Abstract
Cysteine residues are concentrated at key functional sites within proteins, performing diverse roles in metal binding, catalysis, and redox chemistry. Chemoproteomic platforms to interrogate the reactive cysteinome have developed significantly over the past 10 years, resulting in a greater understanding of cysteine functionality, modification, and druggability. Recently, chemoproteomic methods to examine reactive cysteine residues from specific subcellular organelles have provided significantly improved proteome coverage and highlights the unique functionalities of cysteine residues mediated by cellular localization. Here, the diverse physicochemical properties of the mammalian subcellular organelles are explored in the context of their effects on cysteine reactivity. The unique functions of cysteine residues found in the mitochondria and endoplasmic reticulum are highlighted, together with an overview into chemoproteomic platforms employed to investigate cysteine reactivity in subcellular organelles.
Introduction
Cysteine is one of the least abundant, yet most highly conserved, amino-acid residues in proteins and perform diverse functional roles including regulating catalysis, structure, redox sensitivity, and metal trafficking. Importantly, cysteine, with its highly ionizable and oxidation sensitive sulfur atom, is responsive to the different pH and redox conditions of eukaryotic sub cellular organelles. The functional regulation of cysteine-containing proteins in these subcellular environments is poorly characterized. Chemoproteomic platforms, such as isoTOP-ABPP, have greatly expanded our knowledge of cysteine reactivity, druggability, and susceptibility to reactive small molecules. However, these analyses have limitations including; (1) analysis of cysteine reactivity in cell lysates that disrupt subcellular environments; and, (2) analysis of unfractionated cellular proteomes resulting in signal suppression by highly abundant cytosolic proteins. Therefore, novel chemoproteomic strategies are required to target low-abundance cysteine residues localized in subcellular organelles within the context of their native pH and redox environments. This review provides; (1) an overview of the properties of eukaryotic organelles that impact cysteine function; (2) select examples that highlight the diverse roles of functional cysteine residues within subcellular compartments; and, (3) a survey of strategies to probe these subcellular compartments for reactive and functional cysteine residues.
Definition of Cysteine Reactivity and Functionality
Despite its low abundance compared to other amino acids, cysteine residues are highly conserved and enriched at functionally important sites on proteins [1,2]. The cysteine thiol is both redox-active and highly nucleophilic due to the large atomic radius of the sulfur atom and the low dissociation energy of the thiol S—H bond. Functional cysteines can be categorized by activity; 1) structural disulfides, 2) redox-active disulfides, 3) active-site nucleophiles and proton donors, 4) metal ligands, and 5) regulatory sites (ie. post-translationally modified) [3] (Figure 1A). Cysteine reactivity has been shown to be a strong indicator of functionality, particularly for catalytic, redox-active, and regulatory cysteine residues [4]. The nucleophilicity of a cysteine residue is governed by the pKa and ionization state of the thiol [5], while oxidative capacity is determined by the redox potential of a disulfide pair [6][7,8]. While the pKa of a surface-exposed cysteine is ~8.0 [9], cysteine pKa values range from 3.5 to 12 depending on the local protein microenviroment [10–13]. Similarly, redox potentials of cysteine residues can vary significantly from −470 to −90 mV [14]. The propensity for cysteine ionization and/or oxidation is sensitive to variations in pH and reduction potential within the local environment.
Figure 1:
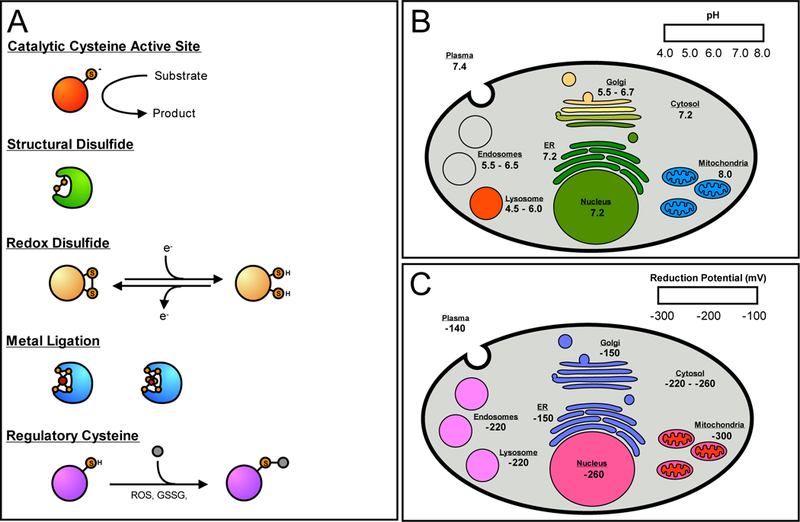
The functional roles of reactive cysteine residues and the pH and redox ranges of the mammalian organelle. (A) Cysteine residues can be grouped into several functional classes; catalytic, structural, redox-active, metal ligating, and regulatory. (B) The pH ranges found in the organelle of a typical mammalian cell. Red/orange is indicative of a low pH, green of a neutral pH, and blue of a higher, more alkaline pH. The mitochondrial pH is referring to the pH found in the mitochondrial matrix. (C) The reduction potentials found in the various organelles, where red indicates a low potential, and blue a higher relative reduction potential. The mitochondrial reduction potential refers to the mitochondrial matrix. The intermembrane space has a higher potential of ~ −255 mV.
Cellular Determinants of Cysteine Reactivity
The eukaryotic cell relies on subcellular organelles, such as the mitochondria, nucleus, and endoplasmic reticulum (ER), to create distinct cellular environments for incompatible physiological functions (ie. protein folding in the oxidizing environment of the ER versus biosynthesis of iron-sulfur clusters in the reducing environment of the mitochondrial matrix). These subcellular organelles maintain distinct pH and redox potentials, as well as concentrations of reactive metabolites [15,16]. Cysteine residues have evolved to exploit these unique subcellular conditions to generate functional diversity across the subcellular proteome. The pH and redox states of the various organelles that influence cysteine function are summarized below.
pH:
The pH of the cytosol is ~ 7.2 (Figure 1B), which is slightly more acidic than the pH of the extracellular environment (7.4 for plasma) [17,18]. Passive transport of protons across organellar membranes of the nucleus, ER, and peroxisomes, results in pH profiles similar to that of the cytosol [15]. In contrast, some organelles generate pH gradients across their membranes through the proton pumping action of intermembrane ATPases [19]. In the secretory pathway, a progressively acidic gradient from 6.7 to 5.5 is generated from the Golgi to secretory vesicles. Similarly, the endocytic pathway has a progressively acidic pH gradient from 6.5 to 4.5 in the fully mature lysosome [20]. In both systems, reduced pH is essential to organelle activities, such as posttranslational processing in the Golgi, and biomolecule degradation and ligand dissociation from receptors in the endocytotic pathway [15,21]. These pH ranges regulate the activity of cysteine-containing proteins, such as lysososomal cysteine cathepsins, which have active-site cysteine pKa values lower than those of cysteine proteases in other cellular compartments [22]. These depressed pKa values allow lysosomal cathepsins to retain the active thiolate form under acidic conditions. In contrast, the mitochondrial matrix has an alkaline pH of 8.0, due to the proton pumping activity of the respiratory pathway [23,24]. This increased pH should theoretically stabilize thiolates of mitochondrial matrix proteins to produce more reactive cysteine residues, but this phenomena has not been rigorously examined to date. While a number of methods exist to measure the pH of individual organelle, the use of pH-sensitive green fluorescent protein (GFP) is widely employed and amenable to genetically tractable organisms [23,24].
Reduction Potential:
Reduction potentials of subcellular organelles are difficult to calculate due to the presence of different redox couples including thioredoxin (TXN), glutathione (GSH/GSSG), as well as the cysteine/cystine couple (Cys/CySS) [16,25]. Furthermore, midpoint potentials for each of these couples can differ significantly within and across different organelles. The best characterized redox couple is GSH/GSSG, which is calculated to vary between −220 to −260 mV in the cytosol of resting and proliferating cells, respectively [26] (Figure 1C). Secretory and endocytic organelles have more oxidizing potentials with GSH/GSSG couples for the ER and plasma calculated to be −150 and −140 mV, respectively [27,28]. This oxidizing environment is essential for the formation and retention of structural disulfide bonds on secretory proteins [29]. In contrast, the mitochondrial matrix maintains the most reducing potential of all organelles with a GSH/GSSG couple of −300 to −330 mV [25], with the mitochondrial intermembrane space displaying a more oxidizing GSH/GSSG couple of only −255 mV [30]. TXN redox couples are generally more reducing than their GSH/GSSG counterparts, but TXN is only found in the cytosol, nucleus, and mitochondria. TXN1, found in the cytosol and nucleus, has redox couples of −280 and −300 mV, respectively, while mitochondrial TXN2 has a redox couple of −340 to −360 mV [16,25]. These more reducing GSH/GSSG and TXN2 mitochondrial redox couples likely indicate that mitochondrial cysteine residues are maintained in their reduced state, unless exposed to oxidative stress conditions. As such, mitochondrial cysteine residues may be primed to participate in redox signaling which is not possible in the more oxidizing secretory organelles. Measurements of organelle reducing potential can be technically challenging, but most current studies accomplish this by organelle-targeted redox-sensitive variants of GFP (roGFP) [30].
Cysteine Functions Unique to the Mitochondria - Redox and Cofactor Biosynthesis
Mitochondria are the site of aerobic metabolism and oxidative phosphorylation, as well as fatty- acid oxidation and amino-acid breakdown. Additionally, mitochondria are the primary site of iron- sulfur biosynthesis, providing many of the redox cofactors for the electron transport chain. Cysteine residues are essential to many of these unique mitochondrial functions (Figure 2A).
Figure 2:
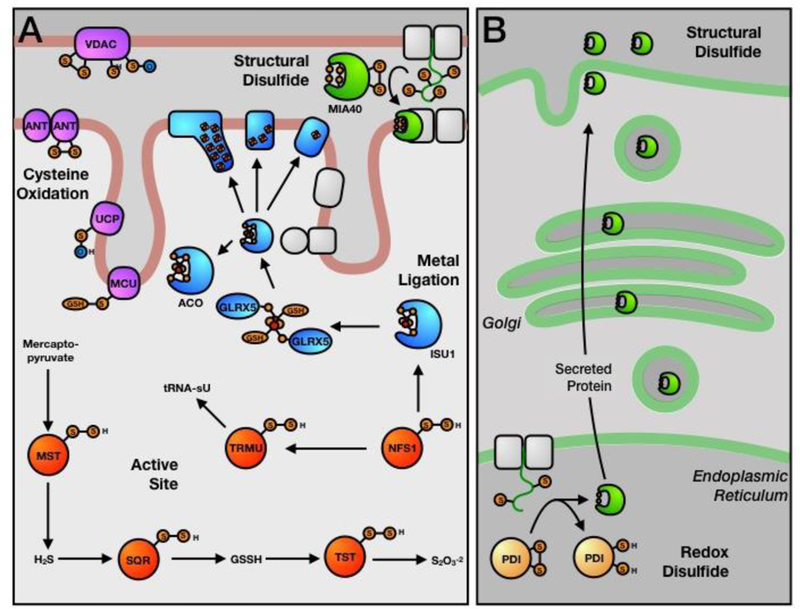
Functional cysteine residues of the mitochondria and endoplasmic reticulum (ER). (A) Mitochondrial cysteine residues involved in persulfide chemistry (red -active site), iron-sulfur trafficking (blue - metal ligation), disulfide bond formation (green - structural disulfide), and oxidative regulation of membrane transporters (purple - regulatory). (B) ER cysteine residues involved in redox-dependent processes (yellow - redox disulfide).
Iron-sulfur Clusters (Metal-Ligation):
One of the main functions of the mitochondria is the biosynthesis of cellular iron-sulfur (Fe-S) clusters [31,32]. [2Fe-2S] clusters are assembled on the scaffold protein ISU1, from the input of iron, sulfide, and electron-reducing equivalents [33]. ISU1 then transfers these clusters to proteins such as glutaredoxin 5 (GLRX5) [34], for eventual delivery to downstream target proteins or further maturation to [4Fe-4S] and [3Fe-4S] clusters [35] (Figure 2A) via cysteine ligands on transfer proteins. In addition to a complete biosynthetic machinery for Fe-S synthesis, the mitochondria harbors approximately half of the known mammalian Fe-S clusters. These include the [4Fe-4S] clusters of metabolic proteins such as aconitase and lipoyl synthase, but also include the ~15 clusters of the respiratory complexes, essential for aerobic respiration and ATP synthesis within the mitochondria. This enrichment in Fe-S ligation and trafficking is likely the result of the highly reducing conditions within the mitochondrial matrix. Up to 4 cysteine residues are required to ligate an Fe-S cluster, which are often in close sequence and spatial proximity to one another. These presumably must remain reduced before insertion of Fe-S clusters, since oxidation would likely prevent cluster delivery. In agreement with this idea, Fe-S proteins also exist within the relatively reducing environments of the nucleus and cytosol, but are not known to exist within the oxidizing organelles of the secretory pathway [36].
Cysteine Persufides (Active-Site):
Oxidation of a cysteine thiol by a sulfur species, results in the formation of a cysteine persulfide (-SSH). This can occur through a number of mechanisms, including; 1) enzymatic activation; 2) transpersulfidation from another protein cysteine persulfide; or, 3) non-enzymatic persulfidation by GSSH [37,38]. In the mitochondria, two important pathways generate cysteine persulfides from sulfur containing substrates. In the hydrogen sulfide (H2S) oxidation pathway [39], mercaptopyruvate sulfur transferase (MST) generates free H2S from mercaptopyruvate. In the most parsimonious pathway, H2S is oxidized by sulfide:quinone oxidoreductase (SQR), generating the product GSSH. The sulfane sulfur of GSSH has a number of fates, including; 1) further oxidation to thiosulfate by rhodenase (TST); 2) oxidation to sulfite and sulfate by persulfide dioxygenase (PDO) and sulfite oxidase, respectively; or, 3) non- enzymatic persulfidation of protein cysteines as a potential regulatory signaling mechanism. The catalytic mechanisms of MST [40], SQR [41], and TST [42] all proceed through cysteine persulfide intermediates (Figure 2A). The second primary mechanism of persulfide generation in the mitochondria is via the cysteine desulferase, NFS1, which generates a cysteine persulfide from the conversion of cysteine to alanine [32]. The persulfide of NFS1 has a number of fates, including providing a source of sulfide for the generation of iron-sulfur clusters, and for mitochondrial tRNA-specific 2-thiouridylase 1 (TRMU), an enzyme responsible for the essential thiouridinylation of mitochondrial tRNAs (Figure 2A) [43]. Thiouridinylation of cytosolic tRNA is also dependent on the activity of mitochondrial NFS1, through the transfer of an as yet unidentified sulfur species from the mitochondria into the cytosol [44]. The mitochondria appear to be an essential regulator of cellular sulfur metabolism, which may again be related to the relatively reducing conditions of the mitochondrial matrix.
Mitochondrial Transporter Oxidation (Regulatory):
Mitochondria have long been considered a primary generator of reactive oxygen species (ROS) through the activity of the electron transport chain [45]. These ROS are important second messengers for the cell, primarily through their ability to reversibly oxidize protein cysteine residues [46]. Many mitochondrial and non-mitochondrial proteins have been shown to harbor redox-regulated cysteine residues. For example, a number of mitochondrial membrane pores and transporters are regulated through cysteine oxidation [47]. In the outer mitochondrial membrane, voltage-dependent anion channels (VDACs 1, 2, and 3) transport small metabolites into the mitochondrial intermembrane space. VDAC2 and 3 have a large number of cysteine residues at the pore opening, which regulate pore activity upon oxidation [48–50]. In the inner mitochondrial membrane, adenine nucleotide transporters (ANTs) [51], uncoupling proteins (UCPs) [52], and mitochondrial calcium uniporter (MCU) [53] have all been shown to be regulated by cysteine oxidation. As all of these transporters control the transfer of essential metabolites for the respiratory chain, the regulation of these transport events directly impacts mitochondrial function. These regulatory events are also likely a response to ROS production by the respiratory chain, resulting in a complex and as yet poorly understood feedback mechanism within this organelle.
Twin CxC Proteins (Structural Disulfides):
Due to the reducing environment of the mitochondria, the presence of structural disulfides in this organelle is unexpected. However, several mitochondrial proteins including TIM proteins of the IMM [54,55], and COX17, a copper chaperone [56], are known to incorporate structural disulfide bonds. These proteins are imported into the intermembrane space (IMS), the most oxidizing mitochondrial compartment (−255 mV), and undergo disulfide-bond formation via the MIA40 pathway (Figure 2A) [57]. Substrates of the Mia40 pathway contain a twin CX3C motif (small TIM proteins) or a twin CX9C motif (i.e. COX17 and CMC1P) [58]. MIA40, itself a twin CX9C motif containing protein, contains a catalytic CPC motif that transfers disulfide bonds via thiol-disulfide exchange reactions yielding inter-twin CXnC motif disulfide bonds and a reduced MIA40 CPC active site. The CPC active site can then be reoxidized by ERV1, a quiescin-sulfhydryl oxidase (QSOX) [59]. Proteins with twin CxnC motifs are not imported into the highly reducing environment of the mitochondrial matrix.
Cysteine Functions Unique to the ER and Secretory Pathway - Disulfide Formation
The ER, along with the Golgi apparatus and secretory vesicles, is responsible for the synthesis, folding, glycosylation, trafficking and secretion of all proteins destined for the extracellular space or plasma membrane. In addition, the ER is important for lipid biosynthesis, steroid metabolism, detoxification, and calcium storage and signaling. Cysteine residues in this organelle support these processes, particularly the folding and trafficking of proteins (Figure 2B).
Protein Disulfide Isomerase (Redox Disulfides):
The ER, as one of the most oxidizing cellular organelles, is ideally suited for the oxidative folding of proteins destined for the equally oxidizing extracellular environments. Native disulfide-bond formation in the ER is essential to maintaining the structural and functional integrity of secreted proteins, and is catalyzed by the 22-member protein disulfide isomerase (PDI) family [60,61]. The most abundant family member, PDIA1, contains two catalytic CGHC motifs whose redox state determines whether oxidase (disulfide) or isomerase (dithiol) activity is exerted on the substrate protein [62,63]. Oxidase activity yields reduced PDIA1 and requires reoxidation of PDIA1 by oxidase ERO1 prior to further rounds of substrate oxidation [64]. With the exception of the small number of previously described twin CxnC proteins of the mitochondrial IMS, cellular systems for disulfide-bond formation do not exist in the more reducing environment of the cytosol or other organelles.
Identifying Functional Cysteine Residues by Organelle
To explore the role of subcellular cysteine residues, an ideal proteomic platform should fulfill the following criteria; 1) preservation of the native subcellular environment, including pH, redox state, and organelle functionality during cysteine labeling; and, 2) efficient enrichment of proteins from the targeted organelle to prevent contamination from abundant cytosolic proteins. To date, two general strategies have been employed to achieve these goals; 1) enrichment and isolation of intact organelles for chemoproteomic labeling [65]; and, 2) organelle-targeted cysteine-reactive probes for live-cell labeling [66,67].
Organelle Isolation and Labeling:
Subcellular isolation procedures utilize a combination of differential and density gradient centrifugation steps (Figure 3A) [68,69], whereby eukaryotic/mammalian cells are; 1) gently lysed in a buffer designed for isolation of a specific organelle; 2) centrifuged to remove un-lysed cells, debris, and intact nuclei; 3) separation of organelles with significantly different densities by differential centrifugation; and, 4) purification of the organelle from those of similar density by density gradient centrifugation. Intact and functional organelles can then be labeled with cysteine-reactive probes, such as iodoacetamide- alkyne (IA-alkyne), for chemoproteomic analysis of cysteine reactivity (Figure 3B). Labeling of intact organelles will maintain the subcellular pH and redox environment, thereby decreasing non- physiological cysteine oxidation or loss of highly reversible cysteine modifications during lysis. Equal levels of cysteine labeling could be achieved whether intact mitochondria or mitochondrial lysates were used for cysteine labeling experiments [65]. A potential limitation of this strategy is the contamination of organelles with similar physico-chemical properties, making this approach non-ideal for experiments dependent on organelle purity.
Figure 3:
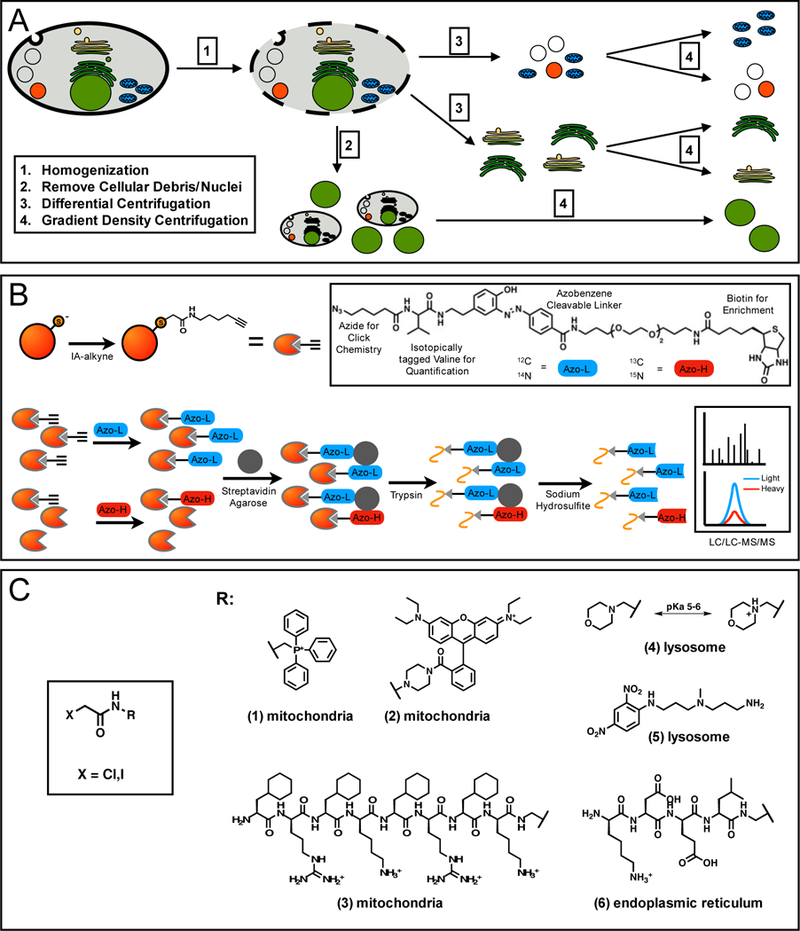
Chemoproteomic platforms to study organelle-specific reactive and functional cysteine residues. (A) Enrichment and isolation of subcellular organelles is most commonly achieved through differential and gradient centrifugation. Cells are gently lysed by dounce homogenization (1), heavy cellular debris and intact nuclei are removed by low speed centrifugation (2), differential centrifugation at moderate and high-speed centrifugation can isolate fractions of organelle (3), which can be fully separated and enriched by density gradient centrifugation (4). (B) Cysteine residues from intact organelles can be quantified by labeling with IA-alkyne, followed by click-chemistry with a chemically cleavable biotin-azide tag. For quantification, an isotopic label is incorporated into either the protein (SILAC), the IA-alkyne probe, or the biotin-azide tag. Tagged proteins are enriched on streptavidin, reduced and alkylated, cleaved by trypsin on-bead, and labeled peptides released by chemical cleavage of the biotin-azide tag. These peptides are then analyzed by LC/LC-MS/MS, followed by isotopic quantification to determine peptide L/H ratios. (C) Cysteine-reactive groups can be appended to organelle-targeting groups. These groups include mitochondrial targeting lipophilic cations, such as rhodamine (1) and triphenylphosphonium (2), mitochondrial-targeting peptide, FxRFxKFxRFxK (3), lysosomal-targeting groups, such as morpholine (4) and 3-(2,4,-dinitroanilino)-3’-amino-N-methyldipropyl-amine (DAMP) (5), and ER-targeting peptide, KDEL (6)
Organelle-Targeted Probes:
An alternative strategy for labeling sub-cellular cysteine residues is the use of organelle-targeted cysteine-reactive electrophiles. Several small molecule and peptide-based targeting agents are capable of localizing to the various sub-cellular compartments within a cell (Figure 3C), and have been used to deliver various cargoes, such as therapeutics, antioxidants, and fluorophores. Organelle-targeting small molecules take advantage of the unique chemical environments of the target organelle. Examples include; 1) lipophilic cations, such as rhodamine or triphenylphosphonium, which localize to the mitochondrial matrix due to the proton gradient across the inner membrane [70,71 ]; 2) weakly basic amine-containing molecules, such as morpholine and 3-(2,4-dinitroanilino)-3,-amino-N-methyldipropylamine (DAMP) [72,73], which are protonated and retained specifically in the acidic environment of the lysosome; and, 3) DNA- binding Hoechst dyes, which localize to the nucleus [80]. Small signaling peptides, endogenously found as terminal organelle-targeting sequences on proteins, can also be adapted for similar applications. Examples include FxRFxKFxRFxK for mitochondrial targeting [74] and NLS peptides (eg. VQRKRQKLMP) for nuclear localization [75]. Recent work has demonstrated success in the coupling of rhodamine or Hoechst with haloacetamides to achieve highly specific mitochondrial and nuclear cysteine labeling [66,67], as well as DAMP with ethyl succinate epoxide to target lysosomal cathepsins [76]. A potential limitation of this strategy is the large bulky size of targeting groups, which can affect the population of cysteine residues that may be targeted.
Both strategies result in the enrichment of mitochondrial cysteine-containing peptides as a percentage of total peptides, ~55% for mitochondrial isolation [65] and ~80% mitochondrial- targeted probes [66] (Figure 4A). In contrast, when whole-cell lysates were labeled with IA- alkyne, only ~10% of the cysteine-containing peptides were mitochondrial [4]. Mitochondrial isolation resulted in the identification of over 500 mitochondrial cysteine-containing peptides (compared to 290 and 131 for mitochondria-targeted probes and IA-alkyne whole cell lysate labeling, respectively), many of which represented cysteine functionalities unique or important to the mitochondria (ie. Fe-S trafficking, persulfides, and oxidation-sensitive residues on membrane transporters) (Figure 4B). In contrast, labeling of whole-cell lysates with IA-alkyne did not result in the identification of any of these functional cysteine residues.
Figure 4:
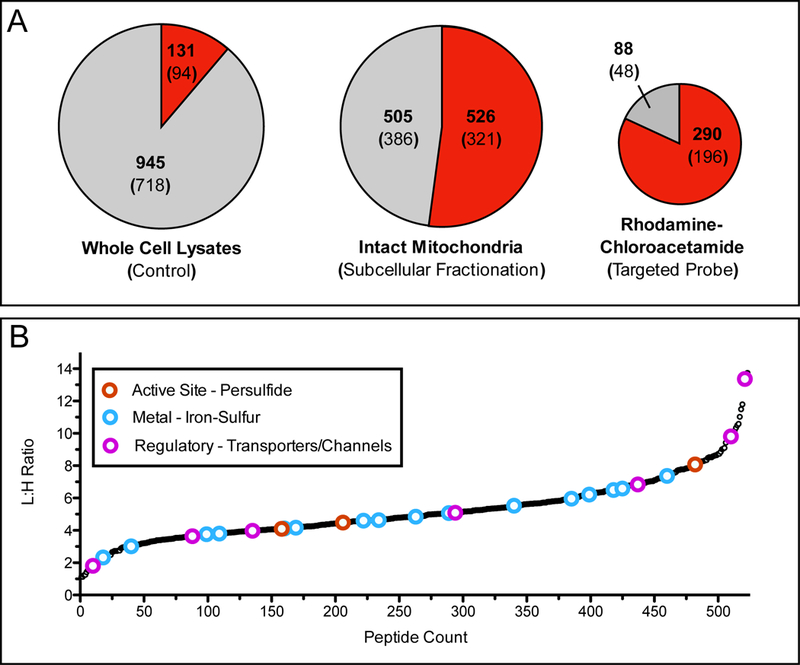
Application of organelle-specific proteomics to examine mitochondrial reactive cysteine residues. (A) Pie charts demonstrate the fraction of mitochondrial peptides identified through probe labeling and proteomic analysis of whole cell lysates by iodoacetamide (IA)-alkyne (left), enriched mitochondria labeled with IA-alkyne (center) [65], and live cells labeled with rhodamine-chloroacetamide (right) [66]. Red shading indicates fraction of total peptides annotated as mitochondrial, while the gray shading indicates peptides from other subcellular organelle or those without annotation. Bold number indicates identified peptides, number within parentheses is the total number of proteins those peptides were identified from. (B) L/H ratios for mitochondrial cysteine-containing peptides identified by labeling of enriched mitochondria with either 100 μM (light) or 10 μM (heavy) IA-alkyne. Lower L/H ratios indicate more reactive cysteine residues. Colored open circles indicate cysteine residues that are annotated with functions described previously as important and somewhat specific to the mitochondria (red - active site cysteine persulfide, blue - iron-sulfur ligating, and pink - regulatory oxidation). Importantly these annotations are generally not observed during whole cell lysate analysis.
Summary and Future Directions
The proteomic analysis of reactive cysteine residues has been employed to better understand the breadth of cysteine functionality, as well as harness their reactivity for the development of covalent inhibitors and therapeutics. Recently, greater coverage of the mammalian cysteinome has been achieved through the enrichment of organelle-specific cysteine residues, by subcellular enrichment or organelle-targeted cysteine reactive probes. Further utilization and development of these enrichment strategies will allow for greater analysis of cysteine residues within their native cellular environment, as well as highlight the unique functions of cysteine residues within organelles. To date organelle-targeted cysteine proteomic studies have been mostly limited to identifying novel mitochondrial reactive cysteine residues, but further analysis of cysteine reactivity across the subcellular space will likely reveal new avenues of cysteine reactivity to explore. As many of the highlighted sub-cellular cysteine functions have implications for disease, including cancer, heart disease, and aging, a better understanding of how these organelle function through the activities of essential cysteine residues could greatly improve future therapeutic strategies.
Acknowledgments
This work was financially supported by NIH grants 1R01GM117004 and 1R01GM118431–01A1 to E. W., by the Smith Family Foundation, the Damon Runyon Cancer Research Foundation (DRR-18–12) and Boston College.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
•• of outstanding interest
References
- 1.Marino SM, Gladyshev VN: Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 2010, 404:902–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pe’er I, Felder CE, Man O, Silman I, Sussman JL, Beckmann JS: Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins 2004, 54:20–40. [DOI] [PubMed] [Google Scholar]
- 3.Pace NJ, Weerapana E: Diverse functional roles of reactive cysteines. ACS Chem. Biol. 2013, 8:283–296. [DOI] [PubMed] [Google Scholar]
- 4.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MBD, Bachovchin DA, Mowen K, Baker D, Cravatt BF: Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468:790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bulaj G, Kortemme T, Goldenberg DP: Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry 1998, 37:8965–8972. [DOI] [PubMed] [Google Scholar]
- 6.Jensen KS, Hansen RE, Winther JR: Kinetic and thermodynamic aspects of cellular thiol disulfide redox regulation. Antioxid. Redox Signal. 2009, 11:1047–1058. [DOI] [PubMed] [Google Scholar]
- 7.Wilson JM, Bayer RJ, Hupe DJ: Structure-reactivity correlations for the thiol-disulfide interchange reaction. J. Am. Chem. Soc. 1977, 99:7922–7926. [Google Scholar]
- 8.Harris TK, Turner GJ: Structural Basis of Perturbed pKa Values of Catalytic Groups in Enzyme Active Sites. IUBMB Life 2002, 53:85–98. [DOI] [PubMed] [Google Scholar]
- 9.Cannan RK, Knight BC: Dissociation Constants of Cystine, Cysteine, Thioglycollic Acid and a-Thiolactic Acid. Biochem. J. 1927, 21:1384–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinitglang S, Watts AB, Patel M, Reid JD, Noble MA, Gul S, Bokth A, Naeem A, Patel H, Thomas EW, et al. : A classical enzyme active center motif lacks catalytic competence until modulated electrostatically. Biochemistry 1997, 36:9968–9982. [DOI] [PubMed] [Google Scholar]
- 11.Zhang ZY, Dixon JE: Active site labeling of the Yersinia protein tyrosine phosphatase: The determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry 2002, 32:9340–9345. [DOI] [PubMed] [Google Scholar]
- 12.Lohse DL, Denu JM, Santoro N, Dixon JE: Roles of aspartic acid-181 and serine-222 in intermediate formation and hydrolysis of the mammalian protein-tyrosine- phosphatase PTP1. Biochemistry 1997, 36:4568–4575. [DOI] [PubMed] [Google Scholar]
- 13.Bulaj G, Kortemme T, Goldenberg DP: Ionization-reactivity relationships for cysteine thiols in polypeptides. Biochemistry 1998, 37:8965–8972. [DOI] [PubMed] [Google Scholar]
- 14.Wouters MA, Fan SW, Haworth NL: Disulfides as redox switches: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 2010, 12:53–91. [DOI] [PubMed] [Google Scholar]
- 15.Casey JR, Grinstein S, Orlowski J: Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 2010, 11:50–61. [DOI] [PubMed] [Google Scholar]
- 16.Jones DP, Go YM: Redox compartmentalization and cellular stress. Diabetes Obes, Metab. 2010, 12 Suppl 2:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb BA, Chimenti M, Jacobson MP, Barber DL: Dysregulated pH: a perfect storm for cancer progression. Nat. Rev. Cancer 2011, 11:671–677. [DOI] [PubMed] [Google Scholar]
- 18.Grinstein S, Furuya W, Biggar WD: Cytoplasmic pH regulation in normal and abnormal neutrophils. Role of superoxide generation and Na+/H+ exchange. J. Biol. Chem. 1986, 261:512–4. [PubMed] [Google Scholar]
- 19.Forgac M: Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8:917–929. [DOI] [PubMed] [Google Scholar]
- 20.Paroutis P, Touret N, Grinstein S: The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 2004, 19:207–215. [DOI] [PubMed] [Google Scholar]
- 21.Weisz OA: Organelle acidification and disease. Traffic 2003, 4:57–64. [DOI] [PubMed] [Google Scholar]
- 22.Turk B, Stoka V: Protease signalling in cell death: caspases versus cysteine cathepsins. FEBS Lett. 2007, 581:2761–2767. [DOI] [PubMed] [Google Scholar]
- 23.Llopis J, McCaffery JM, Miyawaki A, Farquhar MG, Tsien RY: Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 1998, 95:6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abad MF, Di Benedetto G, Magalhäes PJ, Filippin L, Pozzan T: Mitochondrial Ph monitored by a new engineered GFP mutant. J. Biol. Chem. 2003, 279:11521–11529. [DOI] [PubMed] [Google Scholar]
- 25.Kemp M, Go Y-M, Jones DP: Nonequilibrium thermodynamics of thiol/disulfide redox systems: A perspective on redox systems biology. Free Radic. Biol. Med. 2008, 44:921–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirlin WG, Cai J, Thompson SA, Diaz D, Kavanagh TJ, Jones DP: Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic. Biol. Med. 1999, 27:1208–18. [DOI] [PubMed] [Google Scholar]
- 27.Hwang CJ, Sinskey AJ, Lodish HF: Oxidized redox state of glutathione in the endoplasmic reticulum. Science 1992, 257:1496–502. [DOI] [PubMed] [Google Scholar]
- 28.Jones DP, Carlson JL, Mody VC Jr, Cai J, Lynn MJ, Sternberg P Jr: Redox state of glutathione in human plasma. Free Radic. Biol. Med. 2000. 28:625–35. [DOI] [PubMed] [Google Scholar]
- 29.Depuydt M, Messens J, Collet J-F: How proteins form disulfide bonds. Antioxid. Redox Signal. 2011, 15:49–66. [DOI] [PubMed] [Google Scholar]
- 30.Hu J, Dong L, Outten CE: The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J. Biol. Chem. 2008, 283:29126–29134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.⦁Rouault TA, Maio N: Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 2017, 292:12744–12753.• Review of the mitochondrial Fe-S cluster biogenesis pathway, with an emphasis on the protein factors which drive cluster transfer and delivery to target mitochondrial proteins.
- 32.⦁Braymer JJ, Lill R: Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292:12754–12763.• Comprehensive review of both the early and late stages of mitochondrial Fe-S cluster biogenesis and a catalogue of the proteins which are known or predicted to be involved in this process.
- 33.⦁⦁Cory SA, Van Vranken JG, Brignole EJ, Patra S, Winge DR, Drennan CL, Rutter J, Barondeau DP: Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc. Natl. Acad. Sci. U.S.A. 2017, 114:E5325–E5334.•• Crystalographic and electron microscopy structures of the Nfs1-Isd11-ACP complex involved in the generation of sulfur for Fe-S cluster synthesis. These structures help explain the role of accessory subunits, Isd11 and ACP, in regulating Nfs1 desulferase activity and Fe-S cluster synthesis.
- 34.Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, Delft von F, Holmgren A, Oppermann U, Kavanagh KL: The crystal structure of human GLRX5: iron- sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 2011, 433:303–311. [DOI] [PubMed] [Google Scholar]
- 35.Maio N, Kim KS, Singh A, Rouault TA: A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell Metab. 2017, 25:945–953.e6. [DOI] [PubMed] [Google Scholar]
- 36.Paul VD, Lill R: Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 2015, 1853:1528–1539. [DOI] [PubMed] [Google Scholar]
- 37.Mishanina TV, Libiad M, Banerjee R: Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat. Chem. Biol. 2015, 11:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.⦁Filipovic MR, Zivanovic J, Alvarez B, Banerjee R: Chemical Biology of H2S Signaling through Persulfidation. Chem. Rev. 2018, 118:1253–1337.• Comprehensive review of H2S chemistry within the cell, including a complete description of the sulfur oxidation pathway and a description of the role of protein persulfidation in biology.
- 39.Libiad M, Yadav PK, Vitvitsky V, Martinov M, Banerjee R: Organization of the human mitochondrial hydrogen sulfide oxidation pathway. J. Biol. Chem. 2014, 289:30901–30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav PK, Yamada K, Chiku T, Koutmos M, Banerjee R: Structure and kinetic analysis of H2S production by human mercaptopyruvate sulfurtransferase. J. Biol. Chem. 2013, 288:20002–20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson MR, Melideo SL, Jorns MS: Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 2012, 51:6804–6815. [DOI] [PubMed] [Google Scholar]
- 42.Blumenthal KM, Heinrikson RL: Structural studies of bovine liver rhodanese I. Isolation and characterization of two active forms of the enzyme. J. Biol. Chem. 1971, 246:2430–7. [PubMed] [Google Scholar]
- 43.Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K, Suzuki T: Mitochondria- specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005, 280:1613–1624. [DOI] [PubMed] [Google Scholar]
- 44.⦁⦁Pandey A, Pain J, Dziuba N, Pandey AK, Dancis A, Lindahl PA, Pain D: Mitochondria Export Sulfur Species Required for Cytosolic tRNA Thiolation. Cell Chem. Biol. 2018, 25:738–748.e3.•• Report that thio-modification of cytosolic tRNAs is dependent on a mitochondrial generated sulfur species. Specifically, the mitochondrial cysteine desulferase, Nfs1, and the ABC transporter, Atm1, are required to deliver a undefined low molecular weight activated sulfur species to the cytosol.
- 45.⦁Bak DW, Weerapana E: Cysteine-mediated redox signalling in the mitochondria. Mol. BioSys. 2015, 11 678.- .• Comprehensive review of ROS production and signaling within the mitochondria. Particular focus is given to the role of cysteine oxidation as a regulatory mechanism of mitochondrial biology.
- 46.Reczek CR, Chandel NS: ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015, 33:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O-Uchi J, Ryu S-Y, Jhun BS, Hurst S, Sheu S- S: Mitochondrial ion channels/transporters as sensors and regulators of cellular redox signaling. Antioxid. Redox Signal. 2014, 21:987–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reina S, Checchetto V, Saletti R, Gupta A, Chaturvedi D, Guardiani C, Guarino F, Scorciapino MA, Magri A, Foti S, Ceccarelli M : VDAC3 as a sensor of oxidative state of the intermembrane space of mitochondria: the putative role of cysteine residue modifications. Oncotarget 2016, 7:2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saletti R, Reina S, Pittalà MGG, Magri A, Cunsolo V, Foti S, De Pinto V: Post-translational modifications of VDAC1 and VDAC2 cysteines from rat liver mitochondria. Biochim. Biophys. Acta 2018, 1859:806–816. [DOI] [PubMed] [Google Scholar]
- 50.De Pinto V, Reina S, Gupta A, Messina A, Mahalakshmi R: Role of cysteines in mammalian VDAC isoforms’ function. Biochim. Biophys. Acta 2016, 1857:1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McStay GP, Clarke SJ, Halestrap AP: Role of critical thiol groups on the matrix surface of the adenine nucleotide translocase in the mechanism of the mitochondrial permeability transition pore. Biochem. J. 1911, 367:541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.⦁⦁Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik- Bogoslavski D, Vetrivelan R, Clish CB, et al. : Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 2016, doi: 10.1038/nature17399.•• First report that cysteine sulfenylation of UCP1 occurs upon increased ROS production during thermogenesis. This cysteine oxidation drives activation of UCP1 and increased respiration and heat production in brown adipose tissue.
- 53.⦁⦁Dong Z, Shanmughapriya S, Tomar D, Siddiqui N, Lynch S, Nemani N, Breves SL, Zhang X, Tripathi A, Palaniappan P, et al. : Mitochondrial Ca2+ Uniporter Is a Mitochondrial Luminal Redox Sensor that Augments MCU Channel Activity. Mol. Cell 2017, 65:1014– 1028.e7.•• Glutathionylation of MCU1, the mitochondrial calcium uniporter, increases channel activity and regulates mitochondrial metabolism in response to ROS levels.
- 54.Leuenberger D, Bally NA, Schatz G, Koehler CM: Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 1999, 18:4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Curran SP, Leuenberger D, Schmidt E, Koehler CM: The role of the Tim8p-Tim13p complex in a conserved import pathway for mitochondrial polytopic inner membrane proteins. J. Cell Biol. 2002, 158:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnesano F, Balatri E, Banci L, Bertini I, Winge DR: Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding. Structure 2005, 13:713–722. [DOI] [PubMed] [Google Scholar]
- 57.Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM: A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 2005, 121:1059–1069. [DOI] [PubMed] [Google Scholar]
- 58.Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K: Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 2007, 282:37461–37470. [DOI] [PubMed] [Google Scholar]
- 59.Lee J-E, Hofhaus G, Lisowsky T : Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000, 477:62–66. [DOI] [PubMed] [Google Scholar]
- 60.Kozlov G, Määttänen P, Thomas DY, Gehring K: A structural overview of the PDI family of proteins. FEBS J. 2010, 277:3924–3936. [DOI] [PubMed] [Google Scholar]
- 61.Xu S, Sankar S, Neamati N: Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov. Today 2014, 19:222–240. [DOI] [PubMed] [Google Scholar]
- 62.Chivers PT, Prehoda KE, Raines RT: The CXXC motif: a rheostat in the active site. Biochemistry 1997, 36:4061–4066. [DOI] [PubMed] [Google Scholar]
- 63.Walker KW, Gilbert HF: Oxidation of Kinetically Trapped Thiols by Protein Disulfide Isomerase. Biochemistry 1995, 34:13642–13650. [DOI] [PubMed] [Google Scholar]
- 64.Frand AR, Kaiser CA: Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 1999, 4:469–477. [DOI] [PubMed] [Google Scholar]
- 65.•• Bak DW, Pizzagalli MD, Weerapana E: Identifying Functional Cysteine Residues in the Mitochondria. ACS Chem. Biol. 2017, 12:947–957.•• Isolation of intact mitochondria followed by IA-alkyne labeling, achieved significant enrichment and identification of over 500 reactive mitochondrial cysteine residues, involved in diverse functional processes, including Fe-S cluster binding, oxidative signaling, and eznyme activity.
- 66.•• Yasueda Y, Tamura T, Fujisawa A, Kuwata K, Tsukiji S, Kiyonaka S, Hamachi I: A Set of Organelle-Localizable Reactive Molecules for Mitochondrial Chemical Proteomics in Living Cells and Brain Tissues. J. Am. Chem. Soc. 2016, 138:7592–7602.•• The coupling of rhodamine to diverse electrophiles resulted in the highly specific live-cell labeling of mitochondrial nucleophilic amino acids, including cystiene.
- 67.Yasueda Y, Tamura T, Hamachi I: Nucleus-selective Chemical Proteomics Using Hoechst- tagged Reactive Molecules. Chem. Lett. 2016, 45:265–267. [Google Scholar]
- 68.Frezza C, Cipolat S, Scorrano L: Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nat. Protoc. 2007, 2:287–295. [DOI] [PubMed] [Google Scholar]
- 69.Cox B, Emili A: Tissue subcellular fractionation and protein extraction for use in mass- spectrometry-based proteomics. Nat. Protoc. 2006, 1:1872–1878. [DOI] [PubMed] [Google Scholar]
- 70.Dickinson BC, Srikun D, Chang CJ: Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr. Opin. Chem. Biol. 2010, 14:50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murphy MP: Targeting lipophilic cations to mitochondria. Biochim. Biophys. Acta 2008, 1777:1028–1031. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Zhou L, Qiu L, Lu D, X-Wu Y, Zhang B: An efficient ratiometric fluorescent probe for tracking dynamic changes in lysosomal pH. Analyst 2015, 140:5563–5569. [DOI] [PubMed] [Google Scholar]
- 73.Anderson RG, Falck JR, Goldstein JL, Brown MS: Visualization of acidic organelles in intact cells by electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 1984, 81:4838–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jean SR, Ahmed M, Lei EK, Wisnovsky SP, Kelley SO: Peptide-Mediated Delivery of Chemical Probes and Therapeutics to Mitochondria. Acc. Chem. Res. 2016, 49:1893–1902. [DOI] [PubMed] [Google Scholar]
- 75.Wen Y, Liu K, Yang H, Li Y, Lan H, Liu Y, Zhang X, Yi T: A highly sensitive ratiometric fluorescent probe for the detection of cytoplasmic and nuclear hydrogen peroxide. Anal. Chem. 2014, 86:9970–9976. [DOI] [PubMed] [Google Scholar]
- 76.•Wiedner SD, Anderson LN, Sadler NC, Chrisler WB, Kodali VK, Smith RD, Wright AT: Organelle-specific activity-based protein profiling in living cells. Angew. Chem. Int. Ed. 2014, 53:2919–2922. • The application of a lysosomal-targeting DAMP group coupled to a broad spectrum cystiene protease inhibitor, ethyl succinate epoxide, for identification of lysosomal Cathepsins.


