Acta 30: 12–20. [DOI] [PubMed] [Google Scholar]
- Beck CF, Eisenhardt AR and Neuhard J (1975). “Deoxycytidine triphosphate deaminase of Salmonella typhimurium. Purification and characterization.” J. Biol. Chem 250: 609–616. [PubMed] [Google Scholar]
- Bhattacharjee SB and Das J (1973). “Strand breaks in deoxyribonucleic acid of thymine-starved bacteria.” Int. J. Radiat. Biol 23: 167–174. [DOI] [PubMed] [Google Scholar]
- Birnboim HC (1983). “A rapid alkaline extraction method for the isolation of plasmid DNA.” Methods Enzymol. 100: 243–255. [DOI] [PubMed] [Google Scholar]
- Bochner BR and Ames BN (1982). “Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography.” J. Biol. Chem 257(16): 9759–9769. [PubMed] [Google Scholar]
- Bousque JL and Sicard N (1976). “Size and transforming activity of deoxyribonucleic acid in Diplococcus pneumoniae during thymidine starvation.” J. Bacteriol 128: 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman TR, Finkleman A and Rabinovitz M (1971). “Methionineless death in Escherichia coli.” J. Bacteriol 108: 1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman TR, Maury PB and Toal JN (1972). “Loss of deoxyribonucleic acid-thymine during thymine starvation of Escherichia coli.” J. Bacteriol 112(1): 646–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H and Dennis PP (1996). Modulation of chemical composition and other parameters of the cell by growth rate Escherichia coli and Salmonella. Neidhardt FC Washington, D.C., ASM Press: 1553–1569. [Google Scholar]
- Buick RN and Harris WJ (1975). “Thymineless death in Bacillus subtilis.” J. Gen. Microbiol 88: 115–122. [DOI] [PubMed] [Google Scholar]
- Cerritelli SM and Crouch RJ (2009). “Ribonuclease H: the enzymes in eukaryotes.” FEBS J. 276: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SS (1971). “On the nature of thymineless death.” Ann. N. Y. Acad. Sci 186: 292–301. [DOI] [PubMed] [Google Scholar]
- Cohen SS and Barner HD (1954). “Studies on unbalanced growth in Escherichia coli.” Proc. Natl. Acad. Sci. USA 40: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DL and Lovett ST (2011). “Toxicity and tolerance mechanisms for azidothymidine, a replication gap-promoting agent, in Escherichia coli.” DNA Repair 10: 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan GE, Kouzminova EA and Kuzminov A (2019). “Near-continuously synthesized leading strands in Escherichia coli are broken by ribonucleotide excision.” Proc. Natl. Acad. Sci. U S A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA and Wanner BL (2000). “One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products.” Proc. Natl. Acad. Sci. USA 97(12): 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornfeld K and Johnson M (2005). “AP endonuclease deficiency results in extreme sensitivity to thymidine deprivation.” Nucleic Acids Res. 33: 6644–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hajj HH, Wang L and Weiss B (1992). “Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth.” J. Bacteriol 174(13): 4450–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hajj HH, Zhang H and Weiss B (1988). “Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli.” J. Bacteriol 170(3): 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville NC, Bates D, Hastings PJ, Hanawalt PC and Rosenberg SM (2010). “Role of RecA and the SOS response in thymineless death in Escherichia coli.” PLoS Genet 6: e1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville NC, Vaksman Z, Denapoli J, Hastings PJ and Rosenberg SM (2011). “Pathways of resistance to thymineless death in Escherichia coli and the function of UvrD.” Genetics 189: 23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederico LA, Kunkel TA and Shaw BR (1990). “A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy.” Biochemistry 29: 2532–2537. [DOI] [PubMed] [Google Scholar]
- Freifelder D (1969). “Single-strand breaks in bacterial DNA associated with thymine starvation.” J. Mol. Biol 45: 1–7. [DOI] [PubMed] [Google Scholar]
- Freifelder D and Katz G (1971). “Persistence of small fragments of newly synthesized DNA in bacteria following thymidine starvation.” J. Mol. Biol 57: 351–354. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA and Ellenberger T (2006). DNA Repair and Mutagenesis. Washington, D.C., ASM Press. [Google Scholar]
- Gallant J and Suskind SR (1961). “Relationship between thymineless death and ultraviolet inactivation in Escherichia coli.” J. Bacteriol 82: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M, Bleile B and Tseng BY (1980). “The effect of methotrexate on levels of dUTP in animal cells.” J. Biol. Chem 255(22): 10630–10637. [PubMed] [Google Scholar]
- Goulian M, Bleile B and Tseng BY (1980). “Methotrexate-induced misincorporation of uracil into DNA.” Proc. Natl. Acad. Sci. USA 77(4): 1956–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino E, Salguero I, Jiménez-Sánchez A and Guzmán EC (2007). “Double-strand break generation under deoxyribonucleotide starvation in Escherichia coli.” J. Bacteriol. 189: 5782–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G and Weiss B (1998). “Endonuclease V (nfi) mutant of Escherichia coli K-12.” J. Bacteriol. 180(1): 46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S and Kugelberg E (2014). “Bacterial persisters: formation, eradication, and experimental systems.” Trends Microbiol. 22: 417–424. [DOI] [PubMed] [Google Scholar]
- Hill WE and Fangman WL (1973). “Single-strand breaks in deoxyribonucleic acid and viability loss during deoxyribonucleic acid synthesis inhibition in Escherichia coli.” J. Bacteriol 116(3): 1329–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhauser SJ and Weiss B (1978). “Escherichia coli mutants deficient in deoxyuridine triphosphatase.” J. Bacteriol 134(1): 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R, Wierzbicki A and Abremski K (1985). “Formation of small circular DNA molecules via an in vitro site-specific recombination system.” Gene 40: 325–329. [DOI] [PubMed] [Google Scholar]
- Hong Y, Li L, Luan G, Drlica K and Zhao X (2017). “Contribution of reactive oxygen species to thymineless death in Escherichia coli.” Nat. Microbiol 2: 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O and D’Ari R (1981). “An inducible DNA replication-cell division coupling mechanism in E. coli.” Nature 290: 797–799. [DOI] [PubMed] [Google Scholar]
- Itsko M and Schaaper RM (2014). “dGTP starvation in Escherichia coli provides new insights into the thymineless-death phenomenon.” PLoS Genet. 10: e1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsko M and Schaaper RM (2016). “Transcriptome Analysis of Escherichia coli during dGTP Starvation.” J. Bacteriol 198: 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Beuning PJ, Cohen SE and Walker GC (2007). “Y-family DNA polymerases in Escherichia coli.” Trends Microbiol. 15: 70–77. [DOI] [PubMed] [Google Scholar]
- Kabir MS, Sagara T, Oshima T, Kawagoe Y, Mori H, Tsunedomi R and Yamada M (2004). “Effects of mutations in the rpoS gene on cell viability and global gene expression under nitrogen starvation in Escherichia coli.” Microbiology 150: 2543–2553. [DOI] [PubMed] [Google Scholar]
- Khan SR and Kuzminov A (2012). “Replication forks stalled at ultraviolet lesions are rescued via RecA and RuvABC protein-catalyzed disintegration in Escherichia coli.” J. Biol. Chem 287: 6250–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky A, Guzmán EC and Hanawalt PC (2015). “Thymineless Death Lives On: New Insights into a Classic Phenomenon.” Annu. Rev. Microbiol 69: 247–263. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA, Kadyrov FF and Kuzminov A (2017). “RNase HII Saves rnhA Mutant Escherichia coli from R-Loop-Associated Chromosomal Fragmentation.” J. Mol. BIol 429: 2873–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova EA and Kuzminov A (2004). “Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair.” Mol. Microbiol 51(5): 1279–1295. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA and Kuzminov A (2006). “Fragmentation of replicating chromosomes triggered by uracil in DNA.” J. Mol. Biol 355(1): 20–33. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA and Kuzminov A (2008). “Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-strand breaks.” Mol. Microbiol 68(1): 202–215. [DOI] [PubMed] [Google Scholar]
- Kouzminova EA and Kuzminov A (2012). “Chromosome demise in the wake of ligase-deficient replication.” Mol. Micorbiol 84: 1079–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova EA, Rotman E, Macomber L, Zhang J and Kuzminov A (2004). “RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure.” Proc. Natl. Acad. Sci. USA 101(46): 16262–16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz BA and Haynes RH (1982). “DNA repair and the genetic effects of thymidylate stress in yeast.” Mutat. Res 93: 353–375. [Google Scholar]
- Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J and Schär P (2009). “Base excision by thymine DNA glycosylase mediates DNA-directed cytotoxicity of 5-fluorouracil.” PLoS Biol. 7: e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ and Kuzminov A (2009). “Cyanide, peroxide and nitric oxide formation in solutions of hydroxyurea causes cellular toxicity and may contribute to its therapeutic potency.” J. Mol. Biol 390: 845–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ and Kuzminov A (2010). “Stalled replication fork repair and misrepair during thymineless death in Escherichia coli.” Genes Cells 15: 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuong KJ and Kuzminov A (2012). “Disintegration of nascent replication bubbles during thymine starvation triggers RecA- and RecBCD-dependent replication origin destruction.” J. Biol. Chem 287: 23958–23970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A (1999). “Recombinational repair of DNA damage in Escherichia coli and bacteriophage l.” Microbiol. Mol. Biol. Rev 63(4): 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A (2011). “Homologous Recombination—Experimental Systems, Analysis, and Significance.” EcoSal Plus 4: doi: 10.1128/ecosalplus.1127.1122.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner RD (2001). “The role of dUTPase and uracil-DNA repair in cancer chemotherapy.” Curr. Protein Pept. Sci 2(4): 361–370. [DOI] [PubMed] [Google Scholar]
- Maaloe O and Hanawalt PC (1961). “Thymine deficiency and the normal DNA replication cycle. I.” J. Mol. Biol 3: 144–155. [DOI] [PubMed] [Google Scholar]
- Madsen CT, Mengel-Jørgensen J, Kirpekar F and Douthwaite S (2003). “Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry.” Nucleic Acid Res. 31: 4738–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino F and Munakata N (1978). “Deoxyuridine residues in DNA of thymine-requiring Bacillus subtilis strains with defective N-glycosidase activity for uracil-containing DNA.” J. Bacteriol 134(1): 24–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF and Sambrook J (1982). Molecular cloning. A laboratory manual. Cold Spring Harbor, Cold Spring Harbor Laboratory. [Google Scholar]
- Martín CM and Guzmán EC (2011). “DNA replication initiation as a key element in thymineless death.” DNA Repair 10: 94–101. [DOI] [PubMed] [Google Scholar]
- McCann MP, Kidwell JP and Matin A (1991). “The putative sigma factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli.” J. Bacteriol. 173: 4188–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY, Cold Spring Harbor Laboratory Press. [Google Scholar]
- Nakayama H, Nakayama K, Nakayama R and Nakayama Y (1982). “Recombination-deficient mutations and thymineless death in Escherichia coli K12: reciprocal effects of recBC and recF and indifference of recA mutations.” Can. J. Microbiol 28: 425–430. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Kusano K, Irino N and Nakayama H (1994). “Thymine starvation-induced structural changes in Escherichia coli DNA. Detection by pulse field gel electrophoresis and evidence for involvement of homologous recombination.” J. Mol. Biol 243: 611–620. [DOI] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J and Fuchs RP (2000). “All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis.” EMBO J. 19: 6259–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J and Munch-Petersen A (1966). “Studies on the acid-soluble nucleotide pool in thymine-requiring mutants of Escherichia coli during thymine starvation. II. Changes in the amounts of deoxycytidine triphosphate and deoxyadenosine triphosphate in Escherichia coli 15 T-A-U.” Biochim. Biophys. Acta 114: 61–71. [DOI] [PubMed] [Google Scholar]
- Neuhard J and Nygaard P (1987). Purines and pyrimidines. Escherichia coli and Salmonella typhimurium Cellular and molecular biology. Neidhardt FC. Washington, D.C., American Society for Microbiology: 445–473. [Google Scholar]
- Neuhard J and Thomassen E (1971). “Deoxycytidine triphosphate deaminase: identification and function in Salmonella typhimurium.” J. Bacteriol 105: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundström EB, Burgers PM, Johansson E, Chabes A and Kunkel TA (2010). “Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases.” Proc. Nat. Acad. Sci. U.S.A 107: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa T (1975). “Studies of intracellular thymidine nucleotides. Thymineless death and the recovery after re-addition of thymine in Escherichia coli K 12.” Eur. J. Biochem 60: 57–66. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M and Kanaya S (1999). “Molecular diversities of RNases H.” J. Biosci. Bioeng 88: 12–19. [DOI] [PubMed] [Google Scholar]
- Rinken R, Thoms B and Wackernagel W (1992). “Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding.” J. Bacteriol 174(16): 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp WD and Howard-Flanders P (1968). “Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation.” J. Mol. Biol 31: 291–304. [DOI] [PubMed] [Google Scholar]
- Sangurdekar DP, Hamann BL, Smirnov D, Srienc F, Hanawalt PC and Khodursky AB (2010). “Thymineless death is associated with loss of essential genetic information from the replication origin.” Mol. Micorbiol 75: 1455–1467. [DOI] [PubMed] [Google Scholar]
- Schroeder JW, Randall JR, Matthews LA and Simmons LA (2015). “Ribonucleotides in bacterial DNA.” Crit. Rev. Biochem. Mol. Biol 50: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiple L, Jaruga P, Dizdaroglu M and Stivers JT (2006). “Linking uracil base excision repair and 5-fluorouracil toxicity in yeast.” Nucleic Acids Res. 34: 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva OV, Bogdanov AA and Sergiev PV (2015). “What do we know about ribosomal RNA methylation in Escherichia coli?” Biochimie 117: 110–118. [DOI] [PubMed] [Google Scholar]
- Sivaramakrishnan P, Sepúlveda LA, Halliday JA, Liu J, Núñez MAB, Golding I, Rosenberg SM and Herman C (2017). “The transcription fidelity factor GreA impedes DNA break repair.” Nature 550: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB and Kushner SR (2012). “RNAsnap™: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria.” Nucleic Acids Res. 40: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanoi F and Okazaki T (1978). “Uracil incorporation into nascent DNA of thymine-requiring mutant of Bacillus subtilis 168.” Proc. Natl. Acad. Sci. USA 75(5): 2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AF and Weiss B (1982). “Role of exonuclease III in the base excision repair of uracil-containing DNA.” J. Bacteriol 151(1): 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur AR and Poddar RK (1977). “Growth and reactivation of single stranded DNA phage phiX174 in E. coli undergoing “thymineless death”.” Mol. Gen. Genet 151: 313–318. [DOI] [PubMed] [Google Scholar]
- Ting H, Kouzminova EA and Kuzminov A (2008). “Synthetic lethality with the dut defect in Escherichia coli reveals layers of DNA damage of increasing complexity due to uracil incorporation.” J. Bacteriol 190: 5841–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman A, Kuban W, McDonald JP, Karata K, Yang W, Goodman MF and Woodgate R (2012). “Critical amino acids in Escherichia coli UmuC responsible for sugar discrimination and base-substitution fidelity.” Nucleic Acids Res. 40: 6144–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsman JT, Kemp S and Kogg L (1964). “Thymineless death in Bacillus megaterium.” J. Bacteriol 87: 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JR (1970). “Thymine starvation and single-strand breaks in chromosomal deoxyribonucleic acid of Escherichia coli.” J. Bacteriol 104: 1391–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner HR, Duncan BK, Garrett C and Neuhard J (1981). “Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli.” J. Bacteriol 145(2): 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Hatahet Z, Melamede RJ and Kow YW (1994). “Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3’ endonuclease, from Escherichia coli.” J. Biol. Chem 269: 16260–16268. [PubMed] [Google Scholar]
- Yao M and Kow YW (1997). “Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA.” J. Biol. Chem 272(49): 30774–30779. [DOI] [PubMed] [Google Scholar]
- Yao NY, Schroeder JW, Yurieva O, Simmons LA and O’Donnell ME (2013). “Cost of rNTP/dNTP pool imbalance at the replication fork.” Proc. Natl. Acad. Sci. U S A 110: 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K (1973). “Double-strand scission of DNA involved in thymineless death of Escherichia coli 15 TAU.” Biochim. Biophys. Acta 294: 204–213. [PubMed] [Google Scholar]
- Zahradka K, Buljubasić M, Petranović M and Zahradka D (2009). “Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli.” J. Bacteriol 191: 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.