Abstract
Two-cysteine peroxiredoxins (Prx) have a three-step catalytic cycle consisting of (1) reduction of peroxide and formation of sulfenic acid on the enzyme, (2) condensation of the sulfenic acid with a thiol to form disulfide, also known as resolution, and (3) reduction of the disulfide by a reductant protein. By following changes in protein fluorescence, we have studied the pH dependence of reaction 2 in human peroxiredoxins 1, 2, and 5 and in Salmonella typhimurium AhpC and obtained rate constants for the reaction and pKa values of the thiol and sulfenic acid involved for each system. The observed reaction 2 rate constant spans 2 orders of magnitude, but in all cases, reaction 2 appears to be slow compared to the same reaction in small-molecule systems, making clear the rates are limited by conformational features of the proteins. For each Prx, reaction 2 will become rate-limiting at some critical steady-state concentration of H2O2 producing the accumulation of Prx as sulfenic acid. When this happens, an alternative and faster-resolving Prx (or other peroxidase) may take over the antioxidant role. The accumulation of sulfenic acid Prx at distinct concentrations of H2O2 is embedded in the kinetic limitations of the catalytic cycle and may constitute the basis of a H2O2-mediated redox signal transduction pathway requiring neither inactivation nor posttranslational modification. The differences in the rate constants of resolution among Prx coexisting in the same compartment may partially explain their complementation in antioxidant function and stepwise sensing of H2O2 concentration.
Graphical Abstract
Peroxiredoxins (Prx) are enzymes capable of reducing hydroperoxides in remarkably rapid reactions using one or two cysteine residues as reductants. Reported rate constants for the reduction of H2O2 range from 9 × 103 M−1 s−1 for bacterioferritin comigrating protein1 (now known as PrxQ) to 1 × 108 M−1 s−1 for human peroxiredoxin 2 (PRDX2).2 It has been repeatedly pointed out that this is 4−8 orders of magnitude faster than the same reaction with a low-molecular weight thiol as a reductant,3 and explanations for the accelerated reaction have been provided on the basis of structural approaches4,5 and quantum mechanics/molecular mechanics calculations.3,6,7
Since the discovery of the enzymatic activity of Prx,8,9 the emphasis on its catalytic characterization has been placed mostly on the exceptionally large rate constants of their reactions with hydroperoxides and peroxynitrous acid,10–14 with less attention being paid to the rest of the reactions completing the catalytic cycle.
Among Prx, those that use two cysteine residues (2Cys Prx) have a three-step catalytic cycle described by reactions 1–3. Reduction of peroxide, albeit rapid, is only one of the steps. Situations in which peroxide formation is sustained for a long period of time require the enzymes to go through the whole catalytic cycle repeatedly. In reaction 1, a hydroperoxide is reduced and the peroxidatic cysteine (CP) is oxidized to sulfenic acid, usually with a very high rate constant (kROOH). Reaction 2 is the condensation between the newly formed CP sulfenic acid and a second cysteine thiol (the resolving cysteine or CR). It is called the resolution reaction and is always first-order, even in the case of typical 2Cys Prx, in which CP and CR of the disulfide belong to different subunits, because of the extreme stability of the dimers. Finally, reaction 3 is the reduction of the disulfide formed in the previous step, and the typical reductant is a thioredoxin or a similar thiol oxidoreductase.
| (1) |
| (2) |
| (3) |
One aspect that is often overlooked in the study of Prx is their diversity, which is further complicated by the incomplete quantitative information about their reaction kinetics. As mentioned, the reaction rate constant with H2O2 in the Prx family spans 4 orders of magnitude. The rate constant of reaction 2 (kres) is also different, even between very similar proteins. Finally, reaction 3 is largely understudied with very few direct determinations. The scenario is further complicated if additional phenomena like hyperoxidation, posttranslational modification, and oligomerization equilibria and dynamics are considered. Take for instance the six Prx present in human cells, for which the reported kinetic data, summarized in Table 1, has major gaps in the rate constants of reduction.
Table 1.
Reaction Rate Constants of the Catalytic Cycle of Human Peroxiredoxins
| reaction 1 | reaction 2 | reaction 3 | |||
|---|---|---|---|---|---|
| Prx | localization | kres (s−1) | reductanta | k3(M−1 s−1) | |
| PRDX1 | cytosolic | 3.8 × 107 15 | 915 | Trx1 | ND |
| 1.1 × 108 b | 12.9b | ||||
| PRDX2 | cytosolic | 0.13 to 1 × 108 2 | 2;16 0.2515 | Trx1 | ND |
| 1.6 × 108b | 0.64b | ||||
| PRDX3 | mitochondrial | 2 × 107 17 | 2016 | Trx2 | ND |
| PRDX4 | endoplasmic reticulum | 2.2 × 107 18 | ND | PDI19 | ND |
| PRDX5 | mitochondrial, cytosolic, peroxisomal | 3–4 × 105 3,20 | 1520 | Trx2 | 2 × 106 20 |
| 18.7b | |||||
| PRDX6 | cytosolic, lysosomal | 3.4 × 107 21,c | not applicabled | π-GST + GSH22 | ND |
The reductant usually invoked as part of the catalytic thiol peroxidase cycle. Other potential redox partners have been identified but not kinetically characterized.
The values in italics were determined in the work presented here (see Results).
This value was obtained with the rat protein.
PRDX6 is a 1Cys Prx and as such does not undergo a resolution reaction.
Because of their extreme rate of reduction of H2O2, Prx have often been implicated in signaling mediated by this peroxide. The large reaction rate constants and high concentrations make Prx the preferred target, accounting for the reduction of nearly all H2O2 formed or otherwise present in the same compartment as long as upstream reductants can keep up with demand.23–25 Once oxidized, the Prx may relay the signal via thiol−disulfide exchange reactions with other proteins.26–30 Some researchers consider that the signaling can be modulated via posttranslational modifications that affect the peroxidase activity, such as hyperoxidation31 of the CP, phosphorylation,32 and glutathionylation.33 In addition, transient inactivation of Prx is viewed by some authors as a means of allowing H2O2 to react with slower targets that would become the receptors of the H2O2 signal.31,34 The two mechanisms (direct relay or modulation/inactivation via posttranslational modifications to permit the oxidation of slower targets) are not mutually exclusive, and they are theoretically possible but, again, suffer from a lack of quantitative information about the chemical and enzyme kinetics involved. Additionally, the apparent redundancy of different Prx coexisting in the same compartment evidenced in Table 1 is often disregarded.
The center of attention in this article is reaction 2, or “resolution”, for which some rate constant (kres) values have been published: 15 s−1 in human PRDX5,20 2 and 20 M−1 s−1 in human PRDX2 and PRDX3, respectively,16 9 s−1 in human PRDX1,15 and 75 s−1 in Salmonella typhimurium AhpC.35 One of the remarkable aspects of this reaction is that it requires an important conformational change, called the fully folded to locally unfolded or FF−LU transition, in which both CP and CR move to bridge the gap of 13−15 Å that separates them in the dithiol state according to the X-ray structures. The conformational dynamics of this process have been studied in Arabidopsis thaliana PrxQ, belonging to another subfamily of Prx. The authors reported a relatively fast conformational exchange rate (1650 s−1) that, according to them, is not likely to be limiting for catalysis.36 The chemical reaction itself, the condensation of sulfenic acid and thiol moieties, is known to be fast when low-molecular weight sulfenic acids and thiols are involved37,38 but appears to be limited by the protein structure in Prx.
In this work, we study the pH dependence of the resolution process for four 2Cys Prx and find that all four of them show similar patterns, consistent with two ionizable reacting groups. Nevertheless, the rate constants are different, spanning 2 orders of magnitude that cannot be easily explained in terms of differences in reactivity of the sulfenic acids and thiols involved. We then explore how such evolutionarily conserved differences in the rates of the resolution step would allow differential accumulation of the sulfenic acid of certain Prx so that they could play direct roles in peroxide signaling.
EXPERIMENTAL PROCEDURES
Proteins.
Human PRDX2 was purified from red blood cells, as previously described.2 Recombinant human PRDX5 and S. typhimurium AhpC were expressed and purified as previously described.20,39 The coding sequence of PRDX1 (GenBank accession number NM_001202431), including an N-terminal His tag and TEV protease cleavage site, was synthesized and cloned into a pET28a plasmid with NcoI and HindIII by Genscript.
BL21(DE3) cells, harboring the PRDX1pET-28a vector, were grown in Luria broth (LB) medium containing kanamycin (50 μg/mL) at 37 °C to an OD600 of ~0.6. Expression was induced by adding isopropyl β-D-1-thiogalactopyranoside to a final concentration of 0.5 mM at 20 °C. The next morning cultured cells were harvested by centrifugation and suspended in buffer A [50 mM sodium phosphate (pH 7.4) and 150 mM NaCl] with 5 mM phenylmethanesulfonyl fluoride and 1 mg/mL lysozyme. The cell suspension was sonicated, and the supernatant was separated by centrifugation. Histidine-tagged PRDX1 was purified using a HisTrap immobilized metal ion affinity chromatography (IMAC) column (GE Healthcare) equilibrated with buffer A with 20 mM imidazole. Elution was achieved with an imidazole gradient from 20 to 500 mM. To remove the His tag, 2 mg of TEV protease was added to the collected fractions, and they were extensively dialyzed against buffer A with 0.5 mM EDTA and 1 mM dithiothreitol (DTT) at 4 °C. The cleaved His tag and TEV protease were removed with a second IMAC column. Finally, the protein was concentrated, reduced with DTT for 30 min at room temperature, and injected onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with buffer A with0.1 mM diethylenetriamine pentaacetic acid (DTPA). Fractions with PRDX1 were pooled, concentrated, and stored at −80 °C. Purity was evaluated by sodium dodecyl sulfate−polyacrylamide gel electrophoresis.
Buffer System.
Unless otherwise indicated, for kinetic determinations we used a wide-range buffer solution at a constant ionic strength (I = 0.15) independent of the pH as proposed by Ellis and Morrison,40 consisting of 30 mM Tris, 15 mM MES, and 15 mM acetic acid; the buffer also contained 120 mM NaCl and 0.1 mM DTPA.
Kinetic Studies.
The change in fluorescence of PRDX1, PRDX2, and AhpC upon oxidation with excess H2O2 was used to measure the rate constant of the oxidation (, reaction 1) and the resolution reaction (kres, reaction 2). Time courses are biphasic, with a faster phase that is first-order in H2O2 concentration and a slower phase independent of H2O2 concentration and assigned to reaction 2 in the case of AhpC.35 Thus, kres was measured in the presence of excess H2O2 (5 μM for PDRX1 and PRDX2 and 50 μM for AhpC) as the slower phase of the fluorescence time course, which was fitted to a single-exponential function. In some instances, a much slower decay in fluorescence was apparent at longer times, which was at least partially due to photolysis of the tryptophans; this was considered but essentially affected neither the fitting process nor its results (Figure S1).
The behavior of PRDX5 fluorescence upon oxidation is different, likely because it has only one tryptophan (W85) <6 Å from the CP and none near CR. Additionally, the oxidation to sulfenic acid causes an increase in fluorescence emission, and the resolution reaction apparently does not produce any change in fluorescence, as evidenced by the study of the CR mutant.20 Therefore, kres was measured as previously described, as the asymptotic limit of the rate constant of oxidation at high H2O2 concentrations.20 We studied the dependence of k on H2O2 concentration at three different pHs in the range 5.84−8.94 and chose 200 μM H2O2 as a concentration close enough to the asymptotic limit.
Cysteine pKa Determination.
The pKa values of CP and CR of PRDX2 were determined through the pH dependence of the rate of alkylation by monobromobimane as previously described.41
RESULTS AND DISCUSSION
Kinetics of the resolution reaction were previously reported at single pH values (pH 7−7.4) for PRDX5,20 AhpC,35 and PRDX115 measured through the changes in fluorescence of the proteins caused by oxidation, but independent of H2O2 concentration. The reported rate constant for resolution of PRDX2 was previously obtained through a combination of competition kinetics of reaction 2 with the hyperoxidation reaction (reaction 4) monitored by SDS−PAGE and competition kinetics between reaction 4 and catalase.16
| (4) |
In this work, we monitored reaction 2 directly through changes in the intrinsic fluorescence of the Prx. As seen for AhpC and PRDX1, when PRDX2 reacts with H2O2 the time course of fluorescence changes shows two distinct phases (Figure 1). The rate constant of the faster phase depends linearly on H2O2 concentration, and the calculated second-order rate constant[(1.59 ± 0.06) × 108 M−1 s−1] agrees reasonably well with previous values of obtained by HRP.2,14 competition On the other hand, the rate constant of the slower phase is independent of H2O2 concentration in the range used and is interpreted here as the kres.
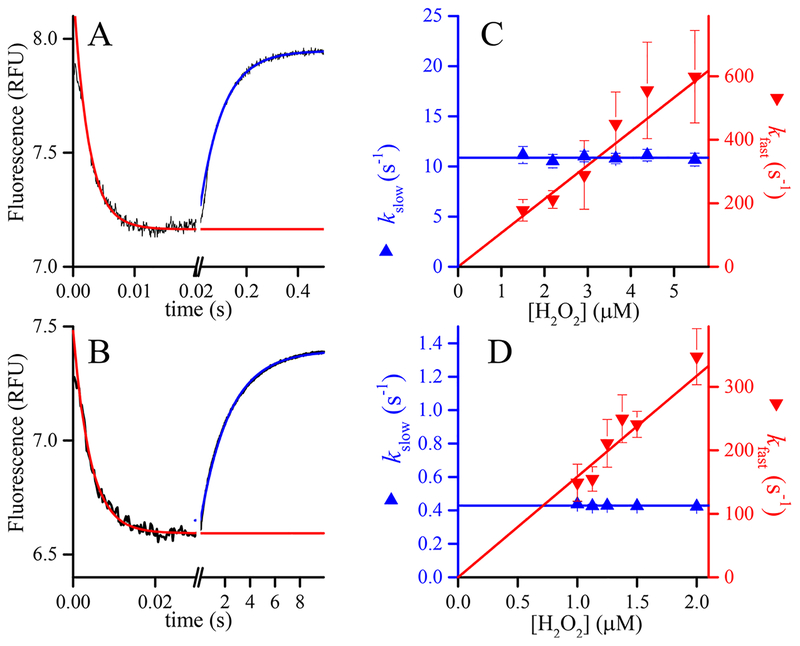
Figure 1.
Kinetics of oxidation of PRDX1 and PRDX2 by H2O2. Time courses of the changes in intrinsic fluorescence of (A) 0.25 μM reduced PRDX1 upon reaction with 2.9 μM H2O2 at pH 6.7 and (B) 0.2 μM reduced PRDX2 upon reaction with 1.5 μM H2O2 at pH 7.4. Note the different time scales. The two phases were fitted independently to single-exponential functions to obtain first-order rate constants. The dependence of the rate constants on H2O2 concentration, kfast, corresponding to the descending phase of the reaction (down triangles, right y axis) depends linearly on H2O2 concentration, whereas the rate constant of the ascending phase (kslow, up triangles, left y axis) is apparently independent of H2O2 concentration for both (C) PRDX1 and (D) PRDX2.
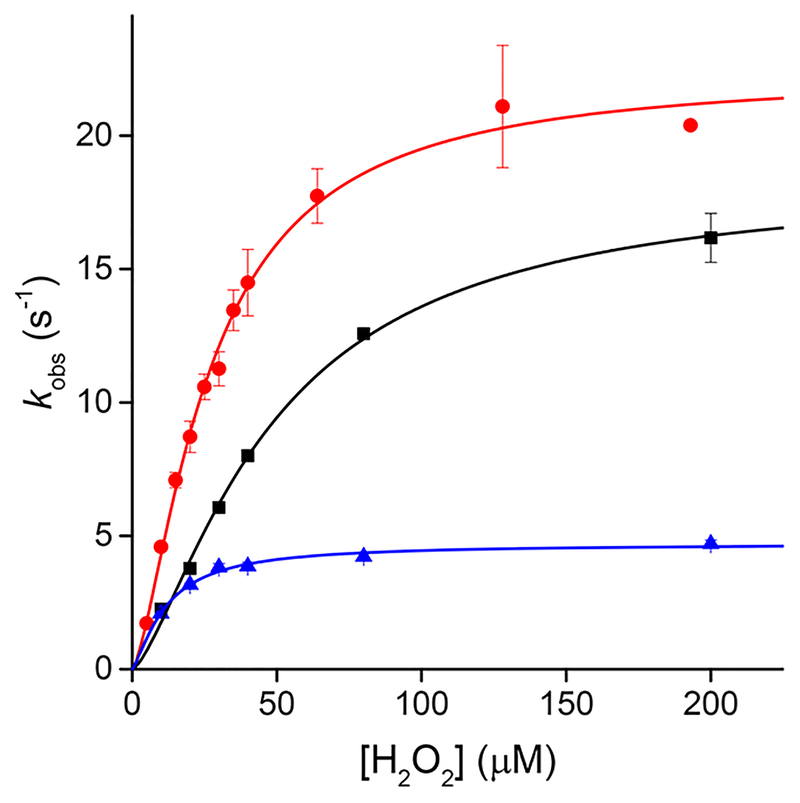
The time courses of oxidation of PRDX5 by H2O2 are single exponentials, in which the rate constant tends asymptotically to the value of kres at high H2O2 concentrations.3 Because the oxidation and the resolution cannot be discriminated from the time courses alone, as they can be for AhpC and PRDX2, we studied the kinetics of oxidation at different pHs and in a range of H2O2 concentrations to make sure we could select a concentration close enough to the plateau in rate constant. Experiments performed at pH 5.84, 7.08, and 8.94 showed that with 200 μM H2O2 the rate constant was practically indistinguishable from the asymptote (Figure 2).
Figure 2.
Kinetics of oxidation of PRDX5 by H2O2. The reaction rate constants were measured through the change in intrinsic fluorescence of the protein at different concentrations of H2O2, at 25 °C and pH5.84 (black squares), pH 7.08 (red circles), and pH 8.94 (blue triangles).
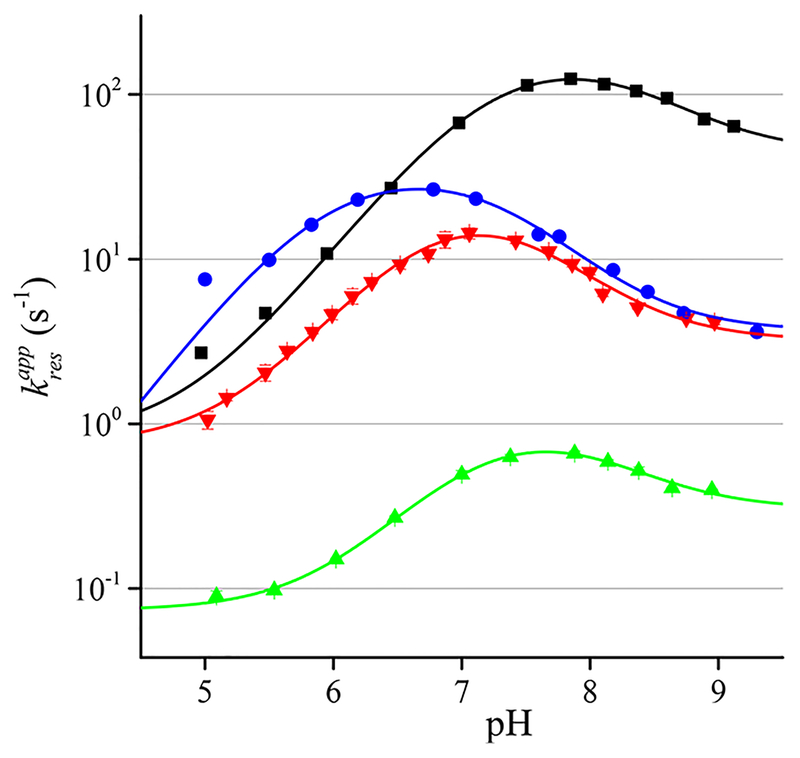
We then measured the apparent rate constants of resolution of each peroxiredoxin at a single excess concentration of H2O2 in the pH range from 5 to 9. The pH profiles of are bell-shaped as expected for reactions with two acid−base groups involved. We posit that the CR thiol and CP sulfenic acid would be the groups whose pKa values could most affect the rate constant (Figure 3).
Figure 3.
pH profiles of the rate constant of resolution of AhpC (black squares), PRDX1 (red triangles), PRDX2 (green triangles), and PRDX5 (blue circles). The lines represent the fits to eq 5.
Taking into account the protonation states of the two reacting groups, reaction 2 can be considered as four possible combinations. Considering that RS− is the best nucleophile and RSOH is the best electrophile, we can order the combinations of protonation states in reactions from faster to slower as
| (2A) |
| (2B) |
| (2C) |
| (2D) |
In this order, we have assumed that the nucleophilicity of the thiolate is more important than the electrophilicity of the sulfenic acid in determining the rate constant.
The general rate law of the reaction would be
where [RS(H)]T and [RSO(H)]T are the total concentrations of thiol plus thiolate and sulfenic acid plus sulfenate, respectively. The rate of the reaction is the sum of the rates of the four possible combinations (2A−2D) that are pH-dependent according to the pH distribution of the species involved. For instance, the rate law of reaction 2A would be
Or
where and are the ionization constants of the CR thiol and the CP sulfenic acid, respectively. Thus, the apparent rate constant would be
The apparent rate constants of reactions 2B−2D can be analogously derived (see the Supporting Information). As the four combinations happen simultaneously, the overall rate constant of the resolution would be the sum of the apparent rate constants:
Or
| (5) |
The resolution reaction is first-order, because CP and CR are contained in the same protein chain (PRDX5) or belong to dimers that do not dissociate (PRDX1, PRDX2, and AhpC). Nevertheless, the pH dependence is the same as in a bimolecular reaction.
The pH profiles of Figure 3 were fitted to eq 5 using Origin8.6, and to simplify the fitting, the value of the least likely rate constant (k2D) was fixed to 0. Results of the best fits are listed in Table 2.
Table 2.
Best Fit Values of eq 5 to the Apparent Rate Constants of Resolution Shown in Figure 3 and Values of Cysteine pKa Obtained by Other Experimental Approaches for Comparison
| AhpC | PRDX1 | PRDX2 | PRDX5 | |
|---|---|---|---|---|
| k2A | 2410 ± 725 | 113 ± 49 | 10.5 ± 5.5 | 1032 ± 425 |
| k2B | 44 ± 4.7 | 3.2 ± 0.3 | 0.30 ± 0.03 | 3.7 ± 0.5 |
| k2C | 1.1 ± 1.2 | 0.7 ± 0.2 | 0.06 ± 0.01 | 0 ± 3.1 |
| 7.22 ± 0.06 | 6.8 ± 0.1 | 7.0 ± 0.1 | 5.9 ± 0.1 | |
| 8.3 ± 0.1 | 7.4 ± 0.2 | 8.0 ± 0.2 | 7.35 ± 0.09 | |
| 5.84 ± 0.0242 | not determined | 4.8 ± 0.1 | 5.2 ± 0.220 | |
| 8.7 ± 0.142 | not determined | 8.5 ± 0.2 | 8.8 ± 0.141 |
The values in Table 2 show, as expected, that reaction 2A is the main contributor to the overall reaction rate; reaction 2B has some minor relevance, mostly because its relative contribution to will be perceived only when . The rate constant of reaction 2C makes a negligible contribution to the overall reaction rate.
There are two pKa values reported in the literature for the sulfenic acid of CP in ; a value of 6.6 was determined for Mycobacterium tuberculosis AhpE,43 and a value of 6.1 was determined for S. typhimurium AhpC C165S.44 The latter is significantly lower than the value determined herein for wild-type AhpC (7.22 ± 0.06). Remarkably, the values of for CR determined through fitting of the resolution reaction are significantly lower than those previously measured by alkylation in refs 41 and 42 or in this work (Figure S1). The shift in the pKa values of both sulfenic acid and thiol points to a potential alteration of the protein environment of both CP and CR upon formation of the sulfenic acid that may occur in a manner independent of the conformational change that enables disulfide bond formation.
In comparison with the CP thiol/thiolate present in the reduced form of these enzymes, the sulfenic acid/sulfenate is bulkier, significantly less acidic,20,42 and potentially much better at forming hydrogen bonds because of its oxygen atom. These steric, electrostatic, and interaction properties will alter the environment of the active site and could result in a destabilization of the FF conformer and may accelerate the FF → LU transition.
Reactivity of the Residues versus Encounter Frequency of the Reacting Moieties.
Sulfenic acids in the presence of thiols are short-lived. They react to yield a disulfide that is much more stable than the reactants; thus, measuring the kinetics of condensation through partial oxidation of a thiol is difficult. This difficulty was evident in a recent debate in the literature37,38 about the rate constant of reaction of Cys with Cys sulfenic acid at pH 6.0 to form a disulfide (kCysSS). One group reported a value of 720 M−1 s−1,37 and the other set a lower limit of 105 M−1 s−1;38 both rate constants indicate relatively fast reactions. There is nevertheless a detailed kinetic study of a sulfenic acid reacting with thiols in aqueous solution; the proton pump inhibitor omeprazole produces a sulfenic acid by a rearrangement in acidic medium in the absence of thiols, and the sulfenic acid is in fast equilibrium with a cyclic sulfenylamide. The mixture of sulfenic and cyclic sulfenylamide reacts with β-mercaptoethanol with a diffusion-limited rate,45 in line with the highest rate constant obtained in the cysteine studies. In any case, both reported rate constants of the condensation of cysteine with cysteine sulfenic acid can be compared to the resolution reactions of the four Prx studied here. To make the comparison, we need to calculate effective molarities as (Table 3). A simple model assuming that both CP sulfenic acid and CR thiol are constrained to move in a sphere with a 30 Å radius (twice the distance between the residues in the reduced enzyme) produces an effective molarity of 0.68 M, much higher than those calculated even with the smallest rate constant. It follows that the encounter probability of the two residues is restricted by the protein structure; i.e., the Prx structure limits the resolution reaction rate and thus may limit the overall turnover of the enzyme. One of the secondary effects of the slow resolution is the differential propensity to hyperoxidation previously observed among different Prx.31
Table 3.
Resolution Rate Constants at pH 6.0 and Effective Molarities Estimated as the Resolution Rate Constant Divided by the Reported Bimolecular Rate Constant of the Condensation between Cys and Cys Sulfenic Acid ()
| effective molarity | |||
|---|---|---|---|
| Prx | (s−1 at pH 6.0)a | kCysSS = 720 M−1 s−1 | kcysSS > 105 M−1 s−1 |
| AhpC | 11.8 | 2.71 × 10−2 | <1.95 × 10−4 |
| PRDX1 | 4.61 | 6.4 × 10−3 | <4.61 × 10−5 |
| PRDX2 | 0.15 | 2.08 × 10−4 | <1.50 × 10−6 |
| PRDX5 | 19.5 | 1.64 × 10−2 | <1.18 × 10−4 |
Values interpolated from Figure 3, using pH 6 to compare the results with those of small-molecule studies.
The FF active site of Prx is an extremely conserved structure;5,46,47 thus, the sulfenic acids of the CP are formed in similar environments in the four Prx studied. Also, the CR thiol appears to be a regular cysteine in terms of reactivity, with the expected pKa for a surface Cys and not particularly fast in any of the reactions studied so far. However, 2 orders of magnitude separate the apparent rate constants of PRDX2 and AhpC. Thus, it appears, particularly in the case of PRDX2, that the resolution rate is limited not by the reactivity of the thiol−sulfenic acid pair but by a structural restriction that hinders the access to the LU conformation.
Furthermore, according to the estimates in Table 2, the resolution reaction is slower than expected in all four cases. This evidence supports the hypothesis that the protein structure restricts the rate of resolution, not necessarily by affecting the reactivity of the groups involved but by sequestering or stabilizing them in the FF conformation and limiting their productive encounters.
Stabilization of the LU conformation happens in nitrated PRDX248 and results in faster overall catalysis. Additional evidence of this structural limitation of the rate comes from another thiol peroxidase from the GPx family. In a recent report on a mycothiol peroxidase from Corynebacterium glutamicum (Mpx),49 the authors show that the resolution reaction happens at a rate on par with that of AhpC (kres = 111 s−1) but a C64S mutation makes the reaction 5-fold faster (kres = 559 s−1). Because C64 is not involved in the reaction and seems to be too far from either CP or CR (according to structural models) to affect the reactivity of either, the best hypothesis is that the C64S mutation helps stabilize the conformation where the reaction is possible.
The sole example of the dynamics of the FF−LU transition studied by nuclear magnetic resonance in a Prx indicates a rather fast process,36 but even if the conformational transition is fast, it could limit the overall rate of resolution. The reaction sequence, including the conformational transition, would be
| (6) |
The rate law of the system can be approximated using any of the three simplified approaches, namely, steady state, prior equilibrium, or improved prior equilibrium,50 described in Table 4.
Table 4.
Approximate Solutions for the Kinetic System of Resolution Described in eq 6
| steady state | prior equilibrium | improved prior equilibrium | |
|---|---|---|---|
| kres ≈ |
In all three cases, the approximate rate constant of product buildup (kres) includes kLU and kFF, with kFF always being in the denominator. Then, regardless of the approximation, assuming that the LU conformation is less stable than FF (kLU ≪ kFF),51 the conformational change will limit the rate of resolution by restricting the available concentration of the LU conformer. In other words, the FF → LU transition does not need to be slow to decelerate the resolution reaction; as long as the transition in the LU direction is slower than its reverse, kres will be lower than the rate constant of the chemical process (k2).
Stoichiometric versus Catalytic Consumption of H2O2 and the Sensing of Peroxide Flux.
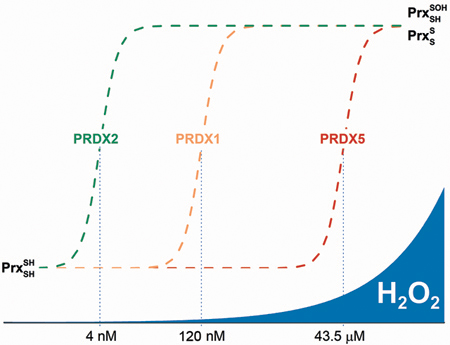
Limitation of the catalytic turnover of Prx by the resolution reaction could have profound consequences in terms of the sensing of peroxides. The rate of peroxide consumption by Prx depends crucially on the concentration of enzyme and the rate of H2O2 generation. If the enzyme is in the dithiol state and in excess, and H2O2 is introduced suddenly into the system, as in a bolus addition, the consumption of peroxide will obey the rate law of reaction 1 (Table 5). This stoichiometric consumption of H2O2 is extremely rapid, particularly by PRDX1, PRDX2, and AhpC that are abundant and have large rate constants. As a result, under these conditions, the enzymes will consume H2O2 very fast. Nevertheless, if H2O2 is produced constantly, the excess reduced Prx will be eventually depleted, and further H2O2 reduction can be possible only if the enzyme goes through the complete catalytic cycle. In this case, reaction 2 or 3 may become rate-limiting. Using the values of Prx rate constants, we calculated the steady-state concentrations of the oxidant ([H2O2]ss) and reductant (either Trx or AhpF, [Red]ss) that make all three steps in the catalytic cycle proceed at the same rate (Table 5). The calculated [H2O2]ss in the case of PRDX2 is remarkably low (4 nM) and separated from that of PRDX1 and PRDX5, enzymes that coexist in the cytosolic compartment. Interestingly, the range of [H2O2]ss between the two main cytosolic Prx (PRDX1 and PRDX2) coincides with the range described recently as physiological, or “oxidative eustress”.52
Table 5.
Calculated Steady-State Concentrations of Substrates Making All Three Steps in the Catalytic Cycle of Prx Equally Fast at pH 7.4a
| reaction 1 | reaction 2 | reaction 3 | |||
|---|---|---|---|---|---|
| v = kred[red][PrxS2] | |||||
| [H2O2]ss | kres (s−1) | kres (M−1 s−1) | [Red]ss (μM) | ||
| AhpC | 1.36 × 108b | 0.77 μM | 104.8 | 2.00 × 107b | 5.24 |
| PRDX1 | 1.1 × 108 | 0.12 μM | 12.9 | not determined | not determined |
| PRDX2 | 1.6 × 108 | 4 nM | 0.64 | not determined | not determined |
| PRDX5 | 4.3 × 105 | 43.5 μM | 18.7 | 2.00 × 106 | 9.35 |
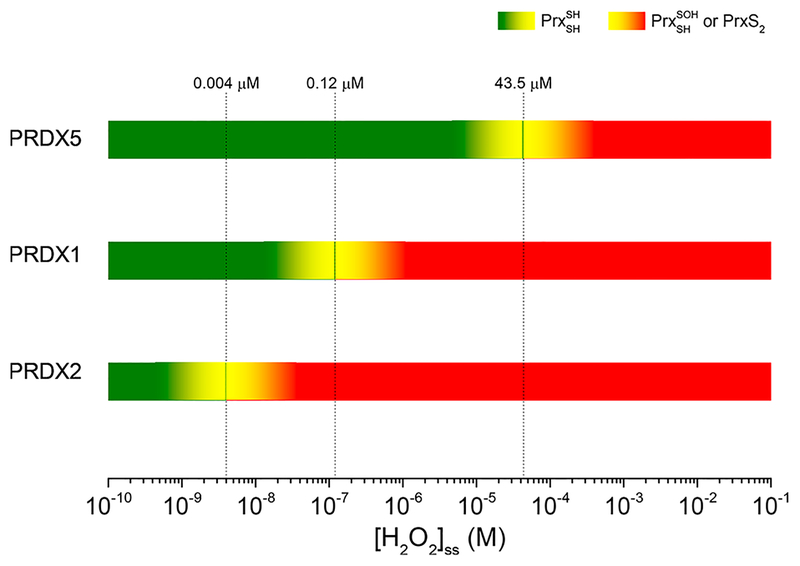
In the case in which a sustained influx of H2O2 oxidizes the Prx and forces it to consume the peroxide catalytically through multiple turnovers (i.e., when [H2O2]ss exceeds the critical values of Table 5 for each Prx), the actual rate of consumption of H2O2 by said Prx will not increase with an increasing [H2O2]ss, as reaction 2 or 3 will become rate-limiting. For instance, if the H2O2 influx is sufficient to sustain [H2O2]ss > 4 nM, PRDX2 will accumulate as either sulfenic acid or disulfide and will not be able to consume H2O2 any faster, thus forfeiting its antioxidant function. An alternative, faster Prx present in the same compartment can then perform the catalytic reduction of H2O2 below their respective critical [H2O2]ss, namely up to 120 nM for PRDX1 and up to 43.5 μM for PRDX5. In summary, in a cytosolic compartment where PRDX1, PRDX2, and PRDX5 coexist, in the range of [H2O2]ss of ≤4 nM all three Prx contribute to the catalytic reduction of peroxide. From 4 to 120 nM, PRDX1 and PRDX5 will be the catalytic antioxidant whereas PRDX2 will accumulate as sulfenic acid and/or disulfide (depending on the rate of reaction 3). When [H2O2]ss > 120 nM, only PRDX5 will continue functioning as the antioxidant. Of course, in the same compartment there may be other peroxidases (glutathione peroxidase, PRDX6, and catalase) sharing the burden of reducing H2O2; nevertheless, the ranges of steady-state concentrations are embedded in each 2Cys Prx through the rate-limiting step that reaction 2 may become (Figure 4).
Figure 4.
Ranges of H2O2 concentration and PRDX roles. Below the [H2O2]ss threshold imposed for each PRDX by their resolution reaction, the enzymes act as fast peroxidases consuming most of the peroxide and keeping its concentration low. Once the flux of H2O2 overcomes the maximal flux of consumption by a Prx, resolution or reduction reactions become rate-limiting, and the enzyme accumulates as sulfenic acid or disulfide and may acquire the role of reporting the increased H2O2 flux (i.e., as a threshold sensor). The other Prx continue the peroxidase function until the [H2O2]ss reaches their respective critical points. Other peroxidases such as catalase and GPx also contribute to the consumption of H2O2 and may prove to be important for preventing hyperoxidation, a reaction that needs a rather high [H2O2]ss.
There is the additional possibility of reaction 3 becoming the rate-limiting step, for instance, because of low reducing partner concentrations. In those cases, the critical [H2O2]ss will be lower than those in Table 5 and the Prx will accumulate as the disulfide, instead of as the sulfenic acid.
Above each critical [H2O2]ss, the catalytic capability of one Prx is overwhelmed and the next one takes over with the possibility of setting a stepwise sensing system with hard-wired detection points at specific concentrations of the oxidant. At each critical point, and in a very narrow range of concentrations, the Prx will switch from mostly reduced to mostly oxidized, providing a recognizable target in a signaling pathway. As each 2Cys Prx has a unique combination of rate constants, several such switches are possible in compartments where multiple 2Cys Prx exist.
One reaction that is usually invoked in systems where Prx turnover occurs with excess H2O2 is hyperoxidation of the CP to render the catalytically inactive sulfinic and sulfonic forms53,54 (reaction 4). To gain a better quantitative perspective on the importance of hyperoxidation, we can make some simple calculations at one of the critical [H2O2]ss values listed in Table 5. What fraction of PRDX2, the most sensitive Prx, will be inactivated through hyperoxidation at 120 nM, the point at which PRDX1 switches from a catalytic antioxidant to a signaling sulfenic acid or disulfide? The reported rate constant of reaction 4 is 12000 M−1 s−1;16 then at 120 nM, the rate of reaction 4 would be
Meanwhile, with the turnover of PRDX2 limited by the resolution reaction, its rate would be
The quotient v4/vres tells us that under these conditions PRDX2 is inactivated at a rate of once every 440 turnovers. Of course, because v4 increases with increasing H2O2 concentrations, it may become more prevalent under conditions of higher H2O2 concentrations, and hyperoxidized protein could accumulate under such conditions.
Other reactions of the sulfenic acid may be even more important than hyperoxidation, e.g., glutathionylation of the CP. The rate constant of that reaction has been reported as 500 M−1 s−1;55 then, assuming a conservative glutathione concentration of 1 mM, the rate would be
In this case, a vGSH/vres ratio of 1.28 indicates that a very important fraction of PRDX2 may become glutathionylated in the system and its return to the basic catalytic cycle will depend on the action of glutaredoxins or other enzymatic systems.55
We still do not know whether or under what circumstances the Prx present as sulfenic acid, as disulfide, gluthionylated, or as the hyperoxidized sulfinic acid (or all of them) would serve as the recognizable transducer of the signal for H2O2 concentration. Recent results point to the involvement of CP sulfenic acid in the formation of mixed disulfides of target protein thiols with PRDX1 and PRDX2 in cells subject to mild H2O2 exposure.27 The involvement of the reduction reaction, by thioredoxin or alternative reductants, in the modulation of the distribution of the three catalytic species of Prx (thiol, sulfenic acid, and disulfide) requires further research. Detailed kinetic studies are needed, beginning with the systems of reduction of Prx by thioredoxins and exploring further the alternative redox partners that may relay the signal. In any case, we currently know that the Prx properties (i.e., structures, dynamics, and kinetic rate constants) are clearly tunable and have been adapted to fulfill their biological niche in ways that we are just beginning to understand.
In summary, by evaluating the disulfide bond formation rate constant across a pH range among a set of Prx with distinct features and roles in biology, we have determined the functional pKa values of both the CP sulfenic acid and CR thiol for each. As expected, disulfide bond formation proceeding by the nucleophilic attack of the deprotonated CR on the protonated, electrophilic CP sulfenic acid is the most efficient (by >30-fold) versus the next fastest reaction (between the CR thiolate and CP sulfenate), while the contributions of reactions involving the protonated CR are negligibly small. In all four Prx studied (PRDX1, PRDX2, PRDX5, and AhpC), the maximal potential rate of disulfide bond formation is never reached because of the higher pKa of the CR thiol relative to that of the CP sulfenic acid (limiting the maximal observed rate constant to approximately 12−30-fold lower than the rate constant of the optimal redox forms of CP and CR). Where the disulfide bond formation rate is slowest, in PRDX2, stoichiometric amounts of H2O2 can still be rapidly reduced (comparable to the case for the other major cytosolic Prx, PRDX1), but as H2O2 concentrations increase and turnover of the system is required, disulfide bond formation rapidly becomes a bottleneck that favors flux through PRDX1 rather than PRDX2, even if reductant availability becomes an issue. This supports the possibility that PRDX2 redox status, which is strongly shifted at [H2O2]ss values of >4 nM, is used by the cell as a threshold sensor of H2O2 levels for the purpose of regulating cell signaling processes.
Supplementary Material
Funding
Financial support was provided by Universidad de la República (CSIC C632–348 to A.D.), National Institutes of Health Grant R01 GM119227 to L.B.P. and P.A.K., and Centro Argentino-Brasileño de Biotecnologiá CABBIO 2014–05 to G.F.-S. S.P.-L., L.M.R., and J.D.R. were partially supported by CAP UdelaR and PEDECIBA, Uruguay.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00188.
Derivation of kinetic equations for fitting Figure 3, alternative fittings to the fluorescence time courses (Figure S1), and determination of the pKa of PRDX2 using the reaction with mBBr (Figure S2) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Reeves SA, Parsonage D, Nelson KJ, and Poole LB (2011) Kinetic and thermodynamic features reveal that Escherichia coli BCP is an unusually versatile peroxiredoxin. Biochemistry 50, 8970–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Manta B, Hugo M, Ortiz C, Ferrer-Sueta G, Trujillo M, and Denicola A (2009) The peroxidase and peroxynitrite reductase activity of human erythrocyte peroxiredoxin 2. Arch. Biochem. Biophys 484, 146–154. [DOI] [PubMed] [Google Scholar]
- (3).Portillo-Ledesma S, Sardi F, Manta B, Tourn MV, Clippe A, Knoops B, Alvarez B, Coitiño EL, and Ferrer-Sueta G (2014) Deconstructing the Catalytic Efficiency of Peroxiredoxin-5 Peroxidatic Cysteine. Biochemistry 53, 6113–6125. [DOI] [PubMed] [Google Scholar]
- (4).Hall A, Nelson K, Poole LB, and Karplus PA (2011) Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signaling 15, 795–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Perkins A, Parsonage D, Nelson KJ, Ogba OM, Cheong PH, Poole LB, and Karplus PA (2016) Peroxiredoxin Catalysis at Atomic Resolution. Structure 24, 1668–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zeida A, Reyes AM, Lebrero MC, Radi R, Trujillo M, and Estrin DA (2014) The extraordinary catalytic ability of peroxiredoxins: a combined experimental and QM/MM study on the fast thiol oxidation step. Chem. Commun. (Cambridge, U. K.) 50, 10070–10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nagy P, Karton A, Betz A, Peskin AV, Pace P, O’Reilly RJ, Hampton MB, Radom L, and Winterbourn CC (2011) Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: a kinetic and computational study. J. Biol. Chem 286, 18048–18055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Jacobson FS, Morgan RW, Christman MF, and Ames BN (1989) An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J. Biol. Chem 264, 1488–1496. [PubMed] [Google Scholar]
- (9).Chae HZ, Chung SJ, and Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem 269, 27670–27678. [PubMed] [Google Scholar]
- (10).Bryk R, Griffin P, and Nathan C (2000) Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215. [DOI] [PubMed] [Google Scholar]
- (11).Ogusucu R, Rettori D, Munhoz DC, Soares Netto LE, and Augusto O (2007) Reactions of yeast thioredoxin peroxidases I and II with hydrogen peroxide and peroxynitrite: rate constants by competitive kinetics. Free Radical Biol. Med 42, 326–334. [DOI] [PubMed] [Google Scholar]
- (12).Jaeger T, Budde H, Flohe L, Menge U, Singh M, Trujillo M, and Radi R (2004) Multiple thioredoxin-mediated routes to detoxify hydroperoxides in Mycobacterium tuberculosis. Arch. Biochem. Biophys 423, 182–191. [DOI] [PubMed] [Google Scholar]
- (13).Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, and Poole LB (2005) Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44, 10583–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, and Winterbourn CC (2007) The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem 282, 11885–11892. [DOI] [PubMed] [Google Scholar]
- (15).Carvalho LAC, Truzzi DR, Fallani TS, Alves SV, Toledo JC Jr., Augusto O, Netto LES, and Meotti FC (2017) Urate hydroperoxide oxidizes human peroxiredoxin 1 and peroxiredoxin 2. J. Biol. Chem 292, 8705–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Peskin AV, Dickerhof N, Poynton RA, Paton LN, Pace PE, Hampton MB, and Winterbourn CC (2013) Hyper-oxidation of peroxiredoxins 2 and 3: rate constants for the reactions of the sulfenic acid of the peroxidatic cysteine. J. Biol. Chem 288, 14170–14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Cox AG, Peskin AV, Paton LN, Winterbourn CC, and Hampton MB (2009) Redox potential and peroxide reactivity of human peroxiredoxin 3. Biochemistry 48, 6495–6501. [DOI] [PubMed] [Google Scholar]
- (18).Wang X, Wang L, Wang X. e., Sun F, and Wang C. c. (2012) Structural insights into the peroxidase activity and inactivation of human peroxiredoxin 4. Biochem. J 441, 113–118. [DOI] [PubMed] [Google Scholar]
- (19).Tavender TJ, Springate JJ, and Bulleid NJ (2010) Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 29, 4185–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Trujillo M, Clippe A, Manta B, Ferrer-Sueta G, Smeets A, Declercq JP, Knoops B, and Radi R (2007) Pre-steady state kinetic characterization of human peroxiredoxin 5: taking advantage of Trp84 fluorescence increase upon oxidation. Arch. Biochem. Biophys 467, 95–106. [DOI] [PubMed] [Google Scholar]
- (21).Toledo JC Jr., Audi R, Ogusucu R, Monteiro G, Netto LE, and Augusto O (2011) Horseradish peroxidase compound I as a tool to investigate reactive protein-cysteine residues: from quantification to kinetics. Free Radical Biol. Med 50, 1032–1038. [DOI] [PubMed] [Google Scholar]
- (22).Manevich Y, Feinstein SI, and Fisher AB (2004) Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc. Natl. Acad. Sci. U. S. A 101, 3780–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, and Denicola A (2011) Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol 24, 434–450. [DOI] [PubMed] [Google Scholar]
- (24).Cox AG, Winterbourn CC, and Hampton MB (2010) Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J 425, 313–325. [DOI] [PubMed] [Google Scholar]
- (25).Randall LM, Ferrer-Sueta G, and Denicola A (2013) Peroxiredoxins as preferential targets in H2O2-induced signaling. Methods Enzymol. 527, 41–63. [DOI] [PubMed] [Google Scholar]
- (26).Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, and Dick TP (2015) Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol 11, 64–70. [DOI] [PubMed] [Google Scholar]
- (27).Stocker S, Maurer M, Ruppert T, and Dick TP (2018) A role for 2-Cys peroxiredoxins in facilitating cytosolic protein thiol oxidation. Nat. Chem. Biol 14, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Winterbourn CC (2018) Biological Production, Detection and Fate of Hydrogen Peroxide. Antioxid. Redox Signaling, DOI: 10.1089/ars.2017.7425. [DOI] [PubMed] [Google Scholar]
- (29).Jarvis RM, Hughes SM, and Ledgerwood EC (2012) Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radical Biol. Med 53, 1522–1530. [DOI] [PubMed] [Google Scholar]
- (30).Stocker S, Van Laer K, Mijuskovic A, and Dick TP (2018) The Conundrum of Hydrogen Peroxide Signaling and the Emerging Role of Peroxiredoxins as Redox Relay Hubs. Antioxid. Redox Signaling 28, 558–573. [DOI] [PubMed] [Google Scholar]
- (31).Wood ZA, Poole LB, and Karplus PA (2003) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653. [DOI] [PubMed] [Google Scholar]
- (32).Woo HA, Yim SH, Shin DH, Kang D, Yu DY, and Rhee SG (2010) Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell 140, 517–528. [DOI] [PubMed] [Google Scholar]
- (33).Park JW, Piszczek G, Rhee SG, and Chock PB (2011) Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry 50, 3204–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Rhee SG, Woo HA, and Kang D (2018) The Role of Peroxiredoxins in the Transduction of H2O2 Signals. Antioxid. Redox Signaling 28, 537–557. [DOI] [PubMed] [Google Scholar]
- (35).Parsonage D, Nelson KJ, Ferrer-Sueta G, Alley S, Karplus PA, Furdui CM, and Poole LB (2015) Dissecting peroxiredoxin catalysis: separating binding, peroxidation, and resolution for a bacterial AhpC. Biochemistry 54, 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Aden J, Wallgren M, Storm P, Weise CF, Christiansen A, Schroder WP, Funk C, and Wolf-Watz M (2011) Extraordinary μs-ms backbone dynamics in Arabidopsis thaliana peroxiredoxin Q. Biochim. Biophys. Acta, Proteins Proteomics 1814, 1880–1890. [DOI] [PubMed] [Google Scholar]
- (37).Luo D, Smith SW, and Anderson BD (2005) Kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J. Pharm. Sci 94, 304–316. [DOI] [PubMed] [Google Scholar]
- (38).Ashby MT, and Nagy P (2007) Revisiting a proposed kinetic model for the reaction of cysteine and hydrogen peroxide via cysteine sulfenic acid. Int. J. Chem. Kinet 39, 32–38. [Google Scholar]
- (39).Poole LB, and Ellis HR (1996) Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry 35, 56–64. [DOI] [PubMed] [Google Scholar]
- (40).Ellis KJ, and Morrison JF (1982) Buffers of constant ionic strength for studying pH-dependent processes. Methods Enzymol. 87, 405–426. [DOI] [PubMed] [Google Scholar]
- (41).Sardi F, Manta B, Portillo-Ledesma S, Knoops B, Comini MA, and Ferrer-Sueta G (2013) Determination of acidity and nucleophilicity in thiols by reaction with monobromobimane and fluorescence detection. Anal. Biochem 435, 74–82. [DOI] [PubMed] [Google Scholar]
- (42).Nelson KJ, Parsonage D, Hall A, Karplus PA, and Poole LB (2008) Cysteine pK(a) values for the bacterial peroxiredoxin AhpC. Biochemistry 47, 12860–12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hugo M, Turell L, Manta B, Botti H, Monteiro G, Netto LE, Alvarez B, Radi R, and Trujillo M (2009) Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry 48, 9416–9426. [DOI] [PubMed] [Google Scholar]
- (44).Poole LB, and Ellis HR (2002) Identification of cysteine sulfenic acid in AhpC of alkyl hydroperoxide reductase. Methods Enzymol. 348, 122–136. [DOI] [PubMed] [Google Scholar]
- (45).Brändström A, Lindberg P, Bergman N, Tekenbergs-Hjelte L, and Ohlson K (1989) Chemical Reactions of Omeprazole and Omeprazole Analogues. IV. Reactions of Compounds of the Omeprazole System with 2-Mercaptoethanol. Acta Chem. Scand 43, 577–586. [Google Scholar]
- (46).Hall A, Parsonage D, Poole LB, and Karplus PA (2010) Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J. Mol. Biol 402, 194–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, and Nelson KJ (2011) PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 39, D332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Randall LM, Manta B, Hugo M, Gil M, Batthyany C, Trujillo M, Poole LB, and Denicola A (2014) Nitration transforms a sensitive peroxiredoxin 2 into a more active and robust peroxidase. J. Biol. Chem 289, 15536–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Pedre B, Van Molle I, Villadangos AF, Wahni K, Vertommen D, Turell L, Erdogan H, Mateos LM, and Messens J (2015) The Corynebacterium glutamicum mycothiol peroxidase is a reactive oxygen species-scavenging enzyme that shows promiscuity in thiol redox control. Mol. Microbiol 96, 1176–1191. [DOI] [PubMed] [Google Scholar]
- (50).Espenson JH (1995) Chemical Kinetics and Reaction Mechanisms, McGraw-Hill, New York. [Google Scholar]
- (51).Perkins A, Nelson KJ, Williams JR, Parsonage D, Poole LB, and Karplus PA (2013) The sensitive balance between the fully folded and locally unfolded conformations of a model peroxiredoxin. Biochemistry 52, 8708–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Sies H (2017) Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 11, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Rabilloud T, Heller M, Gasnier F, Luche S, Rey C, Aebersold R, Benahmed M, Louisot P, and Lunardi J (2002) Proteomics analysis of cellular response to oxidative stress. Evidence for in vivo overoxidation of peroxiredoxins at their active site. J. Biol. Chem 277, 19396–19401. [DOI] [PubMed] [Google Scholar]
- (54).Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, and Rhee SG (2002) Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J. Biol. Chem 277, 38029–38036. [DOI] [PubMed] [Google Scholar]
- (55).Peskin AV, Pace PE, Behring JB, Paton LN, Soethoudt M, Bachschmid MM, and Winterbourn CC (2016) Glutathionylation of the Active Site Cysteines of Peroxiredoxin 2 and Recycling by Glutaredoxin. J. Biol. Chem 291, 3053–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.