Abstract
Antibody diversity is initiated by activation-induced deaminase (AID), which deaminates cytosine to uracil in DNA. Uracils in the Ig gene loci can be recognized by uracil DNA glycosylase (UNG) or mutS homologs 2 and 6 (MSH2-MSH6) proteins, and then processed into DNA breaks. Breaks in switch regions of the heavy chain locus cause isotype switching and have been extensively characterized as staggered and blunt double-strand breaks. However, breaks in V regions that arise during somatic hypermutation are poorly understood. Here we characterize AID-dependent break formation in JH introns from mouse germinal center B cells. We used a ligation-mediated PCR assay to detect single-strand breaks and double-strand breaks that were either staggered or blunt. In contrast to switch regions, V regions contained predominantly single-strand breaks, which peaked 10 days after immunization. We then examined the pathways utilized to generate these breaks in UNG- and MSH6-deficient mice. Surprisingly, both DNA repair pathways contributed substantially to break formation, and in the absence of both UNG and MSH6, the frequency of breaks was severely reduced. When the breaks were sequenced and mapped, they were widely distributed over a 1,000-bp intron region downstream of JH3 and JH4 exons and were unexpectedly located at all four nucleotides. These data suggest that during DNA repair, nicks are generated at distal sites from the original deaminated cytosine, and these repair intermediates could generate both faithful and mutagenic repair. During mutagenesis, single-strand breaks would allow entry for low-fidelity DNA polymerases to generate somatic hypermutation.
Introduction
In the iconic 1966 publication on “Origin of antibody variation” (1), Brenner and Milstein proposed that somatic hypermutation (SHM) starts with a cleaving enzyme. Exonuclease trimming then makes a gap at the nick, which is filled in by an error-prone polymerase. This prescient paper thus places the crux of mutation on a DNA break (2). DNA breaks are observed during SHM and class switch recombination (CSR), two processes that diversify antibodies. Both processes are initiated by the enzyme activation-induced deaminase (AID) (3), which is not the cleaving enzyme, but instead deaminates cytosine to uracil in both the V and switch (S) regions of the immunoglobulin loci (4). Breaks then occur when the uracils are recognized by proteins in the base excision repair and mismatch repair pathways (5). Although breaks in S regions have been extensively studied and mostly characterized as staggered or blunt double-strand breaks (6–14), breaks in V regions are poorly understood (15, 16). DNA breaks have long been assumed to occur in V genes during the process of gene conversion in chickens, when pseudogenes recombine with a rearranged gene by homologous recombination (17, 18). During SHM, breaks were first detected by Sale and Neuberger (19), who used terminal deoxynucleotidyl transferase to catalyze the addition of non-templated nucleotides at the ends of breaks in VH genes from a Burkitt lymphoma cell line that constitutively undergoes mutation. However, it was not determined if the breaks were an intermediate or a by-product of the mutation process. Kong and Maizels (20) used ligation mediated-PCR (LM-PCR) to detect both single- and double-strand breaks in mouse Vλ genes from germinal center B cells, but it was not known if they were dependent on AID. Bross et al. (21) and Papavasiliou and Schatz (22) independently used LM-PCR to observe blunt double-strand breaks in V genes from mouse germinal center B cells, and a large proportion of these breaks occurred at AID target motifs. However, later experiments by the same investigators nullified the significance of these breaks by showing that they also occurred in Aid −/− mice (23–25). Similarly, LM-PCR was used on a V gene in the human BL-2 cell line, and single- and double-strand breaks were detected in the absence of AID, implying no obvious correlation of breaks with SHM (26). Soon after, a study by Zan et al. (27) detected blunt double-strand breaks that did not require AID, and concluded that AID acted downstream of break formation to form staggered breaks. These numerous conflicting reports raise essential questions about whether AID is necessary to induce breaks, what kind of breaks are made in V genes, how the breaks are generated, and where they are located.
In view of the certainty that breaks must occur to allow low-fidelity DNA polymerases to synthesize from free 3’-hydroxyl groups, we sought to characterize the types of breaks in rearranged JH introns from germinal center B cells following immunization. Three types of breaks could develop: a single-strand break, which would consist of a sole break on either strand; a staggered break, where two nearby bases located on opposing strands would be cleaved producing either a 5’ or 3’ overhang; or a blunt double-strand break, where breaks on opposing strands would be located directly across from each other. We used a LM-PCR strategy to demonstrate that AID-induced breaks are predominantly single-stranded and are generated through the activity of either UNG or MSH2-MSH6 DNA repair pathways.
Materials and Methods
Mice
C57BL/6, Aid −/−, Ung −/−, and Msh6 −/− mice were previously described (3, 28, 29); Ung −/− Msh6 −/− double-knockout mice were bred in our colony. Mice were co-housed, and males and females were used at 2–6 months of age. All animal protocols were reviewed and approved by the Animal Care and Use Committee of the National Institute on Aging.
LM-PCR assay on plasmid DNA
A LM-PCR protocol (20) was adapted to detect single-strand, staggered, and blunt double-strand breaks simultaneously on the nontranscribed DNA strand (30). Using universal primer 1st 5’J558 positioned in framework 3 of the J558 family of V genes and primer 1 (extension JH4 1) located 803 bp downstream of JH4, a 902-bp fragment was amplified from splenic B cell DNA using Herculase II enzyme (Agilent) and cloned into the pSC-A-amp/kan vector (Agilent). Primer sequences (Integrated DNA Technologies) are listed in Supplemental Table I. Sequential quick-change mutagenesis reactions were performed to introduce StuI (primers: VDJ4-StuI Fwd and VDJ4-StuI Rev) and BbvCI restriction sites (primers: VDJ4-BbvCI Fwd and VDJ4-BbvCI Rev) at the 3’ end of the JH4 gene segment. The BseR1 site was naturally encoded in the intron. Experimental templates were generated by cutting plasmid with StuI (New England Biolabs) to produce a blunt-ended linear plasmid, BbvCI (New England Biolabs) to yield a linear plasmid with a staggered 5’ overhang of 3 bases, BseR1 (New England Biolabs) to generate a linear plasmid with a staggered 3’overhang of 2 bases, or with Nt.BbvCI (New England Biolabs) which is a mutant version of BbvCI that only cuts the top strand to generate a nicked circular template. One fg of cut plasmid was then added to 100 μg of genomic splenic DNA from AID-deficient mice for the LM-PCR assay. All the restriction enzyme breaks were detected by denaturing the DNA, annealing JH4 extension primer 1, and extending with Fidelitaq (Affymetrix) to convert single-strand breaks to blunt double-strand breaks. For staggered breaks, nondenatured DNA was incubated with Fidelitaq; 5’ overhang breaks were extended from the bottom strand without primer 1, and 3’ overhang breaks were excised back to blunt ends by the 3’ to 5’ exonuclease activity in Fidelitaq. Blunt double-strand breaks were detected without denaturation and extension. A double-stranded linker containing top and bottom strands (Supplemental Table I) was ligated to all products and amplified by PCR with Taq polymerase (Takara) using nested primers. Thus, LL4 linker primer was paired with JH4 2–1, and nested LL2 linker primer was paired with nested JH4 2–2. DNA was separated on an agarose gel and stained with ethidium bromide. Break sites were confirmed by sequencing.
LM-PCR assay on JH intron DNA from germinal center B cells
Mice were immunized i.p. with 100 μg phycoerythrin (Prozyme) in CFA (Sigma Aldrich). Splenocytes were isolated and stained with FITC-labeled anti-B220 (clone RA3–6B2) and Alexa Fluor 647-labeled anti-GL7 (clone GL-7) antibodies (Biolegend) to detect germinal center B cells. B220+GL7+ cells were separated via fluorescence-activated cell sorting with doublet exclusion, and genomic DNA was obtained by proteinase K digestion (Ambion) and phenol/chloroform extraction. There was no difference in amplification if the DNA was incubated in the presence or absence of DNA polymerase β-lyase (a generous gift from Sam Wilson, NIEHS, NIH), which removed the break site sugar moiety to produce a 5’ hydroxyl end. The DNA concentration for all three break assays was standardized by PCR amplification of a Gapdh control gene (Supplemental Table I) using two-fold dilutions of input DNA (30). Breaks were then detected as described in the plasmid assay. Breaks from all assays were visualized by Southern blotting and hybridization with probe 3 (JH4 3), which was labeled with 32P-γATP. ImageQuant TL software was used to measure relative break intensity.
Analysis of DNA alterations
PCR products were TA-cloned (Strataclone) and sequenced (Macrogen) to identify where the linker had ligated in the JH introns spanning 1100 bp from primer 2. Since the PCR assay detected breaks on the nontranscribed strand, the base where the break occurred was the first 5’ nucleotide that was replaced by the linker. 15–60 amplifications were performed on DNA from 3 mice per genotype to generate enough unique sequences for analysis.
Results
Assays to detect DNA breaks in the JH4 intron
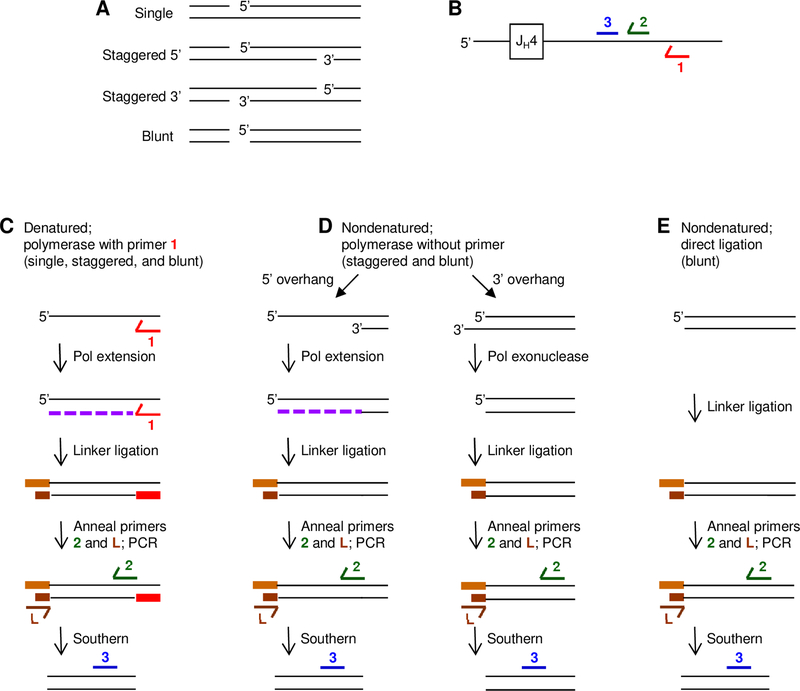
Single, staggered, and blunt breaks could occur in the V region (Fig. 1A). We mainly focused on the intronic region downstream of rearranged JH4 gene segments (Fig. 1B) as a surrogate for heterogeneous V genes, because it sustains a high level of mutations at a frequency of 10−2 per bp (31), and the somatic mutation events are not biased by selection for codon changes. A strategy using LM-PCR was devised to detect all 3 types of breaks in the 492-bp region. Although breaks and mutagenesis occur on both transcribed and nontranscribed strands (5, 20), in this assay we used primers to detect events on the nontranscribed strand, because primer extension was more specific originating in the single copy intron than it was originating in the multiple V genes on the transcribed strand. Single, staggered, and blunt breaks were simultaneously detected by DNA denaturation, followed by primer extension using Fidelitaq polymerase with intron primer 1 annealing to the top 5’ strand (Fig. 1C). Staggered and blunt breaks were detected without denaturation and primer annealing steps (Fig. 1D). For staggered breaks with a 5’ overhang, polymerase extension occurred from the bottom 3’-OH strand, and for breaks with a 3’ overhang, the 3’ nucleotides were excised to a blunt end using the 3’ exonuclease activity of Fidelitaq. This assay could also detect blunt breaks. Blunt breaks alone were detected without denaturation and extension steps (Fig. 1E). For all breaks, a double-stranded linker was ligated to the blunt ends made after polymerase extension or excision, or pre-existing ends in the case of blunt double-strand breaks. Intron primer 2 and a linker primer L were used in PCR reactions with Taq polymerase to amplify the DNA, and amplified products were separated on an agarose gel. Bands were detected after Southern hybridization with intron probe 3.
FIGURE 1.
Detection of DNA breaks in the JH4 intron on the nontranscribed strand. (A) Schematic drawing of 4 types of breaks, including staggered with either a 5’ or 3’ overhang. (B) Location of primers and probes: 1, primer for extension (red); 2, primer for PCR (green); and 3, probe for Southern blot (blue). (C) Single, staggered, and blunt. DNA was denatured and extended (purple dashed line) by polymerase from primer 1 to produce a blunt double-strand break substrate for ligation to a double-stranded blunt linker (orange). PCR amplification used a linker primer L (brown) and primer 2. (D) Staggered and blunt. For staggered breaks with a 5’overhang, nondenatured DNA was extended by polymerase from the 3’ hydroxyl on the bottom strand in the absence of a primer to produce a blunt end. For staggered breaks with a 3’ overhang, nondenatured DNA was trimmed by the exonuclease function of Fidelitaq to produce a blunt end. Blunt breaks were ligated in the absence of polymerase. (E) Blunt. DNA was nondenatured, and the linker was ligated directly to the break site, followed by PCR amplification. For all assays, PCR-amplified DNA was separated on an agarose gel and analyzed by Southern blot with probe 3.
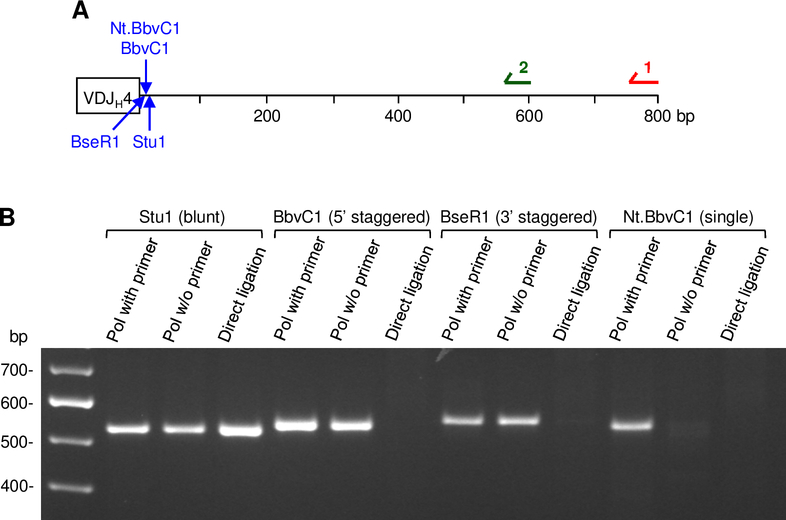
To confirm the validity of our strategy to detect the different breaks, breaks were artificially generated in a plasmid containing the JH4 intron using different restriction enzymes. Nt.BbvC1 produces a single-strand break on the top strand; BbvC1 generates a staggered break with a 5’ overhang of 3 bases; BseR1 generates a staggered break with a 3’ overhang of 2 bases, and Stu1 makes blunt cuts (Fig. 2A). The cut plasmids were mixed with genomic DNA from AID-deficient mice, and then assayed for breaks. As shown in Fig. 2B, the data indicate that the assay using polymerase extension with a primer can detect blunt, staggered, and single breaks, whereas extension without a primer detected blunt and staggered breaks, and direct ligation alone recognized only blunt breaks. The detection of all 3 breaks established that the assay was sensitive enough to detect staggered and blunt breaks as well as single-strand breaks in the presence of genomic DNA.
FIGURE 2.
Validation of LM-PCR break assay using plasmid DNA containing a rearranged VDJH4 gene and intron DNA. (A) Location of restriction enzymes and primers. (B) DNA was digested, and breaks were assayed as described in Fig. 1. Ligation and PCR steps were performed in the presence of Aid −/− genomic DNA. Amplified products were separated on an agarose gel and visualized with ethidium bromide.
To further verify that the 3 assays can detect breaks in a switch region that is affected during CSR, we examined DNA from splenic B cells stimulated ex vivo with lipopolysaccharide and IL-4 for 2 days. Using primers specific for Sμ (32), breaks were readily detected in all three assays, with the staggered and blunt assays being more efficient than the all break assay (data not shown).
JH4 intron breaks are predominantly single-stranded and depend on AID
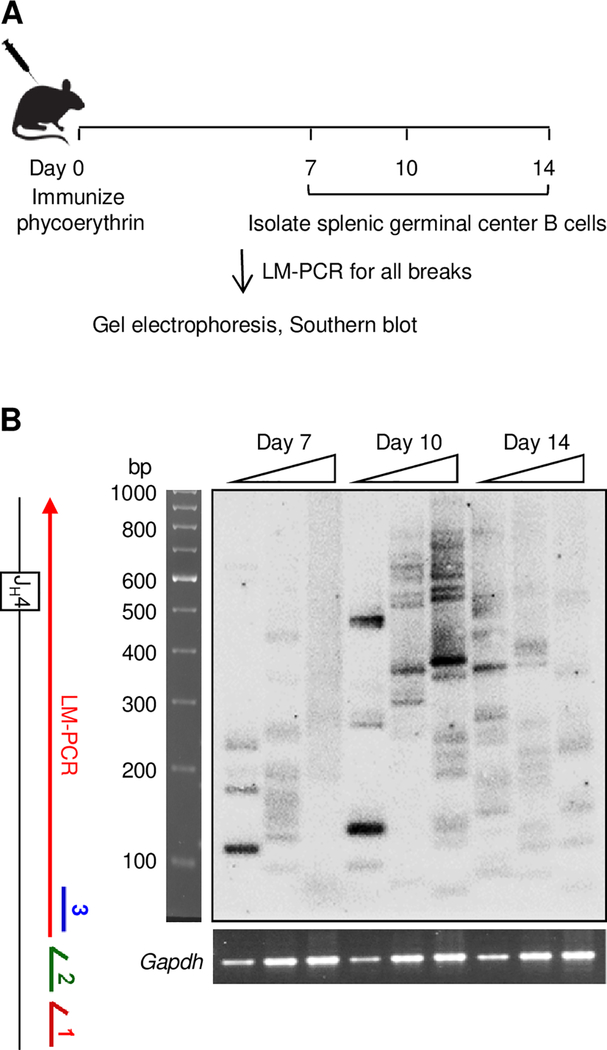
We then performed a time-course study to determine when the maximum number of breaks occurred after immunization in germinal center B cells (B220+GL7+) from spleens of C57BL/6 mice. Cells were collected 7, 10, or 14 days after i.p. immunization with phycoerythrin in adjuvant to induce SHM (Fig. 3A). The assay to detect all breaks by primer extension (Fig. 1C) was used to examine the JH4 intron region on the nontranscribed strand. Thus, primer 1 was annealed to the strand and extended until it hit a break. As shown in Fig. 3B, Southern blotting detected multiple distinct bands per sample, indicative of multiple cells having different break sites. Breaks were present at all 3 time points, but the day 10 time point was chosen for further studies because it exhibited the most breaks.
FIGURE 3.
Timing of breaks following immunization. (A) C57BL/6 mice were immunized, and splenic germinal center B cells were isolated 7, 10, and 14 days later. LM-PCR was then performed using the assay to detect all breaks. (B) The map shows location of primers on the nontranscribed strand and the direction of LM-PCR (red arrow). Representative Southern blot using probe 3 with two-fold increasing dilutions of DNA (triangles) from one mouse for each day. Below, input DNA was standardized by amplification and ethidium bromide staining of a Gapdh control.
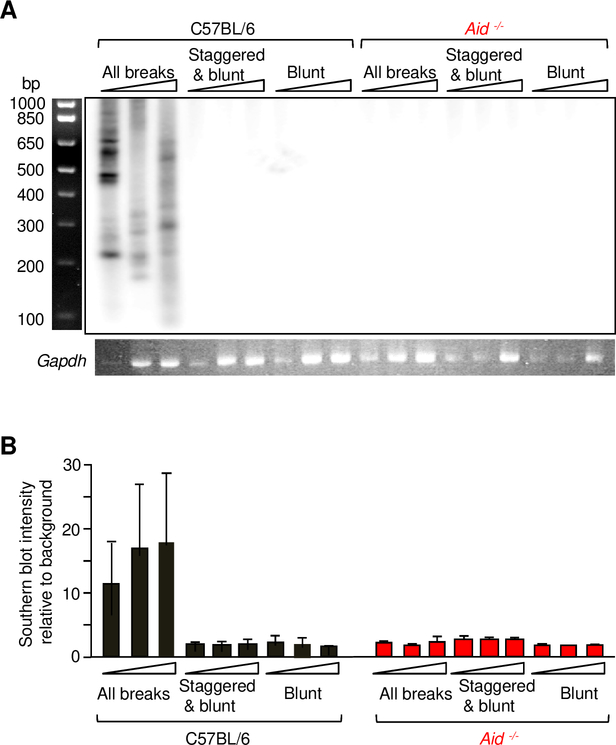
We next characterized the type of breaks that occurred in the JH introns. Breaks were assigned in DNA from germinal center B cells by performing separate assays for all breaks, staggered and blunt ends, and blunt ends only. As shown in Fig. 4A, samples using the primer extension step to detect all breaks, which included single-strand breaks, showed robust accumulation of breaks, whereas staggered and blunt breaks appeared at background levels. No breaks were detected in Aid −/− cells (Fig. 4B). In contrast, in the Sμ region, we and others have identified staggered and blunt breaks during CSR in B cells activated ex vivo with mitogen (11, 32). Thus, the majority of DNA breaks in the JH introns were single-stranded and depended on expression of AID.
FIGURE 4.
Characterization of types of breaks in DNA from day 10 germinal center B cells to detect single, staggered, and blunt breaks from C57BL/6 and Aid −/− mice (red). (A) Representative Southern blot of DNA using the three different assays with two-fold increasing dilutions of DNA (triangles). Lower panel, ethidium bromide staining of a Gapdh loading control, which shows that DNA was present in all lanes. (B) Relative break intensity. Values were calculated from 2–4 independent PCR reactions per mouse with 4–14 mice per genotype. Error bars represent SD.
Breaks are generated by both UNG and MSH6 proteins
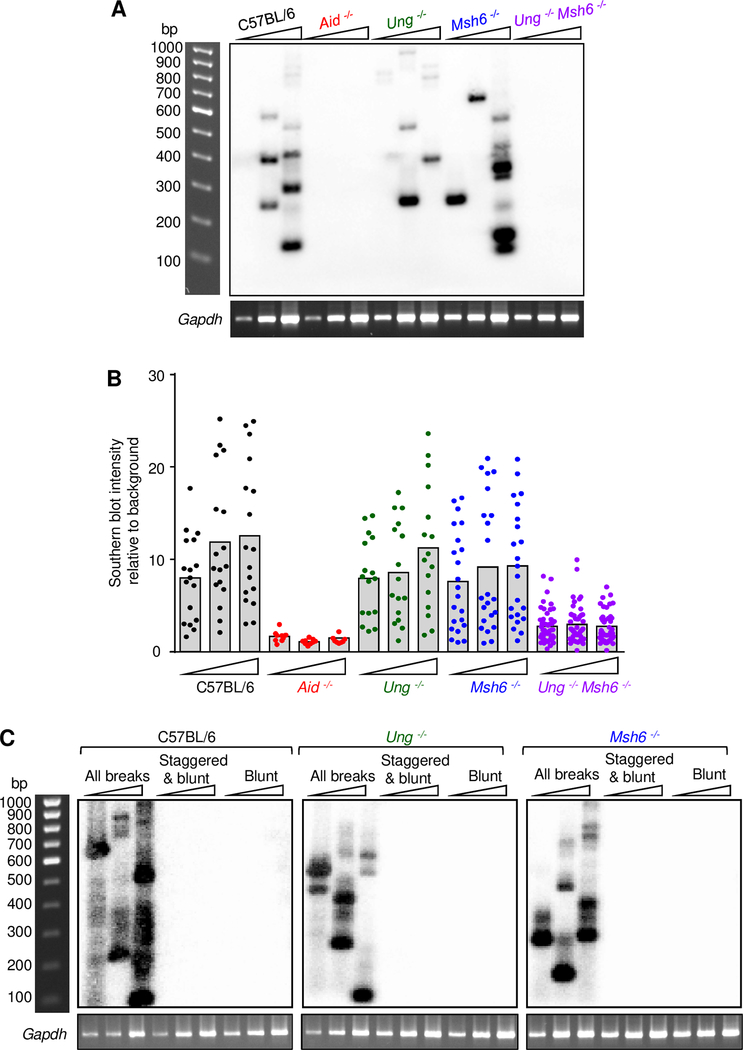
The role of specific proteins in break formation was examined in several repair-deficient mouse models to determine which repair pathway generated the ruptures. C57BL/6 and Aid −/− mice were used as positive and negative controls. In Ung −/− mice, uracils can be removed by mismatch repair proteins. Conversely, in Msh2 −/− or Msh6 −/− mice, uracils can be processed by base excision repair proteins. We used Msh6 −/− mice in these studies, because they have the same phenotype in SHM and a longer lifespan than Msh2 −/− mice (29, 33, 34). The absence of one repair pathway in Ung −/− or Msh6 −/− mice could potentially lead to a decrease in breaks. Alternatively, the number of breaks in these single knockouts would be unchanged if the remaining pathway is able to compensate. As shown in Fig. 5A-B, breaks were detected in C57BL/6, Ung −/−, and Msh6 −/− mice, but were absent in Aid −/− mice. Substantial numbers of breaks were found in the absence of either UNG or MSH6, indicating that both pathways contribute to the generation of breaks. Although V region break formation utilizes both UNG and MSH6, other proteins could be involved. To test this possibility, Ung −/− Msh6 −/− double knockout mice, which do not have functional base excision or mismatch repair pathways to remove uracils, were examined. DNA from these mice contained fewer breaks relative to C57BL/6 (Fig. 5B), indicating that UNG and MSH6 comprise the major pathways for break formation in the JH4 intron, although other minor activities appear to exist. Finally, to determine if staggered or blunt breaks were increased in the absence of DNA repair, we performed the corresponding assays in DNA from UNG- and MSH6- deficient mice. As seen in Fig. 5C, the only breaks that were detected were with the assay that detected all breaks, confirming that single-strand breaks predominate in the V region in the absence of DNA repair.
FIGURE 5.
Breaks from UNG- and MSH6-deficient mice. (A) Representative Southern blot of breaks from C57BL/6 (black), Aid −/− (red), Ung −/− (green), Msh6 −/− (blue), and Ung −/− Msh6 −/− (purple) mice, using the LM-PCR assay to detect all breaks. Below, two-fold increasing dilutions of DNA were standardized with a Gapdh loading control, which shows that DNA was present in all lanes. (B) Relative break intensity at 3 DNA dilutions. Each point represents a single PCR reaction. Data are from 2–4 independent PCRs per mouse, with 4–14 mice per genotype. (C) Nature of breaks in UNG and MSH6-deficient mice. Representative Southern blot of the three break assays from repair-deficient mice. Input DNA was standardized with a Gapdh loading control.
Break site locations suggest nicks are made during DNA repair
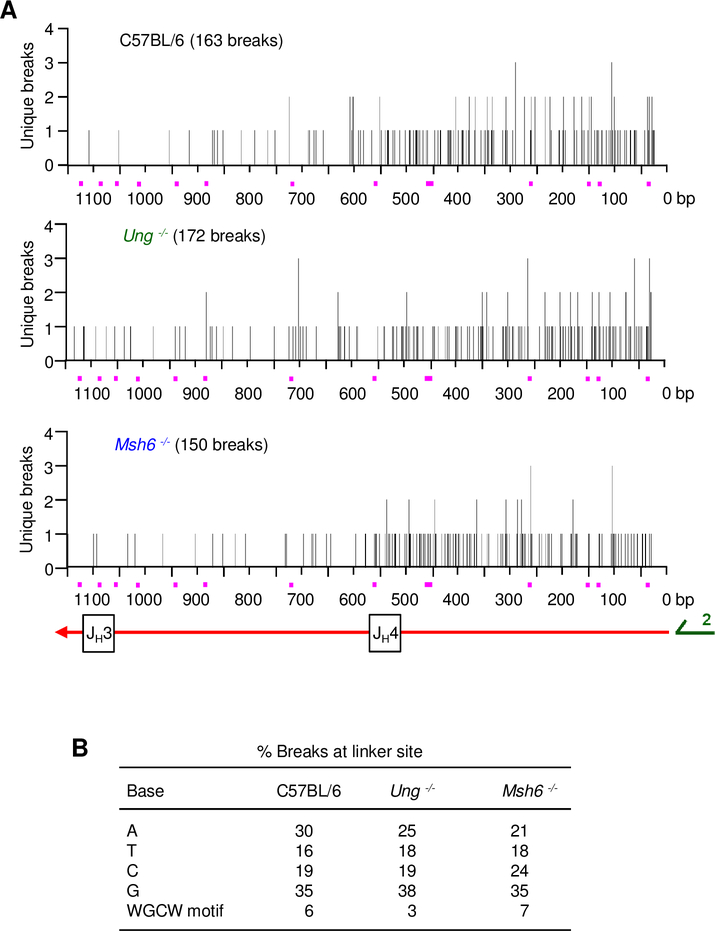
The break site spectrum and/or hotspot specificity may depend on whether UNG or MSH6 proteins are involved. In both cases, AID will deaminate cytosine, frequently in a WGCW (W = A/T) hotspot motif, to produce uracil. In the absence of UNG, breaks could be detected away from the U:G mismatch due to generation of a gap by exonuclease 1 (EXO1). In the absence of MSH6, breaks would seemingly be concentrated around uracil after incision of the abasic site by apurinic/apyrimidinic endonuclease 1 and /or 2 (APE1/2). LM-PCR products from C57BL/6, Ung −/−, and Msh6 −/− mice were cloned and sequenced to determine where the linker attached to a break was located. The resulting spread of ~150 breaks from each genotype in 1 kb of DNA containing JH3 and JH4 introns is shown in Fig. 6A. The distribution was similar across all genotypes and did not show any prominent clustering in WGCW hotspots. Although breaks were detected in both introns and occasional V genes rearranged to JH3 or JH4, we only scored those in introns to compare the genotypes. Most of the breaks were distributed in the JH4 intron, which is closer to primer 2 used for PCR, and would be preferentially amplified. The base that was replaced by the linker was recorded for each of these sequences (Fig. 6B). The percentage of breaks at a given base was comparable between genotypes; the small increase in breaks at C in Msh6 −/− clones was not statistically significant (p = 0.57, Fisher’s exact test), and there was no significant difference in breaks located in the 14 WGCW motifs. The unexpected lack of predominant selection for C suggested that single-strand nicks were made at distal sites away from the original uracil, and we had captured the breaks in the middle of DNA repair.
FIGURE 6.
Location of break sites in JH3 and JH4 introns. (A) Break sites from C57BL/6, Ung −/−, or Msh6 −/− mice were mapped by sequencing the PCR products shown in Fig. 5B. Y-axis shows the number of unique sites detected at each base. Bottom schematic diagram shows the relative locations of the introns, with the red arrow showing the direction of extension from PCR primer 2 towards the J exons. 14 WGCW hotspots are represented by red squares on the X-axis. (B) Percentage of break sites at a base or within WGCW motifs. Data were corrected for the sequence’s nucleotide composition and were from 3 independent experiments with 1 mouse per experiment per genotype.
Discussion
As mentioned in the introduction, previous studies found that many DNA breaks in V genes were independent of AID expression. Collateral break formation may have occurred from apoptotic cell debris during cell isolation or DNA preparation. Accordingly, we eliminated samples that had random, double-strand breaks in a non-immunoglobulin gene, CD19, which could have arisen during apoptosis or from DNA shearing during isolation. Here, we analyzed breaks in JH introns that were captured in splenic germinal center B cells 7–14 days after immunization by stringent gating for B220+GL7+ by flow cytometry without prior column purification. Using a comprehensive LM-PCR assay design, the breaks were characterized as predominantly single-strand breaks with negligible contributions from staggered and blunt double-strand breaks. These breaks were present in mice expressing AID, UNG, and MSH6, and were significantly reduced in Aid −/− mice, and Ung −/− Msh6 −/− double-deficient mice. By these biologic and genetic criteria, the breaks are likely to be intermediates in the SHM pathway.
What causes the breaks? Both UNG and MSH2-MSH6 pathways have been shown to affect SHM and CSR by altering the types of nucleotide substitutions and reducing isotype switching (5, 35). The direct candidates for an endonuclease in the UNG pathway are APE1/2, which cleave at the abasic site left by UNG removal of uracil (36). Mice that are deficient for these enzymes have reduced switching (37), and mice deficient for APE2 have greatly reduced frequencies of hypermutation with altered mutational spectra (38, 39). The candidate for an endonuclease in the MSH2-MSH6 pathway has recently been identified as PMS2 (40, 41). Recognition of the U:G mismatch by the MSH2-MSH6 heterodimer (42) recruits the PMS2-MLH1 heterodimer to produce nicks in the vicinity, and creates a gap using EXO1 (43). We show here that the cleaving enzymes for breaks in V regions come from both UNG and MSH2-MSH6 pathways, which is similar to S regions. However, it was surprising that both UNG-deficient and MSH6-deficient cells had similar amounts of breaks, because the two mouse models behave differently in terms of SHM and CSR. The frequency of SHM is reduced in Msh6 −/− mice relative to Ung −/− mice (33, 35), likely due to the lack of polymerase η error-prone synthesis, and there is substantially reduced CSR in Ung −/− mice (4) compared to Msh6 −/− mice (34), due to lack of robust cleavage by APE enzymes. Double knockout Ung −/− Msh6 −/− mice had greatly reduced breaks, suggesting that in the absence of these two proteins, other uracil glycosylases, e.g., single-strand-selective monofunctional-uracil-DNA glycosylase and thymine-DNA glycosylase (41, 44), make only minor contributions to the formation of DNA breaks.
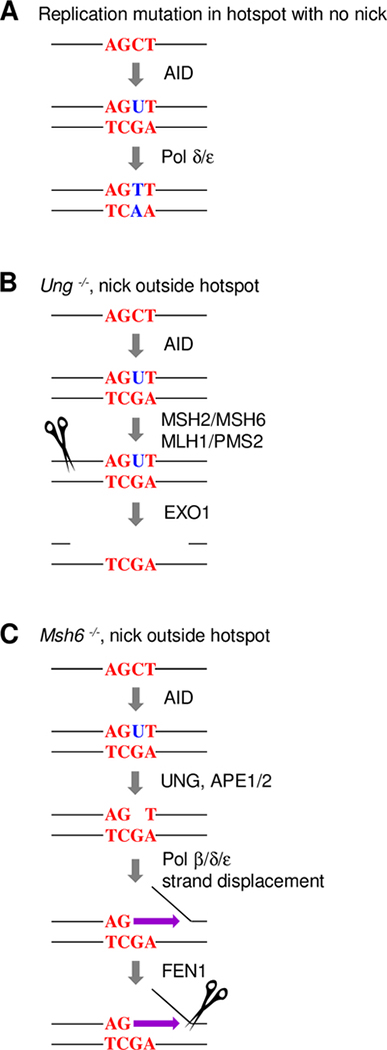
We previously used an analogous LM-PCR assay to detect uracils in JH introns from germinal center B cells, after in vitro treatment of DNA with UNG, APE1, and β-lyase (30). The uracil-associated breaks were mapped at multiple positions over the JH3 and JH4 introns, and they were detected predominantly at cytosine residues, showing that they were generated by AID. Therefore, the lack of focused breaks at C in the in vivo assays described here indicates that nicks were made later in B cells during DNA repair of the uracils. We therefore posit that the location of breaks does not predict the location of mutations. For example, as shown in Fig. 7A, the majority of hotspot-focused mutations of C could occur by direct DNA replication over AID-generated uracil in the absence of nicks. In contrast, breaks occur during DNA repair. Fig. 7B describes the generation of nicks in UNG-deficient B cells, which occurs during mismatch repair. After recognition of the U:G mismatch by the MSH2-MSH6 duplex, PMS2 in conjunction with MLH1 makes a nick at an unknown distance 5’ or 3’ of the mismatch (45). This provides entry for EXO1 to generate a gap for DNA synthesis. Fig. 7C illustrates the generation of nicks in MSH6-deficient B cells, which occurs during base excision repair. UNG and APE1/2 produce a nicked abasic site which allows entry for polymerases β, δ, or ε to synthesize (46). The efficiency of amplification and location of breaks was not affected by the presence or absence of β-lyase in the reaction, which implies that the breaks were not captured immediately after APE cleavage, but after additional processing. The resulting strand displacement during long-patch repair produces a flap, which is nicked by flap endonuclease 1 (FEN1). The nucleotide specificity of cleavage by PMS2, EXO1, and FEN1 is unknown, and could result in the variety of break sites reported in this study. This interpretation proposes that error-free DNA repair is occurring side by side with error-prone repair of AID-induced uracils during SHM in both mismatch and base excision pathways (47–49). Indeed, the major tipping point for mutagenesis appears to be fueled by the use of low-fidelity DNA polymerases, predominantly Rev1 instead of polymerase β in base excision repair, and polymerase η instead of polymerases δ/ε in mismatch repair. Thus, both error-free and error-prone pathways use the same repair proteins and can generate DNA breaks, but mutagenesis uniquely relies on the substitution of error-prone polymerases for replicative polymerases.
FIGURE 7.
Single-strand breaks made during DNA repair. (A) Replication mutation in hotspot with no nick. Mutations accumulate at C in AGCT (WGCW) hotspots (red) following replication over AID-generated uracil (blue). (B) Nicks in Ung −/− cells generated during mismatch repair outside of hotspots by PMS2 (scissor) and EXO1. (C) Nicks generated in Msh6 −/− cells during base excision repair outside of hotspots by FEN1 cleavage (scissor) during polymerase strand displacement (purple arrow).
DNA breaks in V regions are clearly handled differently than those in S regions. First, the widespread distribution of linker locations away from the original deaminated cytosine suggests that many breaks are made during faithful DNA repair of uracil. In contrast, linker locations in S regions were predominantly located at G:C bp in a plethora of hotspots (11, 50) to catalyze a program of recombination using double-strand breaks. Second, the predominance of single- versus staggered/double-strand breaks in V regions correlates with the paucity of WGCW motifs relative to the S region. WGCW sites are the main target for AID deamination (34, 51, 52), which initiate pathways leading to break formation. For example, there are 14 WGCW sites spanning 5% of the 1.1 kb JH3-JH4 intronic region (Fig. 6A), whereas 204 WGCW sites cover 25% of the 3.2 kb Sμ region (53). Cleavage at two nearby WGCW motifs on opposite strands in the S region would generate a staggered break, which can be processed into a blunt break. Third, breaks in S regions are funneled into nonhomologous end-joining programs for CSR, whereas those in V regions are seemingly untouched by these proteins. Mice deficient for recombination proteins DNA-PKCS (54), H2AX (55), 53BP1 (56), and ATM (57, 58) have normal SHM but crippled CSR. Fourth, translocations involving double-strand breaks to other chromosomes rarely occur in the V region, but are far more frequent in the S region (59).
Although staggered and blunt double-strand breaks were below the level of detection in our LM-PCR assays, they do exist, since large insertion and deletion mutations require double-strand breaks. These lesions are infrequent, and they have been reported in both coding and intron regions (60–63). Therefore, we surveyed the JH4 intron from multiple references to determine a frequency for substitutions, insertions and deletions. The non-coding intron is particularly useful for this analysis since addition or subtraction of nucleotides would not cause frameshifts and be negatively selected, as they would in an exon. As shown in Supplemental Table II, in a sequence analysis of over 2600 mutations in the 492-bp intron, 98.7% were base substitutions, 0.3% were small insertions of 1 bp, and 1% were deletions of 1–164 bp. In recent reports, a higher frequency (~15%) of mutated passenger alleles from Peyer’s patches were reported to contain deletions in VDJ exons, as measured by high-throughput sequencing (64, 65). This discrepancy in the frequency of double-strand breaks compared to our results could reflect the timing of the LM-PCR assay, which captures breaks as they occur 10 days after immunization, versus the sequence assay where deletions could progressively build up over time in Peyer’s patch cells.
An alternative proposal to explain small insertions and deletions rather than double-strand breaks is to consider the role of DNA polymerases. Polymerase η, which is predominantly used during SHM (66), has been reported to generate insertions and deletions in repetitive sequences (67), which would not require double-strand breaks. Polymerase η could enter the DNA site at a single-strand break, synthesize the template, and occasionally slip and re-align, generating an insertion or deletion (60). These mutation events might also result from imperfect alignment of direct repeats or palindromes in the V region, again without a strand break (15).
In summary, endonucleases in two repair pathways, UNG and MSH2-MSH6, generate a minefield of DNA lesions in V and S regions, following AID deamination of cytosine. In V regions, breaks traverse the region and are predominantly single-stranded due to infrequent numbers of WGCW motifs. These allow entry by low-fidelity DNA polymerases to synthesize mutations for affinity maturation. In S regions, breaks are clustered at multiple WGCW motifs on both strands and are processed into blunt double-strand ends for CSR and potential translocations to other loci.
Supplementary Material
Acknowledgments
The authors thank C. Nguyen, T. Wallace, J. Scheers, C. Dunn, G. McDonough, and R. Wersto of the National Institute on Aging flow core facility for cell sorting. David M. Wilson provided thoughtful comments on the manuscript.
This research was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging (AG000732).
Abbreviations used in this article:
- AID
activation-induced deaminase
- CSR
class switch recombination
- LM-PCR
ligation mediated-PCR
- MSH
mutS homolog
- SHM
somatic hypermutation
- S
switch
- UNG
uracil DNA glycosylase
References
- 1.Brenner S, and Milstein C 1966. Origin of antibody variation. Nature 211: 242–243. [PubMed] [Google Scholar]
- 2.Gearhart PJ 2006. Antibody wars: extreme diversity. J Immunol 177: 4235–4236. [DOI] [PubMed] [Google Scholar]
- 3.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, and Honjo T 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102: 553–563. [DOI] [PubMed] [Google Scholar]
- 4.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, and Neuberger MS 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12: 1748–1755. [DOI] [PubMed] [Google Scholar]
- 5.Rada C, Di Noia JM, and Neuberger MS 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16: 163–171. [DOI] [PubMed] [Google Scholar]
- 6.Wuerffel R, Du J, Thompson R, and Kenter AL 1997. Ig Sγ3 DNA-specific double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J Immunol 159: 4139–4144. [PubMed] [Google Scholar]
- 7.Chen X, Kinoshita K, and Honjo T 2001. Variable deletion and duplication at recombination junction ends: implication for staggered double-strand cleavage in class-switch recombination. Proc Natl Acad Sci U S A 98: 13860–13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalan N, Selz F, Imai K, Revy P, Fischer A, and Durandy A 2003. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J Immunol 171: 2504–2509. [DOI] [PubMed] [Google Scholar]
- 9.Rush JS, Fugmann SD, and Schatz DG 2004. Staggered AID-dependent DNA double strand breaks are the predominant DNA lesions targeted to S mu in Ig class switch recombination. Int Immunol 16: 549–557. [DOI] [PubMed] [Google Scholar]
- 10.Arudchandran A, Bernstein RM, and Max EE 2004. Single-stranded DNA breaks adjacent to cytosines occur during Ig gene class switch recombination. J Immunol 173: 3223–3229. [DOI] [PubMed] [Google Scholar]
- 11.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, and Stavnezer J 2005. Inducible DNA breaks in Ig S regions are dependent on AID and UNG. J Exp Med 202: 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, and Alt FW 2007. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449: 478–482. [DOI] [PubMed] [Google Scholar]
- 13.Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, and Nussenzweig MC 2010. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vuong BQ, Herrick-Reynolds K, Vaidyanathan B, Pucella JN, Ucher AJ, Donghia NM, Gu X, Nicolas L, Nowak U, Rahman N, Strout MP, Mills KD, Stavnezer J, and Chaudhuri J 2013. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol 14: 1183–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golding GB, Gearhart PJ, and Glickman BW 1987. Patterns of somatic mutations in immunoglobulin variable genes. Genetics 115: 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacob J, Przylepa J, Miller C, and Kelsoe G 1993. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J Exp Med 178: 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S, Humphries EH, Tjoelker L, Carlson L, and Thompson CB 1990. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol Cell Biol 10: 3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buerstedde JM, Reynaud CA, Humphries EH, Olson W, Ewert DL, and Weill JC 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J 9: 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sale JE, and Neuberger MS 1998. TdT-Accessible Breaks Are Scattered over the Immunoglobulin V Domain in a Constitutively Hypermutating B Cell Line. Immunity 9: 859–869. [DOI] [PubMed] [Google Scholar]
- 20.Kong Q, and Maizels N 2001. DNA breaks in hypermutating immunoglobulin genes: evidence for a break-and-repair pathway of somatic hypermutation. Genetics 158: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, and Jacobs H 2000. DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity 13: 589–597. [DOI] [PubMed] [Google Scholar]
- 22.Papavasiliou FN, and Schatz DG 2000. Cell-cycle-regulated DNA double-strand breaks in somatic hypermutation of immunoglobulin genes. Nature 408: 216–221. [DOI] [PubMed] [Google Scholar]
- 23.Bross L, Muramatsu M, Kinoshita K, Honjo T, and Jacobs H 2002. DNA double-strand breaks: prior to but not sufficient in targeting hypermutation. J Exp Med 195: 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papavasiliou FN, and Schatz DG 2002. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med 195: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bross L, and Jacobs H 2003. DNA double strand breaks occur independent of AID in hypermutating Ig genes. Clinical & Developmental Immunology 10: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill JC, and Reynaud CA 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat Immunol 3: 815–821. [DOI] [PubMed] [Google Scholar]
- 27.Zan H, Wu X, Komori A, Holloman WK, and Casali P 2003. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity 18: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, and Barnes DE 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell 5: 1059–1065. [DOI] [PubMed] [Google Scholar]
- 29.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane M, and Lipford J 1997. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell 91: 467–477. [DOI] [PubMed] [Google Scholar]
- 30.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM 3rd, and Gearhart PJ 2011. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol 12: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jolly CJ, Klix N, and Neuberger MS 1997. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res 25: 1913–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saribasak H, Maul RW, Cao Z, McClure RL, Yang W, McNeill DR, Wilson DM 3rd, and Gearhart PJ 2011. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. J Exp Med 208: 2209–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiesendanger M, Kneitz B, Edelmann W, and Scharff MD 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J Exp Med 191: 579–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martomo SA, Yang WW, and Gearhart PJ 2004. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med 200: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen HM, Tanaka A, Bozek G, Nicolae D, and Storb U 2006. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J Immunol 177: 5386–5392. [DOI] [PubMed] [Google Scholar]
- 36.Stivers JT, and Drohat AC 2001. Uracil DNA glycosylase: insights from a master catalyst. Archives of biochemistry and biophysics 396: 1–9. [DOI] [PubMed] [Google Scholar]
- 37.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, and Schrader CE 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med 204: 3017–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabouri Z, Okazaki IM, Shinkura R, Begum N, Nagaoka H, Tsuchimoto D, Nakabeppu Y, and Honjo T 2009. Apex2 is required for efficient somatic hypermutation but not for class switch recombination of immunoglobulin genes. Int Immunol 21: 947–955. [DOI] [PubMed] [Google Scholar]
- 39.Stavnezer J, Linehan EK, Thompson MR, Habboub G, Ucher AJ, Kadungure T, Tsuchimoto D, Nakabeppu Y, and Schrader CE 2014. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci U S A 111: 9217–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Oers J, Roa S, Werling U, Liu Y, Genschel J, Hou H, Sellers R, Modrich P, Scharff MD, and Edelmann W 2010. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci U S A 107: 13384–13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girelli Zubani G, Zivojnovic M, De Smet A, Albagli-Curiel O, Huetz F, Weill JC, Reynaud CA, and Storck S 2017. Pms2 and uracil-DNA glycosylases act jointly in the mismatch repair pathway to generate Ig gene mutations at A-T base pairs. J Exp Med 214: 1169–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, and Gearhart PJ 2005. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med 201: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bardwell PD, Woo CJ, Wei K, Li Z, Martin A, Sack SZ, Parris T, Edelmann W, and Scharff MD 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol 5: 224–229. [DOI] [PubMed] [Google Scholar]
- 44.Dingler FA, Kemmerich K, Neuberger MS, and Rada C 2014. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A:T substitutions during somatic mutation. Eur J Immunol 44: 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kadyrov F, Dzantiev L, Constantin N, and Modrich P 2006. Endonucleolytic function of MutLalpha in human mismatch repair. Cell 126: 297–308. [DOI] [PubMed] [Google Scholar]
- 46.Sung JS, and Demple B 2006. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J 273: 1620–1629. [DOI] [PubMed] [Google Scholar]
- 47.Storb U, Shen HM, and Nicolae D 2009. Somatic hypermutation: processivity of the cytosine deaminase AID and error-free repair of the resulting uracils. Cell Cycle 8: 3097–3101. [DOI] [PubMed] [Google Scholar]
- 48.Roa S, Li Z, Peled JU, Zhao C, Edelmann W, and Scharff MD 2010. MSH2/MSH6 complex promotes error-free repair of AID-induced dU:G mispairs as well as error-prone hypermutation of A:T sites. PLoS One 5: e11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saribasak H, and Gearhart PJ 2012. Does DNA repair occur during somatic hypermutation? Semin Immunol 24: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrader CE, Guikema JE, Linehan EK, Selsing E, and Stavnezer J 2007. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol 179: 6064–6071. [DOI] [PubMed] [Google Scholar]
- 51.Bransteitter R, Pham P, Calabrese P, and Goodman MF 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem 279: 51612–51621. [DOI] [PubMed] [Google Scholar]
- 52.Yu K, Huang FT, and Lieber MR 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem 279: 6496–6500. [DOI] [PubMed] [Google Scholar]
- 53.Wu X, and Stavnezer J 2007. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med 204: 1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bemark M, Sale JE, Kim HJ, Berek C, Cosgrove RA, and Neuberger MS 2000. Somatic hypermutation in the absence of DNA-dependent protein kinase catalytic subunit (DNA-PK(cs)) or recombination-activating gene (RAG)1 activity. J Exp Med 192: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, and Nussenzweig MC 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med 197: 1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, and Carpenter PB 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol 5: 481–487. [DOI] [PubMed] [Google Scholar]
- 57.Lumsden JM, McCarty T, Petiniot LK, Shen R, Barlow C, Wynn TA, Morse HC 3rd, Gearhart PJ, Wynshaw-Boris A, Max EE, and Hodes RJ 2004. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med 200: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reina-San-Martin B, Chen HT, Nussenzweig A, and Nussenzweig MC 2004. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med 200: 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorsett Y, Robbiani DF, Jankovic M, Reina-San-Martin B, Eisenreich TR, and Nussenzweig MC 2007. A role for AID in chromosome translocations between c-myc and the IgH variable region. J Exp Med 204: 2225–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson PC, de Bouteiller O, Liu YJ, Potter K, Banchereau J, Capra JD, and Pascual V 1998. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med 187: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goossens T, Klein U, and Kuppers R 1998. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci U S A 95: 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bemark M, and Neuberger MS 2003. By-products of immunoglobulin somatic hypermutation. Genes, chromosomes & cancer 38: 32–39. [DOI] [PubMed] [Google Scholar]
- 63.Briney BS, Willis JR, and Crowe JE Jr. 2012. Location and length distribution of somatic hypermutation-associated DNA insertions and deletions reveals regions of antibody structural plasticity. Genes Immun 13: 523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, Wang JH, and Alt FW 2015. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell 163: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwang JK, Wang C, Du Z, Meyers RM, Kepler TB, Neuberg D, Kwong PD, Mascola JR, Joyce MG, Bonsignori M, Haynes BF, Yeap LS, and Alt FW 2017. Sequence intrinsic somatic mutation mechanisms contribute to affinity maturation of VRC01-class HIV-1 broadly neutralizing antibodies. Proc Natl Acad Sci U S A 114: 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, and Gearhart PJ 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol 2: 537–541. [DOI] [PubMed] [Google Scholar]
- 67.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, and Kunkel TA 2001. Error rate and specificity of human and murine DNA polymerase eta. J Mol Biol 312: 335–346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.