Abstract
Objective:
To investigate the expression profile of circular RNAs (circRNAs) and proposed circRNA–microRNA (miRNA) regulatory network in atrial fibrillation (AF).
Methods:
Atrial tissues from patients with persistent AF with rheumatic heart disease and non-AF myocardium with normal hearts were collected for circRNA differential expression analyses by high-throughput sequencing. The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed to predict the potential functions of the differentially expressed genes and AF-related pathways. Co-expression networks of circRNA–miRNA were constructed based on the correlation analyses between the differentially expressed RNAs. Quantitative reverse transcription polymerase chain reaction (PCR) was performed to validate the results.
Results:
A total of 108 circRNAs were found to be differentially expressed in AF. Among them, 51 were up-regulated, and 57 were down-regulated. Dysregulated circRNAs were validated by quantitative real-time PCR. The GO and KEGG pathway enrichment analyses were executed to determine the principal functions of the significantly deregulated genes. Furthermore, we constructed correlated expression networks between circRNAs and miRNAs. circRNA19591, circRNA19596, and circRNA16175 interacted with 36, 28, and 18 miRNAs, respectively; miR-29b-1-5p and miR-29b-2-5p were related to 12 down-regulated circRNAs, respectively.
Conclusion:
Our findings provide a novel perspective on circRNAs involved in AF due to rheumatic heart disease and establish the foundation for future research of the potential roles of circRNAs in AF.
Keywords: circular RNAs, non-coding RNAs, atrial fibrillation, gene expression profile
Introduction
Growing evidence demonstrates an increased incidence and prevalence of atrial fibrillation (AF) (1). According to its pathogeny, AF can be divided into two categories: pulmonary vein (PV)-related AF and non-PV-related AF. Despite advances in medications and ablation technologies, the efficacy of current strategies for non-PV-related AF is suboptimal, reflecting that an improved understanding of arrhythmia mechanisms is urgently needed (2, 3). Currently, atrial dilatation, cellular hypertrophy, atrial fibrosis, inflammation, oxidative stress, apoptosis, calcium overload, loss of cell–cell contacts, altered autonomic tone, deposition of amyloid, protein catabolism, ion channel deficiency, posttranscriptional changes, and epigenetic factors are all thought to be involved in the electrophysiological and structural remodeling of AF (4-12). However, critical and initial mechanisms of AF are still poorly understood.
Non-coding RNAs (ncRNAs) comprise a class of RNA molecules that do not encode proteins but regulate protein expression (13), such as microRNAs (miRNAs), Piwi-interacting RNAs, long ncRNAs, circular RNAs (circRNAs), and endogenous siRNAs, and so on. It has been speculated that these ncRNAs are emerging key regulators of gene expression under physiological and pathological conditions (14, 15). Moreover, emerging data have shown that circRNAs, a novel type of endogenous non-coding RNAs, are involved in the pathophysiology of cardiovascular diseases (16, 17). However, their expression profile and circRNA–miRNA network in cardiac arrhythmia remains unclear. In the present study, we analyzed and predicted circRNA expression profiles in AF using whole transcriptome resequencing techniques.
Methods
Adult heart sample collection
The study was conducted in accordance with the Declaration of Helsinki guidelines. The Institutional Ethics Review Board of our hospital approved the study. Tissue samples were collected from the removed left atrial appendages of nine adult patients with rheumatic heart disease and persistent AF undergoing mitral valve replacement. Control samples of the left atrial appendages were obtained from organ donors with six normal hearts collected at the time of organ procurement with consent provided for research tissue collection. The consent to donate to research was obtained through the Transfer of Tissue Agreement of our institution. Patients with cardiac or pulmonary diseases were excluded from the study (Table 1). Each sample was preserved in an RNA stabilization reagent (RNA Safety International) and was subsequently stored at −80°C until use.
Table 1.
Baseline characteristics of the subjects
| Variable | AF group (n=9) | Non-AF group (n=6) |
|---|---|---|
| Age | 50.1±7.2 | 47.3±12.1 |
| Gender (%) | ||
| Female | 6 (66.7%) | 3 (50%) |
| Male | 3 (33.3%) | 3 (50%) |
| Left atrial diameter (mm) | 70.1±25.1† | 35.2±2.6 |
| Ejection fraction | 51.7±3.2 | 55.1±4.9 |
| Rheumatic heart disease | Yes | None |
| Hypertension | None | None |
| Hyperlipidemia | None | None |
| Diabetes mellitus | None | None |
| Coronary heart disease | None | None |
| Infectious disease | None | None |
| Connective tissue disease | None | None |
| Other autoimmune diseases | None | None |
| Other cardiovascular diseases | None | None |
Data are presented as mean±standard deviation and n (%).
P<0.01 (AF group vs. non-AF group).
AF - atrial fibrillation
RNA extraction and qualification
Total RNA was extracted from the atrial samples using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. RNA integrity was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The samples with an RNA integrity number ≥7 were subjected to the subsequent analysis. Total RNA was quantified by the NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA).
Library preparation and RNA-Seq
The cDNA libraries were constructed using TruSeq Stranded Total RNA with Ribo-Zero Gold according to the manufacturer’s instructions. Then, these libraries were sequenced on the Illumina HiSeq X Ten platform, and 150 bp paired-end reads were generated.
Detection, annotation, and quantification of circRNAs
RNA sequencing (RNA-Seq) data were analyzed using CircRNAs Identifier (CIRI), an algorithm for de novo circRNA identification (18). All alignment records in SAM file were generated by BWA-MEM40 and then analyzed by CIRI for searching the potential back-spliced junction reads that are made up of two segments that align to the reference genome in chiastic order. Junction reads and circRNA candidates in SAM files were scanned twice by CIRI. Finally, the identified circRNAs are output with annotation information.
Quantitative real-time PCR validation
The first strand of cDNA was synthesized by Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, UK). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using an iCycler iQ system (Bio-Rad, CA, USA) as described previously (19). The primer sequences were designed in the laboratory and synthesized by Generay Biotech (Generay, Shanghai, China) based on the mRNA sequences obtained from the National Center for Biotechnology Information database (Table 2). BLAST was used to verify the specificity of the PCR primers. Melting curve analysis was performed to validate the specific generation of the expected PCR product. The expression levels of circRNAs were normalized to ACTB and were calculated using the 2−ΔΔCt method.
Table 2.
Primers designed for qRT-PCR validation of selected lncRNAs, circRNAs, and mRNAs
| Gene symbol | Forward primer | Reverse primer | Product length (bp) |
|---|---|---|---|
| circRNA_20118 | CTTCAAGGCAAGATGCTCC | GCTATGAAAGTCCTCGTTGG | 94 |
| circRNA_17558 | CCAGGAGTGTTCAAGATGC | GGTACGGTACTTGATGTCG | 133 |
| circRNA_16688 | GTCACAAC G CATG CAACA | CTGAAAGGGTTGGGTTCATAG | 109 |
| circRNA_11058 | ACCACCAGCTAAAGTGTCA | ACTTTGGAGGTTCTTTGGC | 95 |
| circRNA_11017 | AAGGAAGTGGTCCCAGAAA | CACAATTCTTGAAGGTTCTAGC | 114 |
| circRNA_11109 | CCAAGAAGCTCATCCCAGA | CAG G CTTG ATGTCAAAGAAG G | 108 |
| ACTB | CCATCATGAAGTGTGACG | GCCGATCCACACGGAGTA | 185 |
GO and KEGG pathway enrichment analyses
Each circRNA was first annotated to linear host mRNA according to their position relationship on the chromosome. Then, using the linear host mRNA as the proxy of its related circRNAs, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were applied to investigate the potential functions of differentially expressed circRNAs. GO analysis was applied to annotate the genes with terms under biological process (BP), cellular component (CC), and molecular function (MF) (http://www.geneontology.org). KEGG pathway analysis was performed to explore the significant pathways of the differentially expressed genes (http://www.genome.jp/kegg/).
circRNA–miRNA co-expression network
We constructed a circRNA–miRNA network to reveal the interactions between circRNAs and miRNAs in AF pathogenesis. miRNA-targeted circRNAs were predicted through the miRanda software. Then, the interaction network was built and visually displayed using the Cytoscape software based on the screening of circRNA–miRNA gene pairs. A diamond node represents circRNA, and a circle node represents miRNA. Red and green colors represent up- and down-regulation, respectively. The significant nodes in a core position of the regulated network are potentially more associated with AF.
Statistical analysis
Data are presented as mean±standard error of the mean or n (%), unless otherwise indicated. Student’s t-test was used for analyzing two-group differences. DESeq package (version 1.18.0) of R language was used to determine the differential expression of circRNAs (20). |log2 Fold Change| >1.0 and p<0.05 were considered to indicate a statistically significant difference on sequence analysis.
Results
Expression profile of circRNAs
The genes with |log2 Fold Change| >1.0 and p<0.05 were considered to be up-regulated, and those with |log2 Fold Change| <−1.0 and p<0.05 were considered to be down-regulated. A total of 108 circRNAs were detected to be differentially expressed. Among them, 51 circRNAs were up-regulated, and 57 circRNAs were down-regulated in AF tissues compared with controls, respectively, of which the top 40 differently expressed circRNAs were listed in Table 3. Differentially expressed circRNAs with statistical significance between the two groups were identified using a volcano plot filtering (Fig. 1).
Table 3.
Top 40 differently expressed circRNAs in the AF group
| circRNA ID | |log2 fold change| | P-value | Regulation | Transcript_position | Gene |
|---|---|---|---|---|---|
| circRNA_00949|chr1:94458607_94491247_+ | 4.089 | 0.002 | Up | chr1:94418086_94518663_+ | ABCD3 |
| circRNA_13172|chr3:69287752_69313517_- | 3.405 | 0.008 | Up | chr3:69168782_69386304_- | FRMD4B |
| circRNA_15620|chr5:79396823_79447145_- | 3.259 | 0.042 | Up | chr5:79373824_79513836_- | HOMER1 |
| circRNA_14245|chr4:38089932_38118192_+ | 3.101 | 0.020 | Up | chr4:37891084_38139173_+ | TBC1D1 |
| circRNA_04241|chr12:19253940_19287556_+ | 3.090 | 0.006 | Up | chr12:19129692_19376400_+ | PLEKHA5 |
| circRNA_01452|chr1:180003174_180024582_+ | 2.876 | 0.039 | Up | chr1:179954773_180114875+ | CEP350 |
| circRNA_20118|chr9:111786793_111787947_+ | 2.818 | <0.001 | Up | chr9:111686175_111794992_- | C9orf84 |
| circRNA_08942|chr18:35846281_35852268_- | 2.701 | 0.037 | Up | ||
| circRNA_02905|chr10:95336521_95367699_- | 2.683 | 0.038 | Up | chr10:95311773_95389791_- | SORBS1 |
| circRNA_03391|chr11:22221097_22276199_+ | 2.621 | 0.019 | Up | chr11:22192513_22283357_+ | ANO5 |
| circRNA_02637|chr10:68142940_68161752_+ | 2.618 | 0.046 | Up | chr10:68106117_68212017_+ | MYPN |
| circRNA_11035|chr2:178670218_178688224_- | 2.500 | 0.048 | Up | chr2:178525989_178807423_- | TTN |
| circRNA_16688|chr6:54202105_54230917_+ | 2.267 | 0.004 | Up | chr6:54010979_54262761_+ | MLIP |
| circRNA_17648|chr7:18585281_18648683_+ | 1.907 | 0.038 | Up | chr7:18086942_18999521_+ | HDAC9 |
| circRNA_15410|chr5:50399107_50411383_- | 1.803 | 0.036 | Up | chr5:50396197_50441400_- | EMB |
| circRNA_06639|chr15:42827928_42878684_- | 1.71 4 | 0.044 | Up | chr15:42744338_42920809_- | TTBK2 |
| circRNA_11090|chr2:178678125_178678830_- | 1.71 1 | 0.025 | Up | chr2:178525989_178807423_- | TTN |
| circRNA_03059|chr10:113876521_113884380_+ | 1.607 | 0.044 | Up | chr10:113854632_113907974_+ | NHLRC2 |
| circRNA_17558|chr7:5641154_5652510_- | 1.499 | <0.001 | Up | chr7:5620041_5781730_- | RNF216 |
| circRNA_01695|chr1:219179147_219211752_+ | 1.329 | 0.014 | Up | chr1:219173878_219212863_+ | LYPLAL1 |
| circRNA_11174|chr2:178694599_178721202_- | -2.003 | 0.001 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_10998|chr2:178654445_178715774_- | -2.185 | 0.001 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_06953|chr15:63924816_63926093_- | -2.192 | 0.003 | Down | chr15:63907036_64046322_- | DAPK2 |
| circRNA_11058|chr2:178672635_178721202_- | -2.365 | 0.021 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_11040|chr2:178670218_178715774_- | -2.400 | 0.001 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_19591|chr9:13939661 _14021355_- | -2.520 | 0.013 | Down | ||
| circRNA_14783|chr4:113174416_113199109_+ | -2.559 | 0.013 | Down | chr4:112818083_113383740_+ | ANK2 |
| circRNA_16183|chr5:146254943_146258593_+ | -2.587 | 0.037 | Down | chr5:146203550_146289223_+ | RBM27 |
| circRNA_11109|chr2:178678125_178722134_- | -2.598 | 0.006 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_03961|chr11:115209574_115240420_- | -2.684 | 0.001 | Down | chr11:115173625_115504523_- | CADM1 |
| circRNA_11081|chr2:178674314_178715774_- | -2.714 | 0.002 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_11156|chr2:178689813_178722134_- | -2.950 | 0.049 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_11103|chr2:178678125_178715774_- | -3.010 | <0.001 | Down | chr2:178525989_178807423_- | TTN |
| circRNA_02368|chr10:24495147_24545103_+ | -3.013 | <0.001 | Down | chr10:24042336_24547840_+ | KIAA1217 |
| circRNA_16170|chr5:145866501_145935763_- | -3.173 | 0.002 | Down | chr5:145858387_145937176_- | GRXCR2 |
| circRNA_17137|chr6:123438063_123464983_- | -3.263 | 0.018 | Down | chr6:123216339_123637093_- | TRDN |
| circRNA_01283|chr 1:155926676_155927156_- | -3.408 | 0.041 | Down | chr1:155913043_155934442_- | KIAA0907 |
| circRNA_16169|chr5:145866501_145931677_- | -3.590 | 0.019 | Down | chr5:145858387_145937176_- | GRXCR2 |
| circRNA_18020|chr7:79652499_79671000_+ | -3.942 | 0.002 | Down | ||
| circRNA_11017|chr2:178662966_178715774_- | -4.367 | 0.001 | Down | chr2:178525989_178807423_- | TTN |
Figure 1.
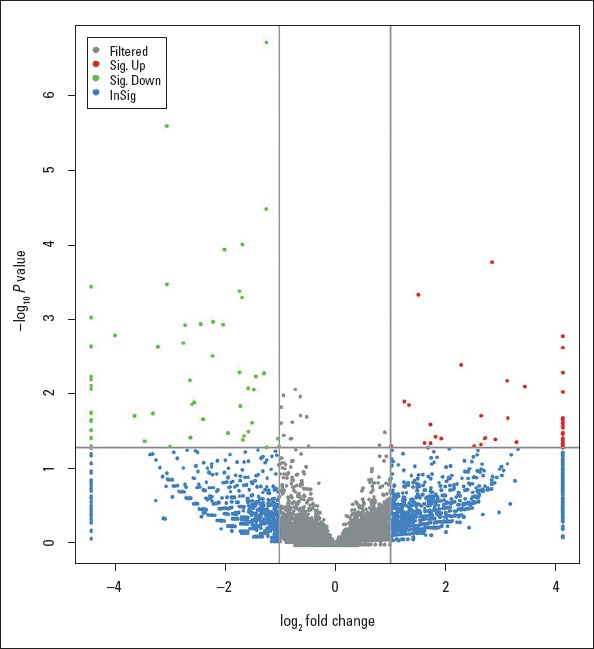
Volcano plot of circRNAs between AF and controls. Green plots represent down-regulated circRNAs. Red plots represent up-regulated circRNAs with absolute |log2 Fold Change| >1.0 and corrected P-value <0.05. Gray plots represent circRNAs with no significant difference. Blue plots represent circRNAs with |log2 Fold Change| >1.0 but with no significant difference
Validation of differentially expressed circRNAs
Six circRNAs (circRNA_20118, circRNA_17558, circRNA_16688, circRNA_11109, circRNA_11017, and circRNA_11058) were randomly selected for qRT-PCR validation and Sanger sequencing to validate the reliability of the sequencing results. As expected, the expression of the first three circRNAs was up-regulated, and the last three circRNAs were down-regulated in the AF samples versus control samples (Fig. 2), consistent with the sequencing results. Furthermore, the sequence of the circRNAs was identified by Sanger sequencing results (data not shown).
Figure 2.
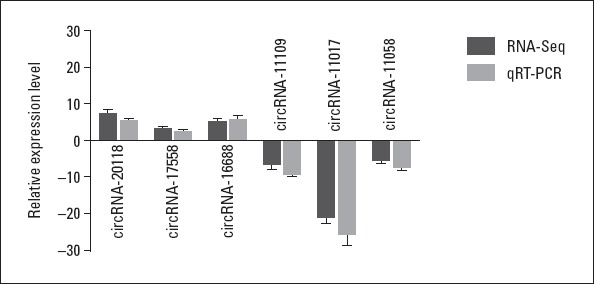
Comparison of circRNA expression levels between sequencing and qRT-PCR results. The Y-axis of the columns in the chart represents the log2-transformed fold changes computed from the sequencing and qRT-PCR data
GO and KEGG pathway analyses
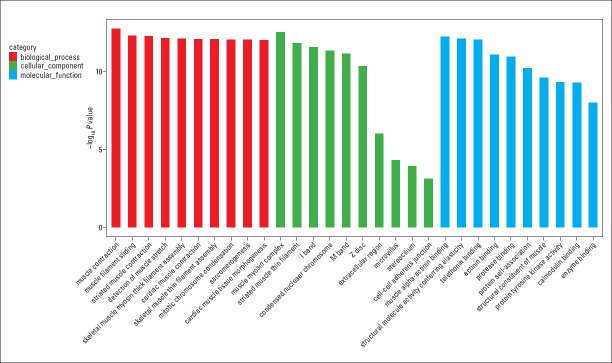
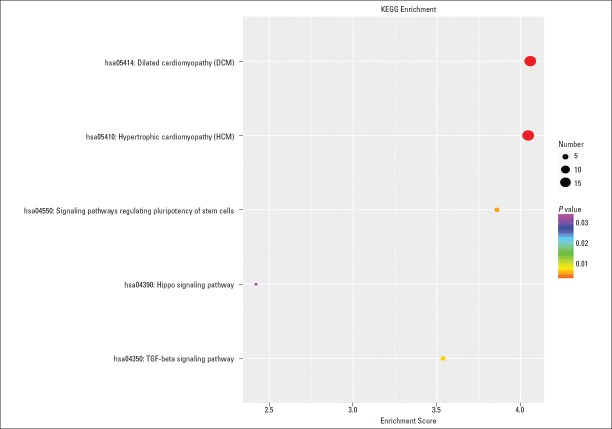
We conducted the GO and KEGG pathway analyses to predict the potential functions of circRNAs. The predicted functional terms with p-value <0.05 were selected and ranked by enrichment score [−log10 (p-value)]. The top 10 generally changed GO terms in all comparison groups were classified by BP, CC, and MF (Fig. 3). We found that the most significantly enriched BP term was muscle contraction (GO: 0006936). The most significantly enriched CC term was muscle myosin complex (GO: 0005859). The most significantly enriched MF term was muscle alpha-actinin binding (GO: 0051371). The pathway analysis indicated that five pathways might be involved in AF pathogenesis (Fig. 4). The most significantly involved pathways were dilated cardiomyopathy (DCM) (path: hsa05414) and hypertrophic cardiomyopathy (HCM) (path: hsa05410).
Figure 3.
GO enrichment analysis for dysregulated circRNA gene symbols. Most significantly enriched [−log10 (p-value)] GO terms of circRNA gene symbols according to biological process (red bar), cellular component (green bar), and molecular function (blue bar)
Figure 4.
KEGG pathway enrichment analysis of up- and down-regulated circRNAs with the top five enrichment score
Construction of the circRNA–miRNA network
We subsequently constructed a circRNA–miRNA network (Fig. 5) based on the sequencing results. In the network, a diamond node represents circRNA, and a circle node represents miRNA. There was a relatively intensive relationship; circRNA19591, circRNA19596, and circRNA16175 interacted with 36, 28, and 18 miRNAs, respectively; miR-29b-1-5p and miR-29b-2-5p were related to 12 down-regulated circRNAs, respectively (Table 4).
Figure 5.
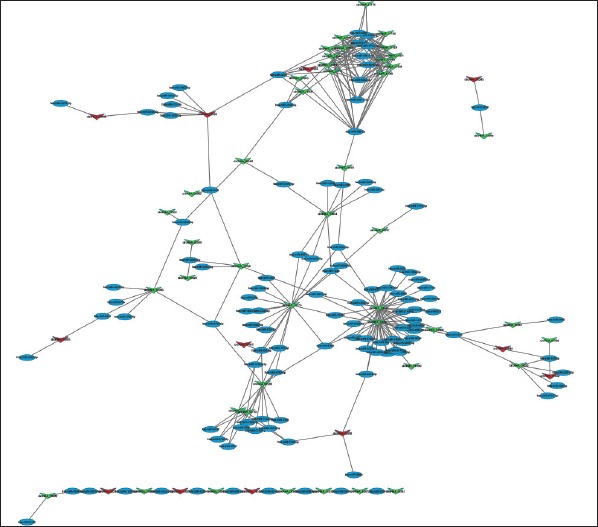
circRNA–miRNA regulatory network analysis of ncRNAs in patients with AF. Red diamonds represent up-regulated circRNAs. Green diamonds represent down-regulated circRNAs. Blue dots represent miRNA
Table 4.
Supposed circRNA-miR-29 axes
| miRNA ID | Term | List Hits | P-value | circRNA ID | Regulation |
|---|---|---|---|---|---|
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_10998 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11017 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11040 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11044 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11058 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11071 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11074 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11081 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11103 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11109 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11156 | Down |
| hsa-miR-29b-1-5p | 24 | 12 | <0.001 | circRNA_11108 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_10998 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11017 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11040 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11044 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11058 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11071 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11074 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11081 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11103 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11109 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11156 | Down |
| hsa-miR-29b-2-5p | 22 | 12 | <0.001 | circRNA_11108 | Down |
Discussion
AF is a heterogeneous disease, and its incidence is influenced by epidemiological factors and genetic predisposition (21). Despite the broad exploration of pathogeny in AF, (22-27) its cellular and biological mechanisms remain largely unknown. At present, PV isolation with cryoballoon and radiofrequency ablation is effective in the therapy of AF initiated by premature atrial contractions originated from PV and distribution of the muscle fascicle within the PV antrum. However, optimal clinical treatment for non-PV-related AF due to elusive pathogenesis is still lacking, such as AF in rheumatic heart disease.
circRNAs, a recently discovered new form of RNA, have been found to regulate transcription, which expanded our knowledge in understanding the complexity of non-coding RNA. Emerging evidence uncovered that endogenous circRNAs might regulate miRNA function as miRNA sponges to inhibit miRNA activity and be involved in transcriptional control (28, 29). circRNAs associated with related miRNAs or “circRNA–miRNA axes/network” are involved in multiple physiological and pathological processes, including the development of cardiovascular diseases (30-34). For example, heart-related circRNA acts as an endogenous miR-223 sponge to modulate the expression of miR-223 and apoptosis repressor with CARD domain, through which it regulates cardiomyocyte hypertrophy and heart failure (22, 23). In addition, Cdr1as, one of the circRNAs, plays proapoptotic roles during the development of myocardial infarction via function as miR-7 sponges (35). Moreover, circRNA circ-Foxo3 can promote cardiac senescence (34). However, to our knowledge, circRNA–miRNA axes/network in AF has not yet been reported.
In the present study, we investigated that circRNA expression profiles are significantly different between patients with AF and no AF. Fifty-one up-regulated and fifty-seven down-regulated circRNAs were significantly differentially expressed in patients with AF. We also predicted the potential functions of significant differential circRNAs using the GO and KEGG pathway analyses in patients with AF. GO analysis revealed that the main BPs are correlated with the structure or function of muscle contraction, such as cytoskeleton of cardiomyocytes. Interestingly, KEGG pathway analysis also indicates that there is molecular crosstalk between AF and cardiomyopathy, especially DCM and HCM, which may reveal that these three groups of patients possibly share a common circRNA-target network. Moreover, according to the KEGG enrichment scores, signaling pathway regulating pluripotency of stem cells was detected, which indicated that circRNAs may contribute to the homeostatic mechanisms of AF.
Furthermore, we investigated the possible circRNA–miRNA axes/network in AF. A network of significantly dysregulated circRNAs with their adjacent miRNA was delineated based on the binding capacity of circRNAs on miRNAs, which might provide a new clue for elucidating the underlying mechanism of AF. Figure 5 shows that the 36, 28, and 18 nearby miRNAs corresponding to circRNA19591, circRNA19596, and circRNA16175, respectively, were identified, and these three circRNAs were all down-regulated and might be relatively potential regulators of gene expressions by interacting with the corresponding endogenous miRNAs in AF. In addition, accumulating studies have demonstrated a functional role for miRNAs in the pathophysiology of AF (36, 37). Among them, miR-29 is considered to be a biomarker and/or therapeutic target of AF due to the contribution to atrial fibrotic remodeling (38). Intriguingly, for the first time, our network displayed that miR-29b-1-5p and miR-29b-2-5p have interactions with 24 down-regulated circRNAs. Therefore, it is hypothesized that these 24 circRNAs may be directly or indirectly involved in structural remodeling in AF. However, the detailed mechanisms still need to be explored, and functional studies are required to elucidate their roles in AF. In our study, circRNA–miRNA network possibly provides a new perspective for competitiveness of AF. Further research on these circRNA–miRNA axes/network is being conducted in our laboratory.
Study limitations
Our study had a limited number of patients analyzed. Moreover, we just preliminarily investigated the expression profile of circRNAs in AF, and functional protein structures, protein–protein interactions, and detailed molecular pathways in the AF process should be further explored.
Conclusion
The incidence of AF is increasing. The curative effect of non-PV-related AF may not be desirable due to its unclear mechanism. We gain a landscape of circRNA expression and constructed a circRNA–miRNA network that might be associated with the development of AF. These results suggest that specific circRNAs could be valuable for AF therapy due to rheumatic heart disease. These studies might enrich our understanding of the pathogenesis of AF and enable further research on the pathogenesis of AF.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 81703407 and 31370996) and scientific and technological project in Shaanxi province (no. 2016SF-289).
Footnotes
Conflict of interest: None declared.
Peer-review: Externally peer-reviewed.
Authorship contributions: Concept – H.C., Y.Y.; Design – H.C., Y.Y.; Supervision – M.H., M.L., L.T., L.W., M.Z., H.C., Y.Y.; Fundings – X.W., Y.Y.; Materials – M.H., M.L., L.T., L.W., M.Z.; Data collection &/or processing – M.H., M.L., L.T., L.W., M.Z.; Analysis &/or interpretation – M.H., M.L., L.T., L.W.; Literature search – M.H., H.C., Y.Y.; Writing – M.H., X.W., Y.Y.; Critical review – M.H., M.L., L.T., L.W., M.Z.
References
- 1.Staerk L, Sherer JA, Ko D, Benjamin EJ, Helm RH. Atrial Fibrillation: Epidemiology, Pathophysiology, and Clinical Outcomes. Circ Res. 2017;120:1501–17. doi: 10.1161/CIRCRESAHA.117.309732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuck KH, Fürnkranz A, Chun KR, Metzner A, Ouyang F, Schlüter M, et al. FIRE AND ICE Investigators. Cryoballoon or radiofrequency ablation for symptomatic paroxysmal atrial fibrillation: reintervention, rehospitalization, and quality-of-life outcomes in the FIRE AND ICE trial. Eur Heart J. 2016;37:2858–65. doi: 10.1093/eurheartj/ehw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morillo CA, Verma A, Connolly SJ, Kuck KH, Nair GM, Champagne J, et al. Radiofrequency ablation vs antiarrhythmic drugs as first-line treatment of paroxysmal atrial fibrillation (RAAFT-2): a randomized trial. JAMA. 2014;311:692–700. doi: 10.1001/jama.2014.467. [DOI] [PubMed] [Google Scholar]
- 4.Nattel S, Dobrev D. Electrophysiological and molecular mechanisms of paroxysmal atrial fibrillation. Nat Rev Cardiol. 2016;13:575–90. doi: 10.1038/nrcardio.2016.118. [DOI] [PubMed] [Google Scholar]
- 5.Schotten U, Verheule S, Kirchhof P, Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev. 2011;91:265–325. doi: 10.1152/physrev.00031.2009. [DOI] [PubMed] [Google Scholar]
- 6.Jalife J, Kaur K. Atrial remodeling, fibrosis, and atrial fibrillation. Trends Cardiovasc Med. 2015;25:475–84. doi: 10.1016/j.tcm.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reumann M, Bohnert J, Osswald B, Hagl S, Doessel O. Multiple wavelets, rotors, and snakes in atrial fibrillation--a computer simulation study. J Electrocardiol. 2007;40:328–34. doi: 10.1016/j.jelectrocard.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Haïssaguerre M, Jaïs P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–66. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 9.Tsai CT, Lai LP, Hwang JJ, Lin JL, Chiang FT. Molecular genetics of atrial fibrillation. J Am Coll Cardiol. 2008;52:241–50. doi: 10.1016/j.jacc.2008.02.072. [DOI] [PubMed] [Google Scholar]
- 10.Rossi A, Enriquez-Sarano M, Burnett JC, >Jr, Lerman A, Abel MD, Seward JB. Natriuretic peptide levels in atrial fibrillation: a prospective hormonal and Doppler-echocardiographic study. J Am Coll Cardiol. 2000;35:1256–62. doi: 10.1016/s0735-1097(00)00515-5. [DOI] [PubMed] [Google Scholar]
- 11.Hancox JC, James AF, Marrion NV, Zhang H, Thomas D. Novel ion channel targets in atrial fibrillation. Expert Opin Ther Targets. 2016;20:947–58. doi: 10.1517/14728222.2016.1159300. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Choi E, Cha MJ, Hwang KC. Looking into a conceptual framework of ROS-miRNA-atrial fibrillation. Int J Mol Sci. 2014;15:21754–76. doi: 10.3390/ijms151221754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 14.Daniel C, Lagergren J, Öhman M. RNA editing of non-coding RNA and its role in gene regulation. Biochimie. 2015;117:22–7. doi: 10.1016/j.biochi.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 15.Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177–86. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in Cardiovascular Disease: An Overview. Biomed Res Int. 2017;2017 doi: 10.1155/2017/5135781. 5135781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Ding W, Sun T, Tariq MA, Xu T, Li P, et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018;285:220–32. doi: 10.1111/febs.14191. [DOI] [PubMed] [Google Scholar]
- 18.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Li YL, Zhang CC, Cui W, Wang X, Xia Y, et al. Inhibition of Toll-like receptor 2 reduces cardiac fibrosis by attenuating macrophage-mediated inflammation. Cardiovasc Res. 2014;101:383–92. doi: 10.1093/cvr/cvt258. [DOI] [PubMed] [Google Scholar]
- 20.Anders S, Huber W. European Molecular Biology Laboratory (EMBL) Heidelberg, Germany: 2012. Differential expression of RNA-Seq data at the gene level–the DESeq package. [Google Scholar]
- 21.Khaji A, Kowey PR. Update on atrial fibrillation. Trends Cardiovasc Med. 2017;27:14–25. doi: 10.1016/j.tcm.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–11. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 23.He F, Xu X, Yuan S, Tan L, Gao L, Ma S, et al. Oxidized Low-density Lipoprotein (ox-LDL) Cholesterol Induces the Expression of miRNA-223 and L-type Calcium Channel Protein in Atrial Fibrillation. Sci Rep. 2016;6:30368. doi: 10.1038/srep30368. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Wang T, Wang B. Identification of microRNA-mRNA interactions in atrial fibrillation using microarray expression profiles and bioinformatics analysis. Mol Med Rep. 2016;13:4535–40. doi: 10.3892/mmr.2016.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Liu L, Hu J, Song L. MicroRNA Regulatory Network Revealing the Mechanism of Inflammation in Atrial Fibrillation. Med Sci Monit. 2015;21:3505–13. doi: 10.12659/MSM.895982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu XJ, Zou LH, Jin JH, Xiao F, Li L, Liu N, et al. Long noncoding RNAs and novel inflammatory genes determined by RNA sequencing in human lymphocytes are up-regulated in permanent atrial fibrillation. Am J Transl Res. 2017;9:2314–26. [PMC free article] [PubMed] [Google Scholar]
- 27.Tribulova N, Egan Benova T, Szeiffova Bacova B, Viczenczova C, Barancik M. New aspects of pathogenesis of atrial fibrillation: remodelling of intercalated discs. J Physiol Pharmacol. 2015;66:625–34. [PubMed] [Google Scholar]
- 28.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–15. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–8. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035–45. doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao YJ, Wang JH, Fu B, Ma MX, Li BX, Huang Q, et al. Effects of 3-aminobenzamide on expressions of poly (ADP ribose) polymerase and apoptosis inducing factor in cardiomyocytes of rats with acute myocardial infarction. Chin Med J (Engl) 2009;122:1322–7. [PubMed] [Google Scholar]
- 33.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 36.Masè M, Grasso M, Avogaro L, D'Amato E, Tessarolo F, Graffigna A, et al. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep. 2017;7:41127. doi: 10.1038/srep41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulte C, Karakas M, Zeller T. microRNAs in cardiovascular disease - clinical application. Clin Chem Lab Med. 2017;55:687–704. doi: 10.1515/cclm-2016-0576. [DOI] [PubMed] [Google Scholar]
- 38.Dawson K, Wakili R, Ordög B, Clauss S, Chen Y, Iwasaki Y, et al. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127:1466–75. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]