Aspergillus fumigatus is the primary cause of aspergillosis, a devastating ensemble of diseases associated with severe morbidity and mortality worldwide. A. fischeri is a close relative of A. fumigatus but is not generally observed to cause human disease. To gain insights into the underlying causes of this remarkable difference in pathogenicity, we compared two representative strains (one from each species) for a range of pathogenesis-relevant biological and chemical characteristics. We found that disease progression in multiple A. fischeri mouse models was slower and caused less mortality than A. fumigatus. Remarkably, the observed differences between A. fischeri and A. fumigatus strains examined here closely resembled those previously described for two commonly studied A. fumigatus strains, AF293 and CEA10. A. fischeri and A. fumigatus exhibited different growth profiles when placed in a range of stress-inducing conditions encountered during infection, such as low levels of oxygen and the presence of chemicals that induce the production of reactive oxygen species. We also found that the vast majority of A. fumigatus genes known to be involved in virulence are conserved in A. fischeri, whereas the two species differ significantly in their secondary metabolic pathways. These similarities and differences that we report here are the first step toward understanding the evolutionary origin of a major fungal pathogen.
KEYWORDS: comparative genomics, evolution of virulence, fungal pathogenesis, laeA, secondary metabolism, specialized metabolism
ABSTRACT
Aspergillus fischeri is closely related to Aspergillus fumigatus, the major cause of invasive mold infections. Even though A. fischeri is commonly found in diverse environments, including hospitals, it rarely causes invasive disease. Why A. fischeri causes less human disease than A. fumigatus is unclear. A comparison of A. fischeri and A. fumigatus for pathogenic, genomic, and secondary metabolic traits revealed multiple differences in pathogenesis-related phenotypes. We observed that A. fischeri NRRL 181 is less virulent than A. fumigatus strain CEA10 in multiple animal models of disease, grows slower in low-oxygen environments, and is more sensitive to oxidative stress. Strikingly, the observed differences for some traits are of the same order of magnitude as those previously reported between A. fumigatus strains. In contrast, similar to what has previously been reported, the two species exhibit high genomic similarity; ∼90% of the A. fumigatus proteome is conserved in A. fischeri, including 48/49 genes known to be involved in A. fumigatus virulence. However, only 10/33 A. fumigatus biosynthetic gene clusters (BGCs) likely involved in secondary metabolite production are conserved in A. fischeri and only 13/48 A. fischeri BGCs are conserved in A. fumigatus. Detailed chemical characterization of A. fischeri cultures grown on multiple substrates identified multiple secondary metabolites, including two new compounds and one never before isolated as a natural product. Additionally, an A. fischeri deletion mutant of laeA, a master regulator of secondary metabolism, produced fewer secondary metabolites and in lower quantities, suggesting that regulation of secondary metabolism is at least partially conserved. These results suggest that the nonpathogenic A. fischeri possesses many of the genes important for A. fumigatus pathogenicity but is divergent with respect to its ability to thrive under host-relevant conditions and its secondary metabolism.
IMPORTANCE Aspergillus fumigatus is the primary cause of aspergillosis, a devastating ensemble of diseases associated with severe morbidity and mortality worldwide. A. fischeri is a close relative of A. fumigatus but is not generally observed to cause human disease. To gain insights into the underlying causes of this remarkable difference in pathogenicity, we compared two representative strains (one from each species) for a range of pathogenesis-relevant biological and chemical characteristics. We found that disease progression in multiple A. fischeri mouse models was slower and caused less mortality than A. fumigatus. Remarkably, the observed differences between A. fischeri and A. fumigatus strains examined here closely resembled those previously described for two commonly studied A. fumigatus strains, AF293 and CEA10. A. fischeri and A. fumigatus exhibited different growth profiles when placed in a range of stress-inducing conditions encountered during infection, such as low levels of oxygen and the presence of chemicals that induce the production of reactive oxygen species. We also found that the vast majority of A. fumigatus genes known to be involved in virulence are conserved in A. fischeri, whereas the two species differ significantly in their secondary metabolic pathways. These similarities and differences that we report here are the first step toward understanding the evolutionary origin of a major fungal pathogen.
INTRODUCTION
Aspergillosis is a major cause of human morbidity and mortality, resulting in over 200,000 life-threatening infections each year worldwide, and is primarily caused by the filamentous fungus Aspergillus fumigatus (1). Multiple virulence traits related to invasive aspergillosis (IA) are known for A. fumigatus, including growth at human body temperature and under low-oxygen conditions, the ability to acquire micronutrients such as iron and zinc in limiting environments, and the production of a diverse set of secondary metabolites (1).
Growth at human body temperature is a key trait for the survival of A. fumigatus inside mammalian hosts and may have arisen through adaptation to the warm temperatures present in decaying compost piles, one of the organism’s ecological niches (2–4). The primary route of A. fumigatus colonization and infection is through the lung, where oxygen levels have been observed to be as low as 2/3 of atmospheric pressure during infection. A successful response to this hypoxic environment is required for fungal pathogenesis (5, 6). Moreover, A. fumigatus produces a diverse set of small, bioactive molecules, known as secondary metabolites, which are biosynthesized in pathways that are often encoded by biosynthetic gene clusters (BGCs) (7). Some of these secondary metabolites and their regulators have been shown to contribute to disease in mouse models (8–10). Furthermore, a master regulator of secondary metabolism, laeA, is required for full virulence in IA mouse model studies (11, 12).
Species closely related to A. fumigatus are also capable of causing disease, but they are rarely observed in the clinic (1, 13–15). For example, Aspergillusfischeri is the closest evolutionary relative to A. fumigatus for which a genome has been sequenced (16, 17), but it is rarely reported to cause human disease (1). Recent evolutionary genomic analyses suggest that A. fischeri and A. fumigatus last shared a common ancestor approximately 4 million years ago (95% credible interval: 2 to 7 million years ago) (17). Why A. fischeri-mediated disease is less common than A. fumigatus-mediated disease remains an open question. Non-mutually exclusive possibilities include differences in ecological abundance, lack of species-level diagnosis of disease-causing strains in the clinic, and innate differences in pathogenicity and virulence between the two species.
Previous studies have suggested that the difference in the frequencies with which the two species cause disease is unlikely to be solely due to ecological factors, as both can be isolated from a variety of locales, including soils, fruits, and hospitals (18–20). For example, approximately 2% of the fungi isolated from the respiratory intensive care unit at Beijing Hospital were A. fischeri compared to approximately 23% of fungal species identified as A. fumigatus (20). While A. fischeri is easily isolated from a variety of environments, only a few cases of human infections have been reported (21–24). Furthermore, numerous recent epidemiological studies from multiple countries that used state-of-the-art molecular typing methods were able to identify several rarely isolated pathogenic species closely related to A. fumigatus, such as Aspergilluslentulus and Aspergillusudagawae, as the source of 10 to 15% of human infections but did not identify A. fischeri in any patient sample (13, 14, 25–27).
If ecological factors and lack of precision in species identification do not convincingly explain why A. fischeri is nonpathogenic and A. fumigatus is pathogenic, other factors must be responsible. An initial genomic comparison between strains of A. fumigatus, A. fischeri, and the more distantly related Aspergillus clavatus identified 818 genes that were A. fumigatus specific (16). These genes were enriched for functions associated with carbohydrate transport and catabolism, secondary metabolite biosynthesis, and detoxification (16), raising the possibility that the observed differences in pathogenicity observed between A. fischeri and A. fumigatus have a molecular basis.
To gain further insight into why A. fischeri-mediated disease is less abundant than A. fumigatus-mediated disease, we took a multipronged approach to investigate phenotypic, genomic, and chemical differences between A. fischeri strain NRRL 181 and A. fumigatus strain CEA10. We observed that while A. fischeri is able to cause fatal disease in multiple animal models, its disease progression and response to multiple host-relevant stresses are markedly reduced compared to A. fumigatus CEA10. We also found that while the two organisms’ genomes are in general very similar, the sets of secondary metabolite pathways in each of them exhibit a surprisingly low level of overlap. Examination of the secondary metabolite profile of A. fischeri identified both previously isolated and novel compounds. Finally, construction of a mutant A. fischeri strain that lacked the laeA gene, a master regulator of secondary metabolism, and examination of its chemical profile suggested that LaeA-mediated regulation of secondary metabolism in A. fischeri closely resembles that of A. fumigatus. These results begin to reveal the molecular differences between A. fischeri and A. fumigatus related to fungal pathogenesis and suggest that a functional evolutionary genomic comparison between pathogenic and nonpathogenic species closely related to A. fumigatus harbors great promise for generating insights into the evolution of fungal disease.
RESULTS
A. fischeri is less virulent than A. fumigatus in multiple animal models of invasive pulmonary aspergillosis (IPA).
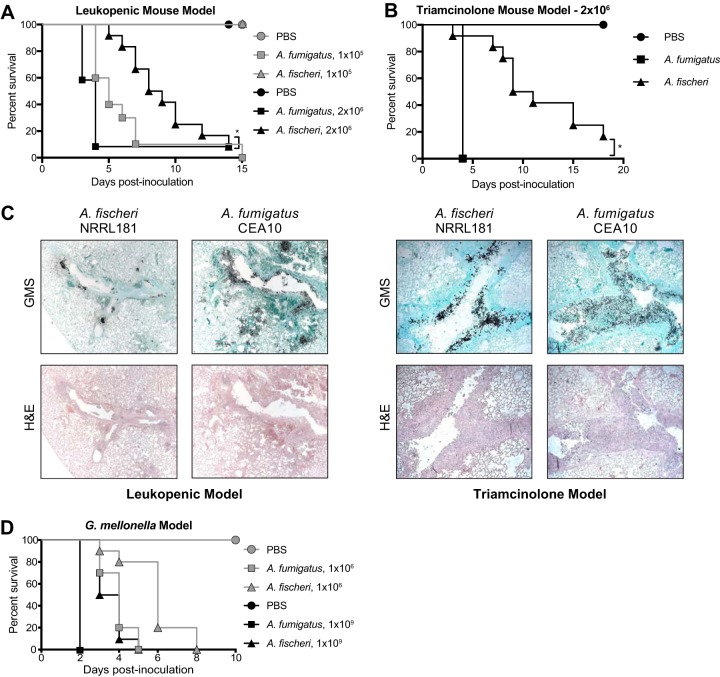
In contrast to A. fumigatus-mediated disease, only a few cases of invasive fungal infections have been reported to be caused by A. fischeri (21–24). Given this contrast, we utilized two immunologically distinct murine IPA models to assess differences in pathogenicity and virulence between the two species. In a leukopenic murine model, A. fischeri NRRL 181 was significantly less virulent than A. fumigatus CEA10, in a dose-dependent manner (Fig. 1A). Using an inoculum of 1 × 105 conidia, A. fischeri was completely attenuated in virulence, with 100% murine survival by day 15 post-fungal challenge. In contrast, inoculation with A. fumigatus resulted in 100% murine mortality by day 15 (Fig. 1A). Using a higher dose (2 × 106) of conidia, both strains caused 90% mortality by day 14; however, the disease progression was markedly different. Eighty percent of mice inoculated with A. fumigatus succumbed to infection by day 4, whereas in mice inoculated with A. fischeri, mortality started occurring on day 5, and then one or two mice succumbed each day until day 14 (Fig. 1A). Thus, despite the similar overall mortality at higher fungal challenge doses, A. fischeri is substantially less virulent than A. fumigatus in a leukopenic murine IPA model.
FIG 1.
A. fischeri is significantly less virulent than A. fumigatus in multiple murine models of invasive pulmonary aspergillosis. (A) Cumulative survival of mice inoculated with 1 × 105 (gray) or 2 × 106 (black) conidia in a leukopenic model of IPA. For the inoculations of 1 × 105 conidia, 10 mice were infected per group, and for the inoculations of 2 × 106 conidia, 12 mice were infected per group. For each PBS control, 4 mice were inoculated. *, P = 0.0098 by log rank test and P = 0.0002 by Gehan-Breslow-Wilcoxon test. (B) Cumulative survival of mice inoculated with 2 × 106 conidia in a steroid model of IPA. n = 12/group, 4 for PBS. *, P = <0.0001 by log rank and Gehan-Breslow-Wilcoxon tests. (C) Histological sections from 3 days postinoculation in a leukopenic (left) and steroid (right) model of IPA, stained with H&E and GMS. Images were acquired at 100×. (D) Cumulative survival of G. mellonella larvae inoculated with 1 × 106 (gray) or 1 × 109 (black) conidia. Ten larvae were used per condition in all assays. Survival curves for A. fischeri and A. fumigatus were significantly different (P < 0.003) in both log rank and Gehan-Breslow-Wilcoxon tests for both inocula.
To better understand what is happening in vivo during disease progression with A. fischeri NRRL 181 and A. fumigatus CEA10, we performed histological analyses on lungs from the leukopenic model 3 days postinoculation. Histological sections were stained with Gomori methenamine silver (GMS) to visualize fungal burden and with hematoxylin and eosin (H&E) stain to visualize host-related pathology (Fig. 1C). Leukopenic mice inoculated with A. fumigatus had an overall greater number of fungal lesions per animal than mice inoculated with A. fischeri. These A. fumigatus lesions contained a greater amount of fungus, which coincided with increased tissue necrosis surrounding the lesions. There was also a greater number of A. fumigatus lesions with fungi invading from the larger airways into the lung parenchyma. Meanwhile, the A. fischeri lesions were largely maintained within the larger airways. A striking difference between the lesions caused by the two species was in the appearance of the hyphae of the two species, where A. fischeri filaments were much shorter than those of A. fumigatus. We hypothesize that in the leukopenic mouse, A. fischeri is unable to grow as well as A. fumigatus, resulting in smaller and fewer fungal lesions, shorter fungal filaments, and attenuation of virulence.
As the patient population at risk for IA continues to change and grow (28), we next tested a nonleukopenic, triamcinolone (steroid)-induced immune suppression model and observed a significant reduction in virulence of A. fischeri compared to A. fumigatus (P < 0.0001 by log rank and Gehan-Breslow-Wilcoxon tests). All mice inoculated with A. fumigatus succumbed to infection by day 4; however, similar to the leukopenic model, mice inoculated with A. fischeri had significantly slower disease progression as monitored by Kaplan-Meier analyses (Fig. 1B).
Histological analyses were also carried out on lungs from the steroid model 3 days post-fungal inoculation (Fig. 1C). Overall, mice inoculated with A. fischeri in the steroid model had similar numbers of fungal lesions as those inoculated with A. fumigatus, but the lesions caused by the two species were phenotypically distinct (Fig. 1C). In larger terminal bronchioles infected with A. fumigatus, there was greater fungal growth per lesion, and the growth was observed throughout the bronchiole itself, extending well into the lumen. These lesions were accompanied by substantial granulocytic inflammation and obstructed the airways surrounding the hyphae (Fig. 1C). In the lesions containing A. fischeri, the fungal growth was contained to the epithelial lining of the bronchioles. This pattern of growth was accompanied by inflammation at the airway epithelia, leaving the airway lumen largely unobstructed (Fig. 1C). The lack of airway obstruction during A. fischeri infection and reduced inflammation may contribute to the reduced virulence compared to A. fumigatus in this murine model.
Although the distribution of the fungal lesions varied, there was still significant fungal growth in the steroid immunosuppressed mice infected with A. fischeri, suggesting that A. fischeri is capable of growing within the immunocompromised murine host. Indeed, we tested the growth rate of A. fischeri and A. fumigatus in lung homogenate as a proxy for growth capability within the nutrient environment of the host and observed no difference between the two strains (see Fig. S1 in the supplemental material). These experiments show that in multiple models of fungal disease, A. fischeri is less virulent than A. fumigatus even though it is able to grow within the immunocompromised murine lung.
A. fumigatus grows slower than A. fischeri in glucose minimal medium (GMM) but at the same speed as A. fischeri in lung homogenate medium. A. fumigatus CEA10 or A. fischeri NRRL 181 was cultured in flat-bottom 96-well plates at 2 × 104 conidia per well. Conidia were added in 20 µl of 0.01% Tween 80, and medium was carefully pipetted over the inoculum into each well. Lung homogenate was generated according to reference 29. Plates were incubated for 7 hours at 37°C before measurements at 405 nm were taken every 10 min. Mean and SEM for eight technical replicates; data are representative of three biological replicates. Download FIG S1, PDF file, 0.2 MB (181.8KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We observed similar pathogenicity and virulence results when using the Galleria mellonella insect larva model of aspergillosis (Fig. 1D). Both low (1 × 106 conidia)- and high (1 × 109 conidia)-inoculum experiments showed significant differences between the disease progression of A. fischeri (slower) and A. fumigatus (faster) in this insect model of fungal pathogenicity.
Compared to A. fumigatus, A. fischeri differs in its response to several host-relevant stresses.
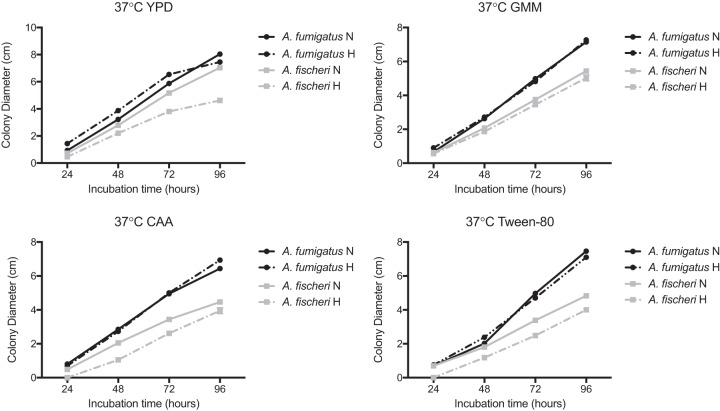
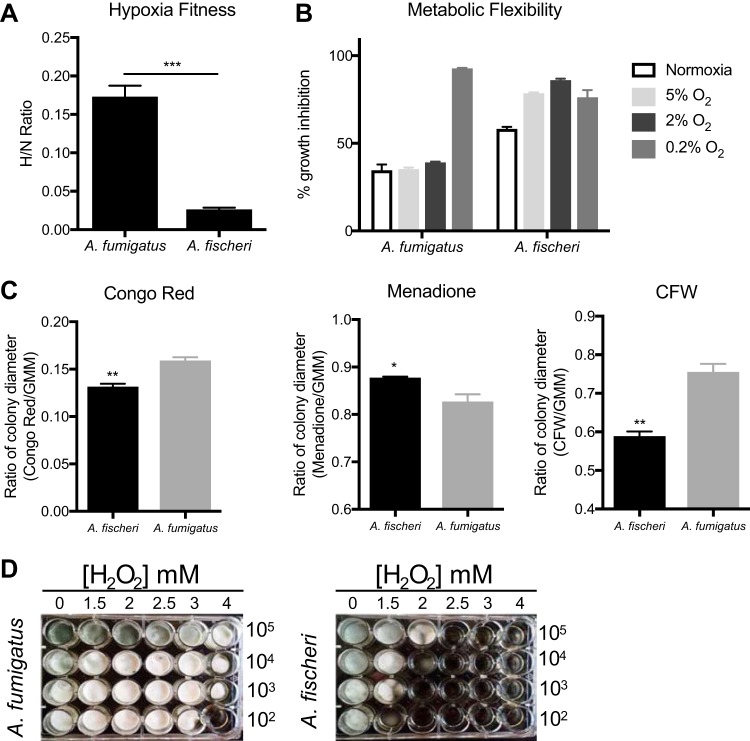
Our in vivo experiments suggested that the lower virulence of A. fischeri is not the result of an inability to grow within the host per se. Therefore, we hypothesized that A. fischeri is unable to mitigate stresses encountered in the host as effectively as its close evolutionary relative A. fumigatus. Nutrient fluctuation is a stress encountered in vivo during A. fumigatus infection (29), and to assess differences in metabolic plasticity between the two species, we measured the two organisms’ growth on media supplemented with glucose, fatty acids (Tween 80), or Casamino Acids. Because low oxygen tension is a significant stress encountered during infection (5), and recently, fitness in low oxygen has been correlated with virulence of A. fumigatus (30), we measured growth of both species at 37°C under both normoxic (ambient air) and hypoxia-inducing (0.2% O2, 5% CO2) conditions. In normoxia with glucose, fatty acids (Tween 80), or Casamino Acids supplied as the carbon source, radial growth of A. fischeri was reduced compared to A. fumigatus (Fig. 2). However, on rich media both organisms grew equally well (Fig. 2). We also observed a lower growth rate of A. fischeri than A. fumigatus in the first 16 h of culture in liquid media supplied with glucose at 37°C. At 30°C, A. fischeri grew the same as, or better than, A. fumigatus except on Tween 80, where A. fumigatus had a slight advantage (Fig. S2). Also, A. fischeri grew substantially worse than A. fumigatus at 44°C (Fig. S2), a temperature rarely observed in the patient but easily observed in compost piles. To determine relative fitness in hypoxic liquid environments, we measured the ratio of biomass in liquid culture in ambient air (normoxia) versus hypoxic (0.2% O2, 5% CO2) conditions. A. fischeri showed significantly lower fitness under hypoxic conditions, with about an 8.5-fold-lower biomass than A. fumigatus (Fig. 3A). These data suggest that A. fischeri is less fit than A. fumigatus at 37°C and under low-oxygen conditions, both of which have been shown to impact fungal virulence.
FIG 2.
A. fischeri is unable to thrive under suboptimal metabolic conditions at 37°C. A total of 1 × 103 conidia were point inoculated on each plate, and then plates were incubated at 37°C in normoxia (N; ∼21% oxygen, 5% CO2) or hypoxia (H; 0.2% O2, 5% CO2); colony diameter was measured every 24 h. Mean and SEM for triplicates. CAA, Casamino Acids; GMM, glucose minimal medium.
FIG 3.
A. fischeri is more susceptible to multiple host-relevant stresses than A. fumigatus. (A) Fitness ratio of A. fumigatus or A. fischeri during hypoxic versus normoxic growth (measured as the dry weight of cultures). Data represent mean and SEM for biological triplicates; ***, P = 0.0006 by Student’s t test. (B) Growth inhibition of strains grown on 1% lactate minimal medium with 0.1% 2-deoxyglucose (2-DG) under a range of low-oxygen conditions. (C) A. fumigatus and A. fischeri were grown in the presence of the cell wall-perturbing agent Congo red (0.5 mg/ml), the oxidative stressor menadione (20 µM), or the chitin-perturbing agent calcofluor white (CFW; 25 µg/ml). Plates were grown for 96 h at 37°C and 5% CO2. For all plates except Congo red and its GMM control, 1 × 103 spores were plated. For Congo red and the control GMM plate, 1 × 105 spores were plated. Student’s t test was performed, and significance was as follows: *, P < 0.05; **, P < 0.01. (D) Strains were grown for 48 h at 37°C in liquid complete medium supplemented with increasing concentrations of hydrogen peroxide.
A. fischeri and A. fumigatus exhibit similar growth patterns at 30°C but not at 44°C. (A to D) Conidia at 1 × 103 were point inoculated on each plate, and plates were then incubated at 30°C in normoxia (∼21% oxygen, 5% CO2); colony diameter was measured every 24 hours. Mean and SEM for triplicates. Tween 80, 1% Tween 80 provided as sole carbon source; CAA, Casamino Acids; GMM, glucose minimal medium. (E) Error bars indicate standard deviations between biological duplicates (**, P value < 0.005 in a paired, equal-variance Student t test). Download FIG S2, PDF file, 0.7 MB (723.5KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic flexibility, or the ability for an organism to utilize multiple carbon sources simultaneously, has been suggested to provide a fitness advantage to Candida albicans during in vivo growth (31). Metabolic flexibility can be characterized using the glucose analog 2-deoxyglucose (2-DG), in combination with an alternative carbon source available in vivo, such as lactate. 2-DG triggers carbon catabolite repression, which shuts down alternative carbon utilization pathways. However, in C. albicans this shutdown is delayed and growth occurs on lactate with 2-DG (31, 32). We tested the metabolic flexibility of both A. fumigatus and A. fischeri and observed that while both species can grow in the presence of 2-DG on lactate, the growth inhibition of A. fischeri is higher (∼60%) than that of A. fumigatus (∼35%; Fig. 3B). Even under low-oxygen conditions (5% and 2%), A. fumigatus maintains this metabolic flexibility except under extremely low oxygen conditions (0.2%), whereas A. fischeri shows even greater inhibition at all oxygen tensions of 5% or below. These data suggest that while both species exhibit some level of metabolic flexibility, A. fumigatus appears more metabolically flexible under a wider range of conditions than A. fischeri.
Next, we measured the susceptibility of A. fischeri to oxidative stress, cell wall stress, and antifungal drugs as they are stresses that are encountered during infection and treatment (33). Interestingly, we observed that A. fischeri is more resistant to the intracellular oxidative stress agent menadione than A. fumigatus but more susceptible to the external oxidative stress agent hydrogen peroxide (Fig. 3C and D). As the in vivo levels of inflammation caused by the two species appeared different, we indirectly tested for differences in cell wall pathogen-associated molecular patterns using the cell wall-perturbing agents Congo red and calcofluor white. A. fumigatus was significantly more resistant to both agents than A. fischeri (Fig. 3C), suggesting differences in the response to cell wall stress or in the composition and organization of the cell wall between the two species. These differences are likely important for host immune cell recognition and interaction, which in turn influence pathology and disease outcome.
Last, A. fischeri showed enhanced resistance relative to A. fumigatus for three of the four antifungal drugs tested (Table 1), consistent with previous experiments (34). Overall, our phenotypic data show that the response of A. fischeri to host-related stresses and antifungals is substantially different from that of A. fumigatus. Furthermore, our results suggest that increased growth capability of A. fumigatus in low oxygen and in high temperatures are two important attributes that likely contribute to its pathogenic potential compared to A. fischeri.
TABLE 1.
A. fischeri shows enhanced resistance relative to A. fumigatus for several antifungal drugs
| Strain | MIC of drug (µg/ml) |
MEC of caspofungin (µg/ml) | ||
|---|---|---|---|---|
| Posaconazole | Voriconazole | Itraconazole | ||
| A. fumigatus | 0.7 | 0.8 | 5 | 0.09 |
| A. fischeri | 2.4 | >4 | >24 | 0.06 |
The proteomes of A. fumigatus and A. fischeri are highly similar, but their secondary metabolic pathways show substantial divergence.
The large differences in virulence and virulence-related traits we observed between A. fumigatus and A. fischeri led us to investigate the genotypic differences that could be responsible. To describe the genomic similarities and differences between A. fumigatus and A. fischeri, we determined how many orthologous proteins and how many species-specific proteins were present in each genome using a reciprocal best BLAST hit approach (35). We identified 8,737 proteins as being shared between the two species (Fig. S3), representing 88% and 84% of the A. fumigatus and A. fischeri proteomes, respectively, and 1,684 A. fischeri-specific proteins (16% of its proteome) and 1,189 A. fumigatus-specific proteins (12% of its proteome). These results are similar to what had previously been reported in an early analysis of the A. fischeri genome (16).
The genomes of A. fumigatus and A. fischeri are largely similar, but their secondary metabolic pathways are quite divergent. Left, Venn diagram showing the sets of A. fischeri-specific proteins, shared orthologous proteins, and A. fumigatus-specific proteins encoded in each genome. Numbers below each species name indicate the total number of proteins encoded in that genome. Right, Venn diagram showing the sets of A. fischeri-specific BGC proteins, shared BGC proteins, and A. fumigatus-specific BGC proteins. Numbers below each species name indicate the total number of BGC proteins encoded in that genome. In each diagram, circles are proportional to the number of proteins they contain. Download FIG S3, PDF file, 0.1 MB (129.3KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To narrow our search for genes that are absent in A. fischeri but are important for A. fumigatus disease, we compiled a list of 49 A. fumigatus genes considered to be involved in virulence (see Table S1 in the Figshare document at https://doi.org/10.6084/m9.figshare.7149167) based on two previously published articles (36, 37) and extensive literature searches of our own. We observed that all but one of these virulence-associated genes were also present in A. fischeri, a surprising finding considering the substantial differences observed between the two species in our animal models of infection. The virulence-associated gene not present in A. fischeri is pesL (Afu6g12050), a nonribosomal peptide synthase that is essential for the synthesis of the secondary metabolite fumigaclavine C and required for virulence in the Galleria model of A. fumigatus infection (38).
Since the only previously described A. fumigatus virulence-associated gene not present in the A. fischeri genome (i.e., pesL) is involved in secondary metabolism and a previous study suggested that secondary metabolism is not conserved between these two species (39), we investigated the differences between the repertoire of secondary metabolic pathways present in A. fumigatus and A. fischeri. Using the program antiSMASH (40), we identified 598 genes distributed among 33 BGCs in A. fumigatus (see Table S2 in the Figshare document at https://doi.org/10.6084/m9.figshare.7149167) and 786 genes spread out over 48 BGCs in A. fischeri (see Table S3 in the Figshare document at https://doi.org/10.6084/m9.figshare.7149167). Of these 598 A. fumigatus genes, 407 (68%) had an orthologous gene that was part of an A. fischeri BGC. This level of conservation of BGC genes (68%) is much lower than the amount of conservation observed for the rest of the proteome (88%), illustrating the high rate at which fungal metabolic pathways evolve (41, 42).
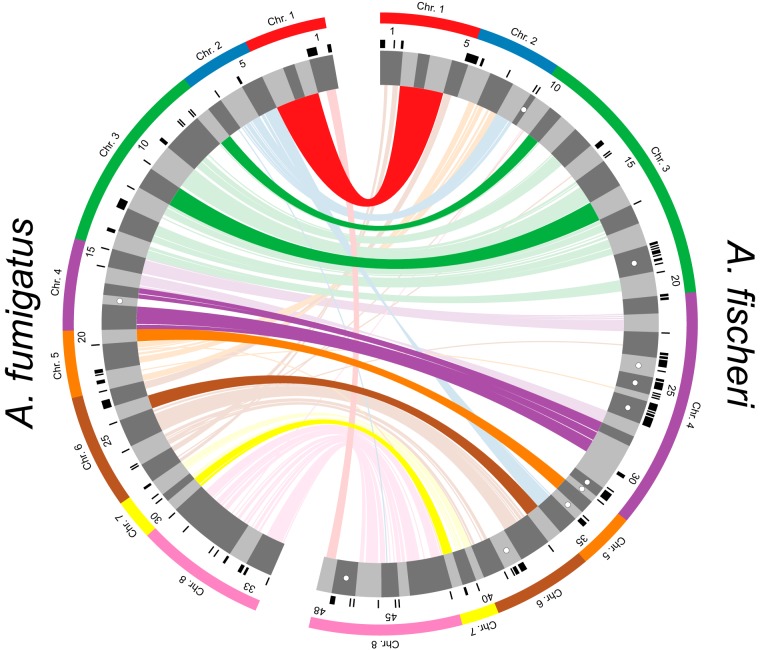
We next directly compared the BGCs of the two organisms. An A. fumigatus BGC was considered conserved in A. fischeri if ≥90% of its genes were also present in an A. fischeri BGC and vice versa. We found that only 10/33 A. fumigatus BGCs are conserved in A. fischeri and only 13/48 A. fischeri BGCs are conserved in A. fumigatus (Fig. 4), a finding consistent with previous results (39) and the low conservation of individual secondary metabolic genes between the two species. While only 10 A. fumigatus BGCs were conserved in A. fischeri, many other BGCs contained one or more orthologs of genes in A. fischeri BGCs.
FIG 4.
Biosynthetic gene clusters (BGCs) of A. fumigatus and A. fischeri show substantial evolutionary divergence. Predicted BGCs are shown in the inner track and are alternatively colored dark and light gray, and their size is proportional to the number of genes in them. Black ticks on the exterior of the cluster track indicate genes that possess an ortholog in the other species but are not in a BGC in the second species. White dots indicate species-specific clusters. Solid bars on the exterior correspond to the chromosome on which the BGCs below them reside. Genes are connected to their orthologs in the other species with dark lines if >90% of the BGC genes in A. fumigatus are conserved in the same BGC in A. fischeri. Lighter lines connect all other orthologs that are present in both species’ sets of BGCs. The image was made using Circos version 0.69-4 (86).
Only one BGC (cluster 18) was completely A. fumigatus specific. Interestingly, our previous examination of the genomes of 66 A. fumigatus strains showed that this BGC was a “jumping cluster,” as it was found to be present in only 5 strains and to reside in three distinct genomic locations (42). Conversely, there are 10 A. fischeri-specific BGCs that do not have orthologs in BGCs in A. fumigatus. One of these BGCs is responsible for making helvolic acid (a gene cluster known to be absent from the A. fumigatus strain CEA10 but present in strain Af293 [42]), but the other 9 have not been biochemically connected to any metabolite.
All the genes required for the production of the mycotoxin gliotoxin are located in a BGC in A. fischeri (Fig. S4) and are in fact similar to their A. fumigatus orthologs (43), even though A. fischeri is not known to produce this mycotoxin (44). Both the gliotoxin and acetylaszonalenin BGCs are adjacent to one another in the A. fischeri genome (Fig. S4). In A. fumigatus, the gliotoxin BGC is immediately next to what appears to be a truncated version of the acetylaszonalenin BGC that lacks portions of the nonribosomal peptide synthase and acetyltransferase genes as well as the entire indole prenyltransferase gene required for acetylaszonalenin production. The close proximity of these two BGCs is noteworthy, as it is similar to previously reported “superclusters” in A. fumigatus and A. fumigatus-related strains (45). These superclusters have been hypothesized to be “evolutionary laboratories” that may give rise to new compounds and pathways (42).
The acetylaszonalenin and gliotoxin biosynthetic gene clusters in A. fumigatus and A. fischeri are located immediately next to one another. The portions of clusters 37 and 25 from A. fischeri and A. fumigatus, respectively, that are known to contain the previously characterized acetylaszonalenin (96) and gliotoxin (43) BGCs are shown. Genes colored in shades of green are involved in the acetylaszonalenin biosynthetic pathway. Dark green, anaPS (nonribosomal peptide synthase); light green, anaAT (acetyltransferase); green, anaPT (prenyltransferase); orange, gliotoxin biosynthetic genes. Gray arrow, syntenic gene in both species not involved in gliotoxin synthesis. Sequences that are similar to one another (based on blastn scores) are marked by gray parallelograms. Image was made using Easyfig version 2.2.2 (88). Download FIG S4, PDF file, 0.1 MB (118.2KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Isolation and characterization of three new compounds from A. fischeri.
The relatively low level of conservation of BGCs we observed between A. fumigatus and A. fischeri led us to characterize the secondary metabolites produced by A. fischeri (Fig. S5) (46–50). The one strain-many compounds (OSMAC) approach was used to alter the secondary metabolites being biosynthesized in order to produce a diverse set of molecules (51–54). Depending on the medium on which it was grown, A. fischeri produced as few as 4 (yeast extract soy peptone dextrose agar [YESD]) or as many as 10 (oatmeal agar [OMA]) compounds (Fig. S6). These results showed that culture medium influences the biosynthesis of secondary metabolites in A. fischeri, a phenomenon observed in many other fungi (52, 55).
A custom chemical analysis protocol was developed for studying the metabolites produced by A. fischeri. Approximately 60 ml of 1:1 CH3OH-CH3Cl was added to cultures of Aspergillus fischeri grown on solid-state fermentation for 2 weeks. The cultures were then chopped thoroughly with a large scalpel and shaken for 16 hours using an orbital shaker. The liquid culture was then vacuum filtered and concentrated using 90 ml CH3Cl and 150 ml water and transferred into a separatory funnel. The organic (bottom) layer was drawn off and evaporated to dryness. The dried, desugared extract was reconstituted in 100 ml of 1:1 CH3OH-CH3CN and 100 ml of hexane. The biphasic solution was shaken vigorously and transferred to a separatory funnel. The CH3OH-CH3CN layer was evaporated to dryness under vacuum, producing a defatted extract. The extract was then subdivided into several peaks or fractions using flash chromatography. The subfractions were further separated using HPLC until pure compounds were isolated. The pure compounds were subjected to UPLC-MS analysis to establish the molecular formula and fragmentation patterns. Finally, pure compounds were identified using both NMR analysis and information from UPLC-MS data. Download FIG S5, PDF file, 0.9 MB (899.7KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. fischeri produces different numbers of metabolites, depending on the medium it is grown on. Base peak chromatograms as measured by LC-MS, illustrating how the chemistry profiles varied based on growth conditions. PDA + ab was used as the chemical control to observe the differences in the secondary metabolites, due to it being the medium in which A. fischeri is stored. There were overall no chemical differences observed between the different variations of PDA medium. Each peak (which indicates different chemical entities) was observed in the three PDA variations, albeit at fluctuating intensities. SDA, PYG, and YESD produced the majority of the peaks observed in PDA, but it also lacked some observed peaks, indicating that these growth conditions were not chemically favored. CYA produced the majority of the peaks, as well as an additional peak that was observed at a much lower intensity in PDA. However, this peak was similarly observed in OMA. OMA produced similar peaks as those observed in PDA, but with higher intensity. Due to this, OMA was selected for further study. The gray boxes indicate differences in the observed peaks from PDA. See the Figshare document at https://doi.org/10.6084/m9.figshare.7149167 for more information. Download FIG S6, PDF file, 0.6 MB (670.6KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
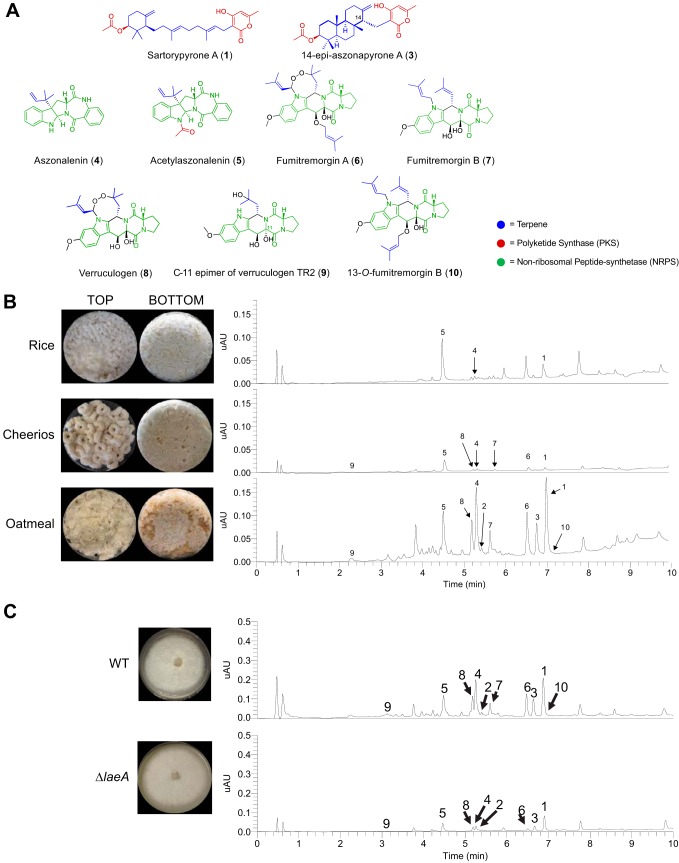
To characterize the peaks of interest we observed when A. fischeri was grown on OMA, we increased the size of our fungal cultures; doing so yielded seven previously isolated compounds (sartorypyrone A [compound 1], aszonalenin [compound 4], acetylaszonalenin [compound 5], fumitremorgin A [compound 6], fumitremorgin B [compound 7], verruculogen [compound 8], and the C-11 epimer of verruculogen TR2 [compound 9]) and three newly biosynthesized secondary metabolites (sartorypyrone E [compound 2], 14-epiaszonapyrone A [compound 3], and 13-O-fumitremorgin B [compound 10]). Two of the secondary metabolites were new compounds (compounds 2 and 3), and one was a new natural product (compound 10) (Fig. 5B). The structures for all 10 compounds were determined using a set of spectroscopic (1D and 2D NMR) and spectrometric techniques (high-resolution mass spectrometry [HRMS]). Our data for sartorypyrone A (compound 1) (56), aszonalenin (compound 4) (57, 58), acetylaszonalenin (compound 5) (56, 59), fumitremorgin A (compound 6) (60, 61), fumitremorgin B (compound 7) (62–64), verruculogen (compound 8) (65, 66), and the C-11 epimer of verruculogen TR2 (compound 9) (66) correlated well with literature values. The structures of 14-epiaszonapyrone A (compound 3) and 13-O-prenyl fumitremorgin B (compound 10) were fully characterized in this study (see Figshare document at https://doi.org/10.6084/m9.figshare.7149167); the structure elucidation of sartorypyrone E (compound 2) is ongoing and will be reported in detail in a forthcoming manuscript.
FIG 5.
Secondary metabolite production in A. fischeri. (A) Compounds isolated from A. fischeri: 1, sartorypyrone A; 2, sartorypyrone E; 3, 14-epimer aszonapyrone A; 4, aszonalenin; 5, acetylaszonalenin; 6, fumitremorgin A; 7, fumitremorgin B; 8, verruculogen; 9, C-11 epimer verruculogen TR2; and 10, 13-O-prenyl-fumitremorgin B. The color coding indicates which putative class the molecule belongs to, e.g., terpenes, PKS, or NRPS. (B) (Top) Aspergillus fischeri was initially grown on rice for 2 weeks and then extracted using methods outlined in Fig. S5. The rice culture yielded compounds 1, 4, and 5. (Middle) A. fischeri was grown on multigrain Cheerios for 2 weeks, which yielded compounds 1 and 4 to 9. (Bottom) A. fischeri grown on Quaker oatmeal for 2 weeks. All compounds that were previously isolated in rice and multigrain Cheerios cultures in addition to three new compounds (2, 3, and 10) were found in the oatmeal culture. All pictures depict fungi growing in 250-ml Erlenmeyer flasks; the left panel indicates the top view, while the right panel shows the bottom view. All chromatographic profiles have been normalized to the highest μAU value. (C) A. fischeri wild-type and ΔlaeA strains were grown on solid breakfast oatmeal for 2 weeks and extracted using organic solvents as indicated previously. The crude desugared and defatted extracts were run using UPLC-MS at a concentration of 2 mg/ml with 5 μl being injected for analysis. The chromatographic profiles were normalized to the highest μAU value. Mass spectrometry analysis indicated the presence of secondary metabolites 1 to 10 within the wild type, and only compounds 1 to 6, 8, and 9 were seen in the ΔlaeA mutant. All pictures show A. fischeri grown on oatmeal agar in petri plates.
Since four secondary metabolites (compounds 5 to 8) from A. fischeri had also been reported from A. fumigatus (Table 2), we hypothesized that the mechanisms A. fischeri employs to regulate its secondary metabolism would also be similar to those used by A. fumigatus. To test this hypothesis, we constructed a deletion mutant of laeA in A. fischeri (Fig. S7). LaeA is a master regulator of secondary metabolism in A. fumigatus and a variety of other fungi (67–69). Both the wild-type and ΔlaeA strains of A. fischeri were subjected to LC-MS analysis. The chromatographic profile of the ΔlaeA strain showed mass data that corresponded to sartorypyrone A (compound 1), sartorypyrone E (compound 2), 14-epiaszonapyrone A (compound 3), aszonalenin (compound 4), acetylaszonalenin (compound 5), fumitremorgin A (compound 6), verruculogen (compound 8), and the C-11 epimer of verruculogen TR2 (compound 9). However, the relative abundance of compounds present was very low compared to the wild type (Fig. 5C). Fumitremorgin B (compound 7) and 13-O-prenyl-fumitremorgin B (compound 10) were not produced by the ΔlaeA mutant at all.
TABLE 2.
Secondary metabolites isolated from A. fischeri that have been reported from A. fumigatus
| Secondary metabolite (compound no.) | Presence in strain: |
|
|---|---|---|
| A. fischeri | A. fumigatus | |
| Sartorypyrone A (1) | + | − |
| Sartorypyrone E (2) | + | − |
| 14-Epiaszonapyrone A (3) | + | − |
| Aszonalenin (4) | + | − |
| Acetylaszonalenin (5) | + | + |
| Fumitremorgin A (6) | + | + |
| Fumitremorgin B (7) | + | + |
| Verruculogen (8) | + | + |
| C-11 epimer of verruculogen TR2 (9) | + | − |
| 13-O-Fumitremorgin B (10) | + | − |
Southern blotting confirms construction of the ΔlaeA mutant. A 1-kb probe recognizes a single DNA band (∼4.4 kb) in the wild-type strain and a single DNA band (∼2.7 kb) in the ΔlaeA mutant. Download FIG S7, PDF file, 0.2 MB (240.8KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
A. fumigatus is a major human fungal pathogen, yet its close relative A. fischeri is rarely an agent of human disease. A number of traits that contribute to the virulence of A. fumigatus have been characterized, but their distribution and potential role in A. fischeri-mediated disease are largely unknown. In this study, we thoroughly characterized A. fischeri (strain NRRL 181) and compared it to A. fumigatus (strain CEA10) for multiple disease-relevant biological and chemical differences. Our data show that A. fischeri can grow in a mammalian host but is much less fit and causes a disease progression quite different than that observed during A. fumigatus infections (Fig. 1 and 2). Future studies will be needed to determine if these differences between A. fischeri NRRL 181 and A. fumigatus CEA10 are conserved across a range of A. fischeri and A. fumigatus isolates.
Further investigations revealed that secondary metabolic genes are much less conserved than genes in the rest of the genome (see Fig. S3 in the supplemental material), and a chemical analysis of A. fischeri resulted in the identification of both previously identified and new compounds (Fig. 5). While the BGCs producing secondary metabolites in A. fischeri and A. fumigatus appear to be quite different, our data suggest that a master regulator of secondary metabolism in A. fumigatus (laeA) possesses a similar role in A. fischeri (Fig. 5C). This is consistent with previous observations that the global regulation of secondary metabolism is conserved but that the underlying BGCs are not, suggesting that the regulatory circuit involved in Aspergillus secondary metabolism has undergone extensive regulatory rewiring (41). Our analyses are the first to infer the occurrence of regulatory circuit rewiring in A. fischeri and are among the first gene knockout studies of this organism (70).
In order to cause disease, a microbe must be able to respond to the set of diverse and stressful environments presented by its host. Based on our data, A. fischeri strain NRRL 181 is unable to respond to many of these stresses as well as A. fumigatus CEA10 (Fig. 2 and 3). We hypothesize that this inability to thrive under stress contributes to the various disease progressions observed during our animal model experiments (Fig. 1). Some or all of the genetic determinants responsible for this discrepancy in stress response and virulence could reside in the ∼1,200 A. fumigatus-specific genes we identified (Fig. S3); alternatively, some of the ∼1,700 A. fischeri-specific genes we identified may inadvertently facilitate control of A. fischeri in a mammalian host. An additional explanation for the discrepancy in stress response is that A. fumigatus has evolved more efficient regulatory circuits than A. fischeri that allow it to cope with more stringent conditions that are observed in the host.
Even though more than 10% of the genes in each species lack an ortholog in the other species, only ∼2% (1/49) of previously identified genetic determinants of virulence in A. fumigatus are not conserved in A. fischeri (see Table S1 in the Figshare document at https://doi.org/10.6084/m9.figshare.7149167). This result and our observation that many of the pathways of secondary metabolism are quite different between A. fischeri and A. fumigatus support a multifactorial model of A. fumigatus virulence (71–73) and suggest a need to investigate virulence on multiple levels of biological complexity. In order to cause disease in a host, A. fumigatus (and other species closely related to it) must adhere and germinate in the lung (73), survive inherently stressful conditions presented by host environments (e.g., severe lack of metals and oxygen) (5, 74, 75), and modulate or endure actions of the host immune system (76). Given the diversity of these activities, it is unlikely that single genes or pathways will be responsible for the totality of A. fumigatus-derived disease, even though many genes in the genome have not yet been characterized for their role in pathogenicity. We hypothesize that multiple pathways (including those involved in secondary metabolism) have changed during the evolution of A. fischeri and A. fumigatus, resulting in their differing ability to cause disease.
As mentioned, an important caveat to our experiments is that we analyzed only a single, representative strain from each species, and further studies are needed to determine how typical these observed trait and genomic differences are across multiple strains from each species. Several recent studies have identified a wide variety of differences between A. fumigatus strains, which have in turn been shown to contribute to physiological differences, including but not limited to secondary metabolism and virulence (30, 72, 73). While the genome of only one isolate of A. fischeri has so far been sequenced (16) and the organism has been reported to cause human disease only a few times (21–24), it would be of great interest to compare patient-derived and environment-derived isolates at the genomic, phenotypic, and chemical levels. Although it appears that clinical and environmental isolates do not stem from separate lineages in A. fumigatus (77), whether this is also the case for largely nonpathogenic species, such as A. fischeri, or for rarely isolated pathogenic species, such as A. lentulus or A. udagawae, remains largely unknown.
Our mouse infection studies revealed that A. fischeri strain NRRL 181 was less virulent than A. fumigatus strain CEA10 in both leukopenic and steroid-induced immune suppression models of disease. The difference in disease outcome in the steroid model closely matches what was observed in comparisons of two A. fumigatus strains, CEA10 and Af293 (30). It is intriguing that the difference between pathogenic (A. fumigatus CEA10) and nonpathogenic (A. fischeri NRRL 181) species in disease outcomes in this model is similar to that between two clinically derived pathogenic strains (A. fumigatus CEA10 and Af293). Interestingly, Kowalski et al. (30) did not observe any statistically significant difference in disease progression between CEA10 and Af293 in the leukopenic model, whereas we found that A. fischeri is less virulent in this second murine model of disease (Fig. 1). These strain-, species-, and infection model-specific differences in disease outcomes are consistent with our hypothesis that A. fumigatus virulence is a complex genetic trait and highlight the need for further studies using wider sets of strains, species, and virulence-related traits.
A. fumigatus and A. fischeri are members of Aspergillus section Fumigati, a clade that includes multiple closely related species, some of which are pathogens (e.g., A. fumigatus, A. lentulus, and A. udagawae) and some of which are considered nonpathogens (e.g., A. fischeri, A. aureolus, and A. turcosus) (1, 44, 78, 79). The ability to cause disease in humans appears to have either arisen or been lost (or both) multiple times independently during the evolution of this lineage, as pathogenic species are spread throughout the phylogeny (17, 80). A broader, phylogenetically informed comparison of pathogenic and nonpathogenic species in section Fumigati would provide far greater resolution in identifying (or dismissing) factors and pathways that may contribute to or prevent the ability of these organisms to cause disease. Also, leveraging the diversity of section Fumigati would give researchers a better understanding of the nature and evolution of human fungal pathogenesis as the appreciation for the health burden caused by fungi increases (81).
MATERIALS AND METHODS
Strains and growth media.
A. fischeri strain NRRL 181 was acquired from the ARS Culture Collection (NRRL). A. fumigatus strain CEA10 (CBS 144.89) was obtained from the Westerdijk Fungal Biodiversity Institute (CBS). All strains were grown on glucose minimal medium (GMM) from conidial glycerol stocks stored at −80°C. All strains were grown in the presence of white light at 37°C. Conidia were collected in 0.01% Tween 80 and enumerated with a hemocytometer.
Murine virulence studies.
For the chemotherapeutic (leukopenic) murine model, outbred CD-1 female mice (Charles River Laboratories, Raleigh, NC, USA), 6 to 8 weeks old, were immunosuppressed with intraperitoneal (i.p.) injections of 150 mg/kg (of body weight) cyclophosphamide (Baxter Healthcare Corporation, Deerfield, IL, USA) 48 h before and 72 h after fungal inoculation, along with subcutaneous (s.c.) injections of 40 mg/kg triamcinolone acetonide (Kenalog-10; Bristol-Myers Squibb, Princeton, NJ, USA) 24 h before and 6 days after fungal inoculation. For the murine triamcinolone model, outbred CD-1 female mice, 6 to 8 weeks old, were treated with 40 mg/kg triamcinolone acetonide by s.c. injection 24 h prior to fungal inoculation.
Unless otherwise noted, conidial suspensions of 2 × 106 conidia were prepared in 40 μl sterile PBS and administered to mice intranasally while under isoflurane anesthesia. Mock mice were given 40 μl PBS. Mice were monitored three times a day for signs of disease for 14 or 18 days postinoculation. Survival was plotted on Kaplan-Meier curves, and statistical significance between curves was determined using Mantel-Cox log rank and Gehan-Breslow-Wilcoxon tests. Mice were housed in autoclaved cages at 4 mice per cage with HEPA-filtered air and autoclaved food and water available ad libitum.
Galleria mellonella virulence studies.
G. mellonella larvae were obtained by breeding adult moths (82). G. mellonella larvae of a similar size were selected (approximately 275 to 330 mg) and kept without food in glass containers (petri dishes) at 37°C in darkness for 24 h prior to use. A. fumigatus and A. fischeri conidia were obtained by growth on YAG medium for 2 days. The conidia were harvested in PBS and filtered through Miracloth (Calbiochem). The concentration of conidia was estimated by using a hemocytometer, and conidia were resuspended at a concentration of 2.0 × 108 conidia/ml. The viability of the conidia was determined by incubation on YAG medium at 37°C for 48 h. Inocula (5 µl) of conidia from both strains were used to investigate the virulence of A. fumigatus and A. fischeri against G. mellonella. Ten G. mellonella larvae in the final (sixth) instar larval stage of development were used per condition in all assays. The control group was the larvae inoculated with 5 μl of PBS to observe the killing due to physical trauma. The inoculation was performed by using a Hamilton syringe (7000.5KH) and injecting 5 μl into the hemocoel of each larva via the last left proleg. Afterward, the larvae were incubated in a glass container (petri dishes) at 37°C in the dark. Larval killing was scored daily. Larvae were considered dead by presenting the absence of movement in response to touch.
Histopathology.
Outbred CD-1 mice, 6 to 8 weeks old, were immunosuppressed and intranasally inoculated with 2 × 106 conidia as described above for the chemotherapeutic and corticosteroid murine models. Mice were sacrificed 72 h postinoculation. Lungs were perfused with 10% buffered formalin phosphate before removal and then stored in 10% buffered formalin phosphate until embedding. Paraffin-embedded sections were stained with hematoxylin and eosin (H&E) and Gomori methenamine silver (GMS). Slides were analyzed microscopically with a Zeiss Axioplan 2 imaging microscope (Carl Zeiss Microimaging, Inc., Thornwood, NY, USA) fitted with a QImaging Retiga-SRV Fast 1394 RGB camera. Analysis was performed in Phylum Live 4 imaging software.
Ethics statement.
We carried out our mouse studies in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (83). The mouse experimental protocol was approved by the Institutional Animal Care and Use Committee (IACUC) at Dartmouth College (Federal-wide assurance number A3259-01).
Growth assays.
Radial growth was quantified by point inoculation of 1 × 103 conidia in 2 μl on indicated media; plates were incubated at 37°C in normoxia (∼21% O2, 5% CO2) or hypoxia (0.2% O2, 5% CO2). Colony diameter was measured every 24 h for 4 days and reported as the average from three biological replicates per strain.
For 2-DG experiments, 1 × 103 conidia in 2 μl were spotted on 1% lactate minimal medium with or without 0.1% 2-deoxyglucose (2-DG; Sigma, D8375). Plates were incubated for 3 days at 37°C in normoxia or hypoxia with 5% CO2. Percent inhibition was calculated by dividing radial growth on 2-DG plates by the average radial growth of biological triplicates on plates without 2-DG.
Fungal biomass was quantified by measuring the dry weight of fungal tissue from 5 × 107 conidia grown in 100 ml liquid GMM with shaking at 200 rpm for 48 h in normoxia (∼21% O2) and hypoxia (0.2% O2, 5% CO2). Liquid biomass is reported as the average from three biological replicates per strain. Hypoxic conditions were maintained using an InvivO2 400 hypoxia workstation (Ruskinn Technology Limited, Bridgend, United Kingdom) with a gas regulator and 94.8% N2.
Liquid growth curves were performed with conidia adjusted to 2 × 104 conidia in 20 μl 0.01% Tween 80 in 96-well dishes, and then 180 μl of medium (GMM or lung homogenate) was added to each well. Plates were incubated at 37°C for 7 h, and then A405 measurements were taken every 10 min for the first 16 h of growth with continued incubation at 37°C. Lung homogenate medium was prepared as follows: lungs were harvested from healthy CD-1 female mice (20 to 24 g) and homogenized through a 100-μm cell strainer in 2 ml PBS/lung. Homogenate was diluted 1:4 in sterile PBS, spun down to remove cells, and then filter sterilized through 22-μm PVDF filters.
Cell wall and oxidative stresses.
Congo red (0.5 mg/ml), menadione (20 µM), or calcofluor white (CFW; 25 µg/ml) was added to GMM plates. Conidia at 1 × 103 (calcofluor white and menadione) or 1 × 105 (Congo red) were point inoculated, and plates were incubated for 96 h at 37°C with 5% CO2.
Orthology determination and analyses.
Genomes for A. fumigatus CEA10 and A. fischeri NRRL 181 were downloaded from NCBI (accession numbers GCA_000150145.1 and GCF_000149645.1, respectively). To identify putative orthologous genes between A. fischeri and A. fumigatus, a reciprocal best BLAST hit (RBBH) approach was used. We performed a BLAST search on the proteome of A. fischeri to A. fumigatus and vice versa using an E value cutoff of 10−3 and then filtered for RBBHs according to bitscore (84). A pair of genes from each species was considered orthologous if their best BLAST hit was to each other. Species-specific and orthologous protein sets were visualized using version 3.0.0 of eulerAPE (85).
Biosynthetic gene cluster (BGC) prediction and analyses.
Version 4.2.0 of antiSMASH (40) was used with its default settings to identify BGCs. Orthologous cluster genes were identified using our RBBH results and visualized using version 0.69 of Circos (86). Chromosomes were identified for A. fischeri NRRL1 and A. fumigatus CEA10 using NUCMER (87) and chromosomal sequences from A. fumigatus strain AF293 from NCBI (accession number GCA_000002655.1). Syntenic clusters were visualized using Easyfig version 2.2.2 (88).
Secondary metabolite extraction and identification.
Secondary metabolites were extracted from A. fischeri using techniques well established in the natural product literature (89, 90). This was done by adding a 1:1 mixture of CHCl3-CH3OH and leaving the mixture to shake overnight. The resulting slurry was partitioned twice, first with a 4:1:5 CHCl3-CH3OH-H2O solution, with the organic layer drawn off and evaporated to dryness in vacuo, and second, reconstituting 1:1:2 CH3CN-CH3OH-hexanes, where the organic layer was drawn off and evaporated to dryness. The extract then underwent chromatographic separation (flash chromatography and HPLC) using varied gradient systems. The full structural characterization of the new secondary metabolites is provided in a Figshare document (https://doi.org/10.6084/m9.figshare.7149167).
Construction of the A. fischeri ΔlaeA mutant.
The gene replacement cassettes were constructed by in vivo recombination in Saccharomycescerevisiae as previously described (91, 92). Approximately 2.0 kb from the 5′-UTR and 3′-UTR flanking regions of the targeted ORF were selected for primer design. The primers pRS NF010750 5′fw (5′-GTAACGCCAGGGTTTTCCCAGTCACGACGCAGTCTAACGCTGGGCCCTTCC-3′) and pRS NF010750 3′rv (5′-GCGGTTAACAATTTCTCTCTGGAAACAGCTACGGCGTTTGACGGCACAC-3′) contained a short homologous sequence to the multicloning site (MCS) of the plasmid pRS426. Both the 5′- and 3′-UTR fragments were PCR amplified from A. fischeri genomic DNA (gDNA). The prtA gene, conferring resistance to pyrithiamine, which was placed within the cassette as a dominant marker, was amplified from the pPRT1 plasmid by using the primers prtA NF010750 5′rv (5′-GTAATCAATTGCCCGTCTGTCAGATCCAGGTCGAGGAGGTCCAATCGG-3′) and prtA NF010750 3′fw (5′-CGGCTCATCGTCACCCCATGATAGCCGAGATCAATCTTGCATCC-3′). The deletion cassette was generated by transforming each fragment along with the plasmid pRS426, cut with BamHI/EcoRI, into the S. cerevisiae strain SC94721 using the lithium acetate method (93). The DNA from the transformants was extracted by the method described by Goldman et al. (94). The cassette was PCR amplified from these plasmids utilizing TaKaRa Ex Taq DNA polymerase (Clontech TaKaRa Bio) and used for A. fischeri transformation according to the protocol described by Malavazi and Goldman (92). Southern blotting and PCR analyses were used to demonstrate that the cassette had integrated homologously at the targeted A. fischeri locus. Genomic DNA from A. fischeri was extracted by grinding frozen mycelia in liquid nitrogen, and then gDNA was extracted as previously described (92). Standard techniques for manipulation of DNA were carried out as described previously (95). For Southern blot analysis, restricted chromosomal DNA fragments were separated on a 1% agarose gel and blotted onto Hybond N+ nylon membranes (GE Healthcare). Probes were labeled using [α-32P]dCTP using the Random Primers DNA labeling system (Life Technologies). Labeled membranes were exposed to X-ray films, which were scanned for image processing. Southern blotting and PCR schemes are shown in Fig. S7 in the supplemental material.
ACKNOWLEDGMENTS
Computational infrastructure was provided by The Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University.
M.E.M., J.L.S., and A.R. were supported by a Vanderbilt University Discovery Grant. Research in A.R.’s lab is also supported by the National Science Foundation (DEB-1442113), the Guggenheim Foundation, and the Burroughs Wellcome Fund. R.A.C. holds an Investigator in the Pathogenesis of Infectious Diseases Award supported by the Burroughs Wellcome Fund and is also supported by National Institute of Allergy and Infectious Diseases award 1R01AI130128. S.R.B. was supported, in part, by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM008704. S.L.K. was supported by the National Center for Complementary and Integrative Health, a component of the National Institutes of Health, under award number T32 AT008938. G.H.G. was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), both from Brazil.
REFERENCES
- 1.Lamoth F. 2016. Aspergillus fumigatus-related species in clinical practice. Front Microbiol 7:683. doi: 10.3389/fmicb.2016.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tekaia F, Latgé J-P. 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr Opin Microbiol 8:385–392. doi: 10.1016/j.mib.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 3.Bhabhra R, Miley MD, Mylonakis E, Boettner D, Fortwendel J, Panepinto JC, Postow M, Rhodes JC, Askew DS. 2004. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun 72:4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagener J, Echtenacher B, Rohde M, Kotz A, Krappmann S, Heesemann J, Ebel F. 2008. The putative α-1,2-mannosyltransferase AfMnt1 of the opportunistic fungal pathogen Aspergillus fumigatus is required for cell wall stability and full virulence. Eukaryot Cell 7:1661–1673. doi: 10.1128/EC.00221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grahl N, Puttikamonkul S, Macdonald JM, Gamcsik MP, Ngo LY, Hohl TM, Cramer RA. 2011. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog 7:e1002145. doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willger SD, Puttikamonkul S, Kim K-H, Burritt JB, Grahl N, Metzler LJ, Barbuch R, Bard M, Lawrence CB, Cramer RA. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog 4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keller NP. 2018. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rokas A, Wisecaver JH, Lind AL. 2018. The birth, evolution and death of metabolic gene clusters in fungi. Nat Rev Microbiol 16:731–744. doi: 10.1038/s41579-018-0075-3. [DOI] [PubMed] [Google Scholar]
- 9.Keller NP, Turner G, Bennett JW. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol 3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 10.Bignell E, Cairns TC, Throckmorton K, Nierman WC, Keller NP. 2016. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos Trans R Soc B 371:20160023. doi: 10.1098/rstb.2016.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugui JA, Pardo J, Chang YC, Müllbacher A, Zarember KA, Galvez EM, Brinster L, Zerfas P, Gallin JI, Simon MM, Kwon-Chung KJ. 2007. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot Cell 6:1552–1561. doi: 10.1128/EC.00140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bok JW, Balajee SA, Marr KA, Andes D, Nielsen KF, Frisvad JC, Keller NP. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot Cell 4:1574–1582. doi: 10.1128/EC.4.9.1574-1582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balajee SA, Kano R, Baddley JW, Moser SA, Marr KA, Alexander BD, Andes D, Kontoyiannis DP, Perrone G, Peterson S, Brandt ME, Pappas PG, Chiller T. 2009. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. J Clin Microbiol 47:3138–3141. doi: 10.1128/JCM.01070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alastruey-Izquierdo A, Mellado E, Peláez T, Pemán J, Zapico S, Alvarez M, Rodríguez-Tudela JL, Cuenca-Estrella M, FILPOP Study Group. 2013. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study). Antimicrob Agents Chemother 57:3380–3387. doi: 10.1128/AAC.00383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Linden JWM, Arendrup MC, Warris A, Lagrou K, Pelloux H, Hauser PM, Chryssanthou E, Mellado E, Kidd SE, Tortorano AM, Dannaoui E, Gaustad P, Baddley JW, Uekötter A, Lass-Flörl C, Klimko N, Moore CB, Denning DW, Pasqualotto AC, Kibbler C, Arikan-Akdagli S, Andes D, Meletiadis J, Naumiuk L, Nucci M, Melchers WJG, Verweij PE. 2015. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg Infect Dis 21:1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, Angiuoli S, Bussey H, Bowyer P, Cotty PJ, Dyer PS, Egan A, Galens K, Fraser-Liggett CM, Haas BJ, Inman JM, Kent R, Lemieux S, Malavazi I, Orvis J, Roemer T, Ronning CM, Sundaram JP, Sutton G, Turner G, Venter JC, White OR, Whitty BR, Youngman P, Wolfe KH, Goldman GH, Wortman JR, Jiang B, Denning DW, Nierman WC. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet 4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steenwyk JL, Shen X-X, Lind AL, Goldman GG, Rokas A. 2018. A robust phylogenomic timetree for biotechnologically and medically important fungi from Aspergillaceae (Eurotiomycetes. Ascomycota). bioRxiv doi: 10.1101/370429. [DOI] [PMC free article] [PubMed]
- 18.Hong S-B, Kim D-H, Park I-C, Samson RA, Shin H-D. 2010. Isolation and identification of Aspergillus section Fumigati strains from arable soil in Korea. Mycobiology 38:1. doi: 10.4489/MYCO.2010.38.1.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frąc M, Jezierska-Tys S, Yaguchi T. 2015. Occurrence, detection, and molecular and metabolic characterization of heat-resistant fungi in soils and plants and their risk to human health. Adv Agron 132:161–204. doi: 10.1016/bs.agron.2015.02.003. [DOI] [Google Scholar]
- 20.Tong X, Xu H, Zou L, Cai M, Xu X, Zhao Z, Xiao F, Li Y. 2017. High diversity of airborne fungi in the hospital environment as revealed by meta-sequencing-based microbiome analysis. Sci Rep 7:39606. doi: 10.1038/srep39606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summerbell RC, de Repentigny L, Chartrand C, St Germain G. 1992. Graft-related endocarditis caused by Neosartorya fischeri var. spinosa. J Clin Microbiol 30:1580–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lonial S, Williams L, Carrum G, Ostrowski M, McCarthy P. 1997. Neosartorya fischeri: an invasive fungal pathogen in an allogeneic bone marrow transplant patient. Bone Marrow Transplant 19:753–755. doi: 10.1038/sj.bmt.1700715. [DOI] [PubMed] [Google Scholar]
- 23.Gerber J, Chomicki J, Brandsberg JW, Jones R, Hammerman KJ. 1973. Pulmonary aspergillosis caused by Aspergillus fischeri var. spinosus: report of a case and value of serologic studies. Am J Clin Pathol 60:861–866. doi: 10.1093/ajcp/60.6.861. [DOI] [PubMed] [Google Scholar]
- 24.Coriglione G, Stella G, Gafa L, Spata G, Oliveri S, Padhye AA, Ajello L. 1990. Neosartorya fischeri var fischeri (Wehmer) Malloch and Cain 1972 (anamorph: Aspergillus fischerianus Samson and Gams 1985) as a cause of mycotic keratitis. Eur J Epidemiol 6:382–385. doi: 10.1007/BF00151712. [DOI] [PubMed] [Google Scholar]
- 25.Escribano P, Peláez T, Muñoz P, Bouza E, Guinea J. 2013. Is azole resistance in Aspergillus fumigatus a problem in Spain? Antimicrob Agents Chemother 57:2815–2820. doi: 10.1128/AAC.02487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negri CE, Gonçalves SS, Xafranski H, Bergamasco MD, Aquino VR, Castro PTO, Colombo AL. 2014. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. J Clin Microbiol 52:3633–3640. doi: 10.1128/JCM.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabino R, Verissimo C, Parada H, Brandao J, Viegas C, Carolino E, Clemons KV, Stevens DA. 2014. Molecular screening of 246 Portuguese Aspergillus isolates among different clinical and environmental sources. Med Mycol 52:519–529. doi: 10.1093/mmy/myu006. [DOI] [PubMed] [Google Scholar]
- 28.Abers MS, Ghebremichael MS, Timmons AK, Warren HS, Poznansky MC, Vyas JM. 2016. A critical reappraisal of prolonged neutropenia as a risk factor for invasive pulmonary aspergillosis. Open Forum Infect Dis 3:ofw036. doi: 10.1093/ofid/ofw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beattie SR, Mark KMK, Thammahong A, Ries LNA, Dhingra S, Caffrey-Carr AK, Cheng C, Black CC, Bowyer P, Bromley MJ, Obar JJ, Goldman GH, Cramer RA. 2017. Filamentous fungal carbon catabolite repression supports metabolic plasticity and stress responses essential for disease progression. PLoS Pathog 13:e1006340. doi: 10.1371/journal.ppat.1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowalski CH, Beattie SR, Fuller KK, McGurk EA, Tang Y-W, Hohl TM, Obar JJ, Cramer RA. 2016. Heterogeneity among isolates reveals that fitness in low oxygen correlates with Aspergillus fumigatus virulence. mBio 7:e01515-16. doi: 10.1128/mBio.01515-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Childers DS, Raziunaite I, Mol Avelar G, Mackie J, Budge S, Stead D, Gow NAR, Lenardon MD, Ballou ER, MacCallum DM, Brown AJP. 2016. The rewiring of ubiquitination targets in a pathogenic yeast promotes metabolic flexibility, host colonization and virulence. PLoS Pathog 12:e1005566. doi: 10.1371/journal.ppat.1005566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandai D, Yin Z, Selway L, Stead D, Walker J, Leach MD, Bohovych I, Ene IV, Kastora S, Budge S, Munro CA, Odds FC, Gow NAR, Brown AJP. 2012. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 3:e00495-12. doi: 10.1128/mBio.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown NA, Goldman GH. 2016. The contribution of Aspergillus fumigatus stress responses to virulence and antifungal resistance. J Microbiol 54:243–253. doi: 10.1007/s12275-016-5510-4. [DOI] [PubMed] [Google Scholar]
- 34.Houbraken J, Weig M, Groß U, Meijer M, Bader O. 2016. Aspergillus oerlinghausenensis, a new mould species closely related to A. fumigatus. FEMS Microbiol Lett 363:fnv236. doi: 10.1093/femsle/fnv236. [DOI] [PubMed] [Google Scholar]
- 35.Salichos L, Rokas A. 2011. Evaluating ortholog prediction algorithms in a yeast model clade. PLoS One 6:e18755. doi: 10.1371/journal.pone.0018755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abad A, Fernández-Molina JV, Bikandi J, Ramírez A, Margareto J, Sendino J, Hernando FL, Pontón J, Garaizar J, Rementeria A. 2010. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev Iberoam Micol 27:155–182. doi: 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 37.Kjærbølling I, Vesth TC, Frisvad JC, Nybo JL, Theobald S, Kuo A, Bowyer P, Matsuda Y, Mondo S, Lyhne EK, Kogle ME, Clum A, Lipzen A, Salamov A, Ngan CY, Daum C, Chiniquy J, Barry K, LaButti K, Haridas S, Simmons BA, Magnuson JK, Mortensen UH, Larsen TO, Grigoriev IV, Baker SE, Andersen MR. 2018. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc Natl Acad Sci U S A 115:E753–E761. doi: 10.1073/pnas.1715954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Hanlon KA, Gallagher L, Schrettl M, Jöchl C, Kavanagh K, Larsen TO, Doyle S. 2012. Nonribosomal peptide synthetase genes pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl Environ Microbiol 78:3166–3176. doi: 10.1128/AEM.07249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. 2010. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, de los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lind AL, Wisecaver JH, Smith TD, Feng X, Calvo AM, Rokas A. 2015. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet 11:e1005096. doi: 10.1371/journal.pgen.1005096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lind AL, Wisecaver JH, Lameiras C, Wiemann P, Palmer JM, Keller NP, Rodrigues F, Goldman GH, Rokas A. 2017. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol 15:e2003583. doi: 10.1371/journal.pbio.2003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gardiner DM, Howlett BJ. 2005. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett 248:241–248. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 44.Frisvad JC, Larsen TO. 2015. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Front Microbiol 6:1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiemann P, Guo C-J, Palmer JM, Sekonyela R, Wang CCC, Keller NP. 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A 110:17065–17070. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El-Elimat T, Raja HA, Day CS, Chen W-L, Swanson SM, Oberlies NH. 2014. Greensporones: resorcylic acid lactones from an aquatic Halenospora sp. J Nat Prod 77:2088–2098. doi: 10.1021/np500497r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.El-Elimat T, Raja HA, Figueroa M, Falkinham JO, Oberlies NH. 2014. Isochromenones, isobenzofuranone, and tetrahydronaphthalenes produced by Paraphoma radicina, a fungus isolated from a freshwater habitat. Phytochemistry 104:114–120. doi: 10.1016/j.phytochem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 48.El-Elimat T, Raja HA, Day CS, McFeeters H, McFeeters RL, Oberlies NH. 2017. α-Pyrone derivatives, tetra/hexahydroxanthones, and cyclodepsipeptides from two freshwater fungi. Bioorg Med Chem 25:795–804. doi: 10.1016/j.bmc.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 49.Raja HA, Paguigan ND, Fournier J, Oberlies NH. 2017. Additions to Lindgomyces (Lindgomycetaceae, Pleosporales, Dothideomycetes), including two new species occurring on submerged wood from North Carolina, USA, with notes on secondary metabolite profiles. Mycol Prog 16:535–552. doi: 10.1007/s11557-017-1282-4. [DOI] [Google Scholar]
- 50.Rivera-Chávez J, Raja HA, Graf TN, Gallagher JM, Metri P, Xue D, Pearce CJ, Oberlies NH. 2017. Prealamethicin F50 and related peptaibols from Trichoderma arundinaceum: validation of their authenticity via in situ chemical analysis. RSC Adv 7:45733–45751. doi: 10.1039/c7ra09602j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez JF, Somoza AD, Keller NP, Wang CCC. 2012. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep 29:351–371. doi: 10.1039/c2np00084a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bode HB, Bethe B, Höfs R, Zeeck A. 2002. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem 3:619–627. doi:. [DOI] [PubMed] [Google Scholar]
- 53.Vandermolen KM, Raja HA, El-Elimat T, Oberlies NH. 2013. Evaluation of culture media for the production of secondary metabolites in a natural products screening program. AMB Express 3:71. doi: 10.1186/2191-0855-3-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemphill CFP, Sureechatchaiyan P, Kassack MU, Orfali RS, Lin W, Daletos G, Proksch P. 2017. OSMAC approach leads to new fusarielin metabolites from Fusarium tricinctum. J Antibiot (Tokyo) 70:726–732. doi: 10.1038/ja.2017.21. [DOI] [PubMed] [Google Scholar]
- 55.Frisvad JC, Andersen B, Thrane U. 2008. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol Res 112:231–240. doi: 10.1016/j.mycres.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 56.Eamvijarn A, Gomes NM, Dethoup T, Buaruang J, Manoch L, Silva A, Pedro M, Marini I, Roussis V, Kijjoa A. 2013. Bioactive meroditerpenes and indole alkaloids from the soil fungus Neosartorya fischeri (KUFC 6344), and the marine-derived fungi Neosartorya laciniosa (KUFC 7896) and Neosartorya tsunodae (KUFC 9213). Tetrahedron 69:8583–8591. doi: 10.1016/j.tet.2013.07.078. [DOI] [Google Scholar]
- 57.Kimura Y, Hamasaki T, Nakajima H, Isogai A. 1982. Structure of aszonalenin, a new metabolite of aspergillus zonatus. Tetrahedron Lett 23:225–228. doi: 10.1016/S0040-4039(00)86791-X. [DOI] [Google Scholar]
- 58.Ruchti J, Carreira EM. 2014. Ir-catalyzed reverse prenylation of 3-substituted indoles: total synthesis of (+)-aszonalenin and (−)-brevicompanine B. J Am Chem Soc 136:16756–16759. doi: 10.1021/ja509893s. [DOI] [PubMed] [Google Scholar]
- 59.Ellestad GA, Mirando P, Kumstmann MP. 1973. Structure of the metabolite LL-S490 beta from an unidentified Aspergillus species. J Org Chem 38:4204–4205. doi: 10.1021/jo00963a024. [DOI] [PubMed] [Google Scholar]
- 60.Yamazaki M, Fujimoto H, Kawasaki T. 1980. Chemistry of tremorogenic metabolites. I. Fumitremorgin A from Aspergillus fumigatus. Chem Pharm Bull (Tokyo) 28:245–254. doi: 10.1248/cpb.28.245. [DOI] [PubMed] [Google Scholar]
- 61.Feng Y, Holte D, Zoller J, Umemiya S, Simke LR, Baran PS. 2015. Total synthesis of verruculogen and fumitremorgin A enabled by ligand-controlled C–H borylation. J Am Chem Soc 137:10160–10163. doi: 10.1021/jacs.5b07154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pohland AE, Schuller PL, Steyn PS, Van Egmond HP. 1982. Physicochemical data for some selected mycotoxins. Pure Appl Chem 54:2219–2284. doi: 10.1351/pac198254112219. [DOI] [Google Scholar]
- 63.Afiyatullov SS, Kalinovskii AI, Pivkin MV, Dmitrenok PS, Kuznetsova TA. 2005. Alkaloids from the marine isolate of the fungus Aspergillus fumigatus. Chem Nat Compd 41:236–238. doi: 10.1007/s10600-005-0122-y. [DOI] [Google Scholar]
- 64.Mundt K, Wollinsky B, Ruan H-L, Zhu T, Li S-M. 2012. Identification of the verruculogen prenyltransferase FtmPT3 by a combination of chemical, bioinformatic and biochemical approaches. Chembiochem 13:2583–2592. doi: 10.1002/cbic.201200523. [DOI] [PubMed] [Google Scholar]
- 65.Fayos J, Lokensgard D, Clardy J, Cole RJ, Kirksey JW. 1974. Letter: structure of verruculogen, a tremor producing peroxide from Penicillium verruculosum. J Am Chem Soc 96:6785–6787. doi: 10.1021/ja00828a054. [DOI] [PubMed] [Google Scholar]
- 66.Fill TP, Asenha HBR, Marques AS, Ferreira AG, Rodrigues-Fo E. 2013. Time course production of indole alkaloids by an endophytic strain of Penicillium brasilianum cultivated in rice. Nat Prod Res 27:967–974. doi: 10.1080/14786419.2012.701210. [DOI] [PubMed] [Google Scholar]
- 67.Bok JW, Keller NP. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot Cell 3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoff B, Kamerewerd J, Sigl C, Mitterbauer R, Zadra I, Kürnsteiner H, Kück U. 2010. Two components of a velvet-like complex control hyphal morphogenesis, conidiophore development, and penicillin biosynthesis in Penicillium chrysogenum. Eukaryot Cell 9:1236–1250. doi: 10.1128/EC.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wiemann P, Brown DW, Kleigrewe K, Bok JW, Keller NP, Humpf H-U, Tudzynski B. 2010. FfVel1 and FfLae1, components of a velvet-like complex in Fusarium fujikuroi, affect differentiation, secondary metabolism and virulence. Mol Microbiol 77:972–994. doi: 10.1111/j.1365-2958.2010.07263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyatt TT, van Leeuwen MR, Wösten HAB, Dijksterhuis J. 2014. Mannitol is essential for the development of stress-resistant ascospores in Neosartorya fischeri (Aspergillus fischeri). Fungal Genet Biol 64:11–24. doi: 10.1016/j.fgb.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 71.Latgé JP. 1999. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev 12:310–350. doi: 10.1128/CMR.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dagenais TRT, Keller NP. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin Microbiol Rev 22:447–465. doi: 10.1128/CMR.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ben-Ami R, Lewis RE, Kontoyiannis DP. 2010. Enemy of the (immunosuppressed) state: an update on the pathogenesis of Aspergillus fumigatus infection. Br J Haematol 150:406–417. doi: 10.1111/j.1365-2141.2010.08283.x. [DOI] [PubMed] [Google Scholar]
- 74.Hissen AHT, Wan ANC, Warwas ML, Pinto LJ, Moore MM. 2005. The Aspergillus fumigatus siderophore biosynthetic gene sidA, encoding L-ornithine N5-oxygenase, is required for virulence. Infect Immun 73:5493–5503. doi: 10.1128/IAI.73.9.5493-5503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moreno MA, Ibrahim-Granet O, Vicentefranqueira R, Amich J, Ave P, Leal F, Latgé J-P, Calera JA. 2007. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol Microbiol 64:1182–1197. doi: 10.1111/j.1365-2958.2007.05726.x. [DOI] [PubMed] [Google Scholar]
- 76.Cramer RA, Rivera A, Hohl TM. 2011. Immune responses against Aspergillus fumigatus: what have we learned? Curr Opin Infect Dis 24:315–322. doi: 10.1097/QCO.0b013e328348b159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC. 2015. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. mBio 6:e00536-15. doi: 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mellado E, Alcazar-Fuoli L, Cuenca-Estrella M, Rodriguez-Tudela JL. 2011. Role of Aspergillus lentulus 14-α sterol demethylase (Cyp51A) in azole drug susceptibility. Antimicrob Agents Chemother 55:5459–5468. doi: 10.1128/AAC.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sugui JA, Vinh DC, Nardone G, Shea YR, Chang YC, Zelazny AM, Marr KA, Holland SM, Kwon-Chung KJ. 2010. Neosartorya udagawae (Aspergillus udagawae), an emerging agent of aspergillosis: how different is it from Aspergillus fumigatus? J Clin Microbiol 48:220–228. doi: 10.1128/JCM.01556-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubka V, Barrs V, Dudová Z, Sklenář F, Kubátová A, Matsuzawa T, Yaguchi T, Horie Y, Nováková A, Frisvad JC, Talbot JJ, Kolařík M. 2018. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia 41:142. doi: 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nature Microbiology. 2017. Stop neglecting fungi. Nat Microbiol 2:17120. doi: 10.1038/nmicrobiol.2017.120. [DOI] [PubMed] [Google Scholar]
- 82.Fuchs BB, O’Brien E, El Khoury JB, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482. doi: 10.4161/viru.1.6.12985. [DOI] [PubMed] [Google Scholar]
- 83.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC. [Google Scholar]
- 84.Madden T. 2013. The BLAST sequence analysis tool In The NCBI handbook, 2nd ed National Center for Biotechnology Information, Bethesda, MD: https://www.ncbi.nlm.nih.gov/books/NBK153387/. [Google Scholar]
- 85.Micallef L, Rodgers P. 2014. eulerAPE: drawing area-proportional 3-Venn diagrams using ellipses. PLoS One 9:e101717–e101718. doi: 10.1371/journal.pone.0101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Elimat T, Figueroa M, Ehrmann BM, Cech NB, Pearce CJ, Oberlies NH. 2013. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J Nat Prod 76:1709–1716. doi: 10.1021/np4004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Paguigan ND, El-Elimat T, Kao D, Raja HA, Pearce CJ, Oberlies NH. 2017. Enhanced dereplication of fungal cultures via use of mass defect filtering. J Antibiot (Tokyo) 70:553–561. doi: 10.1038/ja.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A 103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malavazi I, Goldman GH. 2012. Gene disruption in Aspergillus fumigatus using a PCR-based strategy and in vivo recombination in yeast. Methods Mol Biol 845:99–118. doi: 10.1007/978-1-61779-539-8_7. [DOI] [PubMed] [Google Scholar]
- 93.Schiestl RH, Gietz RD. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 94.Goldman GH, dos Reis Marques E, Duarte Ribeiro DC, de Souza Bernardes LA, Quiapin AC, Vitorelli PM, Savoldi M, Semighini CP, de Oliveira RC, Nunes LR, Travassos LR, Puccia R, Batista WL, Ferreira LE, Moreira JC, Bogossian AP, Tekaia F, Nobrega MP, Nobrega FG, Goldman MHS. 2003. Expressed sequence tag analysis of the human pathogen Paracoccidioides brasiliensis yeast phase: identification of putative homologues of Candida albicans virulence and pathogenicity genes. Eukaryot Cell 2:34–48. doi: 10.1128/EC.2.1.34-48.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sambrook J, Russell DW. 2001. Molecular cloning, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 96.Yin W-B, Grundmann A, Cheng J, Li S-M. 2009. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem 284:100–109. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. fumigatus grows slower than A. fischeri in glucose minimal medium (GMM) but at the same speed as A. fischeri in lung homogenate medium. A. fumigatus CEA10 or A. fischeri NRRL 181 was cultured in flat-bottom 96-well plates at 2 × 104 conidia per well. Conidia were added in 20 µl of 0.01% Tween 80, and medium was carefully pipetted over the inoculum into each well. Lung homogenate was generated according to reference 29. Plates were incubated for 7 hours at 37°C before measurements at 405 nm were taken every 10 min. Mean and SEM for eight technical replicates; data are representative of three biological replicates. Download FIG S1, PDF file, 0.2 MB (181.8KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. fischeri and A. fumigatus exhibit similar growth patterns at 30°C but not at 44°C. (A to D) Conidia at 1 × 103 were point inoculated on each plate, and plates were then incubated at 30°C in normoxia (∼21% oxygen, 5% CO2); colony diameter was measured every 24 hours. Mean and SEM for triplicates. Tween 80, 1% Tween 80 provided as sole carbon source; CAA, Casamino Acids; GMM, glucose minimal medium. (E) Error bars indicate standard deviations between biological duplicates (**, P value < 0.005 in a paired, equal-variance Student t test). Download FIG S2, PDF file, 0.7 MB (723.5KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The genomes of A. fumigatus and A. fischeri are largely similar, but their secondary metabolic pathways are quite divergent. Left, Venn diagram showing the sets of A. fischeri-specific proteins, shared orthologous proteins, and A. fumigatus-specific proteins encoded in each genome. Numbers below each species name indicate the total number of proteins encoded in that genome. Right, Venn diagram showing the sets of A. fischeri-specific BGC proteins, shared BGC proteins, and A. fumigatus-specific BGC proteins. Numbers below each species name indicate the total number of BGC proteins encoded in that genome. In each diagram, circles are proportional to the number of proteins they contain. Download FIG S3, PDF file, 0.1 MB (129.3KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The acetylaszonalenin and gliotoxin biosynthetic gene clusters in A. fumigatus and A. fischeri are located immediately next to one another. The portions of clusters 37 and 25 from A. fischeri and A. fumigatus, respectively, that are known to contain the previously characterized acetylaszonalenin (96) and gliotoxin (43) BGCs are shown. Genes colored in shades of green are involved in the acetylaszonalenin biosynthetic pathway. Dark green, anaPS (nonribosomal peptide synthase); light green, anaAT (acetyltransferase); green, anaPT (prenyltransferase); orange, gliotoxin biosynthetic genes. Gray arrow, syntenic gene in both species not involved in gliotoxin synthesis. Sequences that are similar to one another (based on blastn scores) are marked by gray parallelograms. Image was made using Easyfig version 2.2.2 (88). Download FIG S4, PDF file, 0.1 MB (118.2KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A custom chemical analysis protocol was developed for studying the metabolites produced by A. fischeri. Approximately 60 ml of 1:1 CH3OH-CH3Cl was added to cultures of Aspergillus fischeri grown on solid-state fermentation for 2 weeks. The cultures were then chopped thoroughly with a large scalpel and shaken for 16 hours using an orbital shaker. The liquid culture was then vacuum filtered and concentrated using 90 ml CH3Cl and 150 ml water and transferred into a separatory funnel. The organic (bottom) layer was drawn off and evaporated to dryness. The dried, desugared extract was reconstituted in 100 ml of 1:1 CH3OH-CH3CN and 100 ml of hexane. The biphasic solution was shaken vigorously and transferred to a separatory funnel. The CH3OH-CH3CN layer was evaporated to dryness under vacuum, producing a defatted extract. The extract was then subdivided into several peaks or fractions using flash chromatography. The subfractions were further separated using HPLC until pure compounds were isolated. The pure compounds were subjected to UPLC-MS analysis to establish the molecular formula and fragmentation patterns. Finally, pure compounds were identified using both NMR analysis and information from UPLC-MS data. Download FIG S5, PDF file, 0.9 MB (899.7KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A. fischeri produces different numbers of metabolites, depending on the medium it is grown on. Base peak chromatograms as measured by LC-MS, illustrating how the chemistry profiles varied based on growth conditions. PDA + ab was used as the chemical control to observe the differences in the secondary metabolites, due to it being the medium in which A. fischeri is stored. There were overall no chemical differences observed between the different variations of PDA medium. Each peak (which indicates different chemical entities) was observed in the three PDA variations, albeit at fluctuating intensities. SDA, PYG, and YESD produced the majority of the peaks observed in PDA, but it also lacked some observed peaks, indicating that these growth conditions were not chemically favored. CYA produced the majority of the peaks, as well as an additional peak that was observed at a much lower intensity in PDA. However, this peak was similarly observed in OMA. OMA produced similar peaks as those observed in PDA, but with higher intensity. Due to this, OMA was selected for further study. The gray boxes indicate differences in the observed peaks from PDA. See the Figshare document at https://doi.org/10.6084/m9.figshare.7149167 for more information. Download FIG S6, PDF file, 0.6 MB (670.6KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Southern blotting confirms construction of the ΔlaeA mutant. A 1-kb probe recognizes a single DNA band (∼4.4 kb) in the wild-type strain and a single DNA band (∼2.7 kb) in the ΔlaeA mutant. Download FIG S7, PDF file, 0.2 MB (240.8KB, pdf) .
Copyright © 2019 Mead et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.