Abstract
In today’s radiology workflow, free-text reporting is established as the most common medium to capture, store, and communicate clinical information. Radiologists routinely refer to prior radiology reports of a patient to recall critical information for new diagnosis, which is quite tedious, time consuming, and prone to human error. Automatic structuring of report content is desired to facilitate such inquiry of information. In this work, we propose an unsupervised machine learning approach to automatically structure radiology reports by detecting and normalizing anatomical phrases based on the Systematized Nomenclature of Medicine—Clinical Terms (SNOMED CT) ontology. The proposed approach combines word embedding-based semantic learning with ontology-based concept mapping to derive the desired concept normalization. The word embedding model was trained using a large corpus of unlabeled radiology reports. Fifty-six anatomical labels were extracted from SNOMED CT as class labels of the whole human anatomy. The proposed framework was compared against a number of state-of-the-art supervised and unsupervised approaches. Radiology reports from three different clinical sites were manually labeled for testing. The proposed approach outperformed other techniques yielding an average precision of 82.6%. The proposed framework boosts the coverage and performance of conventional approaches for concept normalization, by applying word embedding techniques in semantic learning, while avoiding the challenge of having access to a large amount of annotated data, which is typically required for training classifiers.
Keywords: Radiology reports, Concept normalization, Anatomical classification, word2vec, Semantic learning, SNOMED CT
Introduction
In today’s clinical care practice, radiology reports are established as one of the most common formats of capturing and communicating patient health information across the care team. Radiologists transcribe imaging observations, and the corresponding diagnosis followed by appropriate follow-up actions. As a result, abundant critical information is captured in a radiology report. During the last couple of decades, there has been an increased interest in using machine-powered intelligence to automate the information extraction and structuring of the report content to improve the quality and consistency of clinical workflow.
A common approach for structuring radiology report content is to sort information based on anatomy of reference. Extracting the anatomical reference helps with classifying radiological observations, correlating clinical events longitudinally, and identifying relevant prior studies. Radiology reports contain descriptions of radiological observations (e.g., nodule in lung, lesion in liver). Often, the anatomy of reference for each observation is explicitly mentioned in the same sentence using a standard terminology such as Systematized Nomenclature of Medicine—Clinical Terms (SNOMED CT) [1]. Sometimes, the radiologist uses elongated phrases to describe the anatomical site of an observation (e.g., inferior aspect of the glenohumeral joint space, adjacent to the scapular body). On other occasions, the anatomical phrase describes part of an anatomical region without explicitly mentioning the site itself, such as wall, left lobe, or right segment. Furthermore, radiologists often use custom abbreviations that do not exist in standard ontologies to refer to anatomical regions (e.g., pulm. referring to pulmonary; semi. vesc. referring to seminal vesicle). As a result, inferring the anatomical site of every observation is a challenging task for machine automated processes.
Concept recognition in radiology reports has previously been tackled using a variety of natural language processing (NLP) and machine learning techniques. Previous efforts can be categorized under three main approaches: (1) dictionary and ontology look-up; (2) handcrafted rules, grammars, and pattern matching; and (3) statistical machine learning. Dictionary-based approaches rely on existing domain knowledge resources such as ontologies to build dictionaries for string-based look-ups. In grammar matching approaches, rules are extracted from morphology and semantics of phrases. To provide standard concept mapping, grammar matching approaches are combined with ontologies. Finally, machine learning approaches are data-driven statistical prediction algorithms that rely on feature-based representation of the observed data.
Among dictionary-based approaches, one can refer to Radiology Analysis tool (RADA) [2], Clinical Text Analysis and Knowledge Extraction System (cTAKES) based on Unified Medical Language System (UMLS) Metathesaurus terms [3, 4], and Health Information Text Extraction (HITEx) [5].
One of the earliest grammar-based approaches is the Medical Language Extraction and Encoding System (MedLEE) [6, 7] used by Friedman et al. to extract UMLS concepts from medical text documents [8–10]. Aronson used an NLP parser, known as MetaMap, for mapping medical concepts to UMLS concepts [11]. A major drawback of dictionary-based and grammar-based approaches is the dependency of the performance on limited hand-crafted dictionaries and rules based on observed data in hand. As a result, such methods often yield low recall compared to their precision.
A pioneer work on using machine learning for concept recognition was proposed by Taira et al. [12] for extracting and labeling of medical information in radiology reports. Pyysalo and Anaiadou proposed a machine learning-based anatomy tagger based on integration of a variety of techniques such as UMLS- and Open Biomedical Ontologies (OBO)-based lexical resources, statistical and nonlocal features [13]. Campos et al. presented a survey of machine learning tools for the biomedical Named Entity Recognition (NER) [14]. The success of machine learning techniques in NER applications heavily relies on the availability of a large-scale high-quality training data [12, 14], which is difficult to obtain. Another major drawback of most machine learning approaches is that they require feature engineering, meaning selecting features that yield the best performance, which may result in overfitting. As a result, the adoption of such data-driven approaches to a new domain or a different clinical site is a costly process with no guarantee of success.
Recently, there has been significant interest in using word embedding techniques for NLP applications. Word embedding refers to a set of unsupervised language modeling techniques in which every word/phrase from the vocabulary is transformed into a lower dimensional real-number vector space. Word embedding techniques are well known for their ability to capture similarity between words and infer the relationship purely from unlabeled data. Tang et al. investigated the use of three different types of word representation for biomedical NER [15]. Wu et al. explored the word embedding for clinical NER using a relatively large unlabeled corpus [16]. Limsopatham and Collier proposed two neural network-based approaches, convolutional and recurrent neural network (RNN) architectures, for medical concept normalization of layman’s language used in social media messages to formal medical language [17]. Authors demonstrated significant improvement in performance compared to state-of-the-art approaches.
Word embedding techniques have become popular because they do not require expensive manual labeling of the training data. Nevertheless, a key limitation of the original form of word embedding is the inability to provide recognition due to its unsupervised learning nature [18]. To overcome this challenge, Ferré et al. proposed to create two separate vector spaces: one for the domain-specific data such as clinical notes and the other for the ontology concepts [19]. In order to compare similarities between vectors from two different spaces, authors proposed to train a prediction model to transform vectors from domain corpus to the ontology vector space, and finally, cosine similarity between phrase vector and ontology concepts vectors is used to determine the most appropriate label. A major shortcoming of such approach is that it still requires labeled data to be able to train the prediction model to transform vectors from domain space to ontology space.
In this work, we propose a solution for anatomical phrase labeling and normalization in free-text radiology reports using unsupervised semantic learning [18]. The proposed solution is designed and tested to overcome major shortcomings of state-of-the-art approaches as follows: (1) consistent performance across multiple corpora from different clinical sites without requiring a large corpus of prelabeled training data from each clinical site, (2) labeling capability beyond exact match with ontologies and interpretations based on contextual learning, and (3) ability to identify and label elongated phrases corresponding to the whole human anatomy using 56 labels from SNOMED CT.
The proposed framework uses word embedding to create a low-dimensional feature space in which contextually related terms are transformed to a common neighborhood in the feature space. Furthermore, the proposed framework provides normalization of phrases to a standard ontology space, known as SNOMED CT [1]. Normalization is achieved based on measuring the similarity between the word embedding representation of the unlabeled phrases and anatomical class representatives defined in SNOMED CT ontology.
Materials and Methods
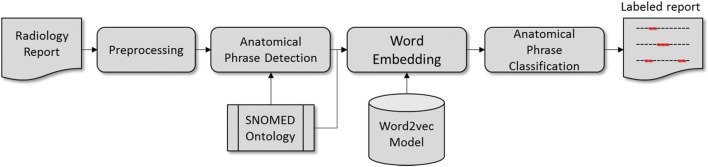
Figure 1 depicts an overview of the proposed framework.
Fig. 1.
An overview of the proposed framework for anatomical phrase labeling in radiology reports
In summary, anatomical phrases are automatically identified using a previously proposed anatomical phrase detection tool [20]. Next, a low-dimensional vector representation of the detected phrase is derived using a trained word embedding model. Concept normalization is achieved via a naïve classifier that determines the label by calculating the similarity between SNOMED CT class representative vectors and the unlabeled phrase vector and selecting the class label yielding maximum similarity. The following sections describe the individual steps in detail.
Preprocessing
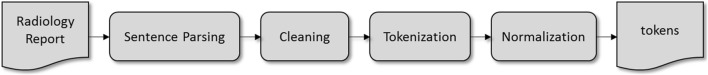
The preprocessing pipeline is illustrated in Fig. 2. Sentence boundary extraction (sentence segmentation) is performed using the Natural Language Toolkit, known as NLTK [21]. Text cleaning includes the removal of special characters and unnecessary white space and new lines and, finally, lower-casing. Sentence-to-word tokenization is achieved using an in-house built tool in Python. The tokenizer is configured to partition each text string into contiguous segments of various types, such as special notations (time, date, number, number-unit values), lexical forms (alphabetic runs, hyphenated terms), punctuation (other ink characters), and whitespace (vertical, horizontal). The final step of preprocessing is normalization. Normalization is applied to specific types of segments, such as case (lowercase alphabetics), dates and times (make all identical), and numerical values (replace each labeled token with the character 9).
Fig. 2.
An overview of the preprocessing pipeline
Anatomical Phrase Detection
The goal of anatomical phrase detection is to identify strings in a body of text that describes an anatomical location. These strings can be short, such as abdominal or in the liver, or longer, such as inferior aspect of the glenohumeral joint space, adjacent to the scapular body. Many of the single words or short phrases can be labeled by finding an exact match in a dictionary or an ontology such as SNOMED CT. Nevertheless, the detection of longer phrases requires recognizing lengthy, potentially noncontiguous, sequences of words. Previously, we proposed an anatomical phrase detection framework based on SNOMED CT [20]. The proposed method uses SNOMED CT as an initial look-up dictionary for anatomical concepts, and if not found in the ontology, it uses grammar-based patterns learned from SNOMED CT to be able to extract longer and more complex anatomical phrases. Details of the anatomical phrase detection algorithm are given in the Appendix.
Word Embedding
In this work, four of the most popular word embedding approaches are compared:
Skip-gram (SG) word2vec: A single hidden layer fully-connected neural network with linear neurons [22].
Continuous Bag-of-words (CBOW) word2vec: The architecture of CBOW is the same as SG with the exception that, in CBOW, the goal is to predict a word given a context [22].
Global Vectors (GloVe): GloVe is a log-bilinear model with a weighted least-squares objective [23].
Swivel (SW): SW is a highly efficient word2vec algorithm [24]. SW is somewhat similar to GloVe, since they both include global co-occurrence data. Besides, SW takes into account the fact that many co-occurrences are unobserved in the corpus.
The implementation of the word embedding techniques is available from different sources as given in Table 1.
Table 1.
Link to libraries for implementation of the word embedding techniques
| Technique | Library | Reference |
|---|---|---|
| Skip-gram | FastText | https://github.com/facebookresearch/fastText |
| CBOW | FastText | https://github.com/facebookresearch/fastText |
| GloVe | GloVe | https://nlp.stanford.edu/projects/glove/ |
| Swivel | Swivel | https://github.com/tensorflow/models/ree/master/swivel |
A mixture of two corpora is used for training the word embedding model: (1) a large corpus of radiology reports and (2) abstracts from SNOMED CT (terms). The reason for including SNOMED CT in the training corpus was to extend the vocabulary being learned in word2vec training beyond the terms covered in the training corpus of radiology reports from a specific site. All sentences and phrases in the training corpus are preprocessed and broken down to tokens.
Parameter Search
It has been shown that parameter tuning has a significant impact on the quality of the word embedding model training [25]. In this work, the impact of two types of hyper-parameters (vector dimension and context window size) on the performance of the anatomical classification is evaluated.
Word vector dimensionality: as the name implies, this parameter defines the size of the word vectors (defined by the size of the hidden layer in the neural network).
Context/window size: this parameter defines the number of words in context that the training algorithm should take into account.
In order to determine the best performing model for the anatomical classification problem, all combinations from mixing four word embedding techniques (SG, CBOW, GloVe, and SW), three vector dimension sizes (100, 300, and 500), and three context window size options (2, 5, and 10) are implemented and compared.
Phrase2vec
The output of anatomical phrase detection is either one word (e.g., thyroid) or multiple-word phrases (e.g., structure of lymphatics of mammary gland). To derive a vector representation for phrases, referred to as phrase2vec, five different aggregation techniques are defined and compared: minimum, maximum, median, average, and weighted average.
Assume phrase S consists of m words, S = {S1 S2. . Sm}, each word represented by an n-dimensional vector: v1 × n. Matrix V is created by row-wise concatenation of word vectors constituting the phrase.
| 1 |
Phrase2vec is derived by column-wise operations motivated by the following [26]: (1) minimum (), (2) maximum (), (3) median (), (4) average: (), and (5) weighted average (). Weights are calculated based on term frequency—inverse document frequency (TF-IDF) [27] as follows:
| 2 |
| 3 |
| 4 |
Here, tf(t, d) refers to the term frequency of term t in document d and idf(t, ℂ) refers to the inverse document frequency of term t in corpus ℂ. ft, d refers to frequency of the term t in document d, N refers to the number of documents in corpus ℂ, and nt refers to the number of documents in corpus ℂ containing term t.
Anatomical Phrase Classification
The final step of the proposed framework is the phrase classification. Word embedding representation of the detected anatomical phrases is created using the word2vec model. The anatomical classification is achieved by calculating pairwise similarity (dot product) between the vector representations of the unlabeled phrase and SNOMED CT class representatives; choosing the class label that yields the maximum similarity:
| 5 |
Here, vt denotes the vector of the unlabeled phrase and denotes the representative vector for class Ci.
Anatomical Class Labels
A set of 47 anatomical concepts extracted from SNOMED CT definitions under body organ structure (SNOMED ID: 113343008) is considered as representatives for 47 classes of anatomy. This set consists of a mixture of body organs (e.g., liver) and anatomical systems (e.g., digestive system) to cover the whole human anatomy. To avoid overlaps between organ and system classes, the following rule is applied: if an anatomical term can be categorized as both organ- and system-level classes, it is only included in the organ-level class. The list of 47 classes is further complemented with nine regional labels, abdomen, back, head, lower limb, neck, pelvis, thorax, upper limb, and whole body, making a total of 56 class labels. This helped provide a class label for generic anatomical regional terms that are also found in the radiology reports. The corresponding SNOMED CT concepts and IDs for all 56 classes are provided in the Appendix.
In SNOMED CT, concepts are logically defined through their relationships to other concepts. One example is the parent-child relation IsA. We define a concept to be member of a class if it is a descendent of the concept that represents that class. We include descendants up to a certain maximum tree depth. A class representative feature vector is created by using one of the aforementioned phrase2vec methods on all terms within a class including all descendent terms up to a desired depth. The optimal maximum depth level is determined empirically, comparing depth levels of 1 up to 10.
Clinical Data
Large corpora of radiology reports from the University of Washington (UW) and the University of Chicago (UC) were obtained. Radiology reports were collected with Institutional Review Board (IRB) approvals. All reports were de-identified by offsetting dates with randomly generated, patient-specific integers. All other HIPAA patient health information was removed. In addition, 100 radiology reports were randomly obtained from the MIMICIII corpus as unseen (test) data for performance evaluation. The breakdown of the data as training, validation, and testing sets is described below:
Word Embedding Training
word2vec model was trained using 1,566,921 full radiology reports from the UW dataset plus 334,486 unique terms extracted from SNOMED CT (version 20150731). After applying preprocessing, the training corpus contained 418,761,995 tokens with a vocabulary size of 270,015. Moreover, 560 radiology reports (referred to as UW560) were randomly selected from the remaining reports in UW corpus as the validation set to determine the best performing word2vec model for the anatomical labeling task. UW560 was manually annotated with seven anatomical labels (brain, breast, kidney, liver, lung, prostate, and thyroid) by two annotators (RvO, PP: not domain experts). The BRAT Annotation tool [28] was used to generate manually labeled ground truth data. Anatomical phrases not belonging to these seven classes were labeled as other and excluded from the study. Manual classifications yield 5,066 anatomical phrases with distribution across seven classes as shown in Table 2. No Inter Annotator Agreement (IAA) [29] was measured for this round. The main reason for considering seven class labels rather than 56 labels is to simplify manual labeling in ground truth generation for a relatively large corpus of radiology reports to be used as the validation set. This enables performing a meaningful evaluation and comparison of word embedding models using such large corpus.
Table 2.
Distribution of manually generated annotations for 560 reports from the UW (UW560)
| Anatomical class | No. of terms |
|---|---|
| Brain | 625 |
| Breast | 268 |
| Kidney | 826 |
| Liver | 1,197 |
| Lung | 1,886 |
| Prostate | 145 |
| Thyroid | 115 |
Testing Set
The testing set consists of 300 reports: a set of 100 radiology reports from the UW excluded from the word2vec training and the validation set (UW100), a set of 100 radiology reports from UC (UC100), and finally, a set of 100 radiology reports from the MIMICIII dataset (MIMICIII100). Anatomical phrases corresponding to 56 classes were manually labeled by four annotators (AT, GM, PP, and RvO: not domain experts). The annotations were generated and refined in three rounds. During the first round of annotation, the first 20% reports of each set were manually annotated by every annotator. In the second round, annotations with half or more disagreements between annotators were further reviewed and corrected by a radiologist. Most disagreements among four annotators were related to the system- and organ-level labels. During the third and last round, the remaining 80% of the reports were annotated by the same four annotators independently following the feedback learned from the first and second rounds. After the third round, there was no case of disagreement with more than half of annotators. The average IAA measured as average Kappa [31] for the first and third rounds were UW100: 89.5% and 86.5%; UC100: 85.3% and 90.1%; and MIMICIII100: 92.6% and 87.4%, respectively. Manual annotations yielded 1,616, 2,028, and 1,579 labeled anatomical phrases for UW100, UC100, and MIMICIII100, respectively.
Evaluation
The performance is measured and reported in the form of the precision, recall, and F1 score for each class label, separately, as the following:
where TP, FP,and FN stand for the number of true positives, false positives, and false negatives, respectively.
The performance of the proposed approach was compared against a number of state-of-the-art supervised and unsupervised classification techniques. The input to all selected methods is the anatomical phrases from the ground truth to avoid combining phrase detection and phrase labeling errors. Furthermore, training of the selected supervised approaches was achieved using all SNOMED CT terms from 56 anatomical classes as described before. The word embedding of phrases were used as input feature vectors. Anatomical phrases from UW560 set with seven anatomical labels (referred to as the validation set) were used for parameter tuning consistently across all approaches. A list of the selected approaches and the corresponding parameters are given in Table 3.
Table 3.
list of the selected approaches and corresponding parameters
| Method | Parameters/range | Optimal parameters |
|---|---|---|
| K nearest neighbor (kNN) | K = 5,…,1000 | K = 80 |
| RNN [30] | RNN cell type: LSTM, GRU; RNN depth: 1, 2; number of hidden layers: 32, 64, and 128; learning rate: 1e−3. Batch size of 128; 200 epochs | Cell type: GRU, hidden layers = 128, depth = 1, dropout = 0.7 |
| Bidirectional RNN conditional random field (bidirectional-RNN-CRF) [31] | RNN cell type: LSTM, GRU; RNN depth: 1, 2; number of hidden layers: 32, 64, and 128; learning rate: 1e−3. Batch size of 128; 200 epochs | Cell type: GRU, hidden layers = 128, depth = 1, dropout = 0.9 |
| Support Vector Machine (SVM) | Kernel: RBF/LINEAR; penalty parameter C: 1e−5, 1e−4,…, 1e5 | C = 1, kernel = Linear |
Results
In search of the best performing word embedding model, several options for the following parameters were considered: (1) SNOMED CT term depth level: from 1 to 10; (2) phrase2vec aggregation method: min, max, median, average, and weighted average; (3) word embedding technique: CBOW, SG, GloVe, and SW; (4) word vector size: 100, 300, and 500; and (5) context window size: 2, 5, and 10. This resulted in training and comparing 1980 models. The followings were concluded from the comparisons: (1) Increasing SNOMED CT term depth level results in improving classification performance, up to depth of 7 and after that, does not result in a significant change; (2) weighted averaging technique yields the best performance for the classification; (3) increasing both the vector size and the window size result in higher precision (see Table 4); and (4) SG-based model outperforms the rest by at least 2%. Table 4 compares classification precision between word embedding techniques while altering the vector dimension, and window size, but fixing the depth level to 7 and phrase2vec method to weighted averaging.
Table 4.
Comparing classification precision between word embedding techniques while altering the embedding dimension, and context window size. SNOMED CT term depth and phrase2vec method are fixed to 7 and weighted averaging, respectively
| (vec_size, win_size) | Precision (weighted average) | |||
|---|---|---|---|---|
| CBOW (%) | SG (%) | GloVe (%) | SW (%) | |
| (100, 2) | 90.4 | 94.1 | 85.7 | 90.4 |
| (100, 5) | 91.9 | 95.0 | 89.9 | 92.1 |
| (100, 10) | 92.7 | 95.0 | 91.6 | 92.9 |
| (300, 2) | 90.2 | 94.2 | 87.0 | 89.2 |
| (300, 5) | 92.1 | 94.8 | 90.0 | 89.7 |
| (300, 10) | 93.1 | 95.1 | 92.0 | 92.1 |
| (500, 2) | 90.3 | 93.9 | 86.7 | 90.7 |
| (500, 5) | 92.2 | 95.2 | 90.0 | 91.0 |
| (500, 10) | 92.9 | 95.1 | 91.7 | 91.0 |
Precision, recall, and F1 score per anatomical class for the best performing model (SG, vec_size: 500, window_size: 10) is given in Table 5.
Table 5.
Precision, recall, and F1 score for seven anatomical classes using the best performing model: skip-gram, vector size = 500, window size = 10
| Anatomical class | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|
| Brain | 97.0 | 83.4 | 89.7 |
| Breast | 94.5 | 96.3 | 95.4 |
| Kidney | 96.2 | 96.2 | 96.2 |
| Liver | 98.6 | 93.8 | 96.1 |
| Lung | 92.1 | 99.5 | 95.7 |
| Prostate | 100.0 | 97.2 | 98.6 |
| Thyroid | 96.4 | 93.0 | 94.7 |
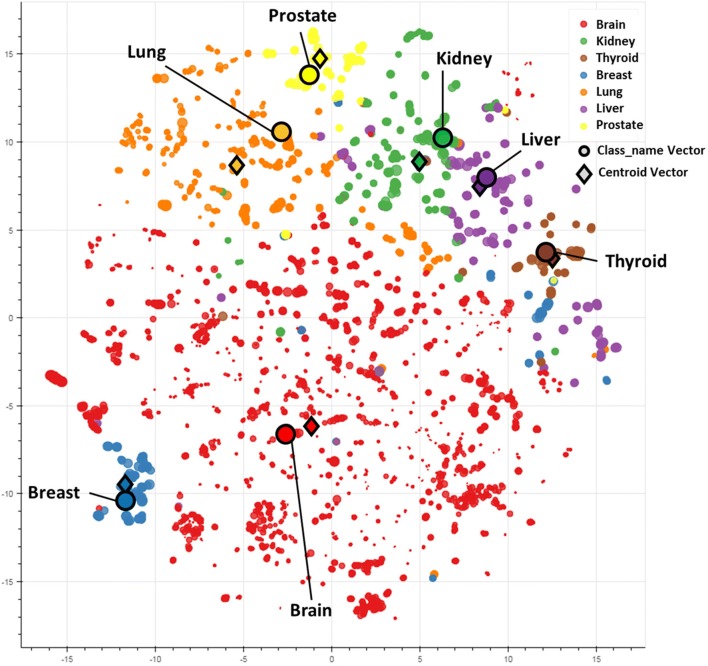
We used t-distributed stochastic neighbor embedding (t-SNE) to derive a visual understanding of the separation of the seven anatomical classes based on the best performing word embedding model [32]. Figure 3 depicts the t-SNE plot of SNOMED CT terms from each of the seven anatomical classes. Each term is shown with a circle. The color represents the corresponding anatomical class. The size of the circle decreases as the depth of the term in SNOMED CT increases. The centroid vectors (determined by averaging all vectors from all terms within the class) and the parent node representative vectors for each class (e.g., lung, liver) are shown with diamond shape and black edge circle, respectively. As can be seen from the figure, there is a clear separation between neighboring classes with a few exceptions.
Fig. 3.
t-SNE plot of SNOMED CT terms from each of the seven anatomical classes. Each term is shown with a circle. The size of the circle indicates its level in SNOMED CT tree, and the color indicates the anatomical class. Circles with black edge depict the spatial location of the category term itself, and the diamond shapes correspond to the centroid of the corresponding category (average of all terms in the class)
Next, the best performing model (SG, vec_size 500, win_size 10) was used on the testing set for the anatomical classification based on 56 anatomical class labels. The classification precision on the 300 reports was measured and compared to other state-of-the-art approaches as described before (see Table 6). Furthermore, precision, recall, and F1 score of the proposed approach for each anatomical class are given in Table 7 (UW100), Table 8 (UC100), and Table 9 (MIMICIII100).
Table 6.
Comparing the performance of the proposed anatomical classification with state-of-the-art approaches
| Method | UW100 (%) | UC100 (%) | MIMICIII100 (%) | Overall (%) |
|---|---|---|---|---|
| Proposed | 83.5 | 83.4 | 80.6 | 82.6 |
| kNN | 63.7 | 68.5 | 60.2 | 64.1 |
| RNN | 74.7 | 66.6 | 63.3 | 68.1 |
| Bi-RNN-CRF | 76.2 | 74.6 | 68.5 | 73.3 |
| SVM | 69.8 | 62.3 | 58.6 | 63.5 |
Table 7.
Performance of the anatomical classification per class label on 100 reports from the UW. NaN: Not a Number
| Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) | Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|---|---|---|---|---|---|
| Abdomen | 70 | 92.6 | 90.0 | 91.3 | Nasal sinus | 25 | 62.5 | 100.0 | 76.9 |
| Adrenal gland | 18 | 100.0 | 100.0 | 100.0 | Neck | 11 | 60.0 | 81.8 | 69.2 |
| Back | 4 | 100.0 | 75.0 | 85.7 | Nervous system | 17 | 38.9 | 82.4 | 52.8 |
| Bile Duct | 17 | 85.0 | 100.0 | 91.9 | Nose | 0 | 0.0 | NaN | NaN |
| Bladder | 16 | 81.3 | 81.3 | 81.3 | Ovary | 14 | 100.0 | 100.0 | 100.0 |
| Brain | 92 | 68.7 | 50.0 | 57.9 | Pancreas | 13 | 100.0 | 100.0 | 100.0 |
| Breast | 41 | 100.0 | 92.7 | 96.2 | Pelvis | 62 | 98.3 | 95.2 | 96.7 |
| Cardiovascular system | 131 | 77.2 | 80.2 | 78.7 | Penis | 0 | NaN | NaN | NaN |
| Diaphragm | 10 | 66.7 | 80.0 | 72.7 | Pericardial sac | 10 | 100.0 | 100.0 | 100.0 |
| Digestive system | 3 | 0.0 | 0.0 | NaN | Peritoneal sac | 12 | 68.8 | 91.7 | 78.6 |
| Ear | 0 | NaN | NaN | NaN | Pharynx | 0 | 0.0 | NaN | NaN |
| Esophagus | 3 | 75.0 | 100.0 | 85.7 | Pleural sac | 35 | 100.0 | 100.0 | 100.0 |
| Eye | 6 | 100.0 | 16.7 | 28.6 | Prostate | 5 | 100.0 | 100.0 | 100.0 |
| Fallopian tube | 0 | NaN | NaN | NaN | Retroperitoneal | 3 | 50.0 | 100.0 | 66.7 |
| Gallbladder | 19 | 100.0 | 84.2 | 91.4 | Seminal vesicle | 4 | 100.0 | 100.0 | 100.0 |
| Head | 49 | 75.0 | 36.7 | 49.3 | Spleen | 13 | 100.0 | 100.0 | 100.0 |
| Heart | 37 | 50.9 | 78.4 | 61.7 | Stomach | 9 | 60.0 | 100.0 | 75.0 |
| Integumentary system | 15 | 100.0 | 93.3 | 96.6 | Testis | 1 | 100.0 | 100.0 | 100.0 |
| Intestine | 51 | 95.7 | 86.3 | 90.7 | Thorax | 67 | 89.9 | 92.5 | 91.2 |
| Kidney | 56 | 96.4 | 96.4 | 96.4 | Thyroid | 6 | 85.7 | 100.0 | 92.3 |
| Laryngeal | 0 | NaN | NaN | NaN | Tracheobronchial | 10 | 100.0 | 80.0 | 88.9 |
| Liver | 54 | 96.1 | 90.7 | 93.3 | Upper limb | 29 | 78.9 | 51.7 | 62.5 |
| Lower limb | 64 | 84.1 | 57.8 | 68.5 | Urethra | 0 | NaN | NaN | NaN |
| Lung | 114 | 86.2 | 98.2 | 91.8 | Uterus | 9 | 69.2 | 100.0 | 81.8 |
| Lymphatic system | 29 | 76.3 | 100.0 | 86.6 | Vagina | 2 | NaN | 0.0 | NaN |
| Mediastinum | 14 | 100.0 | 71.4 | 83.3 | Vas deferens | 0 | NaN | NaN | NaN |
| Mouth | 2 | 33.3 | 50.0 | 40.0 | Vulva | 0 | NaN | NaN | NaN |
| Musculoskeletal system | 342 | 89.1 | 86.3 | 87.7 | Whole body | 2 | 28.6 | 100.0 | 44.4 |
Table 8.
Performance of the anatomical classification per class label on 100 reports from the UC. NaN: Not a Number
| Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) | Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|---|---|---|---|---|---|
| Abdomen | 136 | 97.4 | 82.4 | 89.2 | Nasal sinus | 49 | 66.2 | 100.0 | 79.7 |
| Adrenal gland | 22 | 100.0 | 100.0 | 100.0 | Neck | 40 | 87.8 | 90.0 | 88.9 |
| Back | 4 | 100.0 | 50.0 | 66.7 | Nervous system | 9 | 14.0 | 77.8 | 23.7 |
| Bile Duct | 40 | 88.9 | 100.0 | 94.1 | Nose | 3 | 100.0 | 100.0 | 100.0 |
| Bladder | 20 | 90.0 | 90.0 | 90.0 | Ovary | 12 | 100.0 | 100.0 | 100.0 |
| Brain | 120 | 77.1 | 61.7 | 68.5 | Pancreas | 24 | 95.8 | 95.8 | 95.8 |
| Breast | 80 | 100.0 | 97.5 | 98.7 | Pelvis | 53 | 96.1 | 92.5 | 94.2 |
| Cardiovascular system | 53 | 65.2 | 81.1 | 72.3 | Penis | 0 | NaN | NaN | NaN |
| Diaphragm | 4 | 100.0 | 25.0 | 40.0 | Pericardial sac | 17 | 100.0 | 100.0 | 100.0 |
| Digestive system | 9 | 0.0 | 0.0 | NaN | Peritoneal sac | 26 | 66.7 | 100.0 | 80.0 |
| Ear | 13 | 92.9 | 100.0 | 96.3 | Pharynx | 7 | 50.0 | 100.0 | 66.7 |
| Esophagus | 6 | 75.0 | 100.0 | 85.7 | Pleural sac | 41 | 100.0 | 100.0 | 100.0 |
| Eye | 6 | 50.0 | 16.7 | 25.0 | Prostate | 6 | 100.0 | 100.0 | 100.0 |
| Fallopian tube | 0 | NaN | NaN | NaN | Retroperitoneal | 19 | 82.6 | 100.0 | 90.5 |
| Gallbladder | 21 | 100.0 | 90.5 | 95.0 | Seminal vesicle | 4 | 100.0 | 100.0 | 100.0 |
| Head | 31 | 69.2 | 58.1 | 63.2 | Spleen | 27 | 100.0 | 96.3 | 98.1 |
| Heart | 27 | 51.4 | 70.4 | 59.4 | Stomach | 18 | 60.0 | 100.0 | 75.0 |
| Integumentary system | 14 | 100.0 | 92.9 | 96.3 | Testis | 0 | NaN | NaN | NaN |
| Intestine | 80 | 97.5 | 96.3 | 96.9 | Thorax | 143 | 90.7 | 88.8 | 89.8 |
| Kidney | 87 | 92.9 | 89.7 | 91.2 | Thyroid | 18 | 73.7 | 77.8 | 75.7 |
| Laryngeal | 1 | 100.0 | 100.0 | 100.0 | Tracheobronchial | 11 | 87.5 | 63.6 | 73.7 |
| Liver | 84 | 100.0 | 85.7 | 92.3 | Upper limb | 31 | 56.8 | 80.6 | 66.7 |
| Lower limb | 27 | 84.6 | 81.5 | 83.0 | Urethra | 1 | 100.0 | 100.0 | 100.0 |
| Lung | 123 | 83.9 | 93.5 | 88.5 | Uterus | 7 | 100.0 | 100.0 | 100.0 |
| Lymphatic system | 46 | 53.5 | 100.0 | 69.7 | Vagina | 0 | NaN | NaN | NaN |
| Mediastinum | 68 | 100.0 | 57.4 | 72.9 | Vas deferens | 0 | NaN | NaN | NaN |
| Mouth | 15 | NaN | 0.0 | NaN | Vulva | 0 | NaN | NaN | NaN |
| Musculoskeletal system | 316 | 91.7 | 73.7 | 81.8 | Whole body | 9 | 75.0 | 66.7 | 70.6 |
Table 9.
Performance of the anatomical classification per class label on 100 reports from MIMICIII. NaN: Not a Number
| Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) | Anatomical class | Freq. | Precision (%) | Recall (%) | F1 score (%) |
|---|---|---|---|---|---|---|---|---|---|
| Abdomen | 53 | 95.6 | 81.1 | 87.8 | Nasal sinus | 21 | 87.5 | 100.0 | 93.3 |
| Adrenal gland | 6 | 100.0 | 100.0 | 100.0 | Neck | 7 | 85.7 | 85.7 | 85.7 |
| Back | 2 | 0.0 | 0.0 | NaN | Nervous system | 3 | 10.0 | 66.7 | 17.4 |
| Bile duct | 6 | 100.0 | 100.0 | 100.0 | Nose | 0 | 0.0 | NaN | NaN |
| Bladder | 8 | 63.6 | 87.5 | 73.7 | Ovary | 2 | 100.0 | 100.0 | 100.0 |
| Brain | 168 | 94.6 | 73.2 | 82.6 | Pancreas | 5 | 100.0 | 100.0 | 100.0 |
| Breast | 2 | 66.7 | 100.0 | 80.0 | Pelvis | 55 | 92.5 | 89.1 | 90.7 |
| Cardiovascular system | 151 | 70.6 | 79.5 | 74.8 | Penis | 0 | NaN | NaN | NaN |
| Diaphragm | 5 | 44.4 | 80.0 | 57.1 | Pericardial sac | 8 | 54.5 | 75.0 | 63.2 |
| Digestive system | 30 | 33.3 | 3.3 | 6.1 | Peritoneal sac | 11 | 64.3 | 81.8 | 72.0 |
| Ear | 0 | NaN | NaN | NaN | Pharynx | 7 | 75.0 | 85.7 | 80.0 |
| Esophagus | 8 | 100.0 | 100.0 | 100.0 | Pleural sac | 65 | 100.0 | 90.8 | 95.2 |
| Eye | 5 | 100.0 | 60.0 | 75.0 | Prostate | 6 | 100.0 | 83.3 | 90.9 |
| Fallopian tube | 0 | 0.0 | NaN | NaN | Retroperitoneal | 4 | 66.7 | 100.0 | 80.0 |
| Gallbladder | 8 | 88.9 | 100.0 | 94.1 | Seminal vesicle | 1 | 100.0 | 100.0 | 100.0 |
| Head | 37 | 86.5 | 86.5 | 86.5 | Spleen | 5 | 100.0 | 100.0 | 100.0 |
| Heart | 52 | 63.2 | 82.7 | 71.7 | Stomach | 19 | 33.3 | 89.5 | 48.6 |
| Integumentary system | 9 | 88.9 | 88.9 | 88.9 | Testis | 0 | NaN | NaN | NaN |
| Intestine | 21 | 80.0 | 95.2 | 87.0 | Thorax | 153 | 98.0 | 94.1 | 96.0 |
| Kidney | 69 | 96.7 | 86.6 | 91.3 | Thyroid | 0 | 0.0 | NaN | NaN |
| Laryngeal | 1 | 12.5 | 100.0 | 22.2 | Tracheobronchial | 30 | 84.6 | 73.3 | 78.6 |
| Liver | 100 | 96.8 | 60.0 | 74.1 | Upper limb | 32 | 81.5 | 68.8 | 74.6 |
| Lower limb | 36 | 52.4 | 30.6 | 38.6 | Urethra | 0 | NaN | NaN | NaN |
| Lung | 149 | 79.7 | 97.3 | 87.6 | Uterus | 7 | 75.0 | 100.0 | 85.7 |
| Lymphatic system | 2 | 33.3 | 100.0 | 50.0 | Vagina | 0 | NaN | NaN | NaN |
| Mediastinum | 24 | 94.7 | 75.0 | 83.7 | Vas deferens | 0 | NaN | NaN | NaN |
| Mouth | 3 | 0.0 | 0.0 | NaN | Vulva | 0 | NaN | NaN | NaN |
| Musculoskeletal system | 181 | 84.8 | 83.9 | 84.4 | Whole body | 2 | 20.0 | 50.0 | 28.6 |
Reviewing the confusion matrices revealed the following as the most confused class pairs (i.e., confusions with more than 10 occurrences are only reported): (abdomen and upper limb: e.g., right upper quadrant), (brain and heart: e.g., ventricles), (brain and lung: e.g., right lobe), (brain and nervous system: e.g., intracranial), (brain and nasal sinus: e.g., extra-axial), (cardiovascular system and heart and vice-versa: e.g., coronary artery), (digestive system and stomach: e.g., GI), (head and brain: e.g., vertex), (head and musculoskeletal system and vice-versa: e.g., skull base), (liver and cardiovascular system: e.g., portal vein), (mediastinum and lymphatic system: e.g., hila), (musculoskeletal system and brain: e.g., skull base), (musculoskeletal system and neck: e.g., femoral neck), (musculoskeletal system and nervous system: e.g., spinal), (musculoskeletal system and nasal sinus: e.g., calvarium), (musculoskeletal system and thorax: e.g., thoracic spine), (thorax and lung: e.g., retrocardiac), and (upper limb and cardiovascular system: e.g., brachial).
The performance of an ontology-based exact match classifier was also measured for the same classification task. The ontology-based approach yielded a precision of 48% on the same testing set. Combining the ontology-based exact match with the proposed framework only slightly improved the classification performance: First, every identified phrase was compared against the ontology terms. An exact match was considered as true positive. Otherwise, the labeling process continues using the proposed framework. The overall precision of the combined approach was 84.4% (UW 85.5%, UC 83.6%, and MIMICIII 81.3%).
Discussion
Determining the anatomical context of sentences is a crucial step towards automatically interpreting radiology reports. Nevertheless, free-text reports often suffer from inconsistent terminology and telegraphic presentation, which leads to ambiguity and uncertainty in interpretation [33]. As a result, conventional supervised NER approaches have not been able to deliver satisfactory performance unless provided with sufficiently large labeled training data.
The proposed framework overcomes such limitations by using unsupervised learning of semantic relations between anatomical concepts from a large corpus of radiology reports combined with SNOMED CT ontology without requiring any manual labeling. As demonstrated in Table 6, the proposed approach significantly outperformed some of the state-of-the-art supervised approaches such as RNN and RNN-CRF. The low performance of the supervised approaches is mainly due to the limited size of the training set.
A major strength of the proposed framework is the capability to detect and label anatomical phrases beyond specific language and terminology available through standard ontologies. Other tools, such as cTAKES and MetaMap, fail to correctly label elongated anatomical description such as anterior to the ring of C1 or long head of the biceps tendon. Moreover, most similar frameworks require creating an extensive list of abbreviations and acronyms, whereas the proposed framework, thanks to semantic learning, is capable of correctly labeling abbreviations even beyond what can be found in ontologies (e.g., PEL (pelvis), Pulm (pulmonary), CCA (cardiovascular system), CBD (bile duct)). Another interesting feature of the proposed framework is the ability to detect location descriptors that do not contain organ names (e.g., T12/L1 (musculoskeletal system), left retroareolar region (breast), 12 o’clock position (breast), tail (pancreas)). Contextual learning also helps labeling beyond literal anatomy matching. For example, from the cranial vertex to the foramen magnum was correctly labeled as brain by the proposed framework, whereas other compared approaches labeled the phrase as vertex and/or magnum.
Using the proposed framework, it is straightforward to define new anatomical classes or to merge existing classes for anchoring at a more generic level. For example, in the current implementation, appendix is included within the intestine class as it is a child node of intestine structure; however, appendix and the corresponding terms in SNOMED CT can be excluded from the Intestine class and instead be added as a separate class label. This scenario was tested. The overall performance of the classifier with the new additional class did not change. The classifier yielded 100% precision and recall for labeling four occurrences of appendix in the testing set.
Reviewing incorrect/miss labels (false positives/false negatives) in the confusion matrices revealed that a major source of misclassification is the lack of sufficient context in the detected anatomical phrases. For example, if the detected phrase is ventricles, without considering the context around it, it is impossible to identify the correct label as several organs such as brain or heart contain regions referred to as such. We plan to tackle this shortcoming in the near future using state-of-the-art sequence labeling approaches such as recurrent neural networks [33]. The context information can be extracted from the same or the surrounding sentences or from meta-data included in the report such as the study description (e.g., brain MRI). Another source of error in misclassification is related to the subjective choice of class label in ground truth generation, e.g., should hepatic vein be labeled as Liver or Cardiovascular System? One may argue that both labels are correct depending on what level of specificity is expected from the classification. In order to evaluate the performance of the proposed classifier while avoiding penalization for such subjective cases, a performance metric from Information Retrieval domain was considered, referred to as Success@N [34]: a true positive match is defined as long as the ground truth class label is among top N predicted class labels after ranking similarity values between the unlabeled phrase and all 56 class representatives. For our study, we considered Success@2. The precision of the proposed classifier for 56-classification problem with Success@2 match was measured as 92.1%, 91.9%, and 89.6% for the UW100, UC100, and MIMICIII100, respectively.
Conclusion
In this work, we presented a framework for creating an anatomical phrase classification model for radiology report labeling without requiring a large annotated corpus for training. The proposed framework combines semantic learning through word embedding (i.e., word2vec) with conventional ontology-driven anatomy labeling techniques to overcome shortcomings of the currently available techniques for anatomical phrase labeling in radiology reports. The evaluation of the proposed framework on radiology reports from three different sites demonstrated promising performance. The current implementation of the framework consists of a specific anatomical categorization of the whole body based on SNOMED CT; however, the framework is flexible to be able to incorporate any additional anatomical classification based on any ontology when there is reasonable separation between classes in terms of term scope. The current framework consists of two separate steps for detection and recognition of anatomical phrases. As the next step, a unified approach is under development that combines both tasks (detection and labeling) into one.
Appendix
Anatomical Phrase Detection
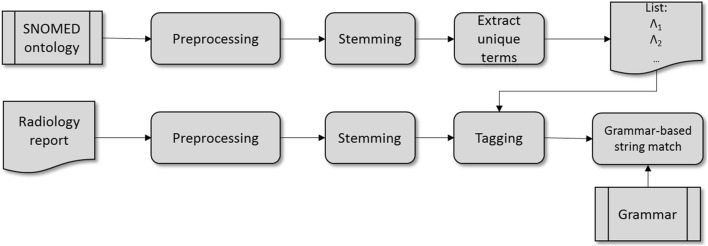
Figure 4 depicts an overview of the anatomical phrase detection pipeline. First, a grammar and a token tagging schema are derived from anatomical descriptions found in SNOMED CT, using tokenization and a simplified version of the Porter stemmer [35, 36]. The input text is processed with the same tokenizer and stemmer. Next, tokens are labeled using the tagging schema and, finally, maximal substrings of labels are identified that match the grammar.
Fig. 4.
Anatomical phrase detection pipeline
Token Tagging
The token labels used in the grammar are of two types: (1) syntactic and (2) anatomical. The syntactic token labels include A for articles and personal pronouns, C for conjunctions, P for prepositions, and X for other words. The anatomical token labels consist of L1 for words that describe an anatomy and L2 for words that are part of an anatomy description but do not specifically describe an anatomy, such as structure, area, or aspect.
An anatomical phrase is defined by a predefined pattern. Examples of predefined patterns include L1, PAL1, and L1CL1, which would match the liver, above the liver, and liver and spleen, respectively.
Grammar Development
The proposed grammar consists of three parts. The lowest level component consists of a sequence of words, each with label L1 or L2, with possible conjunctions C between them. This results in matches like left lung lobe, left and right lung, and 1st, 2nd, and 3rd left rib. Such lowest level components are combined into higher-level components with a word from category P and optionally a word from A between them. This results in matches like inferior aspect of the glenohumeral joint space. These higher-level components are then combined with conjunctions C to form the final grammar, which allows matches like inferior aspect of the glenohumeral joint space, adjacent to the scapular body. The final match needs to contain at least one word with label L1.
SNOMED CT is used to generate the lists Λi containing the words with label Li. All relevant concepts and the corresponding synonyms (i.e., all body structures, specific site descriptors, and anatomical relationship descriptors) are extracted, tokenized, and stemmed. Next, words with tags A, C, or P using the definition given above are excluded. Then, all remaining unique stemmed tokens are collected and assigned to list Λ1, list Λ2, or discarded. The grammar described above is created by starting with an initial grammar (in the form of a regular expression) and, then, further refining it by running the matching method on texts containing anatomical phrases.
Anatomical Categories
The final anatomical classification is determined based on a set of 56 anatomical labels extracted from SNOMED CT as shown in Table 10 in alphabetical order.
Table 10.
Fifty-six anatomical labels extracted from SNOMED CT
| Class | Description | SNOMED ID |
|---|---|---|
| Abdomen | Abdominal structure | 113345001 |
| Adrenal gland | Adrenal structure | 23451007 |
| Back | Back structure, excluding neck | 77568009 |
| Bile duct | Bile duct structure | 28273000 |
| Bladder | Urinary bladder structure | 89837001 |
| Brain | Brain structure | 12738006 |
| Pineal structure | 45793000 | |
| Pituitary structure | 56329008 | |
| Breast | Breast structure | 76752008 |
| Cardiovascular system | Blood vessel structure | 59820001 |
| Diaphragm | Diaphragm structure | 5798000 |
| Digestive system | Structure of digestive system | 86762007 |
| Ear | Ear structure | 117590005 |
| Esophagus | Esophageal structure | 32849002 |
| Eye | Eye region structure | 371398005 |
| Fallopian tube | Fallopian tube structure | 31435000 |
| Gallbladder | Gallbladder structure | 28231008 |
| Head | Head structure | 69536005 |
| Heart | Heart structure | 80891009 |
| Integumentary system | Skin structure | 39937001 |
| Intestine | Intestinal structure | 113276009 |
| Kidney | Kidney structure | 64033007 |
| Ureteric structure | 119220009 | |
| Laryngeal | Laryngeal structure | 4596009 |
| Liver | Liver structure | 10200004 |
| Lower limb | Lower limb structure | 61685007 |
| Lung | Lung structure | 39607008 |
| Lymphatic system | Lymphoid organ structure | 91688001 |
| Mediastinum | Mediastinal structure | 72410000 |
| Mouth | Salivary gland structure | 385294005 |
| Musculoskeletal system | Structure of teeth, gums, and supporting structures | 28035005 |
| Tongue structure | 21974007 | |
| Tooth structure | 38199008 | |
| Bone marrow structure | 14016003 | |
| Structure of bone (organ) | 421663001 | |
| Joint structure | 39352004 | |
| Skeletal and/or smooth muscle structure | 71616004 | |
| Structure of ligament | 52082005 | |
| Nasal sinus | Nasal sinus structure | 2095001 |
| Neck | Neck structure | 45048000 |
| Nervous system | Structure of nervous system | 25087005 |
| Nose | Nasal structure | 45206002 |
| Ovary | Ovarian structure | 15497006 |
| Pancreas | Pancreatic structure | 15776009 |
| Pelvis | Pelvic structure | 12921003 |
| Penis | Penile structure | 18911002 |
| Pericardial sac | Pericardial structure | 76848001 |
| Peritoneal sac | Serous membrane structure | 75858005 |
| Pharynx | Pharyngeal structure | 54066008 |
| Pleural sac | Pleural membrane structure | 3120008 |
| Prostate | Prostatic structure | 119231001 |
| Retroperitoneal | Structure of retroperitoneal space | 699600004 |
| Seminal vesicle | Seminal vesicle structure | 64739004 |
| Spleen | Splenic structure | 78961009 |
| Stomach | Stomach structure | 69695003 |
| Testis | Testis structure | 40689003 |
| Thorax | Thoracic structure | 51185008 |
| Thyroid | Parathyroid structure | 111002 |
| Thyroid structure | 69748006 | |
| Tracheobronchial | Tracheobronchial structure | 91724006 |
| Upper limb | Upper limb structure | 53120007 |
| Urethra | Urethral structure | 13648007 |
| Uterus | Placental structure | 78067005 |
| Uterine structure | 35039007 | |
| Vagina | Vaginal structure | 76784001 |
| Vas deferens | Vas deferens structure | 57671007 |
| Vulva | Vulval structure | 45292006 |
| Whole body | Entire body as a whole | 38266002 |
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no competing interests.
References
- 1.M. Q. Stearns, C. Price, K. A. Spackman, and A. Y. Wang, SNOMED clinical terms: overview of the development process and project status, in Proceedings of the AMIA Symposium, 2001, p. 662. [PMC free article] [PubMed]
- 2.D. B. Johnson, R. K. Taira, A. F. Cardenas, and D. R. Aberle, Extracting information from free text radiology reports, vol. 1, no. 3, pp. 297–308, 1997.
- 3.O. Bodenreider, The unified medical language system (UMLS): integrating biomedical terminology, Nucleic acids research, vol. 32, no. suppl_1, pp. D267–D270, 2004. [DOI] [PMC free article] [PubMed]
- 4.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S. Goryachev, M. Sordo, and Q. T. Zeng, A suite of natural language processing tools developed for the I2B2 project, in AMIA Annual Symposium Proceedings, 2006, vol. 2006, p. 931. [PMC free article] [PubMed]
- 6.G. Hripcsak, C. Friedman, P. O. Alderson, W. DuMouchel, S. B. Johnson, and P. D. Clayton, Unlocking clinical data from narrative reports: a study of natural language processing, vol. 122, no. 9, pp. 681–688, 1995. [DOI] [PubMed]
- 7.C. Friedman, P. O. Alderson, J. H. Austin, J. J. Cimino, and S. B. Johnson, A general natural-language text processor for clinical radiology, vol. 1, no. 2, pp. 161–174, 1994. [DOI] [PMC free article] [PubMed]
- 8.C. Friedman, L. Shagina, Y. Lussier, and G. Hripcsak, Automated encoding of clinical documents based on natural language processing, vol. 11, no. 5, pp. 392–402, 2004. [DOI] [PMC free article] [PubMed]
- 9.J.-D. Kim, T. Ohta, Y. Tsuruoka, Y. Tateisi, and N. Collier, Introduction to the bio-entity recognition task at JNLPBA, in Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and Its Applications, 2004, pp. 70–75.
- 10.M. Gerner, G. Nenadic, and C. M. Bergman, An exploration of mining gene expression mentions and their anatomical locations from biomedical text, in Proceedings of the 2010 Workshop on Biomedical Natural Language Processing, 2010, pp. 72–80.
- 11.A. R. Aronson, Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program, in Proceedings of the AMIA Symposium, 2001, p. 17. [PMC free article] [PubMed]
- 12.Taira RK, Soderland SG, Jakobovits RM. Automatic structuring of radiology free-text reports. Radiographics. 2001;21(1):237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 13.Pyysalo S, Ananiadou S. Anatomical entity mention recognition at literature scale. Bioinformatics. 2013;30(6):868–875. doi: 10.1093/bioinformatics/btt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D. Campos, S. Matos, and J. L. Oliveira, Biomedical named entity recognition: a survey of machine-learning tools, in Theory and Applications for Advanced Text Mining, InTech, 2012.
- 15.B. Tang, H. Cao, X. Wang, Q. Chen, and H. Xu, Evaluating word representation features in biomedical named entity recognition tasks, vol. 2014, 2014. [DOI] [PMC free article] [PubMed]
- 16.Y. Wu, J. Xu, M. Jiang, Y. Zhang, and H. Xu, A study of neural word embeddings for named entity recognition in clinical text, in AMIA Annual Symposium Proceedings, 2015, vol. 2015, p. 1326. [PMC free article] [PubMed]
- 17.N. Limsopatham and N. Collier, Normalising medical concepts in social media texts by learning semantic representation, in ACL (1), 2016.
- 18.Y. Bengio, Deep learning of representations for unsupervised and transfer learning, in Proceedings of ICML Workshop on Unsupervised and Transfer Learning, 2012, pp. 17–36.
- 19.A. Ferré, P. Zweigenbaum, and C. Nédellec, Representation of complex terms in a vector space structured by an ontology for a normalization task, BioNLP 2017, pp. 99–106, 2017.
- 20.Peter Prinsen, Robert van Ommering, Gabe Mankovich, Lucas Oliveira, Vadiraj Hombal, and Amir Tahmasebi, A novel approach for improving the recall of concept detection in medical documents using extended ontologies, in SIIM 2017 Scientific Session, 2017.
- 21.S. Bird, E. Klein, and E. Loper, Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. O’Reilly Media, Inc., 2009.
- 22.T. Mikolov, K. Chen, G. Corrado, and J. Dean, Efficient Estimation of Word Representations in Vector Space, 2013.
- 23.J. Pennington, R. Socher, and C. D. Manning, Glove: global vectors for word representation, in EMNLP, 2014, vol. 14, pp. 1532–1543.
- 24.N. Shazeer, R. Doherty, C. Evans, and C. Waterson, Swivel: improving embeddings by noticing what’s missing, arXiv preprint arXiv:1602.02215, 2016.
- 25.B. Chiu, G. Crichton, A. Korhonen, and S. Pyysalo, How to train good word embeddings for biomedical NLP, Proceedings of BioNLP16, p. 166, 2016.
- 26.Q. V. Le and T. Mikolov, Distributed representations of sentences and documents, in ICML, 2014, vol. 14, pp. 1188–1196.
- 27.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Information processing & management. 1988;24(5):513–523. doi: 10.1016/0306-4573(88)90021-0. [DOI] [Google Scholar]
- 28.P. Stenetorp, S. Pyysalo, G. Topić, T. Ohta, S. Ananiadou, and J. Tsujii, BRAT: a web-based tool for NLP-assisted text annotation, in Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics, 2012, pp. 102–107.
- 29.Artstein R, Poesio M. Inter-coder agreement for computational linguistics. Computational Linguistics. 2008;34(4):555–596. doi: 10.1162/coli.07-034-R2. [DOI] [Google Scholar]
- 30.Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 31.X. Ma and E. Hovy, End-to-end sequence labeling via bi-directional LSTM-CNNs-CRF, 2016.
- 32.L. van der Maaten and G. Hinton, Visualizing data using t-SNE, Journal of Machine Learning Research, vol. 9, no. Nov, pp. 2579–2605, 2008.
- 33.Reiner BI, Knight N, Siegel EL. Radiology reporting, past, present, and future: the radiologist’s perspective. Journal of the American College of Radiology. 2007;4(5):313–319. doi: 10.1016/j.jacr.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 34.C. L. Clarke, N. Craswell, and I. Soboroff, Overview of the TREC 2004 Terabyte Track, in TREC, 2004, vol. 4, p. 74.
- 35.Porter MF. An algorithm for suffix stripping. Program. 1980;14(3):130–137. doi: 10.1108/eb046814. [DOI] [Google Scholar]
- 36.M. F. Porter, Snowball: A Language for Stemming Algorithms. 2001.