Abstract
CD163 facilitates regulation and resolution of inflammation and removal of free hemoglobin, and is highly expressed in myeloid cells from patients with inflammatory disorders such as systemic juvenile idiopathic arthritis (SJIA) and macrophage activation syndrome (MAS). Our recent studies indicate that regulation of CD163 mRNA expression is a key functional property of polarized monocytes and macrophages, and is mediated at the transcriptional and post-transcriptional level including via microRNAs. The goal of the current study is to develop a multi-parameter flow cytometry panel incorporating detection of CD163 mRNA for polarized monocyte and macrophage populations in disorders such as SJIA and MAS. THP-1 cells and CD14+ human monocytes were stained using fluorochrome conjugated antibodies to myeloid surface markers, along with CD163 mRNA. Staining for mRNA could reliably detect CD163 expression, while simultaneously detecting different macrophage populations using antibodies targeting CD14, CD64, CD80, CD163, and CD209. This approach was found to be highly sensitive for increased mRNA expression when macrophages were polarized towards M(IL-10), with a strong signal over a broad range of IL-10 concentrations, and showed distinct kinetics of CD163 mRNA and protein induction upon IL-10 stimulation. Finally, this panel demonstrated clear changes in polarization markers in unstimulated monocytes from patients with SJIA and MAS, including upregulated CD163 mRNA and increased CD64 expression. Together, this approach represents a robust and sensitive system for RNA flow cytometry, useful for studying CD163 expression as part of a multi-marker panel for human monocytes and macrophages, with broad applicability to the pathogenesis of hyperinflammatory diseases.
Keywords: Macrophage activation syndrome, systemic juvenile idiopathic arthritis, macrophage polarization, RNA flow cytometry, IL-10
Introduction
Macrophage activation syndrome (MAS) is a life-threatening episode of systemic hyperinflammation associated with expansion of tissue macrophages and cytotoxic lymphocytes leading to a “cytokine storm”. Although MAS can complicate many rheumatic diseases, in children it is by far most commonly observed in those with systemic juvenile idiopathic arthritis (SJIA), a severe and distinctive form of juvenile arthritis with features of autoinflammation including significant activation of innate immune effector cells(1). The pathologic hallmark of MAS is detection of well-differentiated macrophages or histiocytes with hemophagocytic activity in peripheral tissues such as bone marrow and the reticuloendothelial system. Indeed, MAS bears striking similarity to the rare histiocytic disorder, familial hemophagocytic lymphohistiocytosis (HLH), caused by genetic defects in the perforin cytolytic pathway(2). In both MAS and HLH, activated macrophages are characterized by highly increased expression of cell markers including CD163(3). Recent studies have highlighted the heterogeneity of CD163+ cells and generated great interest in characterizing the specific cellular subtypes in disease (4).
CD163 is an innate pattern recognition and scavenger receptor, binding and engulfing circulating hemoglobin:haptoglobin complexes and mediating cellular anti-inflammatory functions (5, 6). CD163 expression is restricted to the monocyte/macrophage lineage, and increased on tissue resident macrophages. Thus, CD163 is considered a differentiation marker along the path to macrophages (6). CD163 is also upregulated on macrophages involved in later stages of inflammation, such as wound healing and chronic inflammation, supporting a role in resolution of inflammation. Indeed, monocytes and macrophages expressing CD163 have key roles in several inflammatory disorders, including HLH, MAS, and SJIA (7, 8).
Myeloid cells adopt distinct functional states upon ligand stimulation, through a process known as polarization (9). While previously considered a dichotomy between pro-inflammatory “M1” and anti-inflammatory “M2” activation, the contemporary understanding of polarization encompasses a diverse spectrum of phenotypes with specific gene expression profiles (10). Expression of CD163 is largely associated with polarization conditions including IL-10 stimulation (M(IL-10) also referred to as “M2c”) (9, 11–13), and more limited in other conditions including others considered alternative “M2” with immune regulatory properties (14, 15). In SJIA and MAS, circulating monocytes do not align with a single polarization state, but rather have features including both proinflammatory activation and high CD163 expression, consistent with either a mixed phenotype or multiple distinct populations (16–19). Therefore, to further determine the role of monocyte populations in SJIA and MAS, it is crucial to differentiate diverse polarized monocytes and macrophages at the single cell level.
The emergence of techniques to detect mRNA expression at the single cell level allows for analysis of specific genes in the context of various cell markers using either fixed (20–26) or non-fixed (SMARTFLARE) (26) methods, and that the signal is strong enough to detect by flow cytometry (27). The present study focuses on analysis of CD163 at the mRNA and protein level in monocyte and macrophage populations. Our recent findings indicated that regulation of CD163 mRNA expression is mediated at both transcriptional and post-transcriptional levels, including through a network of microRNA (15). Development of tools to define polarization including CD163 expression in specific cell populations remains a key barrier to understanding the pathogenesis of diseases such as MAS and SJIA, and may provide mechanisms for novel therapeutic targets. Here, we demonstrate the detection of CD163 mRNA in monocyte/macrophage populations, and determine a multi-parameter flow cytometry panel that includes analysis of CD163 mRNA at the single cell level for distinction of these cell types in PBMC or bone marrow populations. Our studies indicate distinct changes in CD163 mRNA and protein expression in response to specific stimuli in a time and dose dependent manner. Finally, this mRNA probe can be utilized in conjunction with other monocyte/macrophage polarization markers to examine distinct changes in CD163 mRNA and protein expression at a single cell level using flow cytometry, and aid in defining underlying mechanisms of disease pathogenesis in these hyperinflammatory syndromes.
Materials and Methods
Patient data and study approval:
This study was approved by the Cincinnati Children’s Hospital Institutional Review Board (#2016-2234), and written informed consent was obtained from all patients and/or their legal guardians. Data pertaining to clinical course, laboratory values, and treatment were collected from the electronic medical records. Whole blood was obtained using acid citrate dextrose as an anticoagulant, and peripheral blood mononuclear cells (PBMC) were separated as described below.
Cell isolation and culture:
All cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum. THP-1 monocytic cells were obtained from ATCC (Manassas, VA). Isolation of CD14+ peripheral blood monocytes was as described (28). Monocyte-derived macrophages were generated from peripheral blood from healthy adult donors. Briefly, PBMC were isolated by density gradient centrifugation through Ficoll-Paque Plus (GE Healthcare, Pittsburgh, PA), washed twice and then placed in culture medium supplemented with 5ng/ml monocyte colony stimulating factor (R&D Systems, Minneapolis, MN) at 2×106 cells/ml. After 5-7 days of differentiation, cells were washed 3 times to remove non-adherent lymphocytes prior to cell stimulation.
Cell Stimulation/Polarization:
All ligands were purchased from R&D Systems except for LPS which was from Sigma Aldrich. Prior to stimulation, THP-1 cells were differentiated overnight after addition of 50 ng/mL phorbol myristate acetate (PMA). The cells were left untreated or stimulated with IL-10 (1-50 ng/ml), IFNγ (20 ng/ml), LPS (10-100 ng/ml), or IL-4 (20 ng/ml).
Quantitative PCR:
Cellular mRNA levels were quantified using real-time RT-PCR as described previously (28). Briefly, total RNA was isolated as described using Trizol (Thermo Fisher, Santa Clara, CA) and real-time RT-PCR was performed using Viia7 Realtime PCR Instrument with gene-specific primers and SYBR Green Supermix (Life Technologies). Message copy numbers were normalized against the copy number of the housekeeping gene GAPDH. Primers specific for GAPDH and CD163 were described previously (28).
Antibodies and probes:
All antibodies were from BD Biosciences except for anti-human CD163 protein, which was from Trilium (Brewer, ME), and anti-CD80 and anti-CD163 mRNA probe, which were from ThermoFisher. Antibodies used for these studies were Pacific Blue conjugated anti-human CD14 (clone M5E2), Alexa Fluor 700- or PE-conjugated anti-human CD64 (clone 10.1), BV786- or FITC-conjugated anti-human CD80 (clone L307.4), PE-conjugated anti-human CD163 (clone Mac2.158), FITC-conjugated anti-human CD209 (clone DCN46), and Alexa Fluor 647-conjugated anti-CD163 or anti-CD80 mRNA probes (ThemoFisher Scientific).
Flow Cytometry:
CD163 mRNA transcripts were detected in single cells using the PrimeFlow™ kit according to manufacturer’s directions (Thermo Fisher Scientific). Briefly, approximately 1 million monocytes or macrophages were stained for surface markers for 30 minutes at 4°C. The cells were then washed with FACS buffer (PBS supplemented with 1% fetal calf serum) prior to incubation in fixation buffer 1 at 4ºC for 30 minutes. Samples were then washed 3 times with 1X PrimeFlow RNA Permeabilization Buffer with RNase Inhibitors. 1X PrimeFlow RNA Fixation Buffer 2 was added to each sample at room temperature and left for 1 hour before target probe hybridization. Cells were washed with PrimeFlow RNA Wash Buffer, and CD163 or CD80 Target Probes diluted 1/20 in PrimeFlow RNA Target Probe Diluent were added directly to the samples to incubate for 2 hours at 40ºC. Samples were washed with a preparation of PrimeFlow RNA Wash Buffer with RNase Inhibitor 1 and stored overnight. The next day, PrimeFlow RNA PreAmp Mix was added directly to the cell suspension and incubated for 1.5 hours at 40ºC. Samples were washed 3 times with PrimeFlow RNA Wash Buffer before PrimeFlow RNA Amp mix was added directly for 1.5 hours at 40ºC. Samples were washed again with PrimeFlow RNA Wash Buffer prior to adding diluted Label Probes in PrimeFlow RNA Label Probe Diluent for 1 hour at 40ºC to complete signal amplification. After washing twice with PrimeFlow RNA Wash Buffer, cells were stored overnight at 4ºC in FACS buffer.
Samples were filtered and transferred to 12 × 75 mm polystyrene tubes, and cells were acquired using a BD LSR Fortessa Special Order Research Product analytical cytometer (Becton Dickinson, San Jose, CA) using the following lasers and detectors: 355nm 20 mW; 405 nm 50 mW, PacBlue, 450/50 bandpass filter (BP); BV786, 750 longpass (LP), 780/60 BP; 488nm 50 mW, FITC, 505 LP, 530/30 BP; 640nm 40 mW, Alexa Fluor 647, 670/30 BP; 561nm 50 mW, PE 596/15 BP, or ImageStreamX (Millipore Sigma) 2 camera system using the following camera and laser settings: 40X magnification, 405 nm at 125 mW, 488 nm at 200 mW; 658 nm at 120 mW; 785 nm for SSC at 0.5 mW. Data were analysed using FCS Express (De Novo Software), FACSDiva (Becton Dickinson) or IDEAS (Millipore Sigma).
RESULTS
Single cell detection of CD163 mRNA in monocytes and macrophages using flow cytometry.
CD163 is a key marker of monocyte and macrophage activation and has important roles in immunoregulation. To determine the levels of CD163 mRNA in monocytes and macrophages at the single cell level, we employed an RNA flow cytometry technique, PrimeFlow. THP-1 monocytic cells and primary blood monocytes were analysed using an AF-647-labeled PrimeFlow probe for CD163 mRNA after stimulation with IL-10, known to strongly induce CD163 (15, 29). Analysis showed CD163 mRNA present and strongly increased upon stimulation with human IL-10 in both monocytes from PBMC and THP-1 cells (Figure 1A). The expression of CD163 mRNA increased approximately 3-fold following IL-10 stimulation, as assessed by flow cytometry, which is comparable to that of CD163 protein previously reported for conventional flow cytometry (30).
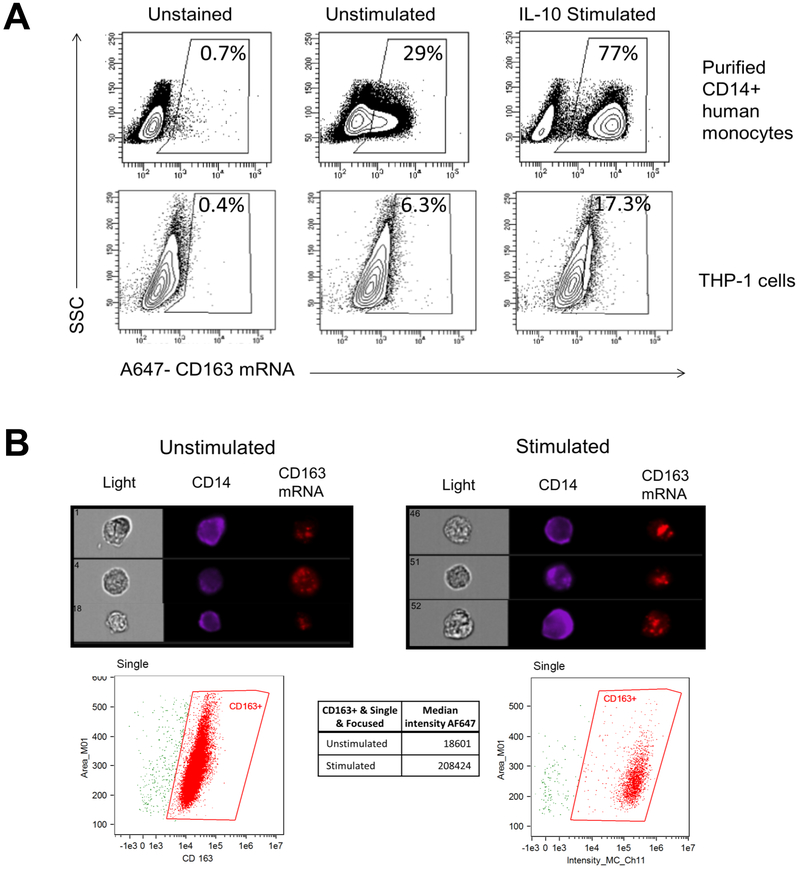
Figure 1. CD163 mRNA is expressed in human blood monocytes and THP-1 monocytic cells and increases in response to IL-10 treatment.
Purified CD14+ human monocytes or THP-1 cells were left unstimulated or stimulated with 50 ng/ml IL-10 for 24 hours. Cells were stained for CD14 expression and then processed for CD163 mRNA detection as described. (A) Density plots indicate percentages of CD163 mRNA signal from gated live singlet CD14+ monocytes (top) or THP-1 cells (bottom) acquired using a LSR Fortessa. MFI values are 332, 665, and 859 for THP-1 unstained, unstimulated, and IL-10 stimulated populations, respectively. Images are representative of three independent experiments. (B) Images are representative of singlet THP-1 cells acquired using an ImageStreamX with median intensity of CD163 gated signal indicated below. Images are representative of two independent experiments.
As a complementary method to quantify cellular CD163 mRNA expression at a single cell level and to visually confirm CD163 mRNA signal within the cell, imaging cytometry was performed on THP-1 cells. In Figure 1B, representative images indicate the presence of CD14 on the outside of the cell and a punctate signal of CD163 mRNA within the cell. Within the CD14+ population, CD163 mRNA signal, as indicated by median intensity, increased approximately 10-fold upon IL-10 stimulation.
Analysis of THP-1 with various drivers of macrophage differentiation indicates that IL-10 specifically induces expression of CD163 protein and mRNA at the single cell level.
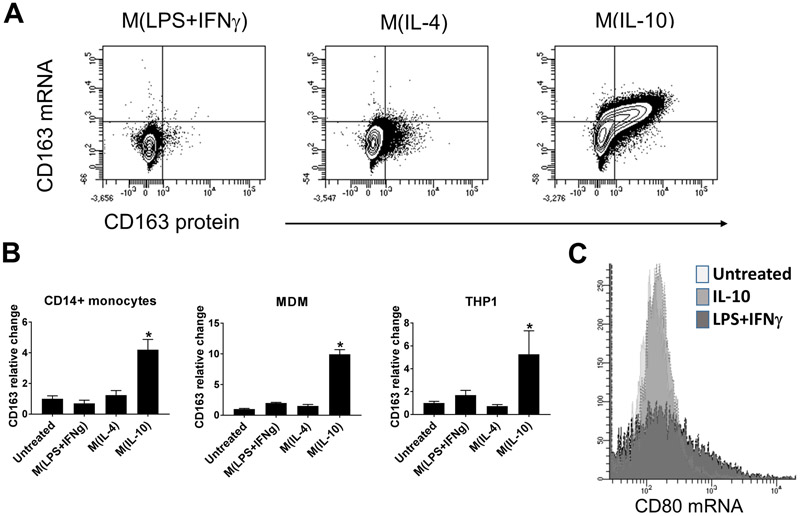
Monocytes and macrophages are implicated as important components of the pathogenesis of both HLH and SJIA-associated MAS (1). CD163 is increasingly recognized as a marker of macrophage activation in disease states, and is induced by several largely immunoregulatory signals (6). To quantify changes in CD163 mRNA and protein expression at the single cell level in response to various drivers of macrophage polarization, THP-1 cells were cultured for 24h under M(LPS+IFNγ) or classical “M1”, M(IL-4) or “M2a”, and M(IL-10) or “M2c” conditions. This 24h time point was chosen as our previous data found it to be optimal for expression of numerous macrophage polarization markers (15, 28). In Figure 2A, flow cytometry analysis confirms (1) no increase in expression of either CD163 mRNA or protein in response to M(LPS+IFNγ) conditions; (2) a slight increase in CD163 surface protein expression in response to M(IL-4) conditions and (3) an increase in both CD163 mRNA and protein in response to M(IL-10) conditions. To compare expression of CD163 mRNA detected by PrimeFlow in response to IL-10 treatment, qRT-PCR was performed on THP-1 cells, primary CD14+ blood monocytes and MDM stimulated with a variety of polarizing conditions. As shown in Figure 2B, we confirm that treatment of these cell types with IL-10 resulted in a significant increase in CD163 mRNA levels above untreated in all cell types; whereas, other treatments did not significantly increase CD163 mRNA levels above that of untreated cells. To analyse the specificity of Primeflow mRNA probes for cytokine-induced gene expression changes in macrophages, an AF-647-labeled probe for CD80 mRNA, which is not significantly upregulated by IL-10 treatment (12, 13) was utilized. As shown in Figure 2C, CD80 mRNA signal was not increased in response to treatment of THP-1 cells with IL-10 . In contrast, classical activation triggered by LPS and IFNγ showed an increase in CD80 mRNA expression in THP-1 cells as expected. Together, this demonstrates the specificity of this system for differential gene expression under distinct inflammatory conditions.
Figure 2. IL-10 specifically increases expression of CD163 mRNA and protein.
(A) Flow cytometry of CD163 mRNA (Alexa647) and protein (PE) expression in THP-1 cells cultured under macrophage polarizing conditions as indicated. Images are representative of two independent experiments. (B) Real time PCR for CD163 mRNA in CD14+ monocytes (left), MDMs (center), or THP-1 cells (right) treated under specific macrophage polarizing conditions for 24 hours as indicated. Bars represent relative fold change to untreated monocytes. Error bars indicate SEM. * = P < 0.05, one-way ANOVA with follow-up Tukey multiple comparisons test for M(IL-10) vs untreated. Data are representative of three independent experiments performed in triplicate. (C) Flow cytometry of CD80 mRNA (Alexa647) levels in THP-1 cells cultured for 24h under the indicated conditions. Data are representative of two independent experiments.
CD163 mRNA and protein increase in response to IL-10 in both a time-dependent and dose-dependent manner.
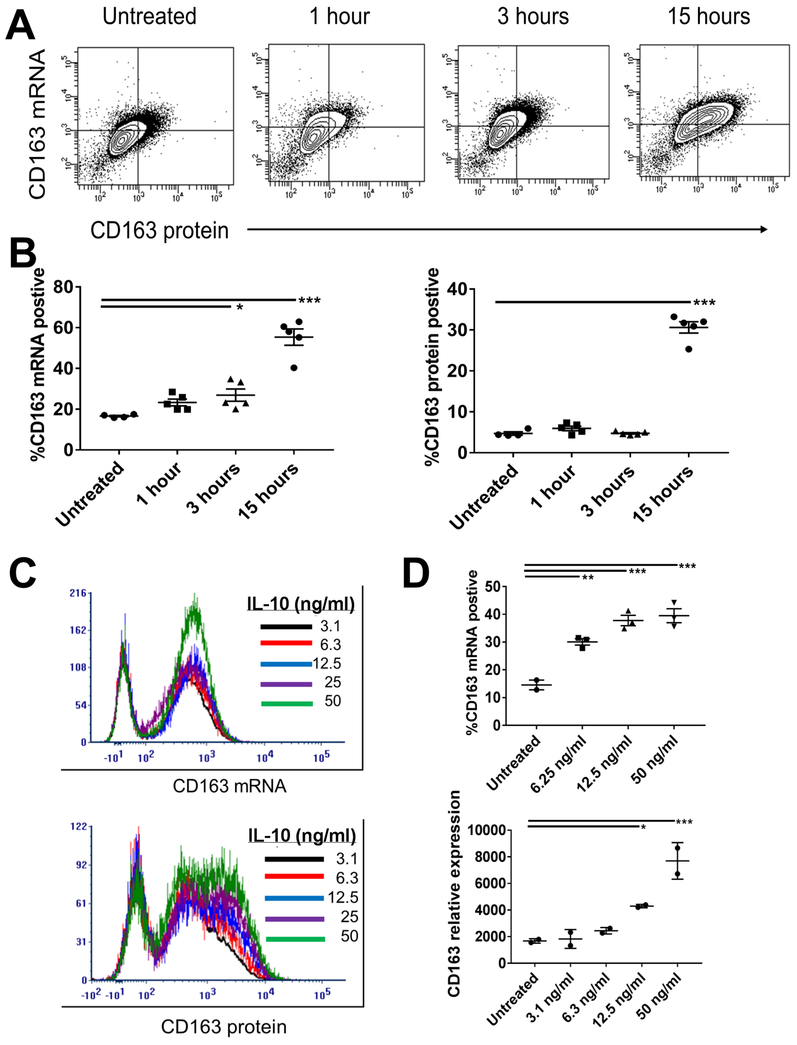
CD163-positive macrophages have been widely reported in inflammatory disease including SJIA, MAS, and HLH. Regulation of CD163 expression is poorly described, but likely occurs at multiple levels, including via microRNA and other post-transcriptional steps (15). To determine the utility of PrimeFlow RNA flow cytometry for studying CD163 regulation, the time course and dose dependence of IL-10-induced CD163 mRNA and protein expression were examined. As shown in Figure 3A-B, using PrimeFlow we found early increased expression of CD163 mRNA following treatment with IL-10, with continued increases up to 15 hours post-stimulation. In contrast, CD163 protein showed no significant changes in expression at early time points (1 and 3 hours post-IL-10), with robust increase only visualized at 15 hours. The percent of cells positive for CD163 mRNA and protein is shown in Figure 3B, demonstrating a trend towards increased mRNA expression at 1 hours and significant increase at 3 hours post-treatment, with a lag in protein expression. This data suggests that PrimeFlow RNA flow cytometry is highly sensitive to changes in mRNA expression, and can detect increased CD163 mRNA levels before significant changes in surface protein expression.
Figure 3. Time and Dose Response of CD163 mRNA and protein to IL-10 stimulations in THP-1 cells.
(A) Representative flow cytometry analysis of CD163 mRNA (AF647) and protein (PE) was performed following IL-10 stimulation of THP-1 cells for the indicated hours. (B) Figures indicate the percent of positive cells for CD163 mRNA (left) or protein (right) after the indicated time of treatment with IL-10. Data are pooled from five experiments. (C) Representative plots of CD163 mRNA (top) and protein (bottom) analysis by flow cytometry following overnight stimulation with IL-10 at the indicated dosages. (D) CD163 mRNA detection by flow cytometry (top) and real time PCR (bottom) from three experiments. Error bars represent SEM. *p<0.05; **p<0.01; ***P<0.001, one-way ANOVA with follow-up Dunnett’s multiple comparisons test vs untreated.
To examine the sensitivity of this system for CD163 mRNA detection further, we also examined the response to increasing concentrations of IL-10. As shown in Figure 3C, RNA flow cytometry showed a small but dose-dependent increase in CD163 mRNA in response to overnight treatment with increasing concentrations of IL-10. In contrast, CD163 protein expression more clearly showed increased surface levels with low concentrations of IL-10 (<10 ng/ml), with continued dose response up to 50 ng/ml. We also compared the sensitivity of CD163 mRNA detection using flow cytometry with real time qPCR. Interestingly, RNA flow cytometry showed significantly increased percentage of CD163 mRNA positive cells at IL-10 doses as low as 6.3ng/ml. In contrast, qPCR analysis did not show significant changes in CD163 expression until doses of 12.5ng/ml (Figure 3D). Collectively, our findings support the use of specific CD163 mRNA PrimeFlow probe as a highly sensitive method to detect changes in gene expression in a variety of cellular conditions, including changes not detected by qPCR, and demonstrate its potential to define distinct kinetics of response compared to surface protein expression.
Development of a multi-marker panel for detection of CD163 mRNA in conjunction with cell surface markers.
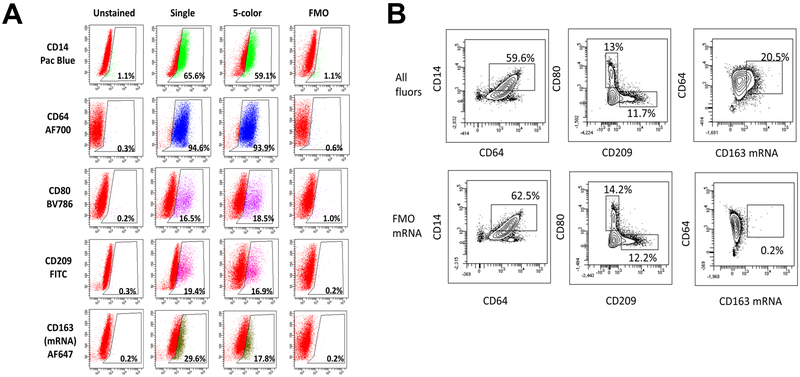
Analysis of CD163 mRNA expression in PBMC or bone marrow cells could serve as an important tool for defining specific monocyte/macrophage populations in translational studies with patient samples. While our above studies demonstrate that IL-10 induces CD163 mRNA and protein expression, disparate stimuli including M(LPS+IFNγ) (also referred to as “M1”) and M(IL-4) (also referred to as “M2a”) induce other well characterized markers of macrophage polarization such as CD64, CD80, and CD209. In order to model a mixed population of polarized macrophages expressing a range of distinct polarization markers, THP-1 were stimulated separately with M(LPS+IFNγ), M(IL-4), or M(IL-10) and then pooled. This mixed population was then utilized to assess the ability of antibodies labelled with specific fluorophores combined with PrimeFlow detection of CD163 mRNA to characterize mixed monocyte/macrophage populations. As shown in Table 1, specific polarization conditions led to variable expression of macrophage markers, in particular with CD80 significantly induced by M(LPS+IFNγ) treatment and CD209 by M(IL-4) treatment. Analyses of all 5 markers is shown in Figure 4, including CD14-Pacific Blue, CD64-AF700, CD80-BV786, and CD209-FITC, as well as the CD163 mRNA AF647 labeled-probe in cells either stained for the marker alone (single), stained with all 5 markers (5-color), or stained with all 5 markers minus a single fluor (fluorescence minus one, FMO) under PrimeFlow conditions. The data were analysed using an FSC vs SSC gate for live cells followed by singlet gates. Shown in Figure 4A are histograms of the singlet gate populations and the percentage of the singlets that were positive for the indicated marker. Unstained tubes are shown for reference of background autofluorescence with the PrimeFlow methodology. Analysis of CD14, CD64, CD80, CD209 and CD163 mRNA showed similar percentages of positive cells that correlated well between the single stain and the 5-color stain. Two dimensional gating shown in Figure 4B indicate similar percentages of positive cells, with a similar distribution of these subpopulations in the presence or absence of CD163 mRNA probe as well. This also clearly distinguishes distinct populations of CD80+CD209− macrophages polarized towards M(LPS+IFNγ) and CD80−CD209+ macrophages polarized towards M(IL-4). Taken together, these findings show that PrimeFlow RNA flow cytometry can be used as part of a multi-marker panel to examine CD163 mRNA and protein expression in mixed functional populations of human myeloid cells.
Table 1:
Expression of macrophage surface markers
| CD14 | CD64 | CD80 | CD163 | CD209 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MFI | ±SD | MFI | ±SD | MFI | ±SD | MFI | ±SD | MFI | ±SD | |
| Untreated | 1360 | 19 | 3249 | 153 | 657 | 37 | 570 | 225 | 129 | 4 |
| M(LPS+IFNγ) | 860*** | 72 | 4132*** | 72 | 2213*** | 211 | 626 | 249 | 111** | 3 |
| M(IL-4) | 1102** | 17 | 3585* | 57 | 566 | 12 | 387 | 29 | 352*** | 4 |
| M(IL-10) | 2263*** | 78 | 6555*** | 166 | 1325 | 540 | 1851*** | 64 | 139 | 3 |
p<0.05.
p<0.01.
p<0.0001 from three replicates and representative of two independent experiments
Figure 4. Several fluorophores are compatible with the use of CD163 mRNA Probe.
THP-1 cells were treated separately with M(LPS+IFNγ), M(IL-4), and M(IL-10) for 24 hours, then pooled together and processed as either unstained, stained with a single fluor, stained for all 5 indicated fluors or stained with all 5 fluors minus the indicated fluor (FMO, fluorescence minus one). (A) Percent of FSC vs SSC live, singlet population for each fluor is depicted in histogram form as SSC vs. Fluor. (B) Gating for subpopulations with all fluors included in staining or all fluors excluding AF647 CD163 mRNA probe (FMO mRNA). Results are representative of three separate experiments.
Primeflow detection of increased CD163 mRNA expression in monocytes from children with SJIA and MAS.
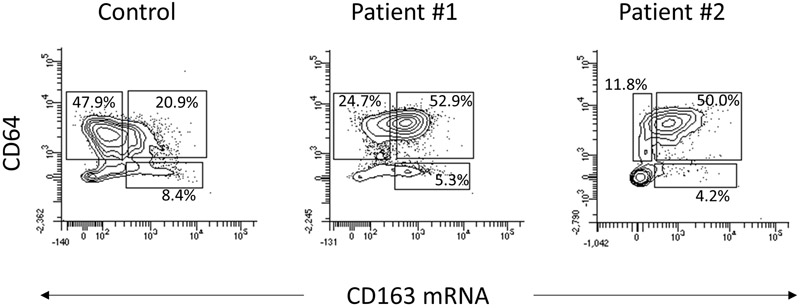
Both active SJIA and MAS are associated with marked innate immune activation and proinflammatory cytokine production, as well as increased expression of CD163 on monocytes and macrophages (3). To determine whether this multi-marker panel including Primeflow can detect changes in monocyte activation markers such as CD163 from clinical samples, we examined fresh, unstimulated PBMC obtained from two children with active SJIA and a healthy donor. Patient 1 had treatment-refractory SJIA, with persistently elevated inflammatory markers, recurrent episodes of MAS and chronic lung disease despite anti-IL-1 therapy. Patient 2 had newly diagnosed SJIA with early/subclinical MAS, and satisfied the 2016 MAS classification criteria(31). Compared to PBMC obtained from healthy donors, monocytes from both patient samples demonstrated markedly elevated CD163 mRNA expression as determined by Primeflow, 58.2% and 54.2% in patients compared to 30.2% in the control (Figure 5). In addition, monocytes from both patients demonstrated enhanced surface CD64 expression, as determined by MFI (4185 and 4296 vs 2084 in controls). Monocyte CD64 expression is known to be enhanced by IFNγ(32), which we have recently shown is linked to emergence of MAS(33, 34), and both patients had elevated serum CXCL9 reflecting IFNγ activity (1900 and 711 pg/ml, normal <121 pg/ml). Surface expression of other monocyte markers including CD14, CD80, and CD209 were not appreciably altered in patients compared with control cells. Interestingly, while CD80 is also typically induced by IFNγ, previous work has shown similarly modest changes in monocyte surface expression in active SJIA (16). Taken together, this demonstrates that a Primeflow multi-marker panel for monocyte and macrophage polarization can be used to examine key functional properties in patient samples from hyperinflammatory states such as SJIA and MAS.
Figure 5.
Expression of CD163 mRNA and CD64 surface protein in unstimulated control and SJIA patient monocytes. PBMC from controls and patients were stained with the 5-color panel as described. Dot plots indicate the percentages of CD64+CD163−, CD64+ CD163+ and CD64−CD163+ populations in control cells, patient 1, and patient 2.
DISCUSSION
Expression of CD163 by monocytes and macrophages serves as both a lineage specific cell marker and a functional pattern recognition receptor with key roles in immunoregulation (6). CD163 directly mediates the binding and uptake of hemoglobin complexed with haptoglobin, limiting oxidative tissue damage. CD163 can also clear some damage-associated molecular patterns such as endogenous high mobility group box 1 (HMGB1), and directly trigger anti-inflammatory functions of tissue macrophages (35). Indeed, mice lacking CD163 displayed enhanced mortality in a mouse model of sepsis (35). Expression of CD163 is also strongly linked to both HLH and MAS associated with SJIA, with high expression on macrophages in tissue and markedly elevated serum sCD163 levels (4, 7, 8, 16, 36). Tools to study polarization properties with complex regulation like CD163 remain a barrier to defining the pathogenesis of these series conditions. Here, we show that RNA flow cytometry is a highly sensitive technique for detecting and quantifying CD163 expression at the single cell level in human monocytes and macrophages. This RNA flow cytometry probe could be combined with fluorescently-conjugated antibodies to allow for simultaneous detection of CD163 surface protein and mRNA expression, along with several other canonical macrophage markers, in myeloid populations with mixed polarization phenotypes. We also demonstrate the utility of this panel to quantify changes in expression of activation markers including CD163 in unstimulated monocytes from children with SJIA and MAS. This multi-parameter panel will support mechanistic characterization of complex monocyte and macrophage populations during systemic inflammatory disorders such as SJIA and MAS.
Historically, analysis of RNA from a biological sample was subject to bulk RNA preparation that reflected heterogeneity of multiple cell types. Until recently detection of mRNA transcripts at a single cell level was limited to in situ hybridization, unless transcripts were in high abundance. Currently, single cell RNA-Seq provides transcriptional profiles at the single cell level, but are not readily correlated with protein expression, although these technologies are rapidly being developed. The advent of branching technology to amplify probe signal has allowed for the detection of transcripts that are not in high abundance (20, 23). Recently, several studies have utilized this technology to examine viral RNA or DNA (37–39) and to determine expression of specific transcripts on a single cell level in various immune contexts(40–42). SmartFlare technology has also recently emerged to detect transcripts and microRNA at the single cell level (26, 43, 44). Experimental design primarily dictates the approach necessary, as SmartFlare technology requires cellular uptake of probe prior to analysis. We utilized Primeflow technology, as our goal is to examine expression of transcripts in fresh PBMC without incubation of cells prior to analysis. This technology is designed to detect signal from as little as 20 RNA copies per cell with high correlation to other mRNA detection systems including PCR (45). Soh and colleagues provide a history of the development of RNA hybridization technique and amplification, as well as limitations, such as fluorophore and antibody combinations that may not work well with the technology (20).
Techniques for RNA cytometry can have crucial roles in defining immune responses, including the regulation of functional phenotypes displayed by macrophages. CD163 is not induced by canonical proinflammatory or “M1” polarization, but rather by several more immunoregulatory or anti-inflammatory polarization conditions broadly termed “M2”, and regulated at both the transcriptional and post-transcriptional levels (9, 15). A more systematic exploration of macrophage polarization strikingly shows that these cells have the capacity for much more phenotypic diversity, and only certain signals induce expression of markers such as CD163 (10). The anti-inflammatory cytokine IL-10 has previously been shown to strongly induce CD163 mRNA expression (12, 29). Our findings using both qRT-PCR on the population level and Primeflow RNA flow cytometry on the single cell level confirm this, showing robust CD163 expression only upon IL-10 treatment compared to other well characterized polarizing conditions.
The precise signaling leading from cytokine stimulation to CD163 gene expression in macrophages are not fully known. Our previous work has demonstrated that microRNA networks including miR-125a-5p and miR-181 family members serve to restrict CD163 expression under some polarization conditions while allowing it in M(IL-10) (15). However, the highly sensitive RNA flow cytometry approach described here provides a useful tool to further study IL-10-induced CD163 expression. Detection of CD163 mRNA with PrimeFlow could visualize changes in gene expression at earlier time points than CD163 protein detection, and at lower concentrations of IL-10 than seen with Real time qPCR. The simultaneous detection of both mRNA and protein at the single cell level is a powerful tool for further studies of CD163 regulation in activated monocytes and macrophages.
While previously considered a dichotomy between classical “M1” and alternative “M2” activation, the understanding of macrophage plasticity and polarization has evolved considerably, with in vitro characterization of numerous functional populations with distinct gene expression profiles (10). However, it remains unclear how these in vitro populations reflect what is observed in vivo during systemic inflammation, including in rheumatic diseases. Macrophages expressing CD163 are increasingly recognized as key effector cells in autoimmune diseases, including discoid lupus skin lesions (46) and lupus nephritis (47). Monocytes and macrophages are also central innate immune effectors in SJIA (16, 48, 49), where they manifest increased CD163 expression which is further enhanced during MAS (8, 36). However, both transcriptional profiling and immunophenotyping of monocytes in SJIA have revealed features reflecting multiple “M1” and “M2” polarization states, which could indicate either a mixed polarization phenotype or multiple distinct cellular populations (16–19). The flow cytometry panel described herein, able to define both CD163 mRNA and multiple surface protein levels at single-cell resolution, represents an ideal tool to detect distinct monocyte and macrophage populations during states of systemic inflammation. Indeed, our analysis of unstimulated PBMC from patients with SJIA and MAS confirm that these monocytes simultaneously show both markedly enhanced IFNγ-induced CD64 expression along with CD163, reflecting the diverse inflammatory signals driving this disease. Further work with these innovative single cell tools will be essential to defining the role of myeloid effectors in SJIA and MAS.
Acknowledgments
This research was supported by the National Institutes of Health P30-AR070549. Dr. Schulert was supported by K08-AR072075 from the National Institute for Arthritis and Musculoskeletal Diseases, National Institutes of Health, a Scientist Development Award from the Rheumatology Research Foundation and a Procter Scholar Award from the Cincinnati Children’s Research Foundation.
Bibliography
- 1.Schulert GS, and Grom AA. 2015. Pathogenesis of macrophage activation syndrome and potential for cytokine- directed therapies. Annu. Rev. Med. 66: 145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang M, Behrens EM, Atkinson TP, Shakoory B, Grom AA, and Cron RQ. 2014. Genetic defects in cytolysis in macrophage activation syndrome. Curr Rheumatol Rep 16: 439. [DOI] [PubMed] [Google Scholar]
- 3.Mellins ED, Macaubas C, and Grom AA. 2011. Pathogenesis of systemic juvenile idiopathic arthritis: some answers, more questions. Nat Rev Rheumatol 7: 416–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canna SW, Costa-Reis P, Bernal WE, Chu N, Sullivan KE, Paessler ME, and Behrens EM. 2014. Brief report: alternative activation of laser-captured murine hemophagocytes. Arthritis Rheumatol. (Hoboken, N.J.) 66: 1666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabriek BO, van Bruggen R, Deng DM, Ligtenberg AJM, Nazmi K, Schornagel K, Vloet RPM, Dijkstra CD, and van den Berg TK. 2009. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113: 887–92. [DOI] [PubMed] [Google Scholar]
- 6.Van Gorp H, Delputte PL, and Nauwynck HJ. 2010. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol. Immunol. 47: 1650–60. [DOI] [PubMed] [Google Scholar]
- 7.Schaer DJ, Schleiffenbaum B, Kurrer M, Imhof A, Bächli E, Fehr J, Moller HJ, Moestrup SK, and Schaffner A. 2005. Soluble hemoglobin-haptoglobin scavenger receptor CD163 as a lineage-specific marker in the reactive hemophagocytic syndrome. Eur. J. Haematol. 74: 6–10. [DOI] [PubMed] [Google Scholar]
- 8.Bleesing J, Prada A, Siegel DM, Villanueva J, Olson J, Ilowite NT, Brunner HI, Griffin T, Graham TB, Sherry DD, Passo MH, V Ramanan A, Filipovich A, and Grom AA. 2007. The diagnostic significance of soluble CD163 and soluble interleukin-2 receptor alpha-chain in macrophage activation syndrome and untreated new-onset systemic juvenile idiopathic arthritis. Arthritis Rheum 56: 965–971. [DOI] [PubMed] [Google Scholar]
- 9.Martinez FO, Sica A, Mantovani A, and Locati M. 2008. Macrophage activation and polarization. Front Biosci 13: 453–461. [DOI] [PubMed] [Google Scholar]
- 10.Xue J, V Schmidt S, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, and Schultze JL. 2014. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 40: 274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang R, Patel D, Morris JJ, Rutschman RL, and Murray PJ. 2002. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169: 2253–63. [DOI] [PubMed] [Google Scholar]
- 12.Williams L, Jarai G, Smith A, and Finan P. 2002. IL-10 expression profiling in human monocytes. J. Leukoc. Biol. 72: 800–9. [PubMed] [Google Scholar]
- 13.Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TRDJ, Reedquist KA, Tak PP, and Baeten DLP. 2012. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J. Immunol. Methods 375: 196–206. [DOI] [PubMed] [Google Scholar]
- 14.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, and Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25: 677–686. [DOI] [PubMed] [Google Scholar]
- 15.Do T, Tan R, Bennett M, Medvedovic M, Grom AA, Shen N, Thornton S, and Schulert GS. 2018. MicroRNA networks associated with active systemic juvenile idiopathic arthritis regulate CD163 expression and anti-inflammatory functions in macrophages through two distinct mechanisms. J. Leukoc. Biol. 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macaubas C, Nguyen KD, Peck A, Buckingham J, Deshpande C, Wong E, Alexander HC, Chang SY, Begovich A, Sun Y, Park JL, Pan KH, Lin R, Lih CJ, Augustine EM, Phillips C, V Hadjinicolaou A, Lee T, and Mellins ED. 2012. Alternative activation in systemic juvenile idiopathic arthritis monocytes. Clin Immunol 142: 362–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fall N, Barnes M, Thornton S, Luyrink L, Olson J, Ilowite NT, Gottlieb BS, Griffin T, Sherry DD, Thompson S, Glass DN, Colbert RA, and Grom AA. 2007. Gene expression profiling of peripheral blood from patients with untreated new-onset systemic juvenile idiopathic arthritis reveals molecular heterogeneity that may predict macrophage activation syndrome. Arthritis Rheum 56: 3793–3804. [DOI] [PubMed] [Google Scholar]
- 18.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, Ardura M, Chung W, Smith E, Wise C, Palucka K, Ramilo O, Punaro M, Banchereau J, and Pascual V. 2007. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med 204: 2131–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogilvie EM, Khan A, Hubank M, Kellam P, and Woo P. 2007. Specific gene expression profiles in systemic juvenile idiopathic arthritis. Arthritis Rheum 56: 1954–1965. [DOI] [PubMed] [Google Scholar]
- 20.Soh KT, Tario JD, Colligan S, Maguire O, Pan D, Minderman H, and Wallace PK. 2016. Simultaneous, Single-Cell Measurement of Messenger RNA, Cell Surface Proteins, and Intracellular Proteins In Current Protocols in Cytometry vol. 75 John Wiley & Sons, Inc., Hoboken, NJ, USA: 7.45.1–7.45.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thaler B, Hohensinner PJ, Krychtiuk KA, Matzneller P, Koller L, Brekalo M, Maurer G, Huber K, Zeitlinger M, Jilma B, Wojta J, and Speidl WS. 2016. Differential in vivo activation of monocyte subsets during low-grade inflammation through experimental endotoxemia in humans. Sci. Rep. 6: 30162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim CC, Nakamura MC, and Hsieh CL. 2016. Brain trauma elicits non-canonical macrophage activation states. J. Neuroinflammation 13: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanley MB, Lomas W, Mittar D, Maino V, and Park E. 2013. Detection of low abundance RNA molecules in individual cells by flow cytometry. PLoS One 8: e57002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalapour S, Font-Burgada J, Di Caro G, Zhong Z, Sanchez-Lopez E, Dhar D, Willimsky G, Ammirante M, Strasner A, Hansel DE, Jamieson C, Kane CJ, Klatte T, Birner P, Kenner L, and Karin M. 2015. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521: 94–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojno EDT, Monticelli LA, V Tran S, Alenghat T, Osborne LC, Thome JJ, Willis C, Budelsky A, Farber DL, and Artis D. 2015. The prostaglandin D₂ receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8: 1313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClellan S, Slamecka J, Howze P, Thompson L, Finan M, Rocconi R, and Owen L. 2015. mRNA detection in living cells: A next generation cancer stem cell identification technique. Methods 82: 47–54. [DOI] [PubMed] [Google Scholar]
- 27.Porichis F, Hart MG, Griesbeck M, Everett HL, Hassan M, Baxter AE, Lindqvist M, Miller SM, Soghoian DZ, Kavanagh DG, Reynolds S, Norris B, Mordecai SK, Nguyen Q, Lai C, and Kaufmann DE. 2014. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat. Commun. 5: 5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulert GS, Fall N, Harley JB, Shen N, Lovell DJ, Thornton S, and Grom AA. 2016. Monocyte microRNA expression in active systemic juvenile idiopathic arthritis implicates miR-125a-5p in polarized monocyte phenotypes. Arthritis Rheumatol. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buechler C, Ritter M, Orsó E, Langmann T, Klucken J, and Schmitz G. 2000. Regulation of scavenger receptor CD163 expression in human monocytes and macrophages by pro- and antiinflammatory stimuli. J. Leukoc. Biol. 67: 97–103. [PubMed] [Google Scholar]
- 30.Sulahian TH, Högger P, Wahner AE, Wardwell K, Goulding NJ, Sorg C, Droste A, Stehling M, Wallace PK, Morganelli PM, and Guyre PM. 2000. Human monocytes express CD163, which is upregulated by IL-10 and identical to p155. Cytokine 12: 1312–21. [DOI] [PubMed] [Google Scholar]
- 31.Ravelli A, Minoia F, Davì S, Horne A, Bovis F, Pistorio A, Aricò M, Avcin T, Behrens EM, De Benedetti F, Filipovic L, Grom AA, Henter J-I, Ilowite NT, Jordan MB, Khubchandani R, Kitoh T, Lehmberg K, Lovell DJ, Miettunen P, Nichols KE, Ozen S, Pachlopnik Schmid J, V Ramanan A, Russo R, Schneider R, Sterba G, Uziel Y, Wallace C, Wouters C, Wulffraat N, Demirkaya E, Brunner HI, Martini A, Ruperto N, and Cron RQ. 2016. 2016 Classification Criteria for Macrophage Activation Syndrome Complicating Systemic Juvenile Idiopathic Arthritis: A European League Against Rheumatism/American College of Rheumatology/Paediatric Rheumatology International Trials Organisation Collaborat. Arthritis Rheumatol. (Hoboken, N.J.) 68: 566–76. [DOI] [PubMed] [Google Scholar]
- 32.Perussia B, Dayton ET, Lazarus R, Fanning V, and Trinchieri G. 1983. Immune interferon induces the receptor for monomeric IgG1 on human monocytic and myeloid cells. J. Exp. Med. 158: 1092–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bracaglia C, de Graaf K, Pires Marafon D, Guilhot F, Ferlin W, Prencipe G, Caiello I, Davì S, Schulert G, Ravelli A, Grom AA, de Min C, and De Benedetti F. 2017. Elevated circulating levels of interferon-γ and interferon-γ-induced chemokines characterise patients with macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 76: 166–172. [DOI] [PubMed] [Google Scholar]
- 34.Cassatella MA, Flynn RM, Amezaga MA, Bazzoni F, Vicentini F, and Trinchieri G. 1990. Interferon gamma induces in human neutrophils and macrophages expression of the mRNA for the high affinity receptor for monomeric IgG (Fc gamma R-I or CD64). Biochem. Biophys. Res. Commun. 170: 582–8. [DOI] [PubMed] [Google Scholar]
- 35.Yang H, Wang H, Levine YA, Gunasekaran MK, Wang Y, Addorisio M, Zhu S, Li W, Li J, V de Kleijn DP, Olofsson PS, Warren HS, He M, Al-Abed Y, Roth J, Antoine DJ, Chavan SS, Andersson U, and Tracey KJ. Identification of CD163 as an antiinflammatory receptor for HMGB1-haptoglobin complexes. JCI insight 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behrens EM, Beukelman T, Paessler M, and Cron RQ. 2007. Occult macrophage activation syndrome in patients with systemic juvenile idiopathic arthritis. J Rheumatol 34: 1133–1138. [PubMed] [Google Scholar]
- 37.Grau-Expósito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocaña I, Ribera E, Torrella A, Planas B, Badía R, Castellví J, Falcó V, Crespo M, and Buzon MJ. 2017. A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4+ T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. MBio 8: e00876–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell JK, Midkiff BR, Israelow B, Evans MJ, Lanford RE, Walker CM, Lemon SM, and McGivern DR. 2017. Hepatitis C Virus Indirectly Disrupts DNA Damage-Induced p53 Responses by Activating Protein Kinase R. MBio 8: e00121–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martrus G, Niehrs A, Cornelis R, Rechtien A, García-Beltran W, Lütgehetmann M, Hoffmann C, and Altfeld M. 2016. Kinetics of HIV-1 Latency Reversal Quantified on the Single-Cell Level Using a Novel Flow-Based Technique. J. Virol. 90: 9018–9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T-T, Gonzalez DG, Cote CM, Kerfoot SM, Deng S, Cheng Y, Magari M, and Haberman AM. 2017. Germinal center B cell development has distinctly regulated stages completed by disengagement from T cell help. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martínez Gómez JM, Periasamy P, Dutertre C-A, Irving AT, Ng JHJ, Crameri G, Baker ML, Ginhoux F, Wang L-F, and Alonso S. 2016. Phenotypic and functional characterization of the major lymphocyte populations in the fruit-eating bat Pteropus alecto. Sci. Rep. 6: 37796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi JS, Russo MA, Massey JM, Juel V, Hobson-Webb LD, Gable K, Raja SM, Balderson K, Weinhold KJ, and Guptill JT. 2017. B10 Cell Frequencies and Suppressive Capacity in Myasthenia Gravis Are Associated with Disease Severity. Front. Neurol. 8: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu L, Kong Y, Zhang J, Claxton DF, Ehmann WC, Rybka WB, Palmisiano ND, Wang M, Jia B, Bayerl M, Schell TD, Hohl RJ, Zeng H, and Zheng H. 2017. Blimp-1 impairs T cell function via upregulation of TIGIT and PD-1 in patients with acute myeloid leukemia. J. Hematol. Oncol. 10: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krönig M, Walter M, Drendel V, Werner M, Jilg CA, Richter AS, Backofen R, McGarry D, Follo M, Schultze-Seemann W, and Schüle R. 2015. Cell type specific gene expression analysis of prostate needle biopsies resolves tumor tissue heterogeneity. Oncotarget 6: 1302–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai C, Stepniak D, Sias L, and Funatake C. 2018. A sensitive flow cytometric method for multi-parametric analysis of microRNA, messenger RNA and protein in single cells. Methods 134–135: 136–148. [DOI] [PubMed] [Google Scholar]
- 46.Chong BF, Tseng L-C, Hosler GA, Teske NM, Zhang S, Karp DR, Olsen NJ, and Mohan C. 2015. A subset of CD163+ macrophages displays mixed polarizations in discoid lupus skin. Arthritis Res. Ther. 17: 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olmes G, Büttner-Herold M, Ferrazzi F, Distel L, Amann K, and Daniel C. 2015. CD163+ M2c-like macrophages predominate in renal biopsies from patients with lupus nephritis. Arthritis Res. Ther. 18: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ling XB, Park JL, Carroll T, Nguyen KD, Lau K, Macaubas C, Chen E, Lee T, Sandborg C, Milojevic D, Kanegaye JT, Gao S, Burns J, Schilling J, and Mellins ED. 2010. Plasma profiles in active systemic juvenile idiopathic arthritis: Biomarkers and biological implications. Proteomics 10: 4415–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Benedetti F, Massa M, Robbioni P, Ravelli A, Burgio GR, and Martini A. 1991. Correlation of serum interleukin-6 levels with joint involvement and thrombocytosis in systemic juvenile rheumatoid arthritis. Arthritis Rheum 34: 1158–1163. [DOI] [PubMed] [Google Scholar]