Abstract
Background
Oral papillomas and verruca vulgaris have been associated with human papillomavirus (HPV) infection. However, approximately half of these have remained HPV‐negative when tested for mucosal HPV genotypes. In this study, we evaluated presence of α‐, β‐, and γ‐HPVs in benign papillary and verrucous lesions.
Methods
Eighty‐three clinical lesions with suspected HPV etiology were analyzed for HPV types of genus α (n = 24), β (n = 46), and γ (n = 52). Immunohistochemistry was used for p16 as a possible surrogate marker of high‐risk HPV, accompanied by Ki‐67 proliferation marker.
Results
Altogether, α‐HPVs were detected in 6.4%, β‐HPVs in 2.4%, and γ‐HPV in 4.8%. The following genotypes were identified: HPV6, 8, 11, 16, 22, 161, and 170. Neither Ki‐67 nor p16 positivity alone were associated with HPV but combined staining showed significant inverse association (P = .042).
Conclusion
HPV infection is found only in a minority of benign verrucous and papillary oral lesions, with the predominance of α‐HPVs.
Level of Evidence
4
Keywords: Oral, HPV, p16, papilloma, benign
INTRODUCTION
Benign epithelial lesions in the oral cavity include squamous cell papilloma, condyloma acuminatum, skin verruca vulgaris, focal epithelial hyperplasia (FEH), and papillary hyperplasia. Comprehensive data on the prevalence of these benign oral mucosal lesions are incomplete, with no systematic population‐based studies. Based on an old survey among 400,000 US children, oral papillomas were the most common (7.5%) oral epithelial lesions/tumors.1 Their prevalence is lower in adults, varying from 0.2% to 0.4%,2, 3, 4 most frequently located on the palate, tongue, lips or gingiva.5
All these benign epithelial lesions have been attributed to human papillomaviruses (HPV). Currently, nearly 200 HPVs are recognized, and their taxonomy is based on the phylogenetic evaluation of HPV L1 alignments.6, 7 HPVs belong to five separate genera: Alpha‐ (α), Beta‐ (β), Gamma‐ (γ), Mu, and Nu papillomavirus, based on five major phylogenetic branches that connect only close to the root of the phylogenetic tree.7 Recently, it has been shown that β‐ and γ‐HPVs can also be detected on the oral mucosa and/or saliva as asymptomatic infections8 but also in head and neck carcinomas.9, 10 HPV association is strongest for FEH, nearly all being caused by HPV13 or 32. Nearly 50% of the 223 papillomas analyzed until 1998 were HPV‐positive, HPV6 and HPV11 being the predominant genotypes.11 A more recent study from Dona et al.12 reported HPV DNA in 70% (22 of 31) of oral papillomas.
Oral condyloma cannot be reliably distinguished from oral papilloma either histologically, clinically or by HPV genotyping, because HPV6/11 are the most prevalent types in both lesions. Based on our limited survey of 116 oral condylomas published in 1988, HPV detection rate was 75%.11 In the genital tract, the terminology was revised in 1970 genital papilloma being replaced by genital condyloma.
Frithiof and Wersäll13 were the first to show viral particles in oral papillomas in 1967 by electron microscopy, followed later by a series of studies where HPV capsid antigen were identified in these lesions by immunohistochemistry.14, 15, 16, 17 Löning et al.18 and Naghasfar et al.19 were the first to describe HPV6/11 DNA with in situ hybridization (ISH), soon confirmed by others20, 21 or PCR.22, 23, 24, 25, 26 Even today, the number of analyzed samples is limited, and the data hampered by the lack of unanimous histological criteria, creating controversy on their etiological association with HPV. Among the 31 samples of Dona et al.,12 of the 22 HPV DNA positive cases, 68% represented mucosal and 32% cutaneous HPV types.
The present study identified 83 randomly selected oral samples in the archives, diagnosed as papilloma, condyloma, verruca vulgaris, or papillary hyperplasia, first assessed for the pathognomonic signs of HPV infection, ie, koilocytosis. In the next step, all samples were analyzed for the presence of α‐, β‐, and γ papillomaviruses. Cell proliferation was analyzed using Ki‐67 immunohistochemistry, also including p16 as a surrogate marker of high‐risk HPV, although anticipated to be rare in these benign lesions.
MATERIALS AND METHODS
Samples
A series of 83 formalin‐fixed paraffin‐embedded samples of oral lesions diagnosed as papilloma, condyloma, verruca vulgaris, or papillary hyperplasia at the Department of Oral Pathology, University of Turku, Finland from 1994 to 2008 were included in this study. No sample of FEH was available for the study. Of the clinical data, only the exclusion criteria for any previous head and neck malignancy were included. Histological diagnoses were reevaluated by an expert oral pathologist (SS). The revised histological diagnoses are shown in Table 1, based on the criteria of the World Health Organization. The study protocol was approved by the ethical committee of South‐Western hospital district (T28/2012).
Table 1.
Samples According to the Histology, Genotypes Detected, and Their Ki‐67 and p16 Results.
| N | Diagnosis | Koilocytes | HPV genotype | Ki67% | p16 |
|---|---|---|---|---|---|
| 1 | Hyperplasia | ‐ | 18.6 | ‐ | |
| 2 | Verruca vulgaris | ‐ | 23.4 | ‐ | |
| 3 | Hyperplasia | ‐ | 11 | 30.8 | ‐ |
| 4 | Papilloma | ‐ | 6 | 32.9 | + |
| 5 | Papilloma | ‐ | 16 | 15.8 | ‐ |
| 6 | Papilloma | ‐ | 27 | + | |
| 7 | Papilloma | ‐ | 28.2 | ‐ | |
| 8 | Hyperplasia | + | 29.5 | ‐ | |
| 9 | Hyperplasia | ‐ | 22.8 | ‐ | |
| 10 | Papilloma | ‐ | 161 | 43.6 | ‐ |
| 11 | Papilloma | + | 31 | ‐ | |
| 12 | Papilloma | ‐ | 31.1 | ‐ | |
| 13 | Verruca vulgaris | ‐ | 38.1 | ‐ | |
| 14 | Hyperplasia | ‐ | 30.2 | ‐ | |
| 15 | Papilloma | ‐ | 40.7 | + | |
| 16 | Hyperplasia | ‐ | 26 | ‐ | |
| 17 | Other* | + | 11 | 22.3 | ‐ |
| 18 | Hyperplasia | + | 16 | 28 | ‐ |
| 19 | Hyperplasia | ‐ | 31.1 | ‐ | |
| 20 | Papilloma | ‐ | 29.7 | ++ | |
| 21 | Papilloma | ‐ | 29.4 | ++ | |
| 22 | Papilloma | ‐ | 34.3 | ‐ | |
| 23 | Hyperplasia | + | 27.9 | ‐ | |
| 24 | Hyperplasia | + | 38 | ‐ | |
| 25 | Verruca vulgaris | ‐ | 8 | 41 | ‐ |
| 26 | Hyperplasia | ‐ | 161 | 49 | ‐ |
| 27 | Hyperplasia | ‐ | 39.1 | ‐ | |
| 28 | Other* | ‐ | 20.6 | + | |
| 29 | Other* | ‐ | 17.1 | ‐ | |
| 30 | Hyperplasia | ‐ | 12 | ‐ | |
| 31 | Hyperplasia | + | 33.6 | ‐ | |
| 32 | Hyperplasia | ‐ | 27.7 | ‐ | |
| 33 | Papilloma | + | 41.2 | ‐ | |
| 34 | Hyperplasia | ‐ | 20.1 | ‐ | |
| 35 | Hyperplasia | + | 36.9 | ‐ | |
| 36 | Papilloma | ‐ | 19.4 | ‐ | |
| 37 | Verruca vulgaris | ‐ | 24.9 | ‐ | |
| 38 | Papilloma | ‐ | 11 | 26.6 | + |
| 39 | Hyperplasia | ‐ | 22.9 | ‐ | |
| 40 | Hyperplasia | ‐ | 11 | 40.1 | ‐ |
| 41 | Hyperplasia | + | 36.4 | ++ | |
| 42 | Papilloma | ‐ | 46.1 | + | |
| 43 | Papilloma | + | 43 | ‐ | |
| 44 | Papilloma | ‐ | 27.5 | + | |
| 45 | Other* | ‐ | 26.6 | ‐ | |
| 46 | Hyperplasia | + | 6 | 12.7 | ++ |
| 47 | Verruca vulgaris | ‐ | 30.9 | ‐ | |
| 48 | Hyperplasia | ‐ | 49.1 | +++ | |
| 49 | Hyperplasia | + | 34.4 | ‐ | |
| 50 | Papilloma | ‐ | 23.9 | + | |
| 51 | Papilloma | ‐ | 30 | + | |
| 52 | Papilloma | ‐ | 41.2 | ‐ | |
| 53 | Papilloma | ‐ | 6 | 30.2 | ‐ |
| 54 | Hyperplasia | + | 16, 170 | 13.4 | ‐ |
| 55 | Hyperplasia | ‐ | 16.1 | ‐ | |
| 56 | Hyperplasia | + | 20.6 | + | |
| 57 | Papilloma | ‐ | 19.5 | ‐ | |
| 58 | Hyperplasia | + | 27.3 | ‐ | |
| 59 | Hyperplasia | ‐ | 26.2 | ‐ | |
| 60 | Papilloma | ‐ | 6 | 16.7 | + |
| 61 | Papilloma | ‐ | 28.8 | + | |
| 62 | Hyperplasia | ‐ | 13.2 | + | |
| 63 | Hyperplasia | ‐ | 170 | 13.6 | + |
| 64 | Papilloma | + | 32 | ‐ | |
| 65 | Hyperplasia | ‐ | 24.9 | ++ | |
| 66 | Hyperplasia | ‐ | 29.3 | + | |
| 67 | Papilloma | ‐ | 28.3 | ‐ | |
| 68 | Papilloma | ‐ | 31.1 | ‐ | |
| 69 | Papilloma | ‐ | 16 | 15 | ‐ |
| 70 | Papilloma | + | 29.5 | ‐ | |
| 71 | Hyperplasia | + | 34.2 | + | |
| 72 | Hyperplasia | ‐ | 38.2 | + | |
| 73 | Papilloma | ‐ | 6 | 21.4 | ++ |
| 74 | Hyperplasia | ‐ | 36.9 | ‐ | |
| 75 | Hyperplasia | ‐ | 45.7 | ‐ | |
| 76 | Hyperplasia | ‐ | 37.1 | ‐ | |
| 77 | Papilloma | ‐ | 39.2 | + | |
| 78 | Hyperplasia | ‐ | 14.5 | ‐ | |
| 79 | Verruca vulgaris | ‐ | 28.1 | ‐ | |
| 80 | Papilloma | ‐ | 22 | 27.4 | ‐ |
| 81 | Hyperplasia | ‐ | 26.2 | ‐ | |
| 82 | Hyperplasia | ‐ | 25.3 | ‐ | |
| 83 | Papilloma | ‐ | 29.8 | + |
Other = condyloma or atypical papilloma or Schneiderian papilloma.
+ = weak, + + = moderate, + ++ = strong positivity.
DNA Extraction
For DNA extraction sequential sections from the paraffin embedded tissue blocks were cut to cover approximately the size of 1 cm2 of the biopsy sample. After deparanifization, the sections were lysed in lysis buffer (10 mM Tris, 400 mM NaCl, 100 mM EDTA, 1% SDS), digested overnight at 37˚C with proteinase K (10 μg/ml) and the proteins were precipitated with saturated NaCl and ethanol.27 DNA was dissolved in 50 μl water, mixed for 15 to 30 min and stored at ‐20˚C for HPV DNA detection.
Detection of α‐ HPVs
Extracted DNA was amplified by nested PCR using general primers MY09 and MY11 and GP5+/bio GP6 + for biotinylation.28 PCR was performed in a 50‐μl reaction mixture using AmpliTaq Gold DNA polymerase. For the first primer set, the following protocol was used: amplification with initial hot start denaturation at 95˚C for 10 min, followed by 30 cycles of denaturation at 95˚C for 30 seconds, annealing for 55 seconds at 55˚C and primer extension for 60 seconds at 72˚C. For the second PCR 1 μl of the first PCR product was taken and PCR was run with the following profile: denaturation at 94˚C for 15 minutes, followed with 40 cycles of denaturation at 94˚C, annealing for 20 seconds at 38˚C for 30 seconds and extension at 71˚C for 4 minutes. HPV genotyping was made with Multimetrix kit (Regensburg, Germany) and Luminex Technology (Luminex Corporation, Austin, TX, USA) according to the manufacturer's instruction.29 Multimetrix kit can detect 24 HPV genotypes (low‐risk HPV6, 11, 42, 43, 44, and 70; high‐risk HPV16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82). Positive control of the beta‐globin gene was included for the quality of the template DNA.
Detection of β‐and γ‐HPV DNAs
β‐ and γ‐HPV detection method has been previously described.30 Shortly, HPV type‐specific E7 PCR was performed with QIAGEN Multiplex PCR kit (Qiagen) targeting type specific primers of 46 β‐HPV types (HPV5, 8, 9, 12, 14, 15, 17, 19, 20, 21, 22, 23, 24, 25, 36, 37, 38, 47, 49, 75, 76, 80, 92, 93, 96, 98, 99, 100, 104, 105, 107, 110, 111, 113, 115, 118, 120, 122, 124, 143, 145, 150, 151, 152, 159, 174) and 52 γ‐HPV types (HPV4, 65, 95, 60, 48, 50, 88,101, 103, 108, 109, 112, 116, 119, 121, 123, 126, 127, 128, 129, 130, 131, 132, 133, 134, 148, 149, 156, 161, 162, 163, 164, 165, 166, 167, 168, 169, 170, 171, 172, 173, 175, 178, 179, 180, 184, 197, 199, 200, 201, 202, and SD2), according to the classification by de Villiers.6 HPV‐SD2 official classification is pending and it was not included in any species.31 Positive control with two primers for amplification of the beta‐globin gene was included for the quality of the template DNA. After PCR amplification, the samples were analyzed with MPG using Luminex Technology (Luminex Corporation, Austin, TX, USA).29
Immunohistochemistry
Ki‐67 and p16 immunohistochemical staining were performed on formalin‐fixed and paraffin‐embedded samples. For monoclonal “Mouse Anti‐Human Ki‐67 Antigen, Clone Mib‐1” (Dako Canada, cat.no. M7240) for Ki‐67 was performed with a TechMate 500 Plus device. For p16 the monoclonal antibody to p16 (Cintec p16 Histology, Roche, cat.no 805‐4713) was used with Ventana Benchmark XT device. For detection Dako REAL Detection System Peroxidase/DAB+, Rabbit Mouse (Dako Canada, cat.no. K5001) and Ultraview Universal DAB (Roche, cat.no.760‐500) were used for Ki‐67 and for p16, respectively.
Ki‐67 positive cells were confirmed under microscopy and each sample was photographed in three different positive areas with x400 magnification and immunostained positive nuclei of the epithelial cells were calculated as the proportion (%) of positive epithelial cells of all epithelial cells in the view.
P16 positive immunostaining was determined as weak positive if some cells of epithelium showed clear cytoplasmic and/or nuclear positive reaction <25%. Staining was considered moderate if one or several parts 25% to 75% of the papilloma/hyperplasia showed strong positive cytoplasmic and/or nuclear reaction, and strong >75%. Samples with no staining or scattered single cells were scored negative.
Statistics
Statistical analyses were carried out using JMP Pro, Version 12 and SAS system for Windows, Version 9.4 (SAS Institute Inc, Cary, NC, USA). Continuous variables were characterized using means and standard deviations (SD) and in case of categorical variables frequencies and percentages were used. The association of histological diagnosis and HPV DNA detection was tested using Pearson's chi‐squared test. The association of Ki‐67 and p16‐staining to α‐HPV or combined HPV positivity was analyzed using logistic regression models, where Ki‐67 and p16 were included in the same model. Also the interactions of Ki‐67 and p16 were tested but if not significant, it was omitted from the model. The numbers for β‐HPV and γ‐HPV were too low for separate analyzes. The results were quantified using odds ratios (OR) with 95% confidence intervals (95% CI). P‐values less than .05 were considered as statistically significant.
RESULTS
Histopathological Diagnosis
Totally, 33 samples of 83 (39.8%) were diagnosed as papilloma (Fig. 1), 40 (48.2%) as papillary hyperplasia, six (7.2%) as verruca vulgaris, and four (4.8%) as other (condyloma, atypical or Schneiderian papillomas). Signs of koilocytes were found in 23% (19 of 83) of the samples (Fig. 1). They were most common in papillary hyperplasia (13 of 40, 32.5%) or papillomas (5 of 33, 15.2%). There was no correlation between HPV positivity and koilocytosis.
Figure 1.
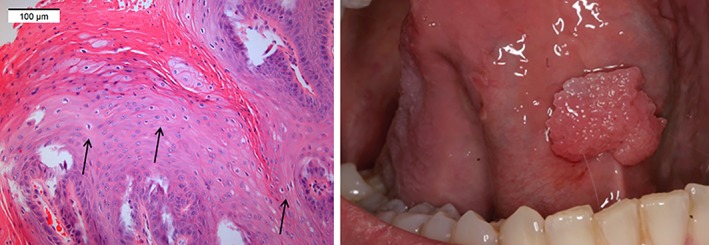
Gross appearance and clinical picture of a papilloma under the mobile tongue. The arrows showing koilocytes.
α‐, β‐ and γ‐HPV DNA Positivity
The overall positivity for HPV DNA was 21.7% (18 of 83). Only 13 of 83 (6.4%) of the samples were positive for α‐HPV, two (2.4%) for β‐HPV and four (4.8%) for γ‐HPV. Among α‐, β, and γ‐HPV, the following genotypes were found 6/11/16, 8/22, and 161/170, respectively (Table 1). Papillomas were most commonly associated with HPV, 9 of 33 (27.3%) samples. Seven papillary hyperplasias and one verruca as well as one in the group of “other” were HPV positive. One papillary hyperplasia was detected as α‐HPV positive (HPV16) and at the same time γ‐HPV positive (HPV170). Otherwise, no other multiple‐type infections were found. There was no statistically significant association between the histological diagnosis and HPV DNA detection (P = .843).
p16 and Ki‐67
Overall, 31.3% (26 of 83) of the samples were p16 positive (Fig. 2); 19 (22.9%) showing weak (<25%) p16 positivity, six (7.2%) moderate (25‐75%) positivity, and one with strong positivity (>75%). Of the α‐HPV positive samples, five were also p16 positive (four weak, one moderate), and of the γ‐HPV positive samples, only one was weakly p16 positive. None of the β‐HPV positive lesions were p16 positive. Of these α‐HPV and p16 positive lesions, four were papillomas and two papillary hyperplasias. The p16 staining (all combined) was not statistically significantly associated with α‐HPV positivity (P = .656). None of the HPV16 positive samples showed p16 positivity.
Figure 2.
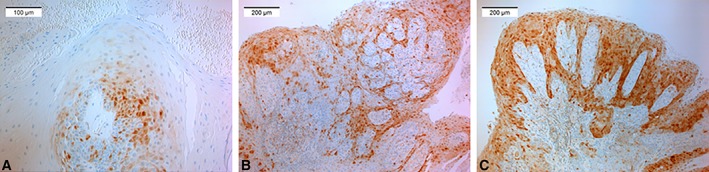
Micrographs showing the grading of p16 immunostaining; A) low (<25 of epithelium), B) moderate (25–75%), and C) strong (>75%) p16 positivity of the epithelia.
The proportion of Ki‐67 positive cells (Fig. 3) varied from 12.0% to 49.1%, with the mean 28.8% (SD 8.8). An increased proliferation rate (Ki‐67) was less likely to be α‐HPV positive (P = .024, OR 0.91, 95% CI 0.85–0.99).
Figure 3.
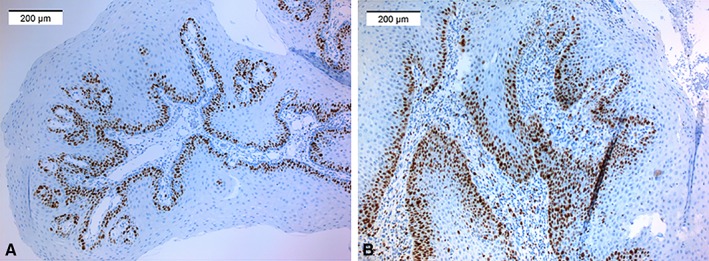
Examples of A) moderate and B) high Ki‐67 stainings.
Any HPV (α+β+γ) positivity was statistically significantly associated with simultaneous p16 and Ki‐67 positivity (P = .042). Among the samples with p16 positivity, the higher proportion (%) of Ki‐67 positive epithelial cells of all epithelial cells was inversely related to HPV positivity (OR 0.85, 95% CI 0.73–0.99).
DISCUSSION
In the present series, only 21.7% of the benign verrucous or papillary oral lesions were HPV DNA positive, dominated by α‐HPVs (68.4%). HPV prevalence in oral and oropharyngeal papillomas, condylomas and verruca vulgaris has been reported to range from 13% to 100% in a review of Castro and Bussoloti Filho.32 In a recent series of 83 squamous cell papillomas from the head and neck region, 48% and 22.6% harbored mucosal and cutaneous HPVs, respectively.12 Of the mucosal low‐risk genotypes, only HPV6 was found, being consonant with series, where HPV6 was also the most prevalent type, accompanied by three HPV11 positive samples. Dona et al.12 found the following mucosal high‐risk genotypes: HPV16, 18, 35, 51, and 74. In our series, only HPV16 was present in four samples. Dona et al.12 found eight different β‐types (5, 12, 23, 93, 96, 98, 110, 120) and six γ‐types (121, 123, 130, 131, 156, SD2). In our study with similar testing panel, these were HPV8, 22, and HPV161, 170, respectively. Not all the skin types were included in the test panel, eg, HPV2 and HPV57, which are known to be common also in oral skin verruca vulgaris.33, 34 Interestingly, we found only one verruca lesion which tested positive for HPV8. HPV8 is the high‐risk genotype traditionally associated with skin cancers associated with epidermodysplasia verruciformis (EV).
Only 2.4% and 4.8%, of our samples contained β‐ and γ‐HPVs, respectively. β‐ and γ‐HPVs have been found in oral mucosa as asymptomatic infections, and they are more prevalent in HIV‐infected than in immunocompetent individuals.8 Recently, β‐ and γ‐HPVs have been found also in head and neck carcinomas.9, 10 The significance of β‐ and γ‐HPVs in oral epithelial lesions needs further studies.
Koilocytosis has been regarded as a pathognomic sign of productive HPV infection. In our study, koilocytosis was not specific enough to identify HPV infection in benign oral epithelial lesions. In the late 1980's, we analyzed the sensitivity of acetic acid application (widely used in colposcopy) of the oral mucosa to predict asymptomatic oral HPV in 262 biopsies from a normal buccal mucosa.35 Koilocytes were found in 8% and 11% of the aceto‐white and aceto‐negative mucosa, while vacuolated cells were present in 63% and 61% of the biopsy samples taken from the aceto‐white and aceto‐negative mucosa. No association was found with the HPV DNA detection.35 Contradictory to that, koilocytosis with dyskeratosis might be a histological marker for HPV presence in oral cancer as reported by Khangura et al.36 This is in line with our original studies from the early 1980s where we first identified the pathognomonic features of HPV in head and neck cancers.15 These results were confirmed by immunohistochemistry using HPV‐specific antibody thus providing the first evidence on HPV as an etiologic agent of a subgroup of head and neck cancers.16 Taken together, the diagnosis of HPV in oral mucosa based on the presence of koilocytosis and/or vacuolated cells only is not reliable. On oral mucosa, continuous mechanical irritations result in degenerative changes in exophytic lesions which closely resemble koilocytosis or vacuolization, in the absence of HPV.
The proliferation rate of normal oral epithelial cells is low as determined from the Ki‐67 immunostaining. In normal mucosa, some 5% of the epithelial cells are Ki‐67 positive and the positivity is mostly located in the nuclei of basal and parabasal cells.37, 38 In our series, 28.8% (mean) of the epithelial cells were Ki‐67 positive, closely corresponding to the proliferation rate in oral squamous cell carcinomas.38 This implicates that oral papillomas and papillary lesions are highly proliferative. Unexpectedly, the proliferation was inversely associated with the presence of HPV DNA. This could suggest that a constant mechanical irritation of the mucosa rather than HPV itself is the leading cause of the cell proliferation in these benign oral lesions.
The overexpression of p16 is considered as a surrogate marker of high‐risk HPV infection. In the uterine cervix, p16 positivity together with Ki‐67 has been shown as a reliable method to predict HPV.39 p16 positivity is also widely used as a marker of HPV infection in oropharyngeal carcinoma. However, nearly 10% of the samples are false negative, while false positivity appears when no strict criteria are followed in grading the intensity and distribution of the p16 positivity.40 Recently, it has been recommended that the cut‐off for p16 positivity should be 75% of the cells in head and neck cancers. We anticipated that up‐regulated of p16 would be rare in our series, consisting of benign proliferative lesions with no progress to malignancy. As expected, the p16 positivity showed a similar staining pattern as found in normal epithelia, ie, diffuse and weak staining locating mostly in the basal layers. Interestingly, p16 positivity did not distinguish between the lesions with low‐risk or high‐risk HPV types. Most likely, this is explained by the fact that only a few samples with moderately strong p16 positivity were included in this series. Furthermore, senescence of the cell can also results in p16 expression.
Simultaneous p16 and Ki‐67 positivity was inversely associated with HPV DNA detection. Prigge et al.41 reported that p16/Ki‐67 co‐expression occurred exclusively in transformed cells of the head and neck region. However, they studied only carcinoma samples while our samples represented proliferative benign mucosal lesions.
To conclude, HPV infection was found only in 21.7% of our benign proliferative oral lesions. HPV genotypes represent mostly the α‐genus, with a few β‐ and γ‐HPVs. The following HPV genotypes were found: HPV6, 11, 16, 8, 22, 161, and 170. We also found that koilocytosis is not a reliable marker for HPV DNA in benign oral lesions. In addition, the use of p16 Ki‐67 immunostaining is of no diagnostic use in these oral lesions.
ACKNOWLEDGEMENTS
We thank Ms. Mariia Valkama and Ms. Tatjana Peskova for their skillful assistance in the laboratory.
BIBLIOGRAPHY
- 1. Jones HJ. Non‐odontogenic tumours in children. Br Dent J 1965:16:439–447. [PubMed] [Google Scholar]
- 2. Knapp MJ. Oral disease in 181,338 consecutive oral examinations. J Am Dent Assoc 1971;83:1288‐1293. [DOI] [PubMed] [Google Scholar]
- 3. Bouquot JE, Gundlach KK. Oral exophytic lesions in 23,616 white Americans over 35 years of age. Oral Surg Oral Med Oral Pathol 1986;62:284–291. [DOI] [PubMed] [Google Scholar]
- 4. Axell T. A prevalence study of oral mucosal lesions in an adult Swedish population. Odontol Revy Suppl 1976;36:1–103. [PubMed] [Google Scholar]
- 5. Abbey LM, Page DG, Sawyer DR. The clinical and histopathologic features of a series of 464 oral squamous cell papillomas. Oral Surg Oral Med Oral Pathol 1980:49:419–428. [DOI] [PubMed] [Google Scholar]
- 6. De Villiers EM. Cross‐roads in the classification of papillomaviruses. Virology 2013;445:2–10. [DOI] [PubMed] [Google Scholar]
- 7. Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of tazonomic amendments. Virology 2010;401:17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bottalico D, Chen Z, Dunne A, et al. The oral cavity contains abundant known and novel human papillomaviruses from the betapapillomavirus and gammapapillomavirus genera. JID 2011;204:787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Agalliu I, Gapstur S, Chen Z, et al. Association of oral α‐, β‐ and γ‐human papillomavirus types with risk of incident head and neck cancer. JAMA Oncol 2016. doi: 10.1001/jamaoncol.2015.5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabol I, Smahelova J, Klozar J, et al. Beta‐HPV types in patients with head and neck pathology and in healthy subjects. J Clin Virol 2016;82:159–165. [DOI] [PubMed] [Google Scholar]
- 11. Syrjänen K, Syrjänen S. Papillomavirus infections in human pathology. 1st ed. Checester, England: Wiley & Sons; 2000; 1‐615. [Google Scholar]
- 12. Dona MG, Pichi B, Rollo F, et al. Mucosal and cutaneous human papillomaviruses in head and neck squamous cell papillomas. Head Neck 2017;39:254–259. [DOI] [PubMed] [Google Scholar]
- 13. Frithiof L, Wersäll J. Virus‐like particles in human oral papilloma. Acta Otolaryngol 1967;64:263–266. [DOI] [PubMed] [Google Scholar]
- 14. Jenson AB, Lancaster WD, Hartmann DP, et al. Frequency and distribution of papillomavirus structural antigens in verrucae, #multiple |papillomas, and condylomata of the oral cavity. Am J Pathol Med 1982:107:212–218. [PMC free article] [PubMed] [Google Scholar]
- 15. Syrjänen K, Syrjänen S, Lamberg M, et al. Morphological and immunohistochemical evidence suggesting human papillomavirus (HPV) involvement in oral squamous cell carcinogenesis. Int J Oral Surg 1983;12:418–424. [DOI] [PubMed] [Google Scholar]
- 16. Syrjänen K, Happonen RP, Syrjänen S, Calonius B. Human papillomavirus (HPV) antigens and local immunologic reactivity in oral squamous cell tumors and hyperplasias. Scand J Dent Res 1984;92:358–370. [DOI] [PubMed] [Google Scholar]
- 17. Padayachee A, Van Wyk CW. Human papillomavirus (HPV) in oral squamous cell papillomas. J Oral Pathol 1987;25:427–432. [DOI] [PubMed] [Google Scholar]
- 18. Löning T, Ikenberg H, Becker J, Gissman L, Hoepfer I, zur Hausen H. Analysis of oral papillomas, leukoplakias, and invasive carcinomas for human papillomavirus type related DNA. J Inverst Dermatol 1985;84:417–420. [DOI] [PubMed] [Google Scholar]
- 19. Naghashfar Z, Sawada E, Kutcher MJ, et al. Identification of genital tract papillomaviruses HPV‐6 and HPV‐16 in warts of the oral cavity. J Med Virol 1985;17:313–324. [DOI] [PubMed] [Google Scholar]
- 20. Syrjänen SM, Syrjänen KJ, Happonen RP, Lamberg MA. In situ DNA hybridization analysis of human papillomavirus (HPV) sequences in benign oral mucosal lesions. Arch Dermatol Res 1987;279:543–549. [DOI] [PubMed] [Google Scholar]
- 21. Bu J, Pang J, Bu R. Study on the role of human papillomavirus in carcinogenesis of oral papillomas by in situ hybridization. Zhonghua Kou Qiang Yi Xue Za Zhi 2001;36:34–36. [PubMed] [Google Scholar]
- 22. Fouret P, Dabit D, Sibony M, et al. Expression of p53 protein related to the presence of human papillomavirus infection in precancer lesions of the larynx. Am J Pathol 1995;146:599–604. [PMC free article] [PubMed] [Google Scholar]
- 23. Ward KA, Napier SS, Winter PC, Maw RD, Dinsmore WW. Detection of human papilloma virus DNA sequences in oral squamous cell papillomas by the polymerase chain reaction. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 1995;80:63–66. [DOI] [PubMed] [Google Scholar]
- 24. Arndt O, Johannes A, Zeise K, Brock J. High‐risk HPV types in oral and laryngeal papilloma and leukoplakia. Laryngorhinotologie 1997;76:142–149. [DOI] [PubMed] [Google Scholar]
- 25. Badaracco G, Venuti A, Di Lonardo A, et al. Concurrent HPV infection in oral and genital mucosa. J Oral Pathol Med 1998;27:130–134. [DOI] [PubMed] [Google Scholar]
- 26. Barzal‐Nowosielska M, Miasko A, Staroslawska E, Sulewska A, Chyczewski L. Detection of human papilloma‐virus in papillomas of the oral cavity. Folia Histochem Cytobiol 2001;39:189–190. [PubMed] [Google Scholar]
- 27. Miller CS, White DK. Human papillomavirus expression in oral mucosa, #premalignant |conditions, and squamous cell carcinoma: a retrospective review of the literature. Oral Med Oral Surg Oral Pathol Endod 1996;82(1):57–68. [DOI] [PubMed] [Google Scholar]
- 28. Snijders PJ, Meijer CJ, Walboomers JM. Degenerate primers based on highly conserved regions of amino acid sequence in papillomaviruses can be used in a generalized polymerase chain reaction to detect productive human papillomavirus infection. J Gen Virol 1991;72:2781–2786. [DOI] [PubMed] [Google Scholar]
- 29. Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead‐based multiplex genotyping of human papillomaviruses. J Clin Microbiol 2006;44:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torres M, Gheit T, McKay‐Chopin S, et al. Prevalence of beta and gamma human papillomaviruses in the anal canal of men who have sex with men is influences by HIV status. J Clin Virol 2015;67:47–51. [DOI] [PubMed] [Google Scholar]
- 31. Mokili JL, Dutilh BE, Lim YW, et al. Identification of a novel human papillomavirus by metagenomics analysis of samples from patients with febrile respiratory illness. PLoS One 2013;8:e58404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Castro TP, Bussoloti Filho I. Prevalence of human papillomavirus (HPV) in oral cavity and oropharynx. Braz J Otorhinolaryngol 2006;72:272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Koning MN, Quint KD, Brukkink SC et al. High prevalence of cutaneous warts in elementry school children and the ubiquitous presence of wart‐associated human papillomavirus on clinically normal skin. Br J Dermatol 2015;172:196–201. [DOI] [PubMed] [Google Scholar]
- 34. Bruggink SC, Gussekloo J, de Koning MN et al. HPV type in plantar warts influences natural course and treatment response: secondary analysis of a randomised controlled trial. J Clin Virol 2013;57:227–232. [DOI] [PubMed] [Google Scholar]
- 35. Kellokoski JK, Syrjänen SM, Syrjänen KJ, Yliskoski M. Oral mucosal changes in women with genital HPV infection. J Oral Pathol Med 1990;19:142–148. [DOI] [PubMed] [Google Scholar]
- 36. Khangura RK, Senzupta S, Sircar K, Sharma B, Singh S, Rastogi V. HPV involvement in OSCC: correlation of PCR results with light microscopic features. J Oral Maxillofac Pathol 2013;17:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Birajdar SS, Radhika MG, Paremala K, Sudhakara M, Soumya M, Gadivan M. Expression of Ki‐67 in normal oral epithelium, leukoplakic oral epithelium and oral squamous cell carcinoma. J Oral Maxillofac Pathol 2014;18:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piatelli A, Rubini C, Fioroni M, Iezzi G, Santinelli A. Prevalence of p53, bcl‐2, and Ki‐67 immunoreactivity and of apoptosis in normal oral epithelium and in premalignant and malignant lesions of the oral cavity. J Oral Maxillofac Surg 2002;60:532–540. [DOI] [PubMed] [Google Scholar]
- 39. Mahajan A. Practical issues in the application of p16 immunohistochemistry in diagnostic pathology. Hum Pathol 2016;51:64–74. [DOI] [PubMed] [Google Scholar]
- 40. Carlander AF, Cronhoj Larsen C, Jensen DH, et al. Continuing rise in oropharyngeal cancer in a high HPV prevalence area: a Danish population‐based study from 2011 to 2014. Eur J Cancer 2017;70:75–82. [DOI] [PubMed] [Google Scholar]
- 41. Prigge E, Toth C, Dyckhoff G, et al. p16INK4a/Ki‐67 co‐expression specifically identifies transformed cells in the head and neck region. Int J Cancer 2015;136:1589–1599. [DOI] [PubMed] [Google Scholar]


