Abstract
Altered expression of long noncoding RNAs (lncRNAs) by environmental chemicals modulates the expression of xenobiotic biotransformation-related genes and may serve as therapeutic targets and novel biomarkers of exposure. The pregnane X receptor (PXR/NR1I2) is a critical xenobiotic-sensing nuclear receptor that regulates the expression of many drug-processing genes, and it has similar target-gene profiles and DNA-binding motifs with another xenobiotic-sensing nuclear receptor, namely, constitutive andronstrane receptor (CAR/Nr1i3). To test our hypothesis that lncRNAs are regulated by PXR in concert with protein-coding genes (PCGs) and to compare the PXR-targeted lncRNAs with CAR-targeted lncRNAs, RNA-Seq was performed from livers of adult male C57BL/6 mice treated with corn oil, the PXR agonist PCN, or the CAR agonist 1, 4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP). Among 125,680 known lncRNAs, 3843 were expressed in liver, and 193 were differentially regulated by PXR (among which 40% were also regulated by CAR). Most PXR- or CAR-regulated lncRNAs were mapped to the introns and 3′-untranslated regions (UTRs) of PCGs, as well as intergenic regions. Combining the RNA-Seq data with a published PXR chromatin immunoprecipitation coupled with high-throughput sequencing; cytochrome P450 (P450; ChIP-Seq) data set, we identified 774 expressed lncRNAs with direct PXR-DNA binding sites, and 26.8% of differentially expressed lncRNAs had changes in PXR-DNA binding after PCN exposure. De novo motif analysis identified colocalization of PXR with liver receptor homolog (LRH-1), which regulates bile acid synthesis after PCN exposure. There was limited overlap of PXR binding with an epigenetic mark for transcriptional activation (histone-H3K4-di-methylation, H3K4me2) but no overlap with epigenetic marks for transcriptional silencing [H3 lysine 27 tri-methylation (H3K27me3) and DNA methylation]. Among differentially expressed lncRNAs, 264 were in proximity of PCGs, and the lncRNA-PCG pairs displayed a high coregulatory pattern by PXR and CAR activation. This study was among the first to demonstrate that lncRNAs are regulated by PXR and CAR activation and that they may be important regulators of PCGs involved in xenobiotic metabolism.
Introduction
Long noncoding RNAs (lncRNAs) are functional transcripts more than 200 nucleotides long whose genes are estimated to constitute at least 62%–75% of the human genome (Djebali et al., 2012; St Laurent et al., 2015). According to the ENCODE Project, lncRNAs represent approximately 80% of functional sequences in the human DNA and are the predominant nonribosomal and nonmitochondrial RNA species in human cells (Kapranov et al., 2010; ENCODE Project Consortium, 2012). LncRNAs transcribed proximally or distally to protein-coding genes (PCGs) can modulate a wide spectrum of biologic events, including chromatin epigenetic remodeling, transcription factor assembly, alternative splicing, mRNA stability, and protein translation efficiency (Geisler and Coller, 2013; Karlsson and Baccarelli, 2016; Dempsey and Cui, 2017). Growing evidence in the literature suggests that lncRNAs are novel biomarkers and/or key contributors during physiologic, pharmacologic, and toxicologic responses, including complex human diseases, developmental disorders, as well as xenobiotic-induced adverse outcomes (Dempsey and Cui, 2017).
Many lncRNAs are highly expressed in the liver, a major organ for xenobiotic biotransformation and nutrient homeostasis. A stringent computational pipeline identified 15,558 lncRNAs expressed in mouse liver, based on an analysis of 186 mouse liver RNA-Seq data sets ranging over 30 biologic conditions (Melia et al., 2016). Interestingly, Melia et al. (2016) also demonstrated greater interspecies conservations within DNA sequences and higher frequency of proximal binding by hepatic transcription factors in the liver-expressed lncRNA gene promoters compared with protein-coding gene (PCG) promoters. The liver-enriched lncRNAs were predicted to be metabolically sensitive regulators with diverse functions in physiologic homeostasis, especially energy metabolism (Yang et al., 2016). Regulation of genes involved in energy metabolism and nutrient homeostasis by lncRNAs found in the literature include gluconeogenesis (Goyal et al., 2017), cholesterol, and bile acid homeostasis (Ananthanarayanan, 2016; Lan et al., 2016; Melia et al., 2016; Zhang et al., 2017), lipid metabolism (Chen, 2015; Li et al., 2015, 2017; Yang et al., 2016; Zhao et al., 2017), and nonalcoholic fatty liver disease (Chen et al., 2017).
In addition to modulating intermediary metabolism in liver, lncRNAs are suggested to modulate hepatic xenobiotic biotransformation. For example, two lncRNAs that are transcribed from the antisense strands of the DNA encoding the hepatocyte nuclear factor 1α (HNF1α) and HNF4α are important in regulating the major drug-metabolizing cytochrome P450s (P450s) in human liver cancer–derived hepatic stem cell line (HepaRG) cells (Chen et al., 2018). Specifically, short hairpin RNA (shRNA) knockdown of the lncRNA gene HNF1α antisense 1 (AS-1) decreased the mRNA expression of PXR and CAR, as well as the expression of seven major P450s (CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2E1, and CYP3A4) in hepaRG cells. In addition, small interfering RNA (siRNA) knockdown of the lncRNA gene HNF4α-AS1 increased the mRNA expression of PXR as well as the expression of six P450s (CYP1A2, CYP2B6, CYP2C9, CYP2C19, CYP2E1, and CYP3A4). This indicated that there is a complex network between transcription factors and lncRNAs that regulate the expression of P450s (Chen et al., 2018).
During postnatal liver maturation, the developmental expression patterns of various lncRNAs have been unveiled at 12 developmental ages in mice (Peng et al., 2014), and the potential role of lncRNAs in regulating the ontogenic expression of the xenobiotic-metabolizing P450s was proposed (Ingelman-Sundberg et al., 2013). LncRNAs have also been suggested to confer drug resistance through modulating the stability and translation of mRNAs that produce proteins involved in cell survival, proliferation, and drug metabolism (Pan et al., 2015); however, very little is known regarding to what extent the hepatic lncRNAs are regulated by exposure to drugs and other xenobiotics.
Regarding xenobiotic biotransformation, in the liver, the nuclear receptors pregnane X receptor (PXR/Nr1i2) and constitutive androstane receptor (CAR/Nr1i3) are major xenobiotic-sensing nuclear receptors that can be activated by a wide spectrum of therapeutic drugs, environmental toxicants, dietary factors, and endogenous chemicals (Kliewer et al., 2002; Willson and Kliewer, 2002; Moore et al., 2003; Pacyniak et al., 2007). Upon activation, PXR and CAR play critical roles in xenobiotic bioactivation and detoxification (Handschin and Meyer, 2003). In addition, recent studies have unveiled novel functions of these drug receptors in various intermediary metabolism pathways, such as lipid and glucose metabolism (Poulin-Dubois and Shultz, 1990; Wada et al., 2009; Gao and Xie, 2010; Mackowiak et al., 2018; Pu et al., 2018). Our research group and others have extensively characterized the effect of pharmacologic activation of PXR and CAR on the regulation of various protein-coding genes, especially the drug-processing genes in liver (Cheng et al., 2005b; Maher et al., 2005; Kiyosawa et al., 2008; Pratt-Hyatt et al., 2013; Oshida et al., 2015; Cui and Klaassen, 2016).
The effect of the the CAR ligand 1, 4-bis[2-(3, 5-dichloropyridyloxy)]benzene (TCPOBOP) has been investigated on the hepatic chromatin assembly and certain lncRNAs that contribute to liver tumor promotion (Lempiäinen et al., 2013; Lodato et al., 2018). TCPOBOP differentially regulated 166 lncRNAs in the liver that are produced from intragenic or antisense strand relative to PCGs that encode CAR-regulated drug-metabolizing enzymes, suggesting that an efficient coregulatory mechanism may exist (Lodato et al., 2017). In addition, two studies have investigated the effect of activating the xenobiotic-sensing transcription factor aryl hydrocarbon receptor (AhR) on the hepatic expression of lncRNAs (Recio et al., 2013; Grimaldi et al., 2018); however, no systematic studies have characterized the effect of PXR activation on the regulation of liver-enriched lncRNAs and how PXR activation differs from CAR activation regarding lncRNA expression profiles.
Therefore, the purpose of the present study was to 1) determine the effect of pharmacologic activation of PXR on hepatic expression of lncRNAs, 2) identify direct PXR-binding sites to the lncRNA gene loci and associated epigenetic signatures under basal and PXR-activated conditions, 3) compare the similarities and differences between PXR- and CAR-targeted lncRNAs, and 4) predict the lncRNA-regulated protein-coding gene networks after PXR/CAR activation in the mouse liver.
Materials and Methods
Chemicals.
Corn oil (vehicle), the mouse PXR ligand pregnenolone-16α-carbonitrile (PCN; ≥97% purity; CAS no. 1434-54-4), and the mouse CAR ligand 1,4-bis-[2-(3,5-dichloropyridyloxy)]benzene,3,3′,5,5′-tetrachloro-1,4-bis(pyridyloxy)benzene (TCPOBOP; ≥98% purity; CAS no. 76150-91-9) were purchased from Sigma-Aldrich (St. Louis, MO).
Animals and Procedures.
As described previously (Cui and Klaassen, 2016), 12-week-old adult male C57BL/6 wild-type mice were purchased from Charles River Laboratories (Wilmington, MA) and housed in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility (Animal Care and Use Program no. 2011-1969) at the University of Kansas Medical Center (KUMC) with ad libitum access to the Laboratory Rodent Chow 8604 (Harlan, Madison, WI) and drinking water. The housing conditions were temperature- and humidity-controlled with a 14-hour light and 10-hour dark cycle in a temperature- and humidity-controlled environment. Mice were acclimated for at least 1 week within the animal facilities before experiments. Mice were administered PCN (200 mg/kg, i.p.), TCPOBOP (3 mg/kg, i.p.), or vehicle (corn oil, 5 ml/kg, i.p.) once daily for 4 consecutive days (n = 5 per group). Twenty-four hours after the final dose, livers were collected, immediately frozen in liquid nitrogen, and stored in a −80°C freezer. All animal experiments were approved by the IACUC at KUMC.
RNA Isolation.
Total RNA was isolated from mouse liver using RNA-Bee reagent (Tel-Test lnc., Friendswood, TX) according to the manufacturer’s protocol and quantified using a Nanodrop 1000 Spectrophotometer (Thermo Scientific, Wilminton, DE). RNA integrity was confirmed using gel electrophoresis and a dual Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). RNA samples used for cDNA library construction and RNA sequencing had RNA integrity values between 7.0 and 10.0.
cDNA Library Construction and RNA-Seq Data Analysis.
The cDNA library preparation from poly-A selection was performed in the KUMC Genome Sequencing Facility, as described previously (Cui and Klaassen, 2016). Samples for sequencing were randomly selected (n = 3 per group) from all biologic replicates (n = 5) for all studies. Sequencing was performed on an Illumina HiSeq2000 sequencer using 100 base-pair paired-end multiplexing strategy. FASTQ files and analyzed data are available at NCBI GEO database (GSE104734), and data were reanalyzed for the present study. Briefly, FASTQ files containing paired-end sequence reads were mapped to the mouse reference genome (GRCm38/mm10) using Hierarchical Indexing for Spliced Alignment of Transcripts (version 2.0.5). The output sequencing alignment/map (SAM) files were converted to binary alignment/map files and sorted using SAMtools (version 1.3.1). The transcript abundance for lncRNAs and PCGs was estimated by Cufflinks (version 2.2.1) using the NONCODE 2016 lncRNA and UCSC mm10 PCG reference databases, respectively. The mRNA abundance was expressed as fragments per kilobase of exon per million reads mapped (FPKM). LncRNAs and PCGs with an average FPKM above 1 in at least one sample were considered expressed. Differential analysis was performed using Cuffdiff, and transcripts with P < 0.05 were considered differentially regulated by chemical exposure. Data were expressed as mean FPKM ± S.E., and asterisks (*) represent statistically significant differences between vehicle and chemical exposure. Two-way hierarchical clustering dendrograms of differentially regulated lncRNAs were generated using the native function of R.
Genomic Annotation of lncRNAs and Proximal lncRNA-PCG Pair Identification.
To annotate and visualize the genomic location of lncRNAs relative to the closest PCGs, the Web-based tool peak annotation and visualization (PAVIS, https://manticore.niehs.nih.gov/pavis2/) was used to identify lncRNAs proximal to PCGs, including 5 kb upstream of the transcription start site (TSS), intronic, exonic, 5′-untranslated region (UTR), 3′-UTR, and up to 1 kb downstream of the transcriptional termination site (TTS). A lncRNA and PCG are considered paired if 1) the lncRNA overlaps with or is within 5 kb upstream of the TSS or 1 kb downstream of TTS of any PCG, and 2) both the lncRNA and the proximal PCG were differentially expressed between vehicle and chemical exposed mice (FPKM >1 at least one sample and P < 0.05). Gene structure and relative genomic location of the lncRNA-PCG pairs were visualized using Integrated Genome Viewer (Broad Institute, Cambridge, MA). Pathways that are associated with PCGs paired with lncRNAs were shown using Ingenuity Pathway Analysis software.
PXR Chromatin Immunoprecipitation Coupled with High-Throughput Sequencing and Motif Analysis for PXR-DNA Binding Near lncRNA Gene Loci.
The PXR chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-Seq) was performed in livers of corn oil– or PCN-exposed adult male C57BL/6 mice as described previously (Cui et al., 2010b), and data were reanalyzed for the present study. The chromosome coordinates of positive PXR DNA binding peaks in either corn oil– or PCN-exposed conditions were retrieved. The chromosome coordinates for the enrichment locations were adjusted to match mm10 using Galaxy to be consistent with the reference genome used in the RNA-Seq data set. The lncRNA genes differentially regulated by PCN were examined for positive enrichment of PXR-DNA binding peaks within 10 kb upstream of the TSS and 10 kb downstream of the TTS. The Motif analysis of PXR binding sites near lncRNA gene loci was performed using Homer (findMotifsGenome.pl). DNA sequences associated with the PXR-DNA binding sites were retrieved using the mouse mm10 genome. The size of the analyzed DNA regions was set as 200 bp, and the motif lengths for de novo motif search were set at 8, 10, and 12 bp.
Epigenetic Marks Near PXR-Regulated lncRNAs.
The positive enrichment of the active gene transcription mark histone H3 lysine 4 di-methylation (H3K4me2), as well as the gene silencing marks H3K27me3 and DNA methylation (5MeC) on chromosomes 5, 12, and 15 in livers of adult C57BL/6 male mice was determined by ChIP-on-chip as described before (Cui et al., 2009, 2010a; Li et al., 2009; Choudhuri et al., 2010). The chromosome coordinates for the epigenetic mark locations that were originally generated were lifted over to mm10 using Galaxy to be consistent with the reference genomes used in the PXR ChIP-Seq and RNA-Seq data sets. The potential colocalizations of PXR and the three epigenetic marks within ±10 kb of the lncRNA gene loci were determined.
Results
Overall Comparison of PCN- and TCPOBOP-Regulated Liver-Expressed lncRNAs.
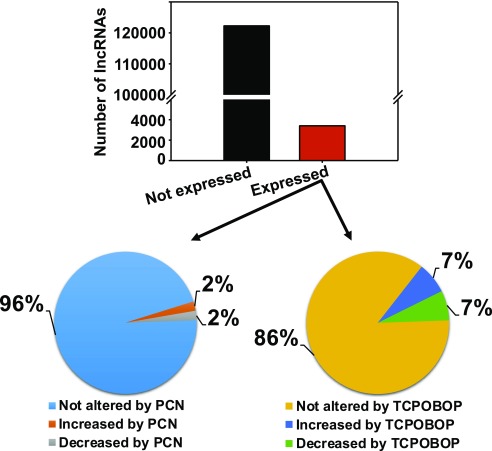
As shown in Fig. 1, most (>120,000) of the lncRNAs were below the detection limit in mouse livers under any exposure condition, whereas approximately 4000 lncRNAs were expressed (average FPKM >1 in at least one group). Among these liver-expressed lncRNAs, most showed stable expression and were not altered after PCN (96%) or TCPOBOP (86%) exposure. PCN upregulated approximately 2% and downregulated 2% of the liver-expressed lncRNAs, whereas TCPOBOP upregulated 7% and downregulated approximately 7% of the liver-expressed lncRNAs.
Fig. 1.
The number of lncRNAs that were not expressed (or not detected using the poly-A selection method) and expressed in liver. An lncRNA is considered to be expressed in liver if the average FPKM is above 1 in at least one exposure group (corn oil, PCN, or TCPOBOP). Among the liver-expressed lncRNAs, the percentages of lncRNAs that were not differentially regulated, increased, or decreased by chemical exposure were calculated and displayed as two pie charts (PCN and TCPOBOP). Differential expression was considered at P < 0.05 (Cuffdiff).
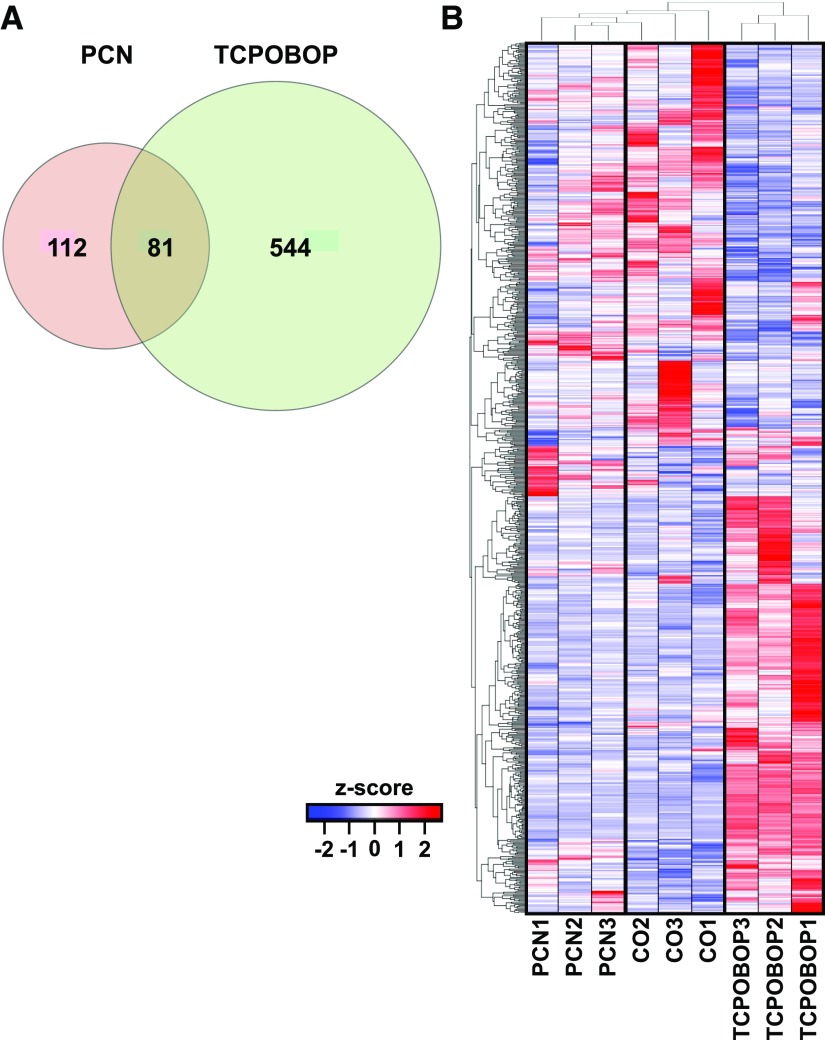
As shown in Fig. 2A, TCPOBOP in general had a more prominent effect than PCN in differentially regulating the liver expressed lncRNAs in that 193 lncRNAs were altered by PCN exposure compared with 625 that were altered by TCPOBOP exposure. There were 81 lncRNAs that were commonly regulated by both PCN and TCPOBOP, suggesting that PXR and CAR have unique lncRNA gene targets in liver. A two-way hierarchical clustering dendrogram showed that biologic replicates from the same exposure groups clustered together for the lncRNA gene expression (Fig. 2B) and confirmed that TCPOBOP had a more profound effect in both upregulation and downregulation of lncRNAs, which may be due to TCPOBOP being a highly potent activator of CAR, whereas the potency of PCN is less toward PXR activation.
Fig. 2.
(A) Common and unique lncRNA targets after PCN and TCPOBOP exposure in mouse liver (P < 0.05 as determined by Cuffdiff). (B) A hierarchical clustering dendrogram showing the relative expression patterns of lncRNAs in liver in corn oil–, PCN-, and TCPOBOP-exposed conditions. Data were standardized and are expressed as z scores.
Genomic Annotation of PCN- and TCPOBOP-Regulated Liver-Expressed lncRNAs Relative to PCGs and Predicted Gene Networks.
As a first step to predict the function of lncRNAs with regard to influencing the transcriptional output of PCGs, the genomic locations of PCN- and TCPOBOP-regulated lncRNAs relative to the PCGs were determined using PAVIS, and the lncRNA-PCG gene pairs were defined if they met all the following criteria: 1) the lncRNA gene overlaps with or is within 5 kb upstream of TSS or 1 kb downstream of TTS of any PCG, and 2) both the lncRNA and the proximal PCG were differentially expressed by PCN or TCPOBOP exposure (average FPKM >1 and P < 0.05). These criteria were set based on the assumption that although exceptions may exist, lncRNAs produced locally around the neighboring PCGs have a more spatial advantage in influencing the transcriptional output of these PCGs compared with lncRNAs produced in distal regions (Ørom et al., 2010; Kim et al., 2012; Villegas and Zaphiropoulos, 2015; Engreitz et al., 2016; Chen et al., 2018).
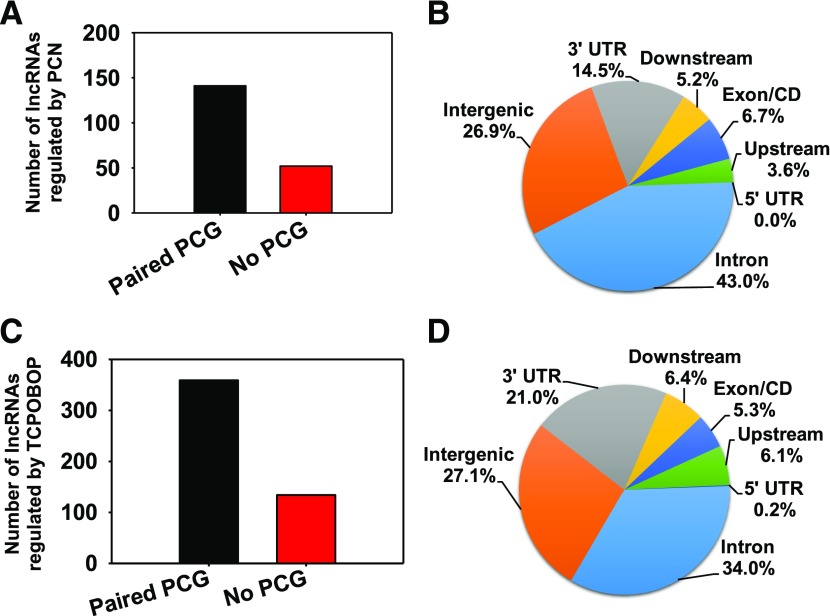
As shown in Fig. 3A and Supplemental Table 1, after PCN exposure, 141 (73.1%) of 193 lncRNAs differentially regulated by PCN paired with distinct PCGs. The remaining 52 (26.9%) were produced from the intergenic regions and did not pair with any PCGs. As shown in Fig. 3B, among the lncRNAs that paired with PCGs, most were produced from the intronic regions of PCGs (43%), followed by the 3′-untranslated regions (3′-UTRs) (14.5%), exonic regions (6.7%), downstream of TTS (5.2%), and upstream of TSS (3.6%). No lncRNAs were codifferentially regulated by PCN and produced within the 5′-UTR of PCGs that were differentially regulated by PCN. The PCGs that paired with lncRNAs formed distinct signaling networks, including lipid metabolism, molecular transport, and small molecular biochemistry centering around extracellular signal regulated kinase 1/2, which is a mitogen-activated protein kinase that catalyzes the phosphorylation of many cytoplasmic and nuclear substrates (Supplemental Fig. 1), as well as cellular growth and proliferation (Supplemental Fig. 2). For example, high-density lipoprotein, as well as its regulator serum amyloid A1, was downregulated, whereas AMPK, which activates glucose and fatty acid uptake and oxidation, was upregulated.
Fig. 3.
(A) Number of lncRNAs that paired with PCGs or not paired with PCGs after PCN exposure. An lncRNA-PCG gene pair is defined as follows:1) the lncRNA gene overlaps with or is within 5 kb upstream of TSS or 1 kb downstream of TTS of any PCG; and 2) both the lncRNA and the proximal PCG were differentially expressed by PCN exposure (average FPKM >1 and P < 0.05). (B) Genomic annotation of PCN-regulated lncRNAs (P < 0.05) relative to the PCGs. Data were analyzed using PAVIS. (C) Number of lncRNAs that paired with PCGs or not paired with PCGs after TCPOBOP exposure. An lncRNA-PCG gene pair is defined as follows:1) the lncRNA gene overlaps with or is within 5 kb upstream of TSS or 1 kb downstream of TTS of any PCG; and 2) both the lncRNA and the proximal PCG were differentially expressed by PCN exposure (average FPKM >1 and P < 0.05). (D) Genomic annotation of TCPOBOP-regulated lncRNAs (P < 0.05) relative to the PCGs. Data were analyzed using PAVIS.
After TCPOBOP exposure, additional lncRNA-PCG pairs were discovered. As shown in Fig. 3C and Supplemental Table 2, 359 (73%) lncRNAs were paired with PCGs, whereas 134 were not paired (27.1%). Similarly to the PCN exposure conditions, most of the paired lncRNAs were produced from the intronic (34%) and 3′-UTR (21.5%) of the PCGs, followed by downstream (6.4%), upstream (6.1%), and exonic (5.3%) regions, whereas mapping to 5′-UTR was minimal (0.2%) (Fig. 3D). Similarly to PCN exposure, the PCGs that paired with lncRNAs after TCPOBOP exposure were also important for lipid metabolism and molecular transport (Supplemental Fig. 3). For example, apoliprotein A4, which is a lipid transporter, was upregulated, whereas serum amyloid A1, which plays an important role in high-density lipoprotein metabolism and cholesterol homeostasis, was downregulated (Supplemental Fig. 3). In addition, the carcinogenesis network was enriched in TCPOBOP-regulated PCGs that paired with lncRNAs (Supplemental Fig. 4).
PXR-DNA Binding to PCN-Regulated Hepatic lncRNAs and Motif Analysis.
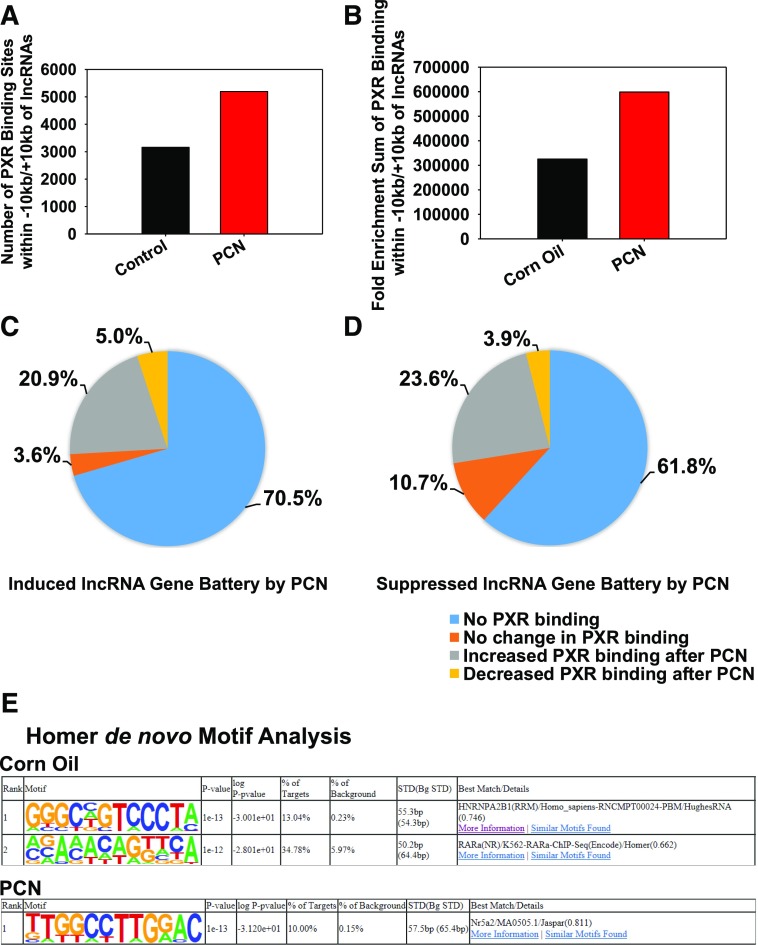
As shown in Fig. 4, both the total numbers of PXR-DNA binding sites and the cumulative PXR-DNA binding fold enrichment near the lncRNA gene loci (±10 kb) increased after PCN exposure. Approximately 70% of the differentially regulated lncRNAs had no PXR binding. Interestingly, among the direct PXR-targeted lncRNA genes, both the upregulated and the downregulated lncRNAs had increased PXR-DNA binding, indicating that PXR has dual functions in the transcription of these lncRNAs—cis-activation and cis-suppression (Fig. 4, C and D)—likely owing to a functional switch of other transcription factors that colocalize with PXR at these sites. To address this, de novo motif analysis was performed in lncRNA gene-associated PXR-DNA binding peaks in corn oil– and PCN- exposed conditions independently (Fig. 4E). Interestingly, in control conditions, HNNRNPA2B1 and RARα were significantly enriched in PXR-DNA binding sites of the lncRNA genes, whereas Nr5a2/liver receptor homolog-1 (LRH-1) was enriched by PCN. The molecular switches of these transcription factors may alter the fate of transcriptional output of PXR-targeted lncRNA genes. Other transcription factors associated with known DNA binding motifs within the PXR-DNA binding intervals around the differentially regulated lncRNA gene loci by PCN are shown in Supplemental Fig. 5A.
Fig. 4.
(A) Number of PXR-DNA binding sites within ±10 kb of lncRNA gene loci in corn oil– and PCN-exposed conditions. (B) Cumulatively PXR-DNA binding fold enrichment within ±10 kb of lncRNA gene loci in corn oil– and PCN-exposed conditions. (C) Percentages of induced lncRNA gene battery by PCN that had no PXR binding, no change in PXR binding, an increase in PXR binding, and decreased PXR binding after PCN exposure. (D) Percentages of decreased lncRNA gene battery by PCN that had no PXR binding, no change in PXR binding, an increase in PXR binding, and decreased PXR binding after PCN exposure. (E) Homer de novo motif analysis of PXR-DNA binding intervals in corn oil– and PCN-exposed groups using findMotifsGenome.pl. Data were reanalyzed from the PXR ChIP-Seq data set as described in Materials and Methods.
Determining the Potential Colocalization of Epigenetic Factors and PXR Near lncRNA Gene Loci.
Distinct PCN exposure-associated permissive and repressive epigenetic marks have previously been identified on mouse chromosomes 5, 12, and 15 [see Materials and Methods]. In a preliminary investigation into the role of epigenetic marks and PXR binding on lncRNA expression, we searched for colocalization of PXR-DNA binding, specific epigenetic marks (H3K4me2, H3K27me3, and 5MeC) near lncRNA loci on the same chromosomes. Most of the lncRNAs associated with these epigenetic marks or PXR were not expressed or not detected on the three chromosomes, likely because of the presence of other suppressive marks that were not investigated in the present study or not detected owing to the method of RNA selection in the cDNA library preparation procedure (i.e., only poly-A-tailed lncRNAs were captured) (Supplemental Table 3). Among the liver-expressed lncRNAs on these three chromosomes, most stably expressed lncRNAs had positive enrichment of the active chromatin epigenetic mark H3K4me2 and PXR. The overlap between H3K4me2 and PXR was also the largest in the stably expressed lncRNA gene category. This indicates that H3K4me2 and PXR may act in concert to maintain the constitutive expression of liver-enriched lncRNAs. Interestingly, DNA methylation (5MeC), commonly thought to silence gene expression, was another epigenetic mark that colocalized with stably expressed lncRNA gene loci and overlapped moderately with PXR binding sites. Paradoxically, although 5MeC is considered a gene-silencing mark, our observation is consistent with the more recent literature report stating that many actively transcribed gene bodies are marked with 5MeC. This observation correlates with transcriptional activity rather than repression, that the basal function of gene body methylation facilitates the establishment of their constitutive expression, and that cytosine-specific methylation may regulate gene expression (Coleman-Derr and Zilberman, 2012; Vyhlidal et al., 2016). After PXR activation, a moderate number of PCN-regulated lncRNAs (either upregulated or downregulated) had positive enrichment of H3K4me2, and most of these lncRNAs had direct PXR-DNA binding sites. In contrast, almost none of the PCN-regulated lncRNAs had positive enrichment of H3K27me3 or 5MeC, and no overlap between these epigenetic marks and PXR were found at these lncRNA gene loci (Supplemental Table 3).
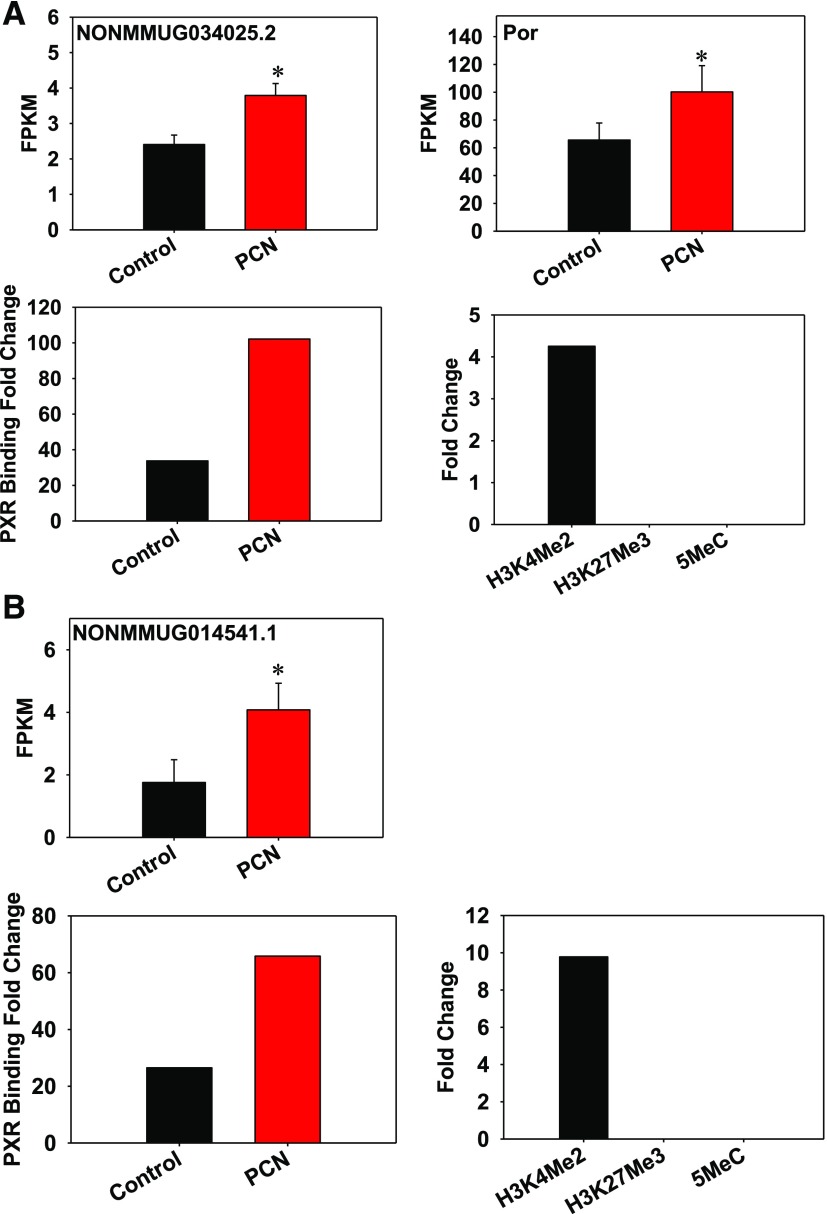
Two examples of lncRNAs that had colocalization of PXR and H3K4me2 are shown in Fig. 5A (a lncRNA that paired with a PCG encoding P450 reductase [Por] that is important for cytochrome P450-mediated hepatic drug metabolism) and Fig. 5B (an intergenic lncRNA) with the gene annotation shown in Supplemental Fig. 6. As shown in Fig. 5A, the lncRNA NONMMUG034025.2 and the neighboring PCG Por were co-upregulated by PCN, and this was associated with increased PXR-DNA binding fold-enrichment around the lncRNA-PGC loci. In addition, this region was marked with the active gene transcription mark H3K4me2, but not H3K27me3 or 5MeC. As shown in Fig. 5B, the PCN-mediated upregulation of the lncRNA NONMMUG014541.1 was also positively associated with increased PXR-DNA binding and positive enrichment of H3K4me2 but was independent of PCGs.
Fig. 5.
(A) Colocalization of PXR and H3K4me2 around the lncRNA NONMMUG034025.2 and the paired PCG Por gene loci. Integrated Genome Viewer (IGV) is a high-performance visualization tool for displaying and exploring large data sets, including RNA-Seq data, and can be accessed at http://software.broadinstitute.org/software/igv/. The genomic locations of lncRNA and PCG are visualized by IGV. Asterisks represent statistically significant differences compared with corn oil control group (P < 0.05, Cuffdiff). FPKM is a unit to express RNA abundance from the RNA-Seq data. Data from RNA-Seq (corn oil– and PCN-exposed groups), ChIP-Seq (for PXR-DNA binding in corn oil– and PCN-exposed conditions), and ChIP-on-chip (for H3K4me2, H3K27me3, and 5MeC on mouse chormosomes 5, 12, and 15) were integrated as described in Materials and Methods. (B) Colocalization of PXR and H3K4me2 around the intergenic lncRNA NONMMUG014541.1 gene locus. The genomic location of lncRNA and PCG is seen by IGV. Asterisks represent statistically significant differences compared with corn oil control group (P < 0.05, Cuffdiff). Data from RNA-Seq (corn oil– and PCN-exposed groups), ChIP-Seq (for PXR-DNA binding in corn oil– and PCN-exposed conditions), and ChIP-on-chip (for H3K4me2, H3K27me3, and 5MeC on mouse chormosomes 5, 12, and 15) were integrated as described in Materials and Methods.
Confirmation of the Literature-Reported CAR-Targeted lncRNAs in Liver after TCPOBOP Exposure and Comparison with the Effect of PCN Exposure.
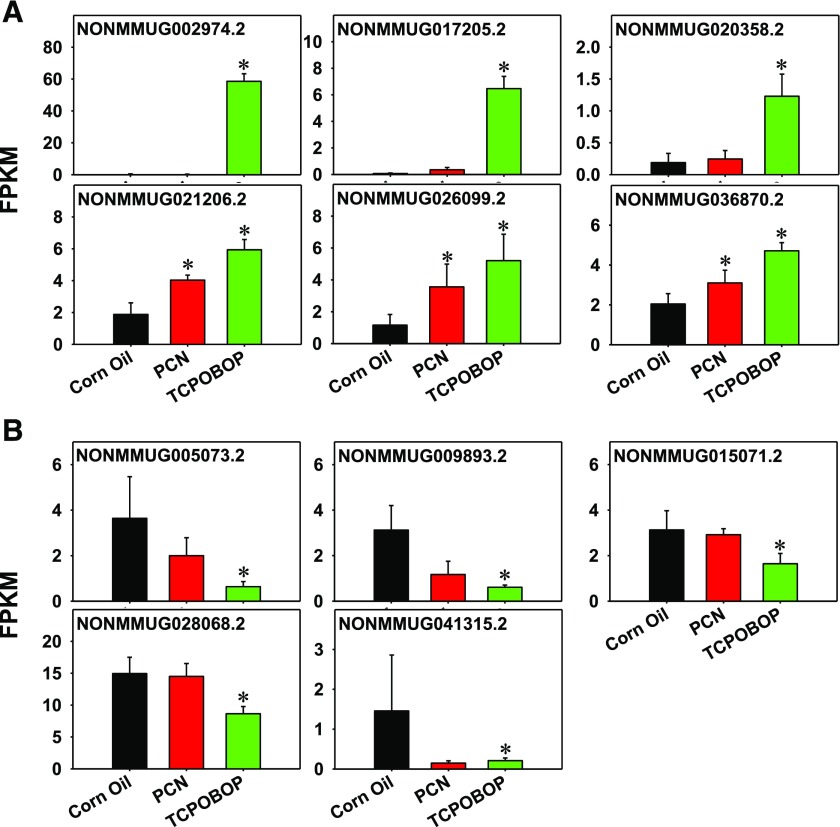
Consistent with the recently published study regarding the effect of the CAR ligand TCPOBOP on the hepatic lncRNA gene expression (Lodato et al., 2017), the present study confirmed the TCPOBOP-mediated increase in six lncRNAs (NONMMUG002974.2, NONMMUG017205.2, NONMMUG020358.2, NONMMUG021206.2, NONMMUG026099.2, and NONMMUG036870.2) (Fig. 6A), as well as the TCPOBOP-mediated decrease in five lncRNAs (NONMMUG005073.2, NONMMUG009893.2, NONMMUG015071.2, NONMMUG028068.2, and NONMMUG041315.2) (Fig. 6B). In comparison, at the given dose, the PXR ligand PCN upregulated three of the same lncRNAs (NONMMUG021206.2, NONMMUG026099.2, and NONMMUG036870.2) as TCPOBOP, albeit to a lesser extent. PCN had only minimal effects on the other TCPOBOP upregulated lncRNAs.
Fig. 6.
Examples of hepatic lncRNAs that were upregulated (A) or downregulated (B) by TCPOBOP (which are consistent with the literature report of Lodato et al., 2017). The effect of PCN on these CAR-targeted lncRNAs is also shown. Asterisks represent statistically significant differences compared with the corn oil group (P < 0.05, Cufdiff). FPKM a unit to express RNA abundance from the RNA-Seq data.
Discussion
The key findings of the present study are 1) at the given doses for the nuclear receptor agonists, CAR activation affects the expression of more lncRNAs than PXR activation in liver, and both commonly and uniquely regulated lncRNAs are observed for the two receptors; 2) most of the differentially regulated lncRNAs are present in introns, intergenic, and 3′-UTRs relative to PCGs; 3) for both receptors, the top differentially regulated PCGs paired with lncRNAs are involved in lipid metabolism and molecular transport, highlighting the potential importance of lncRNAs in fine-tuning this signaling pathway in liver; 4) increased PXR binding by PCN exposure may lead to either transactivation or trans-suppression of direct lncRNA targets, and motif analysis suggests that this context-specific duality may be due to a molecular switch of colocalized transcription factors; and 5) the epigenetic activation mark H3K4me2 may facilitate the PXR recruitment to the genomic regions proximal to lncRNA gene loci for activation of gene transcription.
One technical limitation of this study was the inability to detect nonpolyadenylated lncRNAs, which may also be important for liver functions, owing to the use of poly-A tail selection in RNA-Seq library construction. This likely explains why a previous study identified more liver-enriched lncRNAs than the present study in mice (Fig. 1) (Lodato et al., 2017). The nonpolyadenylated lncRNAs are generally expressed at lower levels than the non-polyadenylated mRNAs and are prevalent in the nucleus, suggesting that they may be involved in the transcriptional regulation of target genes (Cheng et al., 2005a; Furuno et al., 2006). At this time, the ratio between polyadenylated lncRNAs and nonpolyadenylated lncRNAs in liver is not known and most of the well characterized lncRNAs are produced using the same machinery as the PCGs (i.e., transcribed by RNA Pol-II, polyadenylated, and spliced) (Kung et al., 2013). Future studies using whole-transcriptome analysis could use ribosomal depletion as an alternative library construction option, although the tradeoff appears to be lower signal per transcript at the same read depth.
An experimental limitation of the present study is the lack of validations of the findings using PXR and CAR knockout mice. Using these knockout mice would demonstrate the extent to which the regulation of lncRNAs is dependent on PXR and CAR under basal conditions. Previously, one study examined the species differences in gene regulation using human CAR transgenic mice, as well as CAR-null mice (Cheng et al., 2017). This study showed that expression of Cyp3a11 and Cyp2b10 was upregulated by TCPOBOP only in livers of WT mice but not in CAR-null mice. In a separate study, α-tocopherol was shown to be a PXR activator using wild-type and PXR-null mice. PXR-null compared with wild-type mice did not differ greatly in the regulated gene expression of the major P450s (Johnson et al., 2013). The purpose of the current study was to investigate those lncRNAs that are regulated by the pharmacologic activation of PXR or CAR (similarly to a toxicologic response) as a first step. In future directions, it will be important to determine the necessity of the nuclear receptors in modulating the constitutive lncRNA gene expression using knockout mice.
Another interesting finding is that most of the differentially regulated lncRNAs are produced within the intronic region of the PCGs (43%), followed by the intergenic regions (26.9%) and 3′-UTRs (14.5%) (Fig. 3B). Because only the mature mRNAs that are poly-A- tailed are enriched, the intronic transcripts should be separate lncRNA fragments instead of nascent mRNA transcripts, which suggests that the mammalian transcription machinery is highly efficient in that both lncRNAs and PCGs may share the same transcription machinery because of the relative genomic distributions. Intronic lncRNAs are known to initiate their transcription inside introns of PCGs in either direction and terminate without overlapping exons (Rinn and Chang, 2012). Little is known regarding the specific functions of intronic lncRNAs, although their potential involvement in cancer has been suggested (Tahira et al., 2011). Intergenic lncRNAs are encoded completely within intergenic regions between PCG loci and have been suggested to be more stable than intronic lncRNAs (Rinn and Chang, 2012). The differential regulation of intergenic lncRNAs by PXR and CAR ligands indicates the importance of PXR and CAR genomic binding to the intergenic regions. Indeed, we have previously demonstrated that approximately 30% of PXR-genomic DNA binding are present in the intergenic regions (Cui et al., 2010b). CAR ChIP-Seq experiments are challenging because of a lack of a good antibody-targeting endogenous CAR protein, although one could overcome this technical challenge using an adenovirus-based system to direct the expression of YFP-CAR fusion constructs in transgenic mice (Niu et al., 2018); however, it seems reasonable to speculate that a substantial number of differentially regulated intergenic lncRNAs by TCPOBOP may be a result of direct CAR-binding to the intergenic regions. Lastly, lncRNAs produced from the 3′-UTR regions relative to PCGs may be important in protecting the mRNAs from miRNA-mediated degradation and/or inhibition of protein synthesis (Karapetyan et al., 2013; Dempsey and Cui, 2017). Although lncRNAs produced in distal regions may migrate to the 3′-UTRs of PCGs and perform similar functions, the local production of lncRNAs may have a special advantage in this process.
Regarding the potency of the two nuclear receptors, at the given dose, CAR activation by TCPOBOP differentially regulated more lncRNAs in liver than did PXR activation by PCN (Fig. 1). Similarly, as we observed before, CAR activation also differentially regulated more PCGs than PXR activation (Cui and Klaassen, 2016). Therefore, it appears that TCPOBOP-mediated CAR activation is a highly potent stressor leading to an overall greater change in the mouse transcriptome. Among all the differentially regulated PCGs that paired with lncRNAs, two pathways appeared to be affected the most by PXR and CAR activation, namely, lipid metabolism (Fig. 3C; Fig. 4C) and cell proliferation/cancer (Supplemental Figs. 1 and 2). Both nuclear receptors and certain lncRNAs have been implicated in these two pathways independently (Mo et al., 2016; Schmitt and Chang, 2016; Kong and Guo, 2018), whereas the present study is among the first to show that the biologic outcomes (changes in lipid metabolism and cell proliferation status) mediated by PXR/CAR may be coregulated by lncRNAs. The PXR-mediated change in the lipid metabolism pathway is further supported by the motif analysis in that the PXR-targeted ChIP DNA from PCN-treated mouse livers had enrichment in the LRH-1 DNA binding motifs (Fig. 4E). LRH-1 is an important orphan nuclear receptor that regulates cholesterol, bile acid, and steroid hormone synthesis (Lee et al., 2008). The PXR activation by PCN has also been shown to be important for regulating bile acid synthesis (Staudinger et al., 2001). Under basal conditions, there was no enrichment in LRH-1 DNA binding motifs; instead, the DNA-binding motifs for hnRNP2B1, which is involved in alternative splicing of pre-mRNAs, as well as RARα, which is a receptor for retinoic acid, were enriched (Fig. 5E).
Traditionally, PXR is considered a transcriptional activator for various drug- metabolizing enzymes and efflux transporters; however, recent studies using ChIP-Seq and microarrays have suggested that increased PXR binding may result in either transactivation or trans-suppression of bona fide PXR-targeted protein coding genes (Cui et al., 2010b). The present study adds to the existing literature showing that the PXR-targeted lncRNA genes can also be further divided into inducible versus suppressive gene batteries upon increased PXR binding. We propose two potential mechanisms for this phenomenon: 1) the coregulators that interact with PXR may ultimately determine the fate of PXR-target gene transcription. There is evidence in the literature showing that human PXR may interact with either a coactivator (e.g., SRC-1) or a corepressor (e.g., silencing mediator of retinoid and thyroid hormone receptors or nuclear receptor corepressor) (Navaratnarajah et al., 2012). Human PXR activity is repressed by the corepressor silencing mediator of retinoid and thyroid hormone receptors (Johnson et al., 2006). 2) Another possibility is that PXR may compete with other transactivators that are more important in the transcription of the target genes. Previous studies showed that PXR activation inhibits cAMP responsive element binding protein and subsequently downregulates the expression of rate-limiting enzymes for glucose homeostasis, such as glucose-6-phosphatase catalytic subunit (G6Pase) and phosphoenolpyruvate carboxykinase 1 (PEPCK1) (Kodama et al., 2007; Oh et al., 2013). In the present study, in silico analysis suggests that pharmacologic activation of PXR is associated with increased nuclear occupancy of LRH-1 but decreased nuclear occupancy of RARα (Fig. 4E). This molecular switch may also contribute to the transcriptional silencing of a subset of PXR-targeted lncRNA genes. Additional studies using GST pull down assays as well as overlay between PXR cistrome and ChIP-Seq data of other transcription factors will verify our hypotheses.
Regarding the human relevance, it is known that both PXR and CAR both have species differences between mice and humans, especially their contribution in regulating cell proliferation (Kong and Guo, 2018; Niu et al., 2018). However, the regulation of lncRNAs may be more conserved between the two species, because it has been shown that lncRNA promoters are more conserved than lncRNA exons and almost as conserved as those of PCGs (Carninci et al., 2005; Guttman et al., 2009). In addition, there are greater species conservations and higher frequency of proximal binding by hepatic transcription factors in the liver-enriched lncRNA gene promoters than PCG promoters (Melia et al., 2016). Therefore, it is reasonable to speculate that similar regulatory patterns of the lncRNAs may also be present in human livers after PXR and CAR activation.
The present study unveiled PCG-lncRNA pairs based on the positive associations of the neighboring PCG and lncRNAs afterr chemical exposure. Further mechanistic investigations are needed using lncRNA knockdown approach to validate the dependency of lncRNAs in the transcriptional/translational output of the paired PCGs. In humans, a recent study demonstrated that knocking down the neighboring lncRNAs produced from the antisense strand of the transcription factors HNF1α and HNF4α affected the expression of P450s in HepaRG cells (Chen et al., 2018). These observations have demonstrated the critical role of lncRNAs may play in modulating PCG expression.
The rationale for the selection of H3K4me2, H3K27me3, and 5MeC over other epigenetic marks was that these marks were shown to associate with the regulation of important drug-processing genes in liver, as well as the focus of the present study on xenobiotic-sensing nuclear receptors (PXR and CAR). For example, the age-specific enrichment in H3K4me2 around the Cyp3a gene loci positively associates with the age-specific expression of the Cyp3a gene isoforms in mouse liver (Li et al., 2009). In addition, adult-specific enrichment of H3K4me2 positively associated with the adult-specific mRNA expression of other drug-processing genes, including glutathione S-transferase zeta 1 (Gstz1) (Cui et al., 2010a), UDP glucuronosyltransferase 2 and 3 (Ugt2 and Ugt3) (Choudhuri et al., 2010), as well as Ahr gene locus (Cui et al., 2009). Conversely, the presence of H3K27me3 was associated with the downregulation of CYP1A2 mRNA in human embryonic stem cell-derived hepatocytes (hESC-Hep) and primary human hepatocytes (Park et al., 2015). Regarding 5MeC, an investigation of DNA methylation in human liver samples demonstrated variable CpG hyper-methylation of the CYP3A4 promoter region in adults, as well as in other CCAAT-enhancer-binding proteins (C/EBP) and HNF4α binding sites (Kacevska et al., 2012), indicating that DNA methylation contributes to the regulation of CYP3A4 expression and the subsequent modifications in xenobiotic metabolism. It has also been suggested that methylation of gene bodies can serve as a novel therapeutic target for cancer treatment (Yang et al., 2014). Because many drug-processing genes are known PXR and CAR targets, and the present study identified many lncRNAs as being regulated by these drug receptors, the primary goal of the present study was to determine the interactions between these liver genes and the enrichment of these three epigenetic marks related to xenobiotic biotransformation. Many other epigenetic marks have been identified in the literature (Tan et al., 2011; Rivera and Ren, 2013), and it is important to investigate the involvement of these other marks in PXR- and CAR-mediated regulation of PCGs and lncRNAs in future studies.
Previously, it was demonstrated that most lncRNAs share similar epigenetic marks at the promoter regions, such as H3K4me3 and RNA Pol-II binding sites as PCGs; however, a certain fraction of lncRNAs display a high prevalence of H3K4me1, which marks the enhancer region (Kashi et al., 2016). The present study adds to the evidence showing that a subset of mouse chromosomes (5, 12, and 15), H3K4me2, which marks the enhancers and actively transcribed gene bodies, is colocalized with PXR-DNA binding near the PCG-lncRNA gene loci, leading to active gene transcription. H3K4me2 may provide a permissive chromatin environment for PXR-binding and the subsequent gene transcription and may serve as an important mechanism for the coexpression of the PCG-lncRNA pairs.
The present study is among the first to compare systemically the PXR- and CAR-targeted lncRNA profiles in liver, has provided novel insights into the molecular mechanisms underlying the lncRNA gene transcriptional regulation and potential biologic outcomes in liver, and lay the foundation for further decoding the mechanism of the regulation of the gene transcription and the lncRNA-PCG networks in vivo.
Acknowledgments
We thank Dr. Curtis Klaassen and members of the Cui Laboratory for assistance revising the manuscript, as well as Dr. Xiao-bo Zhong and Dr. Hong Lu for their effort and collaboration in the previously published work from the ChIP-on-chip experiments.
Abbreviations
- 5MeC
DNA methylation and cytosine 5th position
- AhR
aryl hydrocarbon receptor
- AS
anti-sense
- CAR
constitutive androstane receptor
- ChIP-Seq
chromatin immunoprecipitation coupled with high-throughput sequencing
- FPKM
fragments per kilobase of exon per million reads mapped
- H3K27me3
H3 lysine 27 tri-methylation
- H3K4me2
H3 lysine 4 di-methylation
- hepaRG
hepatic stem cell line
- HNF
hepatocyte nuclear factor
- KUMC
University of Kanses Medical Center
- lncRNA
long noncoding RNA
- LRH-1
liver receptor homolog-1
- P450
cytochrome P450
- PCG
protein-coding gene
- PCN
pregnenolone-16α-carbonitrile
- PXR
pregnane X receptor
- SAM
sequencing alignment/map
- siRNA
small interfering RNA
- TCPOBOP
1, 4-bis[2-(3, 5-dichloropyridyloxy)]benzene
- TSS
transcription start site
- TTS
transcriptional termination site
- UTR
untranslated region
Authorship Contributions
Participated in research design: Cui.
Conducted experiments: Cui.
Performed data analysis: Dempsey, Cui.
Wrote or contributed to the writing of the manuscript: Dempsey, Cui.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants [GM111381 and T32 ES007032], the University of Washington Center for Exposures, Diseases, Genomics, and Environment [P30 ES0007033], and the Murphy Endowment.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ananthanarayanan M. (2016) A novel long noncoding RNA regulating cholesterol and bile acid homeostasis: a new kid on the block and a potential therapeutic target? Hepatology 64:16–18. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- Chen L, Bao Y, Piekos SC, Zhu K, Zhang L, Zhong XB. (2018) A transcriptional regulatory network containing nuclear receptors and long noncoding RNAs controls basal and drug-induced expression of cytochrome P450s in HepaRG cells. Mol Pharmacol 94:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Huang H, Xu C, Yu C, Li Y. (2017) Long non-coding RNA profiling in a non-alcoholic fatty liver disease rodent Model: new insight into pathogenesis. Int J Mol Sci 18(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. (2015) Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol Metab 5:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. (2005a) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308:1149–1154. [DOI] [PubMed] [Google Scholar]
- Cheng SL, Bammler TK, Cui JY. (2017) RNA sequencing reveals age and species differences of constitutive androstane receptor-targeted drug-processing genes in the liver. Drug Metab Dispos 45:867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Maher J, Dieter MZ, Klaassen CD. (2005b) Regulation of mouse organic anion-transporting polypeptides (Oatps) in liver by prototypical microsomal enzyme inducers that activate distinct transcription factor pathways. Drug Metab Dispos 33:1276–1282. [DOI] [PubMed] [Google Scholar]
- Choudhuri S, Cui Y, Klaassen CD. (2010) Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol 245:378–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. (2012) DNA methylation, H2A.Z, and the regulation of constitutive expression. Cold Spring Harb Symp Quant Biol 77:147–154. [DOI] [PubMed] [Google Scholar]
- Cui JY, Choudhuri S, Knight TR, Klaassen CD. (2010a) Genetic and epigenetic regulation and expression signatures of glutathione S-transferases in developing mouse liver. Toxicol Sci 116:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. (2010b) ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res 38:7943–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui JY, Klaassen CD. (2016) RNA-Seq reveals common and unique PXR- and CAR-target gene signatures in the mouse liver transcriptome. Biochim Biophys Acta 1859:1198–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui YJ, Yeager RL, Zhong XB, Klaassen CD. (2009) Ontogenic expression of hepatic Ahr mRNA is associated with histone H3K4 di-methylation during mouse liver development. Toxicol Lett 189:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JL, Cui JY. (2017) Long non-coding RNAs: a novel paradigm for toxicology. Toxicol Sci 155:3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. (2012) Landscape of transcription in human cells. Nature 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno M, Pang KC, Ninomiya N, Fukuda S, Frith MC, Bult C, Kai C, Kawai J, Carninci P, Hayashizaki Y, et al. (2006) Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet 2:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Xie W. (2010) Pregnane X receptor and constitutive androstane receptor at the crossroads of drug metabolism and energy metabolism. Drug Metab Dispos 38:2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J. (2013) RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14:699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N, Sivadas A, Shamsudheen KV, Jayarajan R, Verma A, Sivasubbu S, Scaria V, Datta M. (2017) RNA sequencing of db/db mice liver identifies lncRNA H19 as a key regulator of gluconeogenesis and hepatic glucose output. Sci Rep 7:8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G, Rajendra S, Matthews J. (2018) The aryl hydrocarbon receptor regulates the expression of TIPARP and its cis long non-coding RNA, TIPARP-AS1. Biochem Biophys Res Commun 495:2356–2362. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Meyer UA. (2003) Induction of drug metabolism: the role of nuclear receptors. Pharmacol Rev 55:649–673. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Zhong XB, Hankinson O, Beedanagari S, Yu AM, Peng L, Osawa Y. (2013) Potential role of epigenetic mechanisms in the regulation of drug metabolism and transport. Drug Metab Dispos 41:1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Bonzo JA, Cheng J, Krausz KW, Kang DW, Luecke H, Idle JR, Gonzalez FJ. (2013) Cytochrome P450 regulation by α-tocopherol in Pxr-null and PXR-humanized mice. Drug Metab Dispos 41:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, Li CW, Chen LY, Ghosh JC, Chen JD. (2006) Regulation and binding of pregnane X receptor by nuclear receptor corepressor silencing mediator of retinoid and thyroid hormone receptors (SMRT). Mol Pharmacol 69:99–108. [DOI] [PubMed] [Google Scholar]
- Kacevska M, Ivanov M, Wyss A, Kasela S, Milani L, Rane A, Ingelman-Sundberg M. (2012) DNA methylation dynamics in the hepatic CYP3A4 gene promoter. Biochimie 94:2338–2344. [DOI] [PubMed] [Google Scholar]
- Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, et al. (2010) The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA [published correction appears in BMC Biol (2011) 9:86]. BMC Biol 8:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karapetyan AR, Buiting C, Kuiper RA, Coolen MW. (2013) Regulatory roles for long ncRNA and mRNA. Cancers (Basel) 5:462–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Baccarelli AA. (2016) Environmental health and long non-coding RNAs. Curr Environ Health Rep 3:178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashi K, Henderson L, Bonetti A, Carninci P. (2016) Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta 1859:3–15. [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. (2012) Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa N, Kwekel JC, Burgoon LD, Dere E, Williams KJ, Tashiro C, Chittim B, Zacharewski TR. (2008) Species-specific regulation of PXR/CAR/ER-target genes in the mouse and rat liver elicited by o, p′-DDT. BMC Genomics 9:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Goodwin B, Willson TM. (2002) The nuclear pregnane X receptor: a key regulator of xenobiotic metabolism. Endocr Rev 23:687–702. [DOI] [PubMed] [Google Scholar]
- Kodama S, Moore R, Yamamoto Y, Negishi M. (2007) Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene. Biochem J 407:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Guo GL. (2019) Is This the Time to Reconsider the Names for Xenobiotic Nuclear Receptors? Hepatology 69:16–18. [DOI] [PubMed] [Google Scholar]
- Kung JT, Colognori D, Lee JT. (2013) Long noncoding RNAs: past, present, and future. Genetics 193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan X, Yan J, Ren J, Zhong B, Li J, Li Y, Liu L, Yi J, Sun Q, Yang X, et al. (2016) A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 64:58–72. [DOI] [PubMed] [Google Scholar]
- Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA. (2008) Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol 22:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H, Couttet P, Bolognani F, Müller A, Dubost V, Luisier R, Del Rio Espinola A, Vitry V, Unterberger EB, Thomson JP, et al. (2013) Identification of Dlk1-Dio3 imprinted gene cluster noncoding RNAs as novel candidate biomarkers for liver tumor promotion. Toxicol Sci 131:375–386. [DOI] [PubMed] [Google Scholar]
- Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J, Fan H, Chang Y, Yang W. (2017) Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int J Biol Sci 13:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, Cao H. (2015) A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab 21:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cui Y, Hart SN, Klaassen CD, Zhong XB. (2009) Dynamic patterns of histone methylation are associated with ontogenic expression of the Cyp3a genes during mouse liver maturation. Mol Pharmacol 75:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato NJ, Melia T, Rampersaud A, Waxman DJ. (2017) Sex-Differential Responses of Tumor Promotion-Associated Genes and Dysregulation of Novel Long Noncoding RNAs in Constitutive Androstane Receptor-Activated Mouse Liver. Toxicol Sci 159:25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato NJ, Rampersaud A, Waxman DJ. (2018) Impact of CAR agonist ligand TCPOBOP on mouse liver chromatin accessibility. Toxicol Sci 164:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackowiak B, Hodge J, Stern S, Wang H. (2018) The roles of xenobiotic receptors: beyond chemical disposition. Drug Metab Dispos 46:1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. (2005) Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos 33:956–962. [DOI] [PubMed] [Google Scholar]
- Melia T, Hao P, Yilmaz F, Waxman DJ. (2016) Hepatic Long Intergenic Noncoding RNAs: High Promoter Conservation and Dynamic, Sex-Dependent Transcriptional Regulation by Growth Hormone. Mol Cell Biol 36:50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo L, Shen J, Liu Q, Zhang Y, Kuang J, Pu S, Cheng S, Zou M, Jiang W, Jiang C, et al. (2016) Irisin is regulated by CAR in liver and is a mediator of hepatic glucose and lipid metabolism. Mol Endocrinol 30:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JT, Moore LB, Maglich JM, Kliewer SA. (2003) Functional and structural comparison of PXR and CAR. Biochim Biophys Acta 1619:235–238. [DOI] [PubMed] [Google Scholar]
- Navaratnarajah P, Steele BL, Redinbo MR, Thompson NL. (2012) Rifampicin-independent interactions between the pregnane X receptor ligand binding domain and peptide fragments of coactivator and corepressor proteins. Biochemistry 51:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Coslo DM, Bataille AR, Albert I, Pugh BF, Omiecinski CJ. (2018) In vivo genome-wide binding interactions of mouse and human constitutive androstane receptors reveal novel gene targets. Nucleic Acids Res 46:8385–8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh KJ, Han HS, Kim MJ, Koo SH. (2013) CREB and FoxO1: two transcription factors for the regulation of hepatic gluconeogenesis. BMB Rep 46:567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshida K, Vasani N, Thomas RS, Applegate D, Gonzalez FJ, Aleksunes LM, Klaassen CD, Corton JC. (2015) Screening a mouse liver gene expression compendium identifies modulators of the aryl hydrocarbon receptor (AhR). Toxicology 336:99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak EK, Cheng X, Cunningham ML, Crofton K, Klaassen CD, Guo GL. (2007) The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol Sci 97:94–102. [DOI] [PubMed] [Google Scholar]
- Pan JJ, Xie XJ, Li X, Chen W. (2015) Long non-coding RNAs and drug resistance. Asian Pac J Cancer Prev 16:8067–8073. [DOI] [PubMed] [Google Scholar]
- Park HJ, Choi YJ, Kim JW, Chun HS, Im I, Yoon S, Han YM, Song CW, Kim H. (2015) Differences in the epigenetic regulation of cytochrome P450 genes between human embryonic stem cell-derived hepatocytes and primary hepatocytes. PLoS One 10:e0132992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Paulson A, Li H, Piekos S, He X, Li L, Zhong XB. (2014) Developmental programming of long non-coding RNAs during postnatal liver maturation in mice. PLoS One 9:e114917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Dubois D, Shultz TR. (1990) The infant’s concept of agency: the distinction between social and nonsocial objects. J Genet Psychol 151:77–90. [DOI] [PubMed] [Google Scholar]
- Pratt-Hyatt M, Lickteig AJ, Klaassen CD. (2013) Tissue distribution, ontogeny, and chemical induction of aldo-keto reductases in mice. Drug Metab Dispos 41:1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu S, Wu X, Yang X, Zhang Y, Dai Y, Zhang Y, Wu X, Liu Y, Cui X, Jin H, Cao J, Li R, Cai J, Cao Q, Hu L, Gao Y. (2018) The Therapeutic Role of Xenobiotic Nuclear Receptors against Metabolic Syndrome. Curr Drug Metab. [DOI] [PubMed] [Google Scholar]
- Recio L, Phillips SL, Maynor T, Waters M, Jackson AF, Yauk CL. (2013) Differential expression of long noncoding RNAs in the livers of female B6C3F1 mice exposed to the carcinogen furan. Toxicol Sci 135:369–379. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CM, Ren B. (2013) Mapping human epigenomes. Cell 155:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, Chang HY. (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. (2001) Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane X receptor. Drug Metab Dispos 29:1467–1472. [PubMed] [Google Scholar]
- St Laurent G, Wahlestedt C, Kapranov P. (2015) The landscape of long noncoding RNA classification. Trends Genet 31:239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahira AC, Kubrusly MS, Faria MF, Dazzani B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC, Reis EM. (2011) Long noncoding intronic RNAs are differentially expressed in primary and metastatic pancreatic cancer. Mol Cancer 10:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146:1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas VE, Zaphiropoulos PG. (2015) Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci 16:3251–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyhlidal CA, Bi C, Ye SQ, Leeder JS. (2016) Dynamics of cytosine methylation in the proximal promoters of CYP3A4 and CYP3A7 in pediatric and prenatal livers. Drug Metab Dispos 44:1020–1026. [DOI] [PubMed] [Google Scholar]
- Wada T, Gao J, Xie W. (2009) PXR and CAR in energy metabolism. Trends Endocrinol Metab 20:273–279. [DOI] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. (2002) PXR, CAR and drug metabolism. Nat Rev Drug Discov 1:259–266. [DOI] [PubMed] [Google Scholar]
- Yang L, Li P, Yang W, Ruan X, Kiesewetter K, Zhu J, Cao H. (2016) Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab 24:627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. (2014) Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yang Z, Trottier J, Barbier O, Wang L. (2017) Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology 65:604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wu J, Liangpunsakul S, Wang L. (2017) Long non-coding RNA in liver metabolism and disease: current status. Liver Res 1:163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]