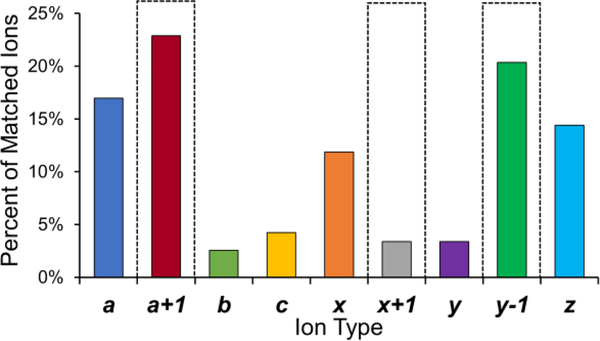Abstract
The ability to map combinatorial patterns of post-translational modifications (PTMs) of proteins remains challenging for traditional bottom-up mass spectrometry workflows. There are also hurdles associated with top-down approaches related to limited data analysis options for heavily modified proteoforms. These shortcomings have accelerated interest in middle-down MS methods that focus on analysis of large peptides generated by specific proteases in conjunction with validated bioinformatics strategies to allow quantification of isomeric histoforms. Mapping multiple PTMs simultaneously requires the ability to obtain high sequence coverage to allow confident localization of the modifications, and 193 nm ultraviolet photodissociation (UVPD) has been shown to cause extensive fragmentation for large peptides and proteins. Histones are an ideal system to test the ability of UVPD to characterize multiple modifications as the combinations of PTMs are the underpinning of the biological significance of histones and at the same time create an imposing challenge for characterization. The present study focuses on applying 193 nm UVPD for identification and localization of PTMs on histones by UVPD and comparison to a popular alternative, electron-transfer dissociation (ETD), via a high throughput middle-down LC-MS/MS strategy. Histone Coder and IsoScale, bioinformatics tools for verification of PTM assignments and quantification of histone peptides, were adapted for UVPD data and applied in the present study. In total, over 300 modified forms were identified and the distributions of PTMs were quantified between UVPD and ETD. Significant differences in patterns of PTMs were found for histones from HeLa cells prior to and after treatment with a deacetylase inhibitor. Additional fragment ion types generated by UVPD proved essential for extensive characterization of the most heavily modified forms (> 5 PTMs).
Graphical Abstract
Introduction
Post-translational modifications (PTMs) of proteins are implicated in an ever-expanding number of crucial biological processes, ranging from gene expression to tumorigenesis to cell death.1–4 Not only the type of modification but also the number, sites, and pattern, collectively known as combinatorial modifications, create an elaborate diversity of protein structure and function. Even minor variations in the distribution of PTMs can significantly influence the outcomes of myriad of cellular processes. Key examples of landmark proteins in which combinatorial modifications have been found to be essential for triggering and regulating downstream effects include p53,5–7 histones (chromatin structural units),8,9 and the C-terminal domain of RNA polymerase.10,11 Hundreds of types of PTMs are now recognized to contribute to the coding of protein function,12,13 and the interplay between different PTMs has created an enormous need for methodologies that can characterize PTMs and allow the cross-talk arising from combinatorial modifications to be deciphered. Significant effort has focused on improving analytical tools, particularly advanced mass spectrometry (MS), to characterize PTMs.14,15 Routine characterization of proteins which are modified at multiple residues remains a challenge,16–18 owing to the dynamic nature of PTMs, their low abundances and the large variation in stoichiometries.19 Other techniques are available to identify PTMs, such as modification-specific antibodies20 and gel electrophoresis (limited applicability).21 However, these methods cannot identify unknown modifications nor co-existing combinatorial PTMs.
Mass spectrometry (MS) offers special attributes in the realm of high-throughput PTM analysis.22–24 In recent years, advances in MS analysis of PTMs have facilitated identification of ever greater numbers and types of PTMs in a single experiment.25–27 Chief among the innovations enabling PTM analysis by MS are the development of selective enrichment methods25,28,29 and the introduction of new ion activation methods which reduce the loss of labile PTMs16,30–33 or enhance localization of modifications via greater sequence coverage.34 For example, collisional activation methods (CID, HCD) result in preferential cleavage of labile modifications such as phosphorylation and sulfation, thus impeding the ability to localize the sites of these modifications.30,35 Implementation of electron-based activation methods, such as electron captured dissociation (ECD)36 and electron transfer dissociation (ETD),37 and 193 nm ultraviolet photodissociation (UVPD)38 alleviates the loss of these labile modifications, enabling their identification and localization for high throughput applications. Another new option for MS/MS analysis is EThcD which is a hybrid method combining higher energy collisional dissociation (HCD) and ETD, to more efficiently generate two series of diagnostic fragment ions (b/y, c/z type ions).39 Moreover, activated ion electron dissociation (AI-ETD) is a hybrid method which uses concurrent infrared laser irradiation during ETD to counteract the charge state dependency inherent to electron activation and overcome “ET-no-D” events which limit sequence coverage.40 UVPD is an alternative to electron- or collisional-based or hybrid methods. UVPD stands out among these methods in that it uses photons for ion excitation and affords retention of labile modifications.30,38,41 UVPD typically generates a larger array of ion types than other activation methods, including a, a+1, b, c, x, x+1, y, y-1, and z type ions.
The development of alternative ion activation methods that allow retention and thus localization of PTMs has been particularly important for the characterization of some of the most heavily modified classes of protein, such as histones.19,42 Histones act as a structural scaffold for packaging of DNA as it wraps into chromatin. Importantly, the N-terminal and C-terminal regions of histones extend beyond the coils of DNA and act as key coding substrates for modification in a way that modulates DNA-protein interactions. Acetylated N-terminal tails promote loose histone-DNA association that guide interactions with transcription factors, whereas N-terminal methylation hinders DNA transcription.43 The extent of modifications along the N-terminal histone tails (e.g. first 50 residues) can be quite complex with many possible co-existing modification patterns with different biological ramifications. This complex relationship between modification states and biological outcomes has been termed the “histone code.”8
When mapping histone PTMs by tandem mass spectrometry (MS/MS), there are two notable hurdles that limit the success of traditional bottom-up MS/MS methods. Deciphering the contextual network of modifications among heterogeneous mixtures of histones is virtually insurmountable based on analysis of small proteolytic peptides. In addition, the prevalence of lysine and arginine residues of the N-terminal tails result in small tryptic peptides, often ones that are poorly separated by reversed phase chromatographic methods and for which combinatorial patterns are lost owing to the short lengths of the peptides.42 In an effort to directly characterize combinatorial PTMs, several groups have developed methods to characterize large peptides (middle-down) or intact proteins (top-down), enabling observation of all PTMs in a key region of a protein or in the entire protein sequence.42,44–46 Top-down workflows analyze even heavily modified proteins as intact species, thus offering an unsurpassed opportunity for mapping all PTMs. However, top-down analysis has greater technical challenges with respect to effective separation of intact proteins, ion activation methods that perform adequately for large ions, and bioinformatics needed to interpret very complicated spectra of proteins.44 The bioinformatics issue becomes particularly challenging when analyzing heavily modified proteins, such as histones, owing to the exponential increase in number of potential modification sites of intact proteins.47 Top-down analysis of histones has been extensively developed and successfully evaluated by several groups, including in a high throughput format for complex mixtures of histones.48,49 Despite the advantages of direct analysis of intact proteins, the need for excellent ion activation methods and the limited software for assignment, confident scoring and quantification of proteoforms with multiple PTMs have impeded widespread successful implementation of large scale LC-MS analyses of intact, heavily modified proteins.
Middle-down strategies offer an intermediate compromise between top-down and bottom-up methodologies, typically achieved via enzymatic or chemical procedures which limit the extent of protein digestion, thus producing peptides that are typically larger than those generated in conventional bottom-up workflows.19,45 MS/MS analysis of middle-down sized peptides has the added advantage of having fewer fragmentation channels in which to distribute ion current compared to analysis of intact proteins, thus affording better S/N in the resulting spectra.40 Furthermore, database searches of middle-down sized peptides is accommodated by the multitude of robust informatics platforms currently available for analysis of bottom-up sized peptides.18,42,46 A majority of PTMs found on histones exist on the first 50–60 amino acids, and this N-terminal stretch may be covered by a single long peptide generated by using GluC or AspN to cleave the histones. One of the most used approaches was developed by Hunt et al., who introduced the derivatization of lysine and N-terminal amines with propionic anhydride.50 Derivatization blocked the ɛ-amino groups of unmodified and monomethyl lysine residues, meaning that conventional trypsin proteolysis occurred only C-terminal to arginine residues instead of at both arginine and lysine residues, ultimately resulting in longer peptides. Moreover, N-terminal derivatization increased peptide hydrophobicity and thus retention on reversed phase media, affording better chromatographic separation.51 Recently a selective protease, neprosin, has also been successfully utilized for middle-down analysis of histones.52 Neprosin cleaves C-terminal to proline providing 3–4 kDa size peptides, thus offering a promising option for histone characterization.52
Despite the isolation of the most heavily modified region of the histone, the issue of separating hundreds of modified species has remained challenging. The Garcia lab pioneered the use of a mixed bed weak cation exchange hydrophilic interaction liquid chromatography (WCX-HILIC) resin to enable separation of N-terminal histone peptides based on number of modifications.17 Significant strides have been made to further develop this method for robust usage, including the introduction of software for filtering results obtained from canonical database searches and quantification.18,53 For example, false positive modification localization is a common problem encountered when analyzing MS/MS spectra of heavily modified peptides. The problem arises when one of two modifiable sites is modified, and upon MS/MS no backbone cleavage occurs between them to unambiguously assign the location of the modification. Often both possible sites will be reported despite one being a false positive.18 Recently developed software (e.g., Histone Coder and isoScale) has addressed this issue by ensuring that each reported modified site was confirmed by the presence of fragment ions that unambiguously localize modifications, thus allowing curation of false positives and quantification of more than 700 combinatorial histone marks.18
Here, we report the use of 193 nm UVPD for characterization of middle-down sized histone peptides. We used the canonical middle-down MS workflow, including GluC proteolysis followed by histone tail separation with WCX-HILIC coupled online to MS.17,42 We have previously shown that UVPD results in extensive fragmentation of proteins and peptides and does not cause loss of labile PTMs.38,41,54 In addition, the performance of UVPD is not strongly dependent on the size of the peptide nor charge state, thus making UVPD well-positioned for the analysis of histones.55 In this study, UVPD performance is benchmarked with attention to number of backbone cleavages, PTM site localization, and characterization of combinatorial PTMs. Having previously evaluated 193 nm UVPD for characterization of modifications on intact histones,56 the advantages discussed above regarding the middle-down approach merited further investigation in order to evaluate the applicability of UVPD for characterization of PTMs. In particular, adaptation of Histone Coder and IsoScale informatics tools for UVPD spectra allow unambiguous localization of modifications and quantification of isobaric peptides.
Experimental
HeLa cell preparation, histone fractionation and digestion.
HeLa S3 cells were treated for 24 hrs with or without 10 mM sodium butyrate (deacetylate inhibitor) and harvested. Histones were extracted as previously described.53 Purified histones (~300 μg) were separated by RP-HPLC as previously described.57 Isolated histones H3 and H4 were submitted to GluC digestion (20:1, w/w – histone to GluC) for 8 hrs in 50 mM ammonium acetate buffer (pH 4) to generate the 50 residue (5.34 kDa) N-terminal tails of H3 proteoforms and the 52 residue (5.59 kDa) N-terminal tail of H4 proteoform prior to LC-MS analysis.
Liquid chromatography mass spectrometry (LC-MS)
The GluC peptides were separated using a Dionex RSLC 3000 nano-LC system (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a column packed in-house with PolyCAT A (3 μm 1500 Å pore size), a weak cation exchange hydrophilic interaction chromatography (WCX-HILIC) resin (PolyLC, Columbia, MD). The nanoLC system was coupled to a Fusion Lumos Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) modified for 193 nm UVPD, as previously described.58 UVPD was performed in the high-pressure linear ion trap using two pulses (2.5 mJ) from a 193 nm Excistar XS excimer laser (Coherent, Santa Clara, CA). ETD was performed in the high pressure linear ion trap with 30 ms reaction time for ETD (2 × 105 reagent AGC) based on optimized conditions reported by previously.18 A typical chromatogram and MS1 spectrum are shown in Figure S1.
Data Analysis
Data processing for the large-scale identification and quantification of histone tails was performed as described previously.18 Briefly, spectra were deconvoluted with Xtract (Thermo) and searched with Mascot (v2.5, Matrix Science, London, UK), including mono- and dimethylation (KR), trimethylation (K) and acetylation (K) as dynamic modifications. Mascot results were filtered for unambiguous identifications and peptides were quantified using a modified version of isoScale18,53 to accept UVPD spectra. Quantitative analysis of histone PTM data and their co-existence patterns was achieved by using tools of the CrossTalkDB resource.59 All spectra are archived and available at: https://repository.jpostdb.org/ and accession numbers are PXD009653 for ProteomeXchange and JPST000416 for jPOST.
Results and Discussion
The performance of ETD and UVPD were evaluated based on metrics including the number of unique peptide forms (including modifications) identified, sequence coverage, and number and position of diagnostic fragment ions; especially modification-localizing ions. For the histone tails in the present study, the 8+ charge state was targeted owing to its consistently high abundance and suitability for ETD (Figure S2). Moreover, selection of the 8+ charge state ensures resolution of the isotopes of precursor and fragment ions at the level necessary for effective deconvolution and ion assignment. Figure S3 shows one representative UVPD mass spectrum for the N-terminal tail of acH4K20me2 (m/z 708.68, the tail is 5661.35 Da, containing 52 residues, 8+ charge state). The spectrum displays the rich fragmentation pattern characteristic of UVPD.
UVPD Optimization
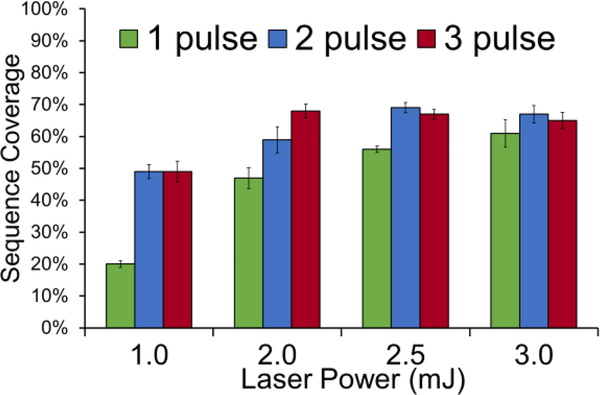
The energy of a single 193 nm photon (6.4 eV) is sufficient to dissociate most peptides; however, other considerations such as photon flux and number of pulses affects the total energy deposition and potential for secondary dissociation.60 A related consideration is the possibility of excessive energy deposition from absorption of multiple photons which can cause secondary fragmentation of ions in a manner that leads to production of un-assignable internal ions or overly small, uninformative sequence ions. Thus, UVPD parameters were optimized for an ideal 5.6 kDa middle-down sized histone peptide originated from acH4K20me2 (52 residues of the N-terminal tail, net 5661.35 Da, 8+ charge state) possessing two modifications. This particular proteoform (acH4K20me2) is one of the most commonly detected and represents an ideal benchmark histone.44,48 Figure 1 shows the dependence of sequence coverage on the N-terminal peptide on laser pulse number and power. Sequence coverage and P-score values were generated using Xtract to deconvolute the raw data and ProSight Lite to match the deconvoluted fragment ions to the theoretical modified sequence of histone H4 (residues 2–53). Using a single laser pulse, the sequence coverage increased with increasing laser power; however, with multiple pulses the increase in sequence coverage peaked or plateaued at 2.5 mJ. Optimal sequence coverage (69%) was obtained using two pulses at 2.5 mJ. Other combinations of laser conditions yielded similar performance, such as 3 pulses at 2 mJ (67% coverage). The P-scores were used to discriminate between the best performing UVPD conditions (Figure S4). The P-score, based on the probability of observed spectra matching theoretical spectra by random chance, is a useful metric as it is often utilized by database searching algorithms such as MASCOT (used in this study) during LC-MS data analysis. Applying 2 pulses at 2.5 mJ gave the lowest P-score (2.6E-66), indicating the highest confidence in the fragment-to-theoretical spectral match. Secondary dissociation and generation of internal fragments occurs if the photon flux is too high or the ions are exposed to multiple pulses. These additional non-diagnostic ions can negatively influence spectral matching confidence, which may be the case for the 3-pulse data, a factor that would explain the high sequence coverage (Figure 1) but non-optimal P-score (Figure S4).
Figure 1.
Dependence of sequence coverage of the N-terminal tail of acH4K20me2 (residues 1–52, 8+) on pulse number and laser power used for UVPD.
Benchmarking UVPD against ETD for mixtures of histones
WCX-HILIC separations followed by high resolution ETD-MS has become the gold standard method for middle-down histone analysis, as originally implemented by Garcia et al.17 Given the large sizes of the N-terminal peptides and their basic nature, they are often multiply protonated and found in charge states ranging from 5+ to 12+ under the acidified conditions utilized in the WCX-HILIC separation. ETD proved to be an efficient means to characterize these multiply charged basic peptides while retaining their abundant modifications.17 While UVPD is similar to ETD with respect to retention of PTMs and the ability to generate excellent sequence coverage, UVPD generates several additional ion types (UVPD: a,a+1,b,c,x,x+1,y,y-1,z compared to predominantly c/z for ETD).60,61 These additional ion types have the potential to add confidence in localization of modification sites and improve sequence coverage, at the expense of potentially reducing the S/N levels of the resulting MS/MS spectra owing to greater dispersion of the ion current.
In order to evaluate the viability of UVPD for LC-MS analysis of the many modified forms of histone H3, a mixture of H3 tails were subjected to WCX-HILIC separation and analyzed by ETD and UVPD. To maximize the sensitivity of the analysis, a narrow mass window bracketing the +8 charge state of the H3 tail and its modified forms was used, followed by data dependent selection of precursors for MS2 analysis.17 The global performance of ETD and UVPD was evaluated based on the number of unique species identified, and detailed evaluation of the fragment ion spectra generated by both methods is discussed later. The number of unique species detected, after filtering out ambiguous matches and non-quantifiable species, was similar for histone H3 (175 proteoforms for ETD and 180 proteoforms for UVPD (Table S1)), thus showing that UVPD is comparable to ETD with respect to number of identifications and is a competitive strategy for identification of heavily modified middle-down sized peptides.
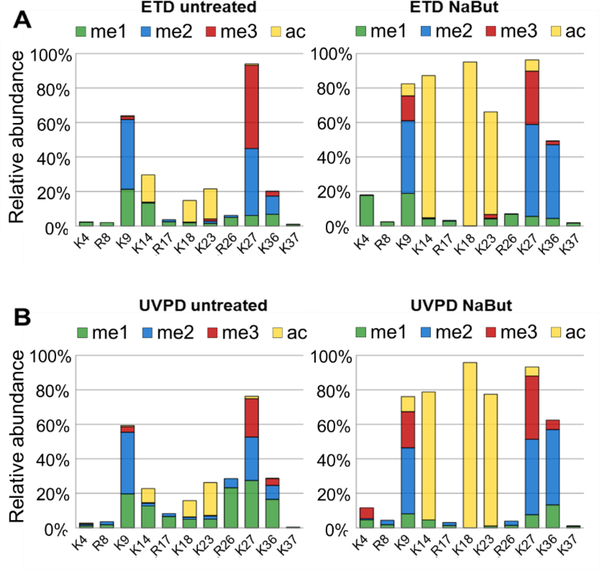
The histone peptides identified by ETD and UVPD were heavily modified. In order to characterize the multitude of modifications, each modifiable site was considered, and the relative contribution of acetylation (ac:yellow), methylation (me1:green), dimethylation (me2:blue), and trimethylation (me3:red) are displayed in Figure 2. Overall, the relative distributions of modifications characterized by UVPD and ETD were similar; however, UVPD of the untreated set resulted in identification of a greater proportion of methylation sites on residues closer to the C-terminus. Several abundant proteoforms identified by UVPD contained K27me1 and K36me1 and contributed to this finding. The fact that UVPD may induce several types of backbone cleavages might explain the ability to better bracket the methylation sites close to the C-terminus, and this finding will be explored in more detail in a larger-scale study. The distributions in Figure 2 can alternatively be displayed based on absolute counts. In fact, the relative abundances are estimated by dividing the absolute counts for a given modified state by the absolute counts of all modified states for a given histone tail. The relative quantification mode used in Figure 2 allows normalization for small biases in amounts of sample injected.
Figure 2.
Distribution of specific modifications detected by ETD (A and C) and UVPD (B and D) based on analysis of the N-terminal peptides of histone H3 (residues 1–50) for untreated (A and B) and butyrate-treated (NaBut) (C and D) cells. The distributions represent the summation of modifications found on all forms of H3.
Among the proteoforms identified, residues K4, K9, K14, K18, K23 and K27 were found to be acetylated. After treatment with NaBut, acetylation of K14, K18 and K23 was detected at significantly increased levels by both UVPD and ETD, whereas acetylation of K9 and K27 increased slightly. NaBut has been shown to block histone deacetylase enzymes (HDACs) leading to hyperacetylation,17 so our results are consistent with this finding. Both UVPD and ETD yielded PTM distributions which were nearly identical and reflected a large increase in acetylation after NaBut treatment, confirming that UVPD should be applicable for relative quantitation of PTMs and is sufficiently sensitive to discriminate different modification distributions based on biological conditions (e.g. NaBut treatment vs. untreated).
Figure 3 shows the log fold change between NaBut-treated and control samples for the individual PTMs of histone H3 resulting from either the ETD or UVPD analysis. The abundance of the PTMs was assessed by summing the relative abundances of all the quantified polypeptides carrying each individual PTM to obtain their total relative abundances. Change in acetylation is highlighted by green data points in Figure 3. Significant increases in acetylation were found by both UVPD and ETD, an outcome consistent with inhibition of HDACs by sodium butyrate.
Figure 3.
Fold change in the relative abundances of modifications for histone H3 identified by: A) ETD and B) UVPD analysis of the N-terminal peptides (8+ charge state) based on triplicate runs. Green markers indicate acetylation, and the five sites with the most prominent changes are labelled. All forms containing each modification contributed to the data in this figure.
In the seminal work of Garcia et al. which entailed the analysis of fractioned NaBut-treated HeLa histones, 205 H3 histoforms were manually identified.17 More recently Paša-Tolić et al.63 evaluated intact HeLa histones by using a 2D RPLC-WCX/HILIC separation method and reported 372 histone isoforms identified by ETD and 44 using CID. However, positive site localization via flanking fragment ions was not mentioned as a requisite for the reported isoforms.63 A 2016 MD study by Yi et al.34 utilizing reversed phase LC-MS and a middle-down strategy for analysis of chemically derivatized Karpas-422 histone N-terminal tails reported up to 311 identified histoforms from El1-treated H3 histones (for ones which surpassed a simple MASCOT ions score of 40). In comparison to these studies, up to 180 histoforms were identified and quantified from NaBut-treated HeLa histones in the present study. The present study utilizes very stringent rules for validating the reported histoforms (i.e. all PTMs must have flanking fragment ions to provide unambiguous site localization), a criterion which explains the discrepancy in the number of histoforms identified between this report and other recent reports in the literature.
Comparison of modified forms found by ETD versus UVPD
The overlap of modified forms identified by both ETD and UVPD accounts for only 15% of the total H3 histoforms, as summarized in the Venn diagrams shown in Figure S5. In fact, the forms identified uniquely by either ETD or UVPD account for over 80% of the total forms identified, demonstrating the complementarity of these two methods. In many cases, a histoform identified uniquely by UVPD diverges from a similar one found by ETD based on a difference in a single modification. For instance, histone H3R8me1K14acR17me1K18acR26me1K36me2 (containing six modifications) identified by UVPD differs in only one position, K23, from H3R8me1K14acR17me1K18acK23acR26me1K36me2 (containing 7 modifications) identified by ETD. For this histone, 6 out of 7 modifications were identified in common by both methods and confirmed by manual interpretation. However, the one identified by UVPD displayed acetylation of Lys18, whereas the one characterized by ETD exhibited acetylation of Lys 18 and Lys23. Despite the large difference in the specific proteoforms identified by each method, the relative distributions of modifications were similar (Figure 2). Inspection of the abundances (found in Table S1) of the modified forms from Figure S5 indicates that the ~17% of modified histones found in common for UVPD and ETD account for approximately 30% of the total abundance of histoforms identified by UVPD and 40% of the total abundance found by ETD.
Because we identify and quantify intact histone tails, it is possible to assess similarities and differences with the estimated co-frequencies of PTMs (i.e. instances where two PTMs occur on the same peptide). Figure 4 shows a web diagram illustrating the co-occurrence of modifications on untreated H4 as indicated by weighted line connections59 (quantified forms are summarized in Table S2). The abundance of these co-occurrences is denoted by the thickness of the line. Similar co-occurrences are observed for both ETD and UVPD, again confirming the reproducibility in PTM quantification despite some differences in the identified combinatorial codes.
Figure 4.
PTM co-existence web for histone H4 (1–52) based on ETD and UVPD analysis. The line connection indicates co-existence, and the line thickness indicates the frequency of co-existence with the thickest line corresponding to 91.1% co-existence frequency. Thicker connecting lines indicate that the two modifications are more often found on the same peptides.
ETD and UVPD fragmentation patterns
One likely factor contributing to the differences in the distribution of PTMs, and both the number and overlap of identified species mentioned above is the significant number of ions generated by UVPD which are not utilized by MASCOT for scoring spectral matches. UVPD consistently generates many diverse ion types, including a, a+ 1, b, c, x, x +1, y, y –1, and z type ions.62 MASCOT has been designed to utilize a, b, c, x, y, z, z+1 and z+2 ions for scoring. Figure 5 shows the distribution of ions generated by UVPD of one typical middle-down sized doubly-modified peptide, representing acH4K20me2, the same species used for the UVPD optimization. The fragment ions were matched at 10 ppm error using ProSight Lite. The results in Figure 5 imply that MASCOT utilizes only 53% of the total number of UVPD fragment ions possible (corresponding to only 35% of the total abundances of identified fragment ions). Moreover, the presence of the diagnostic a+1, x+1, and y-1 ions are not utilized and may be counted as noise, actually depressing the MASCOT scoring metrics for UVPD peptide spectral matches. (For ETD, the fragment ions considered included c, y, z, z+1 and z+2 ions). Figure S6 shows the relative abundances of the ETD fragment ion types for a representative H3 N-terminal histone tail.
Figure 5.
Ion type distribution (by number of matched ions) for the UVPD spectrum of the N-terminal peptide of acH4K20me2 (8+). Boxed bars indicate those ion types which are not used for scoring spectral matches.
Better utilization of the ion types characteristic of UVPD of middle-down size peptides could increase the confidence of UVPD peptide spectral matches, increase the number of overall matched forms, and reconcile some of the differences observed between ETD and UVPD results. Although training MASCOT (or another platform) for UVPD spectra would result in a more ideal performance, the use of isoScale (a custom program currently only compatible with MASCOT output) justifies the workflow used in the present study. isoScale further processes the MASCOT output, specifically focusing on culling false positive modification assignments by virtue of localizing fragment ions. After isoScale processing, the final results are considered unambiguous because each localized modification is supported by assigned fragment ions that bracket the modification site. This feature is crucial for analyzing datasets containing heavily modified peptides with a high level of confidence.
Characterization of the most heavily modified species (> 5 PTMs)
In light of the limitations of the current automated workflow, manual annotation can be used to achieve the greatest sequence coverage and PTM site localization from UVPD spectra. Both ETD and UVPD are effective for characterization of lightly and moderately modified species. UVPD is especially useful for heavily modified forms (i.e. ones containing more than five modifications). In order to highlight the proficiency of UVPD for characterization of highly modified histones, ProSight Lite was used to manually annotate UVPD spectra acquired for the most heavily modified species.64,65 Figure 6 shows deconvoluted ETD and UVPD mass spectra of the hepta-modified peptide H3K4me1K9me2K14acK18acK23acK27acK36me3 (8+ charge state, N-terminal tail containing 50 residues with mono-methylation of residue K4, dimethylation of residue K9, trimethylation of residue K36, and acetylation of residues K14,K18,K23 and K27). Both MS/MS methods adequately localized several of the modifications; however, UVPD was able to achieve higher confidence by virtue of production of multiple PTM-localizing fragment ions. For example, UVPD successfully characterized K14ac and K27ac, generating the greatest number (three or more) of flanking fragment ions containing the modification, including both complementary C-terminal and N-terminal ions. K14ac was localized by a14 + 1, z37, y37 – 1, x37 + 1, and K27ac was localized by a27 + 1, y24 −1, and x24 + 1. By comparison, ETD best characterized K4me, K9me2 and K18, generating only one fragment ion containing the modification and one or more flanking ions facilitating localization.
Figure 6.
Comparison of deconvoluted A) ETD and B) UVPD mass spectra of the N-terminal tail of H3K4me1K9me2K14acK18acK23acK27acK36me3 (50 residues, 8+) showing fragment ion maps, P-score, sequence coverage and labeled PTM site-localizing ions. Residues are shaded to indicate modifications: red indicates acetylation, blue indicates dimethylation, green indicates methylation, and purple indicates trimethylation.
Presence and use of neutral loss ions
Modified peptides can undergo informative neutral losses after activation, often exploited for characterization of phosphorylated and glycosylated peptides.35,66 Traditional bottom-up analysis of histone peptides has utilized neutral losses generated by HCD and ETD of methylated peptides, particularly loss of 59.07 Da from trimethylated Lys residues and loss of 45.06 Da from Arg residues of histones.67 The presence and diagnostic nature of neutral losses upon 193 nm UVPD of methylated species has not been reported previously. These neutral losses can be very useful for determining the specific nature of modified lysines. For instance, the mass difference between a trimethylated lysine and an acetylated lysine is 0.036 Da which for a 5–6 kDa peptide represents a 6–7 ppm mass difference, well within the accepted mass tolerance of 10 ppm. The heavily modified H3 peptide shown in Figure 6 has several acetylated lysines and a trimethylated lysine residue. The presence of a 59.07 Da neutral loss upon UVPD can be used to discriminate between the K36ac and K36me3 forms. Figure S7 shows the occurrence of the 59.07 Da loss, thus confirming the presence of a trimethylation. Conversely, when the trimethylated K36 residue is replaced with acetylated K36 in the search, several scoring metrics degrade, including P-score, number of matched fragments and the ppm mass error. Although other metrics can be used to discriminate between acetylation and trimethylation, the presence of the 59.07 Da mass loss provides further evidence supporting the assignment of trimethylation for this peptide.
Figure S8 shows the sequences and deconvoluted UVPD mass spectra of two nearly isobaric N-terminal peptides of H3K4acR8me2K23acK27me2 (5478.15 Da) and H3K9me3K14acKme2K36me2 (5478.19 Da), differing only by the acetyl-trimethyl mass difference. For each of these proteoforms, the 8+ charge state was subjected to UVPD. The trimethylated species displays the expected 59.07 Da neutral loss which is absent from the UVPD mass spectrum of the acetylated species, confirming the assignment of trimethylation. The presence of this diagnostic neutral loss ion upon UVPD offer notable utility for correctly interpreting ambiguous spectra.
Conclusion
Results from LC-MS analyses of GluC-generated middle-down sized N-terminal tails of histone H3 and H4 demonstrate that 193 nm UVPD is broadly applicable for characterization of heavily modified histones. ETD and UVPD identified largely unique (only 15% overlap) combinatorial species. However, UVPD and ETD led to highly comparable results when assessing the overall abundance of single and co-existing modifications, implying that the high orthogonality in terms of which combinatorial codes are identified in every MS run does not affect significantly the ultimate conclusions. Another promising new ion activation strategy combining ETD and HCD (EThcD) has demonstrated extensive fragmentation of peptides39 and has been used successfully for histone tails.34 Comparison of the performance metrics of EThcD relative to 193 nm UVPD, with both benchmarked to the gold standard ETD, merits further investigation for analysis of histone tails.
UVPD was useful for deciphering changes in modifications related to specific cellular treatments, as shown by the 2-fold to 4-fold up-regulation of acetylation in the NaBut-treated Hela cells. Evaluation of the differences between ETD and UVPD revealed that the automated data processing workflow, which relies on MASCOT, utilizes only approximately 50% of the ions generated by UVPD. Roughly 35% of the total matched fragment ion population from UVPD originates from the a+1 ion series which is not considered by MASCOT. Moving forward, a fully trained search algorithm would extend the capabilities of UVPD for high throughput analysis of modified middle-down sized peptides. In order to evaluate the ability of UVPD to characterize the most heavily modified peptides, manual spectral interpretation facilitated by ProSight Lite was a key to success. The N-terminal peptide of hepta-modified histone H3K4me1K9me2K14acK18acK23acK27acK36me3 was well-characterized and yielded 90% sequence coverage, motivating future investigation of UVPD for interrogating other heavily modified middle-down sized peptides. UVPD also resulted in characteristic neutral loss pathways. For example, loss of 59.07 Da upon UVPD differentiated trimethylation from acetylation for histones H3K4acR8me2K23acK27me2 and H3K9me3K14acKme2K36me2.
We recently reported the application of 193 nm UVPD for top-down shotgun analysis of histones.56 From a crude (non-fractionated) mixture of histones, over 500 modified histoforms encompassing H1, H2, H3, and H4 variants were identified. The present study focused primarily on histones H3 and H4, subsets which were targeted by employing offline pre-fractionation. This fractionation mitigated the problems encountered in the previous top-down study of overlapping proteoforms that led to co-isolation and co-fragmentation, resulting in ambiguous assignments and less depth of identification of histoforms for any particular variant owing to co-elution and disparate abundances of histoforms. The employment of WCX-HILIC in the present study enhanced the performance metrics of the chromatographic separation, allowing a greater number of forms to be analyzed from each histone variant. In essence, identification of 300 histoforms based on middle-down analysis of a single variant of histone H3 or H4 reflects greater depth compared to identification of 500 proteoforms from a collection of H1,H2,H3 and H4 variants. The data analysis and scoring tools for top-down proteomics (and the current state of FDR calculations for high-throughput top-down UVPD spectra) have room for improvement, making middle-down approaches a practical alternative accessible to many laboratories.
Supplementary Material
Acknowledgements:
We acknowledge the following funding sources: NSF (Grant CHE1402753) and the Welch Foundation (Grant F-1155). SMG acknowledges a graduate fellowship from the American Chemical Society Division of Analytical Sciences. Funding from the UT System for support of the UT System Proteomics Core Facility Network is gratefully acknowledged. SS, MC and BAG gratefully acknowledge the NIH grants CA196539, GM110174 and AI118891. The authors acknowledge Dr. Andrew Alpert (PolyLC) for his generous donation of PolyCAT A (WCX-HILIC) resin.
Footnotes
Supporting Information: Available information includes a typical nano WCX-HILIC chromatogram for a mixture of histone tails, typical MS1 and UVPD mass spectra for a histone tail, a histogram of the P-score versus laser power used for UVPD, a Venn diagram showing numbers of modified forms of histone tails for ETD and UVPD, an ion type distribution for ETD, deconvoluted UVPD mass spectra showing neutral losses, and tables summarizing all modified H3 and H4 histones identified by ETD and UVPD.
References
- (1).Venne AS; Kollipara L; Zahedi RP Proteomics 2014, 14 (4–5), 513–524. [DOI] [PubMed] [Google Scholar]
- (2).Jaenisch R; Bird A Nat. Genet 2003, 33, 245–254. [DOI] [PubMed] [Google Scholar]
- (3).Appella E; Anderson CW Eur. J. Biochem 2001, 268 (10), 2764–2772. [DOI] [PubMed] [Google Scholar]
- (4).Kouzarides T Cell 2007, 128 (4), 693–705. [DOI] [PubMed] [Google Scholar]
- (5).Meek DW; Anderson CW Cold Spring Harb. Perspect. Biol 2009, 1 (6), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dai C; Gu W Trends Mol. Med 2010, 16 (11), 528–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bode AM; Dong Z Nat. Rev. Cancer 2004, 4 (10), 793–805. [DOI] [PubMed] [Google Scholar]
- (8).Jenuwein T Science 2001, 293 (5532), 1074–1080. [DOI] [PubMed] [Google Scholar]
- (9).Bannister AJ; Kouzarides T Cell Res. 2011, 21 (3), 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Luo Y; Yogesha SD; Cannon JR; Yan W; Ellington AD; Brodbelt JS; Zhang Y ACS Chem. Biol 2013, 8 (9), 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sims RJ; Rojas LA; Beck DB; Bonasio R; Schüller R; Drury WJ; Eick D; Reinberg D Science 2011, 332 (6025), 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Farriol-Mathis N; Garavelli JS; Boeckmann B; Duvaud S; Gasteiger E; Gateau A; Veuthey A-L; Bairoch A PROTEOMICS 2004, 4 (6), 1537–1550. [DOI] [PubMed] [Google Scholar]
- (13).Garavelli JS PROTEOMICS 2004, 4 (6), 1527–1533. [DOI] [PubMed] [Google Scholar]
- (14).Mann M; Kulak NA; Nagaraj N; Cox J Mol. Cell 2013, 49 (4), 583–590. [DOI] [PubMed] [Google Scholar]
- (15).Olsen JV; Mann M Mol. Cell. Proteomics 2013, 12 (12), 3444–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kjeldsen F; Giessing AMB; Ingrell CR; Jensen ON Anal. Chem. 2007, 79 (24), 9243–9252. [DOI] [PubMed] [Google Scholar]
- (17).Young NL; DiMaggio PA; Plazas-Mayorca MD; Baliban RC; Floudas CA; Garcia BA Mol. Cell. Proteomics 2009, 8 (10), 2266–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Sidoli S; Schwämmle V; Ruminowicz C; Hansen TA; Wu X; Helin K; Jensen ON PROTEOMICS 2014, 14 (19), 2200–2211. [DOI] [PubMed] [Google Scholar]
- (19).Moradian A; Kalli A; Sweredoski MJ; Hess S PROTEOMICS 2014, 14 (4–5), 489–497. [DOI] [PubMed] [Google Scholar]
- (20).Egelhofer TA; Minoda A; Klugman S; Lee K; Kolasinska-Zwierz P; Alekseyenko AA; Cheung M-S; Day DS; Gadel S; Gorchakov AA; Gu T; Kharchenko PV; Kuan S; Latorre I; Linder-Basso D; Luu Y; Ngo Q; Perry M; Rechtsteiner A; Riddle NC; Schwartz YB; Shanower GA; Vielle A; Ahringer J; Elgin SCR; Kuroda MI; Pirrotta V; Ren B; Strome S; Park PJ; Karpen GH; Hawkins RD; Lieb JD Nat. Struct. Mol. Biol 2011, 18 (1), 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pereira Morais MP; Mackay JD; Bhamra SK; Buchanan JG; James TD; Fossey JS; van den Elsen JM H. PROTEOMICS 2010, 10 (1), 48–58. [DOI] [PubMed] [Google Scholar]
- (22).Strack R Nat. Methods 2017, 14 (2), 106–107. [Google Scholar]
- (23).Aebersold R; Mann M Nature 2016, 537 (7620), 347–355. [DOI] [PubMed] [Google Scholar]
- (24).Larance M; Lamond AI Nat. Rev. Mol. Cell Biol 2015, 16 (5), 269–280. [DOI] [PubMed] [Google Scholar]
- (25).Villén J; Gygi SP Nat. Protoc 2008, 3 (10), 1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Carlson SM; Moore KE; Green EM; Martin GM; Gozani O Nat. Protoc 2013, 9 (1), 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Li Y; Silva JC; Skinner ME; Lombard DB In Sirtuins; Hirschey MD, Ed.; Humana Press: Totowa, NJ, 2013; Vol. 1077, pp 81–104. [Google Scholar]
- (28).Wang K; Dong M; Mao J; Wang Y; Jin Y; Ye M; Zou H Anal. Chem 2016, 88 (23), 11319–11327. [DOI] [PubMed] [Google Scholar]
- (29).Zhang L; Liu C-W; Zhang Q Anal. Chem 2017. [Google Scholar]
- (30).Han S-W; Lee S-W; Bahar O; Schwessinger B; Robinson MR; Shaw JB; Madsen JA; Brodbelt JS; Ronald PC Nat. Commun 2012, 3, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Fort KL; Dyachenko A; Potel CM; Corradini E; Marino F; Barendregt A; Makarov AA; Scheltema RA; Heck AJ R. Anal. Chem 2016, 88 (4), 2303–2310. [DOI] [PubMed] [Google Scholar]
- (32).Chi A; Huttenhower C; Geer LY; Coon JJ; Syka JEP; Bai DL; Shabanowitz J; Burke DJ; Troyanskaya OG; Hunt DF Proc. Natl. Acad. Sci 2007, 104 (7), 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wiesner J; Premsler T; Sickmann A Proteomics 2008, 8 (21), 4466–4483. [DOI] [PubMed] [Google Scholar]
- (34).Liao R; Zheng D; Nie A; Zhou S; Deng H; Gao Y; Yang P; Yu Y; Tan L; Qi W; Wu J; Li E; Yi WJ Proteome Res. 2017, 16 (2), 780–787. [DOI] [PubMed] [Google Scholar]
- (35).Boersema PJ; Mohammed S; Heck AJ R. J. Mass Spectrom. JMS 2009, 44 (6), 861–878. [DOI] [PubMed] [Google Scholar]
- (36).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF Proc. Natl. Acad. Sci 2004, 101 (26), 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zubarev RA; Horn DM; Fridriksson EK; Kelleher NL; Kruger NA; Lewis MA; Carpenter BK; McLafferty FW Anal. Chem 2000, 72 (3), 563–573. [DOI] [PubMed] [Google Scholar]
- (38).Syka JEP; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF Proc. Natl. Acad. Sci 2004, 101 (26), 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]; Robinson MR; Taliaferro JM; Dalby KN; Brodbelt JS J. Proteome Res 2016, 15 (8), 2739–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Frese CK; Altelaar AFM; van den Toorn H; Nolting D; Griep-Raming J; Heck AJR; Mohammed S Anal. Chem 2012, 84 (22), 9668–9673. [DOI] [PubMed] [Google Scholar]
- (40).Riley NM; Westphall MS; Coon JJ J. Proteome Res 2017, 16 (7), 2653–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Madsen JA; Kaoud TS; Dalby KN; Brodbelt JS PROTEOMICS 2011, 11 (7), 1329–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Sidoli S; Garcia BA Expert Rev. Proteomics 2017, 14 (7), 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Peters AHFM; Kubicek S; Mechtler K; O’Sullivan RJ; Derijck AAHA; Perez-Burgos L; Kohlmaier A; Opravil S; Tachibana M; Shinkai Y; Martens JHA; Jenuwein T Mol. Cell 2003, 12 (6), 1577–1589. [DOI] [PubMed] [Google Scholar]
- (44).Dang X; Scotcher J; Wu S; Chu RK; Tolic N; Ntai I; Thomas PM; Fellers RT; Early BP; Zheng Y; Durbin KR; LeDuc RD; Wolff JJ; Thompson CJ; Pan J; Han J; Shaw JB; Salisbury JP; Easterling M; Borchers CH; Brodbelt JS; Agar JN; Paša-Tolić L; Kelleher NL; Young NL PROTEOMICS 2014, 14 (10), 1130–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Cannon J; Lohnes K; Wynne C; Wang Y; Edwards N; Fenselau CJ Proteome Res. 2010, 9 (8), 3886–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Cristobal A; Marino F; Post H; van den Toorn HWP; Mohammed S; Heck AJ R. Anal. Chem 2017, 89 (6), 3318–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Park J; Piehowski PD; Wilkins C; Zhou M; Mendoza J; Fujimoto GM; Gibbons BC; Shaw JB; Shen Y; Shukla AK; Moore RJ; Liu T; Petyuk VA; Tolić N; Paša-Tolić L; Smith RD; Payne SH; Kim S Nat. Methods 2017, 14 (9), 909–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Pesavento JJ; Mizzen CA; Kelleher NL Anal. Chem 2006, 78 (13), 4271–4280. [DOI] [PubMed] [Google Scholar]
- (49).Zheng Y; Fornelli L; Compton PD; Sharma S; Canterbury J; Mullen C; Zabrouskov V; Fellers RT; Thomas PM; Licht JD; Senko MW; Kelleher NL Mol. Cell. Proteomics 2016, 15 (3), 776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Syka JEP; Marto JA; Bai DL; Horning S; Senko MW; Schwartz JC; Ueberheide B; Garcia B; Busby S; Muratore T; Shabanowitz J; Hunt DF J. Proteome Res 2004, 3 (3), 621–626. [DOI] [PubMed] [Google Scholar]
- (51).Plazas-Mayorca MD; Zee BM; Young NL; Fingerman IM; LeRoy G; Briggs SD; Garcia BA J. Proteome Res 2009, 8 (11), 5367–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Schräder CU; Ziemianowicz DS; Merx K; Schriemer DC Anal. Chem 2018, 90 (5), 3083–3090. [DOI] [PubMed] [Google Scholar]
- (53).Sidoli S; Lu C; Coradin M; Wang X; Karch KR; Ruminowicz C; Garcia BA Epigenetics Chromatin 2017, 10 (1), 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Cannon JR; Cammarata MB; Robotham SA; Cotham VC; Shaw JB; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS Anal. Chem 2014, 86 (4), 2185–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Greer SM; Holden DD; Fellers R; Kelleher NL; Brodbelt JS J. Am. Soc. Mass Spectrom 2017, 28 (8), 1587–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Greer SM; Brodbelt JS J. Proteome Res. 2018, 17(3) 1138–1145. [DOI] [PubMed] [Google Scholar]
- (57).Lin S; Garcia BA In Methods in Enzymology; Wu C, Allis CD, Eds.; Nucleosomes, Histones & Chromatin Part A; Academic Press, 2012; Vol. 512, pp 3–28. [DOI] [PubMed] [Google Scholar]
- (58).Klein DR; Holden DD; Brodbelt JS Anal. Chem 2016, 88 (1), 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59.) Schwammle V, Aspalter CM, Sidoli S, Jensen ON, Mol. Cell Proteomics, 2014, 13(7), 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Brodbelt JS Chem Soc Rev 2014, 43 (8), 2757–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Brodbelt JS Anal. Chem 2016, 88 (1), 30–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Holden DD; McGee WM; Brodbelt JS Anal. Chem 2016, 88 (1), 1008–1016. [DOI] [PubMed] [Google Scholar]
- (63).Tian Z; Tolić N; Zhao R; Moore RJ; Hengel SM; Robinson EW; Paša-Tolić L Genome Bio. 2012,13 (10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).DeHart CJ; Fellers RT; Fornelli L; Kelleher NL; Thomas PM In Protein Bioinformatics; Methods in Molecular Biology; Humana Press, New York, NY, 2017; pp 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Meng F; Cargile BJ; Miller LM; Forbes AJ; Johnson JR; Kelleher NL Nat. Biotechnol 2001, 19 (10), 952. [DOI] [PubMed] [Google Scholar]
- (66).Yang Y; Liu F; Franc V; Halim LA; Schellekens H; Heck AJ R. Nat. Commun 2016, 7, 13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Zhang K; Yau PM; Chandrasekhar B; New R; Kondrat R; Imai BS; Bradbury ME PROTEOMICS 2004, 4 (1), 1–10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.