Abstract
Durotaxis is a type of directed cell migration in which cells respond to a gradient of extracellular stiffness. Using automated tracking of positional data for large sample sizes of single migrating cells, we investigated 1) whether cancer cells can undergo durotaxis; 2) whether cell durotactic efficiency varies depending on the regional compliance of stiffness gradients; 3) whether a specific cell migration parameter such as speed or time of migration correlates with durotaxis; and 4) whether Arp2/3, previously implicated in leading edge dynamics and migration, contributes to cancer cell durotaxis. Although durotaxis has been characterized primarily in nonmalignant mesenchymal cells, little is known about its role in cancer cell migration. Diffusible factors are known to affect cancer cell migration and metastasis. However, because many tumor microenvironments gradually stiffen, we hypothesized that durotaxis might also govern migration of cancer cells. We evaluated the durotactic potential of multiple cancer cell lines by employing substrate stiffness gradients mirroring the physiological stiffness encountered by cells in a variety of tissues. Automated cell tracking permitted rapid acquisition of positional data and robust statistical analyses for migrating cells. These durotaxis assays demonstrated that all cancer cell lines tested (two glioblastoma, metastatic breast cancer, and fibrosarcoma) migrated directionally in response to changes in extracellular stiffness. Unexpectedly, all cancer cell lines tested, as well as noninvasive human fibroblasts, displayed the strongest durotactic migratory response when migrating on the softest regions of stiffness gradients (2–7 kPa), with decreased responsiveness on stiff regions of gradients. Focusing on glioblastoma cells, durotactic forward migration index and displacement rates were relatively stable over time. Correlation analyses showed the expected correlation with displacement along the gradient but much less with persistence and none with cell speed. Finally, we found that inhibition of Arp2/3, an actin-nucleating protein necessary for lamellipodial protrusion, impaired durotactic migration.
Introduction
Directional cell migration refers to the ability of a cell to polarize and move persistently in a specified direction, generally in response to an extracellular signal that biases the direction of movement. Signals in the extracellular space can take on many forms and can act to either attract or repel the cell. Chemotaxis, the most thoroughly studied and best-characterized mechanism of directed migration, involves a response to diffusible chemicals. Other factors—including substrate-bound gradients of extracellular proteins (haptotaxis), electric fields (galvanotaxis), contact guidance, and changes in substrate rigidity (durotaxis)—have also been shown to direct the movement of cells (for reviews, especially in cancer, see (1, 2, 3, 4, 5, 6, 7, 8, 9, 10)). Directed migration can be contrasted with the chemokinetic, nondirectional migration that cells typically exhibit in homogeneous environments. Cells responding to a chemical stimulus can polarize and temporarily move directionally in an environment that lacks any gradient condition. However, the absence of a sufficiently strong external gradient to “bias” the direction of cell movement results in a population of cells that migrate in random directions.
Durotaxis is a mechanism of directional migration in which a cell responds to an extracellular gradient of stiffness (6, 11). Typically, durotactic migration involves cell movement toward regions of increasing stiffness across steps or up gradients of increasingly stiff substrates (6, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21). Only durotactic migration toward increasing stiffness has been thoroughly documented; however, there is speculation that durotaxis toward increasingly soft substrates may occur (19, 20). Mechanisms proposed to underlie durotaxis of fibroblastic (mesenchymal) cells include contractile mechanosensing, probing of the local substrate by filopodia, and focal adhesion signaling (15, 17, 21, 22, 23, 24).
Cancer cell migration is important for expanding tumor margins and initiating the metastatic cascade. Cells escape from the primary tumor through a variety of migratory mechanisms (25), and they enter the circulatory or lymphatic system. General hallmarks of malignant and normal cell migration on tissue culture substrates can be described as a cyclical series of sequential steps. These steps include 1) protrusion of the leading edge of the cell, 2) adhesion of the leading edge to the extracellular substrate, 3) forward translocation of the cell body, and 4) retraction of the trailing edge (26, 27).
The protrusive leading edge of a migrating cell is often characterized by a broad, sheet-like lamellipodium, which can also contain spike-like filopodia. These protrusions result from globular actin incorporation onto the barbed end of actin filaments during actin polymerization (28, 29). Arp2/3 is a seven-protein complex responsible for initiating the growth of an extensive network of these actin filaments through increased actin branching. This polymerization against the plasma membrane helps push it forward during lamellipodia and filopodia formation (30, 31, 32, 33), thereby playing a fundamental role in this crucial step of migration. Furthermore, Arp2/3 has been implicated in enabling cells to respond by directional migration to chemotactic (EGF but not PDGF) and haptotactic gradients (34, 35). However, the role of Arp2/3 in durotaxis remains unknown.
There is a strong correlation between stiffening of the tumor microenvironment and activation of epithelial-to-mesenchymal transition pathways, tumor growth, and increased malignancy (e.g., see (36, 37, 38, 39)). Consequently, cancer cells might employ durotaxis in the process of metastatic dissemination, but there has been little in vitro or in vivo evidence for the durotactic capacity of cancer cells to date. If they were capable of durotaxis, another unanswered question is whether cancer cells respond to a gradient of stiffness at a physiological range of stiffness associated with the compliance characteristics of different tissues throughout the body (40, 41).
In this study, we first developed software to automate cell tracking to analyze the migratory phenotypes of cells exposed to gradient conditions with sample sizes for each condition of >100–800 cells. We then evaluated commonly used cancer cell lines to determine their capacity to undergo durotactic migration. To gain insight into their mechanisms of durotactic migration, we next compared the efficiency of durotactic migration in regions of different stiffness and discovered that single-cell durotaxis of all lines tested was surprisingly most effective in the softest substrate environments. Nonmalignant, noninvasive human fibroblasts also showed this enhanced durotactic efficiency on the soft region of substrates. We then focused on more mechanistic studies using the glioblastoma cell line U87-MG, which exhibited particularly robust durotaxis. Correlation analyses revealed the expected strong positive correlation with net distance migrated along the gradient but a surprisingly weak correlation with directional persistence of migration (straightness) and absence of any correlation with migration speed. On soft substrates, these cells displayed prominent membrane dynamics at the periphery of migrating cells. Arp2/3 is known to be important for leading edge dynamics, and chemically inhibiting this protein complex was found to disrupt lamellipodia and filopodia and to inhibit durotaxis. Our studies provide new insights into this unique mechanism of directed cell migration.
Materials and Methods
Cell culture
U87-MG (“U87”), T98G, MDA-MB-231, and HT1080 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, high glucose without phenol red or sodium pyruvate, cat. no. SH30284.01; HyClone, Logan, UT) with 10% fetal bovine serum (cat. no. SH30070.03; HyClone), 200 U/mL penicillin/streptomycin, and 200 μg/mL L-glutamine (Gibco, Grand Island, NY) at 37°C with 10% CO2. BJ-5ta normal fibroblasts (CRL-4001; ATCC, Manassas, VA) were cultured in a 4:1 mixture of DMEM and Medium 199 supplemented with 0.01 mg/mL hygromycin B, 10% fetal bovine serum, and 200 U/mL penicillin/streptomycin.
Adenoviral infection
For live-cell confocal imaging, 3 × 105 U87-MG cells were plated into 35-mm cell-culture dishes with TagGFP2-LifeAct adenovirus (Cat. No. 60121; Ibidi, Fitchburg, WI) at a multiplicity of infection of ∼10 overnight. Cells were rinsed with fresh medium the next morning and then replated onto polyacrylamide gel substrates.
Cell migration
Glass-bottomed dishes were seeded sparsely with 1.5 × 104 cells/35-mm dish and cultured overnight. The next day, cells were stained with 1 μg/mL Hoechst for 20 min at 37°C. After transfer to fresh medium, cells were acclimated to the temperature- and humidity-controlled imaging chamber for 1 h before time-lapse imaging. Time-lapse videos were acquired for 24 h at 15-min intervals using a Zeiss Axiovert 135 microscope with a 10 × 0.3 NA phase objective (Carl Zeiss Microscopy, Thornwood, NY). The custom software termed FastTracks (42) was used to automate acquisition of positional coordinates of individual cell nuclei and to assemble cell trajectories. To eliminate analysis of nonmigratory cells in an unbiased manner, the lower quartile of cells for Euclidean displacement was eliminated from the data set before analysis of cell migration.
The metrics for cell migration were calculated separately for each cell that was analyzed, and then all cell values were averaged. To calculate FMI, the displacement value (calculated for the X or Y axis separately) was divided by the total displacement the cell underwent (total track length, not the Euclidean distance). All FMI values for individual cells were then averaged.
Angular displacement was calculated based on the initial and final position of each cell (see Fig. 1 C). Speed was determined as the sum of individual displacements divided by total time, which results in the same final result as determining the speed for each time point and then averaging all values. Persistence was calculated as the Euclidean displacement (determined using first and last positions) divided by total distance.
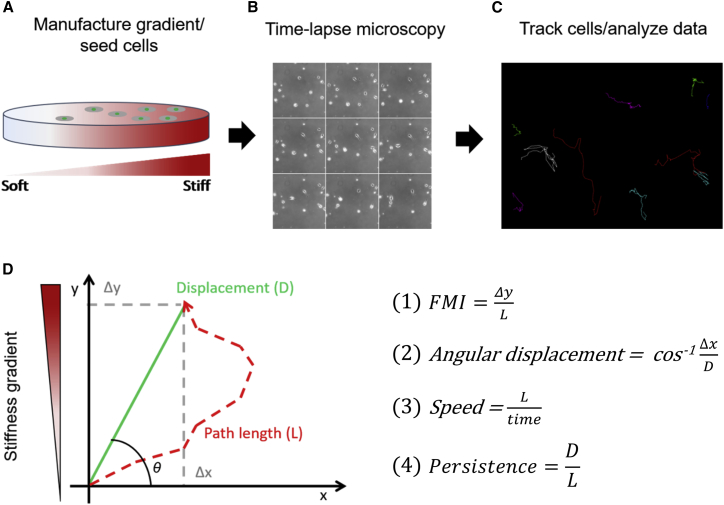
Figure 1.
Procedures for quantifying durotactic migration by a population of single human cancer cells. (A) A polyacrylamide stiffness gradient to which fibronectin is covalently attached to provide an adhesive substrate is seeded with a population of cells at a sufficiently low density to allow (B) imaging of individual migrating cells using phase-contrast or fluorescence time-lapse microscopy for (C) computer-based analysis of migratory track using FastTracks software (42). (D) Graphical schematic of a cell trajectory (red dashed line) and variables used to characterize migratory phenotypes. The equations used to calculate cell migration parameters are listed on the right. To see this figure in color, go online.
Live-cell spinning-disk confocal imaging
For all live-cell fluorescence experiments, DMEM without phenol red or FluoroBrite DMEM (GIBCO) were used, with each supplemented with a 1:100 ratio of Oxyfluor (Oxyrase, Mansfield, OH) and 10 mM DL-lactate (Sigma, St. Louis, MO) to reduce photobleaching and phototoxicity, 10% fetal bovine serum, 200 U/mL penicillin/streptomycin, and 200 μg/mL L-glutamine. U-87 MG cells were imaged with a modified Yokogawa spinning-disk confocal scan head (CSU-21: modified by Spectral Applied Research, Richmond Hill, ON) on an automated Olympus IX-81 microscope using a 30×, 60×, or 100× SAPO-Chromat silicone oil objectives (NA 1.15, 1.3, and 1.35, respectively). A custom laser launch (built by A.D.D.) equipped with 445-nm (80 mW; Vortran Laser Technology, Sacramento, CA), 488-nm (150 mW; Coherent, Santa Clara, CA), 514-nm (150 mW; Coherent), 568-nm (100 mW; Coherent), and 642-nm (110 mW; Vortran Laser Technology) diode lasers supplied excitation wavelengths. A Gooch and Housego AOTF controlled shuttering and intensity for 488, 514, and 568 lines. 445 and 642 lines were shuttered, and intensity was controlled via TTL and direct voltage steps, respectively. The primary dichroics (442/568/647 and 405/488/568/647) were from Semrock (Rochester, NY). Images were captured using a backthinned EM CCD camera in 16-bit format using the 10 MHz digitization setting (Photometrics, Tucson, AZ). EM gain was set at 600–800 (3×) with exposure times 50–100 ms per image acquired every 1 min for 1.5–3 h. A motorized Z-piezo stage was used to rapidly capture Z-stacks every 0.5 microns over a Z-distance of 6–8 microns (ASI Imaging, Eugene, OR). An environmental chamber surrounding the microscope maintained cells at a constant 37°C, with 10% CO2 and ∼50% humidity (Precision Plastics, Beltsville, MD). All components were controlled with MetaMorph imaging software (Molecular Devices, Downington, PA).
Glass substrate treatment
Nunc glass-base 27-mm culture dishes (cat. no. 150682; Thermo Fisher Scientific, Waltham, MA) were treated with 3-(trimethoxysilyl)propyl methacrylate (cat. no. M6514; Sigma-Aldrich), ethanol, and 10% glacial acetic acid in water (ultrapure molecular biology grade water, cat. no. 351-029-131; Quality Biological, Gaithersburg, MD) at a 1:200:6 ratio. This solution was added to glass-bottomed dishes for 5 min, rinsed twice with ethanol, and then allowed to dry.
Polyacrylamide gels
Preparation of gradient gels was based on the protocol of Tse and Engler (43). Briefly, 40% acrylamide monomer (AAm) and 2% N,N′-methylenebisacrylamide (BIS) stock solutions (cat. no. 161-0140 and 161-0142, respectively; Bio-Rad, Hercules, CA) were used to prepare a 10% AAm and 0.3% BIS solution in phosphate-buffered saline (PBS). 0.5% w/v Irgacure 2959 (cat. no. 410896; Sigma) was added to catalyze cross-linking when exposed to UV light. To generate high-stiffness gradient gels, the acrylamide concentrations were increased to 15% AAm and 1.0% BIS. 30 μL of gel solution was added to 3-(trimethoxysilyl) propyl methacrylate-treated dishes and covered with 25 mm glass coverslips that had been presilanized with dichlorodimethylsilane. Dishes were then placed on top of a gradient photomask generated as follows: the photomask template was designed with the Adobe Photoshop Gradient Tool to be the same dimensions as the gel and with a linearly increasing grayscale density of 0–70%. The photomask was generated using a Xerox Phaser 6280 laser printer to print the linear gradient of ink density at 1200 dpi onto laser printer transparency film. The dishes were exposed to 302-nm ultraviolet (UV) light produced by a Maestrogen transilluminator (4 mW/cm2; Hsinchu City, Taiwan; light intensity was measured to be 3.3 joules below the coverslip and 2.7 J above it) for 2.5 min to polymerize the gradient gel, then immediately immersed in PBS, and the top coverslip was removed. Gels were then washed several times with PBS.
Soft and stiff uniform polyacrylamide gels were prepared by combining 5 or 10% AAm, respectively, with 0.1% BIS. 1:1000 N,N,N′,N′-tetramethylethylenediamine and 1:100 10% ammonium persulfate were added to the monomer solutions, and 30 μL was pipetted onto a treated dish and covered with a silanized coverslip. After 25 min of polymerization, PBS was added to the dishes, and the glass coverslip was detached from the gel.
AFM measurements
Microindentations were performed with an atomic force microscopy (AFM) instrument (Bioscope Catalyst; Bruker-Nano, Santa Barbara, CA) positioned on top of an inverted optical microscope (IX-71; Olympus, Tokyo, Japan). We used silicon nitride AFM cantilevers (MSCT; Bruker-Nano) with nominal stiffness of 0.03 and 0.12 nN/nm and silicon cantilevers (FESP; Bruker-Nano) with nominal stiffness of 2.8 nN/nm, depending on the expected gel stiffness range. The stiffness of each probe used was measured using the thermal calibration utility provided with the instrument software (NanoScope v8.15). Glass microspheres, 20 μm in diameter, were attached to each cantilever using UV-curable epoxy and were used as the indentation probes. Gradient gels were positioned on the optical microscope stage, and indentations were performed parallel to the expected stiffness gradient and across the whole width of the gels. Indentations were performed at 30-μm intervals. The acquired force-indentation curves were analyzed using custom-written software (MATLAB; The MathWorks, Natick, MA) to extract the elastic modulus at each point probed and to plot the final results based on n = 3 gradient gels. Regions measuring 160 × 160 μm2 for each uniform gel were probed at points on a 16 × 16 μm grid (10-μm steps in both X and Y directions), and histograms of the distributions of modulus values for individual gels (n = 4 gels for soft and stiff conditions) were plotted to evaluate local elasticity variability and to confirm that the extracellular matrix protein conjugation did not alter elastic modulus.
Extracellular matrix protein conjugation
A 1 mg/mL solution of Sulfo-SANPAH (SS) (cat. no. 22589; Pierce, Waltham, MA, Thermo Fisher Scientific) in PBS was added to each gel and immediately exposed to 365-nm UV light for 8 min using a desktop Spectroline UV lamp (model ENF-280C; Westbury, NY). The same Sulfo-SANPAH treatment and UV illumination were repeated for a second time. Gels were immediately washed twice in PBS, then a 0.1 mg/mL solution of human plasma fibronectin (isolated as described by Akiyama (44)) was added directly to the gels for 4 h. Gels were then washed several times with PBS and incubated with fresh media 30 min before plating cells. Uniformity of conjugation across the gradient gels was confirmed by immunofluorescence staining for fibronectin using a rabbit anti-human plasma fibronectin polyclonal antibody and laser scanning confocal microscopy.
Statistics
Statistical analysis was performed using the MATLAB Statistics and Machine Learning toolbox (The MathWorks) as detailed in each figure legend.
Results
Durotaxis assay
We used the protocol initially established by Tse and Engler (43) to generate gradients that spanned the range of physiological stiffness encountered by cells across many tissues, followed by uniform conjugation of fibronectin protein to the surface of the polyacrylamide gradient gels to mediate cell adhesion (Fig. S1). To evaluate cancer cell durotaxis, cells nuclei were fluorescently labeled with Hoechst, cultured on gradients, and then imaged with time-lapse fluorescence microscopy (Fig. 1, A and B). Fluorescent nuclei were tracked using custom software (42) that enabled unbiased acquisition of cell trajectories for numerous cells (Fig. 1 C). Two different measures of directional migration were then used to quantify cell migration. A forward migration index (FMI) (Fig. 1; Eq. 1) was used to measure the displacement of a cell parallel to the gradient relative to the cell’s total path length. Angular displacement (Fig. 1; Eq. 2) was used to assess the cell’s angular trajectory over the interval that the cell was tracked. Additional statistics such as cell speed (Fig. 1; Eq. 3) and persistence (Fig. 1; Eq. 4), a measure of cell directionality, were also quantified for each cell.
Cancer cells undergo durotaxis
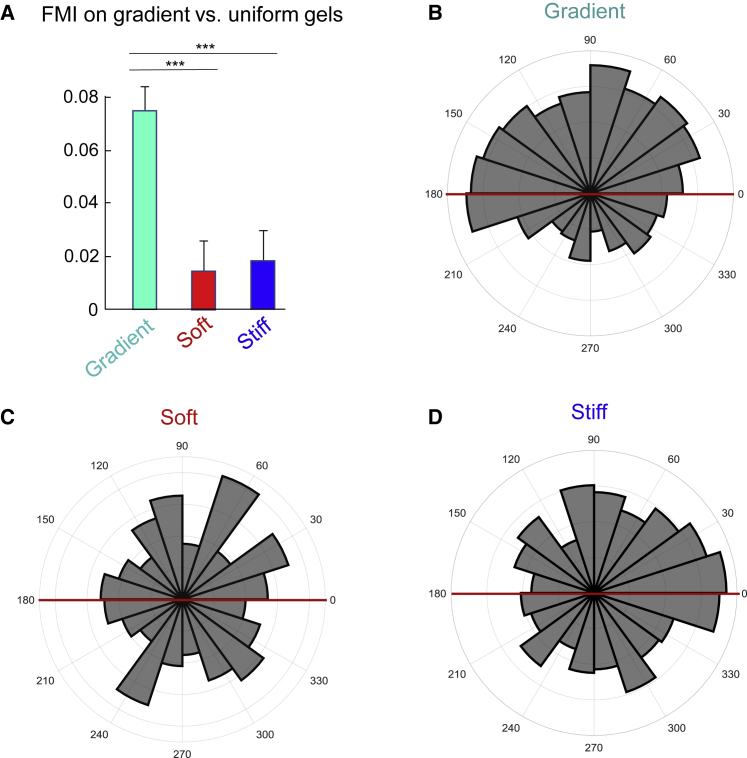
Cancer cell durotaxis was evaluated initially using U87 glioblastoma cells. Representative examples of 8-h migration tracks of cells migrating at locations along the entire gradient but plotted from a single origin are shown in Fig. 2 A. As a control, cells were also cultured on uniform soft and stiff gels that reflect the modulus values associated with the soft (3.5 kPa) and stiff (17.0 kPa) portions of our gradient condition. As can be seen by examination of migratory tracks and by quantification of FMI, U87 cells exhibited a moderate durotactic phenotype in response to the gradient with an average FMI of 0.07 (Fig. 3 A). Although the magnitude of directional migration was not large, it differed substantially from the minimal FMI values obtained for cells cultured on uniformly soft or stiff gels (Fig. 3 A). That is, on uniform gels, U87 cells exhibited a pattern of chemokinetic migration characterized by an absence of biased directional movement, as indicated by an FMI that approximated zero (Fig. 3 A) and uniformly dispersed angular displacements (Fig. 3, C and D). In clear contrast, the angular displacements of cells on a gradient indicated movement toward increasingly stiff regions (Fig. 3 B). To evaluate this qualitative assessment statistically, we employed Rayleigh hypothesis tests to evaluate the significance of the directionality of angular data on a 0–360° scale. We found that cell trajectories were significantly biased toward increasingly stiff regions of the gradient, whereas movement of cells on soft or stiff gels was relatively uniform (Fig. 3 B compared to Fig. 3, C and D).
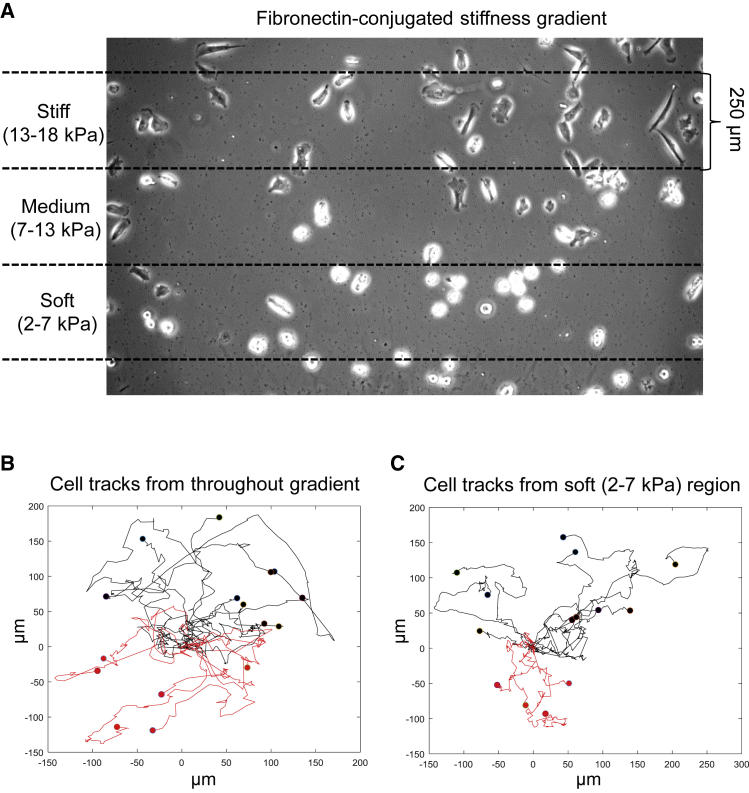
Figure 2.
Tracking migration of cancer cells undergoing durotaxis in response to a stiffness gradient. (A) U87 glioblastoma cells seeded on a polyacrylamide stiffness gradient to which fibronectin had been covalently coupled. Dashed lines indicate the 250-μm-wide soft, medium, and stiff regions; phase-contrast microscopy. (B) Representative tracks of U87 glioblastoma cells migrating on a stiffness gradient, with greater stiffness toward the top. Black tracks indicate cells with net translocation toward the stiffer end of the gradient after 8 h (10 of 16 cells in this example). Red tracks indicate cells with net translocation against the gradient. (C) Representative tracks of U87 cells migrating on the soft (2–7 kPa) region of a stiffness gradient. Note that a higher proportion of cells on this soft region of the gradient migrate toward the stiffer end of the gradient substrate (black tracks; 10 of 14 cells) compared to cells migrating opposite to the gradient (red). To see this figure in color, go online.
Figure 3.
Quantification of cancer cell durotaxis in response to a stiffness gradient. (A) Comparison of FMI values for U87 glioblastoma cells cultured on gradient versus uniform soft or uniform stiff substrates to quantify the effects of a stiffness gradient on directed migration. (B–D) Angular displacement plots for U87 cells cultured on a gradient (B), uniform soft (C), or uniform stiff (D) substrates as a secondary measure of directional migration. Error bars represent mean ± standard error (SE). Data were obtained from n = 572, n = 197, or n = 167 cells from N ≥ 4 biological replicates (independent experiments) for gradient, soft, and stiff conditions, respectively. FMI p-values were calculated using one-way ANOVA and Tukey’s post hoc test (∗∗∗p ≥ 0.001). Angular displacement data were calculated and evaluated statistically using Rayleigh tests for evaluating nonuniformity of circular data (MATLAB) with the assumed mean direction set at 90° (∗∗∗p ≤ 0.001). To see this figure in color, go online.
Local stiffness affects effectiveness of the durotactic response
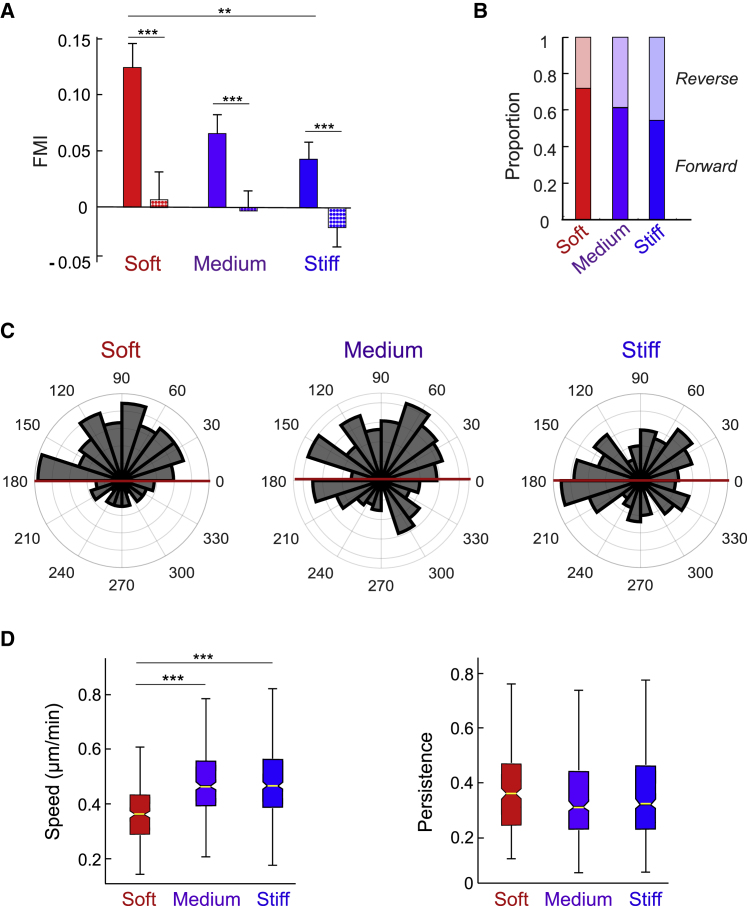
Substrate compliance is known to influence cell morphology and migration (45, 46). Therefore, we hypothesized that the efficiency of cell durotaxis might also be influenced by the specific level of stiffness that a cell encounters on our gradients spanning ∼3–23 kPa. We analyzed cell migration data based on the initial location of each cell on the gradient. Cells were grouped into three 250-μm regions that we defined as soft (2–7 kPa), medium (7–13 kPa), and stiff (13–18 kPa). We observed significant directional migration parallel to (up) the gradient for cells initially occupying each region (Figs. 2 B and 4); control evaluations to rule out lateral directional migration by determining FMI values perpendicular to the gradient confirmed that the only net directionality of migration was up the gradient (Fig. 4 A). However, there was a stepwise decrease in the magnitude of the directed migration up stiffness gradients as cells occupied increasingly stiff regions of the gradient (Fig. 4 A). This trend was further substantiated by evaluating the angular displacements of cells within each of these regions (Fig. 4 C). These durotaxing cells, i.e., “forward”-moving cells that migrated toward increasingly stiff regions, represented 72% of cells evaluated on the soft region and 61 and 55% for cells occupying the medium and stiff regions, respectively (Fig. 4). The remainder of the cells represented a subpopulation that moved in a “reverse” direction relative to increasing stiffness for the interval over which they were tracked. Consequently, durotaxis in these cells is an average population phenomenon.
Figure 4.
Durotaxis efficiency on different regions of the stiffness gradient. (A) FMI for cells moving parallel (bars) versus perpendicular (stippled bars) to the stiffness gradient on the soft, medium, or stiff regions. (B) The proportion of cells moving toward the stiff side of the gradient (forward, darker colored bars) compared to the number of cells moving toward the increasingly soft side (reverse, pastel). (C) Polar histograms indicate the angular displacement of cells within the three different stiffness regions. (D) Speed and persistence (directionality) of cells with respect to position on the gradient. Error bars represent mean ± SE, N = 3, n = 793. Boxplot center lines denote the median, and edges represent the 25th and 75th percentiles, with whiskers incorporating 99.3% of all data. For clarity, nonmigrating outliers were excluded from the graph as described in Materials and Methods, but they were not excluded from the statistical analysis. FMI was evaluated statistically using two-way ANOVA with Tukey’s post hoc test (∗∗∗p ≤ 0.001). Angular displacement was evaluated statistically using a Rayleigh test (∗p ≤ 0.05, ∗∗∗p ≤ 0.001). Speed and persistence (straightness index) were evaluated by one-way ANOVA and Tukey’s post hoc test (∗∗∗p ≤ 0.001). To see this figure in color, go online.
The morphology of these cells in the soft region of gradients was not mesenchymal but was generally rounded in shape and minimally polarized, whereas cells on stiffer parts of the gradient displayed a flattened, polarized morphology (e.g., see Fig. 2 A). In addition, we found that the population undergoing the most robust durotactic migration—on the soft region—moved with speeds significantly slower than cells occupying stiffer regions of the gradient (Fig. 4 D); thus, higher speed of migration does not enhance durotaxis. Moreover, there was no difference in persistence (directionality) of migration between these groups (Fig. 4 D).
A conceptual concern with comparing durotaxis on different regions of a linear stiffness gradient gel is that the rate of change in stiffness becomes progressively less in stiffer regions. Therefore, we examined for the efficiency of durotaxis on high-stiffness gradient gels that rose to 80 kPa, with the softer region 9–25 kPa, medium 25–50 kPa, and stiffest 50–80 kPa. The efficiency of durotaxis as quantified by FMI was lower than gels with the original 2–20 kPa gradients (Fig. S2). Most importantly, there were no significant differences in durotactic efficiency when comparing softer with stiffer regions in these gels possessing much higher absolute stiffness. We conclude that there is a plateau or saturation in the efficiency of durotaxis above the optimal range of absolute stiffness of 2–7 kPa but also that durotaxis can still occur in regions of high stiffness.
Carcinoma, fibrosarcoma, and glioblastoma cell lines all exhibit durotaxis
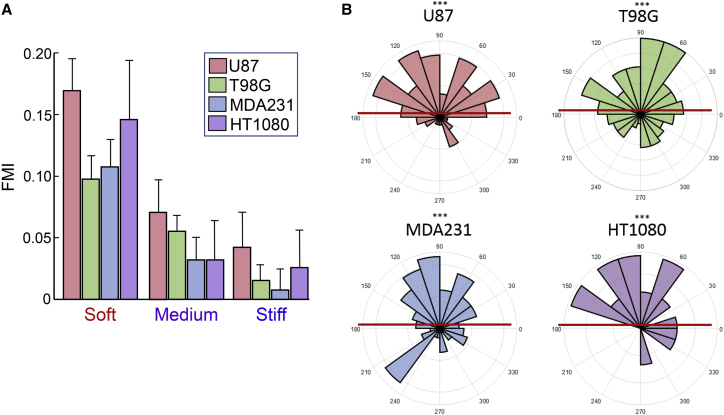
In addition to U87 cells, we evaluated a second glioblastoma cell line (T98G), a metastatic epithelial breast cancer cell line (MDA-MB-231), and a mesenchymal fibrosarcoma cell line (HT1080) (Fig. 5) to determine whether our stiffness gradient condition would elicit a durotactic response in other frequently used cancer cells. Surprisingly, each cell line exhibited a pattern of durotaxis analogous to that seen for U87 cells. Notably, for each of these other cell types, directed movement was also most prominent on the soft region of the stiffness gradient, with an incrementally decreasing durotactic response as the gradient became increasingly stiff. We also compared the durotactic migration of an immortalized nonmalignant fibroblast cell line (BJ-5ta), a normal counterpart to the HT1080 fibrosarcoma line. These nonmalignant fibroblasts displayed the same pattern of enhanced durotactic efficiency on the soft region of gradient gels, with less at intermediate and higher stiffness (Fig. S3).
Figure 5.
Frequently used cancer cell lines undergo durotaxis. (A) FMI values for U87, T98G, MDA-MB-231, and HT1080 cells on soft, medium, and stiff regions of the stiffness gradient. (B) Corresponding angular displacement values for each of the cancer cell populations occupying the soft portion of the gradient. Error bars represent mean ± SE for n = 255, n = 800, n = 495, and n = 136 cells from 3, 3, 2, and 2 biological replicates in independent experiments, respectively. FMI was evaluated statistically by one-way ANOVA and Tukey’s post hoc test (∗p ≤ 0.05, ∗∗∗p ≤ 0.001). Angular displacement was evaluated using Rayleigh tests (∗∗∗p ≤ 0.001). To see this figure in color, go online.
Evaluating directional migration over time
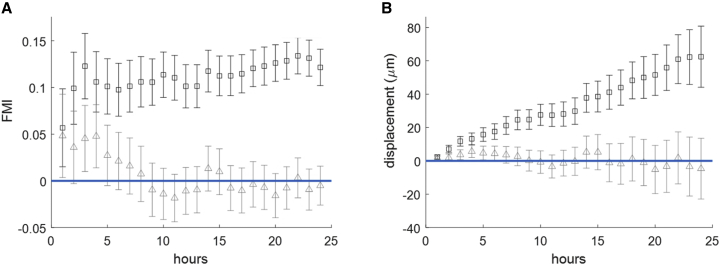
The FMI reported above is a function of the migratory persistence of a cell parallel to the gradient. Evaluating this metric for the population of U87 glioblastoma cells occupying the soft portion of the gradient revealed that this value remains relatively constant over the duration of the experiment (Fig. 6 A). This steady-state FMI suggests a continual increase in the average displacement of cells parallel to the gradient. An inspection of the population’s average displacement confirms persistent movement toward increasingly stiff regions of the gradient (Fig. 6 B). As expected, FMI and displacement values evaluated perpendicular to the axis of increasing stiffness approximated zero (Fig. 6), confirming that there is no directional cue along this axis.
Figure 6.
FMI and displacement calculated over time. (A) The ensemble mean for U87 forward migration index (FMI) parallel (□) and perpendicular (Δ) to the gradient. (B) The average displacement of cells parallel to the gradient was found to increase gradually over time compared to the average displacement perpendicular to the gradient. To see this figure in color, go online.
Correlation analysis confirms contribution to durotaxis from displacement along the gradient but minimally from persistence (straightness) and none from cell speed
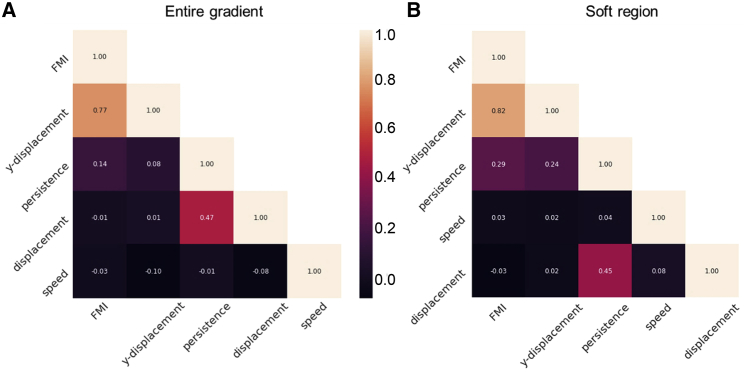
In an effort to identify the cell migration parameter(s) associated most closely with durotaxis, U87 cancer cell data were subjected to correlation analyses. FMI was compared to displacement in the Y direction toward increasing stiffness (which should by definition correlate with FMI), overall cell displacement (net distance traversed over the 8-h assay period), persistence (straightness) of the migration track, and average speed of migration. As shown in Fig. 7 A, evaluation of cells located throughout the gradient (soft through stiff) showed the expected correlation with displacement up the stiffness gradient (correlation coefficient = 0.77). There was only weak correlation of the FMI of each cell with its persistence (correlation coefficient = 0.14 each). There was no correlation with overall displacement and speed (−0.01 and −0.03, respectively).
Figure 7.
Correlation analysis of FMI compared to each of four migration parameters. (A and B) Correlation coefficients between FMI, net cell displacement up the gradient (“Y displacement”), persistence of migration, total cell displacement, and speed of migration. (A) Analysis of all 8-h tracks for 1205 cells on the entirety of the gradients. (B) Analysis of the 8-h tracks for cells located on the soft (2–7 kPa) region of gradients (292 cells). To see this figure in color, go online.
We next focused on cell behavior on the soft region of the gradient, which was the most efficient region for durotaxis (Fig. 7 B). As expected, there was strong positive correlation with displacement up the gradient (0.82) but again only modest correlation with persistence (0.29) and no correlation at all with cell speed and overall displacement (0.03 and −0.03, respectively).
Because it was conceivable that some cells migrating in the soft region of gradients might have initiated migration in a stiffer region, thereby confusing our analysis, we also examined data restricted to the cells that had initiated migration in the soft region (252 out of 292 cells). The results were the same as for the total population of cells in the soft region, i.e., confirming the positive correlation with displacement along the gradient, only modest correlation with persistence, and no correlation with total displacement or speed (data not shown). In addition, even though one tumor cell line showed considerably reduced speed of migration on soft regions, two other tumor cell lines showed less reduction, and a normal human fibroblast line showed no decrease in speed even though durotaxis was also elevated in the soft region. Consequently, even though the capacity of these cells to undergo durotaxis is strongly positively correlated as expected with the overall migratory directional displacement up the gradient, there is relatively minimal correlation with the migratory persistence of a cell and none with individual cell speed or total migratory displacement. Thus, cells that migrate persistently are only slightly more likely to undergo durotaxis, and speed of migration does not affect efficiency of durotaxis.
Arp2/3 inhibition blocks U87 cell durotaxis
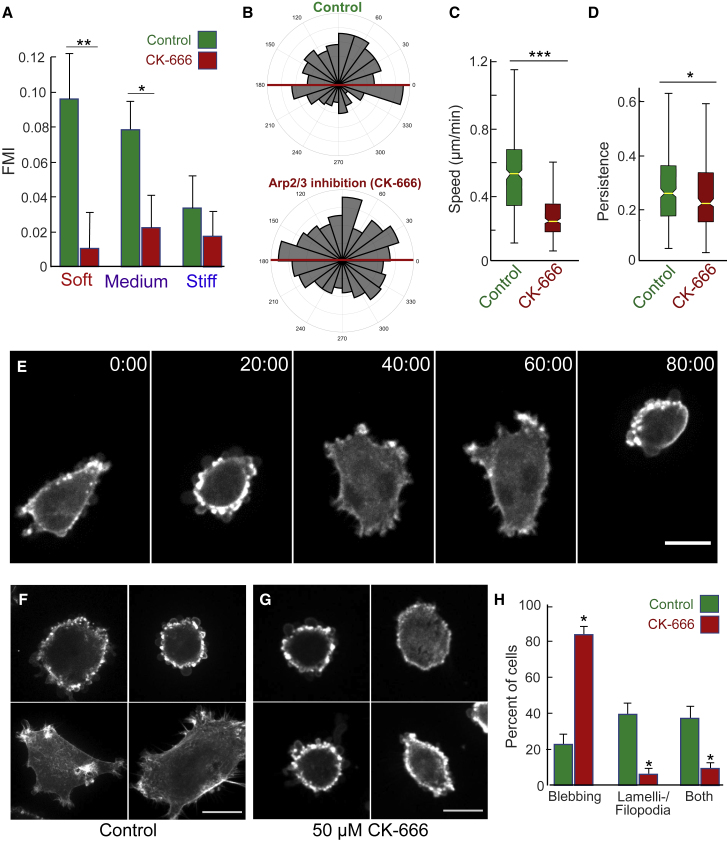
Membrane protrusion via lamellipodial and filopodial dynamics during cell migration depends on Arp2/3-mediated actin polymerization, and this protein complex has been implicated in chemotaxis driven by EGF (35) but not by PDGF (34), as well as in haptotaxis (34). Although they show little or no polarity in soft regions where they display maximal durotactic efficiency (Fig. 8, E and F), U87 cells demonstrate numerous small lamellipodial and filopodial regions around the cell during migration on gradients alternating with a rounded blebbing phenotype (Fig. 8); therefore, we tested whether Arp2/3 plays a role in durotaxis. U87 cells treated with the specific Arp2/3 inhibitor CK-666 (50 μM) displayed substantially lower FMI values compared to DMSO controls. A detailed evaluation of durotaxis in Fig. 8 on soft, medium, and stiff regions of the gradient revealed that CK-666 impairs durotaxis across all regions of our gradients but most strikingly (>9-fold reduction) on the soft region that normally most effectively promotes durotaxis (Fig. 8, A and B). Consistent with previous studies (34, 47), experimentally impairing Arp2/3 activity also resulted in a significant reduction in cell speed (Fig. 8 C). However, correlation analysis had revealed no relationship between cell speed and FMI in untreated cells (Fig. 7). Furthermore, under normal, noninhibited conditions, cells occupying the softest portion of the gradient represent the most durotactic population despite generally migrating significantly more slowly than cells elsewhere on the gradient (Figs. 2, 4, and 5). These findings suggest that the ability to undergo durotaxis is not related to speed of migration but instead depends on some other biological function mediated by Arp2/3.
Figure 8.
Impaired durotaxis resulting from inhibition of Arp2/3. (A) FMI for cells treated with 50 μM CK-666 (red) or DMSO vehicle control (green) moving parallel to the stiffness gradient within the soft, medium, or stiff regions. (B) Angular displacement analysis reveals no evidence for a substrate-directed response of CK-666 treated cells (bottom) cultured on a stiffness gradient compared to vehicle control cells (top). (C) Speed and (D) persistence (directionality) of cell migration are reduced with CK-666 treatment. Data for 320 DMSO- and 391 CK-666-treated cells from N ≥ 3 biological replicates (independent experiments) were collected. (E) Time series of an individual U87 glioblastoma cell (untreated) that spontaneously alternates phenotype periodically between lamellipodial-filopodial and blebbing. (F) Representative fluorescence images of control (untreated) U87 glioblastoma cells expressing TagGFP2-LifeAct to label F-actin showing phenotypes varying from rounded and blebbing to poorly polar cells with multiple small lamellipodia and short spike-like filopodia. (G) Representative fluorescence images of U87 glioblastoma cells treated with 50 μM CK-666, often showing numerous small blebs at the cell periphery. (H) Morphological quantification of control (green bars) versus CK-666-treated (red) U87 cells with blebbing, lamellipodial-filopodial, or both phenotypes in time-lapse videos. (H) FMI p-values were calculated using one-way ANOVA to compare between groups. (∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05). Angular displacement data were analyzed statistically using Rayleigh tests for evaluating circular data with the assumed mean direction set at 90° (∗∗∗p ≤ 0.001). In (A) and (H), error bars represent mean ± SE. In (C) and (D), boxplot center lines denote the median and edges represent the 25th and 75th percentiles, with whiskers incorporating 99.3% of all data. Scale bars, 10 μm for (E) and 20 μm for (F) and (H). To see this figure in color, go online.
We therefore examined for alterations in Arp2/3-dependent lamellipodial and filopodial dynamics. Inhibition of Arp2/3 resulted in morphological alterations in both overall cell morphology and membrane dynamics of treated U87 cells. At any specific time, a majority of U87 cells were observed to display dynamic cell-surface protrusions, with multiple small lamellipodia and filopodia per cell (Fig. 8 E; Video 1). However, roughly 40% of the U87 cells normally switch back and forth from mode of migration characterized by lamellipodia and filopodia to a more rounded, rapidly blebbing phenotype, and more than 20% remain blebbing (Fig. 8 H; Video 2). Inhibition of Arp2/3 converted nearly all nonblebbing cells to the rounded, blebbing morphology (Fig. 8 F). The percentage of such continuously blebbing cells rose from <25% + the 40% that bleb periodically to >80% (Fig. 8 G), with a striking loss of the multiple, highly dynamic small lamellipodia with short filopodia normally characteristic of migration by this cell, which were replaced by numerous, highly active blebs analogous in appearance to the blebbing phenotype of untreated cells (Fig. 8 F; Video 3). This cell blebbing phenotype was reverted back to primarily lamellipodial/filopodial within 1 h after removing the inhibitor (data not shown). These morphological effects and altered dynamics of the membrane protrusions classically associated with cell migration to blebbing resulting from Arp2/3 inhibition are consistent with the accompanying significant loss of cell speed, low persistence of migration, and inability to undergo durotaxis—a process shown here to depend on Arp2/3 function.
U87 cells expressing TagGFP2-LifeAct on the soft (2–7 kPa) side of a gradient polyacrylamide gel demonstrating the lamellipodial form of cell migration. Note the numerous lamellipodia and filopodia forming at the edges, as well as beneath the cell. This cell was imaged every 10 s for 10 min. Time indicates minutes and seconds.
U-87 glioblastoma cell expressing TagGFP2-LifeAct imaged on the soft (2–7 kPa) side of a gradient polyacrylamide gel demonstrating normal switching between lamellipodia and filopodia and blebbing at various times during cell migration. Note that the transition from lamellipodial to blebbing morphology occurs after the cell retracts its trailing edge. This cell was imaged every 1 min for 2 h. Time indicates minutes and seconds.
U-97 glioblastoma cells expressing TagGFP2-LifeAct imaged on the soft (2–7 kPa) side of a gradient polyacrylamide gel. These cells were treated with the specific Arp2/3 inhibitor CK-666 1 min prior to the beginning of the video, demonstrating a switch to continuous blebbing. This cell was imaged every 1 min for 2 h. Time indicates minutes and seconds.
Discussion
Durotaxis is a class of externally directed cell migration in which the direction of cell migration is governed by a local physical parameter: the stiffness of the migratory substrate (11, 12, 13, 14, 15, 16, 17, 18, 19, 20). Durotaxis has been characterized in detail recently in several nonmalignant cell types, generally those derived from a mesenchymal lineage (e.g., see (48, 49)). However, it was not clear whether cancer cells can also undergo effective durotaxis. We applied automated computer tracking of individual human cancer cells in tissue culture to obtain unbiased data sets for characterizing populations of migrating cells. We demonstrate that for all four of the different types of human cancer cells examined, all of them undergo durotaxis on our stiffness gradients using fibronectin as the substrate. Unexpectedly, the efficiency of each of these cancer cell types to undergo durotaxis depended on the local compliance of the stiffness gradients, with maximal durotaxis in the physiologically soft 2–7 kPa range. A comparison with a nonmalignant fibroblast counterpart to the fibrosarcoma cells revealed a similar extent of durotaxis with maximal efficiency on the same soft 2–7 kPa region of the gradient compared to stiffer regions. Time-course and correlation analyses revealed that durotaxis was independent of the time of migration and cell speed. As expected, durotaxis did correlate with the extent of cell migratory displacement along the gradient but only modestly with persistence and not with total extent of displacement. At a molecular level, Arp2/3 function was necessary for both normal lamellipodial and filopodial dynamics and ability to undergo durotaxis.
As will be discussed later, there are theoretical reasons that malignant cells might employ durotaxis during the process of cancer cell spreading during metastasis. However, to our knowledge, this study is the first to document directly the capacity of multiple types of tumor cells—ranging from glioblastoma to breast cancer and fibrosarcoma—to undergo effective durotaxis. In addition, the unexpected shared behavioral characteristic of these various cancer cells and a normal fibroblast line to undergo durotaxis most efficiently on the soft portion of durotactic gradients suggests that physiologically softer regions of the body, e.g., the brain and other soft-tissue regions, may be particularly susceptible to durotaxis-augmented invasion. Interestingly, mesenchymal stem cell chemotaxis toward EGF was recently reported to be more efficient on a soft substrate (50). Taken together, these independent findings on durotaxis and chemotaxis suggest that cells may respond most efficiently to both physical and chemical cues for directional migration in a softer, more compliant microenvironment.
The presence of a polarized, mesenchymal cell morphology and consistent polarity of lamellipodia or lamellae in the direction of migration failed to correlate with efficient durotaxis. In fact, the opposite was observed: cells in soft regions that could mediate durotaxis the most effectively were notably rounded with little or no cell polarity; cells alternated between lamellipodial/filopodial surface phenotype and a blebbing phenotype, but neither cell type showed much polarity (see Figs. 2 A and 8, E and H). Attempts to predict whether a cell would be durotactic by trying to correlate cell morphology with its direction of migration along a stiffness gradient using phase-contrast time-lapse videos completely failed, supporting the observation in static images that the most efficient durotaxis occurred in soft regions where cells were poorly polar. This finding initially appeared nonintuitive because it might be assumed that durotaxis will be the most efficient in polarized cells with lamellipodia confined to the front of a cell to lead it up the stiffness gradient. Instead, our findings suggest that the most efficient durotaxis by single cells (either cancer or nonmalignant fibroblastic cells) involves weak adhesion with rapid sampling of the substrate. Consistent with this view, durotaxis on a much stiffer gradient was approximately half of the FMI for soft regions, suggesting a plateau in durotactic efficiency beyond the 2–7 kPa region but also, importantly, not a total loss of the capacity for durotaxis as predicted in a recent publication (51), even in stiff regions where the cells were generally flattened and polarized (e.g., Fig. 2 A). Although various models will likely be possible, combining a model involving Arp2/3-dependent membrane dynamics with a motor-clutch model (52) to define optimal substrate stiffnesses for migration speed compared to durotactic efficiency could provide an attractive future approach.
We focused on single, individual migrating cells to be able to evaluate durotaxis independent of cell-cell contact. A recent study focusing on durotaxis by confluent monolayers of cells revealed an apparent enhancement of durotactic efficiency in collective cell migration, and actomyosin contractility across the whole population was important for collective cell durotaxis (21). Roles for focal adhesions and probing cell-substrate contacts by filopodia have also been associated with durotaxis (15, 17, 21, 22, 23, 24). To search for a novel contributor to durotaxis, we focused on function of the Arp2/3 complex in the formation of a key sensory organ, the lamellipodium. The selective small-molecule Arp2/3 inhibitor CK-666 blocks conformational activation of the Arp2/3 complex (53). CK-666 is known to inhibit Arp2/3 functions in lamellipodial function in a variety of cell types, including other glioblastoma cell lines besides those examined here (34, 47). Consistent with other studies, inhibition of Arp2/3 with CK-666 severely inhibited normal cell-surface lamellipodial and filopodial dynamics, which were replaced in the glioblastoma cells by rapidly dynamic blebs, with a complete loss of the already minimal extent of cell polarity. Durotaxis was strongly inhibited throughout the stiffness gradient, pointing to a critical role of Arp2/3 in durotaxis. It should also be noted that although Arp2/3 suppressed the speed of cell migration, migration was slower in soft regions of gradient gels where durotaxis is most efficient, and we could not find any correlation between cell speed and durotaxis in untreated cells. Instead, the most striking effect of Arp2/3 inhibition was the complete loss of lamellipodial and filopodial dynamics, suggesting their role in durotaxis.
When considering the potential biological significance of durotaxis in human cancer cells, it is important to note that malignant cells often encounter a diverse microenvironment that provides both chemical and mechanical cues governing cell adhesion that can change over time. To the lists of gradients of chemokines and diffusible growth factors, as well as gradients of extracellular matrix that have been implicated in promoting cancer cell migration (1, 2, 3, 4, 5, 6, 7, 8, 9, 21), we propose adding mechanical gradients of stiffness. As described previously for chemotactic gradients in which high levels of chemoattractant are known to suppress chemotaxis, this study establishes that high levels of mechanical stiffness suppress efficiency of durotaxis. Thus, in both types of cell taxis, key elements are optimal concentrations of the chemical or physical signal. We emphasize that this concept of an optimal level stiffness for durotaxis was also shared by a nonmalignant human fibroblast line, suggesting that normal tissue remodeling by fibroblasts may also involve durotaxis modulated by the local stiffness of extracellular matrix.
The stiffening extracellular stroma surrounding a tumor might offer a durotactic escape route from the potentially threatening hypoxic, necrotic environment of a primary lesion. Inhospitable conditions may trigger innate mechanosensing machinery to be able to initiate efficient directional migration. An analogous phenomenon could be the activation of epithelial-to-mesenchymal transition in response to tumor microenvironmental conditions that enhance extravasation and metastasis (54). Migrating into regions of elevated stiffness is also known to promote the proliferative capacity of cells and enhance tumor growth (55, 56, 57, 58).
The tumor-stroma boundary may represent a “step” gradient in which cells encounter a large difference in extracellular stiffness that prompts movement away from neighboring cancer cells. An intriguing report of breast tumor mastectomies measured with AFM reveals that the cancerous mass itself is more compliant than the invasive front of these tumors (59). Furthermore, a sharp step gradient such as the tumor-stroma boundary may propagate an effective stiffness gradient that can be encountered over longer distances. For example, cells cultured on soft polyacrylamide gels are responsive to the stiff underlying glass if the gel is made sufficiently thin (60, 61). This reveals the concept that as cells exert tension on their surrounding environment, the increased stiffness of the stromal boundary may be felt at a distance that could promote a durotactic response (62).
A great deal of circumstantial evidence also exists for the potential creation of local gradients by stromal cell remodeling of the ECM environment of tumors (63, 64). Fibroblasts strain collagen and fibronectin fibers, thereby transmitting tension to the ECM. As this tension becomes dissipated further from the contractile source, a potential gradient of stiffness can be generated (65). Cancer cells may well escape a primary tumor along these avenues of strained ECM. Although cells have been observed aligning and migrating along strained ECM, cell-induced strain can also align the matrix, making differentiation between migration induced by stiffness or ECM topography difficult (66). It seems likely that a combination of multiple environmental factors ultimately dictates the direction in which a cell moves, just as for any other form of directed migration.
Conclusions
In this study, we have established that human cancer cells of varying tissue origins can undergo durotaxis, i.e., directed cell migration toward regions of increasing stiffness. This response could provide additional biological significance to the alterations in stiffness known to exist at tumor sites. Using an automated tracking system for an unbiased examination of the parameters affecting durotaxis, we found that even though single migrating tumor cells showed the expected correlation between durotaxis and migratory displacement up stiffness gradients, parameters such as cell speed and intrinsic cell directionality did not contribute. Intriguingly, durotaxis occurred most efficiently on a soft substrate and became less effective on stiffer substrates for both cancer cells and normal counterpart nonmalignant fibroblasts. In addition to previously described mechanisms contributing to the process of durotaxis, we established that function of the Arp2/3 complex, which regulates cell-surface dynamics, was also crucial for effective durotaxis. Our studies add durotaxis to a variety of previously described forms of directed migration as a potential contributor to human cancer.
Author Contributions
B.J.D. and K.M.Y. originated the research project. B.J.D., A.D.D., and K.M.Y. designed specific experiments. B.J.D., E.K.D., and A.D.D. performed the research. B.J.D. and E.K.D. contributed analytic tools. B.J.D., A.D.D., and E.K.D. calculated and analyzed quantitative data. All four coauthors contributed to writing the manuscript.
Acknowledgments
This research was supported by the Intramural Research Programs of the National Institutes of Health, National Institute of Dental and Craniofacial Research, and National Institute of Biomedical Imaging and Bioengineering.
Editor: Margaret Gardel.
Footnotes
Four figures and three videos are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30025-6.
Supporting Material
References
- 1.Tweedy L., Susanto O., Insall R.H. Self-generated chemotactic gradients-cells steering themselves. Curr. Opin. Cell Biol. 2016;42:46–51. doi: 10.1016/j.ceb.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Stuelten C.H., Parent C.A., Montell D.J. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat. Rev. Cancer. 2018;18:296–312. doi: 10.1038/nrc.2018.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oudin M.J., Weaver V.M. Physical and chemical gradients in the tumor microenvironment regulate tumor cell invasion, migration, and metastasis. Cold Spring Harb. Symp. Quant. Biol. 2016;81:189–205. doi: 10.1101/sqb.2016.81.030817. [DOI] [PubMed] [Google Scholar]
- 4.Haeger A., Wolf K., Friedl P. Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 2015;25:556–566. doi: 10.1016/j.tcb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Majumdar R., Sixt M., Parent C.A. New paradigms in the establishment and maintenance of gradients during directed cell migration. Curr. Opin. Cell Biol. 2014;30:33–40. doi: 10.1016/j.ceb.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roca-Cusachs P., Sunyer R., Trepat X. Mechanical guidance of cell migration: lessons from chemotaxis. Curr. Opin. Cell Biol. 2013;25:543–549. doi: 10.1016/j.ceb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Lara Rodriguez L., Schneider I.C. Directed cell migration in multi-cue environments. Integr. Biol. 2013;5:1306–1323. doi: 10.1039/c3ib40137e. [DOI] [PubMed] [Google Scholar]
- 8.Bravo-Cordero J.J., Hodgson L., Condeelis J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roussos E.T., Condeelis J.S., Patsialou A. Chemotaxis in cancer. Nat. Rev. Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sander L.M. Modeling contact guidance and invasion by cancer cells. Cancer Res. 2014;74:4588–4596. doi: 10.1158/0008-5472.CAN-13-3294. [DOI] [PubMed] [Google Scholar]
- 11.Lo C.M., Wang H.B., Wang Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isenberg B.C., Dimilla P.A., Wong J.Y. Vascular smooth muscle cell durotaxis depends on substrate stiffness gradient strength. Biophys. J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawano T., Kidoaki S. Elasticity boundary conditions required for cell mechanotaxis on microelastically-patterned gels. Biomaterials. 2011;32:2725–2733. doi: 10.1016/j.biomaterials.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 14.Tse J.R., Engler A.J. Stiffness gradients mimicking in vivo tissue variation regulate mesenchymal stem cell fate. PLoS One. 2011;6:e15978. doi: 10.1371/journal.pone.0015978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plotnikov S.V., Pasapera A.M., Waterman C.M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidoaki S., Sakashita H. Rectified cell migration on saw-like micro-elastically patterned hydrogels with asymmetric gradient ratchet teeth. PLoS One. 2013;8:e78067. doi: 10.1371/journal.pone.0078067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong S., Guo W.H., Wang Y.L. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc. Natl. Acad. Sci. USA. 2014;111:17176–17181. doi: 10.1073/pnas.1412285111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S., Hong J., Lee J. Cell motility regulation on a stepped micro pillar array device (SMPAD) with a discrete stiffness gradient. Soft Matter. 2016;12:2325–2333. doi: 10.1039/c5sm00649j. [DOI] [PubMed] [Google Scholar]
- 19.Löber J., Ziebert F., Aranson I.S. Modeling crawling cell movement on soft engineered substrates. Soft Matter. 2014;10:1365–1373. doi: 10.1039/c3sm51597d. [DOI] [PubMed] [Google Scholar]
- 20.Singh S.P., Schwartz M.P., Anseth K.S. A peptide functionalized poly(ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration. Biomater. Sci. 2014;2:1024–1034. doi: 10.1039/C4BM00022F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunyer R., Conte V., Trepat X. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- 22.Plotnikov S.V., Waterman C.M. Guiding cell migration by tugging. Curr. Opin. Cell Biol. 2013;25:619–626. doi: 10.1016/j.ceb.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raab M., Swift J., Discher D.E. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 2012;199:669–683. doi: 10.1083/jcb.201205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wormer D.B., Davis K.A., Turner C.E. The focal adhesion-localized CdGAP regulates matrix rigidity sensing and durotaxis. PLoS One. 2014;9:e91815. doi: 10.1371/journal.pone.0091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedl P., Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 26.Lauffenburger D.A., Horwitz A.F. Cell migration: a physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 27.Ridley A.J., Schwartz M.A., Horwitz A.R. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 28.Ponti A., Machacek M., Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 29.Svitkina T.M., Borisy G.G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y.L. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 1985;101:597–602. doi: 10.1083/jcb.101.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston S.A., Bramble J.P., Machesky L.M. Arp2/3 complex activity in filopodia of spreading cells. BMC Cell Biol. 2008;9:65. doi: 10.1186/1471-2121-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard T.D., Cooper J.A. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang I., Gorelik R., Gautreau A. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature. 2013;503:281–284. doi: 10.1038/nature12611. [DOI] [PubMed] [Google Scholar]
- 34.Wu C., Asokan S.B., Bear J.E. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suraneni P., Rubinstein B., Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paszek M.J., Zahir N., Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Wei S.C., Fattet L., Yang J. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Provenzano P.P., Inman D.R., Keely P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene. 2009;28:4326–4343. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox T.R., Bird D., Erler J.T. LOX-mediated collagen crosslinking is responsible for fibrosis-enhanced metastasis. Cancer Res. 2013;73:1721–1732. doi: 10.1158/0008-5472.CAN-12-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Discher D.E., Janmey P., Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 41.Swift J., Ivanovska I.L., Discher D.E. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DuChez B.J. Automated tracking of cell migration with rapid data analysis. Current Protoc. Cell Biol. 2017;76:12.12.11–12.12.16. doi: 10.1002/cpcb.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tse J.R., Engler A.J. Preparation of hydrogel substrates with tunable mechanical properties. Current Protoc. Cell Biol. 2010;Chapter 10:Unit 10.16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 44.Akiyama S.K. Purification of fibronectin. Current Protoc. Cell Biol. 2013;60:10.5.1–10.5.13. doi: 10.1002/0471143030.cb1005s60. [DOI] [PubMed] [Google Scholar]
- 45.Yeung T., Georges P.C., Janmey P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 46.Peyton S.R., Putnam A.J. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z., Yang X., Ming H. Expression of the Arp2/3 complex in human gliomas and its role in the migration and invasion of glioma cells. Oncol. Rep. 2013;30:2127–2136. doi: 10.3892/or.2013.2669. [DOI] [PubMed] [Google Scholar]
- 48.Hartman C.D., Isenberg B.C., Wong J.Y. Vascular smooth muscle cell durotaxis depends on extracellular matrix composition. Proc. Natl. Acad. Sci. USA. 2016;113:11190–11195. doi: 10.1073/pnas.1611324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadden W.J., Young J.L., Choi Y.S. Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc. Natl. Acad. Sci. USA. 2017;114:5647–5652. doi: 10.1073/pnas.1618239114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saxena N., Mogha P., Sen S. Matrix elasticity regulates mesenchymal stem cell chemotaxis. J. Cell Sci. 2018;131 doi: 10.1242/jcs.211391. jcs211391. [DOI] [PubMed] [Google Scholar]
- 51.Moriyama K., Kidoaki S. Cellular durotaxis revisited: initial-position-dependent determination of the threshold stiffness gradient to induce durotaxis. Langmuir. 2018 doi: 10.1021/acs.langmuir.8b02529. https://pubs.acs.org/doi/10.1021/acs.langmuir.8b02529 Published online September 25, 2018. [DOI] [PubMed] [Google Scholar]
- 52.Bangasser B.L., Rosenfeld S.S., Odde D.J. Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 2013;105:581–592. doi: 10.1016/j.bpj.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hetrick B., Han M.S., Nolen B.J. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 2013;20:701–712. doi: 10.1016/j.chembiol.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bae Y.H., Mui K.L., Assoian R.K. A FAK-Cas-Rac-lamellipodin signaling module transduces extracellular matrix stiffness into mechanosensitive cell cycling. Sci. Signal. 2014;7:ra57. doi: 10.1126/scisignal.2004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schrader J., Gordon-Walker T.T., Iredale J.P. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53:1192–1205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aragona M., Panciera T., Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 58.Umesh V., Rape A.D., Kumar S. Microenvironmental stiffness enhances glioma cell proliferation by stimulating epidermal growth factor receptor signaling. PLoS One. 2014;9:e101771. doi: 10.1371/journal.pone.0101771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Acerbi I., Cassereau L., Weaver V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buxboim A., Rajagopal K., Discher D.E. How deeply cells feel: methods for thin gels. J. Phys. Condens. Mat. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maloney J.M., Walton E.B., Van Vliet K.J. Influence of finite thickness and stiffness on cellular adhesion-induced deformation of compliant substrata. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2008;78:041923. doi: 10.1103/PhysRevE.78.041923. [DOI] [PubMed] [Google Scholar]
- 62.Buxboim A., Ivanovska I.L., Discher D.E. Matrix elasticity, cytoskeletal forces and physics of the nucleus: how deeply do cells ‘feel’ outside and in? J. Cell Sci. 2010;123:297–308. doi: 10.1242/jcs.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goetz J.G., Minguet S., Del Pozo M.A. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riching K.M., Cox B.L., Keely P.J. 3D collagen alignment limits protrusions to enhance breast cancer cell persistence. Biophys. J. 2014;107:2546–2558. doi: 10.1016/j.bpj.2014.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winer J.P., Oake S., Janmey P.A. Non-linear elasticity of extracellular matrices enables contractile cells to communicate local position and orientation. PLoS One. 2009;4:e6382. doi: 10.1371/journal.pone.0006382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Provenzano P.P., Eliceiri K.W., Keely P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
U87 cells expressing TagGFP2-LifeAct on the soft (2–7 kPa) side of a gradient polyacrylamide gel demonstrating the lamellipodial form of cell migration. Note the numerous lamellipodia and filopodia forming at the edges, as well as beneath the cell. This cell was imaged every 10 s for 10 min. Time indicates minutes and seconds.
U-87 glioblastoma cell expressing TagGFP2-LifeAct imaged on the soft (2–7 kPa) side of a gradient polyacrylamide gel demonstrating normal switching between lamellipodia and filopodia and blebbing at various times during cell migration. Note that the transition from lamellipodial to blebbing morphology occurs after the cell retracts its trailing edge. This cell was imaged every 1 min for 2 h. Time indicates minutes and seconds.
U-97 glioblastoma cells expressing TagGFP2-LifeAct imaged on the soft (2–7 kPa) side of a gradient polyacrylamide gel. These cells were treated with the specific Arp2/3 inhibitor CK-666 1 min prior to the beginning of the video, demonstrating a switch to continuous blebbing. This cell was imaged every 1 min for 2 h. Time indicates minutes and seconds.