Abstract
Intestinal epithelial cells are among the most rapidly proliferating cell types in the human body. There are several different subtypes of epithelial cells, each with unique functional roles in responding to the ever-changing environment. The epithelium’s ability for rapid and customized responses to environmental changes requires multitiered levels of gene regulation. An emerging paradigm in gastrointestinal epithelial cells is the regulation of functionally related mRNA families, or regulons, via RNA-binding proteins (RBPs). RBPs represent a rapid and efficient mechanism to regulate gene expression and cell function. In this review, we will provide an overview of intestinal epithelial RBPs and how they contribute specifically to intestinal epithelial stem cell dynamics. In addition, we will highlight key gaps in knowledge in the global understanding of RBPs in gastrointestinal physiology as an opportunity for future studies.
INTRODUCTION
The intestinal epithelium is one of the most rapidly proliferating organs in the human body and renews itself every 3–5 days (19). Within the epithelial layer, there are several different subtypes of cells, each with unique and/or overlapping genetic markers identified in recent years (13–17). The intestinal epithelium is highly dynamic as a function of its requirement to rapidly adapt to environmental changes. The ability of the epithelium to respond to acute or chronic environmental changes may be disturbed during prolonged intestinal injury, inflammation, irradiation, toxin exposure, or cancer. Therefore, developing a more comprehensive understanding of how key pathways are regulated during homeostasis will lay a foundation for defining new therapeutic strategies in the future.
RNA-binding proteins (RBPs) have long been studied for their roles in fundamental biological processes, including transcription and translation of target RNAs, leading to regulation of various cellular functions. RBPs bind their target RNAs on specific domains and binding motifs within ribonucleoprotein (RNP) complexes and can facilitate mRNA stability, degradation, translation, splicing, polyadenylation, localization, and transport in different contexts (reviewed in Ref. 37). As such, RBPs represent a rapid and efficient mechanism to regulate gene expression and often do so through regulating multiple transcripts within the same pathway, representing posttranscriptional “regulons” (37). From a teleological perspective, RBPs may be particularly critical in rapidly proliferating organs to facilitate cellular adaptation to normal homeostatic stressors as well as during pathological states. It is therefore critical to understand both the basic biology of RBPs and how they may contribute to disease. In this review, we will provide a summary of intestinal epithelial RBPs and how they contribute to normal intestinal epithelial cell dynamics, with a focus on intestinal stem cells and homeostasis.
INTESTINAL RBPs REGULATE CRYPT CELL DYNAMICS
The intestinal epithelial layer functions as conveyor belt of cells that allows for rapid turnover. This phenomenon is dependent on the presence of stem cells residing in the base of intestinal and colonic crypts (1, 62). Two distinct populations of stem cells have been described in the intestinal epithelium. Active stem cells or crypt base columnar cells (CBCs), as their name suggests, reside in the crypt bottoms intermingled with Paneth cells, a differentiated cell type contributing to intestinal immune response and the stem cell niche. CBCs are rapidly cycling (approximately every 24 h), self-renewing, and responsible for maintaining homeostasis during tissue turnover (1). The canonical Wnt target gene Lgr5 was the first protein shown to mark this population, and thus far it has been used extensively to study these cells along with newly discovered markers such as Olfm4 and Ascl2 (1, 71, 72). Reserve intestinal stem cells (rISCs) comprise a functionally distinct population of stem cells in intestinal epithelium, cycling less frequently than CBCs and residing roughly four cell positions from the crypt base (62). Commonly used markers for studying rISCs are Bmi1, Hopx, Lrig1, mTert, among others (25, 50, 56, 62, 65). Studies have shown that during conditions of stress such as whole body irradiation or inflammation, which largely ablate CBCs, rISCs are able to exit their slow-cycling state and acquire stem-like activity, making them essential for intestinal regeneration (58, 67, 79). To regulate stem cell proliferation and differentiation during homeostasis, damage, and disease, intestinal stem cells must be capable of efficient regulation of gene expression. RBPs provide a mechanism by which stem cells can fine-tune or amplify gene expression at the posttranscriptional level via sequestration of newly transcribed RNA molecules into RNP particles where they can be triaged based on context-dependent cellular needs.
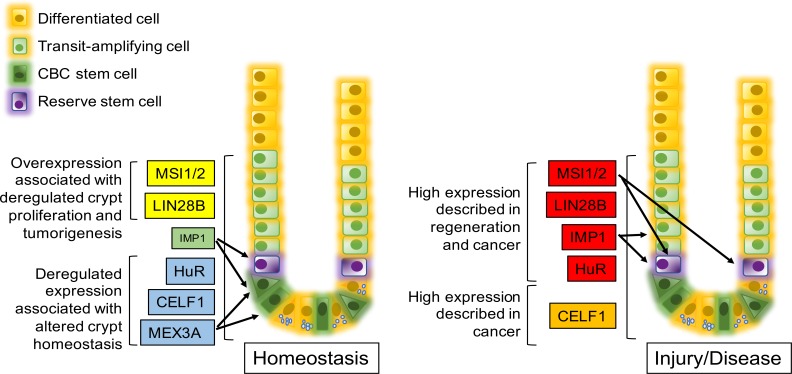
In recent years, several RBPs have been identified with key roles in intestinal homeostasis and response to injury (Fig. 1). Among the most studied RBPs in the intestine are the Musashi (MSI) family of proteins, with particular focus on the highly conserved mammalian isoforms, MSI1 and MSI2. It has been previously reported that the MSI proteins are highly expressed throughout intestinal crypts; however, ablation of these proteins in the intestine using the Villin promoter fails to produce any overt phenotype. Moreover, Msi deletion does not affect stem cell activity or canonical WNT pathway signaling in CBCs. Conversely, Msi ablation specifically in Bmi1/Hopx+ “reserve” stem cells was sufficient to induce G1 arrest during homeostasis and abrogate epithelial regeneration following gamma-irradiation. Msi proteins are further necessary and sufficient to activate rISCs and induce cell cycle reentry, highlighting the critical role of MSI proteins in reserve stem cell dynamics (84). MSI1 and MSI2 are functionally redundant and overexpression of either is sufficient for epithelial transformation. Moreover, loss of both proteins is required to inhibit proliferation in a carcinogen-induced colitis-associated model of intestinal tumorigenesis (40). Together, these studies suggest that despite the dispensability of MSI proteins during homeostasis, their expression in rISCs is critical in regulating crypt dynamics during stress.
Fig. 1.
Schematic of RNA-binding protein (RBP) expression during homeostasis and disease. RBPs are preferentially expressed in intestine and colon crypts. Brackets indicate general area where expression has been observed. Arrows indicate expression described in a specific cell type. Font size indicates relative expression levels. During homeostasis, yellow box indicates that overexpression can lead to deregulated crypt proliferation and tumorigenesis, green box indicates that altered expression of the RBP does not affect homeostasis, and blue indicates that deregulated expression of the RBP affects homeostasis but has not been shown to induce tumorigenesis. During injury or disease, a red box indicates expression of the RBP is increased during intestinal regeneration and cancer, while orange indicates that expression is associated with cancer. CBC, crypt base columnar cell.
LIN28A and LIN28B are RBPs expressed during embryonic development in the intestine. LIN28B is expressed in crypt cells in the adult mouse intestine, with lower expression in villus tip epithelia (48). LIN28B modulates several key pathways in the intestine, both directly by binding to target transcripts in the pathways (26, 68) and indirectly via inhibiting maturation of members of the let-7 miRNA family. LIN28B in turn is regulated by the let-7 microRNA family (30, 31, 55, 59, 75). Intestinal Lin28b overexpression via the murine Vil1 promoter is sufficient to induce intestinal hypertrophy, extensive crypt proliferation, and transformation. This phenotype was attributed mainly to repression of Let7 miRNA through LIN28B, which inhibits intestinal epithelial growth by repressing crypt fission and cell cycle progression (47, 48). Following this study, an independent group also demonstrated enhanced crypt proliferation as well as loss of various differentiated cells following global LIN28A and LIN28B overexpression (68). The same group found that LIN28 overexpression in ApcMin/+ mice accelerated tumor progression and increased invasiveness (68). Finally, studies in Drosophila intestine demonstrate that Lin28 loss leads to decreased intestinal stem cell numbers and slower intestinal stem cell rates of division. Furthermore, Lin28 deficiency significantly decreased symmetric intestinal stem cell division after feeding via reduced insulin-like receptor levels (12). Given the role that LIN28 RBPs play in regulating crypt proliferation, their expression specifically in the stem cell compartment is likely critical to maintain intestinal homeostasis.
Similar to LIN28B, IGF2 mRNA-binding protein 1 (IMP1) is expressed highly during embryonic development in the intestine and its expression is low and limited to the crypts in adult mouse intestine (10, 21). IMP1 has been shown to regulate several developmental signaling pathways such as the IGF2 and WNT/β-CATENIN pathways. IMP1 hypomorphic mice exhibit dwarfism and compromised intestinal development (29). Recent studies demonstrated that IMP1 is upregulated in mice following whole body irradiation, suggesting a possible role during intestinal regeneration. The same study demonstrated that IMP1 is limited or absent in the intestinal epithelium except for crypt cells and is upregulated in reserve stem cells following irradiation (10). Loss of Imp1 in reserve stem cells was associated with reduced weight loss and increased crypt proliferation following whole body irradiation. Furthermore, this study demonstrated that intestinal Imp1 deletion in the context of LIN28B overexpression promoted increased expression of stem cell markers and WNT target genes while increasing crypt proliferation, underscoring a role for IMP1 in regulating stem cell homeostasis (10).
The RBP ELAV-like protein 1 or HuR (human antigen R) is highly expressed in the gut mucosa and has been implicated in intestinal proliferation and regeneration after injury (46). Mice with intestine-specific HuR deletion exhibit increased injury with doxorubicin-induced acute intestinal damage (24). In a separate study, loss of HuR in the intestine was associated with mucosal atrophy as well a robust decrease in crypt proliferation during homeostasis. Additionally, HuR-deficient mice exhibited decreased regenerative potential after exposure to irradiation (43). Interestingly, colonic mucosal growth was unaffected by HuR loss. HuR directly binds to and enhances translation of the WNT coreceptor LRP6 and the WNT target gene MYC. Furthermore, HuR silencing is sufficient to inhibit intestinal epithelial cell proliferation in WNT3A-transfected IEC-6 cells, suggesting an intestinal epithelial cell intrinsic mechanism by which HuR regulates the WNT pathway and intestinal homeostasis (43). Although HuR expression in specific intestinal epithelial cell populations has not yet been addressed, the reliance of the intestinal stem cell compartment on appropriate WNT-signaling regulation to maintain tissue homeostasis suggests HuR may be a key factor in stem cell dynamics. Additionally, it has been shown that deletion of HuR decreases proliferation and tumor burden in multiple models of intestinal tumorigenesis (24).
A member of the CUG-binding protein ELAV-like family of RBPs recently shown to regulate intestinal homeostasis is CELF1 or CUGBP1. CUGBP1 has been associated with upregulation of inhibitor of apoptosis 1 and inhibitor of apoptosis 2 in intestinal epithelial cells, making them more resistant to TNF-α and cyclohexamide-induced apoptosis (20). Furthermore, CUGBP1 regulates tight junction expression and gut barrier function by inhibiting occludin expression at the translational level (85). In the intestine, CUGBP1 is found to compete with HuR for binding to MYC mRNA to repress its translation at homeostasis (44). Moreover, in vitro ectopic CUGBP1 expression in intestinal epithelial cells leads to G1-phase growth arrest and conversely CUGBP1 silencing enhances cell proliferation (44). A transposon-based genetic screen in mice identified CELF1, as a potential promoter of colorectal cancer (64). Beyond the intestine, CUGBP1 effects mRNA stability and is associated with increased proliferation and cell motility and decreased apoptosis in nonsmall cell lung cancer cell lines (23).
Finally, the MEX3 family of RBPs is evolutionarily conserved and differentially recruited to processing bodies in the cytoplasm, suggesting a role in RNA turnover (7). In the intestine, single-cell transcriptome analysis revealed that Mex3a is expressed in a subset of Lgr5+ stem cells that are slow dividing (2). These Mex3a+ cells converted to fast-dividing cell following chemotherapy or radiation-induced toxicity, thus promoting intestinal regeneration. In CaCo2 cells, MEX3A overexpression was associated with enhanced expression of stem cell markers including LGR5, MSI, and BMI1. Furthermore, intestinal MEX3A represses the function of the homeobox transcription factor CDX2 and affects intestinal reprogramming (53). In summary, there are multiple RBPs with emerging roles in intestinal stem cells and the broader crypt compartment, suggesting an essential role for posttranscriptional regulation in stem cell dynamics and plasticity.
RBPs MODULATE WNT AND OTHER KEY SIGNALING PATHWAYS IN INTESTINAL EPITHELIAL CELLS
WNT signaling is essential in intestinal crypt stem cell maintenance. Proliferation and survival of CBC stem cells rely on WNT ligands (54, 70, 88), which are produced in part by Paneth cells, among other nonepithelial cells in the small intestine (63). Indeed, intestinal stem cell growth is enhanced upon culture with Paneth cells, and this can be reversed with WNT inhibition. Inducible β-catenin deletion results in terminal differentiation of intestinal stem cells and subsequent lethality, emphasizing the importance of WNT/β-CATENIN signaling in stem cell maintenance (22).
RBPs play an important role in the regulation of the WNT/β-CATENIN-signaling cascade, emphasizing a significant role of these posttranscriptional regulators in the intestinal stem cell niche (Table 1). The LIN28 family of RBPs has been shown to cooperate with the WNT-signaling axis in tumor growth and progression (8) whereas recent studies demonstrate opposing roles for LIN28B and IMP1 in regulation of WNT signaling during intestinal tumorigenesis (10). During homeostasis, Imp1 deletion is associated with upregulation of WNT target genes including Lgr5, Sox9, and Wnt3, suggesting that IMP1 may normally suppress these genes and enhance intestinal stem cell differentiation in this context (10).
Table 1.
Intestinal epithelial pathways associated with RBP regulation
| Pathway/Function/RBPs | Targets Evaluated* | Reference |
|---|---|---|
| WNT/β-CATENIN | ||
| HuR | LRP6 | (41) |
| KSRP | DSVL3 | (4) |
| RBM3 | β-CATENIN | (74) |
| MSI1 | FRAT1, MYC, CCND, CCND2 | (57) |
| CIRP | LGR5 | (60) |
| LIN28B | Global changes | (10, 68) |
| IMP1 |
C-MYC** LGR5, SOX9, WNT3 |
(77) (10) |
| NOTCH | ||
| MSI1 | NICD, HES1 | (57) |
| mTOR | ||
| MSI1/2 | PTEN, BMPR1A, LRIG1 | (84) |
| Intestinal stem cells | ||
| IMP1 | LGR5, BMI1, SOX9, WNT3 | (10) |
| HUR | OLFM4 | (24) |
| MEX3A | LGR5, MSI, BMI1 | (53) |
| CIRP | BMI1, LGR5, DCLK1 | (60) |
| RBM3 | DCLK1, LGR5, CD44 | (74) |
RBP, RNA-binding protein.
Targets are not necessarily direct binding targets.
Not shown in intestine or colon cells specifically.
While studies evaluating LIN28 and IMP1 suggest global regulation of WNT, studies evaluating mRNA binding are needed to ascertain direct or indirect regulation. The RBP HuR binds to LRP6 mRNA, which is essential for FRIZZLED receptor function and response to WNT (41, 87). HuR can also participate in cell cycle regulation through binding and enhancing translation of CDC42 mRNA, a GTPase that controls cell morphology, migration, and cell cycle progression (45). The RBP K-homology splicing regulator protein (KSRP) has been shown to interact with COOH-terminus of Dishevelled-3 (DVL3), a key component of the FRIZZLED1-LRP-DISHEVELLED pathway. The interaction of KSRP and DVL3 correlated with decreased β-CATENIN mRNA levels, suggesting that KSRP is important for the stability of β-CATENIN mRNA, although the mechanism of this still remains elusive (4).
RBM3, a cold-inducible RBP that is important for cell cycle progression and stress response, increases stability and translation of cyclooxygenase-2, IL-8, and VEGF mRNA. It has also been shown to influence the stability of β-CATENIN. When RBM3 was overexpressed in colon cancer cell lines, both β-CATENIN signaling and expression of LGR5 were increased. Furthermore, overexpression of RBM3 resulted in phosphorylation and attenuation of GSK3B activity, which promotes increased stability and accumulation of β-CATENIN (74). A related protein, Cold-inducible RNA-binding protein (CIRP or hnRNP A18), is also involved in the regulation of β-CATENIN. CIRP has been shown to be upregulated in colitis-associated cancer and inflammatory bowel disease and CIRP levels correlated with levels of BMI1, LGR5, and DCLK1 implicating this protein in stem cell dynamics (60). Although a direct mechanism remains to be demonstrated, in several cell types GSK-3β, which inhibits β-CATENIN, upregulates CIRP transcription and phosphorylates CIRP protein to promote its cytosolic translocation (42, 80, 81). Furthermore, with the use of immunoprecipitation and PCR, it was demonstrated that CIRP maintains expression of β-CATENIN in Xenopus laevis, demonstrating that this protein binds and/or modulates expression of this mRNA (52).
In addition to modulation of WNT signaling, RBPs may also regulate the NOTCH pathway, which is important for postmitotic cell fate decisions in intestinal epithelium (73, 86). Treatment of intestinal epithelial primary cultures with the agonist WNT3A led to an upregulation of Msi1 mRNA, which subsequently increased β-CATENIN expression and nuclear translocation (57). Similarly, overexpression of Msi1 led to an upregulation of the β-CATENIN stabilizer Frat1 and Notch-1 receptor intracellular domain (NICD). Upregulation of Msi1 was found to be due to the binding of TCF/LEF to the Msi1 promoter (57). Finally, the mammalian target of rapamycin (mTOR) pathway is emerging as a key player in intestinal stem cell dynamics (61, 83). Msi proteins have been shown to bind to and inhibit negative regulators of mTORC1 including PTEN, BMPR1A, and LRIG1, leading to induction of this nutrient-sensing protein and ultimately proliferation and differentiation.
PLEIOTROPIC ROLES FOR RBPs
RBPs are rapid and effective regulators since they bind already transcribed targets. Therefore, the functional roles of RBPs depend on the transcripts present within a specific cell type. Even in the same cell type, changes in the transcriptome, due to environmental changes or injury, can lead to different functional outcomes for RBPs. This is also true for different tissues or organs where transcriptomes vary. Furthermore, since RBP binding to target transcripts can result in a number of functional outcomes, it may be difficult to elucidate the functional role from one context to another. For example, although IMP1 is recognized as an oncofetal protein due to its overexpression in colorectal cancers and increased tumor burden with IMP1 expression in xenograft models (28), IMP1 deletion in intestinal mesenchymal cells increased tumor burden in mice (27). Similar, opposing roles for IMP1 have also been reported in breast cancer (51) (76) (66). IMP1 therefore can exhibit both tumor-initiating or tumor suppressive functions. Another example of this paradigm is HuR, which stabilizes and increases translation of several key cancer related pathways (49). In a recent study in pancreatic ductal adenocarcinoma cells, HuR deletion resulted in a decrease in proliferation and an increase in cell death, possibly via regulation of KRAS pathway genes (39). In contrast, other studies have shown that HuR knockdown in myeloid lineage cells in mice caused an enhanced susceptibility to colitis-associated cancer and increases inflammatory mRNAs (82).
Similarly, while CELF1 deletion in several cancer cell lines decreased proliferation and migration (23, 32), and Celf1-deficient mice exhibited growth retardation (38), CELF1 has also been reported to promote apoptotic resistance in esophageal cancer (9). CELF1 exhibits opposing effects on cell proliferation in primary versus malignant T cells due to transcriptomic differences (6). In yet another example, Mex3a is expressed in slow dividing intestinal stem cells (2), but in CRC cell lines the effect of MEX3A on differentiation depends on cell confluence. At low confluency, MEX3A inhibition increased CDX2 expression, but this effect was reverted when the cells were highly confluent (53). Taken together, these studies demonstrate that RBPs represent a complex regulatory network and their functional roles depend on the tissue type, microenvironment, and the transcriptome of cells in which they are expressed. Therefore, understanding the precise, context-dependent mechanisms by which RBPs regulate protein expression in combination with other factors will be essential to understanding their complex roles in gastrointestinal cancers.
EMERGING TECHNIQUES TO EVALUATE RBPs IN THE INTESTINE
There are multiple experimental methods to determine the landscape of RBP biology and their roles in disease. As illustrated above, the functional role of RBPs can vary depending on the cellular context. Therefore, integration of multiple methodologies is required to understand fully the role of these posttranscriptional regulons in the gut. One method for identifying direct roles of RNA-binding proteins has been to determine their target transcripts using crosslinking immunoprecipitation (CLIP). The RBP under investigation is crosslinked in vivo to its target transcripts via a crosslinking agent, such as ultraviolet light, forming covalent bonds between RNA and protein. Cells are then lysed and the target RBP is precipitated using an antibody. Proteinase K is subsequently used to digest the RBP, leaving the bound transcripts intact. After ligating adapters and linkers to RNA, the transcripts are reverse transcribed to cDNA and identified via high-throughput sequencing (69). CLIP techniques have been refined over the years to increase precision and sensitivity as reviewed in Ref. 11. CLIP-sequencing libraries have been generated for several RBPs, including LIN28B (48), IMP1 (18), and MSI1 (3); however, one must consider differences in steady-state transcriptomes in the cell lines in which these experiments were performed before generalizing binding targets from one cell type to another.
In addition to identifying binding targets using CLIP-sequencing, functional effects of RBPs can be studied by overexpressing or deleting RBP genes followed by high-throughput RNA sequencing. This generates a static snapshot of total RNA abundance in the presence or absence of a given RBP. Since RBPs can regulate the stabilization, degradation, localization, and posttranscriptional modification (for example, splicing) of their targets, RNA sequencing provides a robust tool to study these changes. However, as proteins are the functional effectors in cells, RNA sequencing may not serve as a direct indication of the functional role of RBPs. For example, some RBPs may bind to their target transcripts and cage them in RNP granules, rendering these targets inaccessible to the translation machinery (5). In these cases, mRNA abundance does not correlate with translation/functional effects.
In recent years, ribosome footprint profiling has emerged as a powerful tool to obtain a global snapshot of actively translating transcripts within a cell (33–35). This ribosome-centric technique uses binding and density of ribosomes on a transcript as an indicator of active translation. Profiling is done by deep sequencing of ribosome-bound transcripts as well as sequencing total RNA abundance. Polysomes (a cluster of ribosomes held together by a strand of messenger RNA that each ribosome is translating) are stabilized by blocking translation elongation using cycloheximide before cell lysis (36). This technique detects relative changes and therefore relies on cells or tissues with deletion or overexpression of the desired RBP. This technique can help distinguish between genes regulated at the transcriptome and “translatome” levels. The ratio of ribosome footprint profiling to total mRNA abundance for each transcript reveals the translation efficiency for that gene that can be compared across genotypes or conditions (88).
In summary, a comprehensive understanding of RBP functions requires evaluating 1) target transcripts, 2) the context in which binding occurs, 3) downstream effects on transcription and translation of a given transcript or family of transcripts, and 4) the “net effect” of the RBP expression. To address the latter, in vivo models modulating RBP expression are key to understanding the role of RBPs at the whole organisms level. Genetic mouse models of RBP deletion or overexpression in specific cell types have and will continue to be instrumental in understanding RBP roles at homeostasis and injury or disease models.
CONCLUSIONS
Regulation of cellular dynamics via RBPs is becoming recognized increasingly as a key feature in tissue homeostasis and disease. While existing studies evaluating RBPs in intestinal epithelial cell biology are compelling, several questions remain to be addressed in this growing field. For example, how do RBPs cooperate or act in opposition to one another? Our recent study evaluating the LIN28B-IMP1 axis is one example in which genetic mouse models may reveal new and unexpected roles that may be opposing (10). Similarly, miRNAs may also augment the functional roles of RBPs, as reviewed in Ref. 78, and therefore must be accounted for when determining the relative role of posttranscriptional regulation in a given process. In addition, RNA modifications or the “epi-transcriptome,” the outcome of which is largely governed by RBPs, is an emerging field that will shed new light on how RNA regulons determine cellular functions. Finally, the importance of RBPs and posttranscriptional regulation in other gastrointestinal epithelial stem or progenitor cells (e.g., esophageal, stomach) is largely unexplored. Studies defining how RBPs contribute to stem cell dynamics and disease will not only uncover new paradigms in basic gastrointestinal biology but may also hold the promise for new therapeutics as well.
GRANTS
This study was supported by a grant from the University of Pennsylvania Institute for Translational Medicine and Therapeutics (to K. E. Hamilton); National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grants K01-DK-100485 and R03-DK-11446301 (to K. E. Hamilton) and K01-DK-103953 and R03-DK-118304 (to K. A. Whelan); University of Pennsylvania NIDDK Center for Molecular Studies in Digestive and Liver Diseases Grant P30-DK-050306), and Institutional Development Funds from Children’s Hospital of Philadelphia Research Institute.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
L.R.P., P.A.W., and K.E.H. conceived and designed research; L.R.P., P.C., and K.E.H. prepared figures; L.R.P., P.A.W., P.C., K.A.W., and K.E.H. drafted manuscript; P.A.W., P.C., K.A.W., and K.E.H. edited and revised manuscript; P.C. and K.E.H. approved final version of manuscript.
REFERENCES
- 1.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007, 2007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Barriga FM, Montagni E, Mana M, Mendez-Lago M, Hernando-Momblona X, Sevillano M, Guillaumet-Adkins A, Rodriguez-Esteban G, Buczacki SJ, Gut M, Heyn H, Winton DJ, Yilmaz OH, Attolini CS, Gut I, Batlle E. Mex3a marks a slowly dividing subpopulation of Lgr5+ intestinal stem cells. Cell Stem Cell 20: 801–816.e7, 2017. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett CG, Riemondy K, Chapnick DA, Bunker E, Liu X, Kuersten S, Yi R. Genome-wide analysis of Musashi-2 targets reveals novel functions in governing epithelial cell migration. Nucleic Acids Res 44: 3788–3800, 2016. doi: 10.1093/nar/gkw207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bikkavilli RK, Malbon CC. Dishevelled-KSRP complex regulates Wnt signaling through post-transcriptional stabilization of beta-catenin mRNA. J Cell Sci 123: 1352–1362, 2010. doi: 10.1242/jcs.056176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaß M, Möller B, Hüttelmaier S. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res 43: e26, 2015. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohjanen PR, Moua ML, Guo L, Taye A, Vlasova-St Louis IA. Altered CELF1 binding to target transcripts in malignant T cells. RNA 21: 1757–1769, 2015. doi: 10.1261/rna.049940.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchet-Poyau K, Courchet J, Le Hir H, Séraphin B, Scoazec JY, Duret L, Domon-Dell C, Freund JN, Billaud M. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res 35: 1289–1300, 2007. doi: 10.1093/nar/gkm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai WY, Wei TZ, Luo QC, Wu QW, Liu QF, Yang M, Ye GD, Wu JF, Chen YY, Sun GB, Liu YJ, Zhao WX, Zhang ZM, Li BA. The Wnt-β-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci 126: 2877–2889, 2013. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 9.Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J 446: 113–123, 2012. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji P, Hamilton KE, Liang S, Andres SF, Wijeratne HR, Mizuno R, Simon LA, Hicks PD, Foley SW, Pitarresi JR, Klein-Szanto AJ, Mah AT, Van Landeghem L, Gregory BD, Lengner CJ, Madison BB, Shah P, Rustgi AK. The LIN28B-IMP1 post-transcriptional regulon has opposing effects on oncogenic signaling in the intestine. Genes Dev 32: 1020–1034, 2018. doi: 10.1101/gad.314369.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterji P, Rustgi AK. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med 24: 490–506, 2018. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen CH, Luhur A, Sokol N. Lin-28 promotes symmetric stem cell division and drives adaptive growth in the adult Drosophila intestine. Development 142: 3478–3487, 2015. doi: 10.1242/dev.127951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. II. Mucous cells. Am J Anat 141: 481–501, 1974. doi: 10.1002/aja.1001410404. [DOI] [PubMed] [Google Scholar]
- 14.Cheng H. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. IV. Paneth cells. Am J Anat 141: 521–535, 1974. doi: 10.1002/aja.1001410406. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat 141: 461–479, 1974. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- 16.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. III. Entero-endocrine cells. Am J Anat 141: 503–519, 1974. doi: 10.1002/aja.1001410405. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anat 141: 537–561, 1974. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 18.Conway AE, Van Nostrand EL, Pratt GA, Aigner S, Wilbert ML, Sundararaman B, Freese P, Lambert NJ, Sathe S, Liang TY, Essex A, Landais S, Burge CB, Jones DL, Yeo GW. Enhanced CLIP uncovers IMP protein-RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Rep 15: 666–679, 2016. doi: 10.1016/j.celrep.2016.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Creamer B, Shorter RG, Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut 2: 110–116, 1961. doi: 10.1136/gut.2.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell 23: 151–162, 2012. doi: 10.1091/mbc.e11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitriadis E, Trangas T, Milatos S, Foukas PG, Gioulbasanis I, Courtis N, Nielsen FC, Pandis N, Dafni U, Bardi G, Ioannidis P. Expression of oncofetal RNA-binding protein CRD-BP/IMP1 predicts clinical outcome in colon cancer. Int J Cancer 121: 486–494, 2007. doi: 10.1002/ijc.22716. [DOI] [PubMed] [Google Scholar]
- 22.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol 27: 7551–7559, 2007. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao C, Yu Z, Liu S, Xin H, Li X. Overexpression of CUGBP1 is associated with the progression of non-small cell lung cancer. Tumour Biol 36: 4583–4589, 2015. doi: 10.1007/s13277-015-3103-1. [DOI] [PubMed] [Google Scholar]
- 24.Giammanco A, Blanc V, Montenegro G, Klos C, Xie Y, Kennedy S, Luo J, Chang SH, Hla T, Nalbantoglu I, Dharmarajan S, Davidson NO. Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res 74: 5322–5335, 2014. doi: 10.1158/0008-5472.CAN-14-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol 298: G590–G600, 2010. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA 19: 613–626, 2013. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton KE, Chatterji P, Lundsmith ET, Andres SF, Giroux V, Hicks PD, Noubissi FK, Spiegelman VS, Rustgi AK. Loss of stromal IMP1 promotes a tumorigenic microenvironment in the colon. Mol Cancer Res 13: 1478–1486, 2015. doi: 10.1158/1541-7786.MCR-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton KE, Noubissi FK, Katti PS, Hahn CM, Davey SR, Lundsmith ET, Klein-Szanto AJ, Rhim AD, Spiegelman VS, Rustgi AK. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis 34: 2647–2654, 2013. doi: 10.1093/carcin/bgt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464, 2004. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32: 276–284, 2008. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708, 2009. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 32.House RP, Talwar S, Hazard ES, Hill EG, Palanisamy V. RNA-binding protein CELF1 promotes tumor growth and alters gene expression in oral squamous cell carcinoma. Oncotarget 6: 43620–43634, 2015. doi: 10.18632/oncotarget.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell 165: 22–33, 2016. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. Genome-wide annotation and quantitation of translation by ribosome profiling. Curr Protoc Mol Biol 103: 4.18.1–4.18.19, 2013. doi: 10.1002/0471142727.mb0418s103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 8: 1365–1379, 2014. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802, 2011. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543, 2007. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 38.Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol 27: 1146–1157, 2007. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, Romeo C, Stout MC, Londin E, Goetz A, Lowder CY, Nevler A, Yeo CJ, Campbell PM, Winter JM, Dixon DA, Brody JR. CRISPR knockout of the HuR gene causes a xenograft lethal phenotype. Mol Cancer Res 15: 696–707, 2017. doi: 10.1158/1541-7786.MCR-16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li N, Yousefi M, Nakauka-Ddamba A, Li F, Vandivier L, Parada K, Woo DH, Wang S, Naqvi AS, Rao S, Tobias J, Cedeno RJ, Minuesa G, y K, Barlowe TS, Valvezan A, Shankar S, Deering RP, Klein PS, Jensen ST, Kharas MG, Gregory BD, Yu Z, Lengner CJ. The Msi family of RNA-binding proteins function redundantly as intestinal oncoproteins. Cell Rep 13: 2440–2455, 2015. doi: 10.1016/j.celrep.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Chen G, Wang JY, Zou T, Liu L, Xiao L, Chung HK, Rao JN, Wang JY. Post-transcriptional regulation of Wnt co-receptor LRP6 and RNA-binding protein HuR by miR-29b in intestinal epithelial cells. Biochem J 473: 1641–1649, 2016. doi: 10.1042/BCJ20160057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu GP, Zhang Y, Yao XQ, Zhang CE, Fang J, Wang Q, Wang JZ. Activation of glycogen synthase kinase-3 inhibits protein phosphatase-2A and the underlying mechanisms. Neurobiol Aging 29: 1348–1358, 2008. doi: 10.1016/j.neurobiolaging.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Liu L, Christodoulou-Vafeiadou E, Rao JN, Zou T, Xiao L, Chung HK, Yang H, Gorospe M, Kontoyiannis D, Wang JY. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell 25: 3308–3318, 2014. doi: 10.1091/mbc.e14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, Wu J, Donahue JM, Gorospe M, Wang JY. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell 26: 1797–1810, 2015. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Zhuang R, Xiao L, Chung HK, Luo J, Turner DJ, Rao JN, Gorospe M, Wang JY. HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol 37: e00574-16, 2017. doi: 10.1128/MCB.00574-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem 279: 12974–12979, 2004. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- 47.Madison BB, Jeganathan AN, Mizuno R, Winslow MM, Castells A, Cuatrecasas M, Rustgi AK. Let-7 represses carcinogenesis and a stem cell phenotype in the intestine via regulation of hmga2. PLoS Genet 11: e1005408, 2015. doi: 10.1371/journal.pgen.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS, Rustgi AK. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev 27: 2233–2245, 2013. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, Gorospe M, Keene JD, Levenson AS, Gartenhaus RB. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 27: 6151–6163, 2008. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci USA 108: 179–184, 2011. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nwokafor CU, Sellers RS, Singer RH. IMP1, an mRNA binding protein that reduces the metastatic potential of breast cancer in a mouse model. Oncotarget 7: 72662–72671, 2016. doi: 10.18632/oncotarget.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y, Yang PH, Tanner JA, Huang JD, Li M, Lee HF, Xu RH, Kung HF, Lin MC. Cold-inducible RNA binding protein is required for the expression of adhesion molecules and embryonic cell movement in Xenopus laevis. Biochem Biophys Res Commun 344: 416–424, 2006. doi: 10.1016/j.bbrc.2006.03.086. [DOI] [PubMed] [Google Scholar]
- 53.Pereira B, Sousa S, Barros R, Carreto L, Oliveira P, Oliveira C, Chartier NT, Plateroti M, Rouault JP, Freund JN, Billaud M, Almeida R. CDX2 regulation by the RNA-binding protein MEX3A: impact on intestinal differentiation and stemness. Nucleic Acids Res 41: 3986–3999, 2013. doi: 10.1093/nar/gkt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev 17: 1709–1713, 2003. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell 147: 1066–1079, 2011. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149: 146–158, 2012. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci 123: 3256–3265, 2010. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 58.Richmond CA, Rickner H, Shah MS, Ediger T, Deary L, Zhou F, Tovaglieri A, Carlone DL, Breault DT. JAK/STAT-1 signaling is required for reserve intestinal stem cell activation during intestinal regeneration following acute inflammation. Stem Cell Reports 10: 17–26, 2018. doi: 10.1016/j.stemcr.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993, 2008. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 60.Sakurai T, Kashida H, Watanabe T, Hagiwara S, Mizushima T, Iijima H, Nishida N, Higashitsuji H, Fujita J, Kudo M. Stress response protein cirp links inflammation and tumorigenesis in colitis-associated cancer. Cancer Res 74: 6119–6128, 2014. doi: 10.1158/0008-5472.CAN-14-0471. [DOI] [PubMed] [Google Scholar]
- 61.Sampson LL, Davis AK, Grogg MW, Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. FASEB J 30: 1263–1275, 2016. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet 40: 915–920, 2008. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469: 415–418, 2011. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, Gupta M, O’Sullivan MG, Matise I, Dupuy AJ, Collier LS, Powers S, Oberg AL, Asmann YW, Thibodeau SN, Tessarollo L, Copeland NG, Jenkins NA, Cormier RT, Largaespada DA. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323: 1747–1750, 2009. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424, 2011. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res 64: 209–214, 2004. doi: 10.1158/0008-5472.CAN-03-2927. [DOI] [PubMed] [Google Scholar]
- 67.Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478: 255–259, 2011. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu HC, Schwitalla S, Qian Z, LaPier GS, Yermalovich A, Ku YC, Chen SC, Viswanathan SR, Zhu H, Nishihara R, Inamura K, Kim SA, Morikawa T, Mima K, Sukawa Y, Yang J, Meredith G, Fuchs CS, Ogino S, Daley GQ. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev 29: 1074–1086, 2015. doi: 10.1101/gad.256693.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215, 2003. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 70.Valenta T, Degirmenci B, Moor AE, Herr P, Zimmerli D, Moor MB, Hausmann G, Cantù C, Aguet M, Basler K. Wnt ligands secreted by subepithelial mesenchymal cells are essential for the survival of intestinal stem cells and gut homeostasis. Cell Rep 15: 911–918, 2016. doi: 10.1016/j.celrep.2016.03.088. [DOI] [PubMed] [Google Scholar]
- 71.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol 71: 241–260, 2009. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 72.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es JH, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 136: 903–912, 2009. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 73.VanDussen KL, Carulli AJ, Keeley TM, Patel SR, Puthoff BJ, Magness ST, Tran IT, Maillard I, Siebel C, Kolterud Å, Grosse AS, Gumucio DL, Ernst SA, Tsai YH, Dempsey PJ, Samuelson LC. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139: 488–497, 2012. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venugopal A, Subramaniam D, Balmaceda J, Roy B, Dixon DA, Umar S, Weir SJ, Anant S. RNA binding protein RBM3 increases β-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol Carcinog 55: 1503–1516, 2016. doi: 10.1002/mc.22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet 41: 843–848, 2009. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang G, Huang Z, Liu X, Huang W, Chen S, Zhou Y, Li D, Singer RH, Gu W. IMP1 suppresses breast tumor growth and metastasis through the regulation of its target mRNAs. Oncotarget 7: 15690–15702, 2016. doi: 10.18632/oncotarget.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weidensdorfer D, Stöhr N, Baude A, Lederer M, Köhn M, Schierhorn A, Buchmeier S, Wahle E, Hüttelmaier S. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA 15: 104–115, 2009. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xiao L, Wang JY. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr Opin Pharmacol 19: 46–53, 2014. doi: 10.1016/j.coph.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, Sangiorgi E, Capecchi MR, Kuo CJ. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA 109: 466–471, 2012. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang R, Weber DJ, Carrier F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res 34: 1224–1236, 2006. doi: 10.1093/nar/gkj519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang R, Zhan M, Nalabothula NR, Yang Q, Indig FE, Carrier F. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J Biol Chem 285: 8887–8893, 2010. doi: 10.1074/jbc.M109.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yiakouvaki A, Dimitriou M, Karakasiliotis I, Eftychi C, Theocharis S, Kontoyiannis DL. Myeloid cell expression of the RNA-binding protein HuR protects mice from pathologic inflammation and colorectal carcinogenesis. J Clin Invest 122: 48–61, 2012. doi: 10.1172/JCI45021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yilmaz OH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, Nielsen GP, Mino-Kenudson M, Zukerberg LR, Bhan AK, Deshpande V, Sabatini DM. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486: 490–495, 2012. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yousefi M, Li N, Nakauka-Ddamba A, Wang S, Davidow K, Schoenberger J, Yu Z, Jensen ST, Kharas MG, Lengner CJ. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J Cell Biol 215: 401–413, 2016. doi: 10.1083/jcb.201604119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu TX, Rao JN, Zou T, Liu L, Xiao L, Ouyang M, Cao S, Gorospe M, Wang JY. Competitive binding of CUGBP1 and HuR to occludin mRNA controls its translation and modulates epithelial barrier function. Mol Biol Cell 24: 85–99, 2013. doi: 10.1091/mbc.e12-07-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zecchini V, Domaschenz R, Winton D, Jones P. Notch signaling regulates the differentiation of post-mitotic intestinal epithelial cells. Genes Dev 19: 1686–1691, 2005. doi: 10.1101/gad.341705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeng X, Huang H, Tamai K, Zhang X, Harada Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, Wynshaw-Boris A, Hsieh JC, He X. Initiation of Wnt signaling: control of Wnt coreceptor Lrp6 phosphorylation/activation via frizzled, dishevelled and axin functions. Development 135: 367–375, 2008. doi: 10.1242/dev.013540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, Pu WT. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci USA 110: 15395–15400, 2013. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]