Abstract
Sirtuins are highly conserved NAD+-dependent enzymes that are capable of removing a wide range of lipid lysine acyl-groups from protein substrates in a NAD+-dependent manner. These NAD+-dependent activities enable sirtuins to monitor cellular energy status and modulate gene transcription, genome stability, and energy metabolism in response to environmental signals. Consequently, sirtuins are important for cell survival, stress resistance, proliferation, and differentiation. In recent years, sirtuins are increasingly recognized as crucial regulators of stem cell biology in addition to their well-known roles in metabolism and aging. This review highlights our current knowledge on sirtuins in stem cells, including their functions in pluripotent stem cells, embryogenesis, and development, as well as their roles in adult stem cell maintenance, regeneration, and aging.
Keywords: Sirtuins, metabolism, epigenetics, embryonic development, regenerative capacity
Introduction
First discovered in yeast as key components in gene silencing complexes [1], the Silent Information Regulator 2 (Sir2) family of ancient proteins (sirtuins) are present in all kingdoms of life. Sirtuins bear a unique NAD+-dependent protein deacylase activity, through which a variety of lipid acyl-groups, including acetyl, succinyl, malonyl, glutaryl, or long-chain acyl-groups, are transferred from the substrate proteins to the ADP-ribose moiety of NAD+ [2–5]. Given the essential role of NAD+ in cellular metabolism, this exclusive requirement of NAD+ enables sirtuins to monitor the cellular energy status and modulate the function of an array of protein substrates, ranging from histones and transcription factors to metabolic enzymes and cell membrane proteins.
Mammals have seven members of sirtuins, Sirtuin 1 (SIRT1) to Sirtuin 7 (SIRT7) [6]. These seven proteins are localized in distinct subcellular localizations and have different tissue expression patterns with differential substrate specificities [7–11] (Box 1). While a large body of literature is centered on the importance of sirtuins in nutrient sensing, aging, and age-associated conditions/diseases [12, 13], emerging evidence reveals this family of unique enzymes as pivotal regulators of stem cell biology. In this review, we summarize the involvement of sirtuins in metabolic and epigenetic regulation of stem cells, with focus on pluripotent stem cells and embryogenesis, as well as adult stem cell maintenance and tissue regeneration.
Box 1 Mammalian Sirtuins.
Mammals have seven members of sirtuins, Sirtuin 1 (SIRT1) to Sirtuin 7 (SIRT7). Among them, SIRT1, SIRT6, and SIRT7 are nuclear proteins. As the most conserved mammalian sirtuin, SIRT1 predominately resides in the nucleus, but also retains an ability to shuttle between cytosol and nucleoplasm in response to different environmental signals. On the other hand, SIRT6 is a nuclear, chromatin-bound protein, whereas SIRT7 is highly enriched in the nucleolus. Three additional mammalian sirtuins, SIRT3, SIRT4, and SIRT5, are found in the mitochondrial matrix, where they participate in a number of metabolic and survival processes associated with the mitochondrial activity. A substantial proportion of SIRT5 is extramitochondrial/cytosolic, where it functions to regulate glycolysis through demalonylation of key glycolytic enzymes. SIRT2 is primarily a cytosolic protein that can also shuttle between the cytosol and nucleus.
The diverse subcellular localizations of sirtuins, together with their distinct tissue expression patterns and differential substrate specificities, lead to wide-ranging impacts of sirtuins on many fundamental biological processes, including gene transcription, genome stability, energy metabolism, stress responses, inflammation, proliferation, and differentiation. Not surprisingly, dysfunction of this family of crucial regulators has been associated with a number of human diseases, such as metabolic disorders and cancer.
Metabolic and epigenetic regulation of stem cells
Stem cells, including pluripotent stem cells (PSCs) and adult tissue stem cells (ASCs) (Box 2), possess a unique set of metabolic programs and a specific epigenetic status to sustain their unlimited proliferation while maintaining their pluri-/multi-potency [14–17]. Energetically, stem cells share striking similarities with highly proliferating cancer cells, requiring a high glycolytic flux under aerobic conditions (“Warburg effect”) and addicting to exogenous glutamine for rapid growth and maintenance of pluripotency [15, 18–20]. Stem cells, particularly embryonic stem cells (ESCs), also have increased dependence on one-carbon catabolism [21–24]. These metabolic programs provide anabolic precursors required for rapid cell proliferation. They also produce many intermediate metabolites, such as acetyl-CoA, NAD+, α-ketoglutarate, and S-adenosylmethionine (SAM), which function as substrates or cofactors for enzymes that regulate chromatin modification and gene expression [24–29]. Therefore, the unique metabolic state of stem cells directly links to distinctive epigenetics and gene expression profiles, greatly influencing their self-renewal and pluri-/multi-potency [20, 30].
Box 2 Stem Cells.
Stem cells, including pluripotent stem cells (PSCs) and adult tissue stem cells (ASCs), are uniquely characterized by their dual capacity for self-renewal and pluri-/multi-potent differentiation. PSCs are stem cells giving rise to all cells of the tissues of the body. There are two types of PSCs, embryonic stem cells (ESCs) and induced pluripotent stem cell (iPSCs). ESCs are derived from the inner cell mass (ICM) of preimplantation embryos. iPSCs are reprogrammed from adult somatic cells in vitro through simultaneous overexpression of four core pluripotent factors, Oct4, Sox2, Klf4, and cMyc (OSKM). Both ESCs and iPSCs can be indefinitely maintained and expanded in the pluripotent state in vitro, and are capable to differentiate into all the derivatives of the three germ layers.
ASCs are the stem cells characterized by their ability to self-renew and differentiate to generate all the cell types in a tissue. Based on their turnover rates in various tissues/organs, ASCs can be categorized into three types: ASCs that continuously cycle themselves in high-turnover tissues, like intestine stem cells (ISCs) and the short-term hematopoietic stem cells (ST-HSCs); ASCs that are maintained in quiescent state, and proliferation are strongly induced by injury or other environmental/developmental cues, such as muscle satellite cells (SCs), adult neuron stem cells (NSCs), and long-term hematopoietic stem cells (LT-HSCs); and ASCs that periodically alternate between quiescent and proliferative states, like hair follicle stem cells.
Stem cells are essential for embryogenesis, tissue regeneration, and the maintenance of tissue homeostasis throughout the lifetime of an organism. A better understanding of mechanisms underlying stem cell self-renewal and pluri-/multi-potency will therefore improve our knowledge on development, regeneration, and aging.
Sirtuins in metabolic and epigenetic regulation of pluripotent stem cells (PSCs) and development
Different sirtuins have distinct roles in PSCs and development. In mouse ESCs (mESCs), SIRT1 is the most highly expressed sirtuin, followed by SIRT6 and SIRT7 [31]. In contrast, the relative levels of other sirtuins, including mitochondrial sirtuins, are low in mESCs [31]. Consistently, germ-line knocking out SIRT1, SIRT6, or SIRT7, but not other sirtuins, causes significant developmental defects [8, 32–38], highlighting the importance of three nuclear sirtuins in regulation of ESC biology and animal development.
SIRT1 is important for normal embryogenesis and animal development
SIRT1 has numerous functions at different stages of animal life. However, one of the primary functions of SIRT1 is to regulate embryogenesis and animal development. SIRT1 is extremely highly expressed in pre-implantation embryos, particularly at the 2-cell stage when the embryo starts to activate its own genome [39], indicating that SIRT1 is one of the first activated genes during embryogenesis. Consistently, SIRT1 null zygotes generated through in vitro fertilization of SIRT1 deficient sperm and oocytes display a substantially compromised efficiency of developing to the 2-cell stage compared to control zygotes [40]. SIRT1 expression is gradually reduced during development of pre-implantation embryos [39]. However, ESCs isolated from the inner cell mass from the blastocyst still express massively higher amounts of SIRT1 than differentiated cells and tissues [39, 41], further suggesting that SIRT1 is critical for ESC function and embryogenesis. In support of this notion, germ-line deletion of SIRT1 in mice leads to severe developmental defects on various genetic backgrounds, including intrauterine growth retardation, developmental defects of the retina and heart, defective germ cell differentiation, bone developmental delay, and neonatal lethality [32, 33, 42–44]. Although the penetrance and severity of SIRT1 deletion-induced developmental phenotypes vary depending on the genetic background, and mice lacking SIRT1 on the FVB background [45] or a few percentages of SIRT1 null mice on some mixed backgrounds [32, 33] can survive into adulthood, growth retardation and sterility with developmental defects are still observed in these adult mice. Additionally, SIRT1 instability caused by loss of USP22, a deubiquitinating enzyme that stabilizes SIRT1, is associated with the defective embryogenesis in USP22 null mice [46].
Most adult tissues have low levels of SIRT1 compared to pre-implantation embryos and ESCs [32, 39], however, SIRT1 maintains a high expression level in germ cells, particularly in male germ cells [32, 47]. Consistently, whole-body SIRT1 deficiency reduces the spermatogenic stem cell number in E15.5 embryos, abrogating spermatogenesis but not oogenesis [40]. The action of SIRT1 in germ cells is cell-autonomous, as germ cell-specific deletion of SIRT1 disrupts spermatogenesis [43, 47]. Taken together, the high expression of SIRT1 in pre-implantation embryos, ESCs, and germ cells, combined with SIRT1 deficiency-induced defects in embryogenesis and germ cell development, strongly indicate that SIRT1 is a key regulator of embryogenesis and animal development.
SIRT1 maintains pluripotent ESCs through multi-level mechanisms
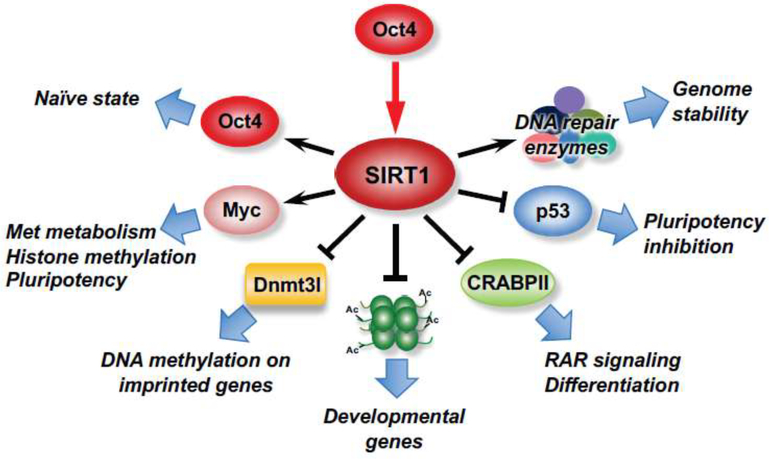
SIRT1 is a multifaceted regulator in the pluripotent ESCs (Figure 1). Firstly, SIRT1 interacts with several master transcription factors to directly modulate the pluripotency of ESCs. Specifically, SIRT1 interacts with Oct4, a key component of the core pluripotency network. In hESCs, SIRT1 is under the transcriptional control of Oct4 and mediates Oct4-dependent pluripotency through repression of p53, a well-known SIRT1 deacetylation substrate [48]. In mESCs, SIRT1 is required for the maintenance of the naïve state through direct deacetylation of Oct4 [49]. During the naive-to-primed transition, the activity of SIRT1 is reduced and Oct4 becomes hyperacetylated. Acetylated Oct4 binds to an enhancer to induce the expression of Otx2, which in turn interacts with acetylated Oct4 to induce the primed pluripotency gene network [49].
Figure 1. SIRT1 is a multifaceted regulator in the pluripotent ESCs.
SIRT1 is critically involved in the maintenance of pluripotent ESCs and animal development through multi-level mechanisms. The expression of SIRT1 in ESCs is under transcriptional control of Oct4, and the interaction between SIRT1, Oct4, and Myc are important to metabolically and epigenetically maintain pluripotent ESCs. SIRT1 also actively represses the expression of developmental and differentiation genes through deacetylation of histones and inhibition of cellular RA signaling. Finally, SIRT1 enhances stress response of ESCs by repressing p53 and promoting DNA repair.
Secondly, SIRT1 directly and indirectly modulates epigenetics of ESCs. We recently reported that SIRT1 interacts with Myc to metabolically and epigenetically maintain pluripotent mESCs [39]. Deletion of SIRT1 in mESCs leads to hyperacetylation of both N-Myc and c-Myc, decreasing their stability or reducing their recruitment to the promoter of methionine adenosyltransferase 2a (mat2a). This defect impairs the conversion of methionine to SAM and markedly decreases methylation levels of histones, especially H3K4me3, thereby altering gene expression profiles, comprising the pluripotency, and sensitizing mESCs to methionine restriction-induced differentiation and apoptosis [39]. This SIRT1-Myc regulated metabolic and epigenetic regulation is physiologically important, as SIRT1 KO embryos are sensitive to maternal methionine restriction-induced lethality, whereas maternal methionine supplementation increases the survival of SIRT1 KO newborn mice [39]. Additionally, SIRT1 interacts with DNA methyltransferase 3-like (Dnmt3l) to modulate DNA methylation, the expression of imprinted and germline genes, and the differentiation potential of mESCs [50]. Heo et al. showed that Dnmt3l, a catalytically inactive DNA methyltransferase that interacts with Dnmt3a and Dnmt3b to stimulate de novo methylation, is regulated by SIRT1 at both transcriptional and posttranslational levels. Consequently, SIRT1 deficiency delayed neurogenesis and spermatogenesis [50]. Therefore, SIRT1 metabolically and epigenetically regulates ESC maintenance and animal development.
Thirdly, SIRT1 actively represses differentiation of ESCs. Highly expressed SIRT1 has been reported to repress the transcription of a number of developmental genes in ESCs through deacetylation of histones [51]. Differentiation of ESCs reduces both SIRT1 mRNA and protein posttranscriptionally [41, 51], either by microRNAs-mediated inhibition of SIRT1 protein translation [41], or by CARM1-HuR-dependent destabilization of SIRT1 mRNA [51, 52]. Reduction of SIRT1 then reactivates key developmental genes [51]. Moreover, SIRT1 also represses mESC differentiation through inhibition of cellular retinoic acid (RA) signaling [44]. In mESCs, SIRT1 interacts with and deacetylates CRABPII, a cellular RA binding protein that is acetylated and translocated into the nucleus to activate RAR signaling upon RA treatment. SIRT1-mediated deacetylation recycles CRABPII back to the cytosol, inactivating the RAR-mediated activation of differentiation genes. Notably, SIRT1 deficiency induced development defects in mice, particularly bone development delay, is associated with increased RA signaling [44].
Finally, SIRT1 is important to determine cell fates in response to stress in ESCs. In response to endogenous reactive oxygen species (ROS), SIRT1 functions to maintain heathy pluripotent ESCs by sensitizing mESCs to mitochondrial p53-induced apoptosis while inhibiting nuclear p53-mediated suppression of Nanog expression [53]. In contrast, SIRT1 protects ESCs from apoptosis induced by high concentrations of exogenous ROS (e.g. 1 mM H2O2) in part through the class III PI3K/Beclin 1 and mTOR mediated autophagy [54]. Therefore, the function of SIRT1 in protection against stress-induced apoptosis in ESCs is dependent on the degree of oxidative stress. Additionally, SIRT1 strongly enhances the survival of hESCs by promoting DNA repair [55], and promotes telomere elongation and genome stability after reprogramming in iPSCs [56]. Collectively, SIRT1 is critically involved in the maintenance of pluripotent ESCs and animal development through regulation of pluripotency factors, metabolism, epigenetics, redox homeostasis, and cellular stress response.
SIRT7 regulates embryogenesis and lifespan through maintenance of genome stability
SIRT7, a nuclear sirtuin enriched in the nucleolus, also plays a role in animal development [38]. SIRT7 deficient pups display defective embryogenesis and growth retardation, and are born at sub-Mendelian ratios. More than 20% of SIRT7 null mice die with the first month of life, and the remaining knockouts show signs of accelerated aging with dramatically reduced lifespan [38]. Mechanistically, SIRT7 is recruited to sites of DNA damage to deacetylate H3K18, which is important for efficient recruitment of damage response factors to DNA double-strand breaks [38]. Therefore, SIRT7 directly promotes DNA non-homologous end joining repair and is important for the maintenance of genome stability.
SIRT6 epigenetically promotes proper lineage commitment of ESCs and animal development
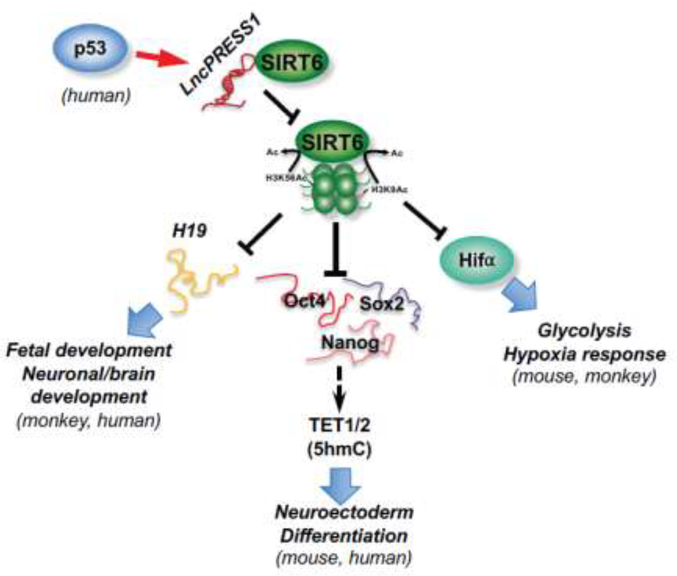
In contrast, SIRT6 is a nuclear sirtuin evolutionally important for proper differentiation of ESCs (Figure 2). SIRT6 knockout mice show developmental delay and premature death, and SIRT6 deficient ESCs form small embryoid bodies (EBs) and exhibit skewed differentiation toward neuroectoderm when induced to differentiation [57]. However, although SIRT6 represses glycolysis that is important for ESC self-renewal [14–17] through deacetylation of histones [58], elevated glycolysis is not accounting for the observed developmental phenotypes in SIRT6 KO ESCs [57]. Etchegaray at al. recently found that SIRT6 epigenetically represses the expression of the core pluripotency genes (OCT4, SOX2, and NANOG) by targeting H3K56ac and H3K9ac. Deletion of SIRT6 in mouse promotes persistent expression of OCT4, SOX2 and NANOG in ESCs and EBs. OCT4 and SOX2 then enhance the expression of TET1 and TET2, increasing 5-hydroxymethylcytosine (5hmC), particularly on neural genes [57]. SIRT6 therefore functions as a chromatin regulator to epigenetically regulate the hierarchical expression of the core pluripotency genes and TET genes, safeguarding the balance between pluripotency and differentiation [57]. Consistent with its role in promoting proper differentiation and development in mice, SIRT6 is inhibited by a p53-targeted lncRNA, lncPRESS1, in hESCs [59]. LncPRESS1 physically interacts with SIRT6 and prevents its chromatin localization, maintaining high levels of H3K56ac and H3K9ac at promoters of pluripotency genes [59]. Importantly, SIRT6 is an evolutionally conserved regulator of ESC differentiation and development in vivo (Figure 2). Deletion of SIRT6 in cynomolgus monkey leads to hyperacetylation of histone, activating the long non-coding RNA H19 and resulting in prenatal developmental retardation and neonatal death [60]. A homozygous SIRT6 inactive mutation in human also leads to hyperacetylation of H3K9 and H3K56, dramatically elevating pluripotent genes and causing severe congenital anomalies and perinatal lethality [61].
Figure 2. SIRT6 epigenetically regulates proper lineage commitment of ESCs and animal development.
In mouse, monkey, and human, SIRT6 represses expression of metabolic genes (particularly glycolytic genes), core pluripotent genes, and maternal imprinted lncRNA H19 through deacetylation of H3K9ac and K3K56ac. These actions of SIRT6 metabolically and epigenetically safeguard the balance between pluripotency and differentiation. In hESCs, a p53-targeted lncRNA, lncPRESS1, physically interacts with SIRT6 and prevents its chromatin localization, which in turn maintains high levels of H3K56ac and H3K9ac at promoters of pluripotency genes. Therefore, in contrast to SIRT1, SIRT6 is an evolutionally conserved regulator of ESC differentiation and animal development both in vitro and in vivo.
SIRT2 promotes differentiation of ESCs in vitro
SIRT2, a predominately cytosolic sirtuin, is not essential for embryonic viability and postnatal development in mice [37]. However, it plays a role in PSCs in vitro, but again in an opposing direction compared to SIRT1. In contrast to SIRT1, the level of SIRT2 is low in hESCs and high in differentiated somatic cells [62], and its induction during RA-induced differentiation of mESCs represses GSK3β and promotes differentiation [63]. Cha et al. recently showed that SIRT2 is the primary deacetylase that deacetylates and inhibits glycolytic enzymes in hESCs [62]. The expression of SIRT2 is under control of miR-200c-5p in hESCs, and its induction/overexpression promotes differentiation through repression of glycolysis. Conversely, inhibition of SIRT2 in fibroblasts promotes the reprogramming of fibroblasts to iPSCs [62]. Therefore, SIRT2 is critical in controlling metabolic switch between PSCs and differentiated cells in vitro. It will be interesting to investigate whether other sirtuin-mediated deacylations that regulate glycolysis, SIRT5 mediated demalonylation and activation of glycolytic enzymes for example [64], also play a role in mediating metabolic transition between PSCs and differentiated cells in response to stress.
Sirtuins in metabolic and epigenetic regulation of adult stem cells (ASCs)
ASCs are the stem cells that persist throughout the life, replacing cells lost to homeostatic turnover, disease and injury [65, 66]. Different tissue ASC populations have varying proliferative plasticity and distinct turn-over patterns (Box2). However, the quiescence, proliferation, self-renewal, and differentiation of ASCs are delicately balanced by common mechanisms [65, 66]. Disruption of ASC homeostasis, either by declines in ASC number and/or functions or by aberrant activation of ASCs, has been linked to a number of human disorders, including degenerative disease, aging, and cancer. Much data has accumulated to show that sirtuins can protect against ASC depletion in response to stress and aging through maintenance of their self-renewal, quiescence, and regenerative capacity.
SIRT1 is important for the maintenance of diverse ASC pools
SIRT1 is an essential factor for preserving ASC pools in multiple tissues. For instance, SIRT1 inactivation has been shown to largely account for the premature phenotype in progeroid syndrome, a group of rare genetic disorders characterized with severe early-onset premature aging [67]. Genetically, progeroid syndrome is caused by progerin, a truncated Lamin A protein. Liu et al showed that both progerin and unprocessed prelamin A protein are defective in binding and activation of SIRT1, leading to development of progeroid or progeroid-like syndrome [67]. A number of ASCs, including mesenchymal stem cells (MSCs), hair follicle progenitor cells, and hematopoietic stem cells (HSCs), are rapidly depleted in mice deficient of Zmpste24, a metalloproteinase responsible for prelamin A maturation. They further showed that resveratrol promotes the interaction between SIRT1 and Lamin, activating SIRT1 and rescuing ASC decline in Zmpste24−/− mice [67]. Although direct in vivo evidence is needed to substantiate the role of SIRT1 in these ASCs, maintaining ASC pool appears to be one of the mechanisms through which SIRT1 promotes longevity.
Mechanistically, SIRT1 maintains different types of ASCs through diverse mechanisms. In ASCs that continuously divide asymmetrically in high-turnover tissues, such as intestinal stem cells (ISCs), SIRT1 is necessary and sufficient to promote their self-renewal and expansion in response to caloric restriction (CR) [68]. It has been previously shown that CR augments ISC functions by reducing mTORC1 signaling thereby increasing the production of a paracrine factor cyclic ADP ribose (cADPR) in Paneth cells, a key constituent of the mammalian ISC niche, but not in ISCs [69]. The increased release of cADPR from Paneth cells then acts on ISCs to activate CaMKK and AMPK [68, 69]. Igarashi and Guarente recently showed that CR-mediated activation of AMPK in ISCs stimulates the transcription of Nampt, the rate-limiting enzyme in a NAD+ salvage pathway, enhancing the activity of SIRT1. Enhanced SIRT1 then deacetylates S6K1, facilitating mTORC1 to phosphorylate S6K1 and stimulating protein synthesis [68]. Although it is surprising that mTORC1 signaling is activated in ISCs in response to CR, this study indicates that mTORC1 inhibition in Paneth cells is cooperated with mTORC1 activation in ISCs to stimulate ISC expansion in response to CR, further supports the notion that SIRT1-mediated preservation of the ASC pool is a part of the longevity response to CR.
In ASCs that are maintained in quiescent state but strongly induced by injury, such as the skeletal muscle stem cells satellite cells (SCs) and adult neuron stem cells (NSCs), SIRT1 is required for the maintenance of their quiescence and regeneration capacity [70, 71, 72]. Ryall et al. showed the transition of SCs from quiescence to proliferation is accompanied with a metabolic switch from fatty acid oxidation to glycolysis despite an increased mitochondrial content [70]. This metabolic switch decreases intracellular NAD+ levels and the activity of SIRT1, leading to elevated H4K16ac and activation of muscle gene transcription. Therefore, decreased activity of SIRT1 is associated with proliferation and differentiation of SCs [70]. Consistently, skeletal muscle specific ablation of SIRT1 deacetylase domain deregulates activation of the myogenic program in SCs, leading to premature differentiation, and impairing muscle growth during development and regeneration upon injury [70]. In adult NSCs, SIRT1 also suppresses their proliferation, as specific deletion of SIRT1 in adult NSCs enhances their self-renewal [71, 72] and promotes lineage specification to oligodendrocyte progenitor cells/oligodendrocytes [73]. This impact of SIRT1 is also sensitive to metabolic switch/redox stress and involves inhibition of Hes1 transcription factor [71], Notch signaling [72], or cell metabolism and growth factor signaling [73]. It would be interesting to determine whether SIRT1 similarly impacts long-term maintenance of the NSC pool and neurogenesis in response to injury as in SCs and skeletal muscle in the future.
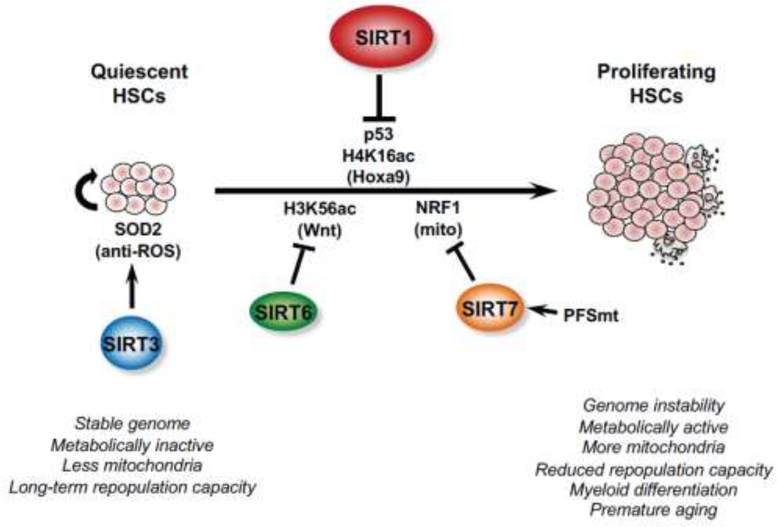
SIRT1 is also important for the maintenance of quiescence and regeneration capacity of HSCs in response to environmental stress and aging [74] (Figure 3). SIRT1 KO mice do not exhibit any abnormalities in their hematopoietic compartment under normal conditions [75]. However, SIRT1 deficient mESCs are compromised in their hematopoietic commitment in vitro and SIRT1 KO mice have decreased survival of hematopoietic progenitors ex vivo, particularly under hypoxia condition or under conditions of delayed growth factor addition [76]. Moreover, conditional ablation of SIRT1 in adult hematopoietic stem/progenitor cells (HSPCs) cell-autonomously induces HSPC expansion and loss of long-term repopulation capacity under stress [77]. This stress-induced loss of HSC function is associated with genomic instability, p53 activation and increased DNA damage in SIRT1 deficient HSPCs [77]. SIRT1 loss also causes a significant increase of H4K16ac and upregulates the expression of Hoxa9, a key regulator of HSPC function and proliferation [77]. Collectively, SIRT1 is critical to maintain diverse ASC pools through maintenance of self-renewal or quiescence and regeneration capacity, particularly in response to stress and injury.
Figure 3. Sirtuins are crucial regulators of HSC functions in response to stress and aging.
Three nuclear sirtuins, SIRT1, SIRT6, and SIRT7 are actively involved in controlling HSC quiescence, which is important to maintain their regenerative capacity and prevent premature aging. Mitochondrial SIRT3 is critical to protect HSCs from age-induced oxidative stress and is therefore indispensable for HSC self-renewal.
SIRT7 regulates quiescence and regenerative capacity of HSCs
SIRT7 is also highly expressed in HSCs to control their quiescence and regenerative capacity through regulating mitochondrial proteostasis [78] (Figure 3). HSCs are maintained in a metabolically inactive state, but are readily transformed into a metabolically active state associated with a dramatic increase of mitochondrial contents. Mohrin et al., showed that mitochondrial protein folding stress (PFSmt) induces SIRT7, which then interacts with nuclear respiratory factor 1 (NRF1), a master regulator of mitochondria, to inhibit the expression of mitochondrial ribosomal proteins and mitochondrial translation factors. Consistently, Sirt7−/− HSCs exhibit phenotypes of aging, including increased PFSmt and apoptosis, loss of quiescence, reduced repopulation capacity, and myeloid-biased differentiation. Conversely, up-regulation of SIRT7 improves the regenerative capacity of aged HSCs [78].
SIRT6 controls regeneration and stress resistance in HSCs and mesenchymal stem cells
SIRT6 is another nuclear sirtuin that inhibits cell proliferation in HSCs (Figure 3). Specific depletion of SIRT6 in adult mouse HSCs leads to a 2-fold expansion of HSPCs due to increased H3K56ac and increased transcription of factors in Wnt pathway [79]. Enhanced proliferation further renders HSCs less quiescent and more prone to depletion. Consequently, SIRT6 deficient HSCs exhibit severe impairment in their long-term repopulation capacity [79]. Together, all three nuclear sirtuins are actively involved in controlling HSC quiescence, which is important to maintain their regenerative capacity and prevent premature aging (Figure 3).
SIRT6 mediated stress resistance also helps to maintain the functionality of mesenchymal stem cells (MSCs) in vitro [80]. SIRT6 deficient human MSCs have increased ROS levels and elevated vulnerability to oxidative injury, and exhibit accelerated cell attrition after implantation to the muscles of immunodeficient mice [80]. This action of SIRT6, again, has been attributed to its deacetylation activity on H3K56ac, which directly upregulates a number of NRF2-regulated antioxidant genes by recruiting RNAP II to their promoters [80]. Although it remains unclear how a deacetylase that usually represses gene expression through histone deacetylation acts as a co-activator to promote expression of antioxidant genes in MSCs, this study indicates that SIRT6 is important for MSC stress resistance and delay of aging.
SIRT3 maintains the pool and regenerative capacity of HSCs during aging
In addition to three nuclear sirtuins, SIRT3, a mitochondrial sirtuin, is also highly enriched in HSCs [81] (Figure 3). Brown et al. showed that deletion of SIRT3 reduces the HSC pool in aged mice and compromises HSC self-renewal upon serial transplantation stress, in part due to hyperacetylation of SOD2 and subsequent increase in oxidative stress [81]. Consistently, SIRT3 is reduced with age, and overexpression of SIRT3 in aged HSCs reduces oxidative stress and rescues their reconstitution capacity [81]. Taken together, by linking diverse cellular stresses to distinct signaling pathways, sirtuins transcriptionally and post-translationally maintain HSC functions in response to stress and aging.
Concluding remarks and future perspectives
Emerging evidence reveals crucial functions of the sirtuin family of metabolic sensors in regulation of self-renewal, maintenance, pluri-/multi-potency, and differentiation of both PSCs and ASCs. Dysfunction and dysregulation of sirtuins in stem cells have been linked to a number of human diseases, including developmental defects, degenerative diseases, and cancer. Therefore, it is clear that stem cell sirtuins are at the crossroads of development, aging, and cancer.
Given the essential role of NAD+ in diverse cellular processes including energy metabolism, redox sensing, DNA repair, and genome stability [82], it is not surprising that stem cells harness sirtuin family of NAD+-dependent enzymes to tightly link their metabolic plasticity to epigenetics, transcription, genome stability, and stress resistance in response to developmental and environmental cues. However, despite their common dependence on cellular NAD+, different sirtuins display cell type-specific and/or stage-dependent impacts on stem cell biology. On the one hand, different sirtuins often have distinct even contrasting functions in the same stem cell type. For example, as nuclear sirtuins in ESCs, SIRT1 maintains pluripotency and represses differentiation genes, whereas SIRT6 suppresses pluripotency genes and is critical for their proper lineage commitment upon differentiation. This phenomenon is likely more consistent with their expression dynamics and/or substrate specificities than NAD+ sensing. On the other hand, the same sirtuin may elicit differential functional impacts in different stem cells. For instance, SIRT6 maintains the quiescent state and longterm regeneration capacity of ASCs, but suppresses the stemness of ESCs. Much work is still needed to comprehend how different members of sirtuins integrate distinct environmental cues via their differential dosages, distinct sensitivities to NAD+, and diverse substrates and interacting partners at various subcellular compartments to coordinately mediate environmental influence on stem cell activities.
It is also worth noting that mitochondrial sirtuins, SIRT3 in particular, are crucial defenders from oxidative stress in various cell types including HSCs during the process of aging, yet evidence for their contribution to the maintenance of PSCs is still lacking. This is likely due to the fact that stem cells have evolved a variety of mechanisms to protect themselves from ROS, including their preferential use of glycolysis instead of oxidative phosphorylation to reduce ROS production [30, 83] and their employments of a number of alterative mechanisms to protect against oxidative stress [84]. Consistently, PSCs such as ESCs have relatively low expression of mitochondrial sirtuins [31]. Nevertheless, given the prevalence of obesity and metabolic syndrome in the reproductive population as well as the increasing parental age in modern society, it will be interesting to study whether mitochondrial sirtuins can regulate the maintenance and survival of PSCs, particularly iPSCs and germ-line stem cells, under these oxidative stress-associated conditions.
Recent advancements in the field have shown that lysine acetylation is just one type of a widespread protein modification known as lipid lysine acylation. These short-chain and long-chain lipids are generated by completely different metabolic pathways, and distinct protein acylations are associated with differential regulation of gene transcription, particularly during germ cell development [85, 86]. Intriguingly, sirtuins have been shown to have deacylation activities to lipid lysine acylations in addition to acetylation [85, 87–89]. Further studies on sirtuin-mediated protein deacylation in stem cells will shed light on the functions of these newly discovered protein modifications in regulating stem cell biology.
Outstanding Questions.
What determines the specificities of different sirtuins at the same stage of a stem cell? How is the expression/activity of a sirtuin regulated at the different stages of a stem cell? How is the expression/activity of a sirtuin regulated in different types of stem cells? Are there any NAD+-insensitive functions of sirtuins in stem cells? How do different members of sirtuins integrate distinct environmental cues via their differential dosages, distinct sensitivities to NAD+, and diverse substrates and interacting partners at various subcellular compartments to coordinately mediate environmental influence on stem cell activities?
Whether mitochondrial sirtuins regulate the maintenance and survival of iPSCs and germ-line stem cells under oxidative stress-associated pathological conditions?
Are there any cell type-specific and/or stage-dependent impacts of protein acylations other than acetylation on stem cell functions? Whether sirtuins mediate these impacts through their deacylase activity?
As cellular metabolic and stress sensors, sirtuin family of NAD+-dependent deacylases are pivotal regulators of stem cell biology in addition to their well-known roles in metabolic diseases and aging.
Despite their common dependence on cellular NAD+, different sirtuins display cell typespecific and/or stage-dependent impacts on stem cell biology in response to various environmental cues.
Nuclear and cytosolic sirtuins modulate pluripotent stem cells and embryogenesis through regulation of pluripotency factors, metabolism, epigenetics, redox homeostasis, and cellular stress response.
Sirtuins maintain self-renewal, quiescence, and regenerative capacity of adult stem cells and protect against adult stem cell depletion in response to stress and aging.
Acknowledgements
We thank Drs. Guang Hu and Raja Jothi, and members of the Li laboratory for critical reading of the manuscript. The work related to this article was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences to X.L. (Z01 ES102205).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shore D et al. (1984) Characterization of two genes required for the position-effect control of yeast mating-type genes. Embo J 3 (12), 2817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He W et al. (2012) Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 23 (9), 467–76. [DOI] [PubMed] [Google Scholar]
- 3.Choudhary C et al. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol 15 (8), 536–50. [DOI] [PubMed] [Google Scholar]
- 4.Wagner GR and Hirschey MD (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 54 (1), 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imai SI and Guarente L (2016) It takes two to tango: NAD(+) and sirtuins in aging/longevity control. NPJ Aging Mech Dis 2, 16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frye RA (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-likeproteins. Biochem Biophys Res Commun 273 (2), 793–8. [DOI] [PubMed] [Google Scholar]
- 7.Tanno M et al. (2007) Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem 282 (9), 6823–32. [DOI] [PubMed] [Google Scholar]
- 8.Mostoslavsky R et al. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124 (2), 315–29. [DOI] [PubMed] [Google Scholar]
- 9.Ford E et al. (2006) Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20 (9), 1075–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verdin E et al. (2010) Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35 (12), 669–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Ven RAH et al. (2017) Mitochondrial Sirtuins and Molecular Mechanisms of Aging. Trends Mol Med 23 (4), 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang HC and Guarente L (2014) SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab 25 (3), 138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh A et al. (2017) The brain, sirtuins, and ageing. Nat Rev Neurosci 18 (6),362–374. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J et al. (2012) Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11 (5), 589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folmes CD et al. (2012) Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11 (5), 596–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito K and Suda T (2014) Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 15 (4), 243–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teslaa T and Teitell MA (2015) Pluripotent stem cell energy metabolism: an update. EMBO J 34 (2), 138–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang CV (2012) Links between metabolism and cancer. Genes Dev 26 (9), 87790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward PS and Thompson CB (2012) Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21 (3), 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey BW et al. (2015) Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518 (7539), 413–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalhan SC and Marczewski SE (2012) Methionine, homocysteine, one carbon metabolism and fetal growth. Rev Endocr Metab Disord 13 (2), 109–19. [DOI] [PubMed] [Google Scholar]
- 22.Locasale JW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13 (8), 572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J et al. (2009) Dependence of mouse embryonic stem cells on threonine catabolism. Science 325 (5939), 435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiraki N et al. (2014) Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19 (5), 780–94. [DOI] [PubMed] [Google Scholar]
- 25.Cai L et al. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42 (4), 426–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellen KE et al. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324 (5930), 1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19 (1), 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shyh-Chang N et al. (2013) Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339 (6116), 222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moussaieff A et al. (2015) Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21 (3), 392–402. [DOI] [PubMed] [Google Scholar]
- 30.Folmes CD et al. (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14 (2), 264–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efroni S et al. (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2 (5), 437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBurney MW et al. (2003) The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23 (1), 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng HL et al. (2003) Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A 100 (19), 10794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haigis MC et al. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126 (5), 941–54. [DOI] [PubMed] [Google Scholar]
- 35.Lombard DB et al. (2007) Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27 (24), 8807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vakhrusheva O et al. (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102 (6), 703–10. [DOI] [PubMed] [Google Scholar]
- 37.Kim HS et al. (2011) SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 20 (4), 487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez BN et al. (2016) SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J 35 (14), 1488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang S et al. (2017) Methionine metabolism is essential for SIRT1-regulatedmouse embryonic stem cell maintenance and embryonic development. EMBO J 36 (21), 3175–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coussens M et al. (2008) Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 3 (2), e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders LR et al. (2010) miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2 (7), 415–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang RH et al. (2008) Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14 (4), 312–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C et al. (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144 (3), 441–451. [DOI] [PubMed] [Google Scholar]
- 44.Tang S et al. (2014) SIRT1-Mediated Deacetylation of CRABPII Regulates Cellular Retinoic Acid Signaling and Modulates Embryonic Stem Cell Differentiation. Mol Cell 55 (6), 843–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satoh A et al. (2010) SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J Neurosci 30 (30), 10220–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Z et al. (2012) USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell 46 (4), 484–94. [DOI] [PubMed] [Google Scholar]
- 47.Bell EL et al. (2014) SirT1 is required in the male germ cell for differentiation and fecundity in mice. Development 141 (18), 3495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang ZN et al. (2014) Oct4 maintains the pluripotency of human embryonic stem cells by inactivating p53 through Sirt1-mediated deacetylation. Stem Cells 32 (1), 15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams EO et al. (2016) Sirtuin 1 Promotes Deacetylation of Oct4 and Maintenance of Naive Pluripotency. Cell Rep 17 (3), 809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heo J et al. (2017) Sirt1 Regulates DNA Methylation and Differentiation Potential of Embryonic Stem Cells by Antagonizing Dnmt3l. Cell Rep 18 (8), 1930–1945. [DOI] [PubMed] [Google Scholar]
- 51.Calvanese V et al. (2010) Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci U S A 107 (31), 13736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abdelmohsen K et al. (2007) Posttranscriptional orchestration of an anti-apoptoticprogram by HuR. Cell Cycle 6 (11), 1288–92. [DOI] [PubMed] [Google Scholar]
- 53.Han MK et al. (2008) SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell 2 (3), 241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ou X et al. (2014) SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells 32 (5), 1183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jang J et al. (2017) SIRT1 Enhances the Survival of Human Embryonic Stem Cells by Promoting DNA Repair. Stem Cell Reports 9 (2), 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Bonis ML et al. (2014) SIRT1 is necessary for proficient telomere elongation and genomic stability of induced pluripotent stem cells. Stem Cell Reports 2 (5), 690706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Etchegaray JP et al. (2015) The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17 (5), 545–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong L et al. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 140 (2), 280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain AK et al. (2016) LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol Cell 64 (5), 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang W et al. (2018) SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560 (7720), 661–665. [DOI] [PubMed] [Google Scholar]
- 61.Ferrer CM et al. (2018) An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev 32 (5–6), 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cha Y et al. (2017) Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol 19 (5), 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Si X et al. (2013) Activation of GSK3beta by Sirt2 is required for early lineage commitment of mouse embryonic stem cell. PLoS One 8 (10), e76699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishida Y et al. (2015) SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell 59 (2), 321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Signer RA and Morrison SJ (2013) Mechanisms that regulate stem cell aging and life span. Cell Stem Cell 12 (2), 152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biteau B et al. (2011) Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9 (5), 402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu B et al. (2012) Resveratrol rescues SIRT1-dependent adult stem cell decline and alleviates progeroid features in laminopathy-based progeria. Cell Metab 16 (6), 738–50. [DOI] [PubMed] [Google Scholar]
- 68.Igarashi M and Guarente L (2016) mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell 166 (2), 436–450. [DOI] [PubMed] [Google Scholar]
- 69.Yilmaz OH et al. (2012) mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486 (7404), 490–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryall JG et al. (2015) The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16 (2), 171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prozorovski T et al. (2008) Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol 10 (4), 385–94. [DOI] [PubMed] [Google Scholar]
- 72.Ma CY et al. (2014) SIRT1 suppresses self-renewal of adult hippocampal neural stem cells. Development 141 (24), 4697–709. [DOI] [PubMed] [Google Scholar]
- 73.Rafalski VA et al. (2013) Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol 15 (6), 614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seita J and Weissman IL (2010) Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med 2 (6), 640–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leko V et al. (2012) SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood 119 (8), 1856–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ou X et al. (2011) SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood 117 (2), 440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh SK et al. (2013) Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. J Exp Med 210 (5), 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohrin M et al. (2015) Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347 (6228), 1374–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H et al. (2016) SIRT6 Controls Hematopoietic Stem Cell Homeostasis through Epigenetic Regulation of Wnt Signaling. Cell Stem Cell 18 (4), 495–507. [DOI] [PubMed] [Google Scholar]
- 80.Pan H et al. (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26 (2), 190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown K et al. (2013) SIRT3 reverses aging-associated degeneration. Cell Rep 3 (2), 319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Canto C et al. (2015) NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab 22 (1), 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simsek T et al. (2010) The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7 (3), 380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tower J (2012) Stress and stem cells. Wiley Interdiscip Rev Dev Biol 1 (6), 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sabari BR et al. (2017) Metabolic regulation of gene expression through histone acylations. Nat Rev Mol Cell Biol 18 (2), 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sabari BR et al. (2015) Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol Cell 58 (2), 203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bheda P et al. (2016) The Substrate Specificity of Sirtuins. Annu Rev Biochem 85, 405–29. [DOI] [PubMed] [Google Scholar]
- 88.Tasselli L et al. (2017) SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol Metab 28 (3), 168–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anderson KA et al. (2017) SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. Cell Metab 25 (4), 838–855 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]