Abstract
Most animals depend upon olfaction to find food, mates, and to avoid predators. An animal’s olfactory circuit helps it sense its olfactory environment and generate critical behavioral responses. The general architecture of the olfactory circuit, which is conserved across species, is made up of a few different neuronal types including first-order receptor neurons, second- and third-order neurons, and local interneurons. Each neuronal type differs in their morphology, physiology, and neurochemistry. However, several recent studies have suggested that there is intrinsic diversity even among neurons of the same type and that this diversity is important for neural function. In this review, we first examine instances of intrinsic diversity observed among individual types of olfactory neurons. Next, we review potential genetic and experience-based plasticity mechanisms that underlie this diversity. Finally, we consider the implications of intrinsic neuronal diversity for circuit function. Overall, we hope to highlight the importance of intrinsic diversity as a previously underestimated property of circuit function.
Keywords: Olfaction, neuronal diversity, olfactory receptor neurons, interneuron, mitral cell, projection neuron
Introduction
A central theme of neuroscience, observed since the early drawings of neurons by Santiago Ramon y Cajal, has been the marked intrinsic variety of cells. In any neural circuit, distinct types of neurons interact in intricate networks to drive functional outputs. Even among neurons of the same type, neurons can differ in their morphologies, synaptic connections, molecular expression, or physiological properties (Mainen and Sejnowski 1996; Root et al. 2008; Schulz et al. 2006; Vetter et al. 2001; Yagi 2013). While individual neuronal properties have been extensively studied and some also appreciated in the context of neuronal diversity, the importance of intrinsic diversity within neurons of the same type to overall brain function has largely been ignored. However, recent studies are beginning to appreciate that this diversity has implications for brain function. Thus, knowledge of intrinsic neuronal diversity is essential to understand information processing within a neural circuit.
Most animals in the animal kingdom depend upon olfaction to find food and mates and to avoid predators. The peripheral olfactory circuit, the general architecture of which is conserved across species, is composed of a few different types of neurons based on their position in the circuit: (i) First-order olfactory receptor neurons (ORNs) expressing the same odor receptor gene project their axons onto the same glomeruli in the olfactory bulb (in mammals) (Mombaerts 2004) or antennal lobe (in insects) (Bhalerao et al. 2003; Gerber et al. 2004; Stocker 1994) (Fig 1) (Lin and Katz 2006; Wang et al. 2003). (ii) Dendrites of second-order mitral and tufted (MT) cells (in mammals) or projection neurons (PNs) (in insects) collect information from the glomeruli and carry it to higher olfactory targets in the brain, including regions that promote associations between odors and rewards or punishments, innate responses to odors etc. (Choi et al. 2011; Parnas et al. 2013; Root et al. 2014; Seki et al. 2017; Sosulski et al. 2011; Stocker et al. 1990). (iii) The peripheral olfactory circuit also includes local interneurons that are essential for information processing (Chou et al. 2010; Christensen et al. 1993; Wachowiak and Shipley 2006) (Fig 1). In general, odors bind to odor receptors expressed in ORNs (Buck and Axel 1991; Clyne et al. 1999). ORNs convert this chemical information into electrical information (Buck and Axel 1991; Couto et al. 2005; Dalton and Lomvardas 2015; Fishilevich and Vosshall 2005; Kreher et al. 2005; Kreher et al. 2008; Manzini and Korsching 2011; Sato et al. 2008; Smart et al. 2008; Wicher et al. 2008; Yao and Carlson 2010). Electrical information is encoded at various levels of an olfactory circuit to eventually affect a behavioral response (Chalasani et al. 2007; Fishilevich et al. 2005; Gomez-Marin and Louis 2014; Gomez-Marin et al. 2011; Turner et al. 2011; Wilson et al. 2004). Considerable instances of neuronal diversity are observed among each class of olfactory neurons.
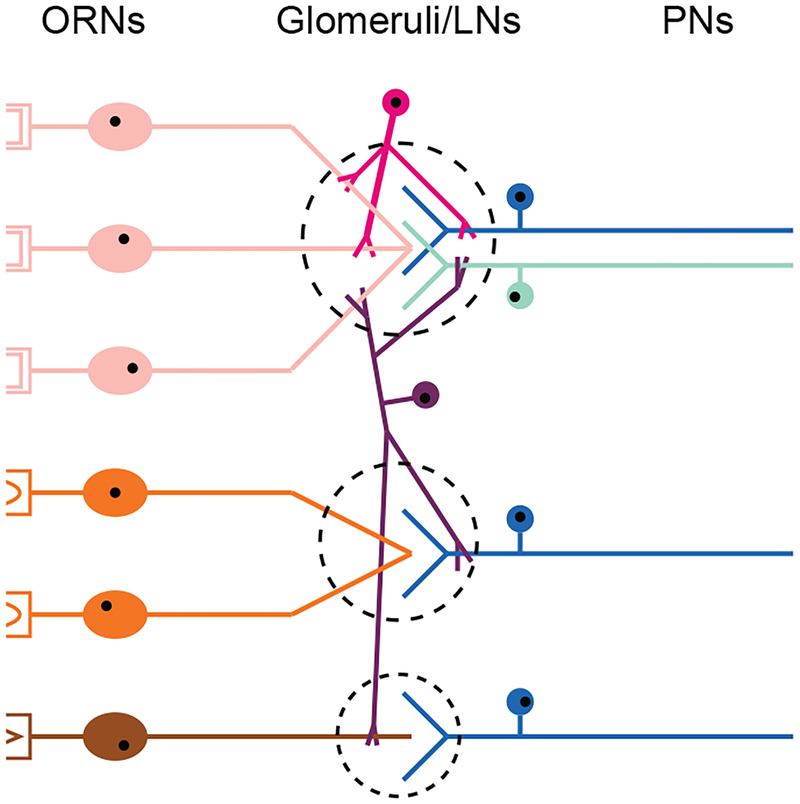
Figure 1. Intrinsic diversity in the peripheral olfactory circuit.
Schematic diagram depicting diversity among neurons of the peripheral olfactory circuit. First-order ORNs expressing the same odor receptors (shades of orange, brown) project their axons into the same glomeruli (dashed circle) within the olfactory bulb (mammals) or antennal lobe (insects). ORNs form excitatory synapses with second-order MT-cells (mammals) or PNs (insects) (shades of cyan). Second-order neurons project their axons to deeper regions in the brain. Interneurons (shades of red, violet) make intra- or inter-glomerular connections with first- and second-order neurons. Diversity is observed in the number, connectivity, and functional properties of neurons of the same type. Diversity in glomerular size is also depicted. ORN, olfactory receptor neuron; MT, mitral and tufted; PN, projection neuron.
Our knowledge of how odors are encoded in olfactory circuits relies on decades of work in different animal species in many laboratories around the world (Boeckh et al. 1987; Carlson 1996; Homberg et al. 1989; Laurent 1996; Menzel 2012; Schneider 1969; Vosshall 2000; Wilson and Mainen 2006). Circuit connections have been mapped and are currently being studied in more detail (Su et al. 2009; Vosshall and Stocker 2007; Wang et al. 1998). For instance, the complete neural connectivity within the Drosophila melanogaster larval antennal lobe, an olfactory neuropil similar to the vertebrate olfactory bulb, was recently mapped by A. Cardona and colleagues (Berck et al. 2016). The components of olfactory signal transduction in first-order ORNs have been described in insect and mammalian systems (Kato and Touhara 2009; Kaupp 2010; Nakagawa and Vosshall 2009; Silbering and Benton 2010). The responses of first-order ORNs, second-order MT cells or PNs, and local interneurons to olfactory inputs have also been described (Hallem and Carlson 2006; Kreher et al. 2008; Mathew et al. 2013; Olsen et al. 2010; Saito et al. 2009; Wang et al. 2003; Wilson and Mainen 2006). Despite this rich history and considerable progress in the cellular, molecular, and physiological understanding of olfaction, contemporary computational models of odor coding do not accurately predict animal behavior and consequently do not provide satisfactory explanations for how olfactory information is organized and processed within a circuit. While there could be several reasons for this, one reason could be the underestimation of intrinsic neuronal diversity among circuit neurons in computational models. Some of this neuronal diversity is appreciated. For instance, extensive work has been done, in various organisms, to understand the diversity of ORN responses as defined by the expression of specific odor receptors. However, the neuronal diversity that result from factors other than the differential expression of odor receptors have largely been ignored. In this review, we list several other examples of neuronal diversity observed in olfactory circuits, the potential mechanisms that underlie this diversity, and the implications of diversity for neuronal function and information coding. This review will focus on neuronal diversity observed in olfactory circuits. We acknowledge that several instances of diversity have been reported in other sensory systems as well, and we will provide few examples throughout the review as appropriate. However, a more detailed analysis of diversity observed in other sensory systems is beyond the scope of this review.
Section 1: Functional diversity among olfactory neurons of the same type
1.1. Diversity observed among first-order olfactory neurons
The olfactory system is astonishingly diverse. In olfactory systems, neuronal diversity is generated primarily by the expression of different odor receptor types (Buck and Axel 1991; Sakano 2010). The mouse olfactory system has 913 odor receptor genes that give rise to approximately a thousand neuronal types among 12 million olfactory neurons, whereas the fruit fly (D. melanogaster) olfactory system has approximately 80 odor receptor genes that give rise to 50 neuronal types among 1300 neurons in the adult (Clyne et al. 1999; Godfrey et al. 2004). In mammals and insects, individual ORNs express only one or a small number of receptors. Thus, ORN activity reflects the activity of specific odor receptors. Ultimately, the perception and discrimination of odors is based on the pattern of odor receptor activity (Hallem and Carlson 2006; Kreher et al. 2008; Malnic et al. 1999; Saito et al. 2009; Xia et al. 2008). These studies also revealed that individual odor receptors are activated by specific subsets of odors. Some receptors are broadly tuned in that they respond to many odors while others are narrowly tuned, responding to one or few odors. The primary responses of ORNs to odors also vary in their temporal dynamics. Some ORN responses to odors may be of short-duration, lasting the duration of the stimulus, while others may be of long-durations, in some cases in the order of minutes after the end of stimulation (Hallem et al. 2004; Montague et al. 2011). Here, we argue that neuronal functional diversity extends beyond the differential expression of odor receptors in ORNs.
Beyond the differential expression of odor receptors, ORNs may differ in the expression levels of neurotransmitter receptors. Such an example of diversity among olfactory neurons was provided by Root and colleagues. They showed that each D. melanogaster ORN has a unique baseline level of GABABR expression. ORNs that sense the aversive odor CO2 do not express GABABRs and do not have significant presynaptic inhibition. In contrast, pheromone-sensing ORNs express high levels of GABABRs and exhibit strong presynaptic inhibition (Root et al. 2008).
An important question in the study of olfactory systems relates to the equivalency with which its primary sensory neurons drive behavioral outputs. One hypothesis is that different sensory neurons expressing different receptors might drive equivalent behavioral responses, especially in a simple sensory system. The alternate hypothesis is that different neurons drive different behavioral responses, particularly if the connectivity and downstream processing are complex (Masse et al. 2009; Su et al. 2009; Wilson and Mainen 2006). Mathew and colleagues showed that each Drosophila larval ORN, upon activation, drove different behavioral outputs. To carry out this experiment, they first performed a large-scale electrophysiology screen to identify “private odors” for each larval ORN. Each private odor elicited specific physiological response from a single ORN. They found that some private odors elicited strong physiological responses but weak behavioral responses while others elicited weak physiological responses but strong behavioral responses. One implication of these results is that each ORN contributes differentially to the olfactory circuit (Mathew et al. 2013). The mechanisms that underlie this functional diversity among ORNs remain unclear. Hernandez-Nunez and colleagues showed that individual ORNs in the D. melanogaster larva, upon activation, could produce behavioral responses with distinct dynamics. The D. melanogaster larva chemosensory system, in addition to encoding attractant and repellent responses, is also capable of shaping the dynamics of behavioral responses in ecologically important ways. So, behavioral priorities may not only include relative degrees of attraction or repulsion but also the speed of the response or adaptation (Hernandez-Nunez et al. 2015). Related to this, Munch and Galizia systematically analyzed temporal dynamics of ORN responses in adult D. melanogaster. Their studies showed that there is temporal diversity in the response of ORNs to various odor combinations. This ability of olfactory neurons to alter the temporal dynamics of their response greatly increases the coding capacity of the system (Munch and Galizia 2017).
1.2. Diversity observed among second-order olfactory neurons
Second-order projection neurons (PNs) in the Drosophila antennal lobe show diversity in their connectivity with ORNs (Tobin et al. 2017). There is a systematic co-variation in synapse number and PN dendrite size. In glomerulus with fewer PNs, PN dendrites are larger but there was also a compensatory increase in synapse numbers (Grabe et al. 2016; Tanaka et al. 2004; Tobin et al. 2017). PNs receive 35% more synapses from ipsilateral ORNs and the connections are 30–40% stronger compared to connections with contralateral ORNs. However, no sizeable difference in synaptic conductance was observed among the ipsilateral and contralateral connections. The authors argue that each connection is composed of random distributions of dendritic filtering properties that filter these conductance and so the average effect is similar across connections (Tobin et al. 2017). While the diversity in PN dendritic connections may provide a structural basis for some phenomenon such as odor lateralization behavior. Homeostatic mechanisms, on the other hand, may counteract this structural diversity and together they likely impact network performance.
Each glomerulus in the mammalian olfactory bulb is a functional unit that collects input from ORNs that express the same type of olfactory receptor gene (Bozza et al. 2002; Mombaerts et al. 1996). Each glomerulus provides exclusive excitatory inputs to a set of 10–20 second-order mitral cells. Mitral cells are equivalent to second-order projection neurons in the insect antennal lobe. Sister MT cells that receive input from the same genetically identified glomerulus exhibit diversity in their temporal patterns of responses (Arneodo et al. 2018; Dhawale et al. 2010; Kikuta et al. 2013). Whether sister MT cells respond stereotypically or in a diverse manner to the stimulus depends on the stimulus. Stimuli with high ligand affinity to the primary sensory neurons elicit stereotypic responses from the sister MT cells. In contrast, ligands with lower affinity elicit diverse temporal responses from the sister MT cells (Arneodo et al. 2018). Thus, information about ligand affinity is potentially encoded in the collective stereotypy or diversity of activity among sister MT cells.
MT cells of the mouse olfactory bulb exhibit intrinsic biophysical diversity (Friedrich and Laurent 2001; Padmanabhan and Urban 2010). Each cell’s spiking response was based on its unique biophysical fingerprint because a neuron’s response to incoming stimuli is shaped by the voltage-gated ion channels expressed in that cell (Taylor et al. 2009). One source of intrinsic diversity in MT cell population is believed to be the differential expression of the voltage-gated potassium channel Kv1.2 subunit (Padmanabhan and Urban 2010). The suggestion is that this intrinsic heterogeneity in responses allows a diverse population of neurons to code for twofold more information than their homogenous counterparts.
1.3. Diversity observed among local interneurons.
Local interneurons are critical for information processing in neural circuits. Olfactory local interneurons (LNs) in the D. melanogaster antennal lobe are highly diverse and variable (Chou et al. 2010; Huang et al. 2010; Nagel and Wilson 2016; Seki et al. 2010; Shang et al. 2007; Wilson and Laurent 2005). Whole cell patch-clamp recordings from adult D. melanogaster LNs revealed several different types of LNs (Chou et al. 2010; Seki et al. 2010). Each type of LN exhibited characteristic electrophysiological properties in terms of spontaneous firing, firing patterns, degree of spike adaptation, and spike amplitudes. LN types also differed in their morphology in terms of dendritic density and distribution within specific antennal lobe glomeruli, in their neurotransmitter types, and odor response properties (Chou et al. 2010; Seki et al. 2010). A more recent study showed that only a subset of larval LNs are selectively reintegrated into the adult circuit via pruning and re-extension mechanisms, while others die during metamorphosis (Liou et al. 2018). Thus, LNs exhibit high levels of plasticity and execute complicated genetic programs with precise temporal control, which could all result in their morphological and functional diversity. Such diversity should translate into variability in the functional connectivity of LNs, ORNs, and PNs. Notably, interneurons in the mammalian olfactory cortex are also remarkably varied (Kay and Brunjes 2014; Miles 2000).
Prolonged exposure to CO2 in fruit flies led to a reversible volume increase in the CO2-specific glomerulus. While neither morphology nor function of ORNs were affected in this case, one type of inhibitory local interneurons showed significantly increased responses to CO2 (Sachse et al. 2007). Thus, when flies are exposed to elevated CO2 levels early in life, the chronic exposure of CO2 sensing neurons promotes functional changes in the LN2 subtype of inhibitory local interneurons, which leads to functional diversity among sub-populations of inhibitory local interneurons. This further impacts the functional output of the downstream projection neurons (Sachse et al. 2007).
Clear examples of functional diversity are also observed among inhibitory local interneurons in other sensory systems (Kim et al. 2017; Markram et al. 2004). The mammalian visual cortex contains various types of GABAergic neurons exerting local inhibition. These neurons show several common features but also considerable diversity in their genetic, morphological, as well as physiological properties. Genetically, most of the cortical GABAergic interneurons express one or more of three different genetic markers: parvalbumin (PV), somatostatin (SST), and 5-HT receptor 3A (5-HT3R). GABAergic neurons expressing each genetic marker show distinct characteristic features and form synapses with specific targets. Morphologically, the interneurons can be classified as large, small, or nest basket cell. Each morphological type can still show varying electrical or molecular properties (Wonders and Anderson 2006). Physiologically, these neurons show several types of response properties including stuttering, irregular spiking, and bursting (Stiefel et al. 2013). The neuronal response properties are believed to arise due to different combinations of ion channel expression as well as morphology. It was suggested that because of diversity in expression of genetic markers, synaptic connections, and response properties, different GABAergic interneurons may play different roles during visual processing. Indeed, selective activation of PV+ GABAergic interneurons resulted in improvement of orientation selectivity of V1 neurons. Selective activation of SST+ GABAergic interneurons did not improve orientation selectivity of V1 neurons (Atallah et al. 2012; Lee et al. 2012). On the other hand, SST+ neurons and not PV+ neurons were found to mediate surround suppression of V1 response to the center of the receptive field (Adesnik et al. 2012). The principles by which this functional diversity of GABAergic interneurons in the visual cortex is organized to help modulate sensory processing remain to be understood.
1.4. Diversity observed deeper in the olfactory circuit
Approximately 2000 third-order olfactory neurons (Kenyon cells) in the D. melanogaster mushroom body converge on to a population of only 34 mushroom body output neurons (MBONs). The MBONs fall in to 21 anatomically distinct cell types (Aso et al. 2014; Tanaka et al. 2008). At the single cell level, uniquely identifiable MBONs show identical tuning across the two hemispheres of an animal but very different tuning across different animals (Hige et al. 2015). The authors also showed that this individualization of MBONs is an active process that requires the learning-related gene, rutabaga. This plasticity-driven individualization of MBONs lies in contrast to the highly stereospecific responses observed in the lateral horn (LH). The LH, which is also part of the olfactory circuit, lies in parallel to the mushroom body and is implicated in the processing of innate responses to odors (Fisek and Wilson 2014; Heimbeck et al. 2001). Functional individuality in MBONs seen at this level of the sensory circuit provides the ability to fine tune an odor representation or even override innate responses driven by the LH and thus allows each individual to have a unique olfactory experience.
Section 2: Mechanisms that contribute to functional diversity among neurons of the same type
2.1. Genetic mechanisms for neuronal diversity
In sensory systems, an obvious mechanism that contributes to functional differences among neurons of the same type is the expression of different receptor types. In the D. melanogaster olfactory system, selective expression of typically a single odor receptor gene from a repertoire of approximately 80 possible genes generates 50 different types of ORNs (Li et al. 2016). Beyond this, other mechanisms that contribute to functional diversity among neurons of the same type include differential expression of cell adhesion molecules and ion channels. Differential expression of genes in individual cells can be generated by independent and stochastic mechanisms. For instance, the cell adhesion proteins, protocadherins (Pcdhs) are stochastically expressed in individual neurons. This random expression is generated by epigenetic regulation in the genome during development (Ribich et al. 2006). Pcdhs have an important role in the precise building of neural circuits. While Pcdhs are only one example, the extensive molecular diversity of neuronal cell-surface proteins affects neuron’s individual properties and connectivity. Differential expression of cell-surface molecules provides a genetic basis for neuronal individuality (Yagi 2013).
Theoretical studies predict different combinations of conductance densities of voltage-dependent currents and synaptic strengths in networks carrying out the same function in different animals (Golowasch et al. 2002; Prinz et al. 2004). One of the implications of these predictions is that the expression of ion channel genes may differ in identified neurons in different animals. Schulz and colleagues tested this hypothesis by measuring multiple ion channel expression levels in single, unambiguously identified neurons in the stomatogastric nervous system of C. borealis crabs, and comparing the values both within and between animals. They found that neurons with similar functional output varied two- to four-fold in terms of both mRNA abundance and current (Schulz et al. 2006). In further support of this hypothesis, differential expression of voltage-gated potassium channel Kv1.2 subunit is believed to underlie the intrinsic diversity observed in MT cells in mice (Padmanabhan and Urban 2010).
The functional and transcriptional complexity of neurons can also be linked to their underlying epigenomic diversity. While the molecular activity of a neuron at any given time is based on its gene expression patterns, information about the neuron’s gene regulatory mechanisms, developmental origins, and potential future responses are based on its epigenomic information. Epigenomic signatures include accessible regions of chromatin, histone modifications, and DNA methylation (Bird 2002; Heintzman et al. 2007; Thurman et al. 2012). A recent study showed that morphological and physiological diversity of neocortical neurons is related to widespread differences in their underlying epigenomes (Mo et al. 2015). Mo and colleagues find relationships between distinctive hyper-methylation patterns and developmentally transient expression of critical transcription factors that shape neuronal subtype identity. They showed that epigenomic states of adult neurons capture long-lasting attributes of neuronal identity including patterns of gene expression and potential experience-dependent responses. Studies in Drosophila have revealed conserved developmental strategies along with epigenetic mechanisms that are used to generate sensory neuron diversity. During development, a network of transcription factors coordinate and pattern the developing olfactory tissue to modulate the eventual level of ORN diversity (Li et al. 2016; Li et al. 2013). In nascent Drosophila ORNs, Hamlet, a proto-oncogene homolog directs locus-specific chromatin modifications, which generates context-dependent responses of Notch signaling (Endo et al. 2012). Notch signaling is known to influence odor receptor gene choice and ORN axon targeting specificity (Endo et al. 2007). Odor receptor gene choice is also influenced by evolutionarily conserved gene regulatory elements that direct the expression of the adjacent odor receptor genes in subsets of ORN classes, by elements that restrict Or gene expression to a single ORN class, and by a combinatorial code of transcription factors that generate the receptor-to-neuron map (Ray et al. 2007; Ray et al. 2008).
While heterogeneity in electrical responsivity of neurons can be attributed to variability in the types and densities of ion channels, the neuronal firing patterns can also be influenced by dendritic geometry. Careful in vitro recordings from somatosensory neurons that each had similar channel distributions in their soma and axons but different dendritic architecture revealed differences in intrinsic firing patterns, adaptation speed (Mainen and Sejnowski 1996), or action potential propagation (Vetter et al. 2001). Thus, altering neuronal geometry can influence neuronal functional diversity and neuronal function (Mainen and Sejnowski 1996; Schaefer et al. 2003; Vetter et al. 2001). A recent study in C. elegans showed that levels of expression of a transmembrane Ig domain protein, OIG-8 can instruct the extent of olfactory cilia membrane elaborations. OIG-8 protein localizes to ciliated endings of olfactory neurons and is expressed at different levels in individual neurons. Low expression levels of oig-8 correspond to less branching patterns of cilia while high expression levels correspond to more elaborate branching patterns (Howell and Hobert 2017).
2.2. Experience-dependent plasticity mechanisms for neuronal diversity
Sensory processing often depends on an individual’s experience. This is clearly evident in studies that showed that certain neurons’ odor responses differ between individual animals but remain consistent across brain hemispheres of the same individual (Hige et al. 2015). We note that past experience impacts gene expression in first-order sensory neurons as well as in second and third order neurons in the circuit (Santoro and Dulac 2012; Xu et al. 2016; Zhao and Reed 2001).
Neuronal diversity can be established due to varying levels of adaptation of sensory neurons to different odors. Adaptation generally occurs after prolonged exposure to an odor, which results in a diminished response to the odor for a prolonged period of time. Adaptation involves a genetically complex mechanism that differs from the mechanisms underlying odor sensation (Zufall and Leinders-Zufall 2000). Chronic exposure to odors has also been shown to activate the Notch signaling pathway in olfactory neurons. Thus, environmental stimuli differentially modulate the pattern of Notch activity in the ORNs. This differential modulation of Notch activity was shown to depend on odor receptor activity, ORN synaptic transmission, as well as activity of local interneurons in the antennal lobe (Lieber et al. 2011). Notch signaling impacts both physiological and morphological plasticity of olfactory neurons. This activity dependent plasticity in ORNs regulated by Notch signaling could establish diversity among neurons (Kidd and Lieber 2016; Kidd et al. 2015). Sensory stimuli fluctuate in a wide range of background conditions. So, the ability of a sensory system to modulate neuron sensitivity becomes critical to expand the dynamic range of sensory input. A recent study identified a conserved phosphorylation site in Orco that is important for modulating odor receptor sensitivity to background odors. Orco is a co-receptor expressed in every insect ORN. In each ORN, an odor receptor and Orco together form a ligand-gated ion channel (Sato et al. 2008; Smart et al. 2008; Wicher et al. 2008). Prolonged odor-exposure leads to a steady-state dephosphorylation at this conserved site in Orco, which reduces odor-sensitivity (Guo et al. 2017). This modulation of sensitivity enables the animal to carry out important food-seeking behaviors in the presence of background odors. Such a mechanism could also explain why mosquitoes pre-exposed to a repellent such as DEET (N,N-diethyl-meta-toluamide) show reduced subsequent repellency to DEET (Stanczyk et al. 2013).
The numbers of specific odor receptor expressing ORNs and their synaptic connections remain plastic in the adult mammalian olfactory system. When mice were trained in a learning paradigm with an odor specific for M71 odor receptor, there was a significant increase in M71-specific ORNs within the nose of trained mice compared to untrained mice. This phenomenon was observed when the odor was paired in both aversive as well as appetitive conditioning paradigms (Jones et al. 2008). In the mouse olfactory bulb, prenatal and early postnatal exposure to odors increases the number, amplitude, and reliability of second-order MT cell responses (Liu and Urban 2017). Similarly, in the D. melanogaster antennal lobe, prolonged exposure to CO2 led to reversible functional changes in the LN2 subtype of inhibitory local interneurons (Sachse et al. 2007). Because second-order neurons and inhibitory local interneurons are important for olfactory bulb/antennal lobe outputs and in mediating lateral inhibitory networks, experience-based changes in the number and response properties of specific groups of olfactory neurons will add to the diversity already observed among them and significantly impact odor representations.
Section 3: Implications of neuronal diversity for circuit function
Although we have laid out several examples of neuronal diversity throughout the olfactory system, its importance remains a source of debate. It is unclear whether intrinsic differences among neurons of the same type or even same molecular type impact a neuron’s function to an extent that it affects the system’s ability to encode information. Are biophysical differences among neurons simply a reflection of the probabilistic nature of gene expression among different cells or could this diversity have functional relevance in neuronal function and coding? Several studies suggest the latter.
Variability in the expression of several channel genes in individual neurons of the same cell type is an important mechanism that leads to neuronal diversity. As described in Section 2 above, the variability in ion channel expression among neurons of the same type could arise in part due to experience-dependent plasticity mechanisms, as well as genetic variability in wild populations. Individual neurons of the same type take advantage of this range of variability in the expression of channel genes and use a variety of tuning rules that enable them to perform adequately in the networks in which they are found (Schulz et al. 2006). One implication of these results is that individual neurons can find different solutions within a multidimensional parameter space that are consistent with their characteristic intrinsic properties. Similar to variability in distributions of ion channels, variability in the distribution of neurotransmitter receptors can also play an important role in neuronal function. A clear example of this is seen in the case of GABABR receptor expression levels at the terminals of D. melanogaster ORNs. GABABR expression expands the dynamic range of ORN transmission. Each ORN has a unique level of GABABR expression levels. ORNs that sense the aversive odor, CO2 do not express GABABRs and do not have significant presynaptic inhibition. This allows flies to rapidly respond and escape from the aversive odor. In contrast, ORNs that sense pheromones express high levels of GABABRs and have significant presynaptic inhibition. This allows flies to be highly selective about pheromone-dependent mate localizations (Root et al. 2008). Thus, different olfactory receptor channels employ different gain control as a mechanism to allow an animal’s innate behavior to match its ecological needs. Could variable expression of GABABRs at the terminals of sensory neurons potentially allow for differential gain control based modulation of individual sensory neurons by the internal state of the animal? This is an important question that needs to be addressed in the future.
While ion channel and neurotransmitter receptor distributions are important factors, several studies concluded that neurons have high levels of variability in morphology and geometry, which are factors that can impact neuronal function (Mainen and Sejnowski 1996; Schaefer et al. 2003; Vetter et al. 2001). Because there is a wide variety in dendritic geometry among neocortical neurons, the ability of different ion channel distributions as well as dendritic shape to impact electrical properties of the neuron support the idea that there is a continuous spectrum of firing patterns rather than discrete categories. Morphological diversity of neurons can also influence the integration of information within neurons. A study conducted on L5 pyramidal neurons revealed that variation in dendritic arborizations is a key factor that tunes coincidence detection in the cells. Simulation studies showed that addition of oblique branches of the main apical dendrite in close proximity to the soma increases the coupling between the apical and axosomatic action potential initiation zones while incorporating distal branches reduced this coupling (Schaefer et al. 2003). Thus, structural plasticity can impact the rules for dendritic integration of multilayer synaptic input to neocortical cells.
Padmanabhan and Urban found that intrinsic diversity among neurons can be important for neural coding. Their study suggests that intrinsic biophysical diversity of neurons influences the correlated activity often observed among a population of neurons (Padmanabhan and Urban 2010). Correlated activity of neurons has been identified as being crucial in a number of systems including the olfactory bulb (Laurent et al. 2001). Intrinsic diversity among mitral cells could reduce the degree to which cell firing is correlated even when the incoming ORN excitatory inputs are very similar and thereby, increase the ability of the animal to discriminate among similar odors. The intrinsic diversity may also influence the extent to which mitral cells can be synchronized by inhibition. Overall, the study found that the coding capacity of populations of biophysically heterogeneous mitral cells was twofold higher than that of their homogenous counterparts (Padmanabhan and Urban 2010).
In natural environments, animals encounter different temporal patterns of odor stimuli. D. melanogaster ORNs show diversity in their temporal response dynamics (Hernandez-Nunez et al. 2015; Mathew et al. 2013; Montague et al. 2011; Munch and Galizia 2017; Nagel and Wilson 2011). A systematic analysis of the temporal dynamics of ORNs revealed that the response dynamics of an ORN depend on the combination of sensory neuron and odor (Munch and Galizia 2017). Several mechanisms can potentially contribute to this diversity in temporal responses. This could be because different odors have different physical properties of adsorption that could naturally occur at the cuticular surface of animals leading to odors that linger for longer, and others that do not (Martelli et al. 2013) or due to the influence of sensillar events that transport odor molecules from the surface to the ORN dendrites (Kaissling 2013; Larter et al. 2016). Properties of the odor and odor receptor interactions such as binding affinity, receptor saturation, allostery etc. could impact response time courses (Kenakin 2017). Elements of the G-protein dependent signal transduction cascade could also be differentially expressed in individual neurons and could potentially contribute to different response dynamics (Raja et al. 2014; Yao and Carlson 2010). It has been theorized that adding information about the temporal development of an odor response across ORNs significantly increases the coding capacity of the system and ability of the olfactory system to identify the stimulus.
Concluding Remarks
In this review, we examined instances of neuronal diversity observed among different types of olfactory neurons, we listed potential mechanisms that could lead to such diversity, and finally we reviewed some of the functional implications that arise because of this diversity. While the list of neuronal diversity examples provided in this review is not exhaustive, we hoped to highlight its importance as a previously underestimated property of circuit function.
Overall, we note that functional diversity exists at various levels of the olfactory circuit. Both genetic mechanisms and experience-dependent plasticity mechanisms contribute to the physical and functional diversity observed among neurons of the same type. While the significance of this type of diversity remains a source of debate, several studies have suggested that neuronal diversity play important roles during brain function, especially in the integration and coding of environmental information in neural circuits. This knowledge challenges contemporary conventions of how olfactory neuron activity is mathematically incorporated in odor coding models and provides opportunities to fine-tune computational models that aim to reliably predict animal behavior. Thus, systematic identification and characterization of diversity among neuronal classes within a circuit are a prerequisite to understanding circuit function. Going forward, this would require innovations in the ability to collect precise data on individual neuronal contributions to the circuit as well as in approaches to incorporate their diversity into computational models. Future studies focused on molecular mechanisms underlying neuronal diversity and their role in ensuring circuit function could also have the potential to impact studies on host-seeking behavior in animals, particularly harmful insects that transmit diseases to humans.
It is only appropriate to start as well as end a review on neuronal diversity by mentioning Ramon y Cajal. The idea of mapping circuits can be traced back to Cajal. Detailed neuronal wiring diagrams are important – although in no way sufficient – prerequisites to understand how brain functions underlie behavior and how brain malfunctions underlie disease (Lichtman and Sanes 2008). Information about diversity among similar neuronal types poses a challenge in how we represent this diversity in wiring diagrams in a way that will impact our basic understanding of neural circuit layout and function.
Acknowledgements:
The authors are supported by a grant from the NIGMS of the National Institute of Health under grant number P20 GM103650 and by startup funds from the University of Nevada, Reno awarded to DM.
Footnotes
Conflict of Interest:
The authors declare no competing financial interests.
References:
- Adesnik H, Bruns W, Taniguchi H, Huang ZJ, Scanziani M (2012) A neural circuit for spatial summation in visual cortex Nature 490:226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneodo EM et al. (2018) Stimulus dependent diversity and stereotypy in the output of an olfactory functional unit Nature communications 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y et al. (2014) The neuronal architecture of the mushroom body provides a logic for associative learning eLife 3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah BV, Bruns W, Carandini M, Scanziani M (2012) Parvalbumin-Expressing Interneurons Linearly Transform Cortical Responses to Visual Stimuli Neuron 73:159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berck ME et al. (2016) The wiring diagram of a glomerular olfactory system eLife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao S, Sen A, Stocker R, Rodrigues V (2003) Olfactory neurons expressing identified receptor genes project to subsets of glomeruli within the antennal lobe of Drosophila melanogaster J Neurobiol 54:577–592 [DOI] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory Genes & development 16:6–21 [DOI] [PubMed] [Google Scholar]
- Boeckh J, Ernst KD, Selsam P (1987) Neurophysiology and neuroanatomy of the olfactory pathway in the cockroach Annals of the New York Academy of Sciences 510:39–43 [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P (2002) Odorant receptor expression defines functional units in the mouse olfactory system Journal of Neuroscience 22:3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition Cell 65:175–187 [DOI] [PubMed] [Google Scholar]
- Carlson JR (1996) Olfaction in Drosophila: from odor to behavior Trends Genet 12:175–180 [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, Goodman MB, Bargmann CI (2007) Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans Nature 450:63–70 [DOI] [PubMed] [Google Scholar]
- Choi GB, Stettler DD, Kallman BR, Bhaskar ST, Fleischmann A, Axel R (2011) Driving Opposing Behaviors with Ensembles of Piriform Neurons Cell 146:1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YH, Spletter ML, Yaksi E, Leong JC, Wilson RI, Luo L (2010) Diversity and wiring variability of olfactory local interneurons in the Drosophila antennal lobe Nature neuroscience 13:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen TA, Waldrop BR, Harrow ID, Hildebrand JG (1993) Local Interneurons and Information-Processing in the Olfactory Glomeruli of the Moth Manduca-Sexta J Comp Physiol A 173:385–399 [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR (1999) A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila Neuron 22:327–338 [DOI] [PubMed] [Google Scholar]
- Couto A, Alenius M, Dickson B (2005) Molecular, anatomical, and functional organization of the Drosophila olfactory system Current biology: CB 15:1535–1547 [DOI] [PubMed] [Google Scholar]
- Dalton RP, Lomvardas S (2015) Chemosensory Receptor Specificity and Regulation Annual Review of Neuroscience, Vol 38:331–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF (2010) Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse Nature neuroscience 13:1404–U1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo K, Aoki T, Yoda Y, Kimura KI, Hama C (2007) Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages Nature neuroscience 10:153–160 [DOI] [PubMed] [Google Scholar]
- Endo K et al. (2012) Chromatin modification of Notch targets in olfactory receptor neuron diversification Nature neuroscience 15:224–233 [DOI] [PubMed] [Google Scholar]
- Fisek M, Wilson RI (2014) Stereotyped connectivity and computations in higher-order olfactory neurons Nature neuroscience 17:280–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E, Domingos AI, Asahina K, Naef F, Vosshall LB, Louis M (2005) Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila Current biology: CB 15:2086–2096 [DOI] [PubMed] [Google Scholar]
- Fishilevich E, Vosshall LB (2005) Genetic and functional subdivision of the Drosophila antennal lobe Current biology: CB 15:1548–1553 [DOI] [PubMed] [Google Scholar]
- Friedrich RW, Laurent G (2001) Dynamic optimization of odor representations by slow temporal patterning of mitral cell activity Science 291:889–894 [DOI] [PubMed] [Google Scholar]
- Gerber B, Scherer S, Neuser K, Michels B, Hendel T, Stocker RF, Heisenberg M (2004) Visual learning in individually assayed Drosophila larvae The Journal of experimental biology 207:179–188 [DOI] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, Buck LB (2004) The mouse olfactory receptor gene family Proceedings of the National Academy of Sciences of the United States of America 101:2156–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowasch J, Goldman MS, Abbott LF, Marder E (2002) Failure of averaging in the construction of a conductance-based neuron model Journal of neurophysiology 87:1129–1131 [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A, Louis M (2014) Multilevel control of run orientation in Drosophila larval chemotaxis Frontiers in behavioral neuroscience 8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Marin A, Stephens GJ, Louis M (2011) Active sampling and decision making in Drosophila chemotaxis Nature communications 2:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe V, Baschwitz A, Dweck HKM, Lavista-Llanos S, Hansson BS, Sachse S (2016) Elucidating the Neuronal Architecture of Olfactory Glomeruli in the Drosophila Antennal Lobe Cell Reports 16:3401–3413 [DOI] [PubMed] [Google Scholar]
- Guo H, Kunwar K, Smith D (2017) Odorant Receptor Sensitivity Modulation in Drosophila Journal of Neuroscience 37:9465–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR (2006) Coding of odors by a receptor repertoire Cell 125:143–160 [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR (2004) The molecular basis of odor coding in the Drosophila antenna Cell 117:965–979 [DOI] [PubMed] [Google Scholar]
- Heimbeck G, Bugnon V, Gendre N, Keller A, Stocker RF (2001) A central neural circuit for experience-independent olfactory and courtship behavior in Drosophila melanogaster Proceedings of the National Academy of Sciences of the United States of America 98:15336–15341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND et al. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome Nat Genet 39:311–318 [DOI] [PubMed] [Google Scholar]
- Hernandez-Nunez L, Belina J, Klein M, Si G, Claus L, Carlson JR, Samuel AD (2015) Reverse-correlation analysis of navigation dynamics in Drosophila larva using optogenetics eLife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hige T, Aso Y, Rubin GM, Turner GC (2015) Plasticity-driven individualization of olfactory coding in mushroom body output neurons Nature 526:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg U, Christensen TA, Hildebrand JG (1989) Structure and function of the deutocerebrum in insects Annual review of entomology 34:477–501 [DOI] [PubMed] [Google Scholar]
- Howell K, Hobert O (2017) Morphological Diversity of C. elegans Sensory Cilia Instructed by the Differential Expression of an Immunoglobulin Domain Protein Current Biology 27:1782-+ [DOI] [PubMed] [Google Scholar]
- Huang J, Zhang W, Qiao WH, Hu AQ, Wang ZR (2010) Functional Connectivity and Selective Odor Responses of Excitatory Local Interneurons in Drosophila Antennal Lobe Neuron 67:1021–1033 [DOI] [PubMed] [Google Scholar]
- Jones SV, Choi DC, Davis M, Ressler KJ (2008) Learning-Dependent Structural Plasticity in the Adult Olfactory Pathway Journal of Neuroscience 28:13106–13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaissling KE (2013) Kinetics of olfactory responses might largely depend on the odorant-receptor interaction and the odorant deactivation postulated for flux detectors J Comp Physiol A 199:879–896 [DOI] [PubMed] [Google Scholar]
- Kato A, Touhara K (2009) Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization Cell Mol Life Sci 66:3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp UB (2010) Olfactory signalling in vertebrates and insects: differences and commonalities Nature reviews Neuroscience 11:188–200 [DOI] [PubMed] [Google Scholar]
- Kay RB, Brunjes PC (2014) Diversity among principal and GABAergic neurons of the anterior olfactory nucleus Front Cell Neurosci 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin T (2017) Theoretical Aspects of GPCR-Ligand Complex Pharmacology Chem Rev 117:4–20 [DOI] [PubMed] [Google Scholar]
- Kidd S, Lieber T (2016) Mechanism of Notch Pathway Activation and Its Role in the Regulation of Olfactory Plasticity in Drosophila melanogaster PloS one 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S, Struhl G, Lieber T (2015) Notch Is Required in Adult Drosophila Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit Plos Genet 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta S, Fletcher ML, Homma R, Yamasoba T, Nagayama S (2013) Odorant Response Properties of Individual Neurons in an Olfactory Glomerular Module Neuron 77:1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kim JH, Song YH, Lee SH (2017) Functional dissection of inhibitory microcircuits in the visual cortex Neurosci Res 116:70–76 [DOI] [PubMed] [Google Scholar]
- Kreher SA, Kwon JY, Carlson JR (2005) The molecular basis of odor coding in the Drosophila larva Neuron 46:445–456 [DOI] [PubMed] [Google Scholar]
- Kreher SA, Mathew D, Kim J, Carlson JR (2008) Translation of sensory input into behavioral output via an olfactory system Neuron 59:110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larter NK, Sun JS, Carlson JR (2016) Organization and function of Drosophila odorant binding proteins eLife 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G (1996) Odor images and tunes Neuron 16:473–476 [DOI] [PubMed] [Google Scholar]
- Laurent G, Stopfer M, Friedrich RW, Rabinovich MI, Volkovskii A, Abarbanel HD (2001) Odor encoding as an active, dynamical process: experiments, computation, and theory Annual review of neuroscience 24:263–297 [DOI] [PubMed] [Google Scholar]
- Lee SH et al. (2012) Activation of specific interneurons improves V1 feature selectivity and visual perception Nature 488:379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY et al. (2016) A Functionally Conserved Gene Regulatory Network Module Governing Olfactory Neuron Diversity Plos Genet 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY et al. (2013) Combinatorial Rules of Precursor Specification Underlying Olfactory Neuron Diversity Current Biology 23:2481–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Sanes JR (2008) Ome sweet ome: what can the genome tell us about the connectome? Current opinion in neurobiology 18:346–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Struhl G (2011) DSL-Notch Signaling in the Drosophila Brain in Response to Olfactory Stimulation Neuron 69:468–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Katz LC (2006) Representation of natural stimuli in the rodent main olfactory bulb Chemical senses 31:A2–A2 [DOI] [PubMed] [Google Scholar]
- Liou NF et al. (2018) Diverse populations of local interneurons integrate into the Drosophila adult olfactory circuit Nature communications 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AN, Urban NN (2017) Prenatal and Early Postnatal Odorant Exposure Heightens Odor-Evoked Mitral Cell Responses in the Mouse Olfactory Bulb Eneuro 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainen ZF, Sejnowski TJ (1996) Influence of dendritic structure on firing pattern in model neocortical neurons Nature 382:363–366 [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB (1999) Combinatorial receptor codes for odors Cell 96:713–723 [DOI] [PubMed] [Google Scholar]
- Manzini I, Korsching S (2011) The peripheral olfactory system of vertebrates: molecular, structural and functional basics of the sense of smell Neuroforum 17:110–118 [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu CZ (2004) Interneurons of the neocortical inhibitory system Nature Reviews Neuroscience 5:793–807 [DOI] [PubMed] [Google Scholar]
- Martelli C, Carlson JR, Emonet T (2013) Intensity Invariant Dynamics and Odor-Specific Latencies in Olfactory Receptor Neuron Response The Journal of neuroscience: the official journal of the Society for Neuroscience 33:6285–6297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse NY, Turner GC, Jefferis GS (2009) Olfactory information processing in Drosophila Current biology: CB 19:R700–713 [DOI] [PubMed] [Google Scholar]
- Mathew D et al. (2013) Functional diversity among sensory receptors in a Drosophila olfactory circuit Proceedings of the National Academy of Sciences of the United States of America 110:E2134–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R (2012) The honeybee as a model for understanding the basis of cognition Nature reviews Neuroscience 13:758–768 [DOI] [PubMed] [Google Scholar]
- Miles R (2000) Neurobiology - Diversity in inhibition Science 287:244–246 [DOI] [PubMed] [Google Scholar]
- Mo A et al. (2015) Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain Neuron 86:1369–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P (2004) Genes and ligands for odorant, vomeronasal and taste receptors Nature reviews Neuroscience 5:263–278 [DOI] [PubMed] [Google Scholar]
- Mombaerts P et al. (1996) Visualizing an olfactory sensory map Cell 87:675–686 [DOI] [PubMed] [Google Scholar]
- Montague SA, Mathew D, Carlson JR (2011) Similar odorants elicit different behavioral and physiological responses, some supersustained The Journal of neuroscience: the official journal of the Society for Neuroscience 31:7891–7899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch D, Galizia CG (2017) Take time: odor coding capacity across sensory neurons increases over time in Drosophila J Comp Physiol A 203:959–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel KI, Wilson RI (2011) Biophysical mechanisms underlying olfactory receptor neuron dynamics Nature neuroscience 14:208–U296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel KI, Wilson RI (2016) Mechanisms Underlying Population Response Dynamics in Inhibitory Interneurons of the Drosophila Antennal Lobe Journal of Neuroscience 36:4325–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Vosshall LB (2009) Controversy and consensus: noncanonical signaling mechanisms in the insect olfactory system Current opinion in neurobiology 19:284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bhandawat V, Wilson RI (2010) Divisive normalization in olfactory population codes Neuron 66:287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Urban NN (2010) Intrinsic biophysical diversity decorrelates neuronal firing while increasing information content Nature neuroscience 13:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnas M, Lin AC, Huetteroth W, Miesenbock G (2013) Odor discrimination in Drosophila: from neural population codes to behavior Neuron 79:932–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E (2004) Similar network activity from disparate circuit parameters Nature neuroscience 7:1345–1352 [DOI] [PubMed] [Google Scholar]
- Raja JSI, Katanayeva N, Katanaev VL, Galizia CG (2014) Role of G(o/i) subgroup of G proteins in olfactory signaling of Drosophila melanogaster European Journal of Neuroscience 39:1245–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters WG, Shiraiwa T, Carlson JR (2007) Mechanisms of odor receptor gene choice in Drosophila Neuron 53:353–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, van Naters WV, Carlson JR (2008) A regulatory code for neuron-specific odor receptor expression PLoS biology 6:1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T (2006) Identification of long-range regulatory elements in the protocadherin-alpha gene cluster Proceedings of the National Academy of Sciences of the United States of America 103:19719–19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Denny CA, Hen R, Axel R (2014) The participation of cortical amygdala in innate, odour-driven behaviour Nature 515:269–U274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root CM, Masuyama K, Green DS, Enell LE, Nassel DR, Lee CH, Wang JW (2008) A presynaptic gain control mechanism fine-tunes olfactory behavior Neuron 59:311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse S, Rueckert E, Keller A, Okada R, Tanaka NK, Ito K, Vosshall LB (2007) Activity-dependent plasticity in an olfactory circuit Neuron 56:838–850 [DOI] [PubMed] [Google Scholar]
- Saito H, Chi QY, Zhuang HY, Matsunami H, Mainland J (2009) Odor coding by a mammalian receptor repertoire Neurosci Res 65:S76–S76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H (2010) Neural Map Formation in the Mouse Olfactory System Neuron 67:530–542 [DOI] [PubMed] [Google Scholar]
- Santoro SW, Dulac C (2012) The activity-dependent histone variant H2BE modulates the life span of olfactory neurons eLife 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall LB, Touhara K (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels Nature 452:1002–1006 [DOI] [PubMed] [Google Scholar]
- Schaefer AT, Larkum ME, Sakmann B, Roth A (2003) Coincidence detection in pyramidal neurons is tuned by their dendritic branching pattern Journal of neurophysiology 89:3143–3154 [DOI] [PubMed] [Google Scholar]
- Schneider D (1969) Insect olfaction: Deciphering system for chemical messages Science 163:1031–1037 [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder E (2006) Variable channel expression in identified single and electrically coupled neurons in different animals Nature neuroscience 9:356–362 [DOI] [PubMed] [Google Scholar]
- Seki Y, Dweck HKM, Rybak J, Wicher D, Sachse S, Hansson BS (2017) Olfactory coding from the periphery to higher brain centers in the Drosophila brain Bmc Biol 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Rybak J, Wicher D, Sachse S, Hansson BS (2010) Physiological and Morphological Characterization of Local Interneurons in the Drosophila Antennal Lobe Journal of neurophysiology 104:1007–1019 [DOI] [PubMed] [Google Scholar]
- Shang Y, Claridge-Chang A, Sjulson L, Pypaert M, Miesenbock G (2007) Excitatory local circuits and their implications for olfactory processing in the fly antennal lobe Cell 128:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Benton R (2010) Ionotropic and metabotropic mechanisms in chemoreception: ‘chance or design’? EMBO reports 11:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart R et al. (2008) Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins Insect biochemistry and molecular biology 38:770–780 [DOI] [PubMed] [Google Scholar]
- Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR (2011) Distinct representations of olfactory information in different cortical centres Nature 472:213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanczyk NM, Brookfield JFY, Field LM, Logan JG (2013) Aedes aegypti Mosquitoes Exhibit Decreased Repellency by DEET following Previous Exposure PloS one 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiefel KM, Englitz B, Sejnowski TJ (2013) Origin of intrinsic irregular firing in cortical interneurons Proceedings of the National Academy of Sciences of the United States of America 110:7886–7891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R (1994) The organization of the chemosensory system in Drosophila melanogaster: a review Cell Tissue Research 275:3–26 [DOI] [PubMed] [Google Scholar]
- Stocker RF, Lienhard MC, Borst A, Fischbach KF (1990) Neuronal architecture of the antennal lobe in Drosophila melanogaster Cell Tissue Res 262:9–34 [DOI] [PubMed] [Google Scholar]
- Su CY, Menuz K, Carlson JR (2009) Olfactory perception: receptors, cells, and circuits Cell 139:45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Awasaki T, Shimada T, Ito K (2004) Integration of chemosensory pathways in the Drosophila second-order olfactory centers Current biology: CB 14:449–457 [DOI] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K (2008) Neuronal assemblies of the Drosophila mushroom body Journal of Comparative Neurology 508:711–755 [DOI] [PubMed] [Google Scholar]
- Taylor AL, Goaillard JM, Marder E (2009) How Multiple Conductances Determine Electrophysiological Properties in a Multicompartment Model Journal of Neuroscience 29:5573–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE et al. (2012) The accessible chromatin landscape of the human genome Nature 489:75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin WF, Wilson RI, Lee WCA (2017) Wiring variations that enable and constrain neural computation in a sensory microcircuit eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SL, Li N, Guda T, Githure J, Carde RT, Ray A (2011) Ultra-prolonged activation of CO2-sensing neurons disorients mosquitoes Nature 474:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter P, Roth A, Hausser M (2001) Propagation of action potentials in dendrites depends on dendritic morphology Journal of neurophysiology 85:926–937 [DOI] [PubMed] [Google Scholar]
- Vosshall LB (2000) Olfaction in Drosophila Current opinion in neurobiology 10:498–503 [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Stocker RF (2007) Molecular architecture of smell and taste in Drosophila Annual review of neuroscience 30:505–533 [DOI] [PubMed] [Google Scholar]
- Wachowiak M, Shipley MT (2006) Coding and synaptic processing of sensory information in the glomerular layer of the olfactory bulb Semin Cell Dev Biol 17:411–423 [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R (1998) Odorant receptors govern the formation of a precise topographic map Cell 93:47–60 [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R (2003) Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain Cell 112:271–282 [DOI] [PubMed] [Google Scholar]
- Wicher D, Schafer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, Hansson BS (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels Nature 452:1007–1011 [DOI] [PubMed] [Google Scholar]
- Wilson RI, Laurent G (2005) Role of GABAergic inhibition in shaping odor-evoked spatiotemporal patterns in the Drosophila antennal lobe The Journal of neuroscience: the official journal of the Society for Neuroscience 25:9069–9079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Mainen ZF (2006) Early events in olfactory processing Annual review of neuroscience 29:163–201 [DOI] [PubMed] [Google Scholar]
- Wilson RI, Turner GC, Laurent G (2004) Transformation of olfactory representations in the Drosophila antennal lobe Science 303:366–370 [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA (2006) The origin and specification of cortical interneurons Nature Reviews Neuroscience 7:687–696 [DOI] [PubMed] [Google Scholar]
- Xia YF, Wang GR, Buscariollo D, Pitts RJ, Wenger H, Zwiebel LJ (2008) The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae Proceedings of the National Academy of Sciences of the United States of America 105:6433–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu PS, Lee D, Holy TE (2016) Experience-Dependent Plasticity Drives Individual Differences in Pheromone-Sensing Neurons Neuron 91:878–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T (2013) Genetic basis of neuronal individuality in the mammalian brain Journal of neurogenetics 27:97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CA, Carlson JR (2010) Role of G-Proteins in Odor-Sensing and CO2-Sensing Neurons in Drosophila Journal of Neuroscience 30:4562–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HQ, Reed RR (2001) X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system Cell 104:651–660 [DOI] [PubMed] [Google Scholar]
- Zufall F, Leinders-Zufall T (2000) The cellular and molecular basis of odor adaptation Chemical senses 25:473–481 [DOI] [PubMed] [Google Scholar]