Abstract
Background
Brain- and lesion-volumes derived from magnetic resonance images (MRI) serve as important imaging markers of disease progression in neurodegenerative diseases and aging. While manual segmentation of these volumes is both tedious and impractical in large cohorts of subjects, automated segmentation methods often fail in accurate segmentation of brains with severe atrophy or high lesion loads. The purpose of this study was to develop an atlas-free brain Classification using DErivative-based Features (C-DEF), which utilizes all scans that may be acquired during the course of a routine MRI study at any center.
Methods
Proton-density, T2-weighted, T1-weighted, brain-free water, 3D FLAIR, 3D T2-weighted, and 3D T2*-weighted images, collected routinely on patients with neuroinflammatory diseases at the NIH, were used to optimize the C-DEF algorithm on healthy volunteers and HIV + subjects (cohort 1). First, manually marked lesions and eroded FreeSurfer brain segmentation masks (compiled into gray and white matter, globus pallidus, CSF labels) were used in training. Next, the optimized C-DEF was applied on a separate cohort of HIV + subjects (cohort two), and the results were compared with that of FreeSurfer and Lesion-TOADS. Finally, C-DEF segmentation was evaluated on subjects clinically diagnosed with various other neurological diseases (cohort three).
Results
C-DEF algorithm was optimized using leave-one-out cross validation on five healthy subjects (age 36 ± 11 years), and five subjects infected with HIV (age 57 ± 2.6 years) in cohort one. The optimized C-DEF algorithm outperformed FreeSurfer and Lesion-TOADS segmentation in 49 other subjects infected with HIV (cohort two, age 54 ± 6 years) in qualitative and quantitative comparisons. Although trained only on HIV brains, sensitivity to detect lesions using C-DEF increased by 45% in HTLV-I-associated myelopathy/tropical spastic paraparesis (n = 5; age 58 ± 7 years), 33% in multiple sclerosis (n = 5; 42 ± 9 years old), and 4% in subjects with polymorphism of the cytotoxic T-lymphocyte-associated protein 4 gene (n = 5; age 24 ± 12 years) compared to Lesion-TOADS.
Conclusion
C-DEF outperformed other segmentation algorithms in the various neurological diseases explored herein, especially in lesion segmentation. While the results reported are from routine images acquired at the NIH, the algorithm can be easily trained and optimized for any set of contrasts and protocols for wider application. We are currently exploring various technical aspects of optimal implementation of CDEF in a clinical setting and evaluating a larger cohort of patients with other neurological diseases. Improving the accuracy of brain segmentation methodology will help better understand the relationship of imaging abnormalities to clinical and neuropsychological markers in disease.
Keyword: Medical imaging
1. Introduction
Quantitative analysis of whole-brain volumes (or those of different brain substructures), from magnetic resonance (MR) images, can provide important markers of disease progression in various neurological diseases, such as Alzheimer's disease and multiple sclerosis [1, 2, 3, 4, 5]. In multiple sclerosis, for example, total, new, and gadolinium-enhancing lesion volumes and counts are widely used to assess disease activity, progression, and efficacy of therapy [4, 5, 6, 7]. While whole-brain atrophy can be measured with minimal processing of tailored imaging sequences [8, 9], classifying substructures and lesions in the brain requires intense computing and prior knowledge of the underlying structures. In these methods, classification is often based on the probability that a tissue class exists at a particular voxel location after registration to an atlas.
Common approaches to performing volumetric segmentation of MR images, such as Statistical Parametric Mapping or SPM [10], SIENA(X) from FMRIB Software Library or FSL [11], 3dseg from Analysis of Functional NeuroImages or AFNI [12], and FreeSurfer [13], are often based on a T1-weighted structural image of the brain. While these types of atlas-based methods generally produce good results in relatively normal-appearing brains, they are usually not robust to segmenting brain images with high lesion loads and/or large atrophy from neurological disease [14]. Algorithms specifically developed for segmenting lesions, on the other hand, typically use T2-weighted or fluid-attenuated inversion recovery (FLAIR), in which white matter lesions generally appear hyperintense, often in addition to T1-weighted images. Early semi-automatic methods have delineated these hyperintense lesions using techniques such as seed-growing or connectivity [15, 16, 17]. However, such methods do not reliably detect lesions in the presence of significant radiofrequency bias fields; they are also inherently tedious to implement in large populations when manual inputs or corrections are required. More recently, fully automated techniques that use bias correction, as well as statistical or topological atlases of lesion location, have been developed to dramatically reduce processing time [18, 19]. However, lesion classification errors in atlas-based methods is compounded not only by the fact that size and distribution of lesions in the brain vary from one individual to next, but also because the likelihood of a lesion being present in any region varies between neurological diseases. This leads to misclassification of lesions and gray matter, which has necessitated disease-specific atlases for reliable lesion segmentation [20].
To overcome these issues, alternative multi-contrast methods have been described. These methods use multiple MRI contrasts, e.g., FLAIR and T1-weighted images, and machine learning algorithms, such as fuzzy c-means, random forest, and support vector machine classifiers, to improve image segmentation. Multiple contrasts are routinely acquired in clinical MRI sessions and are heavily relied upon by radiologists in their clinical assessment. However, the inclusion of any individual contrast, and the protocol used to create it, may be a matter of user preference. There may also be inconsistencies between imaging sessions, which may be tailored for each neurological disease. Nevertheless, brain segmentation algorithms should in principle benefit from the use of multiple imaging contrasts. In addition, not using a priori information about lesion distribution (atlas-free method), should improve its generalizability for application to various neurological disorders.
The purpose of this study was to train and implement an atlas-free brain segmentation algorithm that utilizes multiple MRI contrasts, and that could easily be implemented in a variety of neurological diseases. This atlas-free Brain Classification using Derivative-based Features (C-DEF) algorithm was optimized, and then used to segment brain images of healthy volunteers and individuals clinically diagnosed with a variety of neuroimmunological diseases. The segmentation output from C-DEF was compared qualitatively and quantitatively with popular segmentation algorithms, including Lesion-TOADS (TOpology-preserving Anatomical Segmentation) [18] and FreeSurfer. Lesion-TOADS is a publicly available automatic brain segmentation algorithm used commonly in subjects with lesions, that incorporates information from topological and statistical atlases to an intensity-based segmentation technique using iterative fuzzy classification. FreeSurfer is a very widely used automated brain segmentation tool that registers target images to a probabilistic atlas, and generates tissue class labels using voxel intensity or tissue morphology. T1-hypointensities in the white matter are classified as lesions. We chose subjects infected with HIV for initial training and optimization of C-DEF since they exhibit varying degrees of lesion burden and atrophy. Furthermore, current tissue-segmentation algorithms do not readily delineate lesions in these subjects. The C-DEF algorithm was then evaluated in subjects clinically diagnosed with other neurological diseases, routinely seen at our research clinic, including multiple sclerosis, polymorphism of the cytotoxic T-lymphocyte-associated protein 4 gene (CTLA4), and HTLV-I-associated myelopathy/tropical spastic paraparesis (HAM/TSP).
2. Materials and method
2.1. Recruitment and imaging
The study methods were reviewed and approved by the Institutional Review Boards at the National Institutes of Health (CNS, NIAID IRB at NIH) per regulations. Data were drawn from three cohorts of subjects, who provided informed consent to participate in the study. The first cohort, consisting of healthy volunteers and individuals infected with HIV, was used to train, evaluate, and optimize the C-DEF algorithm. The second cohort, consisting only of subjects infected with HIV, was used to test the final, optimized algorithm in comparison with commonly used segmentation algorithms (FreeSurfer and Lesion-TOADS). Finally, the third cohort, consisting of subjects with MS, HAM/TSP, and CTLA4, was used to determine the robustness and sensitivity of C-DEF classifications to those of Lesion-TOADS and FreeSurfer.
All MR images were collected on a 3T Philips MRI scanner (Philips Medical Systems, Netherlands), with an eight-channel receive head coil. T1-weighted (T1-MPRAGE), T2-weighted, proton-density (PD), brain-free water imaging (BFWI), fluid attenuated inversion recovery (3D FLAIR), post-contrast non-selective fluid attenuated inversion recovery (3D ns-FLAIR), and 3D segmented-EPI T2*-weighted [21] volumes are routinely collected at our center on subjects with neuroinflammatory diseases, and all these contrasts were used in this study. Table 1 contains the MR parameters for these sequences.
Table 1.
Sequence parameters for the MR contrasts used in the C-DEF algorithm.
| 3D-T1 MPRAGE | 3D FLAIR | 2D FSE PD_T2 | 3D BFWI | 3D nsFLAIR | 3D EPI T2* | |
|---|---|---|---|---|---|---|
| Slice thickness (mm) | 1 | 1 | 3 | 0.65 | 1 | 0.55 |
| Inversion time (ms) | 1600 | N/A | 1600 | |||
| Echo time (ms) | 3.2 | 318 | 15.4, 100 | 750 | 318.4 | 29.5 |
| Repetition time (ms) | 7 | 4800 | 3417 | 4500 | 4800 | 54.1 |
| Flip angle (deg) | 9 | 90 | 90 | 90 | 90 | 10 |
| Number of repetitions | 1 | 1 | 2 | 1 | 1 | 1 |
| Total acquisition time (min:sec) | 5:16 | 7:00 | 3:31 | 6:40 | 7:00 | 4:14 |
2.2. Image preprocessing and feature creation
All MR images acquired in a session were registered to the T1-weighted images of the same session using AFNI tools [12]. The bias-field correction was performed using a local statistic function that applies a percentile filter to a sliding window [22]. The filter was chosen to minimize bias field and blurring in the resulting image. This corrected image was then normalized to the intensity at the 90th percentile to generate a scaled image. Next, spatial and edge feature images were generated using the Scipy module in Python [23]. Features were extracted using a Gaussian and Gaussian Gradient filter of four different kernel sizes of 1, 2, 4 and 8 voxels.
2.3. Labeled mask creation
Masks for white matter (WM), gray matter (GM), and globus pallidus (GP) were compiled from FreeSurfer [13] volumetric segmentation of the supratentorial brain, with outputs eroded by one pixel. The GM masks contained both cortical and deep gray matter masks, excluding the GP because its MR properties are substantially different from other gray matter areas. Lesion masks were manually drawn (by PMD, MKS) on the 3D FLAIR images, if they were present, and subsequently verified by a neurologist with specialized training in neuroimaging (MKS). CSF masks were created from the highest intensities (top 3%) in the BFWI, a heavily T2 weighted image acquired at high isotropic spatial resolution of 0.65 mm [9]. A final class, called “other” (OTH), was defined as voxels that did not belong to any of the above tissue classes – e.g., background voxels. Five (WM, GM, CSF, GP, and OTH) or six (WM, GM, CSF, GP, Lesion, and OTH) tissue masks were combined into a “gold standard” labeled mask for training and optimizing the C-DEF algorithm. The cerebellum and brain stem were not included in the labeled masks.
2.4. Training and model selection
The training set consisted of the scaled images of subjects in cohort one, and Gaussian and Gaussian gradient features generated from the scaled images with various kernel sizes (X data), as well as the gold standard tissue masks (Y data). A logistic regression model was implemented using the Python scikit-learn module, with an L2 regularization. For both the healthy volunteers and HIV cases, the regression model was trained on the X and Y image data from four subjects to generate a classifier, which was then applied to X data of the fifth, for leave-one out (LOO) cross-validation. The logistic regression model built from the training data was used to generate the probability mask of various tissue types [24]. In the final step, a segmented image of the brain was created by assigning to each voxel the class label with the highest probability at that location (maximum membership).
Multiple models of C-DEF were trained and evaluated for optimal performance using data from cohort one. C-DEF was trained on healthy volunteers without a lesion class (5-label classifier), and on subjects infected with HIV with a lesion class (6-label classifier). For each classifier, four different feature vectors were tested to evaluate whether features generated from large kernels are necessary for the accuracy of the model. The Raw Model had no additional features and consisted only of scaled images. The 2-Kernel Model included features of kernel sizes 1 and 2, and the 3-Kernel Model included features of kernel sizes 1, 2 and 4 in addition to scaled images. The Full Model included all features (kernel sizes 1, 2, 4 and 8) as well as scaled images. This resulted in feature vectors with a total of 7, 35, 49, and 63 features for the Raw, 2-Kernel, 3-Kernel, and Full Model, respectively.
2.5. Testing
The optimal model from this analysis was then selected to segment the brain images of HIV subjects not included in the training (cohort two), as well as those with other neurological diseases that are routinely seen in our clinic (cohort three). The segmentation outputs from C-DEF were evaluated against those from FreeSurfer and Lesion-TOADS.
2.6. Outcome measures and statistics
The performance of the different models was evaluated qualitatively and quantitatively. Qualitative evaluation was done through a simple visual inspection by experienced radiologists. Quantitative evaluation of the two training sets in cohort one was performed by combining classification results from the five patients in each set to plot the Receiver Operating Characteristic (ROC) curves and calculate the Area Under the Curve (AUC). This was done for every tissue class and for all Models. Repeated-measure ANOVA was used to compare the AUC values and determine the effect of including more features on segmentation quality. P < 0.05 was considered to be statistically significant.
A regression analysis was done to compare tissue class volumes generated from C-DEF, FreeSurfer, and Lesion-TOADS segmentations. The Bland-Altman method was used to calculate the bias and 95% confidence interval between volumes generated from these techniques. A sensitivity analysis was used to compare C-DEF lesion segmentation performance with that of FreeSurfer, and Lesion-TOADS in other neurological diseases, where:
and TP and FN are the true positive and false negative values, respectively.
3. Results
3.1. Subject cohorts
The Institutional Review Board approved the study protocols and all participants provided written informed consent. Healthy volunteers (n = 5, 4 males, average age 36 ± 11 years), as well as subjects infected with HIV (n = 5, 4 males, average age 57 ± 3 years) were included in cohort one. Subjects infected with HIV (n = 49, 30 males, average age 54 ± 6 years), not including the five in the training set, comprised cohort two. To achieve a representative sampling of diseases studied in our neuroimmunology research clinic at NIH, individuals with MS (n = 5, 2 females, average age 42 ± 9 years old), CTLA4 haploinsufficiency (n = 5, 1 female, average age 24 ± 12), and HAM/TSP (n = 5, 5 females, average age 58 ± 7) were studied in cohort three.
3.2. Generation of features and training mask
Representative scaled MR images from a subject infected with HIV are shown in Fig. 1A (56-year-old male). Gaussian and Gaussian gradient features generated from filters of various kernel sizes, applied on the T1-MPRAGE in 1A, are shown in Figs. 1B and 1C, respectively. Combined masks of GM, WM, and GP (from eroded FreeSurfer segmentation), and CSF mask (derived from 97th intensity percentile of the BFWI image), served as gold-standard label masks for the 5-label classifier (Fig. 1D, left). Similar GM, WM, GP, CSF masks, along with manually segmented lesion masks, served as gold-standard training masks for the 6-label classifier (Fig. 1D, right).
Fig. 1.
Training Data for C-DEF: (A) Coregistered and intensity-normalized representative proton density, T2-weighted, T1-MPRAGE, brain free water, 3D FLAIR, 3D-nsFLAIR, and 3D EPI T2* images (left to right) from a subject infected with HIV (male, 56 years old). (B) Gaussian (Gaussian filter) and (C) gradient magnitude (Gaussian Gradient filter) feature images generated from the T1-MPRAGE with kernel sizes 1, 2, 4, and 8 (left to right). (D) The gold standard label mask obtained from healthy volunteer (left) and HIV subject (right), overlaid on the corresponding T1-MPRAGE.
3.3. Model selection
Visual assessment comparing the segmentation outputs from Raw, 2-Kernel, 3-Kernel and Full Models with the uneroded form of the labeled training masks (compiled labels) revealed that increasing the number of features provides better segmentation, especially at the GM-WM and GM-CSF boundaries and in the deep GM structures. A representative FLAIR image and segmentation results from the 5-label classifier (Fig. 2A) and the 6-label classifier (Fig. 2B) on a healthy volunteer show that a focal, nonspecific hyperintensity in the deep WM (red arrow) was classified as GM by the 5-label classifier, but was correctly identified to have the MRI signature of a lesion by the 6-label classifier. However, hyperintensities that typically abut the lateral ventricles, labelled as normal WM in the lesion mask by the neurologist, were consistently classified as lesions in the 6-label classifier. For the Full Model, 0.7% of GM and 0.68% of WM identified in the 5-label classifier was identified as lesion in the 6-label classifier. Segmentation results from 6-label classifier on HIV subjects shows that the 3-Kernel and Full Models were able to accurately segment lesions and produced whole brain segmentations with fewer misclassified areas (Fig. 2C).
Fig. 2.
Qualitative comparison of C-DEF models: Representative 3D FLAIR axial slices, and results from the Raw, 2 Kernel, 3 Kernel, Full model, and compiled FreeSurfer segmentation (left to right). Color-coded segmentation results are overlaid on FLAIR image. (A) Segmentation results from a healthy volunteer (male, 40 years old) for the (A) 5-label and (B) 6-label classifier. (C) Results from a subject infected with HIV (male, 60 years old) for 6-label classifier. The corresponding uneroded forms of GM, WM, CSF, GP masks used in training, combined with manual lesion segmentations (compiled labels), are shown in the far right column.
Quantitative comparison of the four models also reflected the qualitative observation that increasing the number of kernels improved segmentation. ROC curves for the Raw and Full Models from the 5-label classifier (Fig. 3A) and 6-label classifier (Fig. 3B) reveal a significant improvement in AUC with increasing kernel size (GP: p = 0.037, WM: p = 0.037, ANOVA repeated measures). However, quantitative comparison of the 6-label classifier revealed that improvement was shy of statistical significance (GP, p = 0.06; WM, p = 0.06, ANOVA repeated measures). While, the Full Model for the 5-label classifier yielded AUC values >0.98, and for the 6-label classifier >0.99 in all tissue classes, analysis of other structures besides GP and WM did not show statistically significant improvements in AUC from the Raw Model.
Fig. 3.
Quantitative evaluation of C-DEF models: ROC curves of each tissue type for Raw and Full models from (A) training without lesions (5-label classifier) on healthy volunteers (n = 5, age 36 ± 11 years), and (B) training with lesions (6-label classifier) on subjects infected with HIV (n = 5, age 57 ± 2.6 years). Inset: Magnified image of the top left corner on the ROC curves. (C) Tables showing the true positive rates comparing the predicted C-DEF labels with FreeSurfer labels and manual lesion segmentations for the 5-label (top) and 6-label classifiers (bottom).
For the 5-label classifier training with the Full Model, 89% of the voxels identified as GM agreed with those of the compiled labels, while only 30% of the CSF voxels were in agreement between the two methods (Fig. 3C). Most of the CSF from the compiled labels was classified as GM by C-DEF. The agreement of CSF labels to compiled labels increased to 63% for the 6-label classifier. Only 51% of the voxels classified as lesions in C-DEF were deemed to be so by compiled labels. Interestingly, about 19% of all tissue (other than GM) classified by C-DEF was classified as GM by FreeSurfer (Fig. 3C). Taken together, and because of the importance of classifying lesions in neurological disorders, the 6-label Full Model classifier was chosen to the optimal model of C-DEF.
3.4. Evaluation in cohort two
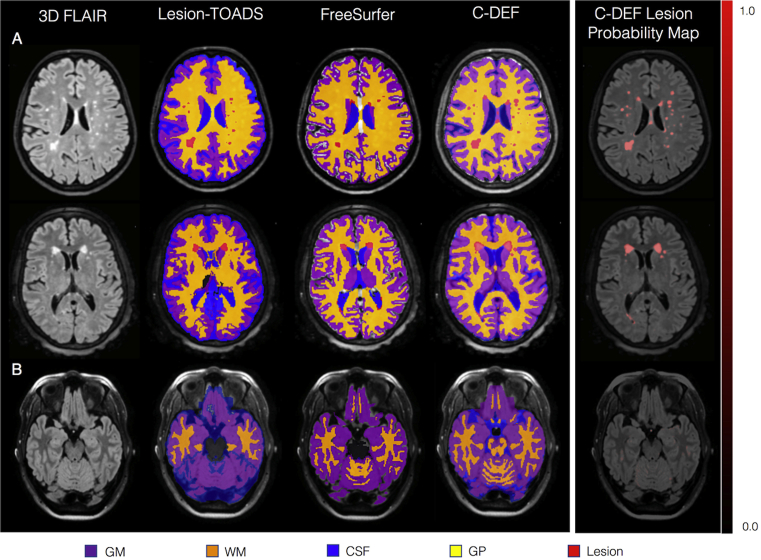
In order to further understand the nature of the mismatch, the performance of C-DEF was compared to FreeSurfer and Lesion-TOADS segmentation in a larger group of subjects infected with HIV (cohort two). A comparison of brain segmentation by Lesion-TOADS, FreeSurfer, and optimized C-DEF revealed regions of misclassification in Lesion-TOADS and FreeSurfer to be the source of the mismatch (Fig. 4A and B). C-DEF was especially adept at picking up lesions, and seemed to perform better than either FreeSurfer or Lesion-TOADS. Furthermore, C-DEF performed well irrespective of the lesion type (periventricular, juxtacortical, or deep white matter lesion), and anatomic location (supratentorial, cerebellum and brainstem). It should be noted that some of the voxels in the temporal lobe and cerebellar regions had relatively high probability of being a lesion, but were not classified as such by the maximum membership criteria.
Fig. 4.
Qualitative comparison of various segmentation algorithms: (A) Maximum membership classification from C-DEF and lesion probability maps of different types of lesions in three different subjects (top: female, 60 years old; middle: female, 56 years old; bottom: female, 44 years old). Lesion types include periventricular, juxtacortical, and deep white matter. (B) C-DEF of the cerebellum and brainstem, without specific training in these structures.
WM (Fig. 5A) and GM (Fig. 5B) volumes derived from C-DEF and FreeSurfer, were significantly correlated (r = 0.87 and 0.92, respectively). However, GM volume from C-DEF was higher than that from FreeSurfer (Bias, -13% 95% CI, -22% to -4%). There was no significant bias noted between WM volumes from C-DEF and FreeSurfer (95% CI, -16% to 9%). The lesion volume from C-DEF was more reliable than the one from FreeSurfer (Fig. 5C) or Lesion-TOADS (Fig. 5D), although there was moderate correlation between the lesion volumes calculated by C-DEF and Free-Surfer (r = 0.51).
Fig. 5.
Quantitative evaluation of various segmentation algorithms: Scatter plots comparing volumes from FreeSurfer and C-DEF of (A) white matter (Bias, -4; 95% CI, -16–9), (B) gray matter (Bias, -13; 95% CI, -22 – -4). Plots comparing lesion volumes obtained from (C) FreeSurfer and C-DEF (Bias, -99; 95% CI, -175 – -22) and (D) Lesion-TOADS and C-DEF (Bias, -33; 95% CI, -182–115). The dotted line represents the X = Y line, or perfect agreement between the two methods (cohort two, n = 49 HIV infected individuals).
The source of this mismatch is better identified in a direct comparison of lesion masks generated by Lesion-TOADS, FreeSurfer, and C-DEF (Fig. 6). Fig. 6A shows that Lesion-TOADS was able to segment lesions with few false positives, but missed the smaller, more punctate deep white matter lesions. Fig. 6B is a case in which Lesion-TOADS incorrectly classified certain cortical gray matter regions as lesions.
Fig. 6.
Qualitative comparison of lesion segmentation: FLAIR images and resulting Lesion-TOADS, FreeSurfer, and C-DEF lesion segmentations from two subjects (top: female, 61 years old; bottom: male, 51 years old) in cohort two (left to right). (A) A case showing relatively good agreement between Lesion-TOADS and C-DEF, (B) a case showing misclassification of lesions by Lesion-TOADS. FreeSurfer was unable to segment most of the lesions. Red arrow points to lesions identified only in C-DEF, and yellow arrows point to false positives identified in other algorithms.
3.5. Segmentation in other neuroimmunological diseases
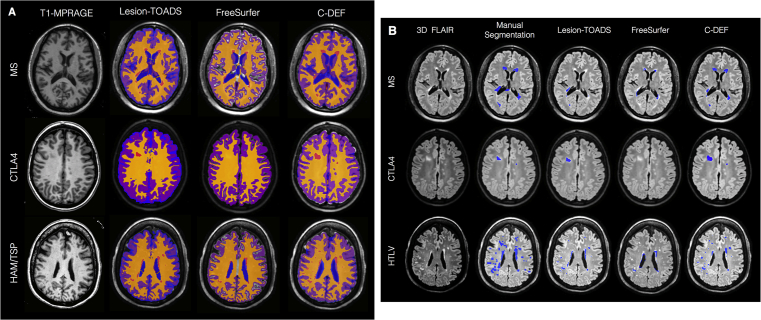
Although it was not trained in other neuroimmunological diseases, the optimized C-DEF algorithm was better at segmenting the brain (Fig. 7A) and lesions (Fig. 7B) than Lesion-TOADS or FreeSurfer. Visual inspection revealed regions of mismatch mainly in lesion and GM segmentation for FreeSurfer, and in CSF-GM boundaries in Lesion-TOADS. Lesion-TOADS and C-DEF performed well in MS brain segmentation. In MS, the sensitivity by volume for lesion segmentation was 49% for C-DEF, and 37% and 21% for Lesion-TOADS and FreeSurfer, respectively. For HAM/TSP, sensitivity was 44% for C-DEF, 30% for Lesion-TOADS, and 16% for FreeSurfer. In CTLA4, the sensitivity was 47% for C-DEF, 45% for Lesion-TOADS and 9% for FreeSurfer.
Fig. 7.
A. Whole brain segmentation applied to other diseases: Qualitative comparison of whole brain segmentation of Lesion-TOADS, FreeSurfer, and C-DEF, on three neurological diseases (top: multiple sclerosis, female, 37 years old; middle: CTLA4, female 14 year old; HTLV, female, 56 years old). Masks have been overlaid on their corresponding T1-MPRAGE images. B. Lesion segmentation applied to other diseases: Qualitative comparison of lesion segmentation methods (manual segmentation, Lesion-TOADS, FreeSurfer, and C-DEF) on three neurological diseases (top: multiple sclerosis, female, 37 years old; middle: CTLA4, female 14 year old; HTLV, female, 56 years old). Lesion masks have been overlaid on their corresponding 3D-FLAIR images. Red arrow points to lesions identified only in C-DEF.
4. Discussion
An atlas-free automatic brain segmentation algorithm (C-DEF) that uses logistic regression on multiple MRI contrasts and derived features was developed and implemented herein. C-DEF outperformed widely used brain segmentation techniques in a variety of neuroimmunological diseases for lesion detection, while still achieving highly accurate brain segmentations. A local image statistics-based approach enabled removal of bias fields associated with high field imaging [22]. Use of 3D-kernels of multiple, isotropic sizes allowed generation of features at various local scales [22, 25]. Overall, C-DEF classification performed consistently in supratentorial, cerebellar, and brainstem regions.
Popular MRI brain segmentation algorithms utilize atlas-based and region growing techniques. Algorithms such as FreeSurfer, SPM, and Lesion-TOADS rely on registration to a standard atlas to determine the likelihood that a voxel at particular location belongs to a particular tissue type. While this approach has been successful in healthy populations, errors tend to arise when the subject's anatomy deviates significantly from the atlas, as is evident in subjects with neurological diseases. Large morphological changes seen in these subjects, such as significant atrophy or severe lesion load, may lead to registration errors and thereby errors in estimation of tissue probability and misclassification. For detecting lesions, depending on spatially derived prior probabilities leads to further classification errors because atlases do not account for the varied size and spatial distribution of lesions in subjects with neuroimmunological diseases. For example, subjects diagnosed with MS often have confluent periventricular lesions, while subjects with chronic, well-controlled HIV infection and HAM/TSP are more likely to have smaller deep WM lesions [26, 27]. Therefore, atlas-based algorithms for lesion segmentation are often disease-specific and may be prone to miss lesions in locations where they are not expected. These segmentation algorithms also use single contrasts, which makes them insensitive to lesions that are not easily visible on that contrast, and studies have shown that the accuracy of such single-contrast-based algorithms are highly influenced by the noise level in an image [28, 29].
An alternative to atlas-based methods are region-growing algorithms such as the one implemented in Lesion Segmentation Tool (LST) [30]. While this technique is robust to morphological variations [31], it is highly sensitive to noise and radiofrequency bias, which can cause disconnections or holes within segmented regions. Adaptive thresholding algorithms [32, 33] performed better by eliminating many of these artifacts, but they suffer from inconsistency and often require significant manual input for lesion segmentation.
C-DEF overcomes these issues by using multiple image contrasts, correcting bias fields, and eliminating the need for an atlas through incorporating image features, thereby resulting in a more robust disease-independent classification. C-DEF minimizes atlas-based registration errors, because registration is performed across the MR contrasts obtained within a single session. Several recent studies have used multi-contrast techniques with machine learning for lesion segmentation [34, 35, 36]; C-DEF extends this idea for segmenting the whole brain by adding derived feature sets to the algorithm. Instead of relying on spatially derived prior probabilities, C-DEF uses local neighborhood information derived from Gaussian and Gaussian gradient filters with multiple kernel sizes. These feature images reinforced the confidence within the model. For the purposes of this classification, we used 3-dimensional kernels and magnitude of the gradient because we hypothesized that directional information would not assist with basic classification, given the variable orientations of both the highly convoluted cortical sheet and the unpredictable location and shape of lesions. In addition, these features are relatively robust to changes in the orientation of the subject's head in the images and to the specific slice orientation with respect to a given structure or lesion.
It should be noted that the classification algorithm was trained on thousands of voxels per patient (an average of about 200,000 for CSF, 500,000 for GM, and 400,000 for WM and 3,000 for GP and lesions per patient) in cohort 1. Given the 63 features used in C-DEF, and the one-in-ten rule of thumb for number of predictors that can be derived from logistic regression, there is no reason to expect overfitting for any of the 6 classes predicted by C-DEF. Therefore it was possible to get robust and reliable results from training on as few as 5 subjects. Indeed, experiments performed to determine optimal size of training set (not reported here) revealed dice similarity coefficient of WM and GM generally increasing when the number of training subjects were increased from 1 to 3, and plateauing between 3-5 subjects (data not shown).
A strength of C-DEF algorithm is its adaptability. The algorithm currently uses seven different MRI contrasts, which are routinely acquired as a part of standard clinical scans for neuroimmunological diseases at the NIH. However, the segmentation algorithm is not heavily dependent on the types of contrasts acquired nor on the protocol used to achieve them. In principle, high-resolution T1-weighted and FLAIR scans (the latter to identify lesions) can be trained to achieve effective brain classification. Using multiple imaging contrasts only increases the confidence of the classification. For example, adding the 3D nsFLAIR images helped the algorithm further differentiate between the GM and lesion class and reduce the partial volume errors (data not shown). Similarly, 3D BFWI (a heavily T2-weighted image) improved the delineation of CSF by the algorithm (data not shown). Additionally, using T2* and nsFLAIR images required the inclusion of a separate GP class, as iron deposition in this brain region resulted in very distinct tissue signatures. While these seven contrasts were readily available to us, C-DEF can be easily adapted and retrained in protocols where fewer or different images are acquired. Along these lines, we have successfully trained and are testing C-DEF on a set of standard clinical images acquired at a collaborator's site to segment brains in patients with MS and small vessel disease (data not shown).
Scanners from different manufacturers and field strengths, and different imaging parameters (some of which may be inaccessible in a non-research setting) can produce subtle to large variations in image contrast, which can directly affect segmentation. It would therefore be important that the training be done on images similar to the ones used in the application. The training is simple to perform and takes just a few hours including registration step, and need only be performed once. However, a limitation to training is that gold-standard masks for the various tissue types would need to be provided. In the present case, the use of images derived from FreeSurfer segmentation on the T1-MPRAGE as the gold-standard for training WM, GM, and GP made this task easier. To reduce training on erroneous voxels, the FreeSurfer segmentation was eroded by one pixel to create the masks, thereby eliminating voxels with partial volume. CSF masks were derived from images with very heavy T2-weighting. Only lesion masks were required to be manually drawn, making the task much simpler. Most of the lesions in HIV-infected individuals were small focal lesions, which are easy to delineate. However, one can envision using outputs of other lesion-classifying algorithm as input for training after elimination or editing of inaccurate voxels in the lesion mask. All masks should be carefully inspected for segmentation errors before training is performed. As an immediate next step, we are exploring imaging parameters and contrasts, as well as optimizing feature and training sets for C-DEF. Such optimized imaging sequences and parameters can be provided along with the C-DEF as recommended imaging protocol for brain segmentation.
Supervised classification algorithms, specifically logistic regression models, have been used in the past to segment brain tissues structures from MRI and CT images [37, 38]. Briefly, logistic regression is a linear classifier that calculates the weighted sum of the input features in order to model the probability that an image voxel belongs to a particular labeled class. While studies have looked at the application of logistic regression in classifying brain tumors [39], annotating traumatic brain injury [38], and segmenting multiples sclerosis lesions [35], this approach has not been previously utilized for whole brain image segmentation of subjects with HIV. Logistic regression has an added advantage over other classifiers, such as support vector machine (SVM) or artificial neural networks (ANN), because it produces coefficients that can be used to interpret and optimize model parameters [40]. SVM and ANN lack transparency in that their results cannot be represented as a parametric function of the input features. These classifiers are also computationally more expensive. Moreover, it has been shown that simpler methods, like the logistic regression model, yield performance that is equivalent to that of other sophisticated methods [41]. For these reasons, we selected logistic regression to be the classifier for the C-DEF algorithm.
We used the Gaussian and Gaussian gradient features, as well as the logistic regression model, to build two different training sets: one without lesions (5-label classifier), and one with lesions (6-label classifier). In general, both sets yielded high performance in their respective cohorts, as seen by the fewer voxels classified as “other” within the brain. This can be attributed to the fact that the predictive models built from the training sets were applied to a test set with similar characteristics; the 5-label classifier was tested on image data from other healthy volunteers. An exception to this was in segmenting images from a healthy volunteer with non-specific deep WM lesions. For this case, we applied the HIV training model and found that it was able to delineate the punctate deep white-matter, with some misclassification of areas around the ventricles. While the tissue signature of these regions around the ventricles matches that of a lesion, they are seldom reported as lesions in a radiological report, as they are commonly seen in healthy volunteers as well [42]. It is possible that the region does indeed undergo very subtle neuroinflammatory changes with aging, but without any noticeable clinical symptoms. Nevertheless, classification of these regions as lesion or normal tissue will depend on the investigator. Since either training set works in healthy volunteers with lesions, it would be up to the user to decide the appropriate model for their study, depending on the question being asked in the specific study.
The Raw, 2-Kernel, 3-Kernel and Full Models were used to test the influence of features generated from larger kernel sizes on segmentation performance. A quantitative evaluation of the models generally showed an increasing trend between number of features and AUC values in all classes, with significant differences between the Raw and Full Models for GP and WM classification. Regardless, a qualitative examination showed a clear improvement in segmentation performance with increasing number of features. Therefore, the Full Model for the C-DEF algorithm was treated as optimal and used for the implementation cohort two and in cases with other neuroinflammatory diseases. As a follow up on these encouraging results, we are in the process of evaluating ways to improve the performance of C-DEF using various other modified feature set such as Laplacian and rotationally invariant features.
Quantitative evaluation showed that C-DEF had a higher sensitivity for delineating lesions in MS and CTLA4 in comparison with widely used segmentation algorithms such as Lesion-TOADS and FreeSurfer, even though the initial training of the C-DEF model was done on subjects with HIV, and the morphology of those lesions were very different. Interestingly, training using lesions in HIV subjects, in which many lesions were small and focal, did not appear to adversely affect detection of large lesions such as those typically seen in other neuroinflammatory diseases.
5. Conclusion
In this study, we use an atlas-free machine learning classification approach utilizing multi-scale local image features generated from multiple MR contrasts to robustly classify both normal brain tissue as well as lesions found in subjects with neuroinflammatory diseases. Since the algorithm performs segmentation based on intensity signatures in image features and contrasts, it outperforms current atlas-based techniques that are susceptible to image noise or not sensitive to structural variations such as high lesion load or atrophy. Future work includes exploring various technical aspects of optimal implementation of CDEF in a clinical setting and application to other neurological diseases. Overall, C-DEF can be reliably used to extract accurate quantitative neurostructural measures, which could serve as biomarkers of disease progression.
Declarations
Author contribution statement
Kartiga Selvaganesan, Emily Whitehead: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Paba M. DeAlwis, Matthew K. Schindler: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Joan E. Ohayon, Irene C.M. Cortese, Bryan Smith, Steven Jacobson, Avindra Nath: Contributed reagents, materials, analysis tools or data.
Daniel S. Reich: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Souheil Inati, Sara Inati, Ziad S. Saad, Govind Nair: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Intramural Research Program at the National Institute of Neurological Disorders and Stroke, and the Office of AIDS Research.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We are grateful to the staff of the NIH Clinical Center Department of Radiology and Imaging Sciences for help with MRI acquisitions. We thank the staff of the National Institute of Neurological Disorders and Stroke Neuroimmunology Clinic for recruitment, care, and collection of clinical data from the study participants. We also thank John Ostuni and the NINDS information technology department for help with servers and software maintenance.
References
- 1.Cash D.M. Imaging endpoints for clinical trials in Alzheimer’s disease. Alzheimer's Res. Ther. 2014;6(9):87. doi: 10.1186/s13195-014-0087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waser M. Neuroimaging markers of global cognition in early Alzheimer's disease: a magnetic resonance imaging-electroencephalography study. Brain Behav. 2018 doi: 10.1002/brb3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols M.J. Atrophic brain signatures of mild forms of neurocognitive impairment in virally suppressed HIV infection. AIDS. 2019;33(1):55–66. doi: 10.1097/QAD.0000000000002042. [DOI] [PubMed] [Google Scholar]
- 4.Harrison D.M. Association of cortical lesion burden on 7-T magnetic resonance imaging with cognition and disability in multiple sclerosis. JAMA Neurol. 2015;72(9):1004–1012. doi: 10.1001/jamaneurol.2015.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hakansson I. Neurofilament levels, disease activity and brain volume during follow-up in multiple sclerosis. J. Neuroinflammation. 2018;15(1):209. doi: 10.1186/s12974-018-1249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Traboulsee A. Effect of interferon beta-1a subcutaneously three times weekly on clinical and radiological measures and no evidence of disease activity status in patients with relapsing-remitting multiple sclerosis at year 1. BMC Neurol. 2018;18(1):143. doi: 10.1186/s12883-018-1145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chitnis T. Trial of fingolimod versus interferon beta-1a in pediatric multiple sclerosis. N. Engl. J. Med. 2018;379(11):1017–1027. doi: 10.1056/NEJMoa1800149. [DOI] [PubMed] [Google Scholar]
- 8.Rudick R.A. Use of the brain parenchymal fraction to measure whole brain atrophy in relapsing-remitting MS. Neurology. 1999;53(8):1698. doi: 10.1212/wnl.53.8.1698. http://n.neurology.org/content/53/8/1698.abstract [DOI] [PubMed] [Google Scholar]
- 9.Gao K.C. Sub-millimeter imaging of brain-free water for rapid volume Assessment in atrophic brains. Neuroimage. 2014;100:370–378. doi: 10.1016/j.neuroimage.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner J., Friston K.J. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Smith S.M. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage. 2002;17(1):479–489. doi: 10.1006/nimg.2002.1040. https://www.ncbi.nlm.nih.gov/pubmed/12482100 [DOI] [PubMed] [Google Scholar]
- 12.Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. https://www.ncbi.nlm.nih.gov/pubmed/8812068 [DOI] [PubMed] [Google Scholar]
- 13.Fischl B. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. https://www.ncbi.nlm.nih.gov/pubmed/11832223 [DOI] [PubMed] [Google Scholar]
- 14.Vrenken H. Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J. Neurol. 2013;260(10):2458–2471. doi: 10.1007/s00415-012-6762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippi M. Semi-automated thresholding technique for measuring lesion volumes in multiple sclerosis: effects of the change of the threshold on the computed lesion loads. Acta Neurol. Scand. 1996;93(1):30–34. doi: 10.1111/j.1600-0404.1996.tb00166.x. https://www.ncbi.nlm.nih.gov/pubmed/8825269 [DOI] [PubMed] [Google Scholar]
- 16.Udupa J.K. Multiple sclerosis lesion quantification using fuzzy-connectedness principles. IEEE Trans. Med. Imag. 1997;16(5):598–609. doi: 10.1109/42.640750. [DOI] [PubMed] [Google Scholar]
- 17.Wicks D.A. Volume measurement of multiple sclerosis lesions with magnetic resonance images. A preliminary study. Neuroradiology. 1992;34(6):475–479. doi: 10.1007/BF00598953. https://www.ncbi.nlm.nih.gov/pubmed/1436453 [DOI] [PubMed] [Google Scholar]
- 18.Shiee N. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage. 2010;49(2):1524–1535. doi: 10.1016/j.neuroimage.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy S. Subject specific sparse dictionary learning for atlas based brain MRI segmentation. IEEE J. Biomed. Health Inf. 2015;19(5):1598–1609. doi: 10.1109/JBHI.2015.2439242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Lorenzo D. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med. Image Anal. 2013;17(1):1–18. doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Sati P. Micro-compartment specific T2* relaxation in the brain. Neuroimage. 2013;77:268–278. doi: 10.1016/j.neuroimage.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vovk A. Segmentation priors from local image properties: without using bias field correction, location-based templates, or registration. Neuroimage. 2011;55(1):142–152. doi: 10.1016/j.neuroimage.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones E., Oliphant T., Peterson P. 2001. SciPy: Open Source Scientific Tools for Python.http://www.scipy.org/ [Google Scholar]
- 24.Sperandei S. Understanding logistic regression analysis. Biochem. Med. 2014;24(1):12–18. doi: 10.11613/BM.2014.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herlidou-Même S. MRI texture analysis on texture test objects, normal brain and intracranial tumors. Magn. Reson. Imag. 2003;21(9):989–993. doi: 10.1016/s0730-725x(03)00212-1. [DOI] [PubMed] [Google Scholar]
- 26.Godoy A.J. Characterization of cerebral white matter lesions of HTLV-I-associated myelopathy/tropical spastic paraparesis in comparison with multiple sclerosis and collagen-vasculitis: a semiquantitative MRI study. J. Neurol. Sci. 1995;133(1–2):102–111. doi: 10.1016/0022-510x(95)00161-t. https://www.ncbi.nlm.nih.gov/pubmed/8583211 [DOI] [PubMed] [Google Scholar]
- 27.Morgan D.J. Brain magnetic resonance imaging white matter lesions are frequent in HTLV-I carriers and do not discriminate from HAM/TSP. AIDS Res. Hum. Retrovir. 2007;23(12):1499–1504. doi: 10.1089/aid.2007.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kazemi K., Noorizadeh N. Quantitative comparison of SPM, FSL, and brainsuite for brain MR image segmentation. J. Biomed. Phys. Eng. 2014;4(1):13–26. https://www.ncbi.nlm.nih.gov/pubmed/25505764 [PMC free article] [PubMed] [Google Scholar]
- 29.Klauschen F. Evaluation of automated brain MR image segmentation and volumetry methods. Hum. Brain Mapp. 2009;30(4):1310–1327. doi: 10.1002/hbm.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt P. An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis. Neuroimage. 2012;59(4):3774–3783. doi: 10.1016/j.neuroimage.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 31.del Fresno M., Venere M., Clausse A. A combined region growing and deformable model method for extraction of closed surfaces in 3D CT and MRI scans. Comput. Med. Imag. Graph. 2009;33(5):369–376. doi: 10.1016/j.compmedimag.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg-Zimring D. Automated detection and characterization of multiple sclerosis lesions in brain MR images. Magn. Reson. Imag. 1998;16(3):311–318. doi: 10.1016/s0730-725x(97)00300-7. [DOI] [PubMed] [Google Scholar]
- 33.Khayati R. Fully automatic segmentation of multiple sclerosis lesions in brain MR FLAIR images using adaptive mixtures method and Markov random field model. Comput. Biol. Med. 2008;38(3):379–390. doi: 10.1016/j.compbiomed.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 34.Su T. White matter hyperintensities in relation to cognition in HIV-infected men with sustained suppressed viral load on combination antiretroviral therapy. AIDS. 2016;30(15):2329–2339. doi: 10.1097/QAD.0000000000001133. [DOI] [PubMed] [Google Scholar]
- 35.Sweeney E.M. OASIS is Automated Statistical Inference for Segmentation, with applications to multiple sclerosis lesion segmentation in MRI. Neuroimage. Clinic. 2013;2(Suppl. C):402–413. doi: 10.1016/j.nicl.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goceri E., Dura E., Gunay M. 2016 15th IEEE International Conference on Machine Learning and Applications (ICMLA) 2016. Review on machine learning based lesion segmentation methods from brain MR images. [Google Scholar]
- 37.Bullmore E.T. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Trans. Med. Imag. 1999;18(1):32–42. doi: 10.1109/42.750253. [DOI] [PubMed] [Google Scholar]
- 38.Dinh T.A. An automated pathological class level annotation system for volumetric brain images. AMIA Annu. Symp. Proc. 2012;2012:1201–1210. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3540549/ [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H.-C. 2017. Hybrid Clustering and Logistic Regression for Multi-Modal Brain Tumor Segmentation. [Google Scholar]
- 40.Dreiseitl S., Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J. Biomed. Inf. 2002;35(5):352–359. doi: 10.1016/s1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 41.Sweeney E.M. A comparison of supervised machine learning algorithms and feature vectors for MS lesion segmentation using multimodal structural MRI. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0095753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller S. Do brain T2/FLAIR white matter hyperintensities correspond to myelin loss in normal aging? A radiologic-neuropathologic correlation study. Acta Neuropathol. Commun. 2013;1 doi: 10.1186/2051-5960-1-14. 14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]