Abstract
Olfactory sensory neurons (OSNs) are generated throughout life from progenitor cells in the olfactory epithelium. OSN axons project in an odorant receptor-specific manner to the olfactory bulb (OB), forming an ordered array of glomeruli where they provide sensory input to OB neurons. The tetracycline transactivator (tTA) system permits developmental stage-specific expression of reporter genes in OSNs and has been widely used for structural and functional studies of the development and plasticity of the mouse olfactory system. However, the cellular ages at which OSNs stop expressing reporters driven by the immature OSN-specific Gγ8-tTA driver line and begin to express reporters driven by the mature OSN-specific OMP-tTA driver line have not been directly determined. We pulse-labeled terminally dividing cells in the olfactory epithelium of 28-day-old (P28) mice with EdU and analyzed EdU labeling in OSNs expressing fluorescent reporter proteins under control of either the Gγ8-tTA or OMP-tTA driver line 5 – 14 days later. Expression of OMP-tTA-driven reporters began in 6-day-old OSNs, while the vast majority of newborn OSNs did not express Gγ8-tTA-driven fluorescent proteins beyond 8 days of cellular age. Surprisingly, we also found a low survival rate for P28-born OSNs, very few of which survived for more than 14 days. We propose that OSN survival requires the formation of stable synaptic connections and hence may be dependent on organismal age.
Keywords: Adult neurogenesis, olfactory sensory neuron, olfactory epithelium, neuronal survival
Introduction
In mammals, volatile odors in inhaled air are detected by odorant receptors expressed on the cilia of olfactory sensory neurons (OSNs), which reside in the olfactory epithelium (OE) at the back of the nasal cavity. OSN axons project through the cribriform plate to provide olfactory sensory input to the olfactory bulb (OB), the first brain processing center for odor information. While the repertoire of odorant receptors is large (~1100 functional genes in mice and ~350 in humans (Godfrey et al. 2004; Malnic et al. 2004; Saraiva et al. 2016)), each OSN expresses a single allele of a single odorant receptor. Furthermore, while OSNs expressing any given odorant receptor are scattered across broad zones of the OE, the axons of OSNs expressing the same receptor coalesce together to form spherical neuropil structures termed glomeruli near the surface of the OB (Strotmann et al. 1992; Ressler et al. 1993, 1994; Vassar et al. 1993, 1994; Mombaerts et al. 1996). Hence, odor input to the OB is highly spatially organized.
Remarkably, despite ongoing OSN neurogenesis, this glomerular map is maintained throughout life. OSN neurogenesis peaks during the second postnatal week, but is maintained at a lower level throughout life in both rodents and humans (Graziadei and Monti Graziadei 1978; Ryba and Tirindelli 1995; Tirindelli and Ryba 1996; Hahn et al. 2005; Kondo et al. 2010). In the healthy OE, the majority of newly-generated OSNs are derived from globose basal cells (GBCs) (Caggiano et al. 1994), whereas horizontal basal cells play a particularly important role in OE regeneration following severe injury (Leung et al. 2007). OSNs are thought to have a half-life of approximately one month (Holl 2018), although a significant fraction has a lifespan of at least three months under standard mouse housing conditions (Mackay-Sim and Kittel 1991) and a minority can survive for at least a year when mice are housed under filtered air or pathogen-free conditions (Hinds et al. 1984; Holl 2018). Synchronous waves of neurogenesis are not seen in the OE of healthy mice, indicating that OSN lifespan is likely to vary (Mackay-Sim and Kittel 1991; Schwob et al. 1992). Nonetheless, individual GBCs follow a clearly defined developmental pathway via a neuronally-committed Ascl1-expressing intermediate to generate nascent OSNs that express CXCR4 and DBN1, which rapidly become immature OSNs that express GAP43 and Gγ8 (Verhaagen et al. 1989; Ryba and Tirindelli 1995; McIntyre et al. 2010; Schwob et al. 2017). As they mature, newly generated OSN somata migrate apically, resulting in a pseudostratified organization of the OE (Farbman and Margolis 1980; Farbman 1992; Roskams et al. 1998; Rodriguez-Gil et al. 2015). Full OSN maturation is defined by the onset of olfactory marker protein (OMP) expression, which is concomitant with the downregulation of GAP43 and Gγ8 expression (Miragall and Graziadei 1982; Verhaagen et al. 1989; Tirindelli and Ryba 1996).
Harnessing these well-established maturation stage-specific markers, the tetracycline transactivator (tTA) system has been widely used to drive specific expression in either immature or mature OSNs. Both the Gγ8-tTA transgenic (Nguyen et al. 2007) and OMP-IRES-tTA knockin [referred to as OMP-tTA] (Yu et al. 2004) driver lines have been crossed with a wide range of tetO promoter-dependent lines to drive maturation stage-specific expression of proteins including but not limited to odorant receptors, fluorescent proteins, optogenetic actuators, genetically encoded calcium indicators, and cytotoxic proteins. Crucially, unlike recombinase-based cell type-specific driver lines, in which reporter expression is irreversible once switched on, continued expression of the tetO reporter gene depends on ongoing tTA expression and hence is downregulated in Gγ8-expressing OSNs when they reach maturity.
However, the precise neuronal ages at which Gγ8-tTA-dependent expression is downregulated, and OMP-tTA-dependent expression begins, have never been directly determined. In this study, we used EdU click chemistry to “birth-date” OSNs expressing a fluorescent reporter protein driven either by Gγ8-tTA or by OMP-tTA, with a particular focus on the transition from immature to mature OSN.
Materials and methods
Experimental Animals
All animal procedures conformed to National Institutes of Health guidelines and were approved by the Carnegie Mellon University and University of Pittsburgh Institutional Animal Care and Use Committees. Mice were bred in-house and were maintained in individually ventilated cages under temperature- and humidity-controlled conditions on a 12 h light/dark cycle with unrestricted access to food and water. Mice were weaned at postnatal day (P) 21.
C57BL/6J mice were bred from mice purchased from The Jackson Laboraotry (strain #000664). Transgenic mouse lines were as described previously (Cheetham et al. 2016). Experimental mice expressed tdTomato or Citrine in either immature OSNs under control of the Gγ8 promoter (Gγ8-tdTomato [Gγ8-tTA+/−;tetO-sypGFP-tdTomato+/−] or Gγ8-Citrine [Gγ8-tTA+/−;tetO-ChIEF-Citrine+/−]) or in mature OSNs under control of the OMP promoter (OMP-tdTomato [OMP-IRES-tTA+/−;tetO-sypGFP-tdTomato+/−] or OMP-Citrine [OMP-IRES-tTA+/−;tetO-ChIEF-Citrine+/−]). All transgenic mice were of mixed 129 × C57BL/6J background. Each experimental group comprised male and female mice. Mice were genotyped by PCR using the primers described previously (Cheetham et al. 2016). A total of 42 mice were used in this study (18 expressing a Gγ8-tTA-driven reporter protein, 18 expressing an OMP-tTA-driven reporter protein, and 6 C57BL/6J mice).
EdU injection
Mice were postnatal day 28 (P28) on the day of EdU injection, and all injections were performed between 8 – 10 am to control for diurnal variations in cell division rates (Hinds et al. 1984). Mice were briefly anesthetized with isoflurane (4% in O2) and received a single intraperitoneal injection of 50 mgkg−1 EdU dissolved in sterile saline.
Perfusion and dissection
EdU-injected mice underwent transcardial perfusion 5, 6, 7, 8, 10 or 14d (120, 144, 168, 192, 240 or 336 h) later. Time points were accurate to ±1 h. C57BL/6J mice underwent perfusion at either P38 or P66. Mice were deeply anesthetized with 200 mgkg−1 ketamine and 20 mgkg−1 xylazine and transcardially perfused with ice-cold PBS followed by 4% paraformaldehyde (PFA). Olfactory epithelia (OEs) were dissected out, post-fixed overnight in 4% PFA at 4 °C then cryopreserved in 30% sucrose for 24 hours at 4°C. OEs were embedded in 10% gelatin, fixed/cryopreserved overnight in 2% PFA/15% sucrose at 4 °C, flash frozen in 2-methylbutane on dry ice and stored at −80 °C.
Tissue sectioning and staining
50 μm coronal sections were cut through each OE using a cryostat (Microm) and stored at −80 °C. Sections 25%, 50% and 75% along the anterior-posterior axis of each OE were selected for staining. Sections were permeabilized and blocked in 5% NDS/0.5% Triton-X100 (T-X100) in PBS for 1 hour. Sections from EdU-injected mice were then incubated in EdU reaction mixture made according to the manufacturer’s instructions using the Click-iT Plus EdU Alexa Fluor 647 Imaging Kit (Thermo Fisher Scientific) for 30 minutes in the dark. Sections were then washed with 0.5% T-X100 in PBS for 3 × 5 minutes in the dark. OE sections from OMP-tdTomato and OMP-Citrine mice were mounted immediately with Vectashield HardSet (Vector Labs). Sections from Gγ8-tdTomato and Gγ8-Citrine mice were next incubated for 4 days at 4°C in 3% NDS/0.2% T-X100/0.01% sodium azide in PBS with 1:5000 anti-OMP primary antibody (catalog #019-22291; Wako Pure Chemical Industries Inc.). Sections were washed for 3 × 5 minutes in PBS, incubated in 3% NDS/0.2% T-X100 in PBS with 1:500 Dylight-405-donkey anti-goat secondary antibody (catalog #705-475-147; Jackson ImmunoResearch) for 1 hour at 23°C, washed for 3 × 5 minutes with PBS, and mounted with Vectashield HardSet (Vector Labs). After permeabilization and blocking as above, OE sections from C57BL/6J mice were incubated for 5 days at 4°C in 3% NDS/0.2% T-X100/0.01% sodium azide in PBS with 1:200 anti-cleaved caspase-3 primary antibody (catalog #9661S; Cell Signaling Technology), washed for 3 × 5 minutes in PBS, incubated in 3% NDS/0.2% T-X100 in PBS with 1:500 AF546-donkey anti-rabbit secondary antibody (catalog #A10040, ThermoFisher), washed 3 × 5 minutes in PBS, and mounted with VectaShield containing DAPI (Vector Labs).
Confocal microscopy
We confined imaging to the septal OE to control for anatomical variations such as concha bullosa. Z-stack images of septal OE were acquired using either a Nikon A1R confocal microscope equipped with an Apo 60x/1.4 NA oil immersion objective or a Zeiss LSM 880 Meta FCS confocal microscope equipped with a 40x/1.4 NA Plan-Apochromat oil immersion objective. Excitation/ bandpass emission wavelengths were (in nm): OMP-Dylight-405 (405/ 410-450); Citrine (514/ 520-555); tdTomato (561/ 580-630); cleaved caspase-3-AF546 (561/ 580-630); Edu-AF647 (633/ 650-700). Confocal image voxel size was 0.13 × 0.13 × 1.00 μm. For colocalization of Gγ8- or OMP-driven fluorescent reporter-expressing cells with EdU or analysis of cleaved caspase-3 staining, z-stacks were 30 μm in depth. For colocalization of Gγ8+EdU+ cells with OMP immunostaining, z-stacks were 10 μm in depth due to the limited tissue penetration of the anti-OMP antibody.
Image analysis
Images were analyzed using Fiji (https://fiji.sc) (Schindelin et al. 2012). Image brightness and contrast were manipulated purely for display purposes. Cell counts were performed blind to genotype and time post-EdU injection (Figures 1-4) or mouse age (Figure 5). For analysis of EdU-labeled OSNs and colocalization with OMP or Gγ8, cells at each time point were manually quantified from ~1mm of OE septum per section from three coronal sections 25%, 50%, and 75% along the anterior-posterior axis for each mouse. For analysis of cleaved caspase-3 labeling, images were collected along the entire left and right septal OE in two coronal sections each taken 25%, 50%, and 75% along the anterior-posterior axis for each mouse. For OMP-tdTomato and OMP-Citrine mice, the number of OMP+, EdU+, and OMP+EdU+ cells were counted. For Gγ8-tdTomato and Gγ8-Citrine mice, the number of Gγ8+, EdU+, Gγ8+EdU+ and Gγ8+EdU+OMP+ cells were counted. The proportion of double-labeled cells is expressed as a percentage of the total Gγ8+ or OMP+ OSNs for each section, to provide an estimate of the contribution of EdU-labeled cells to the total population expressing either Gγ8 or OMP at each time point after EdU injection. All cell counts are reported as linear density per mm of OE. To quantify the position of cells along the basal-apical axis of the OE, the distance from the center of the EdU-labeled or cleaved caspase-3-labeled cell to the border of the basal lamina was measured, as a percentage of the distance from the basal lamina to the apical surface. Figures were prepared using Photoshop (Adobe).
Statistics
The percentages of Gγ8+EdU+ or OMP+EdU+ cells failed tests for normality and/or equal variance and were analyzed using Kruskal-Wallis one-way ANOVA on Ranks and described as ranges. Percentages of Gγ8+EdU+ or OMP+EdU+ cells at different anterior-posterior positions, linear density of cleaved caspase-3-expressing neurons and OE width passed tests for normality and equal variance and were analyzed with two-way ANOVA. Basal-apical locations of EdU-labeled cells also passed tests for normality and equal variance and were compared with unpaired t-tests. Basal-apical locations of cleaved caspase-3-expressing cells failed a normality test and were compared using a Kolmogorov-Smirnov test. A one-phase decay was used to fit the decrease in Gγ8+EdU+ OSNs with time after EdU injection. EdU+ cell linear density with time after EdU injection is described as median (interquartile range) and was analyzed by linear regression. All statistical analyses were performed using GraphPad Prism 7.
Results
Birth-dating of OSNs expressing Gγ8-tTA-driven fluorescent reporter proteins
To determine the neuronal age at which OSNs cease to express Gγ8-driven reporter proteins, we performed EdU injections in mice expressing either tdTomato or Citrine in Gγ8+ OSNs, perfused them 5 – 14 days later, and cut coronal sections through their OEs. Sections were stained for EdU using the Click-iT system, which preserves fluorescent proteins, and the percentage of Gγ8+ OSNs stained with EdU was quantified in the septal OE at each time point. EdU is a thymidine analog that is incorporated into DNA during the S phase of the cell cycle. The time between EdU injection and perfusion can therefore be used to “birth-date” each OSN. Note that we use the term “newborn OSN” to refer to recently generated OSNs, as marked by their incorporation of EdU.
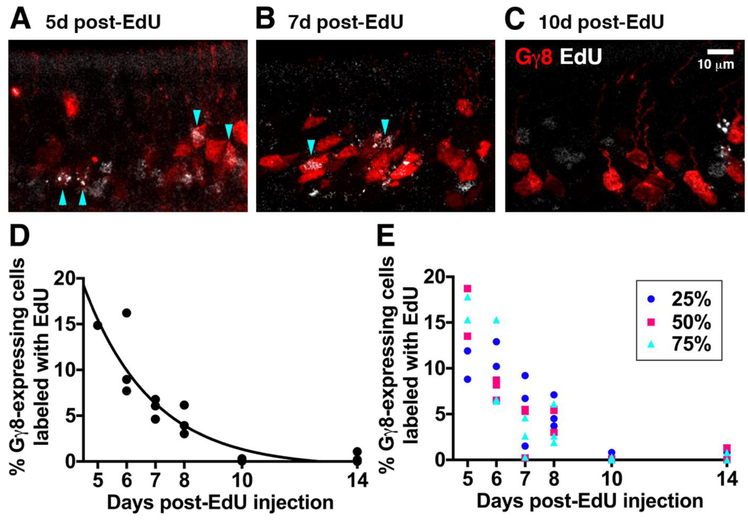
We analyzed a total of 28716 Gγ8+ OSNs in 18 mice (3 mice per time point after EdU injection). Gγ8+EdU+ OSNs were present at all time points analyzed (Fig. 1A-C), but their contribution to the Gγ8+ OSN population depended strongly on neuronal age (Fig. 1D). A one-way ANOVA on Ranks showed a significant effect of time after EdU injection on the proportion of Gγ8+ OSNs that were labeled with EdU (P = 0.013, KW statistic = 14.44). As time after EdU injection increased from 5 to 14 days, the contribution of newborn OSNs to the Gγ8+ OSN population decreased, and this relationship was well fit by a one-phase decay (R2 = 0.87). Notably, EdU-labeled cells comprised just 0.0 – 1.1% of the Gγ8+ OSN population 10-14 days after EdU injection, indicating that the vast majority of OSNs cease to express Gγ8 at these time points.
Figure 1: Gγ8-tTA-driven fluorescent reporters are expressed in 5-8-day old OSNs.
A-C: Single confocal optical sections of the septal OE of mice expressing tdTomato under control of the Gγ8 promoter at 5 (A), 7 (B) or 10 (C) days after EdU injection. D: Percentage contribution of EdU-labeled OSNs to the total Gγ8-driven fluorescent reporter-expressing population at 5–14 days after EdU injection. Data from 3 mice per time point are shown. Total number of cells counted per time point [Gγ8/EdU]: 5d 3730/1171; 6d 4014/1283; 7d 5451/910; 8d 5040/662; 10d 6508/228; 14d 3973/109. E: Percentage of Gγ8+EdU+ OSNs 5-14 days after EdU injection in coronal OE septum sections taken 25%, 50% or 75% along the anterior-posterior axis. Data from 3 mice per time point are shown.
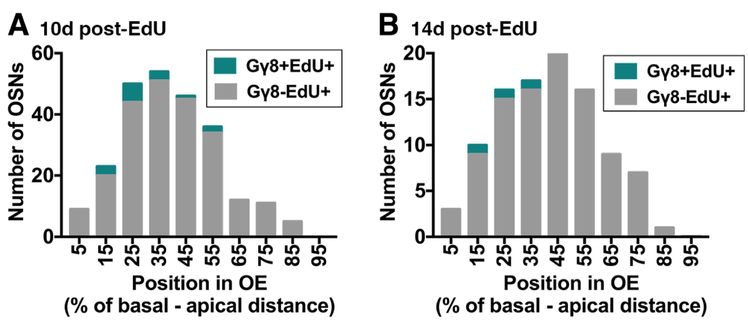
We also performed anti-OMP immunohistochemical staining to determine whether any Gγ8+EdU+ OSNs had already begun to express OMP (Fig. 2). Across all time points after EdU injection, we identified 337 Gγ8+EdU+ OSNs in OMP-stained OE sections, none of which expressed OMP. This suggests that there is a low probability (< 0.3%) that P28-born OSNs expressing a Gγ8-tTA driven reporter will co-express OMP.
Figure 2: Position of EdU labeled cells along the basal-apical axis of the OE 10 – 14 days after EdU injection.
Histograms show number of Gγ8-EdU+ and Gγ8+EdU+ cells (A) 10 days and (B) 14 days post-EdU injection along the basal-apical axis of the septal OE.
We next assessed whether the maturation time course of Gγ8-expressing OSNs varied along the anterior-posterior axis of the OE by comparing the percentage of Gγ8+ OSNs that were labeled with EdU in the septum of coronal OE sections 25%, 50% and 75% along the anterior-posterior axis (Fig. 1E). A 2-way ANOVA showed no effect of OE anterior-posterior position on the proportion of Gγ8+ OSNs labeled with EdU (OE position, P = 0.61, F2,36 = 0.50; days post-EdU, P < 0.001, F5,36 = 24.96; interaction, P = 0.11, F10,36 = 1.75). This indicated that the contribution of newborn OSNs of different ages to the Gγ8+ population is not affected by position along the anterior-posterior axis of the septal OE.
The small number of Gγ8+EdU+ OSNs present at 10-14 days after EdU injection may arise from GBCs that had undergone more than one round of cell division prior to generating these OSNs. The OE has a pseudostratified organization: OSNs migrate apically as they mature (Farbman and Margolis 1980; Roskams et al. 1998; Rodriguez-Gil et al. 2015). Therefore, we reasoned that determining the position along the OE basal-apical axis of the Gγ8+EdU+ OSNs present at 10-14 days after EdU injection would enable us to test the hypothesis that these neurons are younger than the number of days since EdU injection. We first noted that at both 10 and 14 days after EdU injection, there were a small number of EdU-labeled cells in the most basal 10% of the OE (Fig. 2A, B), and concluded that these cells are highly likely to be EdU-labeled GBCs. We then compared the basal-apical distribution of EdU-labeled cells at 10 and 14 days after EdU injection. While EdU labeled cells spanned the OE at both time points, the peak of the distribution shifted to a slightly more apical location at 14 days (44 ± 20%) versus 10 days (40 ± 18%; P = 0.049, t = 1.98, d.f. = 328, unpaired t-test; Fig. 2A, B), consistent with ongoing maturation. Finally, we compared the basal-apical locations of Gγ8+EdU+ cells to those of the total EdU-labeled population, which would be expected also to include mature OSNs occupying more apical locations. Although the number of Gγ8+EdU+ cells was too small to permit statistical analysis, these cells were predominantly confined to more basal regions of the OE than the total population of EdU labeled cells at both 10 and 14 days after injection (Fig. 2A, B). Overall, these data are consistent with the possibility that the small number of Gγ8+EdU+ cells present 10-14 days after EdU injection could have been generated by later GBC cell division and hence are younger than the time elapsed since EdU injection.
Birth-dating of OSNs expressing OMP-tTA-driven fluorescent reporter proteins
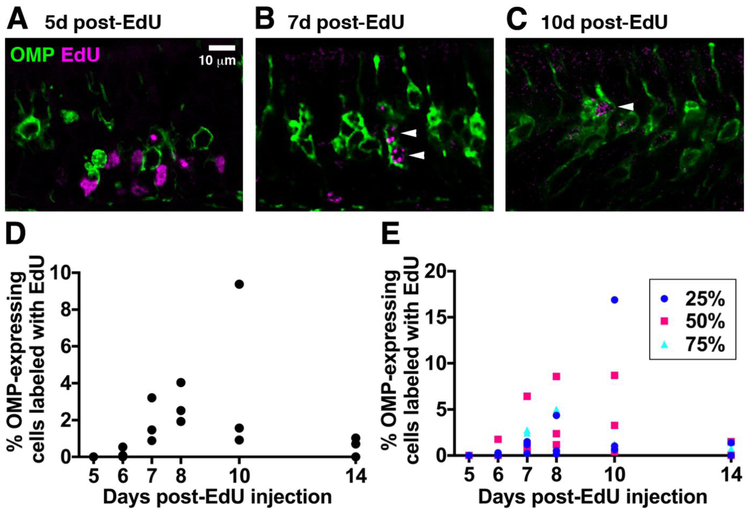
We next determined the contribution of newly-generated OSNs of different ages to the OMP+ OSN population (Fig. 3A-D). We analyzed a total of 35235 OMP+ OSNs in 18 mice (3 mice per time point after EdU injection). We did not detect any 5-day-old OMP+EdU+ OSNs (0/7735 OMP+ OSNs in 3 mice). 6-day-old OSNs accounted for a small subset of OMP+ OSNs in two out of three mice (0%, 0.1% and 0.5% in individual mice). 0.9 – 3.2 % of OMP+ OSNs were 7 days old, 1.9 – 4.0 % were 8 days old and 0.9 – 9.4 % were 10 days old. Surprisingly, 14-day-old OSNs comprised only 0.0 – 1.0% of the OMP+ OSN population. The contribution of newly generated OSNs to the OMP+ population was unaffected by location along the anterior-posterior axis of the septal OE (Fig. 3E; OE position, P = 0.75, F2,36 = 0.29; neuronal age, P = 0.023, F5,36 = 2.99; interaction, P = 0.74, F10,36 = 0.68). Taken together, these data suggest that the minimum age at which an OSN can express an OMP-tTA-driven reporter is 6 days, and that a large proportion of newly mature, OMP-expressing OSNs born at P28 undergo cell death 8-14 days after terminal cell division.
Figure 3: OMP-tTA-driven fluorescent reporters are not expressed in 5-day old OSNs.
A-C: Single confocal optical sections of the septal OE of mice expressing membrane-bound Citrine under control of the OMP promoter at 5 (A), 7 (B) or 10 (C) days after EdU injection. D: Percentage of OSNs that express an OMP-tTA-driven fluorescent reporter that are colabeled with EdU 5-14 days after EdU injection. Data from 3 mice per time point are shown. Total number of cells counted per time point [OMP/EdU]: 5d 7735/1710; 6d 6536/656; 7d 7233/1610; 8d 5521/736; 10d 5366/725; 14d 2844/160. E: Percentage of OMP+EdU+ OSNs 5-14 days after EdU injection in coronal OE septal sections taken 25%, 50% or 75% along the anterior-posterior axis. Data from 3 mice per time point are shown.
Survival of P28-born EdU-labeled cells in the OE
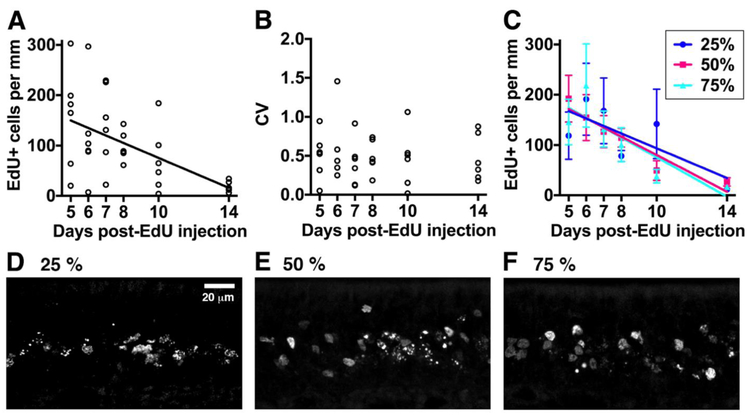
The surprising observation that 14-day-old P28-born OSNs make a smaller contribution to the OMP+ OSN population than their 8-day-old counterparts indicates that many P28-born OSNs have a lifespan of fewer than two weeks. To directly assess the survival of the total population of P28-born OSNs, we determined the linear density of EdU+ OSNs across our entire cohort (6 mice per group, irrespective of whether they expressed a Gγ8-driven or an OMP-driven reporter protein). Overall, there was a strong linear relationship between EdU+ cell linear density and time after EdU injection (Fig. 4A; P = 0.002, R2 = 0.93). EdU+ cell linear density decreased from 182 (115 – 251) per mm at 5 days to 16 (14 – 31) per mm at 14 days [data described as median (interquartile range)]. The slope of the linear regression fit indicated that on average, 15 ± 2 EdU+ OSNs per mm of OE were eliminated per day, while the x-intercept suggested that all EdU+ OSNs would be eliminated 15.1 days after EdU injection.
Figure 4: Most P28-born OSNs survive for less than 14 days throughout the septal OE.
A: EdU+ cell linear density in the septal OE 5-14 days after EdU injection. B: Coefficient of variation (mean/standard deviation) of EdU cell linear density 5-14 days after EdU injection. C: EdU+ cell linear density in the septal OE 5-14 days after EdU injection in coronal sections 25%, 50% and 75% along the anterior-posterior axis of the OE. D-F: Single confocal optical sections of EdU+ cells in the septal OE 7 days after EdU injection in coronal sections 25% (D), 50% (E) or 75% (F) along the anterior-posterior axis of the OE.
However, there was considerable inter-mouse variability in the linear density of EdU+ cells at each individual time point. To determine whether this variability was similar across cellular ages, we calculated the coefficient of variation (CV = standard deviation divided by mean). We found no difference in the CV between individual time points after EdU injection (Fig. 4B; one-way ANOVA on Ranks, P = 0.87, Kruskal-Wallis statistic = 1.87, n = 36 mice in 6 groups). This indicates that the relative variability across experimental time points was similar and hence could not account for the decline in EdU+ cell linear density that we observed with increasing time after EdU injection. A 2-way ANOVA also showed no effect of anterior-posterior location along the septal OE on the linear density of EdU+ cells (Fig. 4C-F; OE location, P = 0.92, F2,88 = 0.1; days post-EdU, P < 0.001, F5,88 = 5.8; interaction, P = 0.76, F10,88 = 0.76). Furthermore, linear regression fits for each anterior-posterior location had similar slopes (Fig. 4C; −15 ± 7 cells per day at 25%, −18 ±3 OSNs per day at 50% and - 20 ± 6 OSNs per day at 75% along the anterior-posterior axis) and x-intercepts (16.3 days at 25%, 14.4 days at 50% and 13.9 days at 75% along the anterior-posterior axis). Overall, we concluded that 5 – 14-day-old P28-born OSNs are eliminated steadily throughout the OE, with very few surviving beyond two weeks after terminal cell division.
Apoptotic cell death in the OE
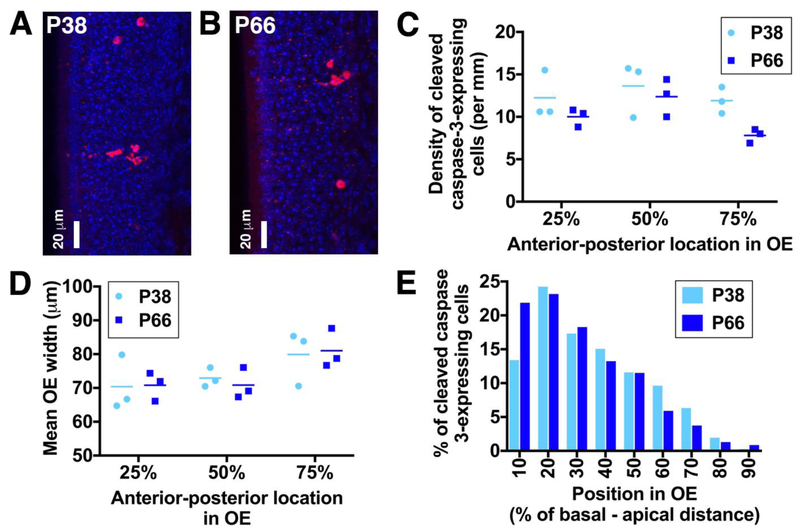
As a first step towards investigating whether the low survival rate of newborn OSNs is specific to those generated at P28, we performed immunohistochemical staining for cleaved caspase-3, an apoptotic marker, in OE sections from P38 and P66 C57BL/6J mice (Fig. 5A,B). These ages were selected to encompass the elimination of OSNs born at ~P28 and ~P56 (Fig. 4A). Cleaved caspase-3 is the active form of this “executioner” caspase, the presence of which leads to protein degradation and DNA fragmentation during programmed cell death.
Figure 5: Apoptotic cells occupy slightly more apical locations in the septal OE of P38 vs. P66 mice.
A,B: Maximum intensity projections of confocal z-stack showing apoptotic cells in the septal OE of P38 (A) and P66 (B) mice. Cleaved caspase-3-expressing cells are shown in red; DAPI-stained nuclei are in blue. Images with a higher than average density of apoptotic cells were selected to illustrate the basal-apical distribution at each age. C: Linear density of cleaved caspase-3-expressing cells along the anterior-posterior axis in P38 and P66 mice. D: Septal OE width in sections along the anterior-posterior axis in P38 and P66 mice. E: Histogram showing percentage of cleaved caspase-3-expressing cells along the basal-apical axis of the septal OE in P38 and P66 mice. Data are from a total of 664 cells in P38 mice and 695 cells in P66 mice.
We first determined the linear density of cleaved caspase-3-expressing cells in the septal OE of P38 and P66 mice (Fig. 5C). We analyzed a total of 664 cells in P38 mice and 695 cells in P66 mice. Very occasional cells with clear sustentacular cell morphology (3 cells in P66 mice) were excluded from our analysis. Although the sample size is small (3 mice per group), the linear density of cleaved caspase-3-expressing cells was significantly higher in the P38 septal OE than in the P66 septal OE (Fig. 5C; two-way ANOVA; P = 0.028, F1,12 = 6.27 for effect of age; P = 0.073, F2,12 = 3.28 for OE position along the anterior-posterior axis; P = 0.53, F2,12 = 0.68 for interaction). This lower rate of apoptotic cell death in P66 mice was not due to ongoing expansion of the OE, as OE width, defined as the distance from the basal lamina to the apical surface, was similar in P38 and P66 mice (Fig. 5D; two-way ANOVA, P = 0.95, F1,12 = 0.003). Furthermore, OE width was greater in the posterior than the anterior OE at both ages (two-way ANOVA, P = 0.029 F2,12 = 4.82 for OE location, P = 0.89 F2,12 = 0.12 for interaction; post-hoc Tukey’s multiple comparisons test, P = 0.036, q = 4.03, d.f. = 12 for 25% vs. 75% along anterior-posterior axis).
We then compared the basal-apical distribution of cleaved caspase-3-expressing cells in the septal OE of P38 and P66 mice. We found that apoptotic cells were present throughout the stem/progenitor cell and neuronal layers of the OE in both age groups (Fig. 5E). However, the distribution of apoptotic cell positions was skewed towards more basal locations in P66 mice (27 [17 – 44] %) relative to P38 mice (32 [19 – 49]; data described as median [interquartile range]; P = 0.006, D = 0.093, Kolmogorov-Smirnov test). This suggests that newly generated OSNs die slightly earlier during their development in P66 mice than in P38 mice.
Discussion
This study has three major conclusions. First, OSNs never expressed OMP-tTA-driven reporter proteins until they were at least six days old. Second, the vast majority of OSNs shut down expression of Gγ8-tTA-driven reporters once they were 8 days old. Finally, we found a very low survival rate for P28-born OSNs in the undamaged OE.
The onset of OMP-tTA-driven reporter protein expression
The initial goal of this study was to define the neuronal age at which newborn OSNs shut down expression of Gγ8-tTA-driven reporters and begin to express OMP-tTA-driven reporters. Our data clearly demonstrate that newborn OSNs do not begin to express OMP-tTA-driven transgenes until at least 6 days after EdU injection in P28 mice on a 129 × C57BL/6J mixed background. This finding is in very good agreement with the general consensus that arises from previous studies of both the healthy OE across the mouse lifespan, and the mouse or rat OE after either direct lesion or olfactory bulbectomy. First, OMP protein expression was shown to begin 7 days after terminal cell division marked by tritiated thymidine incorporation in 3-month-old CD-1 mice (Miragall and Graziadei 1982). Second, using BrdU injections in ICR mice ranging from 10 days to 16 months of age, Kondo et al. (2010) showed that OMP protein was present in 7-day-old but not in 5-day-old OSNs irrespective of mouse age. Third, a recent study that assessed OMP mRNA expression at daily time points after BrdU injection in P7 CD-1 mice concluded that the earliest time point at which an OSN expresses OMP mRNA is 6 days after cell division (Rodriguez-Gil et al. 2015). Fourth, following unilateral olfactory bulbectomy in 2-3-month old Sprague Dawley rats, newborn OSNs began to express OMP between 5 days and 2 weeks of age (Schwob et al. 1992). Finally, newborn OSNs first expressed OMP 7 days after methimazole-mediated ablation (Kikuta et al. 2015). These studies, together with our data, strongly suggest that OSNs follow a very similar maturation time course irrespective of animal age or whether neurogenesis is constitutive or lesion-induced. Furthermore, the concordance of the timing of OMP-tTA-driven transgene expression onset in our study with that of OMP mRNA and protein expression in the undamaged OE across the mouse lifespan in several other studies (Miragall and Graziadei 1982; Kondo et al. 2010; Rodriguez-Gil et al. 2015) indicates that the OMP-tTA driver line provides a faithful readout of the onset of OMP expression.
In marked contrast, a recent study using 10-week-old C57BL/6J x 129S1/Sv1MJ mice found that OMP-stained, EdU+ cells were present as early as 2 days after EdU injection and accounted for 11% of 5-day-old EdU labeled cells in 10-week-old mice (Coleman et al. 2017). It is unclear why this study found OMP expression so much earlier than in other reports, although it is possible that differences in housing type or conditions (Hinds et al. 1984; Holl 2018), analysis of the septal OE vs. the turbinates (Vedin et al. 2009) and/or analysis of confocal vs. widefield images of OE sections could contribute.
We also noted that the contribution of EdU+OMP+ OSNs to the total OMP reporter-expressing population increased across 6 to 8 days after EdU injection. There are two non-mutually-exclusive explanations for this finding. First, the onset of OMP-tTA-driven transgene expression could be asynchronous across the population of newborn OSNs, with expression onset varying by as much as 48 hours. Second, expression onset could be synchronous, but additional cell divisions of EdU-labeled precursors could generate additional cohorts of OSNs that are 1-2 days younger than the time elapsed since EdU injection. Kondo et al. (2010) found that the number of BrdU-labeled OE cells peaked 2-3 days after BrdU injection, supporting the latter possibility. However, in 1-month-old mice, there was a ~50% increase in the number of BrdU-labeled cells between 2 hours and 3 days after injection, which would be insufficient to explain the magnitude of the increase in the percentage of OMP reporter-expressing cells between 6 and 8 days post-EdU injection in our study (Fig. 3). Therefore, it likely that while slightly younger OSNs contribute to the increased number of OMP+EdU+ cells present at 8 vs. 6 days post-EdU injection, the onset of OMP-tTA-driven transgene expression is also somewhat asynchronous across the P28-born OSN population. This effect is also present in previous studies that have assessed OMP mRNA or protein expression onset across a range of time points (Kondo et al. 2010; Rodriguez-Gil et al. 2015; Coleman et al. 2017), again indicating that use of the OMP-tTA line to drive tetO-dependent promoter expression faithfully recapitulates the temporal expression dynamics of OMP itself.
Shut down of Gγ8-tTA-driven reporter expression
The contribution of EdU-labeled cells to the population of OSNs expressing a Gγ8-tTA-driven fluorescent protein decreased as the time post-EdU injection increased from 5 to 10 days. This is as would be expected for an immature marker, expression of which is transient and shuts down as OSNs mature. To our knowledge, no previous study has determined the temporal profile of Gγ8 expression as OSNs mature; however, two studies have investigated the expression of GAP43, another widely-used immature OSN marker, in newborn OSNs. Rodriguez-Gil et al. (2015), using in situ hybridization in BrdU-labeled OE from P7 CD-1 mice, found that GAP43 mRNA expression began in 1-day-old OSNs, peaked at 5 days and then remained relatively stable until 8 days, before abruptly shutting down by 9 days. Coleman et al. (2017), using EdU labeling in 10-week-old C57BL/6J x 129S1/Sv1MJ mice, found that GAP43 protein expression began at 2 days, peaked at 7 days, and then decreased significantly but still accounted for 40% of EdU-labeled cells at 9 days, the latest time point examined. Hence, our results are in broad agreement with these previous studies, especially given that expression of GAP43 and Gγ8 may not follow exactly the same temporal pattern during OSN maturation. Indeed, we have shown previously that Gγ8-tTA-driven reporter-expressing OSNs are slightly more apically located than GAP43-immunostained OSNs in the OE of P21 C57BL/6J x 129 mice, indicating that Gγ8 promoter-driven expression may persist for slightly longer than that of GAP43 as OSNs mature (Cheetham et al. 2016).
The question of precisely when Gγ8-tTA-driven expression shuts down remains incompletely resolved, as we did detect a very small subset of Gγ8+EdU+ cells at both 10 and 14 days after EdU injection. It is possible that a very small subset of OSNs continue to express Gγ8-tTA-driven reporters beyond 8 days of cellular age. Greater stability of fluorescent proteins vs. Gγ8 or GAP43 protein could also extend the time window for Gγ8 and EdU co-labeling by up to 24 hours (Li et al. 1998). Alternatively, these double-labeled cells could have been generated by subsequent cell divisions of EdU-labeled progenitors, and hence be several days younger than the time post-EdU injection. Kondo et al. (2015) provided good evidence that additional labeled cells continue to be generated in the OE for several days after BrdU injection. Furthermore, our analysis of the position of Gγ8+EdU+ cells along the basal-apical axis of the OE, which provides an estimate of OSN maturity (Rodriguez-Gil et al. 2015), indicated that these double-labeled cells were more basally located than many of the Gγ8-EdU+ cells present at 10 or 14 days post-injection. We interpret this to mean that a significant fraction of the Gγ8+EdU+ cells present at the 10- and 14-day post-EdU time points arose from later cell division of EdU-labeled GBCs (Miragall and Graziadei 1982; Schwob et al. 1992), and hence that these cells are younger than the time elapsed since EdU injection. However, we cannot completely exclude the possibility that occasional OSNs older than 8 days continue to express Gγ8-tTA-driven reporters.
We found no OMP staining of Gγ8+EdU+ OSNs at any time point after EdU injection. This suggests that co-expression of Gγ8 and OMP is very rare in P28-born OSNs, which is somewhat surprising given that we previously reported that 6% of OSNs expressing a Gγ8-tTA-driven fluorescent reporter stained for OMP in the OE of 8-week-old mice (Cheetham et al. 2016). The most plausible explanation for this discrepancy is that the very low survival rate of P28-born OSNs means that there are simply very few OSNs in transition to maturity, making them difficult to detect.
Low survival rate of P28-born OSNs
Our data suggest that newborn OSNs in P28 mice are gradually eliminated from the OE between 5 and 14 days of neuronal age. Indeed, the total linear density of 14-day-old EdU+ cells was 88% lower than that of 5-day-old EdU+ cells. This indicates that attaining maturity, as assessed by OMP expression, does not promote survival; that very few P28-born OSNs make a lasting contribution to the OE; and furthermore, that OSNs continue to be generated even under circumstances that are unfavorable to long-term survival.
Is this low survival rate, and the elimination of OSNs that have already begun to express OMP, specific to P28-born OSNs? Similar to a previous report (Kondo et al. 2010), we found that the density of apoptotic cells in the P38 OE was higher than that in the P66 OE, indicating that ~P28-born OSNs are eliminated at a slightly higher rate than those born a month later. Surprisingly, we also found that the distribution of apoptotic cells was skewed slightly apically in P38 vs. P66 mice, indicating that ~P28-born OSNs are dying at slightly later maturational stages than those born a month later. Future studies should further investigate this intriguing result; however, the greater overlap of the P38 vs. P66 apoptotic cell distribution with those of 5-14-day-old OSNs along the basal-apical axis of the OE (Fig 2)(Rodriguez-Gil et al. 2015) may suggest that elimination of newly-matured P28-born OSNs is unusually prevalent.
Previous reports also suggest that OSN survival rates are higher for OSNs generated in P7 CD-1 (Rodriguez-Gil et al. 2015) and in 2-3-month-old CD-1 and outbred Swiss albino (Hinds et al. 1984; Mackay-Sim and Kittel 1991) mice, although comparisons between studies are complicated by differences in mouse strain and – more particularly – housing conditions (Hinds et al. 1984; Holl 2018). Indeed, in a study directly comparing the survival of birth-dated OSNs in mice aged 10 days to 16 months, Kondo et al. (2010) found similar reductions in the number of 14-day-old vs. 5-day-old OSNs across mouse age groups. However, both Kondo et al. (2010) and another study (Brann et al. 2015) reported far greater elimination of OMP-expressing OSNs between 14 and 30 days of neuronal age in mice injected with BrdU at 1-month-old vs. 2-3-months-old. These studies therefore indicate that P28-born OMP-expressing OSNs are less likely to survive long-term than those generated in older mice. Furthermore, the majority of late embryonic- and perinatally-born OMP-expressing OSNs survive for at least three weeks (Holl 2018).
Taking these studies and our results together, the healthy P28 OE may represent an unusual time point, at which postnatal development of the glomerular map is largely complete (Mombaerts et al. 1996; Potter et al. 2001; Zou et al. 2004) and the rate of subventricular zone neurogenesis has declined substantially (Batista-Brito et al. 2008), yet few established mature OSNs have started to undergo apoptosis. In contrast, glomerular maps in younger mice are still being formed and refined, requiring continuous incorporation of newly generated OSNs, while in older mice, the need to replace damaged or dying OSNs promotes the survival of newly-generated OSNs. Indeed, the low survival rate that we observed in the healthy OE is reminiscent of the poor survival of newborn OSNs in the ipsilateral OE of rats that have undergone unilateral olfactory bulbectomy: under these conditions, neurogenesis is upregulated, yet very few OMP-expressing OSNs survive long term (Graziadei 1983; Verhaagen et al. 1990; Schwob et al. 1992). Indeed, Schwob et al. (1992) found that following bulbectomy, 90% of newly-generated OSNs in the ipsilateral OE died between 5 and 14 days of age in adult rats. In contrast, only 40% of a BrdU-labeled cohort of OSNs was lost between 5 and 14 days of age in 2-3-month-old control rats (Schwob 1992). The authors concluded that OSNs are trophically dependent on the OB for prolonged survival (Schwob et al. 1992). In our experiments, the OB is undamaged; however, the reason for lack of OSN survival may be analogous: namely, that P28-born OSNs are very infrequently able to form synaptic connections with OB neurons, which has been suggested to be essential for their survival (Graziadei 1983). Hence, while OSN maturation is independent of animal age, OSN survival may not be. This finding is particularly useful in the context of future studies that will explore how postnatal-born OSNs integrate into the highly ordered glomerular circuits of the OB without disrupting existing function.
Acknowledgements:
This work was supported by grants to CEJC from the National Institute on Deafness and other Communication Disorders (R03DC014788) and the Samuel and Emma Winters Foundation. We thank Gerry Hammond (University of Pittsburgh) for use of the Nikon A1R confocal microscope.
References
- Batista-Brito R, Close J, Machold R, Fishell G (2008) The distinct temporal origins of olfactory bulb interneuron subtypes. The Journal of neuroscience 28:3966–75. doi: 10.1523/JNEUROSCI.5625-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann JH, Ellis DP, Ku BS, et al. (2015) Injury in aged animals robustly activates quiescent olfactory neural stem cells. Frontiers in Neuroscience 9:367. doi: 10.3389/fnins.2015.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano M, Kauer J, Hunter D (1994) Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron 13:339–52. doi: 10.1016/0896-6273(94)90351-4 [DOI] [PubMed] [Google Scholar]
- Cheetham C, Park U, Belluscio L (2016) Rapid and continuous activity-dependent plasticity of olfactory sensory input. Nature communications 7:10729. doi: 10.1038/ncomms10729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JH, Lin B, Schwob JE (2017) Dissecting LSD1-Dependent Neuronal Maturation in the Olfactory Epithelium. Journal of Comparative Neurology 525:3391–3413. doi: 10.1002/cne.24259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbman A (1992) Cell biology of olfaction Cambridge University Press 27 [Google Scholar]
- Farbman A, Margolis F (1980) Olfactory marker protein during ontogeny: immunohistochemical localization. Developmental biology 74:205–15 [DOI] [PubMed] [Google Scholar]
- Godfrey P, Malnic B, Buck L (2004) The mouse olfactory receptor gene family. Proceedings of the National Academy of Sciences of the United States of America 101:2156–61. doi: 10.1073/pnas.0308051100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziadei MG (1983) Experimental studies on the olfactory marker protein. III. The olfactory marker protein in the olfactory neuroepithelium lacking connections with the forebrain. Brain research 262:303–8 [DOI] [PubMed] [Google Scholar]
- Graziadei P, Monti Graziadei G (1978) Continuous nerve cell renewal in the olfactory system In: Development of Sensory Systems. Springer-Verlag [Google Scholar]
- Hahn C, Han L, Rawson N, Mirza N, Borgmann-Winter K, Lenox R, Arnold S (2005) In vivo and in vitro neurogenesis in human olfactory epithelium. Journal of Comparative Neurology 483 154–163 doi: 10.1002/cne.20424 [DOI] [PubMed] [Google Scholar]
- Hinds J, Hinds P, McNelly N (1984) An autoradiographic study of the mouse olfactory epithelium: evidence for long-lived receptors. The Anatomical record 210:375–83. doi: 10.1002/ar.1092100213 [DOI] [PubMed] [Google Scholar]
- Holl A (2018) Survival of mature mouse olfactory sensory neurons labeled genetically perinatally. Molecular and cellular neurosciences 88:258–269. doi: 10.1016/j.mcn.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Kikuta S, Sakamoto T, Nagayama S, et al. (2015) Sensory deprivation disrupts homeostatic regeneration of newly generated olfactory sensory neurons after injury in adult mice. The Journal of neuroscience 35:2657–73. doi: 10.1523/JNEUROSCI.2484-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Suzukawa K, Sakamoto T, et al. (2010) Age-related changes in cell dynamics of the postnatal mouse olfactory neuroepithelium: Cell proliferation, neuronal differentiation, and cell death. Journal of Comparative Neurology 518:1962–1975. doi: 10.1002/cne.22316 [DOI] [PubMed] [Google Scholar]
- Leung C, Coulombe P, Reed R (2007) Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nature neuroscience 10:720–6. doi: 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, et al. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. The Journal of biological chemistry 273:34970–5 [DOI] [PubMed] [Google Scholar]
- Mackay-Sim A, Kittel P (1991) On the Life Span of Olfactory Receptor Neurons. The European journal of neuroscience 3:209–215 [DOI] [PubMed] [Google Scholar]
- Malnic B, Godfrey P, Buck L (2004) The human olfactory receptor gene family. Proceedings of the National Academy of Sciences of the United States of America 101:2584–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Titlow WB, McClintock TS (2010) Axon growth and guidance genes identify nascent, immature, and mature olfactory sensory neurons. Journal of neuroscience research 88:3243–56. doi: 10.1002/jnr.22497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miragall F, Graziadei G (1982) Experimental studies on the olfactory marker protein. II. Appearance of the olfactory marker protein during differentiation of the olfactory sensory neurons of mouse: an immunohistochemical and autoradiographic study. Brain Research 239:245–250. doi: 10.1016/0006-8993(82)90846-0 [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, et al. (1996) Visualizing an olfactory sensory map. Cell 87:675–86 [DOI] [PubMed] [Google Scholar]
- Nguyen M, Zhou Z, Marks C, et al. (2007) Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell 131:1009–17. doi: 10.1016/j.cell.2007.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S, Zheng C, Koos D, et al. (2001) Structure and emergence of specific olfactory glomeruli in the mouse. The Journal of neuroscience 21:9713–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler K, Sullivan S, Buck L (1994) Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell 79:1245–55. doi: 10.1016/0092-8674(94)90015-9 [DOI] [PubMed] [Google Scholar]
- Ressler K, Sullivan S, Buck L (1993) A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73:597–609 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gil DJ, Bartel DL, Jaspers AW, et al. (2015) Odorant receptors regulate the final glomerular coalescence of olfactory sensory neuron axons. Proceedings of the National Academy of Sciences of the United States of America 112:5821–6. doi: 10.1073/pnas.1417955112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskams A, Cai X, Ronnett G (1998) Expression of neuron-specific beta-III tubulin during olfactory neurogenesis in the embryonic and adult rat. Neuroscience 83:191–200 [DOI] [PubMed] [Google Scholar]
- Ryba N, Tirindelli R (1995) A novel GTP-binding protein gamma-subunit, G gamma 8, is expressed during neurogenesis in the olfactory and vomeronasal neuroepithelia. The Journal of biological chemistry 270:6757–67 [DOI] [PubMed] [Google Scholar]
- Saraiva L, Ibarra-Soria X, Khan M, et al. (2016) Hierarchical deconstruction of mouse olfactory sensory neurons: from whole mucosa to single-cell RNA-seq. Scientific Reports 5:srep18178. doi: 10.1038/srep18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nature methods 9:676–82. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J (1992) The biochemistry of olfactory neurons: stages of differentiation and neuronal subsets In: Science of Olfaction. Springer [Google Scholar]
- Schwob J, Jang W, Holbrook E, et al. (2017) Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. The Journal of comparative neurology 525:1034–1054. doi: 10.1002/cne.24105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J, Szumowski K, Stasky A (1992) Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. The Journal of neuroscience 12:3896–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Wanner I, Krieger J, et al. (1992) Expression of odorant receptors in spatially restricted subsets of chemosensory neurones. Neuroreport 3:1053–6 [DOI] [PubMed] [Google Scholar]
- Tirindelli R, Ryba N (1996) The G-protein gamma-subunit G gamma 8 is expressed in the developing axons of olfactory and vomeronasal neurons. The European journal of neuroscience 8:2388–98 [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, et al. (1994) Topographic organization of sensory projections to the olfactory bulb. Cell 79:981–991 [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, Axel R (1993) Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74:309–18 [DOI] [PubMed] [Google Scholar]
- Vedin V, Molander M, Bohm S, Berghard A (2009) Regional differences in olfactory epithelial homeostasis in the adult mouse. Journal of Comparative Neurology 513:375–384. doi: 10.1002/cne.21973 [DOI] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher A, Gispen W, Margolis F (1989) The expression of the growth associated protein B50/GAP43 in the olfactory system of neonatal and adult rats. The Journal of neuroscience 9:683–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaagen J, Oestreicher A, Grillo M, et al. (1990) Neuroplasticity in the olfactory system: differential effects of central and peripheral lesions of the primary olfactory pathway on the expression of B-50/GAP43 and the olfactory marker protein. Journal of neuroscience research 26:31–44. doi: 10.1002/jnr.490260105 [DOI] [PubMed] [Google Scholar]
- Yu C, Power J, Barnea G, et al. (2004) Spontaneous neural activity is required for the establishment and maintenance of the olfactory sensory map [DOI] [PubMed] [Google Scholar]
- Zou D-JJ, Feinstein P, Rivers AL, et al. (2004) Postnatal refinement of peripheral olfactory projections. Science 304:1976–9. doi: 10.1126/science.1093468 [DOI] [PubMed] [Google Scholar]