Abstract
Cpf1, a CRISPR endonuclease discovered in Prevotella and Francisella 1 bacteria, offers an alternative platform for CRISPR-based genome editing beyond the commonly used CRISPR–Cas9 system originally discovered in Streptococcus pyogenes. This protocol enables the design of engineered CRISPR–Cpf1 components, both CRISPR RNAs (crRNAs) to guide the endonuclease and Cpf1 mRNAs to express the endonuclease protein, and provides experimental procedures for effective genome editing using this system. We also describe quantification of genome-editing activity and off-target effects of the engineered CRISPR–Cpf1 in human cell lines using both T7 endonuclease I (T7E1) assay and targeted deep sequencing. This protocol enables rapid construction and identification of engineered crRNAs and Cpf1 mRNAs to enhance genome-editing efficiency using the CRISPR–Cpf1 system, as well as assessment of target specificity within 2 months. this protocol may also be appropriate for fine-tuning other types of CRISPR systems.
INTRODUCTION
The discovery of clustered regularly interspaced short palindromic repeats (CRISPR) from Prevotella and Francisella 1 (Cpf1) expanded the diversity of the CRISPR–Cas family (Fig. 1a)1. Among the 16 Cpf1-family proteins that were originally evaluated, two Cpf1 orthologs, Acidaminococcus sp. Cpf1 (AsCpf1) and Lachnospiraceae bacterium Cpf1 (LbCpf1), were found to display robust genome-editing activity in human cells1 and have been applied for genome editing in a number of organisms, including plants and mice2–12. Most recently, two engineered AsCpf1 variants were shown to be able to recognize TYCV and TATV protospacer adjacent motifs (PAMs; the sequences recognized by the Cpf1 protein to determine the cleavage site), which markedly increased the number of potentially targetable sites13. Compared with Cas9, Cpf1 possesses several unique features1,14: as shown in Figure 1b, Cpf1 induces targeted DNA breaks via a single crRNA without an additional trans-activating crRNA (tracrRNA); a 5′ T-rich PAM is required for protein recognition; the cleavage of the target DNA strand is attributed to the Nuc domain of the protein rather than the HNH domain; Cpf1 creates a staggered end at the PAM-distal region; and Cpf1 precursor crRNA is processed into mature crRNA through Cpf1 protein without the assistance of RNase III. Crystal structures of the Cpf1–crRNA–dsDNA complex validate the PAM recognition and cleavage model for the Cpf1 system15–19. The effector protein Cpf1 binds target sites with high preference in AT-rich genomic regions and cleaves the nontarget and target DNA strands using the RuvC-like domain and the Nuc domain, respectively15–19.
Figure 1 |.
Schematic illustration of the CRISPR–Cpf1 system. (a) AsCpf1-mediated double-stranded DNA breaks1,15. CRISPR–Cpf1 is a two-component genome-editing system consisting of a Cpf1 effector nuclease (colored purple) and a single crRNA (blue strand). A protospacer is located in the genomic DNA immediately downstream of the protospacer-adjacent motif (PAM) (5′-TTTN-3′). The PAM (orange) refers to a DNA sequence recognized by the Cpf1–crRNA complex. RuvC-like and Nuc domains (gray symbols) of the Cpf1 protein are involved in cleaving the nontarget and target DNA strands. (b) Structure of the AsCpf1 crRNA1,15. crRNA is composed of a direct repeat (5′ handle) and a spacer (guide segment). The pseudoknot structure formed by the direct repeat is the prominent Cpf1 recognition region, and the spacer is able to guide Cpf1 to the target region to exert endonuclease activity. The seed region, a crucial sequence that interacts with target DNA sequence, is approximately the first eight nucleotides on the 5′ end of the spacer14. crRNA, CRISPR RNA; NTS, nontarget strand; TS, target strand.
To systematically study the structure–activity relationship of CRISPR–Cpf1 components and maximize genome-editing efficiency, we recently engineered a wide variety of chemically and structurally modified crRNAs and AsCpf1 mRNAs20. The results showed that the combination of the best-performing crRNA and AsCpf1 mRNAs increased genome-editing efficiency by > 300% in comparison with wild-type AsCpf1 crRNA with the AsCpf1 expression plasmid. In the case of LbCpf1, co-delivery of engineered LbCpf1 components induced high-frequency editing at the DNMT1 gene locus, whereas the combination of wild-type LbCpf1 crRNA and LbCpf1 expression plasmid led to undetectable levels of genome editing20. Our studies also showed that AsCpf1 was capable of inducing genome editing with crRNAs from the Cpf1 families of most of the different species tested20. Herein, we describe the design criteria and experimental procedures for engineering the CRISPR–Cpf1 system and using it for genome-editing purposes.
Overview of the procedure
To date, a number of chemically modified nucleotides have been incorporated into synthetic RNA molecules to enhance their chemical stability, nuclease resistance, binding affinity, and the level of protein expression21–25. Figure 2 displays three types of modifications: linkage, ribose, and base modifications. Representative examples of chemically modified nucleotides are shown in Supplementary Figure 1. To rationally design and evaluate engineered CRISPR–Cpf1 crRNAs and mRNAs, we developed the following protocol, facilitating genome editing using the CRISPR–Cpf1 system.
Figure 2 |.
A diagram of chemically modified RNA. Chemically modified nucleotides including linkage-, ribose-, and nitrogenous base (guanine, uracil, adenine, and cytosine)-modified nucleotides can be installed into RNA molecules by an automated solid-phase DNA/RNA synthesizer or in vitro transcription.
Based on the structure of wild-type AsCpf1 crRNA (termed crWT, Fig. 1b), we synthesized a series of engineered crRNAs, including chemically modified and structurally altered crRNAs, and systematically investigated their structure–activity relationships, which enabled us to establish a set of design criteria for engineering Cpf1 crRNAs and identifying the optimal crRNA. This was found to contain five 2′-fluoro ribose modifications at the 3′ terminus (termed cr3′5F). In addition, based on our previous work24, we examined the activity of engineered AsCpf1 mRNAs produced by fully substituting uridines with modified bases or introducing mutations during in vitro transcription. We determined that pseudouridine (Ψ) was a favorable modification as compared with the wild-type AsCpf1 mRNA.
To maximize genome-editing efficiency of the CRISPR–Cpf1 system, we simultaneously delivered the best-performing AsCpf1 crRNA and AsCpf1 mRNAs into human 293T cells. Such a combination gave rise to a marked enhancement in genome-editing activity at the DNMT1 gene locus of human cell lines in comparison with the combination of wild-type crRNA and AsCpf1 plasmid, as determined by the T7 endonuclease I assay and further validated by targeted deep sequencing. This strategy has also been successfully applied for other locus-specific crRNAs, including crRNAs targeting AAVS1 and FANCF (ref. 20). Furthermore, co-delivery of the best-performing crRNA and mRNA did not increase off-target effects, as determined by targeted deep sequencing. For LbCpf1-mediated genome editing, the enhanced efficiency of genome editing was more pronounced when using the best-performing LbCpf1 components. In addition, we also examined the effects of the loop structure of crRNA on Cpf1-mediated genome-editing activity by substituting the loop of AsCpf1 crRNA with those from other crRNAs of the Cpf1 family and demonstrated that AsCpf1 was able to complex with crRNAs from the majority of Cpf1 orthologs to execute genome editing20.
Advantages and limitations
Carrying out genome editing with Cpf1 mRNA offers several advantages in comparison with its plasmid counterpart. Cpf1 mRNA ensures transient expression of Cpf1 endonuclease in order to cleave the gene of interest, reducing unexpected toxicity due to long-term expression of Cpf1. The mRNA does not integrate into the genome of host cells, thus avoiding potential genotoxicity. Moreover, in comparison with the combination of wild-type crRNA and Cpf1 plasmid, the engineered CRISPR–Cpf1 system described here displays a much higher genome-editing efficiency in the three human cell lines tested20. Furthermore, this approach can be adapted to engineer other types of CRISPR–Cas systems. Finally, broad applicability of crRNAs from Cpf1 orthologs may provide more options for guiding the construction of AsCpf1-mediated genome-editing systems.
One limitation of this approach is that extra attention must be paid during experimental procedures, as both mRNA and crRNA are sensitive to ribonuclease. However, precautionary measures can be taken to address this issue (see TROUBLESHOOTING). In this protocol, Lipofectamine 3000 is used to deliver the CRISPR–Cpf1 components. Other types of delivery methods would need to be validated for use with the engineered CRISPR–Cpf1 system. Furthermore, genome-wide analysis of off-target effects may be necessary to provide additional insight into the effects of using the engineered system.
Experimental design
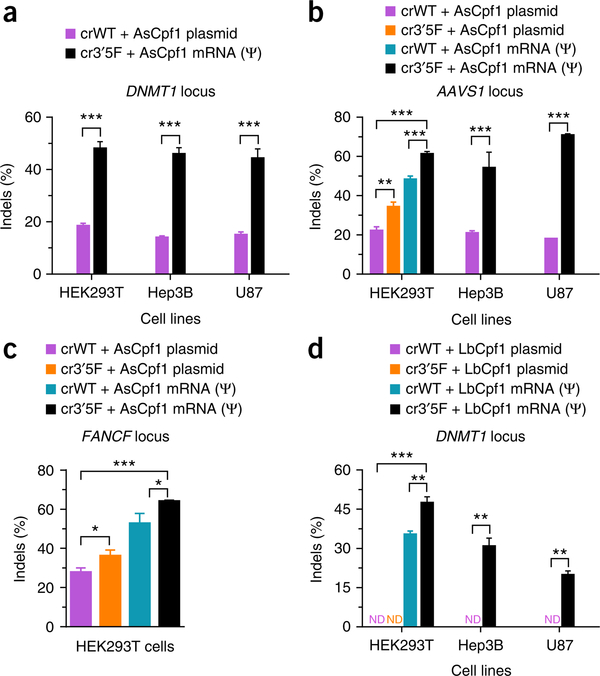
As illustrated in Figure 3, we describe here detailed experimental procedures, including crRNA selection and optimization, engineering of Cpf1 mRNA, and evaluation of biological activity. We use the DNMT1 locus as an example of the target site in human 293T cells. The procedures are compatible with other genomic loci (AAVS1 and FANCF) and cell types (Hep3B and U87), as shown in Figure 4 (ref. 20).
Figure 3 |.
Schematic diagram of the genome-editing workflow to design and assess the engineered CRISPR–Cpf1 system for its use in human cell lines
Figure 4 |.
Assessment of genome-editing efficiency and specificity of the engineered CRISPR–Cpf1 system. (a–c) AsCpf1-mediated genome-editing efficiency at the DNMT1 (a), AAVS1 (b), and FANCF (c) loci in different human cell lines. (d) LbCpf1-mediated genome-editing efficiency at the DNMT1 locus in different human cell lines. Indels at each locus are determined by T7E1 assay and plotted as the mean ± s.d. from three biological replicates (*P < 0.05; **P < 0.01; and ***P < 0.001; two-tailed t test). ND, not detectable. Adapted from ref. 20, Nature Publishing Group. Ψ, pseudouridine.
Selection of Cpf1 crRNAs.
Selection of appropriate Cpf1 crRNAs is an essential step toward engineering the CRISPR–Cpf1 system. crRNAs can be designed via online design tools such as Benchling (https://www.benchling.com) and CHOPCHOP v2 (http://chopchop.cbu.uib.no)26. The Cpf1–crRNA complex recognizes the target DNA region via PAM scanning and Watson–Crick base pairing between crRNA and the target DNA15–19. Previous studies elucidated that PAM recognition of Cpf1 is highly specific to the TTTV sequence27. In addition, crRNA with GC content in the range of 30–70% typically possesses superior activity27. On the basis of these principles, genome-editing activity of designed crRNAs can be scored and ranked by the web tool CINDEL (http://big.hanyang.ac.kr/cindel)27. We used the crRNA that targets the DNMT1 locus (5′-UAAUUUCUACUCUUGUAGAUCUGAUGGUCCAUG UCUGUUACUC-3′)1. To evaluate off-target effects, we applied the web-based tools Cas-OFFinder (http://www.rgenome.net/casoffinder)28 or COSMID (https://crispr.bme.gatech.edu)29. Both consider genomic regions that contain insertions and deletions relative to the crRNA sequence as potential off-target sites28,29.
Engineering of crRNAs.
crRNAs are highly conserved in the Cpf1 orthologs of different bacterial species1. Typically, AsCpf1 crRNA is composed of a 20-nt direct repeat (also known as a 5′ handle) and a 23-nt spacer (guide segment) (Fig. 1b; refs. 1,15). The direct repeat adopts a pseudoknot structure, which contains five Watson–Crick base pairs, one noncanonical U–U base pair, one UCUU tetraloop, one reverse Hoogsteen A–U base pair, and three 5′-end bases15. The spacer is complementary to the target DNA sequence, and the seed region located at the first eight nucleotides of the spacer has a critical role in the target specificity of the CRISPR–Cpf1 system14.
In our work, two strategies were applied to engineer Cpf1 crRNAs: chemical modifications and structural alterations. We first constructed two chemically modified crRNAs according to the modification pattern used for Cas9 guide RNAs30,31. The results indicated that chemical modifications of AsCpf1 crRNA must be different from those used to engineer Cas9 guide RNAs. We subsequently constructed a library of modified AsCpf1 crRNAs by incorporating diverse chemical modifications, including 2′fluoro, 2′-O-methyl, and phosphorothioate (PS) modifications, as well as locked and unlocked nucleotides, into different regions of the wild-type crRNA. We also generated crRNAs with interspersed modifications through the whole crRNA based on the interactions among AsCpf1, crRNA, and the target DNA15.
In addition to chemical modifications, structural alteration is a useful approach for engineering crRNAs32,33. We designed a series of crRNAs through shortening or extending the stem of the crRNA. We also split the single crRNA into two strands in the middle of the loop and rehybridized them to form a hairpin-like structure. However, these structural alterations were found to reduce genome-editing activity20. Finally, we modified the loop structure of AsCpf1 crRNA by replacing its loop with those from Cpf1 orthologs reported previously1. We found that crRNAs containing a three- or five-nucleotide loop impaired genome-editing activity20. For four-nucleotide loop-bearing crRNAs, the base at position −9 (Fig. 1b) substantially affected cleavage potency (the order of genome-editing potency was A > C > G > U)20. U (−10) is a critical position that interacts with A (−18) and A (−19) (ref. 15), and so could not tolerate changes. LbCpf1 was found to be more conservative for recognition of the loop structure as compared with AsCpf1 (ref. 20). After systematic analysis of these data, we established a set of design criteria for future genome-editing applications (Box 1)20. All engineered crRNAs and their mass spectrometry data are shown in Supplementary Table 1. Of note, the purity of the crRNAs described above may greatly affect their efficiencies.
Box 1 | Design criteria for engineered Cpf1 crRNAs.
Analysis of the structure–activity relationship of Cpf1 crRNAs indicates the following design criteria:
A relatively large number of phosphorothioate (PS) modifications usually impairs genome-editing efficiency.
The stem duplex in the 5′ handle does not tolerate splitting, deletion, or insertion of nucleotides tested in the study.
The seed region can accommodate slight chemical modifications.
The 5′ handle is susceptible to chemical modifications, whereas the 3′ spacer tolerates certain modifications; introduction of five 2′-fluoro modifications at the 3′ end yields superior cleavage activity.
Simultaneous chemical modifications tested at both the 5′ and 3′ ends of crRNAs are not favorable.
Engineering of Cpf1 mRNAs.
To reduce immunogenicity and improve translational efficiency of mRNAs, numerous approaches have been investigated24,34–41. Based on our previous studies24, we designed pseudouridine- (Ψ-), N1-methylpseudouridine- (me1Ψ-), and 5-methoxyuridine- (5moU-)modified AsCpf1 mRNAs. In addition, we incorporated both chemical modifications and sequence mutations during mRNA production. For example, we constructed S1228A&Ψ mRNA by substituting the nucleotides that encode serine1228 (S1228) with those encoding alanine (A) and replacing uridines with Ψ in the whole mRNA sequence15,20. These mRNAs were produced via in vitro transcription. In general, Ψ-modified AsCpf1 mRNA showed the highest increase (177%) in genome-editing activity in comparison with AsCpf1 plasmid20.
Co-delivery of the best-performing Cpf1 mRNA and crRNA.
In this section, we formulated Cpf1 mRNA (or plasmid) and crRNA, using Lipofectamine 3000. Subsequently, two CRISPR–Cpf1 components were simultaneously added to 293T cells. After 2 d of treatment, genomic DNA (gDNA) was harvested for downstream assays. Regarding crRNAs with low activity, increase of dosage may be a feasible method as long as no substantial cytotoxicity is observed. Otherwise, new crRNAs must be designed and tested. The combination of wild-type crRNA and an AsCpf1 expression plasmid was included in each independent experiment in order to normalize genome-editing efficiency for other treatment groups20.
Assessment of efficacy and specificity of the engineered CRISPR–Cpf1 system.
T7E1 has been routinely used to quantify the indel (insertion and deletion) percentage induced by the CRISPR system due to its ability to recognize and cleave non-perfectly matched DNA20. T7E1 is compatible with normal PCR buffer and easy to use without the need to further purify the PCR products. Yet it might be challenging to detect low mutagenesis rates and single-base mutations42. Deep sequencing, also known as high-throughput sequencing or next-generation sequencing, is a powerful technology for identifying indels with superior accuracy43. In addition to indel frequency, deep sequencing also provides informative data such as indel patterns, their individual percentage, and size and position distribution44. In this protocol, both T7E1 assay (Fig. 4) and targeted deep sequencing (Fig. 5) are used for assessing efficacy and target specificity of the engineered CRISPR–Cpf1 system. To obtain more accurate genome-editing outcomes, the sequencing data are analyzed by the computational tool CRISPResso (http://crispresso.rocks)44. The detailed data processing and specific parameters are described in Steps 68 and 69.
Figure 5 |.
Targeted deep-sequencing analysis of on-target efficacy and off-target effects of the engineered CRISPR–Cpf1 system. (a) AsCpf1-mediated genome-editing efficiency at the DNMT1 locus and its off-target effects in human 293T cells. (b) LbCpf1-mediated genome-editing efficiency at the DNMT1 locus and its off-target effects in human 293T cells. All data are plotted as the mean ± s.d. from three biological replicates (***P < 0.001, two-tailed t test). Ψ, pseudouridine; NS, not significant; OT, off-target. Adapted from ref. 20, Nature Publishing Group.
MATERIALS
REAGENTS
Acrylamide/bis-acrylamide solution, 40% (wt/vol), 19:1 (Bio-Rad, cat. no. 1610144) ! CAUTION This solution is toxic. Work with it under a fume hood. Wear a lab coat, gloves, and protective goggles.
Ammonium persulfate (Bio-Rad, cat. no. 1610700)
AMPure XP (Beckman Coulter, cat. no. A63881)
Antarctic phosphatase (New England Biolabs, cat. no. M2089S)
Base-modified ribonucleoside triphosphates (TriLink BioTechnologies, 5-methoxyuridine-5′-triphosphate, cat. no. N1093; N1-methylpseudouridine-5′-triphosphate, cat. no. N1081; pseudouridine-5′-triphosphate, cat. no. N1019)
dNTP solution mix (New England Biolabs, cat. no. N0447L)
DMSO (TCI America, cat. no. D0798)
DNA gel loading dye, 6× (Thermo Fisher Scientific, cat. no. R0611)
DNeasy Blood & Tissue Kits (Qiagen, cat. no. 69506) ! CAUTION This kit contains guanidine hydrochloride and proteinase K. When working with this kit, always wear a lab coat, gloves, and protective goggles.
DpnI restriction enzyme (New England Biolabs, cat. no. R0176S)
DMEM (Corning, cat. no. 10–027-CV)
Ethanol, USP grade, 200 proof (Thermo Fisher Scientific, cat no. 04–355-223) ! CAUTION Ethanol is flammable. Handle with care.
EZ-Vision In-Gel Solution, 10,000× (Amresco, cat. no. N391–0.5ML)
FBS, heat inactivated (Thermo Fisher Scientific, cat. no. 10082147)
GeneRuler 100-bp DNA ladder (Thermo Fisher Scientific, cat. no. SM0241)
Hemocytometer (Hausser Scientific, cat. no. 3100)
HiScribe T7 ARCA mRNA Kit (New England Biolabs, cat. no. E2065S)
Human embryonic kidney 293T cell line (American Type Culture Collection, cat. no. CRL-3216) ! CAUTION The cell line should be regularly checked to ensure that the cells are authentic and are not infected with mycoplasma.
Human glioblastoma U-87 MG cell line (American Type Culture Collection, cat. no. HTB-14) ! CAUTION The cell line should be regularly checked to ensure that the cells are authentic and are not infected with mycoplasma.
Human hepatoma Hep3B cell line (American Type Culture Collection, cat. no. HB-8064) ! CAUTION The cell line should be regularly checked to ensure that the cells are authentic and are not infected with mycoplasma.
KAPA HiFi HotStart Ready Mix, 2× (Kapa Biosystems, cat. no. KK2601)
Lipofectamine 3000 (Thermo Fisher Scientific, cat. no. L3000008)
Low-electroendosmosis agarose (BioExpress, cat. no. E-3120–500)
NEBuffer2 (New England Biolabs, cat. no. B7002S)
Nextera XT Index Kit (Illumina, cat. no. FC-131–1002)
Opti-MEM I reduced-serum medium (Thermo Fisher Scientific, cat. no. 31985–070)
PBS, 1×, pH 7.4 (Thermo Fisher Scientific, cat. no. 10010049)
Q5 Hot Start High-Fidelity Master Mix, 2× (New England Biolabs, cat. no. M0494L) ▲ CRITICAL Use a high-fidelity polymerase to minimize error rates during PCR amplification.
Q5 High-Fidelity DNA Polymerase (New England Biolabs, cat. no. M0491L)
QIAquick PCR Purification Kit (Qiagen, cat. no. 28104)
RNA Clean & Concentrator Kit (Zymo Research, cat. no. R1018)
RNA phosphoramidites (Glen Research, Bz-A-CE phosphoramidite, cat. no. 10–3003-02; U-CE phosphoramidite, cat. no. 10–3030-02; Ac-C-CE phosphoramidite, cat. no. 10–3015-02; Ac-G-CE phosphoramidite, cat. no. 10–3025-02)
RNase AWAY surface decontaminant (Thermo Fisher Scientific, cat. no. 7002)
Small-RNA PAGE Recovery Kit (Zymo Research, cat. no. R1070)
T7 Endonuclease I (New England Biolabs, cat. no. M0302L)
TBE-urea sample buffer, 2× (Thermo Fisher Scientific, cat. no. LC6876)
Tetramethylethylenediamine (Bio-Rad Laboratories, cat. no. 1610800)
Tris-acetate-EDTA (TAE) buffer, 10× (Sigma-Aldrich, cat. no. 574797–1L)
Tris-borate-EDTA buffer, 10× (Thermo Fisher Scientific, cat. no. 03500529)
Tris-EDTA buffer, 1×, pH 7.6, nuclease-free (Thermo Fisher Scientific, cat. no. BP2474–1)
Trypsin-EDTA (0.05% (wt/vol)), phenol red (Thermo Fisher Scientific, cat. no. 25300062)
UltraPure dH2O, DNase/RNase-free (Thermo Fisher Scientific, cat. no. 10977015)
Urea (Sigma-Aldrich, cat. no. U5378–1KG)
Plasmids, Cpf1 crRNAs, Cpf1 mRNAs, and primers
Cpf1 crRNAs, PAGE-purified (TriLink BioTechnologies, custom RNA oligonucleotide synthesis) ▲ CRITICAL Cpf1 crRNAs degrade easily. Store at −80 °C in small aliquots for up to 1 year.
Cpf1 mRNAs (TriLink BioTechnologies, custom mRNA synthesis). ▲ CRITICAL Cpf1 mRNAs degrade easily. Store at –80 °C in small aliquots for up to 1 year.
Human codon-optimized AsCpf1 expression plasmid, pcDNA3.1-hAsCpf1 (gift from F. Zhang’s lab at the Broad Institute of MIT and Harvard; this plasmid is currently available from Addgene, plasmid ID 69982)
Human codon-optimized LbCpf1 expression plasmid, pcDNA3.1-hLbCpf1 (gift from F. Zhang’s lab at the Broad Institute of MIT and Harvard; this plasmid is currently available from Addgene, plasmid ID 69988)
Plasmid-encoding Cpf1 protein used for in vitro transcription (TriLink BioTechnologies, custom plasmid synthesis)
Primers (Integrated DNA Technologies, custom DNA oligonucleotide synthesis, standard desalting) for T7E1 assay are listed in Table 1.
Primers with overhang adapter (Eurofins Genomics, custom DNA oligonucleotide synthesis, standard desalting) for targeted deep sequencing are listed in Table 1.
TABLE 1 |.
Primer pairs used to generate PCR amplicons for the T7E1 assay and targeted deep sequencing.
| Step | Primer | Sequence (5’–3’)a | Purpose |
|---|---|---|---|
| 39 | T7E1-ON-Fwd | CTGGGACTCAGGCGGGTCAC | Amplifying the on-target DNMT1 locus for T7E1 assay |
| 39 | T7E1-ON-Rev | CCTCAGCCAGAAGTCCCGTGC | Amplifying the on-target DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT1-Fwd | AGGAAAGCCATGCCAGAGACTCA | Amplifying the off-target site 1 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT1-Rev | CACCGCCACTCTGTTTCCAAG | Amplifying the off-target site 1 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT2-Fwd | GTTGGGACATGAAGGTCAAGTGTG | Amplifying the off-target site 2 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT2-Rev | TTTGTCTCCTGTTGCCTTCAGGCC | Amplifying the off-target site 2 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT3-Fwd | GAGGCATAGCAAGGTCATGCCTTT | Amplifying the off-target site 3 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT3-Rev | TGCTTCCCTTGGTGGAGCTG | Amplifying the off-target site 3 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT4-Fwd | TTTCCATGTAGGCCCATGCCC | Amplifying the off-target site 4 against the DNMT1 locus for T7E1 assay |
| 39 | T7E1-OT4-Rev | CCAGGTTACCAGCAACAGATCTC | Amplifying the off-target site 4 against the DNMT1 locus for T7E1 assay |
| 60 | NGS-ON-Fwd | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTCCCTCACTCCTGCTCGGTGAA | Amplifying the on-target DNMT1 locus for targeted deep sequencing |
| 60 | NGS-ON-Rev | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAAGTCACTCTGGGGAACACGCC | Amplifying the on-target DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT1-Fwd | TCGTCGGCAGCGTCAGATGTGTATAAGAGaCAGACCTTTTGGGCGTGGAGAAGGG | Amplifying the off-target site 1 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT1-Rev | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGGAGGGGGTCAGCATGAAAGG | Amplifying the off-target site 1 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT2-Fwd | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCTCCCCCACCCCCTAGGAAAGT | Amplifying the off-target site 2 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT2-Rev | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCCTTTCTGGTGGAGTGTCCCC | Amplifying the off-target site 2 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT3-Fwd | TCGTCGGCAGCGTCAGAAGTGTATAAGAGACAGTGAAGGTATAGGAGAGGTTTTGGGCT | Amplifying the off-target site 3 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT3-Rev | GTCTCGTGGGCTCGGAGATGTGTaTAAGAGACAGAGACGACCTTAGATGGAGTGTTGTGT | Amplifying the off-target site 3 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT4-Fwd | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTGCCAGTGGAAGGAGGGAGTGT | Amplifying the off-target site 4 against the DNMT1 locus for targeted deep sequencing |
| 60 | NGS-OT4-Rev | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTGCCCAGGAAGTTGCTTCTCCC | Amplifying the off-target site 4 against the DNMT1 locus for targeted deep sequencing |
Bold letters denote the overhang adapter sequences.
EQUIPMENT
Eight-strip PCR tubes (BioExpress, cat. no. T-3135–1)
Axygen microcentrifuge tubes, sterile, 1.5 ml, RNase, DNase, pyrogen-free (Corning, cat. no. MCT-150-C-S)
Biological safety cabinet (Thermo Fisher Scientific, model no. 1300 Series A2)
Cell culture flasks, TC treated, 25 cm2 (Corning, cat. no. 430639)
Cell culture flasks, TC treated, 75 cm2 (Corning, cat. no. 430641U)
Cell culture plates, TC treated, 24 well (Corning, cat. no. 3526)
Bench-top centrifuge (Eppendorf, model nos. 5427 R and 5804 R)
Cryogenic tubes (2 ml; Fisher Scientific, cat. no. 033377D)
Digital water bath (Thermo Fisher Scientific, cat. no. 2320)
Digital dry bath (Thermo Fisher Scientific, cat. no. 88–860-023)
Eppendorf tubes, PCR clean (5 ml; Fisher Scientific, cat. no. 14–282-301)
Falcon conical tubes, polypropylene (15 ml; Corning, cat. no. 430790)
Falcon conical tubes, polypropylene (50 ml; Corning, cat. no. 430828)
Falcon serological pipettes (5 ml; Corning, cat. no. 357543)
Falcon serological pipettes (10 ml; Corning, cat. no. 357551)
Freezing container (Thermo Fisher Scientific, cat. no. 15–350-50)
Gel-imaging systems (ChemiDoc XRS; Bio-Rad Laboratories)
Incubator (Thermo Fisher Scientific, model no. Heracell 150i)
Inverted microscope (Leica, model no. DMi1)
Mini centrifuge (Thermo Fisher Scientific, cat. no. 12–006-901)
MiSeq system (Illumina, cat. no. SY-410–1003)
Nuclease-free pipette tips (Rainin, 20 μl, cat. no. GPS-L10; 200 μl, cat. no. GPS-L250; 1,000 μl, cat. no. GPS-L1000)
Pasteur pipettes (Fisher Scientific, cat. no. 22–042817)
PowerPac basic power supply (Bio-Rad, cat. no. 1645050)
PowerPac HV power supply (Bio-Rad, cat. no. 1645056)
Protean II xi vertical electrophoresis cell (Bio-Rad Laboratories, cat. no. 1651813)
Thermal cycler (Bio-Rad, model no. T100)
UV-visible spectrophotometer (Thermo Fisher Scientific, model no. NanoDrop 2000)
Vacuum filter bottle system, 0.22-μm PES membrane (Corning, cat. no. 431097)
Vortex mixer (Scientific Industries, model no. Genie 2)
Water purification system (Millipore)
Wide Mini-Sub cell GT horizontal electrophoresis system (Bio-Rad, cat. no. 1704468)
REAGENT SETUP
Complete growth medium
Add 50 ml of FBS to 450 ml of DMEM. Filter the mixed solution with a 0.22-μm vacuum filter bottle system. The prepared medium can be stored at 4 °C for up to 1 month. Prewarm the medium to 37 °C before use.
crRNA working solution
Prepare a 15 μM crRNA working solution in Tris-EDTA buffer on ice. Aliquots (50 μl/tube) can be stored at −80 °C for up to 1 year. ! CAUTION Prepare a working solution in an RNase-free area to prevent degradation.
Cryopreservation medium
Mix complete growth medium and DMSO at a ratio of 9:1 and keep the solution at 4 °C for up to 2 months.
Plasmid and mRNA working solution
Prepare a 200 μg/ml working solution in Tris-EDTA buffer on ice. Aliquots (50 μl/tube) can be stored at −80 °C for up to 1 year. ! CAUTION Prepare the working solution in an RNase-free area.
TAE electrophoresis buffer
Dilute 10× TAE buffer to a 1× working solution with ultrapure water from a Millipore water purification system and store it at room temperature (20–25 °C) for up to 2 months.
PROCEDURE
Design of crRNAs targeting the genes of interest ● TIMING 4–6 h
-
1
Design of AsCpf1 crRNAs. crRNAs can be designed via online tools, such as Benchling (https://benchling.com) and CHOPCHOP v2 (http://chopchop.cbu.uib.no) using 5′-TTTV-3′ as the PAM (Experimental design). Alternatively, sequences of crRNAs can be obtained from the literature.
-
2
Identification of potential off-target sites. The potential off-target sites associated with crRNAs can be ranked through the web-based tools Cas-OFFinder (http://www.rgenome.net/cas-offinder) or COSMID (https://crispr.bme.gatech.edu). As COSMID is primarily designed for the CRISPR–Cas9 system, input TTTV plus the protospacer sequence into the ‘Guide Strand Sequence’ text window and leave the PAM suffix empty when using COSMID.
-
3
Rank crRNAs for their indels with an online tool: CRISPR AsCpf1 INDEL Score tool (http://big.hanyang.ac.kr/cindel).
-
4
Select one crRNA with low off-target effects and a high indel score for experimental studies.
-
5
Using COSMID, design primer pairs to amplify genomic segments spanning on-target or off-target sites (used for the T7E1 cleavage assay in Step 39).
▲ CRITICAL STEP A PCR product in the range of 600–1,000 bp—with the cleavage site away from the center of the amplicons to ensure that T7E1 digested products can be easily differentiated on an agarose gel—is optimal. Other tools such as CHOPCHOP v2 and Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast) can be used for primer design as well.
-
6
Check the specificity of the selected primers with Primer-BLAST.
-
7
Order the primers; after obtaining them, resuspend the primers to a stock concentration of 100 μM.
Engineering of the components of the CRISPR–Cpf1 system ● TIMING 30–60 d
-
8
Synthesize engineered Cpf1 crRNAs using an automated solid-phase DNA/RNA synthesizer according to the manufacturer’s instructions. For chemically modified crRNAs, replace unmodified RNA phosphoramidites at the desired positions with corresponding modified phosphoramidites during synthesis (supplementary table 1). Regarding structurally altered crRNAs, substitute only the loop sequences of the wild-type AsCpf1 crRNA with those of crRNAs from other Cpf1 orthologs, while keeping the remaining sequences unchanged during synthesis (supplementary table 1). Alternatively, Cpf1 crRNAs are commercially available from TriLink BioTechnologies.
-
9Cast a 20% (wt/vol) denaturing preparative polyacrylamide gel.
Components Amount Urea 21 g 40% (wt/vol) acrylamide/bis-acrylamide (19:1) 25 ml 5 × Tris-borate-EDTA buffer 10 ml 10% (wt/vol) ammonium persulfate 500 μl Tetramethylethylenediamine 50 μl dH2O to 50 ml ▲ CRITICAL STEP Cast the polyacrylamide gel in a fume hood. Prepare an ammonium persulfate solution shortly before use. Do not add ammonium persulfate or tetramethylethylenediamine solution until the urea has completely dissolved in the other components.
-
10
Allow the gel to polymerize for ~60 min.
-
11
Prerun the gel in Tris-borate-EDTA buffer for 2 h at 10 V/cm.
-
12
Purification of synthetic Cpf1 crRNAs. Denature crRNA samples in an equal volume of TBE-urea sample buffer at 90 °C for 90 s. Load the sample solution onto the gel, and run polyacrylamide urea gel electrophoresis at room temperature for 16 h at 25 V/cm.
-
13
Cut the desired RNA bands out of the gel under UV shadowing.
▲ CRITICAL STEP Wear gloves, and face and eye protection to avoid exposure to UV.
-
14
Extract crRNAs with a Small-RNA PAGE Recovery Kit by following the manufacturer’s instructions.
-
15
Quantify Cpf1 crRNAs (2 μl) with a NanoDrop 2000 spectrophotometer.
-
16
Store the synthetic crRNAs at − 80 °C in small aliquots (20 μl/tube) until use.
∎ PAUSE POINT crRNA stock can be stored at − 80 °C for up to 1 year.
-
17To produce engineered AsCpf1 mRNAs, prepare DNA templates for in vitro transcription from a plasmid-encoding Cpf1 protein from TriLink BioTechnologies, by using a reverse primer containing 120 poly(T) and a corresponding forward primer in the following PCR reaction:
Components Volume (μl) Final concentration Q5 high-fidelity DNA polymerase 1 0.02 U/μl dNTPs, 10 mM 4 400 μM Forward primer, 10 μM 5 0.5 μM Reverse primer, 10 μM 5 0.5 μM Q5 reaction buffer, 5× 20 1× Q5 high GC enhancer, 5× 20 1× AsCpf1 plasmid for in vitro transcription Variable 5 ng Nuclease-free water To 100 Forward primer, TGCAAGGCGATTAAGTTGGGTAAC.
Reverse primer, TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT TTTTTTTTTTTTTTTTTTTTCTTCCTACTCAGGCTTTATTCAAAGACC.
▲ CRITICAL STEP Use high-fidelity DNA polymerase such as Q5 Hot Start enzyme to minimize errors in PCR amplification.
-
18Carry out PCR reactions on a thermal cycler with the following cycling conditions:
Cycle number Denature Anneal Extend Final 1 98 °C, 45 s 2–36 98 °C, 15 s 64 °C, 30 s 72 °C, 150 s 37 72 °C, 150 s 38 4 °C, infinite -
19
Remove AsCpf1 plasmid with 1 μl of DpnI at 37 °C for 30 min.
-
20
Purify DNA templates with a QIAquick PCR Purification Kit, following the manufacturer’s instructions.
-
21
Conduct in vitro transcription of ARCA-capped and poly(A)-tailed mRNA transcripts using a HiScribe T7 ARCA mRNA Kit according to the manufacturer’s protocols. For base-modified mRNA transcripts, replace natural ribonucleoside triphosphates with the corresponding base-modified ribonucleoside triphosphates during in vitro transcription.
-
22
Purify the Cpf1 mRNA transcripts using an RNA Clean & Concentrator Kit according to the manufacturer’s instructions.
-
23
Treat the Cpf1 mRNA transcripts with 2 μl of Antarctic phosphatase at 37 °C for 30 min.
-
24
Purify the Cpf1 mRNA transcripts again using an RNA Clean & Concentrator Kit, following the manufacturer’s instructions.
-
25
Cast a 1% (wt/vol) agarose gel. Add 0.9 g of agarose powder to 90 ml of TAE buffer. Microwave for 2 min and add 9 μl of EZ-Vision In-Gel Solution. Gently mix and slowly pour into a gel tray to avoid bubbles. Cool for ~30 min at room temperature.
-
26
To validate the length of mRNA transcripts, denature the mRNA samples in an equal volume of TBE-urea sample buffer for 90 s at 90 °C. Load the sample solution onto the gel and perform agarose gel electrophoresis at room temperature for 40 min at 6 V/cm.
-
27
Quantify the concentration of Cpf1 mRNAs with a NanoDrop 2000 spectrophotometer.
-
28
Store the synthetic mRNAs at − 80 °C in small aliquots (20 μl/tube) until use. Alternatively, AsCpf1 mRNAs are commercially available from TriLink BioTechnologies.
∎ PAUSE POINT mRNA stock can be stored at − 80 °C for up to 1 year.
Co-delivery of engineered Cpf1 crRNA and Cpf1 mRNA to human cells ● TIMING 3–4 d
-
29
Cell culture. Remove a vial of 293T cells from a liquid nitrogen tank, and rapidly thaw cells in a 37 °C water bath. Add a fivefold volume of prewarmed DMEM supplemented with 10% (vol/vol) FBS, and centrifuge at room temperature for 5 min at 100g to remove the DMSO in freezing medium. Discard the supernatant and resuspend the cell pellets in 5 ml of prewarmed complete growth medium, transfer the cell suspension to a 25-cm2 flask, and then grow the cells at 37 °C in a humidified 5% CO2 incubator.
-
30
Routinely passage 293T cells every 3–4 d at a split ratio of 1:10. Typically, when cells reach 60–80% confluence, rinse them gently with PBS in case cells detach from the flask surface. Add 0.5 ml of prewarmed trypsin-EDTA and wait until the cells are dispersed into small clumps (~10 s). Add 4.5 ml of prewarmed complete growth medium and pipette the solution vigorously to detach the cells. Transfer 0.5 ml of cell suspension to a 15-ml tube containing 4.5 ml of prewarmed complete growth medium, mix well, and transfer to a new 25-cm2 flask.
-
31
Dissociate the cells from a flask with trypsin-EDTA as described in Step 30 ~24 h before addition of the CRISPR–Cpf1 components.
-
32
Seeding of cells. Adjust the cell density to 200,000 cells/ml in fresh culture medium. Transfer 0.5 ml of cell suspension to each well of a 24-well plate. Gently shake the plate to distribute the cells evenly in the plate. If a different size of plate is used, scale up or down the number of cells accordingly based on the relative surface area of the plate. Conditions may vary for different cell types.
▲ CRITICAL STEP The density of the cells may affect genome-editing efficacy. Maintain a similar cell density to ensure experiment reproducibility. Avoid using cells from the first two passages or cells with high passage numbers ( > 20).
-
33
Prepare Lipofectamine 3000-complexed genome-editing components by first prewarming Opti-MEM reduced serum medium and Lipofectamine 3000 reagent to room temperature. To complex crRNA from Step 16 and Cpf1 mRNA from Step 28 or Cpf1 expression plasmid with Lipofectamine 3000 according to the manufacturer’s instructions, typically, to one tube, add 4 μl of crRNA working solution and 50 μl of Opti-MEM reduced serum medium. In a second tube, add 1 μl of Lipofectamine 3000 and 50 μl of opti-MEM medium. Transfer the diluted crRNA solution to the diluted Lipofectamine 3000 solution, mix vigorously by pipetting, and incubate at room temperature for 5 min. Similarly, add 4 μl of Cpf1 mRNA or Cpf1 expression plasmid working solution to 50 μl of Opti-MEM reduced serum medium. Meanwhile, add 1 μl of Lipofectamine 3000 to 50 μl of opti-MEM medium. Combine the two solutions, mix vigorously, and incubate at room temperature for 5 min. For Cpf1 expression plasmid, 1.6 μl of P3000 reagent is also added to the plasmid solution diluted with opti-MEM medium according to the manufacturer’s instructions.
▲ CRITICAL STEP The quality of RNA greatly affects genome-editing efficiency. Biological activity of Cpf1 crRNAs and mRNAs (especially for mRNAs) is reduced after multiple freeze–thaw cycles. Keep the molarity rather than weight consistent for all crRNAs to compare their efficiency because engineered crRNAs have different molecular weights.
-
34
Add Lipofectamine 3000-complexed genome-editing components to the 293T cells from Step 32. Transfer 66 μl of freshly prepared complex containing crRNA to each well dropwise. Next, add 66 μl of complex containing Cpf1 mRNA or Cpf1 plasmid to each well. That is 38 pmol crRNA and 500 ng of Cpf1 mRNA (or plasmid) per well.
▲ CRITICAL STEP It is not necessary to change to fresh medium before and after treatments. For crRNAs with low activity, dosage of formulations may need to be adjusted. Proportionally scale up or down the amount of formulations according to the density of cells, as well as the relative surface area, if a different size of plate is used.
Evaluation of on-target efficacy and off-target effects using the T7E1 assay ● TIMING 1–2 d
-
35
Harvest cells 48 h after treatment with genome-editing components by centrifuging at room temperature for 5 min at 100g. Discard the supernatant and rinse the pellet with PBS.
? TROUBLESHOOTING
∎ PAUSE POINT Cell pellets can be stored at − 20 °C overnight.
-
36
Extract gDNA with a DNeasy Blood & Tissue Kit by following the manufacturer’s instructions.
-
37
Measure the concentration of gDNA using a NanoDrop 2000 spectrophotometer.
-
38
Store the gDNA in a − 20 °C freezer.
∎ PAUSE POINT The isolated gDNA can be stored at − 20 °C for at least 6 months.
-
39Set up PCR reactions for the T7E1 assay. Mix the following components well to amplify genomic regions covering the on-target or off-target site, using primer pairs designed at Step 7 (table 1):
Components Volume (μl) Final concentration Q5 Hot Start High-Fidelity Master Mix, 2× 12.5 1× T7E1-Fwd-primer, 10 μM 1.25 0.5 μM T7E1-Rev-primer, 10 μM 1.25 0.5 μM gDNA template (from Step 38), 100 ng Variable 100 ng Nuclease-free water To 25 ▲ CRITICAL STEP Use high-fidelity DNA polymerase such as Q5 Hot Start enzyme to minimize errors in PCR amplification. Q5 Hot Start master mix can be used at room temperature.
-
40
Spin all liquid down to the bottom of the PCR tubes with a minicentrifuge at room temperature for 20 s.
-
41Carry out PCR reactions on a thermal cycler with the following cycling conditions:
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–36 98 °C, 5 s Variable, 10 s 72 °C, 20 s 37 72 °C, 2 min 38 4 °C, infinite ▲ CRITICAL STEP Unlike other DNA polymerase, high-fidelity Q5 enzyme requires a NEB Tm calculator (http://tmcalculator.neb.com) to determine the optimal annealing temperature for a given set of primers. For example, the annealing temperature for T7E1-ON-Fwd and T7E1-ON-Rev is 72 °C (Table 1). We recommend increasing the cycle number from 35 to 45 for certain genomic loci that are difficult to amplify.
∎ PAUSE POINT PCR products can be stored at − 20 °C for at least 1 month.
-
42
Cast a 2% (wt/vol) agarose gel by adding 1.8 g of agarose powder to 90 ml of TAE buffer. Microwave for 2 min and add 9 μl of EZ-Vision In-Gel Solution. Gently mix and slowly pour into a gel tray to avoid bubbles. Cool for ~30 min at room temperature.
-
43
Check the quality of the amplicons by running 5 μl of the PCR products premixed with 1 μl of 6 × DNA gel loading dye on the 2% (wt/vol) agarose gel for 40 min at 6 V/cm.
-
44
Visualize the gel using a ChemiDoc XRS imaging system.
▲ CRITICAL STEP Ideally, only PCR amplicons will be detected. However, it is acceptable if the size of nonspecific bands does not interfere with the following quantitation of indel frequency. We recommend designing and ordering several primer pairs at Step 7 and selecting the best one (the one that yields a clean and specific band).
? TROUBLESHOOTING
-
45Set up a 19-μl hybridization reaction for the T7E1 assay:
Components Amount (μl) PCR products 10 NEBuffer 2, 10× 7 Nuclease-free water 2 ▲ CRITICAL STEP It is not necessary to purify PCR products before the digestion if T7E1 enzyme is used. The volume of PCR products must be optimized for specific genomic loci.
-
46Re-anneal the PCR products on a thermal cycler with the following program:
Cycle number Denature Anneal Final 1 95 °C, 5 min 2 95 °C85 °C, − 2 °C/s 3 85 °C25 °C, − 0.1 °C/s 4 4 °C, infinite -
47
Cast a 2% (wt/vol) agarose gel as described in Step 42.
▲ CRITICAL STEP Cast a gel beforehand to ensure a constant time for digestion.
-
48
T7E1 digestion. Add 1 μl of T7E1 to the re-annealed PCR products. Mix well and spin down with a minicentrifuge at room temperature for ~20 s.
-
49
Incubate the reaction mixture at 37 °C for 30 min.
▲ CRITICAL STEP Keep the reaction temperature and time constant to ensure experimental reproducibility.
-
50
Add 4 μl of 6 × DNA gel loading dye to each sample and load to the 2% (wt/vol) agarose gel.
-
51
Run the agarose gel in TAE buffer at 6 V/cm for 40 min on a horizontal electrophoresis system.
-
52
Visualize the gel using a ChemiDoc XRS imaging system.
? TROUBLESHOOTING
-
53
Quantify the genome-editing efficiency using Quantity One software downloaded from the Bio-Rad Laboratories website (http://www.bio-rad.com/en-ch/product/quantity-one-1-d-analysis-software).
▲ CRITICAL STEP We occasionally observed that T7E1 induced nonspecific cleavage of amplicons of unedited groups. If the cleaved bands can be differentiated from those of interest, determine editing efficiency and conduct targeted deep sequencing by following Steps 54–69. In some cases, however, nonspecific bands have the same length as that induced by the CRISPR–Cpf1 system after treatment with T7E1, hampering accurate quantification of indels. Under these circumstances, it is essential to conduct a pilot study for T7E1 assays with gDNA extracted from unedited cells before engineering crRNAs in order to avoid false-positive results.
-
54
Determine Cpf1-mediated genome-editing efficiency (indels) according to the formula: 100 × (1–(1–fraction cleaved)1/2) (see the New England Biolabs website (https://www.neb.com/protocols/2014/08/11/determining-genome-targeting-efficiency-using-t7-endonuclease-i)).
-
55
For on-target efficiency, normalize the indel frequency of the combination of engineered cRNAs and AsCpf1 mRNAs by dividing by that of the counterpart (the combination of wild-type crRNA and AsCpf1 expression plasmid).
▲ CRITICAL STEP For off-target effects, the results of the T7E1 assay may have to be considered a preliminary estimate, as it is often difficult to quantify low off-target mutagenesis rates. To obtain more quantitative data for off-target effects, follow Steps 56–69.
? TROUBLESHOOTING
Evaluation of on-target efficacy and off-target effects using targeted deep sequencing ● TIMING 30–40 d
-
56
Design primers using COSMID to amplify genomic segments spanning on-target or off-target sites (corresponding to sites in the T7E1 assay) for targeted deep sequencing.
▲ CRITICAL STEP The PCR product is optimal in the range of 200–300 bp with the predicted cleavage site away from either end of the amplicons to minimize false positives due to imperfect trimming of the reads.
-
57
Check the specificity of the selected primers with Primer-BLAST.
-
58
Add overhang adapter sequences to the 5′ ends of the primers (Table 1). Targeted deep sequencing requires additional overhang adapters in order to append Illumina index and sequencing adapters at Step 64.
-
59
Order and resuspend primers containing overhang adapter sequences to a stock concentration of 100 μM.
-
60Set up PCR reactions for targeted deep sequencing. Mix the following components well to amplify the on-target or off-target sites, using the gDNA extracted in Step 38 and primer pairs containing overhang adapter sequences (Table 1):
Components Volume (μl) Final concentration Q5 Hot Start High-Fidelity Master Mix, 2× 12.5 1× NGS-Fwd-primer, 10 μM 1.25 0.5 μM NGS-Rev-primer, 10 μM 1.25 0.5 μM gDNA template, 1.25 ng/μl 10 12.5 ng -
61Conduct PCR reactions with the following cycling conditions:
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–26 98 °C, 5 s Variable, 10 s 72 °C, 20 s 27 72 °C, 2 min 28 4 °C, infinite ▲ CRITICAL STEP Use the NEB Tm calculator to determine the optimal annealing temperatures for Miseq sequencing primers, without inputting overhang adapter sequences. In other words, only input sequences that are completely complementary to the genomic locus of interest to calculate Tm values. For example, the annealing temperature for NGS-ON-Fwd and NGS-ON-Rev is 72 °C (Table 1).
? TROUBLESHOOTING
▲ PAUSE POINT PCR products can be stored at − 20 °C for 1 month.
-
62
Check the quality of amplicons by running 5 μl of the PCR products on a 2% (wt/vol) agarose as described in Step 43.
-
63
Purify the PCR products with the AMPure XP system, following the manufacturer’s instructions.
-
64Attach index primers (including Illumina index and sequencing adapter sequences) to the pooled amplicons using the Nextera XT Index Kit, by setting up the following reaction:
Components Volume (μl) Purified PCR products 5 Nextera XT index 1 primers 5 Nextera XT index 2 primers 5 2× KAPA HiFi HotStart Ready Mix 25 Nuclease-free water 10 -
65Perform PCR reactions on a thermal cycler with the following cycling conditions:
Cycle number Denature Anneal Extend Final 1 95 °C, 3 min 2–11 95 °C, 30 s 55 °C, 30 s 72 °C, 30 s 12 72 °C, 5 min 13 4 °C, infinite ▲ CRITICAL STEP An increase in PCR cycle number may result in additional nonspecific products.
-
66
Purify the second round of PCR products with AMPure XP beads, following the manufacturer’s instructions.
-
67
Sequence the pooled libraries on an Illumina MiSeq system with appropriate paired-end reads depending on the size of the PCR products.
Data processing ● TIMING 5–7 d
-
68
Analyze the raw data (fastq.gz files) with CRISPResso using the following parameters: ‘experimental design’ (paired-end reads); ‘fastq file R1’ and ‘fastq file R2’ (Miseq paired fastq.gz files); ‘amplicon sequence’ (one strand of the amplicon amplified at Step 61); ‘sgRNA sequence’ (22 nt; T instead of U, immediately 3′ of the TTTV PAM); ‘window size’ (10; 10 bp around each side of the predicted cleavage site); ‘minimum average read quality’ ( > 30); ‘minimum single bp quality’ ( > 20); and other parameters (default). Leave the parameters ‘expected HDR amplicon sequence’ and ‘coding sequence/s’ empty.
▲ CRITICAL STEP The CRISPResso analysis tool is primarily designed for the CRISPR–Cas9 system. Moreover, it contains additional functions such as quantification of homology directed repair (HDR) and frameshift mutations, and analysis of splice sites. Hence, specific parameters are required to precisely quantify the nonhomologous end joining (NHEJ) mutation events induced by the CRISPR–Cpf1 system.
-
69
Export and analyze the genome-editing data. Obtain NHEJ indels from the quantification file (Quantification_of_editing_frequency.txt) generated by CRISPResso. Identify the mutagenesis pattern and corresponding rate from the sequence file (Alleles_frequency_table.txt).
? TROUBLESHOOTING
? TROUBLESHOOTING
Troubleshooting advice can be found in table 2.
● TIMING
Steps 1–7, design of crRNAs targeting the genes of interest: 4–6 h
Steps 8–28, engineering of CRISPR–Cpf1 components: 30–60 d
Steps 29–34, co-delivery of engineered Cpf1 crRNA and Cpf1 mRNA to human cells: 3–4 d
Steps 35–55, evaluation of on-target efficacy and off-target effects using the T7E1 assay: 1–2 d
Steps 56–67, evaluation of on-target efficacy and off-target effects using targeted deep sequencing: 30–40 d
Steps 68 and 69, data processing: 5–7 d
table 2 |.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 35 | Low cell survival | Insufficient quality of cells | Use cells with proper passage number (2 < passage number < 20) |
| Low cell density | Treat cells when the confluency is > 50% | ||
| High dosage of formulations | Lower the dosage of formulated CRISPR–Cpf1 components | ||
| 44 | No or faint PCR bands | Inappropriate primers | Redesign primers and find the optimal one |
| Ineffective DNA polymerase | Divide the DNA polymerase into single-use aliquots before using for the first time | ||
| Inappropriate cycle number | Increase the number of cycles. It is an effective method to enhance the abundance of PCR products without compromising specificity when Q5 Hot Start DNA Polymerase is used | ||
| Unclean electrophoresis buffer | Avoid overusing electrophoresis buffer | ||
| 52 | Nonspecific bands | Primers are not specific enough | Use appropriate primers or try to adjust the annealing temperature |
| Problems stemming from target locus sequence | Use a negative control to eliminate nonspecific interference, or redesign crRNA by looking for a different protospacer located within the same genomic locus | ||
| Inappropriate T7E1 digestion temperature and time | Incubate at 37 °C for 15–30 min | ||
| 55, 61 | Low genome-editing efficiency | Degradation of CRISPR–Cpf1 components before treatment | Thoroughly clean working areas with RNase AWAY solution or equivalent; use a new aliquot after three to four freeze– thaw cycles |
| High cell density | Lower the cell density | ||
| Insufficient transfection reagents | Increase the ratio of transfection reagents to CRISPR–Cpf1 components | ||
| Low dosage | Increase the dosage of formulated CRISPR–Cpf1 components | ||
| Insufficient quality of PCR products | Use PCR products that were stored at − 20 °C for no more than 1 month | ||
| 69 | No reports or abnormal reports | .fastq file is too large | Use the off-line version of CRISPResso if a single .fastq file is > 100 Mb |
| Wrong format of crRNA sequence | Replace uracil (U) with thymine (T) | ||
| Wrong amplicon sequence | Check the sequence of amplicon with BLAST | ||
| Inappropriate window size | Use the recommended window size (10 nt) | ||
| Wrong options for trimming | Confirm the adapter trimming mode |
ANTICIPATED RESULTS
Using this protocol, we previously engineered synthetic AsCpf1 crRNA and AsCpf1 mRNA, and consequently achieved robust genome editing at three genomic loci: DNMT1, AAVS1, and FANCF in different human cell lines tested (293T, Hep3B, and U87) without compromising target specificity (Figs. 4a–c and 5a)20. Such a strategy is also applicable to LbCpf1. For example, 47% genome-editing activity was observed for engineered LbCpf1 components at the DNMT1 locus in 293T cells. Under the same conditions, wild-type LbCpf1 crRNA in combination with LbCpf1 expression plasmid displayed no detectable indels (Figs. 4d and 5b). In addition, we found that AsCpf1 was able to recognize the majority of crRNAs from other Cpf1 families, expanding our understanding of the AsCpf1 system. In summary, this protocol will facilitate engineering of the CRISPR–Cpf1 system and relevant CRISPR platforms in order to maximize genome editing and apply it to diverse applications.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge F. Zhang and his laboratory at the Broad Institute of MIT and Harvard for providing Cpf1 plasmids and technical assistance. This work was supported by the National Institutes of Health through the National Heart, Lung, and Blood Institute (NHLBI; R01HL136652), as well as by the start-up fund from the College of Pharmacy at The Ohio State University.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
References
- 1.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zetsche B et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol 35, 31–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y et al. Generation of knockout mice by Cpf1-mediated gene targeting. Nat. Biotechnol 34, 808–810 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv 3, e1602814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watkins-Chow DE et al. Highly-efficient Cpf1-mediated gene targeting in mice following high concentration pronuclear injection. G3: Genes Genomes Genet. 7, 719–722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo A, Masafumi M, Kaya H & Toki S Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep 6, 38169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim H et al. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun 8, 14406 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang X et al. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 17018 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Wang M, Mao Y, Lu Y, Tao X & Zhu JK Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10, 1011–1013 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Xu R et al. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J 15, 713–717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin X et al. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Hur JK et al. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol 34, 807–808 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Gao L et al. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol 35, 789–792 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonfara I, Richter H, Bratovic M, Le Rhun A & Charpentier E The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Yamano T et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong D et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Gao P, Yang H, Rajashankar KR, Huang Z & Patel DJ Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swarts DC, van der Oost J & Jinek M Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233.e224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stella S, Alcon P & Montoya G Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559–563 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Li B et al. Engineering CRISPR–Cpf1 crRNAs and mRNAs to maximize genome editing efficiency. Nat. Biomed. Eng 1, 0066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corey DR Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest 117, 3615–3622 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deleavey GF & Damha MJ Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol 19, 937–954 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Burnett JC & Rossi JJ RNA-based therapeutics: current progress and future prospects. Chem. Biol 19, 60–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Luo X & Dong Y Effects of chemically modified messenger RNA on protein expression. Bioconjug. Chem 27, 849–853 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Dowdy SF Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol 35, 222–229 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Labun K, Montague TG, Gagnon JA, Thyme SB & Valen E CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HK et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 14, 153–159 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Bae S, Park J & Kim JS Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cradick TJ, Qiu P, Lee CM, Fine EJ & Bao G COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther. Nucleic Acids 3, e214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendel A et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol 33, 985–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahdar M et al. Synthetic CRISPR RNA-Cas9–guided genome editing in human cells. Proc. Natl. Acad. Sci. USA 112, E7110–E7117 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang Y et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 16, 280 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kariko K et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther 16, 1833–1840 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson BR et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kormann MS et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol 29, 154–157 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Zangi L et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol 31, 898–907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahin U, Kariko K & Tureci O mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Andries O et al. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release 217, 337–344 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Thess A et al. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther 23, 1456–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uchida S, Kataoka K & Itaka K Screening of mRNA chemical modification to maximize protein expression with reduced immunogenicity. Pharmaceutics 7, 137–151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vouillot L, Thélie A & Pollet N Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3: Genes Genomes Genet. 5, 407–415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldman D & Domschke K Making sense of deep sequencing. Int. J. Neuropsychopharmacol 17, 1717–1725 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinello L et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol 34, 695–697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.