Abstract
Macrophages exist as innate immune subsets that exhibit phenotypic heterogeneity and functional plasticity. Their phenotypes are dictated by inputs from the tissue microenvironment. G-protein coupled receptors (GPCRs) are essential in transducing signals from the microenvironment, and heterotrimeric Gα signaling links these receptors to downstream effectors. Several Gαi coupled GPCRs have been implicated in macrophage polarization. Here, we use genetically modified mice to investigate the role of Gαi2 on inflammasome activity and macrophage polarization. We report that Gαi2 in murine bone marrow-derived macrophages (BMDMs) regulates IL-1β release after activation of the NLRP3, AIM2, and NLRC4 inflammasomes. We show this regulation stems from the biased polarity of Gαi2 deficient (Gnai2−/−) and RGS-insensitive Gαi2 (Gnai2G184S/G184S) BMDMs. We determined that while Gnai2G184S/G184S BMDMs (excess Gαi2 signaling) have a tendency towards classically activated pro-inflammatory (M1) phenotype, Gnai2−/− BMDMs (Gαi2 deficient) are biased towards alternatively activated anti-inflammatory (M2) phenotype. Finally, we find that Gαi2 deficient macrophages have increased Akt activation and IFN-β production, but defects in ERK1/2 and STAT3 activation after LPS stimulation. Gαi2 deficient macrophages also exhibit increased STAT6 activation after IL-4 stimulation. In summary, our data indicates that excess Gαi2 signaling promotes an M1 macrophage phenotype, while Gαi2 signaling deficiency promotes an M2 phenotype. Understanding Gαi2-mediated effects on macrophage polarization may bring to light insights regarding disease pathogenesis and the reprogramming of macrophages for the development of novel therapeutics.
Keywords: inflammasomes, heterotrimeric G-protein signaling, RGS/AGS proteins, innate immunity, macrophages, M1/M2 phenotype
INTRODUCTION
Macrophages are critical mediators of innate immunity and participate in both the initial propagation of the inflammatory response and later in the suppression of inflammation promoting a return to homeostasis (1). To do so, macrophages have heterogeneous and flexible phenotypes that are dictated by signaling inputs from their microenvironment. Macrophages are broadly classified as pro-inflammatory M1 macrophages implicated in initiating and sustaining inflammation or anti-inflammatory M2 macrophages which produce high levels of IL-10 and promote tissue repair (2). Though progress has been made, the signaling networks responsible for macrophage polarization remain unclear.
G-protein coupled receptors (GPCRs) play a central role in regulating macrophage function, as lipid-based inflammatory mediators, vasoactive amines, complement fragments, and other inflammatory/anti-inflammatory stimuli activate signaling by GPCRs linked to heterotrimeric G-proteins (3). Typically, GPCR activation leads to GTP nucleotide exchange on Gα causing its dissociation from the Gβγ heterodimer; this allows both GTP-bound Gα and Gβγ heterodimers to activate their corresponding downstream effector molecules. Signaling is terminated by the inherent GTPase activity of Gα subunits, which hydrolyze GTP to GDP and cause the re-association of GDP-bound Gα and Gβγ. This cycle highlights the importance of heterotrimeric G-proteins in cell signaling networks, as they transduce environmental stimuli from cell surface receptors to generate a coordinated biological response (4).
Apart from the classical GPCR-G-protein signaling paradigm, there exist several regulatory proteins that modulate G-protein signaling. The regulators of G-protein signaling (RGS) family consists of more than 30 members and is defined by the presence of an RGS domain (5, 6). RGS proteins act as GTPase accelerating proteins (GAPs) for specific Gα subunits. They enhance the intrinsic GTPase activity of Gα by stabilizing the GTPase transition state, accelerating the intrinsic Gα GTPase activity by as much as 100-fold (6, 7). In this way, RGS proteins limit the duration of GPCR-mediated G-protein signaling, and G-proteins genetically modified to be RGS-insensitive proteins show higher basal levels of signaling (6). Conversely, the activators of G-protein signaling (AGS) family broadly functions as receptor independent activators of G-proteins (8, 9). They do so by serving as alternative binding partners to influence subunit interactions independent of nucleotide exchange. Group II AGS proteins, including AGS3 and AGS4, are of interest due to their high expression levels in macrophages (10). They exert their effects on heterotrimeric G-proteins through their G-protein regulatory (GPR) motif, which allows them to bind to GDP-bound Gα thereby regulating Gβγ signaling (8).
Several studies have shown that GPCRs can drive both M1 and M2 macrophage polarization (11). Specifically, Formyl peptide receptor 2 (FPR2), chemokine receptor CXCR3, and Chemerin Receptor 23 (ChemR23) have all have been implicated. FPR2 has multiple cognate ligands that drive either M1 or M2 polarization in a ligand specific manner (12–15). CXCR3 exhibits isoform restricted expression on M1 and M2 subsets, with each isoform functioning to polarize macrophages in opposite directions (16, 17). ChemR23, on the other hand, is only expressed on M1 macrophages and like FPR2 has multiple ligands with opposing proinflammatory/pro-resolving effects. Of note, all three of these receptors couple the inhibitory class of heterotrimeric G-proteins (Gαi), though the isoform/ligand specific effects on Gαi signaling are still unclear (18–22). Given that these receptors have pleiotropic isoform/ligand specific effects, these observations raise the possibility that Gαi signaling functions as a molecular switch regulating macrophage polarization.
Inflammasomes are multimeric complexes that generate IL-1β, an important early mediator of the inflammatory response (23, 24). Each inflammasome consists of a sensor molecular (NLRP3, AIM2, or NLRC4), the adaptor protein ASC, and caspase-1, which cleaves pro-IL-1β and pro-IL-18 upon activation (25, 26). Inflammasome activation is a two-step process. First, an initial priming signal (signal 1) is needed to license inflammasome assembly and mediate the transcriptional upregulation of pro-IL-1β. Next, a sensor specific stimulus (signal 2) initiates inflammasome assembly via activation of the sensor molecule. Macrophage polarization regulates the priming phase of inflammasome activation (27).
Gαi signaling has previously been shown to modulate macrophage chemotaxis, phagocytosis, and activation (6, 28, 29). Though several Gαi-coupled GPCRs show opposing isoform/ligand specific effects on macrophage polarization, the role of Gαi in macrophage polarization is unknown. Here, we use genetically modified mice to directly investigate the physiological role of Gαi and its accessory RGS/AGS proteins on inflammasome activation and macrophage polarization. We find that Gαi2 regulates IL-1β release upon NLRP3, AIM2, and NLRC4 inflammasome activation via modulation of the LPS-priming phase (signal 1). This regulation stems from the biased M1 and M2 polarity of macrophages with excess and deficient Gαi2 signaling respectively. We further establish that these phenotypic differences are built into the macrophage life history, as persistent chemical inhibition of Gαi2 is required to recapitulate the macrophage polarization phenotype seen in genetically modified cells.
MATERIALS AND METHODS
Animals
All animal experiments and protocols used were approved by the NIAID Animal Care and Use Committee (ACUC) at the National Institutes of Health (NIH). C57BL/6J and C57BL/6J vav1-cre mice are from Jackson Laboratory. Gnai2−/−, Gnai3−/−, and Gnai2fl/fl mice were provided by Dr. Lutz Birnbaumer (NIEHS, NIH), and the Gnai2G184S/G184S mice a gift from Dr. Richard Neubig (Michigan State University) (30, 31). Rgs19-deficient GFP knock-in mice were generated by Ozgene. Ric8−/−, Rgs10−/−, Rgs1−/−, and Gpsm1−/− mice have been previously described (32–35). All strains were backcrossed a minimum of 12 times to C57BL/6J and experiments performed using 6–14 week old mice with littermate controls.
Cell isolation and culture
Bone marrow-derived macrophages (BMDMs) were prepared as previously described (36). Gαi2-deficient macrophages were derived from Gnai2−/− or Gnai2fl/flvav1-cre mice, no differences were observed between strains. In certain experiments, PTX (Sigma) was added either on day 0 (50 ng/ml for days 0–2; 100 ng/ml for days 3–7) or on day 7 (200 ng/ml) overnight. Macrophages were M1 polarized using 1 μg/ml LPS from E. coli (Enzo Life Sciences) for 4 hours and towards M2 by treatment with 20 ng/ml recombinant mouse IL-4 (R&D) for 48 hours. CD4+CD25− splenic helper T cells were isolated by negative depletion using biotinylated antibodies to B220 (RA3–6B2, Biolegend), CD8a (53.67, BD Biosciences), CD11b (M1/70, BioLegend), CD11c (HL3, BD Biosciences), and CD25 (PC61, BioLegend) and Dynabeads M280 streptavidin (Thermo). T cell purity was greater than 95%.
Inflammasome activation assays
BMDMs were primed with 1 μg/ml LPS for 4 hours (signal 1), then stimulated with the following activators (signal 2): 4 μM nigericin (Sigma) for 1 h (NLRP3); 1 μg/ml poly(dA:dT) (Sigma) transfected with Lipofectamine 2000 for 4 hours (AIM2); 0.5 μg/ml flagellin from S. typhimurium (tlrl-pstfla, Invivogen) transfected with Profect P1 (Targeting Systems) for 4 hours (NLRC4). Untreated and LPS only treated control groups were included for all inflammasome experiments. BMDM culture supernatant was collected after inflammasome activation and IL-1β levels measured using a commercially available kit following the manufacturer’s protocol (R&D).
Cytokine production assays
BMDMs were stimulated with 1 μg/ml LPS for 4 hours. In certain experiments, BMDMs were concomitantly treated with 10 μg/ml of either an IL-10 neutralizing Ab (JES5–2A5, BioLegend) or a rat IgG1, κ isotype control Ab (RTK2071, BioLegend). Cytokines were measured from BMDM culture supernatant using the following commercial kits: TNF-α (R&D), IL-6 (BioLegend), IL-10 (BioLegend), and IFN-β (PBL Assay Science).
RNA isolation and quantitative RT-PCR
RNA was isolated from BMDMs using RNeasy Mini Kit (Qiagen) and cDNA synthesized using Omniscript RT Kit (Qiagen). RT-PCR was performed using the StepOneTM Real Time PCR System (Applied Biosytems) and the Quantitect SYBR Green PCR kit (Qiagen) according to the manufacturer’s instructions. Primers for TNF-α, IL-12p40, iNOS, Arginase 1 (Arg1), Fizz1, Ym1, and HPRT are from Zhu et al (37) and the GAPDH primer from Qiagen (QT01658692). The results were normalized to HPRT or GAPDH and relative mRNA levels calculated using the 2−ΔΔCt method.
Immunoblotting
For signaling experiments, BMDMs were treated with LPS (250 ng/ml) or IL-4 (20 ng/ml) for the indicated time. BMDM cell lysates were analyzed as described (36). Proteins were transferred onto a nitrocellulose membrane and subject to immunoblotting using the following primary Abs: IL-1β (D4T2D, Cell Signaling), phospho-p65 (Ser536) (93H1, Cell Signaling), total p65 (#06–418, Millipore Sigma), phospho-ERK (Thr202/Tyr204) (E10, Cell Signaling), total ERK (137F5, Cell Signaling), phospho-JNK (Thr183/Tyr185) (#9251, Cell Signaling), total JNK (D-2, Santa Cruz), phospho-Akt (Ser473) (193H2, Cell Signaling), total Akt (40D4, Cell Signaling), phospho-STAT3 (Ser727) (#9134, New England BioLabs), total STAT3 (#9132, New England BioLabs), phospho-STAT6 (Tyr641) (D8S9Y, Cell Signaling), total STAT6 (D-1, Santa Cruz), GAPDH (HRP-60004, Proteintech), β-actin-peroxidase antibody (A3854, Sigma). Membranes were washed and incubated with the appropriate HRP-conjugated secondary Ab (Cell Signaling) prior to detection using Amersham Hyperfilm (GE) or the iBright FL1000 (Thermo). Blots were scanned and imported into Photoshop as unmodified tagged image file format. Quantification of band intensity was performed using standard methods on Image J. Phospho bands were normalized to their respective total protein bands. All western blot quantifications are presented as fold change of the indicated control.
Flow cytometry
For cell surface staining, cells were stained with fluorochrome-conjugated or biotinylated Abs against CD11b (M1/70, BioLegend), F4/80 (BM8, BioLegend), CD169 (3D6.112, BioLegend), SIGN-R1 (eBio22D1, eBiosciences), CD53 (OX-79, BioLegend), CD9 (MZ3, BioLegend), CD37 (Duno85, BioLegend), Tim1 (RMT1–4, BioLegend), Tim 2 (F37–2C4, BioLegend), Tim 3 (RMT3.23, BioLegend), Tim 4 (RMT4–54, BioLegend), CD63 (NVG-2, BioLegend), CD81 (Eat-2, BioLegend), and ADAM10 (139712, R&D Systems). Biotin labeled antibodies were visualized with fluorochrome-conjugated streptavidin (Thermo). LIVE/DEAD® Fixable Aqua Stain (Thermo) was used to exclude dead cells. For phosphorylated protein expression assays, cells were analyzed with Abs against phospho-p38 (Thr180/Tyr182) (28B10, Cell Signaling) and total p38 (#9212, Cell Signaling), and detected using mouse or rabbit Alexa Flour 647 respectively. Data acquisitions were done on a FACSCanto II (BD) flow cytometer and analyzed with FlowJo software version 9 or 10 (Treestar).
T cell suppression assay
96 well round bottom plates were coated with 1 μg/ml CD3 (145–2C11, BD Biosciences) and 0.5 μg/ml CD28 (37.5, BD Biosciences). Macrophages were M1 (250 ng/ml LPS) or M2 (20 ng/ml IL-4) polarized prior to the experiment for 16 h. CD4+CD25− T cells were stained with CFSE proliferation dye (Thermo) and co-cultured with macrophages at a 1:2 ratio for 4 days in RPMI supplemented with 20 ng/ml recombinant IL-2 (R&D Systems). Upon collection, samples were stained with LIVE/DEAD™ Fixable Near-IR Stain Dye (Thermo) and analyzed by flow cytometry. Proliferation modeling was performed using Flow-Jo 10.1 and results normalized to the T cell only control group.
Statistical analysis
Data are expressed as a mean value ± SEM. All results were confirmed in at least three independent experiments. Data were analyzed using Student’s t test or Ordinary One-Way ANOVA with multiple comparisons on Prism software (GraphPad) and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
RESULTS
Gαi2 signaling regulates IL-1β release after inflammasome activation
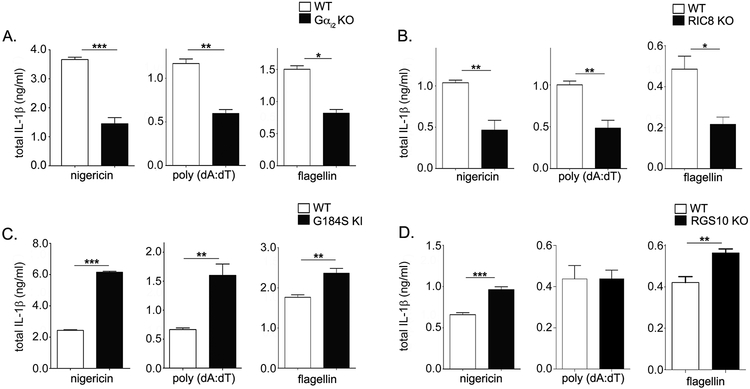
To delineate the role of Gαi and related RGS/AGS proteins on inflammasome activation, we activated the NLRP3, AIM2, and NLRC4 inflammasomes in BMDMs from various genetically modified mice (36). Gαi2 deficient BMDMs show a severe (2.5 fold) reduction in IL-1β release for all inflammasomes assayed (Fig. 1A). To confirm that Gαi2 deficient cells have decreased IL-1β release after inflammasome activation, we looked at Ric-8a deficient BMDMs. Ric-8a is a molecular chaperone critical in targeting Gαi subunits to the plasma membrane (6); thus Ric-8a deficiency leads to a vital loss of Gαi subunits. Like Gαi2 deficient BMDMs, Ric-8a deficient BMDMs demonstrated reduced IL-1β release for all inflammasomes tested (Fig. 1B). Collectively, this suggests that Gαi2 is critical for IL-1β release in macrophages independent of the inflammasome activated.
Figure 1. Gαi2 signaling regulates inflammasome activation in BMDMs.
BMDMs were generated from WT and A) Gαi2 KO (Gnai2−/−), B) Ric-8 KO (Ric8−/−), C) G184S KI (Gnai2G184S/G184S), or D) RGS10 KO (Rgs10−/−) mice. BMDM inflammasomes were activated as indicated in the methods section, and supernatant IL-1β assayed by ELISA. The graphs shown are representative of 5 separate experiments with three replicates. P values were determined using Student’s t-test and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
To determine whether excess Gαi2 signaling reciprocally affects IL-1β release after inflammasome activation, we used Gnai2G184S/G184S knock-in (KI) BMDMs which express the RGS-insensitive Gαi2 G184S allele. The Gαi2 G184S mutation does not affect GPCR-Gαi2 interactions or Gαi2 stability, but results in higher basal levels of Gαi2 signaling due to the inability of the Gαi2 G184S allele to interact with RGS proteins (6). Consistent with our previous results, G184S KI macrophages released significantly more IL-1β than WT macrophages for all inflammasomes assayed (3 fold for NLRP3 and AIM2, 40% for NLRC4; Fig. 1C). This phenomenon was confirmed in BMDMs from mice lacking RGS10, the most highly expressed macrophage RGS protein (38). These BMDMs displayed increased IL-1β release following NLRP3 and NLRC4 activation (Fig. 1D).
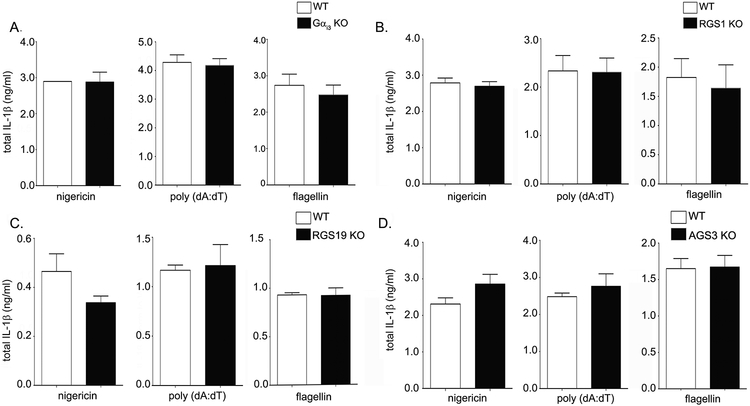
Next, we determined whether Gαi3 deficient BMDMs also show differences in IL-1β release after inflammasome activation. Interestingly, no difference was observed in Gαi3 deficient BMDMs (Fig. 2A). Though both Gαi2 and Gαi3 are expressed in C57BL/6J BMDMs, the Gnai2 transcript is 5 times more abundant, which may explain the relative contributions of Gαi2 and Gαi3 to inflammasome activation (6). Additionally, Gαi3 may have properties different from those of Gαi2, though it is unclear at present which of these properties are important for regulating IL-1β release after inflammasome activation. Mice lacking genes for the less well expressed Gαi2 accessory proteins (RGS1, RGS19, and AGS3) show no phenotypic change after inflammasome activation (Fig. 2B–D), likely due to compensatory overlap from other family members. Untreated and LPS only treated control groups were included for all inflammasome activation experiments, and no IL-1β release was detected for these groups in all cases (data not shown). In sum, our data indicates that deficiencies in Gαi2 but not Gαi3 signaling decrease IL-1β release after inflammasome activation. In contrast, prolonged Gαi2 signaling enhances it.
Figure 2. Gαi3 deficiency does not affect inflammasome activity in macrophages.
BMDMs were generated from WT and A) Gαi3 KO (Gnai3−/−) mice, B) RGS1 KO (Rgs1−/−), C) RGS19 KO (Rgs19 −/−), or D) AGS3 KO (Gpsm1−/−) mice. BMDM inflammasomes were activated as indicated in the methods section, and the supernatant IL-1β assayed by ELISA. The graphs shown are representative of 5 separate experiments with three replicates. P values were determined using Student’s t-test and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Gαi2-mediated signaling plays a role in the priming phase of inflammasome activation
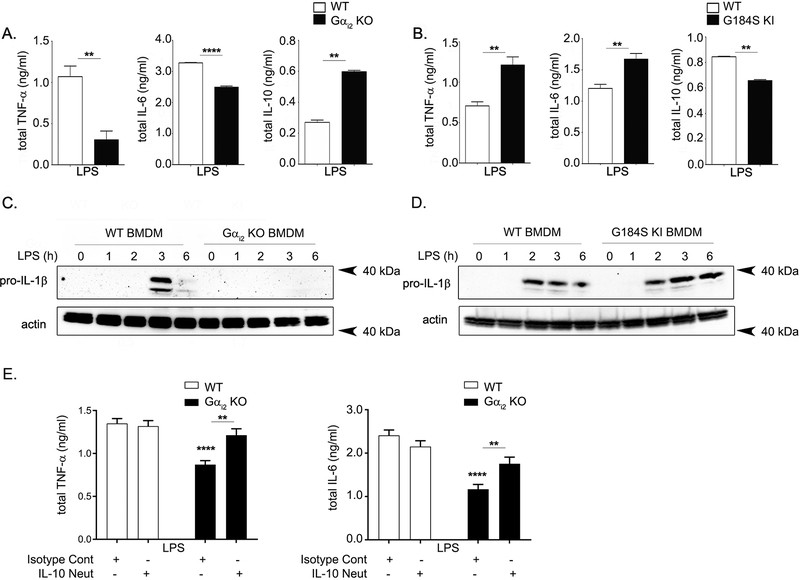
The observation that Gαi2-dependent signaling consistently affects IL-1β release across three types of inflammasomes argues against a role for Gαi2 in inflammasome assembly, as each inflammasome has a different assembly mechanism. Rather, this observation points towards Gαi2 modulating either the LPS priming or the IL-1β secretion phase, which share mechanisms across inflammasomes (39). To determine the phase of Gαi2 involvement, we mimicked inflammasome priming (signal 1) by treatment with LPS and monitored some key downstream cytokines: TNF-α, IL-6, and IL-10 (40). IL-1β is secreted via a non-canonical pathway that utilizes different machinery from standard cytokines; thus changes in supernatant IL-1β without changes in TNF-α and IL-6 secretion suggest IL-1β specific secretion defects. Conversely, changes in IL-1β secretion with accompanying changes in TNF-α and IL-6 point towards global inflammatory changes affecting the LPS priming phase. After LPS stimulation, Gαi2 deficient BMDMs secrete less TNF-α and IL-6 than WT controls, while G184S KI BMDMs secrete more of these cytokines (Fig. 3A–B). These results suggest global inflammatory differences after LPS priming, as opposed to IL-1β specific secretion defects, are responsible for the differences in IL-1β release observed after inflammasome activation in the Gαi2 deficient BMDMs. Consistently, immunoblotting of pro-IL-1β after LPS stimulation showed decreased pro-IL-1β in the Gαi2 deficient BMDMs and increased pro-IL-1β in the G184S KI BMDMs (Fig. 3C–D). Finally, analysis of the anti-inflammatory cytokine IL-10 after LPS stimulation showed an increase in the Gαi2 deficient BMDMs and a decrease in the G184S KI BMDMs (Fig. 3A–B). IL-10 is produced after LPS stimulation in macrophages as an autocrine feedback mechanism to curtail the inflammatory response (41). Thus, we next investigated whether the increased IL-10 production in Gαi2 deficient BMDMs contributes to the IL-6 and TNF-α production defects observed in Gαi2 deficient BMDMs. Neutralization of IL-10 during LPS stimulation rescued both IL-6 and TNF-α production in Gαi2 deficient BMDMs (Fig. 3E), emphasizing the importance of the increased IL-10 production in Gαi2 deficient BMDMs.
Figure 3. Gαi2 deficiency causes global inflammatory changes.
BMDMs generated from WT and A) Gαi2 KO (Gnai2−/−) or B) G184S KI (Gnai2G184S/G184S) mice were stimulated with LPS (1 μg/ml) for 4 hours. The supernatant was collected and assayed for TNF-α, IL-6, and IL-10 by ELISA. The bar graphs shown are representative of five separate experiments with three replicates. C) Immunoblotting for pro-IL-1β after stimulation with LPS (250 ng/ml) in WT and Gαi2 deficient BMDM lysates. D) Immunoblotting for pro-IL-1β after stimulation with LPS (250 ng/mL) in WT and G184S KI BMDM lysates. E) BMDMs generated from WT and Gαi2 KO (Gnai2−/−) mice were stimulated with LPS (1 μg/ml) for 4 hours either in the presence of 10 μg/ml isotype control or IL-10 neutralizing antibody. The supernatant was collected and assayed for TNF-α and IL-6. The graphs shown are representative of five separate experiments with three replicates. P values were determined using Student’s t-test or ordinary one-way ANOVA followed by multiple comparisons, and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Gαi2-mediated signaling is critical in macrophage polarization
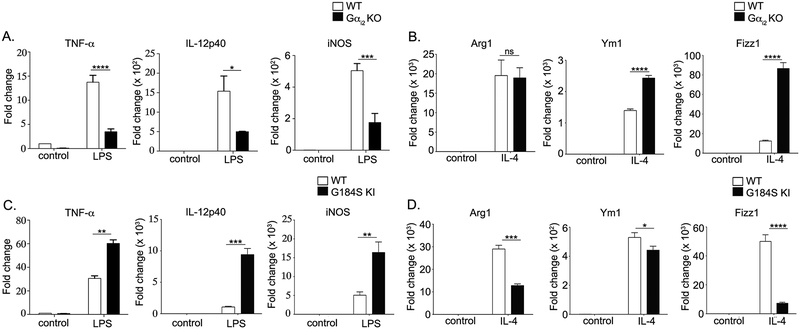
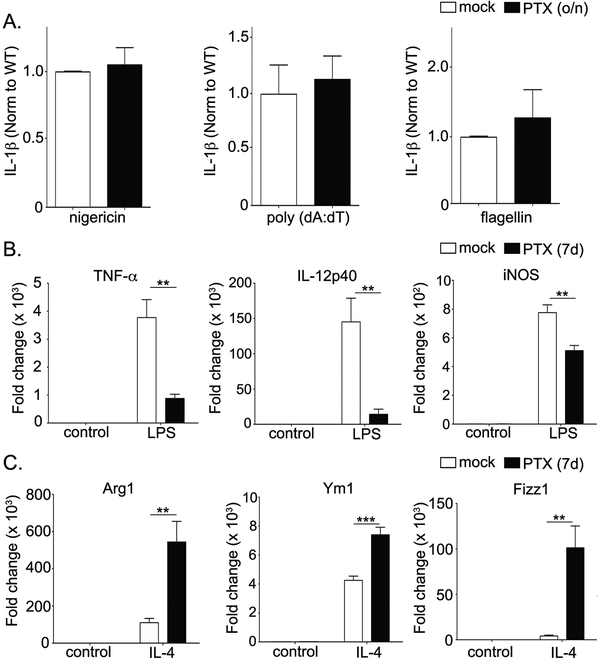
Increased IL-10 despite decreases in TNF-α and IL-6 in Gαi2 deficient BMDMs with reciprocal changes in G184S KI BMDMs suggest alterations in Gαi2 signaling bias macrophage phenotypic determination. To determine the M1/M2 phenotype propensity of Gαi2 deficient and G184S KI BMDMs BMDMs, we examined mRNA levels of standard pro-inflammatory M1 markers (TNF-α, IL-12p40 and iNOS) after LPS stimulation and standard anti-inflammatory M2 markers (Arg1, Ym1, and Fizz1) after IL-4 stimulation. LPS stimulated Gαi2 deficient BMDMs showed decreased TNF-α, IL-12p40, and iNOS mRNA and increased Ym1 and Fizz1 mRNA after IL-4 stimulation (Fig. 4A–B). On the other hand, G184S KI BMDMs had higher levels of pro-inflammatory markers (TNF-α, IL-12p40 and iNOS) after LPS and lower levels of anti-inflammatory markers (Arg1, Ym1, Fizz1) after IL-4 stimulation (Fig. 4C–D). These results are consistent with our observed cytokine and inflammasome changes, indicating that Gαi2 deficiency skews macrophages towards an M2 phenotype while excess Gαi2 signaling results in a tendency towards the pro-inflammatory M1 phenotype. Taken together, this suggests that Gαi2 signaling is critical in macrophage phenotype determination.
Figure 4. Gαi2 signaling is a critical determinant of macrophage pro-inflammatory (M1) vs anti-inflammatory (M2) phenotype.
A) Expression of M1 genes in Gαi2 deficient BMDMs after treatment with LPS (1 μg/ml) for 4 hours. B) Expression of M2 genes in Gαi2 deficient BMDMs after treatment with IL-4 (20 ng/ml) for 48 hours. C) Expression of M1 genes in G184S KI BMDMs after treatment with LPS (1 μg/ml) for 4 hours. D) Expression of M2 genes in G184S KI BMDMs after treatment with IL-4 (20 ng/ml) for 48 hours. Data is presented as fold change relative to the WT control. Experiments were conducted at least 3 times in triplicate. P values were determined using Student’s t-test and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Long term, but not short-term, inhibition of Gαi2 signaling recapitulates inflammatory changes
To investigate the mechanism through which Gαi2 affects macrophage phenotype, we assayed inflammasome activation in BMDMs treated overnight with pertussis toxin (PTX). PTX catalyzes the ADP-ribosylation of Gαi, which blocks receptor-G-protein interaction and inhibits Gαi from undergoing GPCR-mediated nucleotide exchange. Unexpectedly, overnight treatment with PTX showed no differences in IL-1β secretion after inflammasome activation (Fig. 5A). It is unlikely that nucleotide exchange independent effects of Gαi2 are responsible for these changes, given the reciprocal phenotypes of Gαi2 deficient and G184S KI BMDMs. Thus, we investigated whether long-term inhibition of Gαi2 during macrophage development affects phenotype determination. To test this, we eliminated Gαi signaling during the course of macrophage development by growing bone marrow cells in the presence of PTX from day 0 to day 7 (42). On day 7, cells were treated with either LPS or IL-4 followed by transcript analysis of the pro- and anti-inflammatory markers to determine macrophage polarization. Interestingly, BMDMs differentiated in the presence of PTX (long-term) demonstrated a reduction in M1 genes upon LPS stimulation (Fig. 5B) and a corresponding induction M2 genes upon IL-4 treatment (Fig. 5C). Together, this data indicates that prolonged Gαi2 signaling plays a critical role in the establishment of macrophage phenotype, potentially during growth and differentiation.
Figure 5. Long-term, but not short term, inhibition of Gαi2 signaling recapitulates the phenotypes seen in genetically altered BMDMs.
A) BMDMs from WT mice treated at only day 7 with PTX (200 ng/ml) overnight before inflammasome activation. BMDM culture supernatants were collected and IL-1β assayed by ELISA. The bar graphs shown are representative of five separate experiments with three replicates. B) Expression of M1 genes in PTX treated BMDMs (50 ng/ml for days 0–2; 100 ng/ml for days 3–7) after treatment with LPS (1 μg/ml) for 4 hours. C) Expression of M2 genes in PTX treated BMDMs (50 ng/ml for days 0–2; 100 ng/ml for days 3–7) after treatment with IL-4 (20 ng/ml) for 48 hours. RT-PCR experiments were conducted at least 3 times in triplicate. Data is presented as fold change relative to the WT control. P values were determined using Student’s t-test and p < 0.05 was considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Gαi2 modulates signaling downstream of LPS and IL-4
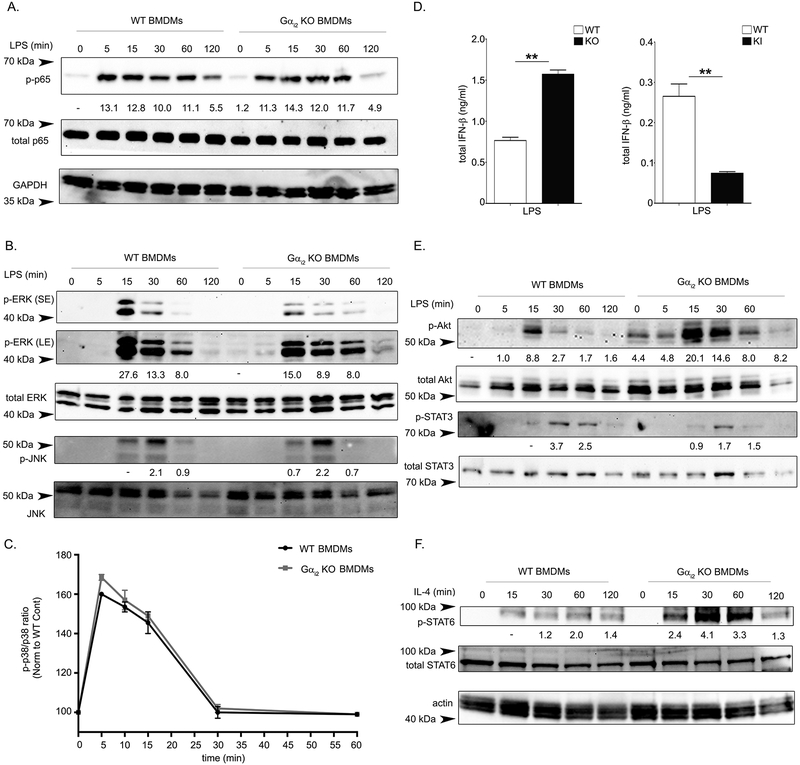
To investigate the mechanisms through which Gαi2 alters macrophage polarization phenotype, we stimulated WT and Gαi2 deficient BMDMs with LPS and monitored critical downstream signaling outputs. NFκB signaling is a major inflammatory pathway downstream of LPS (43), but our results show Gαi2 deficient BMDMs have no alterations in NFκB signaling as assayed by p65 phosphorylation (Fig. 6A). LPS/TLR4 signaling also results in MyD88-dependent activation of the MAPK cascade, activating ERK1/2 (p42/44), JNK1/2 (p46/54), and p38 (43). Activation of MAPK cascade proteins are important in the cytokine production downstream of LPS (44). Gαi2 deficient BMDMs exhibit defects in ERK1/2 activation, but no changes in JNK1/2 or p38 activation after LPS stimulation (Fig. 6B–C). LPS/TLR4 signaling also activates IRF3 in a MyD88-independent manner, resulting in the production of type 1 interferons (43). To monitor this pathway, we stimulated WT, Gαi2 deficient, and G184S KI BMDMs with LPS and monitored IFN-β. Gαi2 deficient BMDMs release significantly more IFN-β after LPS stimulation, while G184S KI BMDMs release less (Fig. 6D). This result suggests that Gαi2 restricts IRF3 mediated signaling downstream of LPS. Finally, both STAT3 and the kinase Akt are activated after LPS stimulation (45–47). Gαi2 deficient BMDMs show clear STAT3 activation defects after LPS stimulation and enhanced Akt activation both basally and after LPS stimulation (Fig. 6E). Collectively, these data indicate that Gαi2 modulates signaling downstream of TLR4/LPS, which may play a role in the observed macrophage polarization phenotypes observed. In order to determine if Gαi2 affects signaling downstream of IL-4, we stimulated WT and Gαi2 deficient BMDMs with IL-4 assayed STAT6 activation. STAT6 is an essential transcription factor downstream of IL-4 that functions as a transcriptional repressor to limit activation of inflammatory enhancers (48). Gαi2 deficient BMDMs show elevated levels of STAT6 activation after IL-4 stimulation, consistent with the M2 skewed phenotype seen in Gαi2 deficient BMDMs (Figure 6F).
Figure 6. Gαi2 modulates signaling downstream of LPS and IL-4.
BMDMs generated from WT and Gαi2 KO (Gnai2−/−) mice were treated with LPS (250 ng/ml) for the indicated time. Whole cell lysates were collected and separated on an SDS PAGE gel. Immunoblots are shown for A) NFκB related protein p-65 (phospho and total) and GAPDH, and B) MAPK cascade proteins ERK1/2 (phospho and total) and JNK1/2 (phospho and total). C) Phosflow analysis of p38 phosphorylation after LPS stimulation (250 ng/ml) for the indicated time in BMDMs generated from WT and Gαi2 KO (Gnai2−/−) mice. Data is presented as the mean fluorescence intensity ratio of phospho p38 to total p38 and normalized to the control. D) BMDMs generated from WT and Gαi2 KO (Gnai2−/−) mice were treated with LPS (1 μg/ml) for 4 hours, cell culture supernatant was collected and assayed for IFN-β. E) Immunoblotting for STAT3 (phospho and total) and Akt (phospho and total) from lysates obtained from LPS stimulated (250 ng/ml) WT or Gαi2 KO (Gnai2−/−) mice. F) BMDMs generated from WT and Gαi2 KO (Gnai2−/−) mice were treated with IL-4 (20 ng/ml) for the indicated time. Immunoblotting for STAT6 (phospho and total) and actin are shown. Phospho protein bands are quantified relative to their respective total protein band. Band intensities are normalized to the indicated control. Experiments were performed a minimum of three times.
Gαi2 deficient macrophages have both functional and qualitative changes
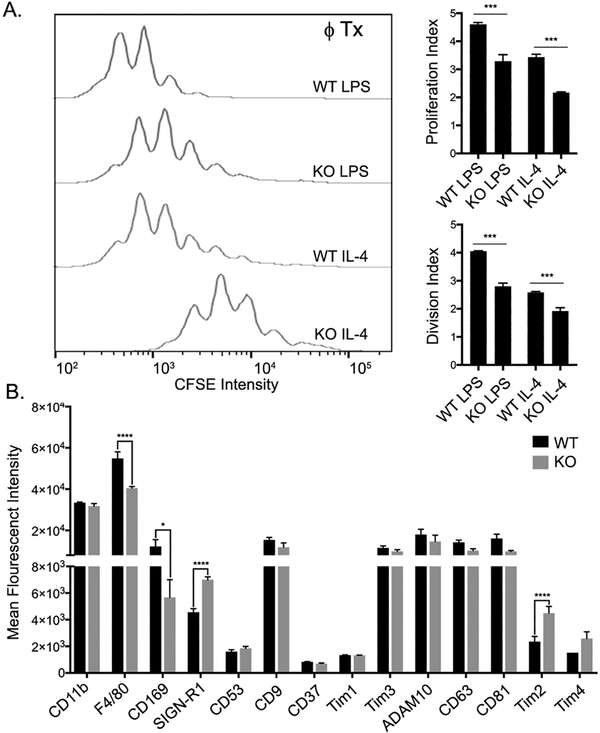
While M1 type macrophages promote T cell proliferation, M2 macrophages are suppressive and inhibit T cells proliferation (49). To test whether Gαi2 deficient macrophages have altered T cell suppression characteristics, we co-cultured naïve T helper cells (CD4+CD25−) with either M1 or M2 polarized WT or Gαi2 deficient BMDMs while stimulating with IL-2 and plate bound anti-CD3/CD28. As expected, co-culture with M1 polarized WT macrophages induced significant proliferation, while M1 polarized Gαi2 deficient macrophages induced significantly less proliferation (Fig. 7A). M2 polarization of WT macrophages significantly suppressed T cell proliferation compared to the M1 macrophages, and this suppression was augmented in M2 polarized Gαi2 deficient BMDMs (Fig. 7A). Together, this data suggests that Gαi2 deficient BMDMs have functional changes that fits the M1/M2 paradigm. Finally, we investigated if Gαi2 deficient BMDMs express a variety of macrophages markers at different levels than WT BMDMs. Interestingly, Gαi2 KO BMDMs had decreased levels of F4/80 and CD169 and increased levels of SIGN-R1 and Tim2 (Fig. 7B). Together, this data suggests that Gαi2 KO BMDMs have functional changes affecting macrophage-T cell interactions, and qualitative phenotypic differences after in vitro culture.
Figure 7. Gαi2 deficient macrophages have functional and qualitative defects.
A) CSFE proliferation peaks of purified T cells stimulated with IL-2 (20 ng/mL), anti-CD3 (1 μg/mL), and anti-CD28 (0.5 μg/mL) and co-cultured with LPS or IL-4 treated WT or Gαi2 deficient BMDMs. Proliferation index and division index are normalized to the T cell only control and shown for each group. B) Mean fluorescent intensity (MFI) of several cell surface markers in day 7 BMDMs from WT or Gαi2 KO mice. Experiments were performed a minimum of three times. Significance was measured by ANOVA with multiple comparisons and p < 0.05 considered statistically significant (* p <0.05; ** p<0.005, *** p<0.001).
Discussion
Our study identifies that Gαi2 signaling regulates IL-1β release after inflammasome activation, likely due to differences in inflammasome priming related to biased M1 and M2 polarization in G184S KI and Gαi2 deficient BMDMs respectively. Previous studies have established that macrophage polarization affects transcriptional upregulation of inflammasome related genes upon LPS priming (27). Specifically, M1 polarized macrophages have higher basal levels and more robustly upregulate pro-IL-1β mRNA after LPS stimulation compared to their M2 counterparts. These transcriptional differences in pro-IL-1β levels affect IL-1β release after inflammasome activation, as the same study showed M1 macrophages release more IL-1β than M2 macrophages after NLRP3 activation (27).
Apart from changes in inflammasome activation, our data shows phenotypic differences in cytokine release after LPS stimulation in Gαi2 deficient and G184S KI BMDMs. Of note, we find that the increased IL-10 production in Gαi2 deficient macrophages is important in limiting production of IL-6 and TNF-α in these macrophages. With regards to the mechanism through which Gαi2 regulates macrophage polarization and cytokine production, many lines of evidence suggest that Gαi-signaling plays a direct role in LPS induced TLR4 signal transduction (50–52). For example, Gαi2 is phosphorylated in response to LPS, co-immunoprecipitates with the LPS co-receptor CD14 (53, 54), and genetic deletion of Gαi2 decreases the activation of signaling pathways downstream of TLR4 (51, 55, 56). While NFκB is an essential transcription factor downstream of LPS responsible for pro-IL-1β, IL-6, and TNF-α transcription (43, 57), our data is in consensus with the literature in indicating that Gαi2 deficient macrophages exhibit no changes in NFκB signaling after TLR4 activation (51, 56).
We observe increased IFN-β production and Akt activation in Gαi2 deficient macrophages after LPS stimulation, and both IFN-β and Akt have previously been implicated in IL-10 production downstream of LPS (41, 47). Increased signaling through these pathways may be responsible for the observed increase in IL-10 in Gαi2 deficient macrophages, which is important in limiting IL-6 and TNF-α production. We also find defects in ERK1/2 and STAT3 activation in Gαi2 deficient macrophages after LPS stimulation. ERK1/2 activation downstream of LPS has previously been shown to contribute to pro-IL-1β, IL-6, and TNF-α production (44, 58), and acute STAT3 activation downstream of TLR4 has been shown to play a role in IL-6 and pro-IL-1β production (46). The altered signaling downstream of TLR4 likely plays an important role in the observed phenotypes after LPS stimulation. In addition to alterations after LPS stimulation, we see greater M2 (alternative) activation after IL-4 signaling in Gαi2 deficient macrophages and less M2 activation in G184S KI macrophages. Signaling studies upon IL-4 stimulation show greater STAT6 activation in Gαi2 deficient macrophages. STAT6 is a critical transcription factor activated by IL-4 that acts as a transcriptional repressor to limit inflammatory enhancer activation in macrophages (48). Differences in M2 activation in Gαi2 deficient and G184S KI macrophages after IL-4 stimulation may be related to signaling downstream of IL-4.
We note that in addition to Gαi2 modulating signaling downstream of LPS and IL-4, other mechanisms may exist that contribute to the observed macrophage polarization phenotypes. For example, M1 polarized macrophages are characterized in part by their dependence on glycolysis, while M2 polarized macrophages are reliant on oxidative metabolism (59). The transcriptional upregulation of pro-IL-1β after LPS stimulation is dependent on this glycolytic switch, which activates the transcription factor HIF-1α enabling its direct binding to the Il1b promoter (60). Our findings warrant further investigation into whether metabolic changes in Gαi2-deficient and G184S KI BMDMs are responsible for alterations in pro-IL-1β upregulation after LPS stimulation. Additionally, epigenetic changes have been implicated in regulating macrophage polarization (61, 62). An interesting observation in our study pointing to the existence of mechanisms in addition to Gαi2 regulating signaling downstream of LPS and IL-4 is that short-term inhibition of Gαi2 using PTX (prior to TLR4 stimulation with LPS) does not recapitulate the LPS dependent inflammasome related changes seen in Gαi2 deficient macrophages, while 7-day inhibition of Gαi2 during differentiation from bone marrow recapitulates the anti-inflammatory phenotype. The mechanisms responsible for this phenomenon remain unclear, but these finding raise the possibility that changes responsible for the macrophage polarization phenotype are built into the macrophage life history.
Our studies generally exhibit a consensus with regards to cytokine response between Gαi2 deficient BMDMs investigated here and Gαi2 deficient peritoneal macrophages previously studied (52, 55). However, G184S KI peritoneal macrophages were previously reported to have decreased cytokine release after LPS stimulation, while our study found increased TNF-α and IL-6 (63). This discrepancy may be explained by developmental differences between peritoneal and BMDMs. Peritoneal macrophages are resident tissue macrophages, which are typically derived prenatally from embryonic precursors as opposed to adult hematopoietic stem cells (64). BMDMs generated in vitro likely share more similarities with in vivo monocyte-derived macrophages, which are of hematopoietic origin (64). Importantly, these types of macrophages have significant developmental differences at the transcriptional level (65), and thus are likely convergently affected by signaling inputs during development. Despite inconsistencies with Li et al. (63), the proinflammatory M1 nature of G184S KI BMDMs revealed in this study are consistent with our previous findings that unprimed G184S KI macrophages have enhanced phagocytic capacity (29, 66). Our data is also consistent with a study on RGS10 deficient macrophages (38). RGS10 is the best expressed RGS protein in macrophages, and absence of RGS10 decelerates GTPase activity of the Gαi subunit (like the Gαi2 G184S KI allele). Lee et al. showed that RGS10 deficient peritoneal and bone-marrow derived macrophages are proinflammatory and M1 skewed (38).
The existing data on Gαi-coupled GPCR signaling remains difficult to interpret, as the functional consequences of specific ligands on Gαi signaling is not well established. FPR2 promotes M1 polarization upon stimulation with the cathelicidin associated microbial peptide LL-37 or serum amyloid protein A (12). On the other hand, this same receptor promotes M2 polarization when bound by Annexin A1, lipid lipoxin A4, or the anti-inflammatory molecule resolvin D1 (12, 15). Studies in FPR2 KO mice show that loss of FPR2 promotes an M2 macrophage phenotype and tumor progression (13, 14), suggesting the physiological role of FPR2 is to promote M1 polarization. As FPR2 couples Gαi proteins and is known to be constitutively active (18, 67), it is possible that the M2 polarization seen in FPR2 KO cells stems from a reduction of Gαi2 signaling. Of note, FPR2 activation of Gαi2 results in concurrent Gβγ dissociation, which may play a role in the observed macrophage polarization phenotype. CXCR3, which also couples Gαi, is similarly implicated in M1 polarization as CXCR3 KO macrophages are M2 biased and promote tumorigenesis (17). Though CXCR3 ligands (CXCL9–11) have similar effects on polarization, recent studies show that CXCR3 has multiple isoforms that are differentially expressed on M1 (CXCR3.1) and M2 (CXCR3.2) macrophages. These isoforms have opposing effects on macrophage polarization, as stimulation of CXCR3.1 promotes an M1 phenotype while CXCR3.2 promotes an M2 phenotype (16). These CXCR3 related phenomena may be related to CXCR3 isoform specific differences in Gαi coupling. Finally, the ChemR23 receptor is expressed only on M1 macrophages; it too couples Gαi and has ligand specific proinflammatory (Chemerin) and proresolving (resolvin E1) affects (68).
Importantly, the question remains as to whether prolonged Gαi2 inhibition itself reprograms macrophages towards the M2 phenotype or whether this inhibition must occur during macrophage differentiation. The former scenario has major therapeutic implications, as prolonged stimulation or inhibition of macrophage specific Gαi2-coupled GPCRs could lay the foundation for the treatment of many inflammatory diseases via macrophage reprograming. Finally, while the M1/M2 paradigm is useful for in vitro evaluation of macrophage phenotype, several macrophage subsets exist in vivo. Notably, Gαi2 deficient BMDMs have alterations in SIGN-R1 and CD169 expression, both of which define specific splenic and lymph node macrophage subsets with unique functions (69). Moving forward, further investigation into the role of Gαi2 in these macrophage subsets and their role in disease pathogenesis is warranted.
Acknowledgements
The authors would like to thank Dr. Anthony Fauci for his long-standing support.
Footnotes
A.V. and N.R.N. contributed equally
A.V. Present address: Department of Pharmacology, Wayne State University School of Medicine, Detroit, Michigan, 48202
The intramural program of the National Institutes of Allergy and Infectious Diseases supported this research. N.R.N. is supported in part by the Thomas Jefferson University MD/PhD training program.
Address correspondence and reprint requests to Neel R. Nabar or John H. Kehrl. B Cell Molecular Immunology Section, Laboratory of Immunoregulation, NIAID, NIH. 10 Center Drive, Rm 11A01, Bethesda MD 20892. Email: neel.nabar@nih.gov or jkehrl@niaid.nih.gov
Abbreviations used in this article: AGS, activator of G protein Signaling; BMDM, bone marrow derived macrophage; GAP, GTPase accelerating protein; GPCR, G-protein coupled receptor; KI, Knock-in; KO, Knockout; PTX, pertussis toxin; RGS, regulator of G protein signaling; WT, wild type
References
- 1.Vural A, and Kehrl JH. 2014. Autophagy in macrophages: impacting inflammation and bacterial infection. Scientifica (Cairo) 2014: 825463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray PJ 2017. Macrophage Polarization. Annual Review of Physiology 79: 541–566. [DOI] [PubMed] [Google Scholar]
- 3.Newton K, and Dixit VM. 2012. Signaling in innate immunity and inflammation Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumer JB, Smrcka AV, and Lanier SM. 2007. Mechanistic pathways and biological roles for receptor-independent activators of G-protein signaling. Pharmacol Ther 113: 488–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodard GE, Jardin I, Berna-Erro A, Salido GM, and Rosado JA. 2015. Regulators of G-protein-signaling proteins: negative modulators of G-protein-coupled receptor signaling. International review of cell and molecular biology 317: 97–183. [DOI] [PubMed] [Google Scholar]
- 6.Kehrl JH 2016. The impact of RGS and other G-protein regulatory proteins on Galphai-mediated signaling in immunity. Biochem Pharmacol 114: 40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srinivasa SP, Watson N, Overton MC, and Blumer KJ. 1998. Mechanism of RGS4, a GTPase-activating Protein for G Protein α Subunits. Journal of Biological Chemistry 273: 1529–1533. [DOI] [PubMed] [Google Scholar]
- 8.Blumer JB, Oner SS, and Lanier SM. 2012. Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol (Oxf) 204: 202–218. [DOI] [PubMed] [Google Scholar]
- 9.Blumer JB, and Lanier SM. 2014. Activators of G Protein Signaling Exhibit Broad Functionality and Define a Distinct Core Signaling Triad. Molecular Pharmacology 85: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vural A, Al-Khodor S, Cheung GY, Shi CS, Srinivasan L, McQuiston TJ, Hwang IY, Yeh AJ, Blumer JB, Briken V, Williamson PR, Otto M, Fraser ID, and Kehrl JH. 2016. Activator of G-Protein Signaling 3-Induced Lysosomal Biogenesis Limits Macrophage Intracellular Bacterial Infection. J Immunol 196: 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin H-H, and Stacey M. 2016. G Protein-Coupled Receptors in Macrophages Microbiology Spectrum 4. [DOI] [PubMed] [Google Scholar]
- 12.Cooray SN, Gobbetti T, Montero-Melendez T, McArthur S, Thompson D, Clark AJ, Flower RJ, and Perretti M. 2013. Ligand-specific conformational change of the G-protein-coupled receptor ALX/FPR2 determines proresolving functional responses. Proceedings of the National Academy of Sciences of the United States of America 110: 18232–18237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Cai L, Wang H, Wu P, Gu W, Chen Y, Hao H, Tang K, Yi P, Liu M, Miao S, and Ye D. 2011. Pleiotropic regulation of macrophage polarization and tumorigenesis by formyl peptide receptor-2. Oncogene 30: 3887–3899. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Chen K, Wang C, Gong W, Yoshimura T, Liu M, and Wang JM. 2013. Cell surface receptor FPR2 promotes anti-tumor host defense by limiting M2 polarization of macrophages. Cancer research 73: 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson JW, Leigh NJ, Mellas RE, McCall AD, Aguirre A, and Baker OJ. 2014. ALX/FPR2 receptor for RvD1 is expressed and functional in salivary glands. American journal of physiology. Cell physiology 306: C178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X-J, Chen Q, Rong Y-J, Chen F, and Chen J. 2017. CXCR3.1 and CXCR3.2 Differentially Contribute to Macrophage Polarization in Teleost Fish. The Journal of Immunology 198: 4692. [DOI] [PubMed] [Google Scholar]
- 17.Oghumu S, Varikuti S, Terrazas C, Kotov D, Nasser MW, Powell CA, Ganju RK, and Satoskar AR. 2014. CXCR3 deficiency enhances tumor progression by promoting macrophage M2 polarization in a murine breast cancer model. Immunology 143: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenzel-Seifert K, Arthur JM, Liu H-Y, and Seifert R. 1999. Quantitative Analysis of Formyl Peptide Receptor Coupling to Giα1, Giα2, and Giα3. Journal of Biological Chemistry 274: 33259–33266. [DOI] [PubMed] [Google Scholar]
- 19.Corminboeuf O, and Leroy X. 2015. FPR2/ALXR Agonists and the Resolution of Inflammation. Journal of Medicinal Chemistry 58: 537–559. [DOI] [PubMed] [Google Scholar]
- 20.Butcher MJ, and Galkina EV. 2015. wRAPping up early monocyte and neutrophil recruitment in atherogenesis via AnnexinA1/FPR2 signaling. Circulation research 116: 774–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson BD, Jin Y, Wu KH, Colvin RA, Luster AD, Birnbaumer L, and Wu MX. 2007. Inhibition of Gαi2 Activation by Gαi3 in CXCR3-mediated Signaling. Journal of Biological Chemistry 282: 9547–9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Henau O, Degroot G-N, Imbault V, Robert V, De Poorter C, McHeik S, Galés C, Parmentier M, and Springael J-Y. 2016. Signaling Properties of Chemerin Receptors CMKLR1, GPR1 and CCRL2. PLoS ONE 11: e0164179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinon F, Mayor A, and Tschopp J. 2009. The inflammasomes: guardians of the body. Annu Rev Immunol 27: 229–265. [DOI] [PubMed] [Google Scholar]
- 24.Nabar NR, and Kehrl JH. 2017. The Transcription Factor EB Links Cellular Stress to the Immune Response. Yale J Biol Med 90: 301–315. [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Callaway JB, and Ting JPY. 2015. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 21: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris J, Lang T, Thomas JPW, Sukkar MB, Nabar NR, and Kehrl JH. 2017. Autophagy and inflammasomes. Mol Immunol 86: 10–15. [DOI] [PubMed] [Google Scholar]
- 27.Awad F, Assrawi E, Jumeau C, Georgin-Lavialle S, Cobret L, Duquesnoy P, Piterboth W, Thomas L, Stankovic-Stojanovic K, Louvrier C, Giurgea I, Grateau G, Amselem S, and Karabina S-A. 2017. Impact of human monocyte and macrophage polarization on NLR expression and NLRP3 inflammasome activation. PLOS ONE 12: e0175336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiege K, Le DD, Syed SN, Ali SR, Novakovic A, Beer-Hammer S, Piekorz RP, Schmidt RE, Nurnberg B, and Gessner JE. 2012. Defective macrophage migration in Galphai2- but not Galphai3-deficient mice. J Immunol 189: 980–987. [DOI] [PubMed] [Google Scholar]
- 29.Huang NN, Becker S, Boularan C, Kamenyeva O, Vural A, Hwang IY, Shi CS, and Kehrl JH. 2014. Canonical and noncanonical g-protein signaling helps coordinate actin dynamics to promote macrophage phagocytosis of zymosan. Mol Cell Biol 34: 4186–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plummer NW, Spicher K, Malphurs J, Akiyama H, Abramowitz J, Nurnberg B, and Birnbaumer L. 2012. Development of the mammalian axial skeleton requires signaling through the Galpha(i) subfamily of heterotrimeric G proteins. Proceedings of the National Academy of Sciences of the United States of America 109: 21366–21371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang X, Fu Y, Charbeneau RA, Saunders TL, Taylor DK, Hankenson KD, Russell MW, D’Alecy LG, and Neubig RR. 2006. Pleiotropic phenotype of a genomic knock-in of an RGS-insensitive G184S Gnai2 allele. Mol Cell Biol 26: 6870–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JK, Chung J, McAlpine FE, and Tansey MG. 2011. Regulator of G-protein signaling-10 negatively regulates NF-kappaB in microglia and neuroprotects dopaminergic neurons in hemiparkinsonian rats. J Neurosci 31: 11879–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boularan C, Hwang IY, Kamenyeva O, Park C, Harrison K, Huang Z, and Kehrl JH. 2015. B Lymphocyte-Specific Loss of Ric-8A Results in a Galpha Protein Deficit and Severe Humoral Immunodeficiency. J Immunol 195: 2090–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang IY, Park C, Harrision KA, Huang NN, and Kehrl JH. 2010. Variations in Gnai2 and Rgs1 expression affect chemokine receptor signaling and the organization of secondary lymphoid organs. Genes Immun 11: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blumer JB, Lord K, Saunders TL, Pacchioni A, Black C, Lazartigues E, Varner KJ, Gettys TW, and Lanier SM. 2008. Activator of G protein signaling 3 null mice: I. Unexpected alterations in metabolic and cardiovascular function. Endocrinology 149: 3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vural A, McQuiston TJ, Blumer JB, Park C, Hwang IY, Williams-Bey Y, Shi CS, Ma DZ, and Kehrl JH. 2013. Normal Autophagic Activity in Macrophages from Mice Lacking Galphai3, AGS3, or RGS19. PLoS One 8: e81886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu L, Yang T, Li L, Sun L, Hou Y, Hu X, Zhang L, Tian H, Zhao Q, Peng J, Zhang H, Wang R, Yang Z, Zhang L, and Zhao Y. 2014. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat Commun 5: 4696. [DOI] [PubMed] [Google Scholar]
- 38.Lee JK, Chung J, Kannarkat GT, and Tansey MG. 2013. Critical role of regulator G-protein signaling 10 (RGS10) in modulating macrophage M1/M2 activation. PLoS One 8: e81785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Broz P, and Dixit VM. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16: 407–420. [DOI] [PubMed] [Google Scholar]
- 40.Arango Duque G, and Descoteaux A. 2014. Macrophage Cytokines: Involvement in Immunity and Infectious Diseases. Frontiers in Immunology 5: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer SS, Ghaffari AA, and Cheng G. 2010. Lipopolysaccharide-Mediated IL-10 Transcriptional Regulation Requires Sequential Induction of Type I IFNs and IL-27 in Macrophages. The Journal of Immunology 185: 6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fields TA, and Casey PJ. 1997. Signalling functions and biochemical properties of pertussis toxin-resistant G-proteins. Biochem J 321 (Pt 3): 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y-C, Yeh W-C, and Ohashi PS. 2008. LPS/TLR4 signal transduction pathway. Cytokine 42: 145–151. [DOI] [PubMed] [Google Scholar]
- 44.Carter AB, Monick MM, and Hunninghake GW. 1999. Both Erk and p38 Kinases Are Necessary for Cytokine Gene Transcription. American Journal of Respiratory Cell and Molecular Biology 20: 751–758. [DOI] [PubMed] [Google Scholar]
- 45.Greenhill CJ, Rose-John S, Lissilaa R, Ferlin W, Ernst M, Hertzog PJ, Mansell A, and Jenkins BJ. 2011. IL-6 Trans-Signaling Modulates TLR4-Dependent Inflammatory Responses via STAT3. The Journal of Immunology 186: 1199. [DOI] [PubMed] [Google Scholar]
- 46.Samavati L, Rastogi R, Du W, Hüttemann M, Fite A, and Franchi L. 2009. STAT3 tyrosine phosphorylation is critical for interleukin 1 beta and interleukin-6 production in response to lipopolysaccharide and live bacteria. Molecular Immunology 46: 1867–1877. [DOI] [PubMed] [Google Scholar]
- 47.Pengal RA, Ganesan LP, Wei G, Fang H, Ostrowski MC, and Tridandapani S. 2006. Lipopolysaccharide-induced production of interleukin-10 is promoted by the serine/threonine kinase Akt. Molecular Immunology 43: 1557–1564. [DOI] [PubMed] [Google Scholar]
- 48.Czimmerer Z, Daniel B, Horvath A, Ruckerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta-Monroy I, Simandi Z, Steiner L, Nagy B Jr., Poliska S, Banko C, Bacso Z, Schulman IG, Sauer S, Deleuze JF, Allen JE, Benko S, and Nagy L. 2018. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity 48: 75–90.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oishi S, Takano R, Tamura S, Tani S, Iwaizumi M, Hamaya Y, Takagaki K, Nagata T, Seto S, Horii T, Osawa S, Furuta T, Miyajima H, and Sugimoto K. 2016. M2 polarization of murine peritoneal macrophages induces regulatory cytokine production and suppresses T-cell proliferation. Immunology 149: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakway JP, and DeFranco AL. 1986. Pertussis toxin inhibition of B cell and macrophage responses to bacterial lipopolysaccharide. Science 234: 743–746. [DOI] [PubMed] [Google Scholar]
- 51.Ferlito M, Romanenko OG, Guyton K, Ashton S, Squadrito F, Halushka PV, and Cook JA. 2002. Implication of Galpha i proteins and Src tyrosine kinases in endotoxin-induced signal transduction events and mediator production. J Endotoxin Res 8: 427–435. [PubMed] [Google Scholar]
- 52.Fan H, Zingarelli B, Peck OM, Teti G, Tempel GE, Halushka PV, Spicher K, Boulay G, Birnbaumer L, and Cook JA. 2005. Lipopolysaccharide- and gram-positive bacteria-induced cellular inflammatory responses: role of heterotrimeric Galpha(i) proteins. American journal of physiology. Cell physiology 289: C293–301. [DOI] [PubMed] [Google Scholar]
- 53.Solomon KR, Kurt-Jones EA, Saladino RA, Stack AM, Dunn IF, Ferretti M, Golenbock D, Fleisher GR, and Finberg RW. 1998. Heterotrimeric G proteins physically associated with the lipopolysaccharide receptor CD14 modulate both in vivo and in vitro responses to lipopolysaccharide. J Clin Invest 102: 2019–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daniel-Issakani S, Spiegel AM, and Strulovici B. 1989. Lipopolysaccharide response is linked to the GTP binding protein, Gi2, in the promonocytic cell line U937. Journal of Biological Chemistry 264: 20240–20247. [PubMed] [Google Scholar]
- 55.Fan H, Williams DL, Zingarelli B, Breuel KF, Teti G, Tempel GE, Spicher K, Boulay G, Birnbaumer L, Halushka PV, and Cook JA. 2007. Differential regulation of lipopolysaccharide and Gram-positive bacteria induced cytokine and chemokine production in macrophages by Galpha(i) proteins. Immunology 122: 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan H, Peck OM, Tempel GE, Halushka PV, and Cook JA. 2004. Toll-like receptor 4 coupled GI protein signaling pathways regulate extracellular signal-regulated kinase phosphorylation and AP-1 activation independent of NFkappaB activation. Shock 22: 57–62. [DOI] [PubMed] [Google Scholar]
- 57.Cogswell JP, Godlevski MM, Wisely GB, Clay WC, Leesnitzer LM, Ways JP, and Gray JG. 1994. NF-kappa B regulates IL-1 beta transcription through a consensus NF-kappa B binding site and a nonconsensus CRE-like site. The Journal of Immunology 153: 712. [PubMed] [Google Scholar]
- 58.Kim SH, Kim J, and Sharma RP. 2004. Inhibition of p38 and ERK MAP kinases blocks endotoxin-induced nitric oxide production and differentially modulates cytokine expression. Pharmacological Research 49: 433–439. [DOI] [PubMed] [Google Scholar]
- 59.Galván-Peña S, and O’Neill LAJ. 2014. Metabolic Reprograming in Macrophage Polarization. Frontiers in Immunology 5: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffery BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, and O’Neill LAJ. 2013. Succinate is a danger signal that induces IL-1β via HIF-1α. Nature 496: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Groot AE, and Pienta KJ. 2018. Epigenetic control of macrophage polarization: implications for targeting tumor-associated macrophages. Oncotarget 9: 20908–20927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivashkiv LB 2013. Epigenetic regulation of macrophage polarization and function. Trends in Immunology 34: 216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li P, Neubig RR, Zingarelli B, Borg K, Halushka PV, Cook JA, and Fan H. 2012. Toll-like receptor-induced inflammatory cytokines are suppressed by gain of function or overexpression of Galpha(i2) protein. Inflammation 35: 1611–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies LC, Jenkins SJ, Allen JE, and Taylor PR. 2013. Tissue-resident macrophages. Nature immunology 14: 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perdiguero EG, and Geissmann F. 2015. The development and maintenance of resident macrophages. Nature Immunology 17: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lam RS, O’Brien-Simpson NM, Holden JA, Lenzo JC, Fong SB, and Reynolds EC. 2016. Unprimed, M1 and M2 Macrophages Differentially Interact with Porphyromonas gingivalis. PLoS ONE 11: e0158629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seifert R, and Wenzel-Seifert K. 2003. The human formyl peptide receptor as model system for constitutively active G-protein-coupled receptors. Life Sciences 73: 2263–2280. [DOI] [PubMed] [Google Scholar]
- 68.Herová M, Schmid M, Gemperle C, and Hersberger M. 2015. ChemR23, the Receptor for Chemerin and Resolvin E1, Is Expressed and Functional on M1 but Not on M2 Macrophages. The Journal of Immunology 194: 2330. [DOI] [PubMed] [Google Scholar]
- 69.Gordon S, Plüddemann A, and Mukhopadhyay S. 2015. Sinusoidal Immunity: Macrophages at the Lymphohematopoietic Interface. Cold Spring Harbor Perspectives in Biology 7: a016378. [DOI] [PMC free article] [PubMed] [Google Scholar]