Abstract
Background
Epidemiologic studies strongly suggest that the pathophysiology of late-onset Alzheimer disease (AD) versus early-onset AD has environmental rather than genetic causes, thus revealing potentially novel therapeutic targets to limit disease progression. Several studies supporting the “pathogen hypothesis” of AD demonstrate a strong association between pathogens and the production of β-amyloid, the pathologic hallmark of AD. Although the mechanism of pathogen-induced neurodegeneration of AD remains unclear, astrocytes, a key player of the CNS innate immune response and producer/metabolizer of β-amyloid, have been implicated. We hypothesized that Chlamydia pneumoniae infection of human astrocytes alters the expression of the amyloid precursor protein (APP)-processing secretases, ADAM10, BACE1, and PSEN1, to promote β-amyloid formation. Utilizing immunofluorescent microscopy, molecular, and biochemical approaches, these studies explore the role of an intracellular respiratory pathogen, Chlamydia pneumoniae, as an environmental trigger for AD pathology. Human astrocytoma cells in vitro were infected with Chlamydia pneumoniae over the course of 6–72 h. The gene and protein expression, as well as the enzymatic activity of non-amyloidogenic (ADAM10), and pro-amyloidogenic (BACE1 and PSEN1) secretases were qualitatively and quantitatively assessed. In addition, the formation of toxic amyloid products as an outcome of pro-amyloidogenic APP processing was evaluated through various modalities.
Results
Chlamydia pneumoniae infection of human astrocytoma cells promoted the transcriptional upregulation of numerous genes implicated in host neuroinflammation, lipid homeostasis, microtubule function, and APP processing. Relative to that of uninfected astrocytes, BACE1 and PSEN1 protein levels were enhanced by nearly twofold at 48–72 h post-Chlamydia pneumoniae infection. The processing of APP in Chlamydia pneumoniae-infected astrocytes favors the pro-amyloidogenic pathway, as demonstrated by an increase in enzymatic activity of BACE1, while that of ADAM10 was decreased. Fluorescence intensity of β-amyloid and ELISA-quantified levels of soluble-APP by products revealed temporally similar increases, confirming a BACE1/PSEN1-mediated processing of APP.
Conclusions
Our findings suggest that Chlamydia pneumoniae infection of human astrocytes promotes the pro-amyloidogenic pathway of APP processing through the upregulation of expression and activity of β-secretase, upregulated expression of γ-secretase, and decreased activity of α-secretase. These effects of astrocyte infection provide evidence for a direct link between Chlamydia pneumoniae and AD pathology.
Electronic supplementary material
The online version of this article (10.1186/s12868-019-0489-5) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer disease, Neuroinflammation, Neurodegeneration, Amyloid, Astrocytes, Secretase, BACE1, Chlamydia pneumoniae, Pathogens
Background
Alzheimer Disease (AD) is a chronic, progressive neurodegenerative illness regarded as the most common cause of dementia, which afflicts 46.8 million people worldwide—a number predicted to quadruple by the year 2050 [1]. AD also ranks among the top 10 causes of death in the US that can be neither prevented nor cured. Several investigations have aimed at deciphering etiologies that suggest the multiple causes or triggers of AD pathology [2, 3]. AD is diagnosed as one of two classifications: early-onset or familial AD, abbreviated EOAD or FAD, or late-onset or sporadic AD, abbreviated LOAD or SAD. Familial AD manifests symptoms at < 65 years of age. These cases are predominantly genetic and account for only 5% of all cases of AD. The majority of AD patients that present with symptoms at > 65 years of age are grouped into the category of late-onset AD. Numerous genome-wide association studies have identified that certain risk-associated alleles are expressed differently in patients suffering from EOAD/FAD versus LOAD/SAD [4, 5]. However, the etiology of SAD pathology, unlike that of FAD, is considered multifactorial rather than strictly genetic.
In 1992, Hardy and Higgins [6] first introduced the amyloid cascade hypothesis to explain the etiology of AD, which proposes that beta-amyloid (Aβ) results from the enzymatic processing of amyloid precursor protein (APP). The amyloid cascade hypothesis claims that the neurotoxicity triggered by Aβ initiates related pathologic processes such as formation of extracellular senile plaques, which is one of the characteristic hallmarks of AD. Senile plaques are composed of oligomerized Aβ and are the pathologic findings in FAD and SAD brains. In its monomeric form, Aβ is a 39–42 amino acid peptide fragment derived from the sequential cleavage of APP, a large, type I transmembrane protein. APP processing can occur in two pathways, the non-amyloidogenic pathway or the pro-amyloidogenic pathway. The initial APP-processing event is catalyzed predominantly by the α-secretase, a disintegrin and metalloproteinase-10 (ADAM10) in the non-amyloidogenic pathway [7], and the β-secretase, β-site APP cleaving enzyme 1 (BACE1), in the pro-amyloidogenic pathway [8, 9]. A second cleavage event is performed by a γ-secretase, a complex containing 4 subunits: presenilin 1 or 2 (PSEN1 or PSEN2), as the catalytic subunit, nicastrin (NCSTN), anterior pharynx defensive phenotype 1 (APH1) and presenilin enhancer-2 (PEN2) [10, 11]. Cleavage of APP by ADAM10 or BACE1 releases the soluble amino terminal products, soluble APP-α (sAPPα) or soluble APP-β (sAPPβ), respectively, and carboxy terminal fragments, C83 and C99, respectively. In the non-amyloidogenic pathway, C83 is further cleaved by PSEN into the APP intracellular domain (AICD) and a p3 peptide, a non-toxic form of amyloid. Whereas in the amyloidogenic pathway, PSEN cleaves C99 into AICD and Aβ fragments, of which the 42 amino acid fragment (Aβ1-42) is considered the most neurotoxic [12]. Due to its hydrophobic characteristics, Aβ1-42 acts as a nidus for seeding additional Aβ peptide fragments, thereby facilitating large, extracellular aggregations of Aβ [13, 14].
In the context of SAD pathogenesis, the well-established mutations of APP and PSEN that promote enhanced pro-amyloidogenic processing of APP in FAD are not implicated; rather, exogenous stimuli, such as environmental toxins or infectious pathogens that may alter their overall expression are implicated [15]. These exogenous stimuli trigger the activation of both neuronal and non-neuronal cells with subsequent release of pro-inflammatory cytokines and activation of intracellular signaling pathways [16, 17]. As a result of these types of stressors, activated glial and neuronal cell models have demonstrated increased transcriptional expression, and/or altered activity of ADAM10, BACE1, and PSEN1 [18–20]. The etiology of neurodegeneration in SAD may therefore result from, at least in part, the effects of exogenous stimuli on the expression of APP-processing secretases.
One such stimulus that has garnered significant support as a potential trigger of SAD pathology is infection of the CNS by various pathogens. Several pathogens that have been implicated in SAD include cytomegalovirus, herpes simplex virus type 1, Borrelia burgdorferi, and Chlamydia pneumoniae (Cpn) [21, 22]. The role of Cpn in SAD pathology has been illustrated at both the epidemiologic and cellular levels. This relationship was first cited in the seminal study by Balin et al. [23] that demonstrated that metabolically active Cpn was found by immunohistochemical, electron microscopic, and PCR techniques to be localized to areas of AD pathology in 17 of 19 post-mortem AD brains compared to 1 of 19 non-AD control brains. Another study validated the presence of viable Cpn in 80% of AD brains (versus 11.1% of age-matched controls) via multiple methods including in situ hybridization and PCR analysis of Cpn-specific targets [24]. Additional evidence for a causal relationship between Cpn and AD was demonstrated through intranasally inoculating the non-genetically manipulated BALB/c mouse with Cpn isolates from AD brains [25]. In that study, Aβ deposits associated with Cpn infection were found in brain areas that are typically affected in AD such as the hippocampus, the dentate gyrus and the amygdala. These plaques were surrounded by reactive astrocytes and, at times, encircled brain vasculature, suggesting the presence of cerebral amyloid angiopathy.
Epidemiologic assessments of Cpn and other infectious burdens in control versus AD brains show a correlation between infection and AD [21, 22, 24]. This evidence supports the hypothesis that the chronic neuronal and glial cell dysfunction visualized in the brains of SAD patients may be derived from early-acquired CNS infection by Cpn and similar intracellular pathogens with the potential to persist over time and reactivate from latency or persistence.
An investigation into aberrant APP metabolism and Aβ accumulation in the setting of inflammation needs to include an analysis of the role of astrocytes, the most abundant glial cells in the CNS. A common observation among studies investigating Cpn in post-mortem AD brains [23] and brains of Cpn-inoculated BALB/c mice [25] was the colocalization of Cpn and GFAP-labeled astrocytes, suggesting astrogliosis in response to Cpn infection. It is interesting to note that glial activation in AD patients is not uncommon, as revealed by PET imaging during the pre-symptomatic stages of AD, and is shown to correlate with the initial signs of Aβ accumulation [26]. Animal models and in vitro studies indicate that astrocytes respond to immune- and AD-associated triggers, such as TNF-α, IFN-γ, IL-1β, bacterial lipopolysaccharide and Aβ by releasing cytokines and modifying the expression and activity of APP processing enzymes, which in turn exacerbate neuroinflammatory and neuropathological changes in the AD brain [19, 20, 27–30]. These findings support the contention that reactive astrocytes contribute to the neurodegeneration and loss of cognition observed in AD. Therefore, investigating the effect of infection by Cpn on the processing of APP by astrocytes is invaluable in modeling potential mechanisms by which Cpn may trigger sporadic AD pathology, especially over time.
This study is aimed at investigating the effects of infection by Cpn on genes and the gene products involved in the processing of APP to produce Aβ, which is a major characteristic of AD pathology. By examining the effect of Cpn infection on validated pathways of astrocytic APP processing, this study provides evidence to support that AD pathology is recapitulated by infection with Cpn. This investigation explores how the expression and activity of APP-processing machinery, as defined by the amyloid cascade hypothesis, is altered as a result of Cpn infection of STTG1 human astrocytoma cells. The STTG1 human astrocytoma cell line has been suggested to be a valuable in vitro model for AD and its experimental therapies. This is due to STTG1’s heterozygous expression of the ApoE ε3/4 gene, its active participation in the pro-inflammatory cascade, and ability to both synthesize and breakdown Aβ [31–34]. Therefore, this in vitro model of Cpn infection of the CNS not only enhances our understanding of pathologic AD mechanisms, but also brings to light new research avenues investigating “the pathogen hypothesis” for early diagnosis and treatment of sporadic AD.
Methods
Cell culture and infection with Chlamydia pneumoniae
The human astrocytoma cell line CCF-STTG1 (CRL-1718) was obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were grown at 37 °C and 5% CO2 as a monolayer in culture medium RPMI-1640 (ATCC, 10-2001) supplemented with 10% (v/v) fetal bovine serum (FBS) in culture flasks (Corning Cell Culture Treated Flasks). Cells were trypsinized (Thermofisher) and transferred to 12 or 6 well polystyrene plates (Corning® CellBIND®) in culture medium for the western blot and RT-PCR experiments. For the immunocytochemistry experiments, cells were grown on 18.5 mm glass coverslips (neuvitro, GG-18-1.5-pre) in sterile 12-well plates. For cell infection experiments, 50% of conditioned growth media was removed and Cpn strain AR39 (ATCC, 52592) at MOI = 1 was added to 5 × 104 to 1 × 105 cells/well. To minimize variability Cpn lot number was held constant throughout experiments and each time point for a given experiment was inoculated on the same day. After centrifugation at 300×g for 30 min at RT, fresh growth media was added and cells were incubated for 6, 24, 48, and 72 h. Uninfected cells used as a negative control were processed in parallel with Cpn-infected cells. This procedure describes the preparation for one biological replicate. Each timepoint was repeated to achieve samples in biological triplicate, for which each was run in at least technical triplicate for western blot analysis, ELISA, immunocytochemistry, and molecular studies. Additional information of sample sizes for each study is included in its respective figure legend.
Immunocytochemistry
Cells grown on sterile 18.5 mm glass coverslips were incubated with the following primary antibodies: anti-Aβ1-42 at 1:500 (Synaptic Systems, 218703); anti-ADAM 10 at 1:100 (abcam ab39180), anti-BACE1 at 1:500 (abcam, ab10716), anti-presenilin-1 at 1:500 (ProSci 4203). Secondary antibody was used at 1:500 (Alexa Fluor® 594); FITC-conjugated chlamydial antibodies (Fitzgerald, 61C75-A and 60C19) were used at 1:100 to visualize the infection. BD Perm/Wash™ was used to as the antibody diluent and cell wash buffer. Coverslips were mounted on glass slides using FLUORO-GEL II with DAPI (EMS, 17985-50). Images were acquired using an Olympus FV1000 laser scanning confocal microscope with a 60×, 1.4NA oil immersion objective lens and FluoView 1000 software. For cell counts, images were acquired at 40× using the Nikon Eclipse 90i epi-fluorescence microscope.
Alzheimer disease RT-PCR array
Cells were harvested and RNA was isolated in biological triplicate from Cpn infected and uninfected astrocytes at each timepoint post-infection. Purified RNA was reverse-transcribed using RT2 First Strand Kit (Qiagen, 330401). To ensure that the comparisons of gene expression were valid for each timepoint post-infection, an equal amount of RNA template from uninfected and Cpn-infected cells within each timepoint was used for cDNA synthesis. cDNA was used to profile 84 different genes included in the Human Alzheimer Disease RT2 Profiler ™ PCR Array (Qiagen, PAHS-057ZC). Web-based PCR array data analysis software provided by Qiagen was used to collectively analyze raw Ct values for each AD-related gene included in the assay [35]. Human β-actin was automatically chosen by the analysis software as the housekeeping gene for standardization. Once each assay was normalized to β-actin, ΔCt values for each gene of interest in Cpn-infected cells were compared to that of uninfected cells to obtain a fold change between gene of interest expression in uninfected cells and that of Cpn-infected cells for each timepoint. Statistical significance in fold change values were determined by the Qiagen online analysis software, which uses a two-tailed student’s t test to compare gene expression in infected and uninfected samples. Statistically significant changes in expression of AD-related genes are listed in Additional File 1.
Western blot analysis
At the indicated timepoints post-infection, uninfected and Cpn-infected cells were lysed using 1 × RIPA lysis buffer (EMD Millipore, 20-188) supplemented with 1× protease inhibitor (Halt ™ Protease Inhibitor 100×, Thermo Scientific, 78430). Cell lysates were homogenized mechanically with mortar and pestle and through ice-cold sonication before resolving on 4–20% precast polyacrylamide gels (Bio-Rad, Mini-PROTEAN® TGX™ gels, 456-1094) using 1× Tris/glycine/SDS running buffer (Bio-Rad, 161-0732). Gels were transferred onto nitrocellulose membrane (iBlot® transfer stack, Life Technologies, IB3010-02). Membranes were then washed with wash buffer (Pierce ® Fast Western Blot Kit, Thermo Scientific, 35050) and labeled (24 h at 4 °C with gentle agitation) with primary antibodies diluted in antibody diluent (Pierce ® Fast Western Blot Kit, Thermo Scientific, 35050). The following primary antibodies were used: anti-ADAM10 at 1:400 (Santa Cruz Biotechnology, sc-48400), anti-BACE1 at 1:500 (abcam, ab108394), anti-presenilin-1 at 1:500 (abcam, ab76083), anti-chlamydial antibody at 1:200 (Fitzgerald 10C27B), and to label the housekeeping protein of interest, anti-β-actin at 1:500 (Santa Cruz Biotechnology, sc-8432). The following secondary antibodies were used: goat anti-mouse and anti-rabbit conjugates at 1:500 (Bio-Rad, 170-5046 and 170-5047, respectively). SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific, 34080) was then applied to the membranes and visualized using Bio-Rad VersaDoc Imaging System 4000MP. Densitometry analysis was conducted using FIJI software [36]. ADAM10, BACE1, and PSEN1 expression were assessed using the same samples, ensuring that the expression of each of these proteins could be accurately compared to each other. The expression of each protein of interest was quantified in 5–7 total samples. To determine statistical significance between protein levels derived from Cpn infected and uninfected cells, student’s t-test was conducted on the optical density values of each protein of interest normalized to that of β-actin as a loading control.
Quantitative analysis of Aβ1-42 using immunofluorescence
Following immunolabeling with anti-Aβ1-42 as previously described, thirty 2 μm Z-stack images were acquired across 3 separate coverslips (approximately 10 cells per coverslip) to ensure that representative populations of cells were captured and that the entire 3-dimensional depth of labeled Aβ was included in the analysis. Using FIJI software [36], each Z-stack image was separated into its three individual channels; DAPI and FITC were removed at this point and analysis was performed on the TRITC channel. From each analyzed image, a single 2D composite image was resolved to represent the maximum fluorescence intensity of each pixel of each 0.2 μm slice. Threshold fluorescence level was defined to include all Aβ labeling within the area of the cell. Mean fluorescence intensity of Aβ fluorescence exceeding the threshold fluorescence level was obtained for each imaged cell and a student’s t-test was conducted to determine statistical significance of Aβ1-42 fluorescence intensity between infected and uninfected cells for 24, 48 and 72 h timepoints.
Quantitative analysis of soluble APPα and APPβ using Meso Scale Discovery (MSD) ELISA
At the indicated timepoints post-infection, conditioned media from Cpn-infected and uninfected cells was removed and stored at − 80 °C. One milliliter of the conditioned media was thawed and concentrated using Eppendorf Vacufuge Plus at 45 °C. The MSD 96-well MULTI-SPOT sAPPα/sAPPβ assay was performed as directed by the manufacturer. Inter- and intra-assay %CV are listed in Additional File 2. Concentration readings of each individual sample (3 for each timepoint post-infection and infection status) of sAPPβ were divided by total sAPP (sAPPα + sAPPβ) to obtain a ratio of sAPPβ to total sAPP in the conditioned media of uninfected and Cpn-infected treatment groups.
ADAM10 and BACE1 Activity Assay
AnaSpec Sensolyte 520 ADAM10 and BACE1 Activity Assay Kits (AS72226 and AS71144, respectively) were used as a fluorimetric method for determining ADAM10 and BACE1 activity in Cpn-infected and uninfected astrocytes. After 48 hpi, cells were counted, (data not shown in results) harvested and placed into pre-chilled microcentrifuge tubes with pre-packed pestle for homogenization. Cells were washed with ice-cold PBS, and pelleted at 4 °C and 800×g for 10 min (4 samples for each treatment group). Mechanical and liquid nitrogen snap-freeze homogenization of fresh lysate allowed for retrieval of lysate without compromising enzymatic activity. Each enzyme assay was performed as directed by the manufacturer. Each sample was assayed in technical duplicate. Completed assays were loaded in a black, clear bottom plate and incubated at 37 °C for 1 h, then analyzed at Ex/Em 490/520 using a Fluoroskan Ascent FL microplate fluorometer. Background fluorescence of assay buffer was subtracted from final fluorescence measurements and each final measurement was normalized based on the protein concentration as determined by BCA protein assay (Pierce, 23225).
Results
Chlamydia pneumoniae infects STTG1 human astrocytoma cells in vitro and is maintained through 72 h post infection
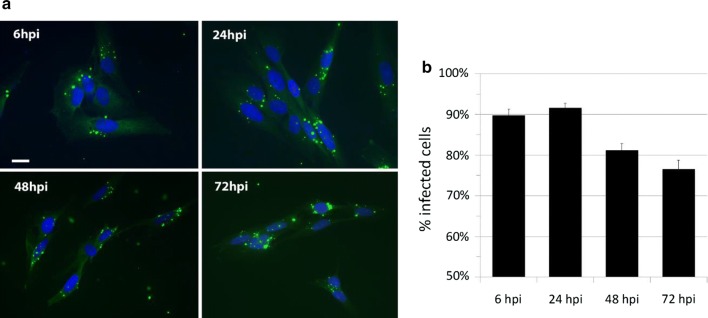
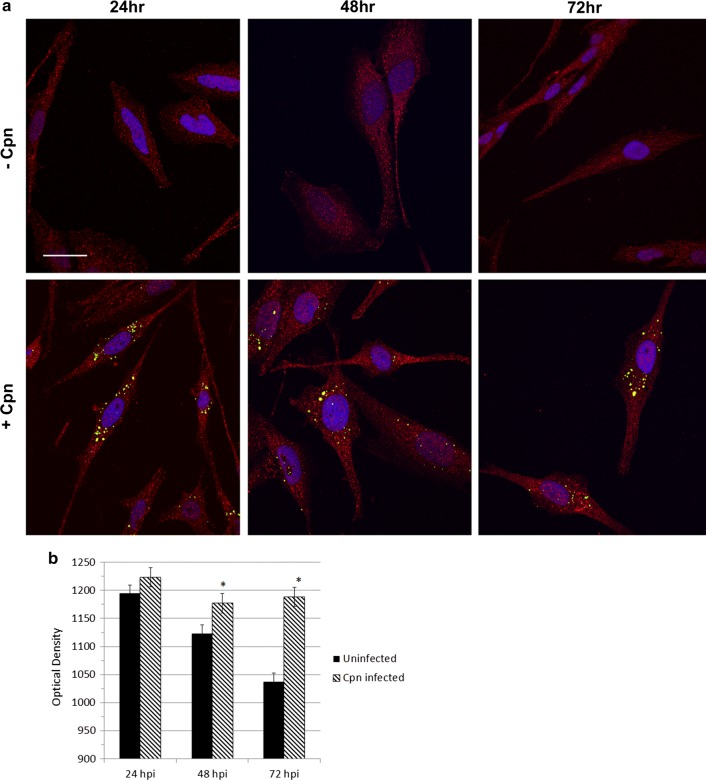
As visualized via confocal microscopy, the respiratory strain of Cpn, strain AR39, robustly infects STTG1 astrocytoma cells in vitro and persists 72 h post-infection (hpi) (Fig. 1a). Percent infected cells, averaged across approximately 2000–2500 cells per timepoint, is shown in Fig. 1b. Percentages of infected cells were significantly different between 6 hpi versus 48 hpi and 72 hpi, 24 hpi versus 48 hpi and 72 hpi. This was determined through conducting a one-way ANOVA, demonstrating p < 0.05, and confirmed with Tukey HSD post hoc analysis. These data indicate that Cpn infects human astrocytoma cells within 6 hpi and appears to remain viable within these cells for at least 72 h, although the numbers of infected cells decrease after 24 hpi.
Fig. 1.
Chlamydia pneumoniae infects human astrocytes in vitro. STTG1 human astrocytes infected with Cpn strain AR39 at an MOI of 1 demonstrated a diffuse punctate labeling of Cpn (green) from 6 to 72 hpi. Nuclei are labeled with DAPI (blue). Scale bar represents 20 μm (a). Infected versus uninfected cell counts were averaged across approximately N = 2000–2500 cells per timepoint and in biological triplicate across two independent infections. Numerical data is expressed as percent infected cells (b). Percentages of infected cells were significantly different between 6 hpi versus 48 hpi and 72 hpi, 24 hpi versus 48 hpi and 72 hpi. Comparisons between populations were determined through one-way ANOVA, where significance was defined as p < 0.05, and confirmed using Tukey HSD post hoc analysis. Error bars represent standard deviation of the mean
Chlamydia pneumoniae infection of human astrocytes alters the transcript expression of AD-related genes
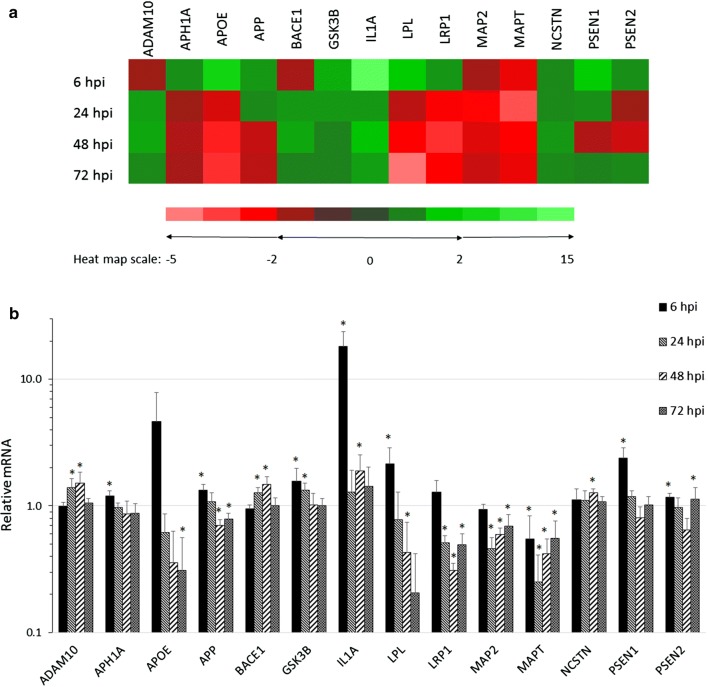
The human Alzheimer disease array revealed that Cpn infection at each timepoint post-infection altered the expression of several genes directly and indirectly involved with the development of AD pathology through APP processing and tau-related mechanisms. The fold changes in expression of the 84 AD-related genes in Cpn-infected cells, compared to that of uninfected cells were standardized to β-actin. At 6, 24, 48, and 72 hpi, significant changes in mRNA expression were observed in 40, 33, 35, and 17 different genes, respectively. The remaining genes were not included in our analysis due to the lack of a significant difference in their expression (p > 0.05). The functional roles of these genes (Fig. 2a) in astrocytes are aberrant in multiple pathways of AD, including lipid metabolism (apolipoprotein E, APOE; lipoprotein lipase, LPL; lipoprotein receptor-related protein 1, LRP1), microtubule organization (microtubule-associated protein 2, MAP2; microtubule-associated protein tau, MAPT; glycogen synthase kinase 3β, GSK3B), and neuroinflammation (interleukin 1-α, IL1A) [29, 37–39]. Moreover, expression of genes associated with several of these pathways have been shown to be altered upon infection with Cpn [40–42]. An additional subset of genes included in this analysis encode the secretases involved in APP processing, as well as APP itself. The transcripts of APP, ADAM10, BACE1, and subunits of the γ-secretase complex (PSEN1, PSEN2 APH1A and NCSTN) were significantly upregulated (p < 0.05) in at least one of the 4 investigated timepoints post-infection (Fig. 2b).
Fig. 2.
Chlamydia pneumoniae infection of human astrocytes alters the transcript expression of AD-related genes. Gene transcripts from Cpn-infected and uninfected cells analyzed at all four timepoints post-infection revealed significant fold changes in genes closely related to AD pathology. The fold changes of fourteen genes implicated in known pathways of AD pathology are presented in a. Histograms of fold changes of these AD-associated genes are presented in b. All expression data was normalized to β-actin and Cpn-infected and uninfected cDNA samples were repeated in biological (N = 3) and technical triplicate for each timepoint. Asterisk indicates p < 0.05. ADAM10, A disintegrin and metalloproteinase 10; APH1A, anterior pharynx defective protein 1A; APOE, apolipoprotein E; APP, amyloid precursor protein; BACE1, βAPP-cleaving enzyme 1; GSK3B, glucogen synthase kinase 3-β; IL1A, interleukin 1α; LPL, lipoprotein lipase; lipoprotein receptor-related protein 1, LRP1; MAP2, microtubule associated protein 2; MAPT, microtubule associated protein tau; NCSTN, nicastrin; PSEN1, presenilin-1, PSEN2, presenilin-2
Cpn had its greatest effect on the transcriptional expression of APP-processing secretases 6, 24, and 48 hpi. The altered gene expression observed as early as 6 hpi indicates that early Cpn entry into the astrocyte host may act as a trigger for the expression of genes required for the processing of APP. The increase in expression of APP (33%), PSEN1 (39%), PSEN2 (17%) and APH1A (20%) observed in infected cells compared to uninfected cells was greatest at 6 hpi, while that of ADAM10 and BACE1 was most elevated at 24 and 48 hpi (approximately 30–50% over that of uninfected cells). PSEN1, PSEN2 and APH1A function in concert with nicastrin, which demonstrated the greatest increase in expression at 48 hpi (approximately 30% over that of uninfected cells), to form the γ-secretase complex (Fig. 2b).
Our data indicate that the greatest increase in expression of secretase genes occurred within the first 48 hpi, though apparently neither the pro- nor the non-amyloidogenic pathway is favored, since expression of both the α-secretase (ADAM10) and β-secretase (BACE1) were similarly increased. These findings suggest that Cpn infection may be enhancing the processing of APP through transcriptional upregulation of secretase-associated genes.
Chlamydia pneumoniae infection of astrocytes alters the expression of the APP processing secretases
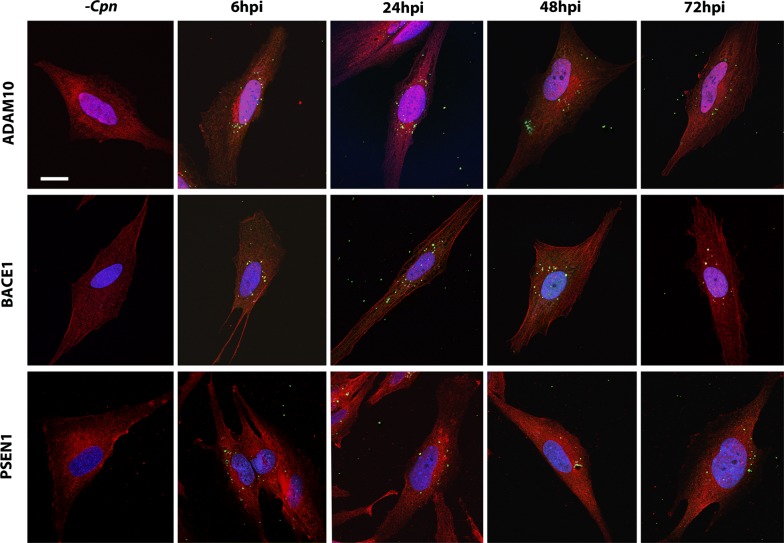
To determine whether Cpn-induced transcriptional changes in ADAM10, BACE1, and PSEN1 expression were consistent at the protein level, these proteins were visualized in Cpn-infected cells via confocal immunofluorescence. As ADAM10, BACE1, and PSEN1 proteins mature, they are recycled between the plasma and endosomal membranes [12], however, the antibodies used to visualize them in this study were chosen to be non-selective for intracellular and plasma membrane-localized populations. For example, BACE1’s C-terminal domain was targeted, which will detect BACE as it is recycled to and from endosomes and the plasma membrane or as it is shuttled through the late endosome/lysosome pathway.
Overall, the total fluorescence of each labeled protein in Cpn-infected cells showed very subtle differences at each timepoint post-infection compared to that of uninfected cells. ADAM10 labeling was not qualitatively different in Cpn-infected versus uninfected cells; BACE1 and PSEN1 labeling did show noticeable variations post-Cpn infection (Fig. 3). In our studies, the pattern of BACE1 labeling differed between uninfected and infected astrocytes, potentially representing localization to cellular membranes in Cpn-infected cells, though the intensity of BACE1 labeling appeared unchanged. Labeling of PSEN1, on the other hand, did appear increased in Cpn-infected relative to uninfected astrocytes.
Fig. 3.
Chlamydia pneumoniae infection of astrocytes alters labeling of secretases. Astrocytes infected with Cpn from 6 to 72 hpi were double labeled for Cpn (green) and secretases ADAM10, BACE1 or PSEN1 C-terminal fragment (red). 10–15 cells per biological replicate were imaged against an equal number of uninfected control cells. Cells were visualized using laser scanning confocal microscopy, maintaining the voltage settings of each color channel identical across biological replicates. DAPI was used to visualize the nucleus. Representative images are included this figure. Scale bar represents 20 μm
Chlamydia pneumoniae infection of astrocytes increases the protein expression of secretases involved in APP processing
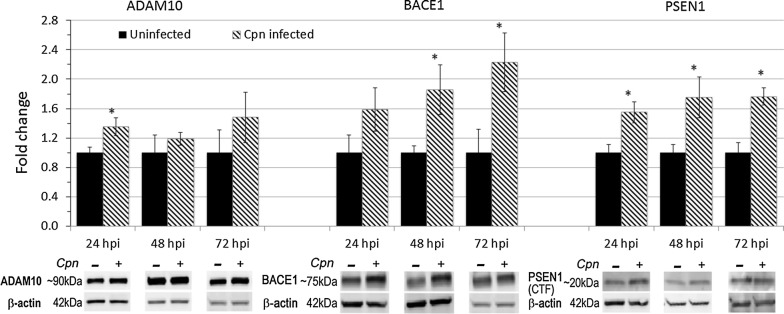
To examine Cpn’s regulatory dynamics on the non-amyloidogenic processing pathway, total cell lysates from Cpn infected and uninfected cells at 24, 48, and 72 hpi were harvested for western blot analysis to semi-quantitatively assess the expression of ADAM10 (α-secretase), BACE1 (β-secretase) and PSEN1 (a component of the γ-secretase complex). Densitometry histograms of fold change of ADAM10, BACE1 or PSEN1 protein levels in Cpn-infected cells represent the average change in protein levels across 5–7 replicates per time point, normalized to that of β-actin within each sample and shown relative to the amount of the respective protein present in uninfected cells for each timepoint (Fig. 4).
Fig. 4.
Protein expression of ADAM10, BACE1 and PSEN1 in Chlamydia pneumoniae-infected and uninfected astrocytes. Whole cell lysate was harvested from Cpn-infected and uninfected astrocytoma cells, resolved via SDS-PAGE gel electrophoresis, and labeled for secretase proteins. Fold change represents densitometry analysis for protein levels of full-length ADAM10, BACE1 and PSEN1 C-terminal fragment in Cpn-infected cells compared to that of uninfected cells at the same timepoint post infection. All densitometry values were normalized to that of β-actin for each biological replicate (N = 5–7). Statistical analysis was conducted using student’s T-test of fold change within each timepoint (asterisk indicates p < 0.05). Error bars represent standard error of the mean
The ADAM10 antibody used for western blots detected an immature, proenzyme form of ADAM10 (~ 90 kDa), the initiating enzyme for this pathway. Full length ADAM10 requires posttranslational modifications to be active, therefore the ADAM10 labeling presented in these analyses detects inactive enzyme [12]. Compared to that of uninfected samples, Cpn-infected astrocyte cell lysates showed a statistically significant 1.4 fold increase in ADAM10 protein levels (p < 0.05) at 24 hpi, but no significant changes were measured at other timepoints, although the trend at 72 hpi indicated increased protein levels in infected cells at this timepoint (Fig. 4).
The protein expression of BACE1 was assessed to determine Cpn’s effect on the pro-amyloidogenic processing of APP. The BACE1 labeling observed at approximately 75 kDa corresponds to the mature form of the protein. Mature BACE1 protein levels detected in Cpn-infected versus uninfected cell lysates significantly increased by 1.9 and 2.2 fold after 48 and 72 hpi, respectively, over that of uninfected cells (Fig. 4). These data suggest that Cpn infection results in a more extensive increase in protein levels of intracellular BACE1 relative to ADAM10, which may promote a pro-amyloidogenic rather than a non-amyloidogenic processing pathway of APP. Additionally, the increase in BACE1 protein expression observed with these studies does not seem transient; as time of infection progresses, BACE1 protein levels continued to increase throughout the 72 h of infection.
Active PSEN1 is localized to numerous subcellular compartments of the cell [43]. The antibody used for PSEN1 labeling via western blot analysis did not detect intracellular levels of full length PSEN1, which appears at 50 kDa, but rather a 20 kDa band, indicating proteolytically cleaved, and therefore active, carboxy terminal fragment (CTF) of PSEN1. At all timepoints tested (24, 48, and 72 hpi), PSEN1 CTF protein levels detected in Cpn-infected cell lysates were significantly increased over those of uninfected controls (p < 0.05) (Fig. 4). Corroborating the observations made via confocal immunofluorescence, these western blot results provide additional evidence for the role of Cpn in facilitating the accumulation of potentially active PSEN1.
Fluorescence intensity of Aβ1-42 is increased in Chlamydia pneumoniae infected astrocytes
Approximately 30 Z-images of uninfected and Cpn-infected cells were acquired for each timepoint and resolved into a single, 2D representation of the maximum Aβ1-42 fluorescence intensity of each 0.2 μm optical section (Fig. 5a). Aβ1-42 was localized within all cells, regardless of treatment group, indicating constitutively active APP-processing. Mean Aβ1-42 fluorescence intensity in Cpn-infected astrocytes, relative to that of uninfected cells, did not differ significantly at 24 hpi; however, it was elevated in infected cells at 48 and 72 hpi relative to uninfected cells from the same timepoint (Fig. 5b), indicating an increase in pro-amyloidogenic processing of APP at later times of infection.
Fig. 5.
Fluorescence intensity of Aβ1-42 is increased in Chlamydia pneumoniae-infected astrocytes. Aβ1-42 (red) and Cpn (green) were visualized by laser scanning, confocal microscopy (a). To analyze the Z-images using FIJI software, a defined threshold subtraction was applied equally to each image to determine Aβ1-42 fluorescence intensity (b); mean fluorescence intensity was calculated for infected and uninfected astrocytes at 24, 48 and 72 hpi. Cells (N = 25–30) were analyzed across three biological replicates to reliably conduct a student’s t-test on the fluorescence intensities of Aβ1-2 in uninfected and Cpn-infected cells. Error bars represent standard error of the mean. Asterisk represents p < 0.05
Chlamydia pneumoniae infection alters the activity of ADAM10 and BACE1 in initiating cleavage of APP
To study Cpn’s effect on ADAM10 and BACE1 activities in the initial cleavage of APP, MSD ELISA was used to quantify the concentration of soluble APPα (sAPPα) and soluble APPβ (sAPPβ) in the conditioned media of uninfected and Cpn-infected cells. Corroborating the significantly increased intracellular Aβ1-42 at 48 and 72 hpi, we observed a significant increase in the ratio of sAPPβ/total sAPP, expressed as a percentage (Fig. 6), at 48 and 72 hpi in Cpn-infected cells when compared to that of uninfected cells (p < 0.05). The levels of sAPPβ relative to total sAPP released by uninfected cells did not vary significantly between time points, suggesting a regulated balance of APP processing by ADAM10 and BACE1.
Fig. 6.
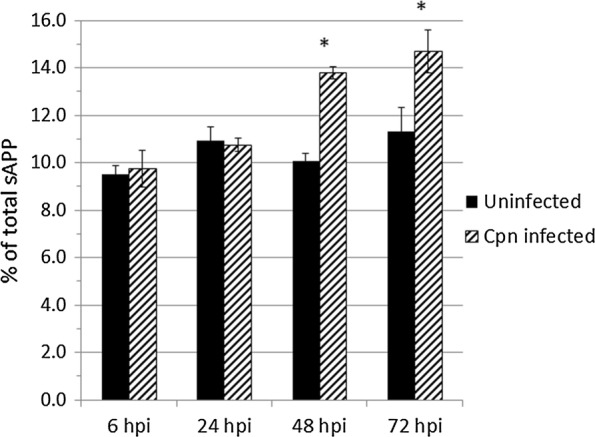
Quantification of sAPPβ/total sAPP in media of uninfected and Chlamydia pneumoniae infected astrocytes using MSD ELISA. Conditioned media of uninfected and Cpn-infected cells at each timepoint post-infection was collected, concentrated, and assayed in equal volumes for sAPPβ and sAPPα levels. Standard curves of known concentrations of sAPPβ and sAPPα was used to determine the concentration of these individual Aβ species. Conditioned media was obtained from three biological replicates and the assay was conducted in technical triplicate. Student’s t-test was calculated using the average sAPPβ/total sAPP ratio of Cpn-infected conditioned media compared to that of uninfected conditioned media. Asterisk represents p < 0.05
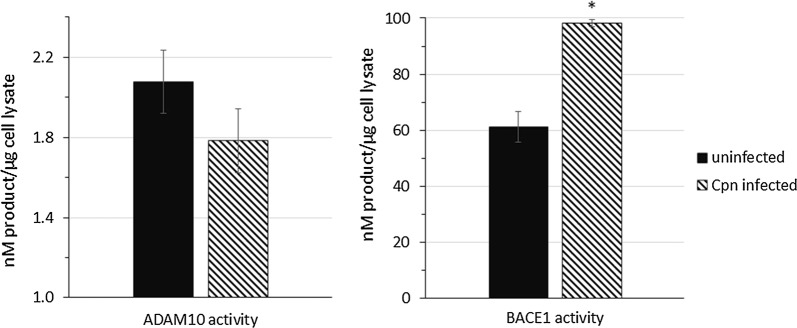
Chlamydia pneumoniae infection of astrocytes results in increased activity of BACE1 and decreased activity of ADAM10
The enzymatic activity of ADAM10 and BACE1 is contingent upon post-translational modifications that affect each enzyme’s trafficking to the optimal subcellular compartments for APP cleavage [12, 44]. To determine if Cpn-induced altered expression of ADAM10 and BACE1 at the protein level correlated to their altered enzymatic activity, fluorimetric FRET-based enzyme assays were conducted on whole cell lysate of uninfected and Cpn-infected cells. Compared to that of uninfected cells, the concentration of ADAM10-cleaved, fluorescent substrate in Cpn-infected cell lysate at 48 hpi exhibited an overall decreased trend in ADAM10 activity (Fig. 7). In contrast, the concentration of BACE1-cleaved, fluorescent substrate generated by Cpn-infected cell lysate 48 hpi was significantly greater than that of uninfected cell lysate (p < 0.05), indicating an overall greater activity of BACE1 in these samples (Fig. 7).
Fig. 7.
BACE1 activity is increased and ADAM10 activity is decreased in Chlamydia pneumoniae infected astrocytes. FRET-based assays were used to quantify the activity of ADAM10 and BACE1 enzyme activity generated by Cpn-infected and uninfected cell lysates at 48 hpi. Fluorescence of the 5-FAM or HiLyte Fluor 488 dyes conjugated to ADAM10 or BACE1-specific substrates was detected as a result of cleavage by the respective enzymes and compared back to fluorescence intensity of known dye concentrations. The quantified dye concentration from ADAM10 and BACE1 fluorescent substrate cleavage from (N = 4) biological replicates of Cpn-infected and uninfected cell lysates is presented as an average in the histograms. Error bars represent standard error of the mean. Asterisk represents p < 0.05
Discussion
By investigating Cpn’s effect on the transcription and localization of APP-processing secretases, this study is the first to propose a mechanism by which a respiratory strain of Cpn alters APP processing in an astrocyte host. Balin et al. [23] identified the Cpn pathogen within neurons, microglia and astroglia of AD brain tissue. Following this initial study, numerous groups have explored the roles of astrocytes as hosts for Cpn [45–47]. While our data indicate that Cpn clearly infects astrocytes, the percent of astrocytes that were infected decreased over time. The life cycle of Cpn is complex and involves transitioning from an infectious, elementary body (EB) to a metabolically active, reticulate body (RB) form of growth before converting back to an EB that can be released to infect new cells. Under conditions of stress, Cpn can persist as viable but noninfectious aberrant bodies (AB), which can revert back to RBs when conditions favor active multiplication [48]. It is possible that if cell division occurs more rapidly than Cpn can complete its life cycle and spread to a new host cell, then the number of infected cells relative to uninfected cells will decrease over time. Alternatively, it is possible that Cpn forms ABs in astrocytes, which would not subsequently spread to astrocytes arising from division of uninfected cells. This also could account for the decrease in % infected astrocytes observed over time.
In this study, we report that having gained access into the astrocyte host, Cpn promotes significant dysregulation of important AD-related genes directly involved with APP processing, pathologic lipid trafficking and microtubule dysfunction. While physiologically abnormal and potentially harmful for a mammalian host, the altered expression of these genes may have distinct benefits for Cpn infecting target cells. For example, Cpn infection of endothelial cells, monocytes, and macrophages has been shown to alter the expression of lipid homeostasis genes [40, 42, 49] and allow the acquisition and utilization of host lipids by Cpn as the pathogen is unable to synthesize them de novo [50]. The increased mRNA levels of apolipoprotien E (ApoE) and lipoprotein lipase (LPL) observed in Cpn-infected astrocytes relative to uninfected cells at 6 hpi may therefore be necessary to allow Cpn to initiate infection within the astrocyte host. In addition, APOE has been shown to play a role in the attachment and internalization of several intracellular pathogens, including Cpn [51–53]. The observed early increase in ApoE transcript expression may enhance internalization of Cpn EB’s during infection. Cpn-induced changes in host gene expression presumably evolved to enhance the infectivity of the bacterium, while potential AD-related pathologic effects associated with altered host gene expression would be an indirect consequence of Cpn infection.
It is conceivable that the initial increase in transcriptional ApoE expression has a direct impact on expression of APP. A recent study noted that the binding of glial ApoE to its receptors enhances the transcription of APP through activating the transcription factor activator protein-1 (AP-1) and its associated family of downstream effectors [54], the consequence of which may account for the 30% increase in APP transcription seen at 6 hpi. Furthermore, infection of endothelial cells by Cpn activates AP-1, and activation of this transcription factor regulates Cpn-induced inflammation [55]. If a similar scenario occurs in Cpn-infected astrocytes, AP-1 may play a role in mediating Cpn-triggered neuroinflammation as well as modulating levels of APP, and indirectly, Aβ levels. Data presented here indicate that Cpn stimulated transcription of the proinflammatory cytokine interleukin 1α (IL1α), suggesting that infection of astrocytes by Cpn promoted an inflammatory response. Moreover, a study by Lim et al. [41] reported that Cpn can activate an inflammatory response in monocytes, which, if occurring in microglial cells of the central nervous system, would reactivate nearby astrocytes. It is likely that Cpn infection in the human brain would trigger an inflammatory response that would exacerbate neurodegeneration associated with AD.
In this study, it is shown that Cpn infection of astrocytes decreased levels of microtubule-associated protein tau (MAPT) and microtubule-associated protein 2 (MAP2) mRNA, and increased that of glycogen synthase kinase 3-β (GSK3β), a kinase that can phosphorylate tau. A decrease in tau protein would destabilize host microtubules, whereas an increase in tau phosphorylation would subsequently decrease tau binding to, and stabilization of, microtubules [56]. An increase in GSK3β activity in Cpn-infected astrocytes could potentially enhance the formation of neurofibrillary tangles, which are composed of hyperphosphorylated tau proteins, thereby contributing to tau-mediated pathology that occurs in AD.
Changes in the expression of ADAM10, BACE1, and PSEN1 in Cpn-infected astrocytes will directly affect the processing of APP. In the present study, we observed significant increases in the expression of ADAM10 mRNA by 40–50% in Cpn-infected astrocytes relative to uninfected cells, as well an increase in the full length ADAM10 protein. However, it is important to note that the ADAM10 proenzyme needs posttranslational processing to be active and thus protein levels determined in this study may not accurately represent enzymatically active ADAM10 [7, 12]. In fact, ADAM10 protein levels tended to increase with Cpn infection while enzymatic activity was diminished at 48 hpi. These data suggest that posttranslational modification and/or trafficking of ADAM10 may differ in infected and uninfected astrocytes, resulting in decreased α-secretase-mediated non-amyloidogenic cleavage of APP in Cpn-infected cells.
Protein levels of both BACE1 and PSEN1 in Cpn-infected astrocytes progressively increased from 24 to 72 hpi relative to uninfected cells. Relative mRNA levels were greatest at 48 hpi for BACE1 and 6 hpi for PSEN1, indicating that the proteins persisted after transcription of these genes returned to levels consistent with that seen in uninfected astrocytes. The progressive increase in the amount of BACE1 and PSEN1 in Cpn-infected astrocytes over time paralleled the observation that Aβ1-42 labeling was greatest in infected cells at 48 and 72 hpi. It has been shown that in the presence of Aβ1-42, BACE1 activity is elevated because of impaired lysosomal degradation of BACE1, indicating that increased pro-amyloidogenic processing of APP favors persistence of this β-secretase [57, 58]. This positive feedback between Aβ1-42 and BACE1 levels may have occurred in our Cpn-infected astrocytes. Furthermore, it has been shown that low-density lipoprotein receptor-related protein 1 (LRP1) regulates BACE1 expression and activity by directing the β-secretase to lysosomes for degradation, while a loss of LRP1 expression correlates with an increase in BACE1 activity [59]. Thus the decreased transcription of LRP1 observed in Cpn-infected astrocytes may contribute to the increase in BACE1 expression and activity observed in infected astrocytes.
The lack of a significant increase in the fluorescence intensity of Aβ1-42 at 24 hpi suggests that prior to 24 hpi, the pro-amyloidogenic pathway has not been upregulated. If the increase in APP mRNA observed in Cpn-infected astrocytes at 6 hpi signals an increase in APP protein, then the surplus APP within the cell is processed by the non-amyloidogenic pathway at an early time of infection. However, at 48 to 72 hpi, the significant increase in Aβ1-42 fluorescence labeling in infected astrocytes suggests that in the presence of Cpn, either pro-amyloidogenic processing of APP is stimulated and/or there is decreased clearance of toxic, intracellular Aβ products. The increased protein concentration of BACE1 and PSEN1 present in infected astrocytes at 48 and 72 hpi supports the conclusion that increased processing of APP may be responsible for the greater intracellular accumulation of Aβ1-42, however doesn’t rule out the possibility of decreased clearance of Aβ. Our data indicating that the ratio of sAPPβ per total sAPP (sAPPα and sAPPβ) was significantly increased in Cpn-infected astrocytes at 48 and 72 hpi further supports the conclusion that APP processing favored the pro-amyloidogenic pathway as the infection progressed. These results could be explained by either an increase in BACE1-mediated cleavage and/or a decrease in ADAM10-mediated cleavage of APP. Interestingly, enzyme activity assays for BACE1 and ADAM10 indicated that at 48 hpi, enzymatic activity in Cpn-infected astrocytes was increased for BACE1 and decreased for ADAM10 relative to that measured in uninfected cells.
Our study supports the postulate that the mechanism by which Cpn induces AD pathology centers on the ability of the pathogen in astrocytes to temporally alter the expression and activity of the α- and β-secretases, and thereby alter the balance between the non- and pro-amyloidogenic APP processing pathways that occurs in uninfected cells. This study is the first to quantify the altered regulation of the predominant α, β-, and γ-secretases in the CNS, namely ADAM10, BACE1, and PSEN1, respectively, and Aβ1-42 in human astrocytes infected with Cpn. We have demonstrated that Cpn infection causes a significant increase in the amyloidogenic processing of APP, which correlates with increased protein levels and activity of the rate-limiting enzyme, BACE1. Whether through upregulating the transcriptional or post-transcriptional expression of BACE1 and the subunits of γ-secretase and/or simultaneously disrupting normal secretase trafficking, severe downstream effects on the CNS may result secondary to Cpn infection. Over time, Cpn-induced astrocyte activation culminating in neuroinflammation, altered APP processing favoring the amyloidogenic pathway, dysregulation of tau expression and function, and eventually neuronal death causes chronic, irreversible damage, resulting in pathology similar to that found in the CNS of AD patients.
Future directions
This investigation explores a potential Cpn-induced mechanism for Aβ formation by focusing on the pathway of APP processing by proamyloidogenic secretases, thereby identifying a putative early event triggering AD-associated pathology. While this study is the first to model a stable, Chlamydia pneumoniae-infection of human astrocytes in vitro and investigate its downstream effects on AD-related secretases, numerous additional studies can be conducted to further support our conclusions. The STTG1 human astrocytoma cell line has been used as a viable astrocyte model for AD [31–34]. Considering Cpn has been found in multiple areas of human cortex and vasculature as reported by Balin et al. [23], reproducing Cpn infection in additional cell types such as other astrocyte and glial cell lines, neuronal cells, and endothelial cells may provide a more thorough understanding of in vivo Cpn infection. Reversing the pathologic effects of Cpn infection with anti-microbial or anti-inflammatory medication may provide a viable therapeutic option for AD. In support of this, Hammond et al. [60] demonstrated decreased cerebral Aβ load in BALB/c mice inoculated with Cpn and subsequently treated with Moxifloxacin. Recent studies implicating pathogens in AD, including this current study, suggest that eliminating infectious triggers for AD pathology may be beneficial as a therapeutic target for preventing the initiation or progression of AD (Additional file 2).
Conclusions
This study examines the effect of Chlamydia pneumoniae infection on astrocytes, a major cell type in the CNS that plays an important role in establishing a state of neuroinflammation and neurodegeneration in the brain. Specifically, the data suggest that infection by Chlamydia pneumoniae promotes the pro-amyloidogenic pathway of APP processing by manipulating the expression and activity of the major secretases involved in generating toxic and nontoxic fragments of APP. Pro-inflammatory processes in the brain, and environmental stimuli that favor the pro-amyloidogenic pathway of APP processing, are emerging as potential triggers for the pathology associated with AD. We present evidence of an association between AD pathology and infection with Chlamydia pneumoniae, supporting the concept of an infectious etiology as a candidate to be considered in the pathogenesis of late onset AD. Furthermore, this study presents a potential target for preventing or slowing the progression of this neurodegenerative disease.
Additional files
Additional file 1: AD array tables. Table S1. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 6 hpi. Table S2. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 24 hpi. Table S3. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 48 hpi. Table S4. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 72 hpi. Description: These tables list the Alzheimer-associated genes whose expression in Cpn-infected cells significantly differed from that of uninfected cells. The genes examined were included in the Human Alzheimer Disease RT2 Profiler ™ PCR Array (Qiagen, PAHS-057ZC) that profiled 84 different Alzheimer-associated genes.
Additional file 2: MSD ELISA %CV values. Table S1. Percent CV for soluble APPα. Table S2. Percent CV for soluble APPβ. Description: These tables list intra- and inter-ELISA % coefficient of variation (CV) for the data generated by MSD ELISA.
Authors’ contributions
ZA and DA conceived and designed experiments. ZA performed all experiments, interpretation and data analysis. DL assisted as technical support for immunocytochemistry, confocal microscopy and was integral in the data analysis for these studies. DA and SH were also involved in data analysis. ZA, DA, and SH were the major contributors in writing the manuscripts. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Dr. Marcus Bell, and the Ph.D. Committee from PCOM, including Drs. Brian Balin and Marina D’Angelo, and at the University of the Sciences, Drs. Peter Berget and C. Nicole Sunnen, for their advice and guidance throughout this research. We especially would like to thank Ahmad Cader and Christine Hammond for their technical contributions and advice as well as Brittany Kane for her assistance with the preparation of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in the body of the manuscript or the additional files. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported from the following foundations: Center for Chronic Disorders of Aging (CCDA-funded through the Osteopathic Heritage Foundation), Division of Research at the Philadelphia College of Osteopathic Medicine; Adolph and Rose Levis Foundation for Alzheimer Disease Research. The Giunta Family Research Scholarship was utilized for Zein Al-Atrache PhD stipend support and lab supplies. These foundations did not have a role in the studies other than financial support.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- Aβ
beta-amyloid
- AD
Alzheimer disease
- ADAM10
a disintegrin and metalloproteinase-10
- AICD
APP intracellular domain
- ANOVA
analysis of variance
- AP-1
activator protein-1
- APH-1
anterior pharynx defective 1
- ApoE
apolipoprotein E
- APP
amyloid precursor protein
- BACE1
β-site APP cleaving enzyme 1
- BCA
bicinchoninic acid
- cDNA
complementary deoxyribonucleic acid
- CNS
central nervous system
- Cpn
Chlamydia pneumoniae
- CTF
carboxy-terminal fragment
- DAPI
4′,6-diamidino-2-phenylindole
- EB
elementary body
- EOAD
early onset Alzheimer disease
- ER
endoplasmic reticulum
- FAD
familial Alzheimer disease
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FRET
fluorescence resonance energy transfer
- GFAP
glial fibrillary acidic protein
- GSK3β
glycogen synthase kinase 3-β
- hpi
hours post-infection
- IFN-y
Interferon-y
- IL-1α
interleukin 1-α
- IL-1β
interleukin 1-β
- LOAD
late onset Alzheimer disease
- LPL
lipoprotein lipase
- LRP1
lipoprotein receptor-related protein-1
- MAP2
microtubule associated protein 2
- MAPT
microtubule associated protein tau
- mRNA
messenger RNA
- MSD
meso scale discovery
- ELISA
enzyme-linked immunosorbent assay
- NCSTN
nicastrin
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PEN2
presenilin enhancer-2
- PET
positron emission tomography
- PSEN1
presenilin-1
- PSEN2
presenilin-2
- RB
reticulate body
- RFU
relative fluorescence unit
- RT-PCR
real time polymerase chain reaction
- SAD
sporadic Alzheimer disease
- sAPPα
soluble APPα
- sAPPβ
soluble APPβ
- SDS-PAGE
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- TGN
trans-Golgi Network
- TNF-α
tumor necrosis factor-α
- TRITC
tetramethylrhodamine
- Tukey’s HSD
Tukey’s honest significance difference
Contributor Information
Zein Al-Atrache, Email: Zeinal@pcom.edu.
Danielle B. Lopez, Email: Daniellelop@pcom.edu
Susan T. Hingley, Email: SusanH@pcom.edu
Denah M. Appelt, Email: DenahA@pcom.edu
References
- 1.Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology. 2013;80:1778–1783. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362:329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 3.Sanabria-Castro A, Alvarado-Echeverría I, Monge-Bonilla C. Molecular pathogenesis of Alzheimer’s disease: an update. Ann Neurosci. 2017;24:46–54. doi: 10.1159/000464422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram L, Lill CM, Tanzi RE. The genetics of Alzheimer disease: back to the future. Neuron. 2010;68:270–281. doi: 10.1016/j.neuron.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Goate A, Hardy J. Twenty years of Alzheimer’s disease-causing mutations. J Neurochem. 2012;120:3–8. doi: 10.1111/j.1471-4159.2011.07575.x. [DOI] [PubMed] [Google Scholar]
- 6.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 7.Vingtdeux V, Marambaud P. Identification and biology of α-secretase. J Neurochem. 2012;120:34–45. doi: 10.1111/j.1471-4159.2011.07477.x. [DOI] [PubMed] [Google Scholar]
- 8.Cole SL, Vassar R. The role of amyloid precursor protein processing by BACE1, the γ-secretase, in Alzheimer disease pathophysiology. J Biol Chem. 2008;283:29621–29625. doi: 10.1074/jbc.R800015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kandalepas PC, Vassar R. Identification and biology of β-secretase. J Neurochem. 2012;120:55–61. doi: 10.1111/j.1471-4159.2011.07512.x. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper B, Iwatsubo T, Wolfe MS. Presenilins and γ-secretase: structure, function, and role in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006304. doi: 10.1101/cshperspect.a006304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurisch-Yaksi N, Sannerud R, Annaert W. A fast growing spectrum of biological functions of γ-secretase in development and disease. Biochim Biophys Acta (BBA) Biomembr. 2013;1828:2815–2827. doi: 10.1016/j.bbamem.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 12.Agostinho P, Pliássova A, Oliveira CR, Cunha RA. Localization and trafficking of amyloid-β protein precursor and secretases: impact on Alzheimer’s disease. J Alzheimers Dis. 2015;45:329–347. doi: 10.3233/JAD-142730. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SIA, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, et al. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc Natl Acad Sci USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarrett JT, Berger EP, Lansbury PT. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 15.Balin B, Hudson A. Etiology and pathogenesis of late-onset Alzheimer’s disease. Curr Allergy Asthma Rep. 2014;14:1–10. doi: 10.1007/s11882-013-0417-1. [DOI] [PubMed] [Google Scholar]
- 16.Ben Haim L, Carrillo-de Sauvage M, Ceyzériat K, Escartin C. Elusive roles for reactive astrocytes in neurodegenerative diseases. Front Cell Neurosci. 2015;9:11–21. doi: 10.3389/fncel.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burda J, Sofroniew M. Reactive gliosis and the multicellular response to CNS damage and disease. Neuron. 2014;81:229–248. doi: 10.1016/j.neuron.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasko I, Stampfer-Kountchev M, Robatscher P, Veerhuis R, Eikelenboom P, Grubeck-Loebenstein B. How chronic inflammation can affect the brain and support the development of Alzheimer’s disease in old age: the role of microglia and astrocytes. Aging Cell. 2004;3:169–176. doi: 10.1111/j.1474-9728.2004.00101.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee JW, Lee YK, Yuk DY, Choi DY, Ban SB, Oh KW, et al. Neuro-inflammation induced by lipopolysaccharide causes cognitive impairment through enhancement of beta-amyloid generation. J Neuroinflamm. 2008;5:37. doi: 10.1186/1742-2094-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto M, Kiyota T, Horiba M, Buescher JL, Walsh SM, Gendelman HE, et al. Interferon-γ and tumor necrosis factor-α regulate amyloid-β plaque deposition and β-secretase expression in swedish mutant APP transgenic mice. Am J Pathol. 2007;170:680–692. doi: 10.2353/ajpath.2007.060378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bu X, Yao X, Jiao S, Zeng F, Liu Y, Xiang Y, et al. A study on the association between infectious burden and Alzheimer’s disease. Eur J Neurol. 2015;22:1519–1525. doi: 10.1111/ene.12477. [DOI] [PubMed] [Google Scholar]
- 22.Harris SA, Harris EA. Herpes simplex virus type 1 and other pathogens are key causative factors in sporadic Alzheimer’s disease. J Alzheimers Dis. 2015;48:319–353. doi: 10.3233/JAD-142853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balin BJ, Gerard HC, Arking EJ, Appelt DM, Branigan PJ, Abrams JT, et al. Identification and localization of Chlamydia pneumoniae in the Alzheimer’s brain. Med Microbiol Immunol (Berl) 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- 24.Gerard HC, Dreses-Werringloer U, Wildt KS, Deka S, Oszust C, Balin BJ, et al. Chlamydophila (Chlamydia) pneumoniae in the Alzheimer’s brain. FEMS Immunol Med Microbiol. 2006;48:355–366. doi: 10.1111/j.1574-695X.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- 25.Little CS, Hammond CJ, MacIntyre A, Balin BJ, Appelt DM. Chlamydia pneumoniae induces Alzheimer-like amyloid plaques in brains of BALB/c mice. Neurobiol Aging. 2004;25:419–429. doi: 10.1016/S0197-4580(03)00127-1. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Vieitez E, Saint-Aubert L, Carter SF, Almkvist O, Farid K, Schöll M, et al. Diverging longitudinal changes in astrocytosis and amyloid PET in autosomal dominant Alzheimer’s disease. Brain. 2016;139:922–936. doi: 10.1093/brain/awv404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blasko I, Veerhuis R, Stampfer-Kountchev M, Saurwein-Teissl M, Eikelenboom P, Grubeck-Loebenstein B. Costimulatory effects of interferon-γ and interleukin-1β or tumor necrosis factor α on the synthesis of Aβ1-40 and Aβ1-42 by human astrocytes. Neurobiol Dis. 2000;7:682–689. doi: 10.1006/nbdi.2000.0321. [DOI] [PubMed] [Google Scholar]
- 28.Garwood CJ, Pooler AM, Atherton J, Hanger DP, Noble W. Astrocytes are important mediators of Aβ-induced neurotoxicity and tau phosphorylation in primary culture. Cell Death Dis. 2011;2:e167. doi: 10.1038/cddis.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennessy E, Griffin ÉW, Cunningham C. Astrocytes are primed by chronic neurodegeneration to produce exaggerated chemokine and cell infiltration responses to acute stimulation with the cytokines IL-1β and TNF-α. J Neurosci. 2015;35:8411–8422. doi: 10.1523/JNEUROSCI.2745-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, O’Connor T, Vassar R. The contribution of activated astrocytes to Aβ production: implications for Alzheimer’s disease pathogenesis. J Neuroinflamm. 2011;8:150. doi: 10.1186/1742-2094-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batarseh YS, Mohamed LA, Al Rihani SB, Mousa YM, Siddique AB, El Sayed KA, et al. Oleocanthal ameliorates amyloid-β oligomers’ toxicity on astrocytes and neuronal cells: in vitro studies. Neuroscience. 2017;352:204–215. doi: 10.1016/j.neuroscience.2017.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidana K, Tatebe T, Ito K, Hara N, Kakita A, Saito T, et al. Loss of kallikrein-related peptidase 7 exacerbates amyloid pathology in Alzheimer’s disease model mice. EMBO Mol Med. 2018;10:n/a. doi: 10.15252/emmm.201708184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim M, Sung M, Seo S, Yoo S, Lim W, Kim H. Water-soluble chitosan inhibits the production of pro-inflammatory cytokine in human astrocytoma cells activated by amyloid beta peptide and interleukin-1β. Neurosci Lett. 2002;321:105–109. doi: 10.1016/S0304-3940(02)00066-6. [DOI] [PubMed] [Google Scholar]
- 34.Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28:11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Data Analysis Center – QIAGEN. https://dataanalysis.qiagen.com/pcr/arrayanalysis.php.
- 36.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Liu C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Hu J, Zhao N, Wang J, Wang N, Cirrito JR, et al. Astrocytic LRP1 mediates brain aβ clearance and impacts amyloid deposition. J Neurosci. 2017;37:4023–4031. doi: 10.1523/JNEUROSCI.3442-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinohara M, Tachibana M, Kanekiyo T, Bu G. Role of LRP1 in the pathogenesis of Alzheimer’s disease: evidence from clinical and preclinical studies. J Lipid Res. 2017;58:1267–1281. doi: 10.1194/jlr.R075796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng B, Wu X, Sun S, Wu Q, Mei C, Xu Q, et al. MAPK–PPARα/γ signal transduction pathways are involved in Chlamydia pneumoniae-induced macrophage-derived foam cell formation. Microb Pathog. 2014;69–70:1–8. doi: 10.1016/j.micpath.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Lim C, Hammond CJ, Hingley ST, Balin BJ. Chlamydia pneumoniae infection of monocytes in vitro stimulates innate and adaptive immune responses relevant to those in Alzheimer’s Disease. J Neuroinflamm. 2014;11:217. doi: 10.1186/s12974-014-0217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu W, He P, Cheng B, Mei C, Wang Y, Wan J. Chlamydia pneumoniae disturbs cholesterol homeostasis in human THP-1 macrophages via JNK-PPARγ dependent signal transduction pathways. Microbes Infect. 2010;12:1226–1235. doi: 10.1016/j.micinf.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Rajendran L, Annaert W. Membrane trafficking pathways in Alzheimer’s disease. Traffic. 2012;13:759–770. doi: 10.1111/j.1600-0854.2012.01332.x. [DOI] [PubMed] [Google Scholar]
- 44.Lundgren J, Ahmed S, Schedin-Weiss S, Gouras G, Winblad B, Tjernberg L, et al. ADAM10 and BACE1 are localized to synaptic vesicles. J Neurochem. 2015;135:606–615. doi: 10.1111/jnc.13287. [DOI] [PubMed] [Google Scholar]
- 45.Boelen E, Steinbusch HWM, van der Ven AJAM, Grauls G, Bruggeman CA, Stassen FRM. Chlamydia pneumoniae infection of brain cells: an in vitro study. Neurobiol Aging. 2007;28:524–532. doi: 10.1016/j.neurobiolaging.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Dreses-Werringloer U, Bhuiyan M, Zhao Y, Gerard HC, Whittum-Hudson JA, Hudson AP. Initial characterization of Chlamydophila (Chlamydia) pneumoniae cultured from the late-onset Alzheimer brain. Int J Med Microbiol. 2009;299:187–201. doi: 10.1016/j.ijmm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond CJ, Hallock LR, Howanski RJ, Appelt DM, Little CS, Balin BJ. Immunohistological detection of Chlamydia pneumoniae in the Alzheimer’s disease brain. BMC Neuroscience. 2010;11:121. doi: 10.1186/1471-2202-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoborg RV. Chlamydia persistence—a tool to dissect Chlamydia–host interactions. Microbes Infect. 2011;13:649–662. doi: 10.1016/j.micinf.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Pietro M, Filardo S, De Santis F, Sessa R. Chlamydia pneumoniae infection in atherosclerotic lesion development through oxidative stress: a brief overview. Int J Mol Sci. 2013;14:15105–15120. doi: 10.3390/ijms140715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elwell CA, Engel JN. Lipid acquisition by intracellular Chlamydiae. Cell Microbiol. 2012;14:1010–1018. doi: 10.1111/j.1462-5822.2012.01794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gérard HC, Wildt KL, Whittum-Hudson JA, Lai Z, Ager J, Hudson AP. The load of Chlamydia pneumoniae in the Alzheimer’s brain varies with APOE genotype. Microb Pathog. 2005;39:19–26. doi: 10.1016/j.micpath.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 52.Gérard HC, Fomicheva E, Whittum-Hudson JA, Hudson AP. Apolipoprotein E4 enhances attachment of Chlamydophila (Chlamydia) pneumoniae elementary bodies to host cells. Microb Pathog. 2008;44:279–285. doi: 10.1016/j.micpath.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Carter CJ. APP, APOE, complement receptor 1, clusterin and PICALM and their involvement in the Herpes Simplex life cycle. Neurosci Lett. 2010;483:96–100. doi: 10.1016/j.neulet.2010.07.066. [DOI] [PubMed] [Google Scholar]
- 54.Huang Y, Mucke L. Alzheimer mechanisms and therapeutic strategies. Cell. 2012;148:1204–1222. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang A, Al-Kuhlani M, Johnston SC, Ojcius DM, Chou J, Dean D. Transcription factor complex AP-1 mediates inflammation initiated by Chlamydia pneumoniae infection. Cell Microbiol. 2013;15:779–794. doi: 10.1111/cmi.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iqbal K, Alonso ADC, Chen S, Chohan MO, El-Akkad E, Gong C, et al. Tau pathology in Alzheimer disease and other tauopathies. Biochim Biophys Acta (BBA) Mol Basis Dis. 2005;1739:198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 57.Guglielmotto M, Monteleone D, Boido M, Piras A, Giliberto L, Borghi R, et al. Aβ1-42-mediated down-regulation of uch-L1 is dependent on NF-κB activation and impaired BACE1 lysosomal degradation. Aging Cell. 2012;11:834–844. doi: 10.1111/j.1474-9726.2012.00854.x. [DOI] [PubMed] [Google Scholar]
- 58.Mamada N, Tanokashira D, Hosaka A, Kametani F, Tamaoka A, Araki W. Amyloid β-protein oligomers upregulate the β-secretase, BACE1, through a post-translational mechanism involving its altered subcellular distribution in neurons. Mol Brain. 2015;8:73. doi: 10.1186/s13041-015-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanokashira D, Motoki K, Minegishi S, Hosaka A, Mamada N, Tamaoka A, et al. LRP1 Downregulates the Alzheimer’s β-Secretase BACE1 by Modulating Its Intraneuronal Trafficking. Eneuro. 2015;2:e0006. doi: 10.1523/ENEURO.0006-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.C. Hammond, C. S. Little, N. Longo, C. Procacci, D. Appelt and B. Balin. Antibiotic Alters Inflammation in the Mouse Brain During Persistent Chlamydia pneumoniae Infection. Paper presented at: 10th International Conference on Alzheimer’s Disease and Related Disorders. International Proceedings for Alzheimer's Disease: New Advances. p.537–540, July 15–20, 2006. Madrid, Spain.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: AD array tables. Table S1. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 6 hpi. Table S2. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 24 hpi. Table S3. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 48 hpi. Table S4. AD-associated genes whose expression was significantly altered in Cpn-infected cells relative to uninfected cells at 72 hpi. Description: These tables list the Alzheimer-associated genes whose expression in Cpn-infected cells significantly differed from that of uninfected cells. The genes examined were included in the Human Alzheimer Disease RT2 Profiler ™ PCR Array (Qiagen, PAHS-057ZC) that profiled 84 different Alzheimer-associated genes.
Additional file 2: MSD ELISA %CV values. Table S1. Percent CV for soluble APPα. Table S2. Percent CV for soluble APPβ. Description: These tables list intra- and inter-ELISA % coefficient of variation (CV) for the data generated by MSD ELISA.
Data Availability Statement
All data generated or analyzed during this study are included in the body of the manuscript or the additional files. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.