Abstract
The partitioning of the proteome between nucleus and cytoplasm affects nearly every aspect of eukaryotic biology. Despite this central role, we still have a poor understanding of which proteins localize in the nucleus and how this varies in different cell types and conditions. Recent advances in quantitative proteomics and high-throughput imaging are starting to close this knowledge gap. Studies on protein interaction are beginning to reveal the spectrum of cargos of nuclear import and export receptors.
We anticipate that it will soon be possible to predict each protein’s nucleocytoplasmic localization based on its importin/exportin interactions and its estimated diffusion rate through the nuclear pore. This insight is likely to provide us with a fundamental understanding of how cells use nucleocytoplasmic partitioning to encode and relay information.
Introduction
The subdivision of the cell into compartments with distinct contents and functions is one of the key hallmarks of eukaryotic biology [1]. A double-layered membrane, the nuclear envelope (NE), separates the nucleus from the rest of the cell. The NE is perforated by thousands of nuclear pore complexes (NPCs), which control the exchange of nuclear and cytoplasmic content by allowing the selective passage of some molecules, while being nearly impermeable to others. This spatial separation of biochemical pathways allows eukaryotes to have additional layers of regulation not available to prokaryotes [2]. Nucleocytoplasmic (NC) partitioning can encode cellular information, similar to protein expression levels or protein phosphorylation. For instance, the activation of the canonical Wnt signaling pathway leads to accumulation of β-catenin in the nucleus, resulting in downstream gene activation. Kinase activity can also be regulated via subcellular localization: cyclin B needs to relocalize into the nucleus to induce NE breakdown, which is required for the transition from G2 to mitosis [3]. Considering the importance of subcellular localization in encoding important cellular information, it is not surprising that mis-regulation of nuclear transport has been associated with multiple diseases, including developmental defects and cancer [4–6]. Targeting mis-regulation of NC partitioning has emerged as a promising therapeutic approach, particularly for cancer treatment [7–11].
Many previous studies have reported this subcellular localization of individual proteins. Recent technological advances in methods such as mass spectrometry (MS) now allow us to look at the entire proteome at once. This global approach allows the observation of protein localization in a wider context. This might reveal emerging principles, which are impossible to observe otherwise. For example, we might discover a set of proteins that localize in an unexpected manner, pointing towards a so far unknown nuclear localization mechanisms. In this review, we will discuss our current knowledge about NC partitioning and highlight the responsible nuclear transport mechanisms. We will focus on the proteome-wide scale and discuss the emerging technologies that might be able to close the considerable gaps that remain in our knowledge.
Which proteins constitute the nuclear proteome?
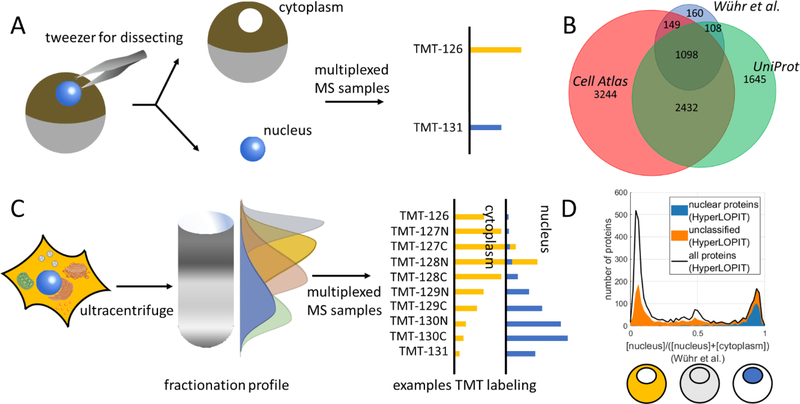
Various resources are currently available that annotate the composition of the nuclear proteome. An emerging source for subcellular localization data comes from quantitative proteomics studies. MS provides a tool to quantify relative protein abundances for thousands of proteins in a single experiment. Quantifying NC distribution with MS relies on the ability to reliably fractionate cells into the nucleus and cytoplasm. Cleanly separating nuclei from the rest of the cell is surprisingly difficult. Once a nucleus is removed from a somatic cell, the time it takes some proteins to diffuse through the nuclear pore is believed to be short compared to the time it takes for standard nuclear isolation protocols e.g., via centrifugation through a sucrose cushion [12,13]. Additionally, ruptures in the nuclear envelope resulting from cell lysis might lead to additional loss of soluble nuclear proteins. The unusually large frog oocyte (~1mm diameter) allows for the rapid and faithful isolation of the nucleus via physical methods and is therefore uniquely suited for proteomics experiments (Fig. 1A).Taking advantage of this system, we previously quantified nucleocytoplasmic partitioning for ~9k proteins in the frog oocyte with two state-of-the-art methods of quantitative proteomics [14–16]. The quantified frog proteins were then matched to human gene symbols for easy comparison to other databases.
Figure 1: What we know about the distribution of the proteome between nucleus and cytoplasm.
A) An MS-based method for proteome-wide quantification of protein partitioning between the nucleus (blue) and cytoplasm (rest of cell) in an amphibian oocyte. The exceptionally large nuclei can be easily and rapidly isolated without loss of material. With proteomics, the relative concentration of proteins in the nucleus and cytoplasm can be quantified. B) Comparison of overlap of nuclear localization prediction of human proteins from three resources. UniProt is curated from the literature, Cell Atlas predictions are based on immunofluorescence in tissue culture cells, and Wühr et al. quantified nucleocytoplasmic partitioning with quantitative proteomics in the frog oocyte, which were then matched to human genes [14,23,24]. A consensus on nuclear localization is emerging for a significant number of proteins but, for many others, the data is contradictory or missing. C) Reconstruction of subcellular localization based on crude fractionation of cell lysate e.g., HyperLOPIT [35]. Proteins from the same organelles are typically not fully separated but often show characteristic fractionation patterns. The elution profile is read out with quantitative proteomics. Based on this profile, proteins are assigned to organelles. D) Comparison of nuclear localization from physical isolation in the frog oocyte [14] or in mouse pluripotent stem cell with HyperLOPIT [35]. Shown is the histogram based on subcellular localization quantification in the frog oocytes for proteins that were measured in both studies (black). The proteins predicted to be nuclear by HyperLOPIT (blue shaded area) show remarkable agreement with the data obtained in frogs. However, for about a third of the frog nuclear proteins quantified in both studies, HyperLOPIT does not make predictions about subcellular localization (orange shaded area on top of the blue).
For many years, studies have determined the localization of individual proteins of interest via immunofluorescence, GFP-fusion proteins, or Western Blotting [17–20]. Various sources like UniProt, Gene Ontology, or LocDB have curated these studies into subcellular localization databases [21–23]. UniProt contains subcellular information for ~17k human proteins (~5k nuclear) (Fig. 1B). Equally large and widely-used as the UniProt database, Gene Ontology has the information of ~5k nuclear proteins among ~20k human entries. Another subcellular localization database, LocDB, provides subcellular localization annotations for ~13k human proteins in which ~5k of them are nuclear. Recently, the Human Cell Atlas Consortium has attempted to generalize the immunofluorescence approach by raising antibodies against every human protein [24]. So far, the consortium has been able to annotate the subcellular localization of ~12k human proteins based on immunofluorescence in human tissue culture cells. Of these, ~7k were identified as nuclear (Fig. 1B). Unlike the categorical information made available by UniProt or Cell Atlas, quantitative proteomics determines the concentration ratio between the nucleus and cytoplasm. Due to the limited sensitivity of proteomics, however, the total number of proteins for which information is available is comparatively small. When comparing the predictions from these three resources, it is apparent that a consensus of proteins constituting the nuclear proteome starts to emerge (Fig. 1B). Surprisingly, however, information about the subcellular localization of many proteins is either unavailable, limited to a single source, or is contradictory between different sources. Some of the discrepancies might be simply due to measurement errors, caused, for instance, by nonspecific antibody staining. Alternatively, it is possible, or even likely, that for many proteins, subcellular localization depends on cell type or environmental conditions. Apparent disagreement might be due to interesting biological differences.
An important question for the field is how much the nuclear proteome composition differs in various cell types and conditions. MS-based proteomics is the technique which will likely be employed to answer such questions, as it is currently the only approach that allows quantification of thousands of proteins in a single experiment at a moderate cost.
While highly valuable as a model system, the frog oocyte is unusually large and rather specialized; hence, the methods for quantification of subcellular localization cannot be fully transferred to small cells, for which nuclei cannot be isolated manually. An elegant approach to overcome the difficulty of isolating individual organelles from small cells is the use of crude fractionation to assign proteins to their compartments (Fig. 1C). This method is compatible with a wide range of cell types and potentially tissue lysates. These approaches typically use density gradient centrifugation to fractionate cell lysates. Protein complexes and organelles are separated based on their size and density. The acquired fractions are differentially labelled with isobaric tags for an MS experiment [25]. Isobaric tags like TMT barcode different conditions e.g., fractions [26]. This allows the co-analysis of multiple samples in a single experiment, which drastically improves measurement precision. Recent progress in multiplexed proteomics technology allows the accurate quantification of ~9k proteins among ~10 conditions with typical coefficients of variation of ~5% [15,27–29]. The centrifugation profile for each protein is read out with quantitative MS and proteins are assigned to compartments based on the similarity of their profiles to organelle-reference proteins [30,31]. This approach has been used to map subcellular localization in multiple eukaryotic cell lines ranging from plants [32], invertebrates [33], vertebrates [34], to mammalian cells such as mouse pluripotent stem cells [35], mouse and rat liver cells [36,37], and HeLa cells [38]. Comparison of the categorical nuclear data of one such study (HyperLOPIT)[35] with the quantitative data from the frog oocyte shows a remarkable agreement for proteins that are predicted to be nuclear (Fig. 1D). However, no predictions can be made for ~1/3 of frog nuclear proteins detected by HyperLOPIT, likely due to unassignable gradient profiles. These unassignable profiles likely result from proteins exhibiting different elution profiles than any of the reference protein sets This is likely due to (partial) disassembly of organelles during the lysis/fractionation, differential behavior of sub-organelle structures, loss of protein-protein interactions due to dilution, proteomics measurement errors, and so forth. Perhaps it will be possible to further improve HyperLOPIT-like approaches by using gentler lysis conditions or by adding complementary fractionation techniques. Another important limitation is that approaches like HyperLOPIT are currently non-quantitative. Nevertheless, it has been shown that, at least in principle, relative abundance between multiple compartments can be inferred [37].
How do nuclear transport pathways give rise to the observed nucleocytoplasmic partitioning of the proteome?
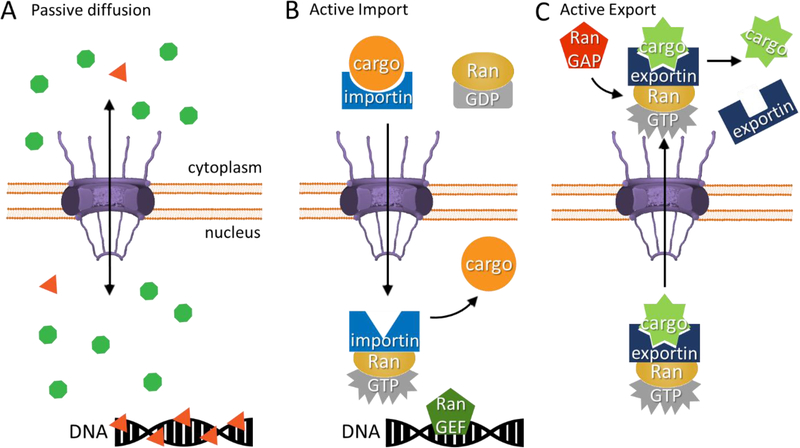
As proteins are exclusively synthesized in the cytoplasm [39], nuclear content is strictly dependent on nuclear pore passage. Based on energy consumption, one can differentiate transport through the NPC into passive diffusion and active transport. Passive diffusion involves the bidirectional and energy-free movement of molecules through NPCs. If diffusion rates are relatively fast compared to protein lifetimes, passive diffusion is expected to lead to equimolar concentrations in the nucleus and cytoplasm. However, if local binding to a structure (e.g., DNA) exclusive to the nucleus or cytoplasm acts as a sink, passive diffusion can lead to asymmetric protein concentrations (Fig. 2A). Einick and Bustin demonstrated that a histone antibody fragment heavily enriches in the nucleus even though it was inert to active nuclear transport [40]. It is unclear to what extent this mechanism is responsible for differential nucleocytoplasmic concentrations of the cell’s proteome.
Figure 2: Schematic of molecular mechanisms responsible for the establishment and maintenance of nucleocytoplasmic partitioning.
A) Small proteins are believed to be able to diffuse rapidly through the nuclear pore. Typically, this would lead to equal concentration in the nucleus and cytoplasm (green). However, with a local sink (e.g., binding to DNA) or localized assembly into large complexes, passive diffusion leads to enrichment of proteins in one compartment (orange). B) Principle of active directional transport over the nuclear pore. The restricted localization of RanGAP in the cytoplasm and RanGEF in the nucleus, in particular on the DNA, maintains a steep concentration gradient of RanGTP across the NE. Importins bind cargo in the cytoplasm. The importin-substrate complex can diffuse through the nuclear pore comparatively rapidly. In the nucleus, the exchange of RanGDP to RanGTP leads to a conformational change in the importin, resulting in the release of its cargo protein. C) Similarly, but in the opposite direction, exportin together with RanGTP binds to its cargo substrate in the nucleus and rapidly diffuses outward through the nuclear pore complex. In the cytoplasm, RanGAP hydrolyzes RanGTP and leads to the exportin’s release of its cargo.
In contrast to passive diffusion, active transport is strictly regulated, directional, and ultimately energy-dependent. This system requires continuous consumption of energy to maintain a RanGTP gradient between the nucleus and cytoplasm [41]. The gradient is controlled by the restricted localization of RanGAP (which hydrolyzes RanGTP into RanGDP) and RanGEF (which converts RanGDP into RanGTP), which leads to a steep concentration gradient of RanGTP across the NE. RanGEF is bound to the DNA to restrict its localization and results in the high concentration of RanGTP in the nucleus [42,43]. Importins and exportins acting as nuclear transport receptors utilize this gradient to selectively transport cargo into or out of the nucleus (Fig. 2B)[44]. The characteristic stretch of amino acids on cargos that allows recognition by importin is called the nuclear localization signal (NLS) and can often be predicted bioinformatically [45].
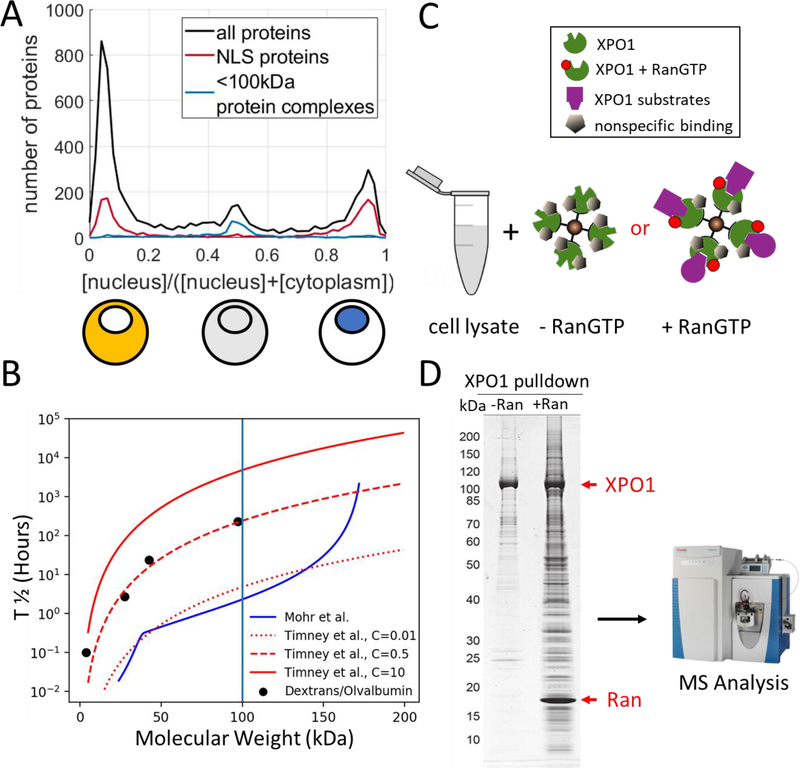
When we overlay current knowledge of nuclear transport pathways with the NC partitioning distribution in the frog oocyte, we can observe some of the expected correlation but also surprising discrepancies (Fig. 3A)[14]: ~17% of the oocyte proteins show roughly the same concentration in nucleus and cytoplasm. The majority of these proteins are in complexes less than ~100 kDa in molecular weight, suggesting that they passively diffuse through the NPC to equidistribute. However, some notable large complexes, like the proteasome, are also equidistributed. Perhaps these complexes diffuse as monomers, or they might be able to permeate through the NPC despite their size. Alternatively, they may be actively transported in both directions. Less than 60% of oocyte nuclear proteins are bioinformatically predicted to carry an NLS [45] (Fig. 3A). It is unclear if the remaining proteins in the nucleus have an undetectable NLS (e.g., cryptic 3D import signals), if there exists a localized binding sink, or if these proteins piggyback into the nucleus with others. Furthermore, nearly as many cytoplasmic proteins contain bioinformatically predicted NLSs as those in the nucleus (Fig. 3A). Among them are ribosomal proteins, which are known to be imported into the nucleus as monomers. Upon assembly into ribosomal subunits, the NLSs are hidden, and exportins transport these subunits to the cytoplasm [46]. It will be exciting to learn why so many other proteins in the cytoplasm appear to carry an apparent NLS. The open questions outlined in figure 3A can be addressed and possibly resolved by studying the contribution of nuclear transport pathways to the nucleocytoplasmic localization of the cell’s proteome.
Figure 3: How does the interplay of nuclear trafficking pathways lead to observed nucleocytoplasmic partitioning?
A) Observed nucleocytoplasmic partitioning in the frog oocyte for all detected proteins and proteins that contain a bioinformatically predicted NLS or for proteins in complexes smaller than 100 kDa [14,45]. Quantification of the concentration in the nucleus over the summed concentration in the nucleus and cytoplasm using the Xenopus oocyte model reveals a trimodal distribution for all ~9k proteins observed (black line). The majority of proteins localize nearly exclusively in either the cytoplasm or the nucleus. The majority of the equidistributed proteins were shown to be in complexes smaller than ~100 kDa and are likely to rapidly diffuse through the nuclear pore (blue line). Interestingly, proteins that are bioinformatically predicted to carry an NLS [45] are nearly as likely to be present in the nucleus or cytoplasm but very unlikely to be equidistributed (red line). B) Comparison of diffusion half-time of a few measured proteins and dextrans to the proposed models for size-dependent diffusion times from separate studies [47–50]. The black dots depict the measured diffusion halftime based on radiography of proteins or dextrans, converted into protein-molecular weight based on matching of stokes radii [47,50]. The curves are the predictions of a protein’s diffusion halftime through the NPCs: in red are the empirical power law relationship between diffusion time constant and protein size with different coefficients presented in different dashed red curves [49]. The blue line depicts how the interplay between the protein’s size and the pore complex affects the protein’s diffusion halftime in the model where the nuclear pore is modelled with three distinct pore sizes with their associated frequencies [48]. The models only poorly predict experimental diffusion times in this system. Furthermore, based on these models it is not apparent why proteins in complexes smaller than ~100 kDa (blue vertical line) are equidistributed while nearly all proteins exclusive to one compartment are in larger assemblies. C, D) Principle of identifying cargos for importin/exportin on the example of exportin 1 (XPO1) [57]. C) Cell lysate is exposed to XPO1 in the binding form (with RanGTP) or to XPO1 which cannot bind substrates. XPO1 is linked to an affinity tag, which allows easy isolation e.g., via magnetic beads. D) Shown is the coomassie gel of eluates of Exportin 1 pulldowns +/− RanGTP. Bands with equal intensities in both lanes are likely due to nonspecific binding. True substrates are expected to show significantly higher abundance in the right lane. With quantitative proteomics, the identity and relative enrichment of each of these bands and many more can be resolved. Reprinted with friendly permission from eLife [57].
Rules for passive diffusion through NPCs have been extrapolated from measurements of a handful of proteins or reporter molecules [47–49]. Based on measurements in HeLa cells, the Görlich group proposed that the passive diffusion can be modeled with a mixture of characteristic pore sizes (Fig 3B) [48]. The Rout group observed that, in yeast, GFP multimers of increasing size passively diffuse with times that relate to a power-law [49]. However, there is no molecular explanation behind this observation and many graphs follow this relationship (Fig. 3B). In figure 3B, we plotted the expected diffusion half-time for spherical proteins as a function of their molecular weight. Interestingly, the models either do not agree with previous halftime measurements, or need to be fitted with a free parameter [47,50]. Even if this free parameter is fitted, it is not obvious why proteins smaller than 100 kDa are nearly always equidistributed (Fig. 3A) whereas nearly all proteins that are exclusive to the nucleus or cytoplasm are larger than ~100 kDa [14]. One possibility is that nuclear pores in different model systems show different permeabilities. An alternative explanation is that deducing diffusion data for all proteins from just a few model substrates might not be appropriate. Recently, the Görlich lab has shown that a protein’s diffusion through NPCs can be altered drastically by modifying its surface properties [51]. Perhaps, the current models for passive diffusion were built on proteins with unusual characteristics. An important task will be to resolve the apparent discrepancies in predicted passive diffusion rate as a function of protein size and potentially incorporate proteins’ surface properties.
Regarding active transport, some studies have already tried to predict which proteins are substrates for particular transport pathways. In humans, ~20 transport receptors are known. For importin pathways, a subset of NLSs can be predicted bioinformatically [45,52,53]. However, it remains a mystery why proteins with predicted NLSs show the observed subcellular distribution (Fig. 3A). Nuclear export signal prediction algorithms have also been developed, but these predictions are even more difficult to make [54–56]. It is possible to infer the substrate transport receptor relationships with differential pulldowns in which the transport receptor is kept in the state where it either binds or does not bind substrates (Fig. 3C, D). Using this approach and quantifying the differential binding, ~1000 substrates were identified for Exportin 1, and ~250 for XPO7 [57,58]. Similar experiments with importins might be able to resolve the surprising NLS distribution shown in figure 3A. In an alternative approach, the nuclear transport receptor-substrate relationship was inferred via proximity labelling [59]. It now seems plausible to map the entire interaction network of all NTRs with their substrates. It should also be possible to estimate relative affinities of all substrates to all NTRs by combining competition studies with quantitative proteomics read-outs. These measurements will be required to predict whether multiple NTRs act on the same substrate(s), or if substrates compete for binding to an NTR. We know of individual examples where differential expression of nuclear transport receptors changes the nuclear proteome composition with drastic consequences for cellular function. For example, the lack of exportin 6 expression in the mature frog oocyte leads to actin accumulation in the nucleus, which provides the stability required for its unusually large size [60]. Knowledge of NTR expression levels could go a long way to predicting differential subcellular protein localization once we understand which NTRs interacts with which cargo.
Summary and Outlook
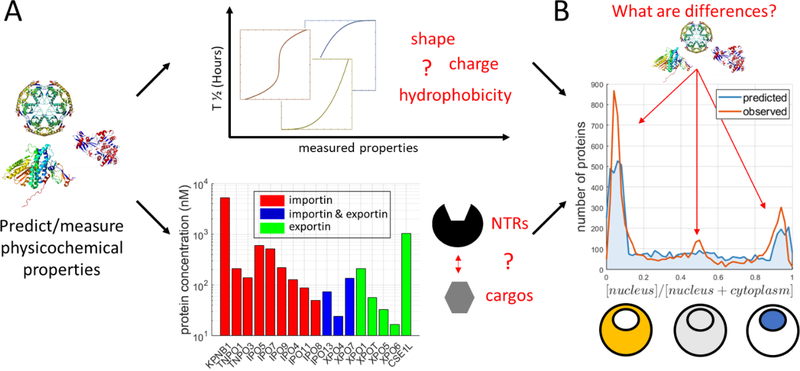
The nucleocytoplasmic distribution of many proteins is highly dynamic and often encodes critical information during development, stress responses, and general cell signaling. Thanks to emerging technologies, which allow the assignment of subcellular localization on a proteome-wide scale, we should soon be able to answer questions such as how nucleocytoplasmic partitioning changes during differentiation or in response to perturbations. These measurements might be just as informative as protein expression and phosphorylation experiments. Besides being able to measure how proteins are distributed between the nucleus and cytoplasm, an exciting challenge is to be able to untangle the underlying mechanisms responsible for this distribution. We discussed approaches to predict the rate of passive diffusion and active transport on a proteome-wide scale with molecular resolution. Data with sufficiently high quality on active transport and passive diffusion through the nuclear pore should allow precise prediction of each protein’s relative concentration between the nucleus and cytoplasm (Fig. 4). Congruence between predictions and observations would be truly satisfying. Perhaps even more exciting would be any discrepancies that could suggest other nuclear transport mechanisms we cannot currently fathom.
Figure 4: Towards an understanding of how the cell partitions its proteome between nucleus and cytoplasm on a global scale with molecular resolution.
A) We can predict many physicochemical properties of proteins from primary sequences and protein structures [61]. These properties can be correlated to their nuclear transport behaviors e.g., TOP - the size/shape of proteins (complexes) and surface properties can be converted to a model that predicts the passive diffusion time through the nuclear pore. BOTTOM - additionally, the expression levels of all importins/exportins along with the identity and affinity of their substrates can be determined bioinformatically or experimentally. B) Together, this should allow us to predict transport for each protein through NPCs and thereby predict their nucleocytoplasmic partitioning. A satisfying model would generate predictions that are very similar to actual measurements. Differences between observed and predicted partitioning could point towards a substantial lack of understanding and provide direction toward new, overlooked mechanisms.
Acknowledgement:
We would like to thank Amanda Amodeo, Thomas Güttler, and members of the Wühr Lab for helpful comments on the manuscript. N.P. was supported by NIH grant T32GM007388.This work was supported by NIH grant 1R35GM128813.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin WF, Garg S, Zimorski V: Endosymbiotic theories for eukaryote origin. Philos Trans R Soc Lond B Biol Sci 2015, 370:20140330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macgillivray AJ, Paul J, Threlfall G: Transcriptional regulation in eukaryotic cells. Adv Cancer Res 1972, 15:93–162. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Meyer AN, Donoghue DJ: Nuclear localization of cyclin B1 mediates its biological activity and is regulated by phosphorylation. Proc Natl Acad Sci U S A 1997, 94:502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paciorkowski AR, Weisenberg J, Kelley JB, Spencer A, Tuttle E, Ghoneim D, Thio LL, Christian SL, Dobyns WB, Paschal BM: Autosomal recessive mutations in nuclear transport factor KPNA7 are associated with infantile spasms and cerebellar malformation. Eur J Hum Genet 2014, 22:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noske A, Weichert W, Niesporek S, Roske A, Buckendahl AC, Koch I, Sehouli J, Dietel M, Denkert C: Expression of the nuclear export protein chromosomal region maintenance/exportin 1/Xpo1 is a prognostic factor in human ovarian cancer. Cancer 2008, 112:1733–1743. [DOI] [PubMed] [Google Scholar]
- 6.van der Watt PJ, Maske CP, Hendricks DT, Parker MI, Denny L, Govender D, Birrer MJ, Leaner VD: The Karyopherin proteins, Crm1 and Karyopherin beta1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. International journal of cancer 2009, 124:1829–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranganathan P, Yu X, Na C, Santhanam R, Shacham S, Kauffman M, Walker A, Klisovic R, Blum W, Caligiuri M, et al. : Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012, 120:1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakakibara K, Saito N, Sato T, Suzuki A, Hasegawa Y, Friedman JM, Kufe DW, Vonhoff DD, Iwami T, Kawabe T: CBS9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood 2011, 118:3922–3931. [DOI] [PubMed] [Google Scholar]
- 9.Turner JG, Dawson J, Emmons MF, Cubitt CL, Kauffman M, Shacham S, Hazlehurst LA, Sullivan DM: CRM1 Inhibition Sensitizes Drug Resistant Human Myeloma Cells to Topoisomerase II and Proteasome Inhibitors both In Vitro and Ex Vivo. J Cancer 2013, 4:614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senapedis WT, Baloglu E, Landesman Y: Clinical translation of nuclear export inhibitors in cancer. Seminars in cancer biology 2014, 27C:74–86. [DOI] [PubMed] [Google Scholar]
- 11.Dickmanns A, Monecke T, Ficner R: Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells 2015, 4:538–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paine PL, Austerberry CF, Desjarlais LJ, Horowitz SB: Protein loss during nuclear isolation. J Cell Biol 1983, 97:1240–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabbi A, Riabowol K: Isolation of Pure Nuclei Using a Sucrose Method. Cold Spring Harbor protocols 2015, 2015:773–776. [DOI] [PubMed] [Google Scholar]
- 14. **.Wühr M, Güttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, Groen AC, Presler M, Erickson BK, Mitchison TJ, et al. : The Nuclear Proteome of a Vertebrate. Current biology : CB 2015, 25:2663–2671. This work quantifies the nucleocytoplasmic partitioning of ~9k proteins in the frog oocyte using two different methods of quantitative proteomics. The unusually large cells allow easy and faithful nuclear isolation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. *.McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP: MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Analytical Chemistry 2014, 86:7150–7158. This work introduced the MultiNotch MS3 quantitative multiplexed proteomics technique, which removes measurement artifacts inherent to standard multiplexed proteomics measurements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. *.Wühr M, Haas W, McAlister GC, Peshkin L, Rad R, Kirschner MW, Gygi SP: Accurate multiplexed proteomics at the MS2 level using the complement reporter ion cluster. Analytical Chemistry 2012, 84:9214–9221. This work introduced a quantitative proteomics multiplexed technique based on the complement reporter ion cluster. Similarly to MultiNotch MS3 it removes measurement artifacts but is compatible with simpler instrumentation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giepmans BN, Adams SR, Ellisman MH, Tsien RY: The fluorescent toolbox for assessing protein location and function. Science 2006, 312:217–224. [DOI] [PubMed] [Google Scholar]
- 18.Stadler C, Rexhepaj E, Singan VR, Murphy RF, Pepperkok R, Uhlen M, Simpson JC, Lundberg E: Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat Methods 2013, 10:315–323. [DOI] [PubMed] [Google Scholar]
- 19.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK: Global analysis of protein localization in budding yeast. Nature 2003, 425:686–691. [DOI] [PubMed] [Google Scholar]
- 20.Yamauchi KA, Herr AE: Subcellular western blotting of single cells. Microsystems & nanoengineering 2017, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rastogi S, Rost B: LocDB: experimental annotations of localization for Homo sapiens and Arabidopsis thaliana. Nucleic Acids Research 2011, 39:D230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Gene Ontology C: Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017, 45:D331–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. *.UniProt Consortium T: UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018, 46:2699 UniProt currently annotates subcellular localization for ~17k human proteins by curating data from various sources. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. **.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. : A subcellular map of the human proteome. Science 2017, 356 The Human Cell Atlas Consortium maps the localization of ~12k human proteins to organelles using immunofluorescence. [DOI] [PubMed] [Google Scholar]
- 25.De Duve C: Tissue fractionation. Past and present. J Cell Biol 1971, 50:20d–55d. [PubMed] [Google Scholar]
- 26.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C: Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 2003, 75:1895–1904. [DOI] [PubMed] [Google Scholar]
- 27.Sonnett M, Yeung E, Wühr M: Accurate, Sensitive, and Precise Multiplexed Proteomics using the Complement Reporter Ion Cluster. Anal Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, Kuhn K, Pike I, Grothe RA, Blethrow JD, et al. : Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem 2012, 84:7469–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakalarski CE, Kirkpatrick DS: A Biologist’s Field Guide to Multiplexed Quantitative Proteomics. Molecular & cellular proteomics : MCP 2016, 15:1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadowski PG, Dunkley TP, Shadforth IP, Dupree P, Bessant C, Griffin JL, Lilley KS: Quantitative proteomic approach to study subcellular localization of membrane proteins. Nat Protoc 2006, 1:1778–1789. [DOI] [PubMed] [Google Scholar]
- 31.Mulvey CM, Breckels LM, Geladaki A, Britovsek NK, Nightingale DJH, Christoforou A, Elzek M, Deery MJ, Gatto L, Lilley KS: Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat Protoc 2017, 12:1110–1135. [DOI] [PubMed] [Google Scholar]
- 32.Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. : Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci U S A 2006, 103:6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan DJ, Dvinge H, Christoforou A, Bertone P, Martinez Arias A, Lilley KS: Mapping organelle proteins and protein complexes in Drosophila melanogaster. J Proteome Res 2009, 8:2667–2678. [DOI] [PubMed] [Google Scholar]
- 34.Hall SL, Hester S, Griffin JL, Lilley KS, Jackson AP: The organelle proteome of the DT40 lymphocyte cell line. Mol Cell Proteomics 2009, 8:1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. **.Christoforou A, Mulvey CM, Breckels LM, Geladaki A, Hurrell T, Hayward PC, Naake T, Gatto L, Viner R, Martinez Arias A, et al. : A draft map of the mouse pluripotent stem cell spatial proteome. Nat Commun 2016, 7:8992 This work uses subcellular protein localization assignment based on crude fractionation and quantification of elution profiles with multiplexed proteomics. The study assigns subcellular localization to ~3k proteins in mouse pluripotent stem cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster LJ, de Hoog CL, Zhang Y, Zhang Y, Xie X, Mootha VK, Mann M: A mammalian organelle map by protein correlation profiling. Cell 2006, 125:187–199. [DOI] [PubMed] [Google Scholar]
- 37.Jadot M, Boonen M, Thirion J, Wang N, Xing J, Zhao C, Tannous A, Qian M, Zheng H, Everett JK, et al. : Accounting for Protein Subcellular Localization: A Compartmental Map of the Rat Liver Proteome. Mol Cell Proteomics 2017, 16:194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itzhak DN, Tyanova S, Cox J, Borner GH: Global, quantitative and dynamic mapping of protein subcellular localization. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D: Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. The EMBO journal 2002, 21:6205–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Einck L, Bustin M: Functional histone antibody fragments traverse the nuclear envelope. The Journal of cell biology 1984, 98:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D: The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. The EMBO journal 1997, 16:6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li HY, Wirtz D, Zheng Y: A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. The Journal of cell biology 2003, 160:635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemergut ME, Mizzen CA, Stukenberg T, Allis CD, Macara IG: Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 2001, 292:1540–1543. [DOI] [PubMed] [Google Scholar]
- 44.Gorlich D, Kutay U: Transport between the cell nucleus and the cytoplasm. Annual review of cell and developmental biology 1999, 15:607–660. [DOI] [PubMed] [Google Scholar]
- 45. *.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM: NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 2009, 10:202 The authors provide an algorithm, based on hidden Markov models, which predicts NLSs based on protein sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melnikov S, Ben-Shem A, Yusupova G, Yusupov M: Insights into the origin of the nuclear localization signals in conserved ribosomal proteins. Nat Commun 2015, 6:7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. *.Paine PL, Moore LC, Horowitz SB: Nuclear envelope permeability. Nature 1975, 254:109–114. In this study, different-size dextrans were injected into amphibian oocytes to measure the permeability of the nuclear envelope. The authors conclude that the envelope is a sieve with pore size of 4.5 nm in radius. [DOI] [PubMed] [Google Scholar]
- 48. **.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D: Characterisation of the passive permeability barrier of nuclear pore complexes. The EMBO journal 2009, 28:2541–2553. Based on studying the diffusion of 10 proteins through the NPCs in HeLa cells, the authors propose to model NPC passage via diffusion through a meshwork with three characteristic pore sizes and abundance frequencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. *.Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP: Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol 2016, 215:57–76. This work used time-resolved fluorescence microscopy to measure the passive diffusion time of GFP multimers of increasing sizes in yeast. The authors found no hard size cut-off for passage, but deduced an empirical power-law relationship between diffusion time constants and proteins sizes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonner WM: Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. The Journal of cell biology 1975, 64:421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. **.Frey S, Rees R, Schunemann J, Ng SC, Funfgeld K, Huyton T, Gorlich D: Surface Properties Determining Passage Rates of Proteins through Nuclear Pores. Cell 2018, 174:202–217 e209 The authors investigate the relationship between the NPC passage rate of proteins and their surface properties. They found that exposed hydrophobic residues, as well as histidine, cysteine, and arginine facilitate, while negative charges and lysine inhibit NPC passage. [DOI] [PubMed] [Google Scholar]
- 52.Chelsky D, Ralph R, Jonak G: Sequence requirements for synthetic peptide-mediated translocation to the nucleus. Molecular and cellular biology 1989, 9:2487–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robbins J, Dilworth SM, Laskey RA, Dingwall C: Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64:615–623. [DOI] [PubMed] [Google Scholar]
- 54.Dong X, Biswas A, Chook YM: Structural basis for assembly and disassembly of theCRM1 nuclear export complex. Nature structural & molecular biology 2009, 16:558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosugi S, Yanagawa H, Terauchi R, Tabata S: NESmapper: accurate prediction of leucine-rich nuclear export signals using activity-based profiles. PLoS computational biology 2014, 10:e1003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu D, Marquis K, Pei J, Fu SC, Cagatay T, Grishin NV, Chook YM: LocNES: a computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 2015, 31:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. **.Kirli K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Gorlich D: A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. Elife 2015, 4 Kirli et al. identified ~1k cargos for Exportin 1 using a differential pulldown in which the exportin is kept in either a binding or non-binding state. Differential binding was assayed with label-free proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aksu M, Pleiner T, Karaca S, Kappert C, Dehne HJ, Seibel K, Urlaub H, Bohnsack MT, Gorlich D: Xpo7 is a broad-spectrum exportin and a nuclear import receptor. J Cell Biol 2018, 217:2329–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mackmull MT, Klaus B, Heinze I, Chokkalingam M, Beyer A, Russell RB, Ori A, Beck M: Landscape of nuclear transport receptor cargo specificity. Mol Syst Biol 2017, 13:962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bohnsack MT, Stuven T, Kuhn C, Cordes VC, Gorlich D: A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nature cell biology 2006, 8:257–263. [DOI] [PubMed] [Google Scholar]
- 61.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE: The Protein Data Bank. Nucleic Acids Research 2000, 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]