Abstract
Glioma-associated oncogene homolog-1 (Gli1)-positive resident mesenchymal stem cell-like cells are the predominant source of kidney myofibroblasts in fibrosis, but investigating Gli1-positive myofibroblast progenitor activation is hampered by the difficulty of isolating and propagating primary cultures of these cells. Using a genetic strategy with positive and negative selection, we isolated Kidney-Gli1 (KGli1) cells that maintain expression of appropriate mesenchymal stem cell-like cell markers, respond to hedgehog pathway activation, and display robust myofibroblast differentiation upon treatment with transforming growth factor-β (TGF-β). Coculture of KGli1 cells with endothelium stabilizes capillary formation. Single-cell RNA sequencing (scRNA-seq) analysis during differentiation identified autocrine ligand-receptor pair upregulation and a strong focal adhesion pathway signal. This led us to test the serum response factor inhibitor CCG-203971 that potently inhibited TGF-β-induced pericyte-to-myofibroblast transition. scRNA-seq also identified the unexpected upregulation of nerve growth factor (NGF), which we confirmed in two mouse kidney fibrosis models. The Ngf receptor Ntrk1 is expressed in tubular epithelium in vivo, suggesting a novel interstitial-to-tubule paracrine signaling axis. Thus, KGli1 cells accurately model myofibroblast activation in vitro, and the development of this cell line provides a new tool to study resident mesenchymal stem cell-like progenitors in health and disease.
Keywords: chronic kidney disease, fibrosis, myofibroblast, pericyte, single-cell RNA sequencing, transcriptomics, transforming growth factor-β
INTRODUCTION
Chronic kidney disease (CKD) is a major health problem that affects 26–30 million adults in the United States (5). Despite current medical therapies, outcomes in CKD remain poor (37). Interstitial fibrosis is the best histologic predictor of renal functional decline in CKD, glomerular diseases, and type I diabetic nephropathy (18, 29, 32). Myofibroblasts, reactive cells present under injury conditions that combine features of fibroblasts and smooth muscle cells, are the cell type primarily responsible for matrix protein secretion during fibrosis. Myofibroblasts are a rational therapeutic target in CKD because of their central role in mediating interstitial fibrosis (14). Proof of principle for this concept derives from the recent observation that an immunoglobulin binding the master regulator of myofibroblast activation, transforming growth factor-β (TGF-β), could both reduce interstitial fibrosis and slow the rate of glomerular filtration rate (GFR) decline in an experimental model (35).
The cellular identification of myofibroblast progenitors remains controversial. Our work indicates that myofibroblasts do not derive from epithelial cells through epithelial-to-mesenchymal transition (15). More recently, we have shown that myofibroblasts also do not derive from circulating bone marrow-derived cells (22). Rather, genetic lineage analysis indicates that the bulk of myofibroblasts are kidney resident mesenchymal stem cell-like pericytes and perivascular fibroblasts (23). These cells express the zinc finger transcription factor glioma-associated oncogene homolog-1 (Gli1), a member of the hedgehog signaling pathway (8), and their ablation reduces fibrosis across organs and improves organ function (21). Key features of kidney Gli1-positive mesenchymal cells are their sensitivity to hedgehog pathway activation, which drives cell cycle progression, and their sensitivity to TGF-β, which drives myofibroblast transformation (20).
Given the known role of Gli1+ kidney myofibroblast progenitors in fibrosis, these cells are a logical choice to isolate and culture in vitro. Because they make up only 0.01% of all kidney cells, however, we have been unable to propagate them in primary culture (23). In this study, we used the Gli1-CreERt2 driver for inducible labeling of kidney Gli1 cells. We crossed bigenic Gli1-CreERt2; R26tdTomato mice with the “immortomouse,” which expresses a temperature sensitive SV40 gene, as well as with the “terminator” mouse, which expresses the diphtheria toxin receptor in all cells except those that undergo Cre-mediated recombination. After tamoxifen-induced recombination in vivo, we could isolate tdTomato-positive Gli1 cells that express the temperature-sensitive SV40 antigen and eliminate all non-recombined cells by administration of diphtheria toxin. The resulting cell line faithfully models myofibroblast activation in vitro, and characterization by single-cell RNA sequencing (scRNA-seq) revealed serum response factor (SRF) as a therapeutic target and an unexpected role for upregulated nerve growth factor (NGF) signaling in renal fibrosis. Upregulation of NGF by TGF-β was not observed in the rat kidney fibroblast cell line NRK49F (16), suggesting that Kidney-Gli1 (KGli1) cells have distinct properties.
MATERIALS AND METHODS
Animals
All mouse experiments were approved and performed according to the animal experimental guidelines issued by the Animal Care and Use Committee at Washington University at St. Louis. Gli1-CreERt2 [Gli1tm3(cre/ERT2)Alj/J, JAX stock no. 007913] and Rosa26-tdTomato [B6-Cg-Gt(ROSA)26Sorttm(CAG-tdTomato)Hze/J JAX stock no. 007909] were purchased from Jackson Laboratories. Immortomouse [CBA;B10-Tg(H2Kb-tsA58)6Kio/Crl] was purchased from Charles River. Terminator mice were a kind gift from the Yale O’Brien Center. For generation of cell lines, 80 mg/kg tamoxifen in corn oil/3% ethanol was administered 2 days and 1 day before euthanizing. Unilateral ureteral obstruction (UUO) and unilateral ischemia-reperfusion injury was performed as previously described (23).
Cell Culture and Cell Line Generation
All Gli1+ cells were cultured in Alpha MEM Glutamax (Thermo Scientific, no. 32571) supplemented with 20% MSC-qualified FBS (Thermo Scientific, no. 12662), 10 ng/ml mouse bFGF (proteintech), 10 ng/ml mouse EGF (proteintech), and 1% penicillin-streptomycin (pen/strep; Sigma). Bone marrow-derived Gli1+ cells were isolated as described previously (39). To generate cell lines of the heart, aorta, liver, lung, and brain, the organs were harvested from triple transgenic Gli1-CreERt2; R26tdTomato/WT; H-2kbSV40tsA58/WT. Organs were minced up and placed in a gentleMACS C tube (Miltenyi Biotec, no. 130-093-237) with 1 mg/ml collagenase II in DMEM-F12 and subjected to program D. The cells were incubated for 30 min in collagenase II (Thermo Scientific, no. 17101015) at 37°C with shaking, before a second round of gentleMACS program D. The cell suspension was then centrifuged at 800 g for 10 min, the supernatant was aspirated, and the pellet was resuspended in Gli1+ media. The whole organ cell suspension was then plated out on 150-cm2 dishes for 24 h. After 24 h, the cells were trypsinized, and FAC sorted for tdTomato. A similar protocol was performed for kidney-derived Gli1+ cells. Kidney cell suspensions from the quadruple transgenic mice (Gli1-CreERt2; R26tdTomato/DTR-LoxP; H-2kbSV40tsA58/WT) were created in a similar fashion and were plated out for 72 h in 150-cm2 dishes. After 72 h, 100 ng/ml diphtheria toxin (List Biological Laboratories, no. 150) was added to the culture media for 7 days. Next, the cells were FAC sorted to remove any non-Gli1 cells. Cells were maintained in Gli1 media and split 1:10. All Gli1 cells were initially cultured at 33°C in the presence of 10 U/ml IFN-γ (Thermo Scientific, no. PMC4034) until a purified polyclonal population of tdTomato+ cells was established. After this, cells were cultured in an unimmortalized state at 37°C without IFN-γ.
For myofibroblast differentiation, Gli1 cells were plated out at 2 × 105 cells into 22-cm2 dishes and incubated overnight. The cells were then serum starved overnight in Alpha MEM GlutaMAX with 0.5% MSC-qualified FBS and 1% pen/strep. The next day, 1 ng/ml TGF-β (Peprotech, no. 100-21) was added to the cells in serum-starved media for 24 h. For smoothened agonist (SAG; Santa Cruz Biotechnology, no. sc-202814) treatment, the cells were similarly starved overnight and treated with either 200 nM or 500 nM SAG, and water control.
For all myofibroblast inhibition assays, cells were cultured in reduced serum conditions (0.5% MSC-qualified FBS) overnight. The next day, media were replaced with reduced serum media containing either vehicle control, TGF-β, inhibitor, or TGF-β + inhibitor. TGF-β was used at a concentration of 1 ng/ml; GANT61 (Selleckchem, no. S-8075) at a concentration of 20 µM in DMSO; rosiglitazone (Rosi) at 40 µM in DMSO (Sigma, no. R-2408); CCG-203971 (R&D systems, no. 5277) at 10 µM in DMSO.
Single-Cell RNA Sequencing
Gli1+ cells were plated at a concentration of 3 × 105 cells into 10-cm3 dishes and allowed to attach overnight in regular media. The following day, cells were starved in serum-free αMEM media containing 1% pen/strep for 2 h. The cells were then treated with 1 ng/ml TGF-β for either 6 h, 12 h, or 24 h. Control cells without TGF-β were harvested after the 2-h starving period. The cells were harvested with TrypLE Select (Thermo Fisher Scientific) for 10 min at 37°C, and after 10 min, cells were further dispersed by gentle pipetting and filtered through a 40-µm cell strainer (pluriSelect). Single-cell suspension was visually inspected under a microscope, counted by hemocytometer (INCYTO C-chip), and resuspended in PBS + 0.01% BSA. Single cells were coencapsulated in droplets with barcoded beads exactly as described (28). Libraries were sequenced on a HiSeq 2500. All sequencing data has been uploaded to Gene Expression Omnibus (GEO series record GSE 108232). We routinely tested our DropSeq setup by running species-mixing experiments before running on actual sample to assure that the cell doublet rate was below 5%.
Computational Data Analysis
Preprocessing of DropSeq data.
Paired-end sequencing reads were processed as previously described using the Drop-Seq Tools v1.12 software available in McCarroll’s laboratory (http://mccarrolllab.org/dropseq/). Briefly, each cDNA read (read2) was tagged with the cell barcode (the first 12 bases in read 1) and unique molecular identifier (UMI; the next 8 bases in read 1), trimmed of sequencing adaptors and poly-A sequences, and aligned to the human (GRCh38) or a concatenation of the mouse and human (for the species-mixing experiment) reference genome assembly using STAR v2.5.3a (28). Cell barcodes were corrected for possible bead synthesis errors using the DetectBeadSynthesisErrors program and then collapsed to core barcodes if they were within an edit distance of 1 as previously described (27). Digital gene expression (DGE) matrix was compiled by counting the number of unique UMIs for a given gene (as row) in a given cell (as column).
Cell clustering and marker gene identification.
Raw DGE matrices from all four time points were combined and loaded into the R package Seurat. For normalization, the DGE matrix was scaled by total UMI counts, multiplied by 10,000, and transformed to log space. Only genes found to be expressing in >10 cells were retained. Cells with a relatively high percentage of UMIs mapped to mitochondrial genes (≥ 0.2) were discarded. Moreover, cells with fewer than 500 or more than 6,000 detected genes were omitted, resulting in ~5,000 cells. Before clustering, variants arising from batch effects, library size, and percentage of mitochondrial genes were regressed out using the function RegressOut in R package Seurat. The highly variable genes were identified using the function MeanVarPlot with the following parameters: 1) x.low.cutoff = 0.0125, x.high.cutoff = 8, and y.cutoff = 0.5, resulting in an output of 1,306 highly variable genes for the organoid from the Little protocol; and 2) x.low.cutoff = 0.0125, x.high.cutoff = 3, and y.cutoff = 2, resulting in 372 highly variable genes for the organoids from the Bonventre protocol. The expression level of highly variable genes in the cells was scaled and centered along each gene and was conducted to principal component (PC) analysis. We then assessed the number of PCs to be included in downstream analysis by 1) plotting the cumulative standard deviations accounted for each PC using the function PCElbowPlot in Seurat to identify the “knee” point at a PC number after which successive PCs explain diminishing degrees of variance, and 2) by exploring primary sources of heterogeneity in the data sets using the PCHeatmap function in Seurat. Based on these two methods, we selected the first 25 PCs for two-dimensional t-distributed stochastic neighbor embedding (tSNE), implemented by the Seurat software with the default parameters. Based on the tSNE map, seven clusters were identified using the function FindCluster in Seurat with the resolution parameter set to 0.6. To identify the marker genes, differential expression analysis was performed by the function FindAllMarkers in Seurat with likelihood-ratio test. Differentially expressed genes that were expressed in at least 25% of cells within the cluster and with a fold change more than 0.25 (log scale) were considered to be marker genes. A heatmap of the top 10 differentially expressed genes was performed using the DoHeatmap function in Seurat. Cell cycle scoring was performed using the CellCycleScoring function in Seurat, with cell cycle genes identified from Kowalczyk et al. (18a). All heatmaps were made using the levelplot function in lattice. For KEGG analysis, gene lists were uploaded to DAVID Bioinformatics Resource 6.8 (22, 30).
Pseudo-temporal analysis.
All clustering data from Seurat were imported into the monocle package in R (31). Expressed genes were classified as having a minimum expression of 0.1 in 10 or more cells. A cell hierarchy was performed based on expression of fibronectin (Fn1) to determine the start and end points of pseudotemporal ordering. Ordering was then performed using a semi-supplemented mode according to publicly available protocols (31–33).
Coculture Assays
Human umbilical vein endothelial cells (HUVEC) were purchased from Lonza (no. C-2519-A) and cultured in EGM-2 media supplemented with EGM-2 bullet kit (Lonza, no. CC-3162). All HUVEC experiments were performed between passages 2–6. Two-dimensional Matrigel assays were performed as previously published (34). HUVEC cells were incubated with green-tracker dye (Thermo Fisher Scientific, no. C-2925) for 30 min before trypsinization. Ten micoliters of GFR Matrigel (Corning, no. 354230) was added to the inner chamber of the μ-angiogenesis slide (ibidi, no. 81506) and put at 37°C for 30 min to solidify. HUVEC and kidney Gli1 cells were mixed at a ratio of 10:1 (7,500 HUVEC, 750 Gli1 cells) per well and incubated for 6 h for quantification and for 24 h for RNA extraction. After 6 h, the cells were fixed for 15 min with 2% cold paraformaldehyde and imaged on a single-plane confocal microscope using Nikon eclipse Ti (Melville, NY). Quantification was performed using ImageJ with the angiogenesis analyzer plugin (35). Average tubule width was obtained from the total area covered divided by the total tubule length. A master segment is a segment adjoined by at least two junctions not associated with end-branches. A master junction is defined as a junction connecting at least three master segments. The mesh area is the total area enclosed by master segments/junctions, thus forming a full mesh without any breaks or branches. Time lapse imaging was performed in 6-well plates at a 10:1 ratio (100,000 HUVEC, 10,000 Gli1 cells) using an inverted microscope (Nikon TE200). Images were obtained with an HQ2 camera (Photometrics, Tucson, AZ). Images were taken every 2 min over a 6-h period.
RNA Isolation, cDNA Synthesis, and Quantitative PCR
Total RNA was isolated using TRIzol (Life Technologies) according to manufacturer’s protocol. For tissue, samples were snap frozen in liquid nitrogen and placed at −80°C. A small piece was cut off the frozen sample and placed in TRIzol. The sample was then homogenized, and extraction continued in the same manner as for cell culture. TRIzol was added to the cells at the end of the experiment. Next, chloroform was added at a ratio of 1:5, and the samples were centrifuged at 12,000 revolutions/min for 18 min. The top layer was removed, and equal parts 2-propanol were added with 1 mg/ml glycogen. The samples were centrifuged again at 12,000 revolutions/min for 18 min and washed twice with 75% ethanol. RNA was suspended in diethyl pyrocarbonate-treated water, and 1 μg cDNA was created using high-capacity cDNA reverse transcriptase kit (Life Technologies, no. 4368813) according to the manufacturer’s protocol. Primer sequences were as follows: Acta2: forward (Fw) 5′-CTGACAGAGGCACCACTGAA-3′, reverse (Rv) 5′-CATCTCCAGAGTCCAGCACA-3′; Fn1: forward: 5′-ATCTGGACCCCTCCTGATAGT-3′, Rv 5′-GCCCAGTGATTTCAGCAAAGG-3′; Col1a2: Fw 5′-AGGAAAGAGAGGGTCTCCCG-3′, Rv 5′-GCCAGGAGGACCCATTACAC-3′; Ctgf: Fw 5′-GGGCCTCTTCTGCGATTTC-3′, Rv 5′-ATCCAGGCAAGTGCATTGGTA-3′; Itga5: Fw 5′-CCTCTCCGTGGAGTTTTACCG-3′, Rv 5′-GCTGTCAAATTGAATGGTGGTG-3′; Itgav: Fw 5′-CCGTGGACTTCTTCGAGCC-3′, Rv 5′-CTGTTGAATCAAACTCAATGGGC-3′; Itgb5: Fw 5′-GAAGTGCCACCTCGTGTGAA-3′, Rv 5′-GGACCGTGGATTGCCAAAGT-3′; Ngf (mouse): Fw 5′-CAAGGACGCAGCTTTCTATACT-3′, Rv 5′-TTGCTATCTGTGTACGGTTCTG-3′; Ngf (rat): Fw 5′-TGCATAGCGTAATGTCCATGTTG-3′, Rv 5′-CTGTGTCAAGGGAATGCTGAA-3′; Pparγ: Fw 5′-GACCTGAAGCTCCAAGAATACC-3′, Rv 5′-TGGCCATGAGGGAGTTAGA-3′; Adrp: Fw 5′-CCTGCCCATCATCCAGAAG-3′, Rv 5′-CTGGTTCAGAATAGGCAGTCTT-3′; Ntrk1: Fw 5′-TCTCGCCAGTGGACGGTAAC-3′, Rv 5′-TGTTGAGCACAAGAAGGAGGG-3′; Gapdh: Fw 5′-AGGTCGGTGTGAACGGATTTG-3′, Rv 5′-TGTAGACCATGTAGTTGAGGTCA-3′. Quantitative PCR (qPCR) was performed using Taq polymerase SYBR Green (Bio-Rad, no. 1725125) on the CFX Connect Real-Time System (Bio-Rad), with GAPDH used as a housekeeping gene. The ΔΔCT method was used to determine fold change compared with control samples.
Immunofluorescence and Western Blotting
Immunostaining was performed as previously described (25). Briefly, mice were anesthetized with isoflurane (Baxter) and subsequently perfused via the left ventricle with 4°C PBS for 1 min. Organs were fixed with 4% paraformaldehyde for 1 h and incubated overnight in 30% sucrose. OCT-embedded (Sakura Finetek) tissues were cryosectioned into 6-µm sections and mounted on Superfrost slides (Fisher Scientific). Cells on chamber slides were fixed for 20 min with 4% paraformaldehyde. Cell membranes were permeabilized with 0.25% Triton-X for 5 min. Blocking was performed with 10% BSA in PBS for 1 h at room temperature and incubated overnight with primary antibody diluted in 10% BSA specific for mouse anti-αSMA (1:250, Sigma, A-2547), rat anti-PDGFRβ (1:50, eBioscience, 16-1402), rabbit anti-NG2 (1:100, Abcam, ab-5320), rabbit anti-CD146 (1:100, Abcam, ab-75769), rat anti-nestin (1:100, Abcam, ab-6142), rabbit anti-calponin (1:100, Abcam, ab-46794), and rat anti-TrkA (1:250, R&D Systems, AF-1056-SP). Samples were washed three times with PBS containing 0.1% Tween, before a 1-h incubation with the corresponding secondary at half the concentration of the primary antibody. Samples were then incubated with DAPI for 5 min and mounted with Prolong Gold antifade reagent (Invitrogen). Confocal microscopy was performed using a Nikon eclipse Ti.
Protein was isolated and run as previously described (25). Briefly, cells were lysed using RIPA buffer [50 mM Tris·HCl (pH 8), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS], protease inhibitor (cOmplete, Mini Protease Inhibitor Cocktail, Roche, 11836153001), and phosphatase inhibitor (phosSTOP, Roche, 4906845001). Protein (5–10 µg) was loaded and separated on a 10% polyacrylamide gel and transferred to a PDVF membrane (Immobilon-P, Millipore). The membranes were blocked for 1 h with 5% milk-TBS with Tween 20 (TBS-T) and incubated overnight with primary antibodies specific to mouse anti-αSMA (1:1,000, Sigma, A2547), mouse anti-SV40 (1:500, EMD Millipore, MABF121), and rabbit anti-GAPDH (1:4,000, Bethyl Laboratories, no. A300-641A). Following incubation, membranes were washed with TBS-T and probed with horseradish peroxidase-linked secondary antibodies (1:4,000, Dako; rabbit secondary, P0448; mouse secondary, P0447) for 1 h at room temperature and then visualized using SuperSignal West Femto (Thermo Fisher, 34095).
Statistical Analysis
Data are presented as mean ± SD. Comparison of two groups was performed using unpaired t-test. Paired t-test was used for comparison of repeated measured in the same group. Statistical analyses were performed using GraphPad Prism 5.0c (GraphPad Software, San Diego, CA). P < 0.05 was considered significant.
RESULTS
Generation and characterization of mouse Gli1+ cell lines
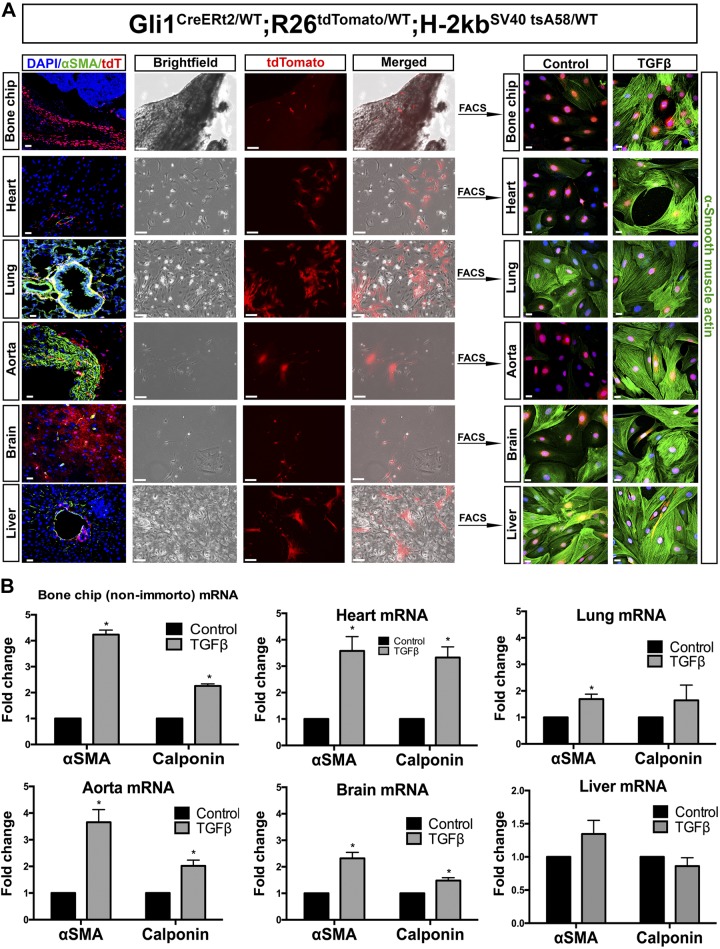
To generate conditionally immortalized pericyte cell lines, we crossed bigenic Gli1-CreERt2; R26tdTomato/WT mice to the “immortomouse” (H-2kbSV40tsA58/WT), which harbors a temperature-sensitive SV40 allele (17). Tamoxifen was administered to 8-wk-old mice, and 2 days later, Gli1-tdTomato expression could be observed predominantly in a perivascular localization in many organs, including bone, heart, aorta, liver, lung, and brain (Fig. 1A). Using the same tamoxifen dosing protocol, another group of mice were euthanized, perfused with cold PBS, and organs were harvested for digestion. After 24 h, many different cell types attached to the plates (Fig. 1A), including cells expressing tdTomato. The tdTomato-positive cells were sorted by fluorescence-activated cell sorting (FACS) to produce a pure cell culture from bone, heart, aorta, liver, lung, and brain. Bone-, heart-, aorta-, and brain-derived Gli1-tdTomato cells all responded to TGF-β by upregulating αSMA, a marker of myofibroblast differentiation (Fig. 1A). Lung- and liver-derived Gli1 cells did not show increased levels of αSMA, due to high basal levels of the protein (Fig. 1A). Despite successful culture of Gli1-tdTomato cells from other organs, kidney Gli1 cells did not survive sorting and could not be passaged, likely because of the very low number of Gli1 cells present in the normal mouse kidney (23). One reason for this is that Gli1-positive cells are predominantly localized to the outer medulla with very few cells in cortex (Fig. 2A).
Fig. 1.
Gli1 labels myofibroblast progenitors across multiple organs. A: tdTomato-labeled Gli1+ cells from Gli1-CreERt2; R26tdTomato/DTR-LoxP; H-2kbSV40tsA58/WT mice were isolated by FACS from bone chip, heart, lung, aorta, brain, and liver. All cells either had endogenous expression of αSMA or could differentiate into myofibroblasts after treatment with TGF-β. Scale bars on organ samples and fluorescent images of cells (columns 1, 5, and 6) represent 20 µm, whereas scale bars on phase contrast (columns 2, 3, and 4) represent 100 µm. B: quantitative PCR analysis of the primary cell cultures show induction of αSMA in all organ-derived Gli1 cells except lung. Each cell type was tested for myofibroblast differentiation by treatment with 10 ng/ml TGF-β for 24 h. *P < 0.05, unpaired Student’s t-test. Gli1, glioma-associated oncogene homolog-1; TGF-β, transforming growth factor-β.
Fig. 2.
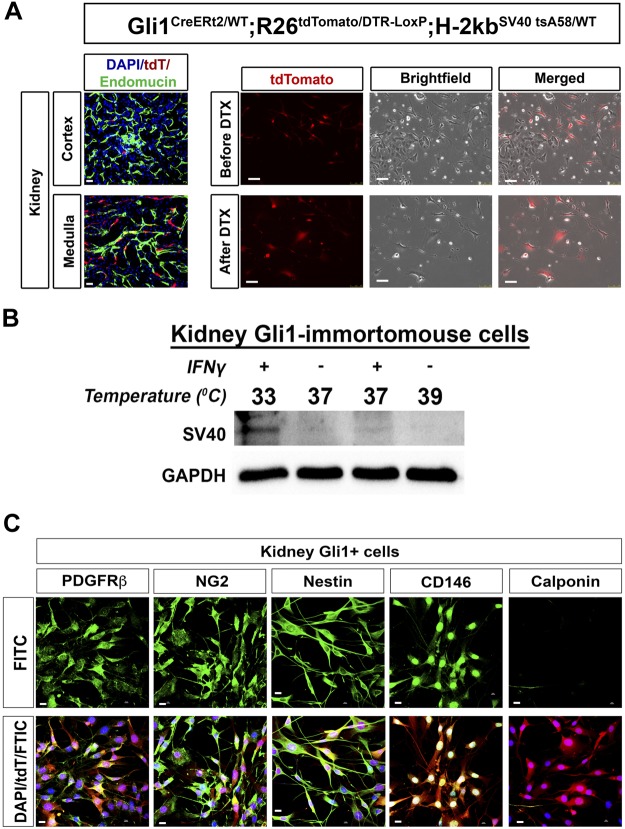
Creation and characterization of Kidney-Gli1 (KGli1) cell lines. A: Gli1 is expressed mainly in the corticomedullary region, adjacent to endomucin-positive endothelial cells. Quadruple transgenic mice (Gli1-CreERt2; R26tdTomato/DTR-LoxP; H-2kbSV40tsA58/WT) were given tamoxifen at 6 wk old to activate cre-recombinase. Mice were then euthanized, and kidneys were harvested, minced up, and plated out on cell culture plates. After treatment with diphtheria toxin, all non-Gli1 expressing cells (non-tdTomato cells) died off, leaving only Gli1 positive cells. Scale bars on tissue samples represent 20 µm, whereas scale bars on cells represent 100 µm. B: SV40 expression can be activated by culturing of cells at 33°C in the presence of IFNγ and turned off at 39°C. Low levels of SV40 remain when cultured at 37°C with IFNγ, with almost no expression seen at 37°C without IFNγ. C: Gli1 cells are positive for PDGFRβ, CD146, nestin, and NG2 and lack expression of calponin, similar to in vivo. DTX, diphtheria toxin; Gli1, glioma-associated oncogene homolog-1.
To circumvent this issue, Gli1-CreERt2; R26tdTomato/WT; H-2kbSV40tsA58/WT mice were crossed to the terminator mouse in which the diphtheria toxin receptor is inserted into the R26 locus preceded by a floxed stop sequence (10). The resulting Gli1-CreERt2; R26tdTomato/DTR-LoxP; H-2kbSV40tsA58/WT mice were treated with tamoxifen and kidney Gli1-tdTomato cells harvested as before but treated with diphtheria toxin (100 ng/ml) in vitro to eliminate non-recombined cells. After diphtheria toxin treatment, all non-Gli1 cells died, and the remaining Gli1+ were allowed to expand. Once enough Gli1+ cells were 50% confluent, they were FACS purified to eliminate any remaining non-Gli1+ cells, resulting in a pure culture of KGli1 cells (Fig. 2A).
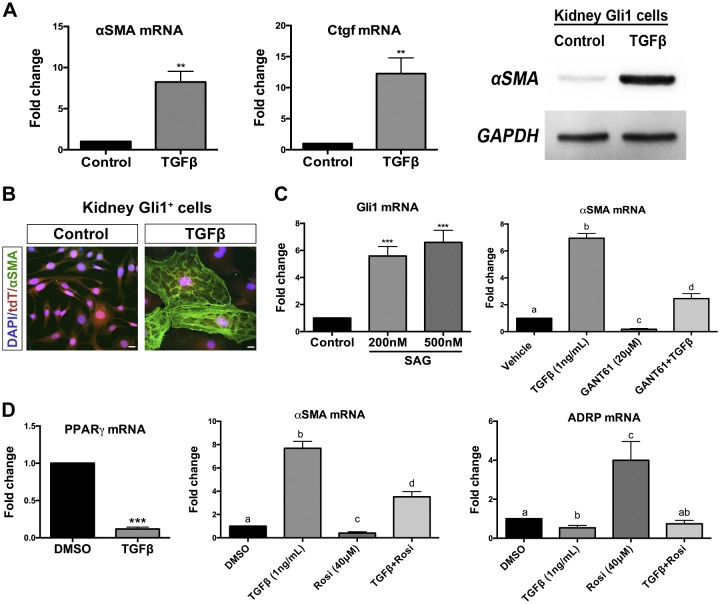
As expected, SV40 was strongly expressed at the permissive temperature of 33°C in the presence of IFN-γ, but it was undetectable at 39°C in the absence of IFN-γ. At 37°C in the absence of IFN-γ, little to no expression of SV40 was observed, and so all experiments were performed under these conditions (Fig. 2B). KGli1 cells maintain expression of mesenchymal stromal cell markers such as PDGFRβ, CD146, nestin, and NG2, while lacking expression of vascular smooth muscle cell marker calponin (Fig. 2C). These cells can be induced to robustly differentiate into myofibroblasts by treatment with TGF-β, with increased mRNA and protein expression of αSMA seen after 24 h and by immunofluorescence after 72 h (Fig. 3, A and B). KGli1 cells also responded to hedgehog agonists, with increased Gli1 mRNA observed after treatment withsmoothened agonist (SAG; Fig. 3C). To test fibrosis inhibition in these cells, we tested two known inhibitors of fibrosis, GANT61 and rosiglitazone. GANT61 is a hedgehog pathway inhibitor previously reported to reduce interstitial fibrosis in UUO models (20). Rosiglitazone is a PPARγ activator and has been shown to attenuate TGF-β-induced lung fibrosis (6). Both inhibitors reduced TGF-β-mediated fibrosis in our cell line, demonstrating that this cell line can be used to test novel antifibrotic therapeutics (Fig. 3, C and D).
Fig. 3.
Kidney Gli1 cells are myofibroblast progenitors. A: kidney Gli1 cells differentiate into myofibroblasts in vitro by treatment with TGF-β. Myofibroblast expression is characterized by increases in αSMA and Ctgf. Increased αSMA is also seen on a proteins level. B: TGF-β induces αSMA protein in association with the actin cytoskeleton. C: kidney Gli1 cells respond to the smoothened agonist, SAG, in vitro after 24 h. A Gli protein inhibitor, GANT61, blocks TGF-β-induced myofibroblast induction, similar to in vivo. D: TGF-β blocks expression of PPARγ, whereas PPARγ agonist rosiglitazone (Rosi) suppresses TGF-β-induced differentiation. ADRP is a PPARγ target gene, which is still repressed by TGF-β. Scale bar, 20 µm. **P < 0.01, ***P < 0.001; statistically significant differences are denoted by letters, with P < 0.05. Gli1, glioma-associated oncogene homolog-1; TGF-β, transforming growth factor-β.
Kidney Gli1 cells stabilize vasculogenesis
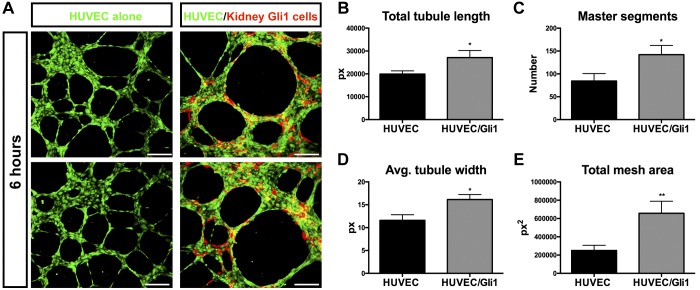
During homeostasis, Gli1 pericyte cells stabilize capillaries. Loss of Gli1 cells leads to capillary rarefaction and tubular injury (24). Therefore, we next asked if KGli1 cells could stabilize endothelial tube formation in a coculture assay with HUVEC. KGli1 cells closely associated with endothelial cells in vitro (Fig. 4A), as they do in the perivascular niche in vivo (Fig. 2A). Indeed, cocultures on two-dimensional Matrigel of HUVEC and kidney Gli1 cells led to increased tubule length and width after 6 h compared with HUVEC alone. Additionally, tubule integrity was also increased. The number of master segments and junctions was significantly increased, as was total mesh area and master segment length (Fig. 4B). During a 6-h time lapse movie of the coculture assay, the tdTomato-positive KGli1 cells can be seen coordinating and recruiting the green fluorescent HUVEC cells (Supplemental Video S1; Supplemental Material for this article is available online at the Journal website). These results provide evidence for a functional role for KGli1 cells in stabilizing capillaries, consistent with their known effects on kidney microvasculature in vivo (24).
Fig. 4.
Kidney Gli1 cells stabilize vasculogenesis. A: two-dimensional cocultures of HUVEC and Gli1 cells form capillary-like structures on Matrigel. Gli1 cells migrate and associate closely with HUVEC cells, similar to in vivo. In addition, Gli1 cells help to stabilize and improve vasculature networks after 6 h of coculturing (B–E, see materials and methods Coculture Assays). *P < 0.05, **P < 0.01, unpaired Student’s t-test between untreated and treated. Scale bar, 200 µm. Gli1, glioma-associated oncogene homolog-1; HUVEC, human umbilical vein endothelial cells; px, pixels.
Single-cell transcriptomic analysis of myofibroblast differentiation
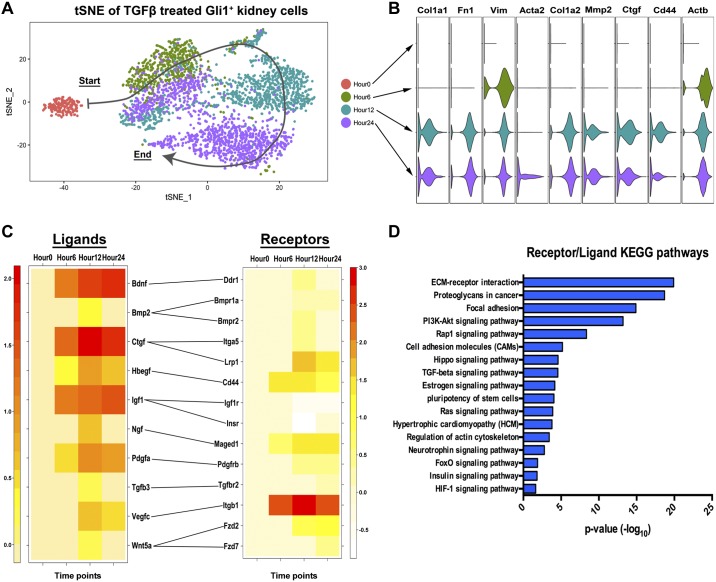
We next sought to assess myofibroblast differentiation in a comprehensive and unbiased manner, so we used single cell RNA-sequencing (scRNA-seq) to track KGli1 cell differentiation after exposure to TGF-β. KGli1 cells were treated with 1 ng/ml TGF-β and the following time points collected for scRNA-seq: 0, 6, 12, and 24 h after exposure. The final cell numbers passing quality filters ranged from 192 (time 0) to 1,238 (24 h). The number of unique molecular identifiers per cell, unique genes per cell, and mitochondrial gene percentage are summarized in Table 1. Using principal component analysis, followed by dimensionality reduction by t-distributed stochastic neighbor embedding (tSNE), weplotted individual cells based on their gene expression profile (Fig. 5A). Differential gene expression analysis of fibrotic genes showed increasing expression over time of Col1a1, Fn1, Vim, Acta2, Col1a2, Mmp2, Ctgf, Cd44, and Actb (Fig. 5B). There were marked differences in expression kinetics. Acta2, which encodes αSMA, was expressed only at 24 h, whereas Vim and Actb were each expressed after only 6 h.
Table 1.
scRNA-seq metrics
| 0 h | 6 h | 12 h | 24 h | |
|---|---|---|---|---|
| Cell, n | 192 | 658 | 1,120 | 1,328 |
| nUMI | 557 | 1,184 | 5,079 | 4,644 |
| nGene | 313 | 509 | 2,086 | 1,826 |
| Mito.percent | 3.4% | 3.2% | 2.4% | 2.2% |
Mito.percent, percentage of mitochondrial genes per cell; nGene, no. of unique genes per cell; nUMI, no. of unique molecular identifiers per cell; scRNA-seq, single-cell RNA sequencing.
Fig. 5.
TGF-β-treated kidney-derived Gli1 cell single-cell RNA sequencing shows increased myofibroblast marker gene expression. A: tSNE plot of hour 0 (red), hour 6 (green), hour 12 (teal) and hour 24 (purple) shows clear separation of hour 0 to all three time points, with a directional differentiation pattern (arrow). B: violin plots of fibrosis-related genes showing an increase in fibrotic gene signature over time. C: heatmap of ligands and receptors upregulated in all TGF-β-treated samples. Receptor list includes receptors, pseudo-receptors, and computer predicted binding receptors. D: KEGG pathway analysis of all receptors and ligands identified from hour 6, hour 12, and hour 24 combined. Scale is represented as the −log10 of the P value. *P < 0.05. ECM, extracellular matrix; Gli1, glioma-associated oncogene homolog-1; TGF-β, transforming growth factor-β; tSNE, t-distributed stochastic neighbor embedding.
We performed a similar differential gene expression analysis on receptor-ligand pairs to assess potential autocrine signaling (Fig. 5C) (30). Several ligands associated with fibrosis were identified (Bmp2, Ctgf, Hbegf, Igf1,Tgfb3), along with novel ligands (Bdnf, Ngf, Vegfc, Pdgfa, Wnt5a). KEGG pathway analysis of the receptor/ligand pair showed that the most altered signaling pathway was extracellular matrix (ECM) signaling and focal adhesion (Fig. 5D). The focal adhesion pathway has been heavily linked to fibrosis and myofibroblast formation (25). Other fibrosis-associated pathways were identified, including the hippo pathway (33), estrogen signaling (26), and insulin/insulin-like growth factor signaling (1) (Fig. 5D). Unexpectedly, the neurotrophin signaling pathway was identified as being activated during differentiation (Fig. 5, C and D). This signaling pathway includes brain-derived neurotropic factor (Bdnf) and neurite growth factor (Ngf), genes with no known role in kidney.
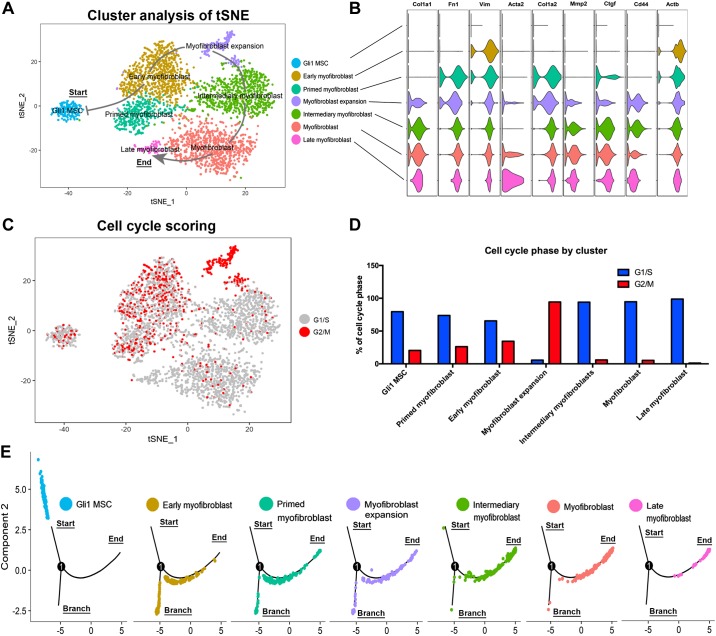
We next performed unsupervised cluster analysis on the combined time points using the Seurat package (3). This yielded seven distinct clusters (Fig. 6A). Based on expression levels of myofibroblast markers (Fig. 6B), we annotated the clusters as “Gli1 mesenchymal stromal cells (Gli1 MSC),” “early myofibroblast,” “primed myofibroblasts,” “myofibroblast expansion,” “intermediary myofibroblasts,” “myofibroblasts,” and “late myofibroblasts” (Fig. 6A). KGli1 cells show little to no expression of fibrotic markers at baseline, and thus are in an undifferentiated state. Early myofibroblasts show increased expression of mesenchymal markers (Vim and Actb) before proceeding to a “primed” state where they express many more of the fibrotic markers, but at low levels. Next, the cells transition through a stage of proliferation termed “myofibroblast expansion.” Cells in this cluster convert to a high level of G2/M phase of the cell cycle, along with expression of proliferation markers. Cells going from Gli1 MSC to myofibroblast expansion show an increasing level of transition from quiescent G1/S to G2/M (Fig. 6, C and D). These results suggest that myofibroblast differentiation proceeds through a proliferative expansion phase before becoming fully differentiated. Once the cells exit the expansion phase, nearly all cells become quiescent in G1/S and continue to differentiate, demonstrating increased levels of all fibrotic markers until they reach the fully differentiated “late myofibroblast” stage (Fig. 6, A and D). Pseudo-temporal ordering validates this progression by showing cells from the individual stages of differentiation advancing towards the end of the pseudo-temporal tree (Fig. 6E). Of note, myofibroblast proliferation in vivo is also characterized by an early peak at 48 h followed by reduced proliferation in the unilateral ureteral obstruction (UUO) model of renal fibrosis (27), suggesting that KGli1 cells accurately model myofibroblast transition.
Fig. 6.
Unsupervised clustering of cells identifies a critical myofibroblast expansion phase before terminal differentiation. A: tSNE plot of the unsupervised cluster analysis. Seven distinct clusters were identified and annotated based on their gene expression. Arrow indicates the directionality of differentiation through the various stages of myofibroblast differentiation. B: fibrotic gene expression of the various clusters. C: bioinformatic cell cycle scoring of the clusters identifies the myofibroblast expansion cluster as being almost completely in a proliferative stage. D: percentage of cells within the cluster that are in either G1/S or G2/M phase. E: breakdown of each cluster in pseudo-time. Differentiation goes from the start (Gli1 MSC) through to the end (late myofibroblasts) with one branch point corresponding to an undifferentiated stage. Gli1, glioma-associated oncogene homolog-1; Gli1 MSC, Gli1 mesenchymal stromal cells; tSNE, t-distributed stochastic neighbor embedding.
Focal adhesion pathway activation drives myofibroblast differentiation
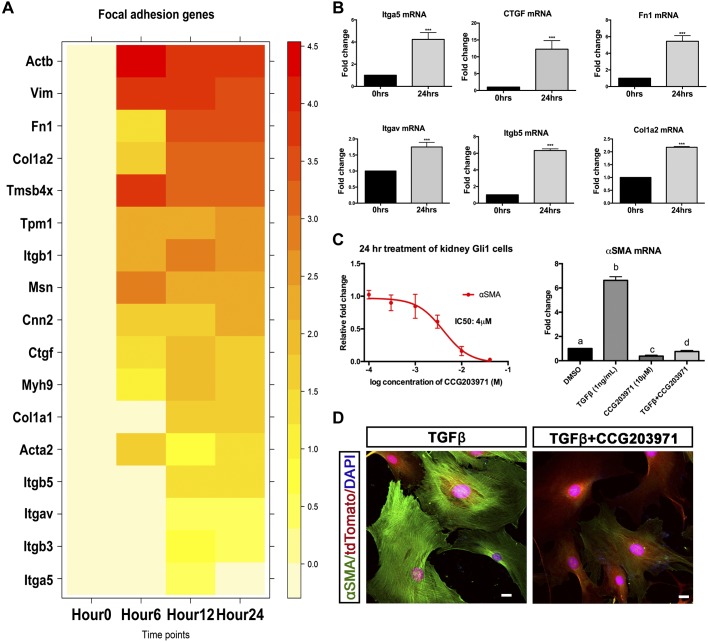
We next asked whether the results from scRNA-seq could be used to identify novel antifibrotic strategies. The most upregulated pathway identified from the receptor/ligand heatmap was ECM receptor interaction and focal adhesion (Fig. 5D). Focal adhesions occur when ECM proteoglycans interact and connect with the internal actin cytoskeleton. Focal adhesion kinase is a key regulator of cell shape, motility, and scaffolding (38). Although this pathway has been linked to fibrosis in lung and other fibrotic diseases, few reports have investigated its role in kidney fibrosis (9, 12). Serum response factor (SRF) is a transcription factor regulating cell growth and survival that integrates focal adhesion signaling (4). Indeed, SRF and myocardin (MRTF) have been hypothesized to be myofibroblast molecular switch in models of skin fibrosis (11). To determine whether this potential master regulator was responsible for renal-derived myofibroblasts, we cross-referenced the differential gene analysis of all 3 points with a list of 196 genes known to be targets of SRF/MRTF transcription (7) (Fig. 7A). One hundred and six out of the 196 genes were differentially expressed at hour 12, with similar results seen for hour 24 (Fig. 7A). qPCR analysis of a subset of these genes verified that focal adhesion is indeed increased in KGli1 myofibroblasts (Fig. 7B). These genes included various integrin receptors (Itga5, Itgav, and Itgb5) along with signaling/ECM proteins (Ctgf, Fn1, and Col1a2) (Fig. 7B). To verify the role of this pathway in fibrosis development, an SRF inhibitor, CCG-203971, was tested in the fibrosis assay. CCG-203971 has been shown to decrease fibrosis in both lung fibrosis and systemic sclerosis models (11, 34). CCG-203971 demonstrated a dose-dependent inhibition of αSMA induction by TGF-β, with an IC50 of 4 µM (Fig. 7C). This decrease in mRNA expression was validated by immunofluorescent staining of cells treated with 1 ng/ml of TGF-β (Fig. 7D). The addition of 10 µM of CCG-203971 blocked TGF-β-induced αSMA protein expression (Fig. 7D). These data implicate SRF or downstream genes as a therapeutic target in kidney fibrosis.
Fig. 7.
Focal adhesion pathway activation drives myofibroblast differentiation. A: heatmap of a select number of genes regulated by serum response factor (SRF) that are involved in focal adhesion from all time points. Expression levels are log fold change compared with hour 0. B: validation of a subset of SRF target genes by quantitative PCR in kidney Gli1 cells. C: dose response of CCG-203971 in kidney Gli1 cells treated with 1 ng/ml of TGF-β to determine therapeutic dosage. A 10 µM concentration of CCG-203971 (−log 2 M) was chosen and had the ability to block TGF-β-induced expression of αSMA. D: fluorescent staining of cells treated with either 1 ng/ml TGF-β alone or 1 ng/ml TGF-β and 10 µM CCG-203971 for 72 h. ***P < 0.001; statistically significant differences are denoted by letters, with P < 0.05. Gli1, glioma-associated oncogene homolog-1; TGF-β, transforming growth factor-β.
Ngf and TrkA receptor expression during fibrosis
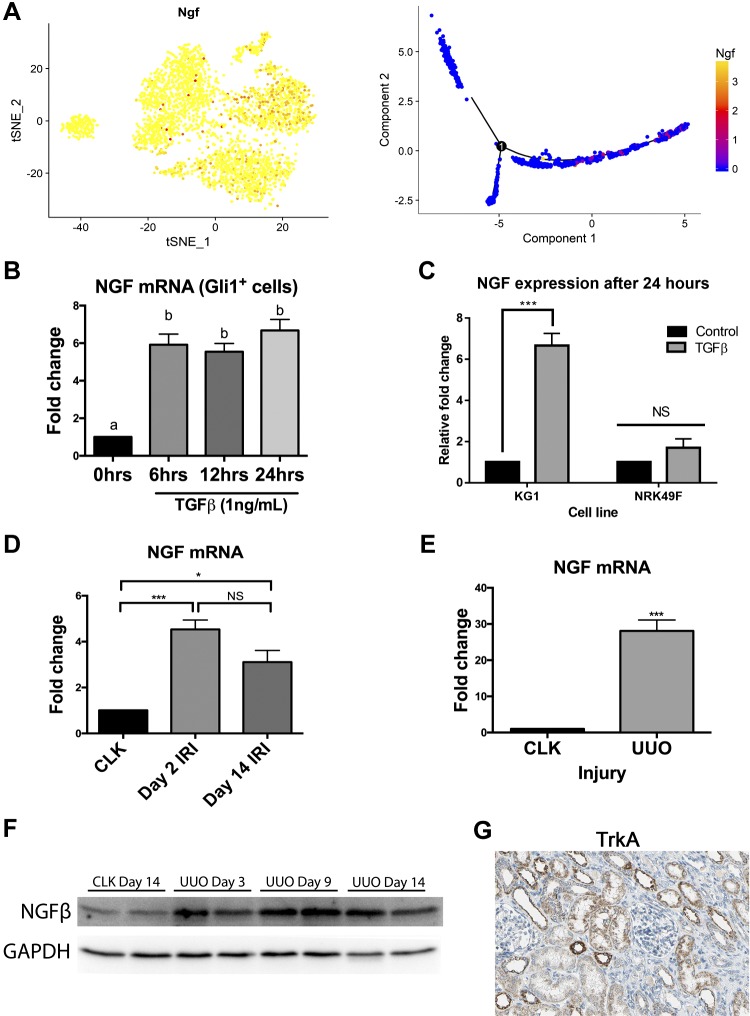
Ngf has never been associated with kidney fibrosis, but our scRNA-seq analysis identified this growth factor as being strongly upregulated in differentiating KGli1 cells. Ngf regulates and promotes neuronal survival (31). Further analysis of Ngf expression kinetics revealed that its expression peaks at hour 12 and continues to be expressed through terminal differentiation (hour 24) (Fig. 8A). Bulk qPCR analysis of KGli1 cells confirmed strong upregulation of Ngf by TGF-β at all three time points (Fig. 8B). The NRK49F cell line is a rat kidney fibroblast cell line that was grown out of a mixed culture of epithelial, mesenchymal, and fibroblast cells that is frequently used to model kidney myofibroblast activation (16). We asked whether Ngf was also upregulated by TGF-β in NRK49F cells, but unexpectedly it was not, despite robust αSMA induction (Fig. 8C). In at least this respect, this result suggests that KGli1 cells more faithfully model myofibroblast transition in kidney. We validated that Ngf is also upregulated in vivo in two different kidney fibrosis models. In unilateral ischemia-reperfusion injury, we observed a fourfold upregulation of Ngf mRNA at day 2, with levels remaining elevated at day 14 (Fig. 8D). In UUO, the upregulation was much more pronounced, with nearly 30-fold induction at day 10 compared with control kidney (Fig. 8E). We could also detect increased Ngf protein levels during the UUO time course (Fig. 8F). Finally, the high affinity Ngf receptor TrkA (13) has been reported to be expressed in tubules (2), and we could verify TrkA expression in healthy human kidney epithelia (Fig. 8G). The upregulation of Ngf in interstitial Gli1-positive mesenchymal progenitors undergoing myofibroblast differentiation, with expression of its receptor TrkA in adjacent epithelia, suggests a possible novel myofibroblast-epithelia Ngf signaling axis in injury.
Fig. 8.
Ngf is upregulated in kidney myofibroblasts after TGF-β in vitro and after fibrotic injury in vivo. A: tSNE plot showing expression levels of Ngf RNA across all time points. Ngf showed the biggest increase at hour 12. Pseudo-temporal ordering shows Ngf expression turning on the most in the latter stages of differentiation. B: quantitative PCR confirmation of increased Ngf mRNA expression after treatment of KGli1 cells with TGF-β. C: rat kidney fibroblast cell line does not upregulate NGF in response to TGF-β. D: increased NGF mRNA expression after unilateral ischemia-reperfusion injury (IRI). E: increased Ngf expression during acute kidney injury after unilateral ureteral obstruction (UUO). F: Ngf protein expression is also increased after UUO. G: TrkA protein is expressed in tubular epithelium in human kidney. *P < 0.05, ***P < 0.001; statistically significant differences are denoted by letters, with P < 0.05. Gli1, glioma-associated oncogene homolog-1; KGli1, kidney-Gli1; NGF, nerve growth factor; NS, not significant; TGF-β, transforming growth factor-β; tSNE, t-distributed stochastic neighbor embedding. [Image from The Human Protein Atlas (version 18; https://www.proteinatlas.org) with permission under Creative Commons Attribution-ShareAlike 3.0 International License.]
DISCUSSION
Recent evidence has focused attention on kidney Gli1-positive mesenchymal stem cell-like cells as the predominant myofibroblast progenitor population in fibrotic disease (14, 15, 22, 23). In this study, we generated a quadruple transgenic mouse to overcome limitations we encountered in isolating and culturing adult Gli1+ kidney pericytes. The resulting cell line, KGli1, maintains similar marker expression to its in vivo counterparts, such as NG2 and PDGFRβ. It also undergoes myofibroblast differentiation in response to treatment with TGF-β. Importantly, KGli1 cells also show vascular stabilizing properties, an important functional property of kidney pericytes (19). Because Gli1+ mesenchymal stem cell-like cells are located in both the pericyte and perivascular niche in kidney, we cannot be sure from which niche KGli1 cells originated. Certainly, the vascular stabilizing property of the line is consistent with a pericyte origin; however, it is unclear whether perivascular Gli1+ fibroblasts might possess this property as well. The absence of any known markers to distinguish between these two cell types, aside from their location (pericyte versus perivascular), makes testing this possibility impossible at present.
To comprehensively describe the transcriptional landscape of myofibroblast differentiation, we performed scRNA-seq of KGli1 cells during TGF-β-induced differentiation. This analysis revealed several insights. It demonstrated that KGli1 cells undergo a discrete phase of proliferative expansion soon after TGF-β exposure, followed by quiescence and upregulation of myofibroblast markers, similar to what has been described in vivo (27). It also revealed that acquisition of myofibroblast markers is a discontinuous process; some markers are upregulated very early, such as Vim and Actb, next are genes such as Col1a1, Fn1, and Mmp2, and finally Acta2 is upregulated very late. This suggests that multiple different signaling pathways regulate expression of myofibroblast genes. Our single-cell analysis also revealed a host of transcription factors, receptors, and ligands whose expression changes during myofibroblast differentiation. The most highly upregulated pathways included extracellular matrix and focal adhesion signaling. Based on this analysis, we validated SRF as a novel therapeutic target in kidney myofibroblasts because blocking SRF-induced signaling could attenuate the fibrotic response induced by TGF-β.
The upregulation of Ngf in KGli1 cells by TGF-β was unexpected. One study of human kidney biopsies with a variety of diseases reported NGF expression in tubular and glomerular cells, but the detection method was limited to immunohistochemistry and has not been subsequently verified (2). Although our study did not evaluate possible expression of Ngf in cell types other than kidney pericytes, the fact that we could confirm Ngf mRNA and protein upregulation in two different mouse models of kidney fibrosis, in addition to our in vitro myofibroblast differentiation model, provides support for our conclusion that this growth factor is strongly induced during injury-induced kidney myofibroblast differentiation. The failure of TGF-β to upregulate Ngf in NRK49F kidney fibroblasts indicates that KGli1 cells are the better cell culture model of Ngf signaling in kidney fibrosis. It also supports the notion that not all kidney mesenchymal cells are equivalent; subsets of interstitial cell types with distinct properties and potential exist within the kidney interstitium. Finally, presence of TrkA receptor in adjacent tubules indicates a possible paracrine Ngf signaling pathway in fibrosis.
In summary, we have developed a new tool for the study of kidney myofibroblast activation. Increasing evidence supports an important role for mesenchymal stem cell-like progenitors in myofibroblast differentiation during CKD and also in mediating tubule-interstitial crosstalk in acute injury (36). We believe this cell line will serve as a useful model to provide molecular insight into roles for kidney myofibroblast progenitors in health and disease.
GRANTS
Primary support for this work was from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Re-Building a Kidney consortium Grant DK107374. Additional support was from the Chan Zuckerberg Initiative Grant 173970, NIH/NIDDK Grant DK103740, and by the NIDDK Diabetic Complications Consortium (DiaComp, http://www.diacomp.org/) Grants DK076169 and DK115255 (all to B. D. Humphreys).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
E.Ó. and B.D.H. conceived and designed research; E.Ó., H.W., Y.M., E.L.D., F.G.M., L.X.F., and M.C.-P. performed experiments; E.Ó., H.W., Y.M., E.L.D., F.G.M., M.C.-P., and B.D.H. analyzed data; E.Ó., H.W., and B.D.H. interpreted results of experiments; E.Ó. and B.D.H. prepared figures; E.Ó. and B.D.H. drafted manuscript; E.Ó. and B.D.H. edited and revised manuscript; E.Ó., H.W., Y.M., F.G.M., M.C.-P., and B.D.H. approved final version of manuscript.
Supplemental Data
REFERENCES
- 1.Bach LA, Hale LJ. Insulin-like growth factors and kidney disease. Am J Kidney Dis 65: 327–336, 2015. doi: 10.1053/j.ajkd.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 2.Bonofiglio R, Antonucci MT, Papalia T, Romeo F, Capocasale G, Caroleo MC, Di Fausto V, Aloe L. Nerve growth factor (NGF) and NGF-receptor expression in diseased human kidneys. J Nephrol 20: 186–195, 2007. [PubMed] [Google Scholar]
- 3.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420, 2018. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai J, Tarnawski AS. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol 53: 147–157, 2002. [PubMed] [Google Scholar]
- 5.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of chronic kidney disease in the United States. JAMA 298: 2038–2047, 2007. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 6.El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Szibor M, Kosanovic D, Schwind F, Schermuly RT, Henneke I, MacKenzie B, Quantius J, Herold S, Ntokou A, Ahlbrecht K, Braun T, Morty RE, Günther A, Seeger W, Bellusci S. Two-way conversion between lipogenic and myogenic fibroblastic phenotypes marks the progression and resolution of lung fibrosis. Cell Stem Cell 20: 571, 2017. [Erratum in Cell Stem Cell 20: P571, 2017]. doi: 10.1016/j.stem.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 28: 943–958, 2014. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian SL, Penchev RR, St-Jacques B, Rao AN, Sipilä P, West KA, McMahon AP, Humphreys BD. Hedgehog-Gli pathway activation during kidney fibrosis. Am J Pathol 180: 1441–1453, 2012. doi: 10.1016/j.ajpath.2011.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan GP, Wang W, Zhao H, Cai L, Zhang PD, Yang ZH, Zhang J, Wang X. Pharmacological inhibition of focal adhesion kinase attenuates cardiac fibrosis in mice cardiac fibroblast and post-myocardial-infarction models. Cell Physiol Biochem 37: 515–526, 2015. doi: 10.1159/000430373. [DOI] [PubMed] [Google Scholar]
- 10.Guo JK, Shi H, Koraishy F, Marlier A, Ding Z, Shan A, Cantley LG. The Terminator mouse is a diphtheria toxin-receptor knock-in mouse strain for rapid and efficient enrichment of desired cell lineages. Kidney Int 84: 1041–1046, 2013. doi: 10.1038/ki.2013.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haak AJ, Tsou PS, Amin MA, Ruth JH, Campbell P, Fox DA, Khanna D, Larsen SD, Neubig RR. Targeting the myofibroblast genetic switch: inhibitors of myocardin-related transcription factor/serum response factor-regulated gene transcription prevent fibrosis in a murine model of skin injury. J Pharmacol Exp Ther 349: 480–486, 2014. doi: 10.1124/jpet.114.213520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao S, Shen H, Hou Y, Mars WM, Liu Y. tPA is a potent mitogen for renal interstitial fibroblasts: role of beta1 integrin/focal adhesion kinase signaling. Am J Pathol 177: 1164–1175, 2010. doi: 10.2353/ajpath.2010.091269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72: 609–642, 2003. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 14.Humphreys BD. Mechanisms of renal fibrosis. Annu Rev Physiol 80: 309–326, 2018. doi: 10.1146/annurev-physiol-022516-034227. [DOI] [PubMed] [Google Scholar]
- 15.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176: 85–97, 2010. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huu Duc-Nguyen, Rosenblum EN, Zeigel RF. Persistent infection of a rat kidney cell line with Rauscher murine leukemia virus. J Bacteriol 92: 1133–1140, 1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jat PS, Noble MD, Ataliotis P, Tanaka Y, Yannoutsos N, Larsen L, Kioussis D. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc Natl Acad Sci USA 88: 5096–5100, 1991. doi: 10.1073/pnas.88.12.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz A, Caramori ML, Sisson-Ross S, Groppoli T, Basgen JM, Mauer M. An increase in the cell component of the cortical interstitium antedates interstitial fibrosis in type 1 diabetic patients. Kidney Int 61: 2058–2065, 2002. doi: 10.1046/j.1523-1755.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- 18a.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 25: 1860–1872, 2015. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramann R, DiRocco DP, Humphreys BD. Understanding the origin, activation and regulation of matrix-producing myofibroblasts for treatment of fibrotic disease. J Pathol 231: 273–289, 2013. doi: 10.1002/path.4253. [DOI] [PubMed] [Google Scholar]
- 20.Kramann R, Fleig SV, Schneider RK, Fabian SL, DiRocco DP, Maarouf O, Wongboonsin J, Ikeda Y, Heckl D, Chang SL, Rennke HG, Waikar SS, Humphreys BD. Pharmacological GLI2 inhibition prevents myofibroblast cell-cycle progression and reduces kidney fibrosis. J Clin Invest 125: 2935–2951, 2015. doi: 10.1172/JCI74929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramann R, Goettsch C, Wongboonsin J, Iwata H, Schneider RK, Kuppe C, Kaesler N, Chang-Panesso M, Machado FG, Gratwohl S, Madhurima K, Hutcheson JD, Jain S, Aikawa E, Humphreys BD. Adventitial MSC-like cells are progenitors of vascular smooth muscle cells and drive vascular calcification in chronic kidney disease. Cell Stem Cell 19: 628–642, 2016. doi: 10.1016/j.stem.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramann R, Machado F, Wu H, Kusaba T, Hoeft K, Schneider RK, Humphreys BD. Parabiosis and single-cell RNA sequencing reveal a limited contribution of monocytes to myofibroblasts in kidney fibrosis. JCI Insight 3: e99561, 2018. doi: 10.1172/jci.insight.99561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramann R, Schneider RK, DiRocco DP, Machado F, Fleig S, Bondzie PA, Henderson JM, Ebert BL, Humphreys BD. Perivascular Gli1+ progenitors are key contributors to injury-induced organ fibrosis. Cell Stem Cell 16: 51–66, 2015. doi: 10.1016/j.stem.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramann R, Wongboonsin J, Chang-Panesso M, Machado FG, Humphreys BD. Gli1+ pericyte loss induces capillary rarefaction and proximal tubular injury. J Am Soc Nephrol 28: 776–784, 2017. doi: 10.1681/ASN.2016030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagares D, Busnadiego O, García-Fernández RA, Kapoor M, Liu S, Carter DE, Abraham D, Shi-Wen X, Carreira P, Fontaine BA, Shea BS, Tager AM, Leask A, Lamas S, Rodríguez-Pascual F. Inhibition of focal adhesion kinase prevents experimental lung fibrosis and myofibroblast formation. Arthritis Rheum 64: 1653–1664, 2012. doi: 10.1002/art.33482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane PH. Estrogen receptors in the kidney: lessons from genetically altered mice. Gend Med 5, Suppl A: S11–S18, 2008. doi: 10.1016/j.genm.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Lin SL, Kisseleva T, Brenner DA, Duffield JS. Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am J Pathol 173: 1617–1627, 2008. doi: 10.2353/ajpath.2008.080433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214, 2015. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nath KA. Tubulointerstitial changes as a major determinant in the progression of renal damage. Am J Kidney Dis 20: 1–17, 1992. doi: 10.1016/S0272-6386(12)80312-X. [DOI] [PubMed] [Google Scholar]
- 30.Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, Forrest AR. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6: 7866, 2015. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci 361: 1545–1564, 2006. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risdon RA, Sloper JC, De Wardener HE. Relationship between renal function and histological changes found in renal-biopsy specimens from patients with persistent glomerular nephritis. Lancet 292: 363–366, 1968. doi: 10.1016/S0140-6736(68)90589-8. [DOI] [PubMed] [Google Scholar]
- 33.Seo E, Kim WY, Hur J, Kim H, Nam SA, Choi A, Kim YM, Park SH, Chung C, Kim J, Min S, Myung SJ, Lim DS, Kim YK. The Hippo-Salvador signaling pathway regulates renal tubulointerstitial fibrosis. Sci Rep 6: 31931, 2016. doi: 10.1038/srep31931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sisson TH, Ajayi IO, Subbotina N, Dodi AE, Rodansky ES, Chibucos LN, Kim KK, Keshamouni VG, White ES, Zhou Y, Higgins PD, Larsen SD, Neubig RR, Horowitz JC. Inhibition of myocardin-related transcription factor/serum response factor signaling decreases lung fibrosis and promotes mesenchymal cell apoptosis. Am J Pathol 185: 969–986, 2015. doi: 10.1016/j.ajpath.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiger S, Grill JF, Ma Q, Bäuerle T, Jordan J, Smolle M, Böhland C, Lech M, Anders HJ. Anti-transforming growth factor β IgG elicits a dual effect on calcium oxalate crystallization and progressive nephrocalcinosis-related chronic kidney disease. Front Immunol 9: 619, 2018. doi: 10.3389/fimmu.2018.00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan RJ, Zhou D, Liu Y. Signaling crosstalk between tubular epithelial cells and interstitial fibroblasts after kidney injury. Kidney Dis (Basel) 2: 136–144, 2016. doi: 10.1159/000446336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.U.S. Renal Data System USRDS 2013 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2013. [Google Scholar]
- 38.Wozniak MA, Modzelewska K, Kwong L, Keely PJ. Focal adhesion regulation of cell behavior. Biochim Biophys Acta 1692: 103–119, 2004. doi: 10.1016/j.bbamcr.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, Guo ZK, Jiang XX, Li H, Wang XY, Yao HY, Zhang Y, Mao N. A protocol for isolation and culture of mesenchymal stem cells from mouse compact bone. Nat Protoc 5: 550–560, 2010. doi: 10.1038/nprot.2009.238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.