Abstract
The most highly studied endocytic pathway, clathrin-dependent endocytosis, mediates a wide range of fundamental processes including nutrient internalization, receptor recycling, and signal transduction. In order to model tissue specific and developmental aspects of this process, CRISPR/Cas9 genomic editing was utilized to fluorescently label the C-terminus of clathrin light chain A (CLTA) within the phenotypically normal, parental CRMi001-A human induced pluripotent stem cell line. Successfully edited cells were isolated by fluorescently activated cell sorting, remained karyotypically normal, and maintained their differentiation potential. This cell line facilitates imaging of endogenous clathrin trafficking within varied cell types.
| Resource Table. | |
|---|---|
| Unique stem cell line identifier | CRMi001-A-1 |
| Alternative name(s) of stem | SANFi002-A-1 |
| cell line | CRMi001-A-1-CLTA-TQ2 |
| NCRM-5-CLTA-TQ2 | |
| Institution | Sanford Research |
| Contact information of distributor | Kevin Francis; Kevin.Francis@SanfordHealth.org |
| Type of cell line | iPSC |
| Origin | Human |
| Additional origin info | Age: Fetal |
| Sex: Male | |
| Ethnicity if known: Unknown | |
| Cell Source | Umbilical cord blood derived CD34 + cells |
| Clonality | Mixed |
| Method of reprogramming | Episomal vectors: Oct4, Sox2, c-Myc, KLF4, Lin28 SV40 Large T antigen |
| Genetic Modification | Yes |
| Type of Modification | Knock-in |
| Associated disease | N/A |
| Gene/locus | CLTA/9p13.3 |
| Method of modification | CRISPR/Cas9 |
| Name of transgene or resistance | Tq2 and puromycin resistance gene |
| Inducible/constitutive system | N/A |
| Date archived/stock date | 05/30/17 |
| Cell line repository/bank | N/A |
| Ethical approval | The CRMi001-A hiPSC line was obtained from the National Heart, Lung, and Blood Institute iPSC Core in Bethesda, MD. All research involving hiPSCs was approved by the Institutional Biosafety Committee at Sanford Research (approval no. 2015101). |
Resource utility
Generation of CRMi001-A-1 human induced pluripotent stem cells enable monitoring and manipulation of endogenous clathrin trafficking dynamics in pluripotent or differentiated cell types.
Resource details
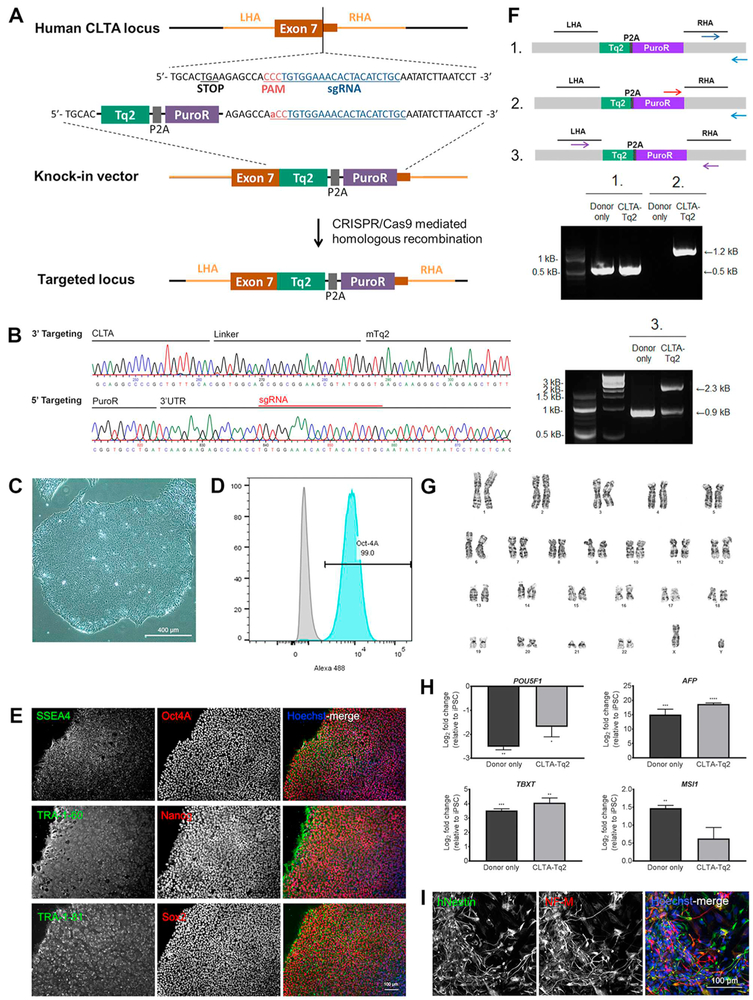
A CRISPR/Cas9 mediated knock-in system was utilized to generate human induced pluripotent stem cells (hiPSCs) expressing CLTA tagged with the fluorescent protein Turquoise2 (Tq2), followed by a self-cleavable P2A peptide sequence and the puromycin N-acetyltransferase gene. 1kB homology arms targeted a Tq2-P2A-puro cassette to the C-terminal end of the CLTA locus (Fig. 1A). A single guide RNA (sgRNA) was designed to target the stop codon in Exon 7 and demonstrated efficient cleavage in a T7 endonuclease assay (Fig. S1). CRMi001-A hiPSCs were transfected with the knock-in vector and sgRNA/Cas9 expression plasmid by nucleofection. Selection for Tq2 positive cells by fluorescence-activated cell sorting (FACS) demonstrated a knock-in efficiency of approximately 1% (Fig. S2A) and co-localized expression with clathrin heavy chain protein CLTC (Fig. S2B). As a control for random integration, CRMi001-A hiPSCs were transfected with the knock-in vector alone by nucleofection. Tq2 positive cells were collected and pooled to minimize potential off-targeting effects. Knock-in to the CLTA locus was validated by PCR and Sanger sequencing (Fig. 1B). Targeted integration to the C-terminus of CLTA was confirmed by PCR analysis of edited versus unedited cells using a primer pair specific to the donor cassette and the genomic region outside of the homology arms (Fig. 1F-1, F-2). PCR amplification with primers designed to span the homology arms suggested predominately heterozygous insert integration (Fig. 1F-3).
Fig. 1.
Targeting of the CLTA locus for genomic insertion and CRMi001-A-1 validation.
CRMi001-A-1 hiPSCs maintained human pluripotent stem cell-like morphology (Fig. 1C, Table 1). Pluripotency of the CRMi001-A-1 line was confirmed by immunofluorescent expression of transcription factors Oct4A, Nanog, and Sox2, as well as surface markers SSEA4, TRA-1-60 and TRA-1-81 (Fig. 1E), and FACS quantitation of Oct4A-Alexa Fluor 488 expression (Fig. 1D). Pluripotency was further confirmed by embryoid body (EB) formation, loss of pluripotent transcription expression (POU5F1) and spontaneous germ layer differentiation exhibited by expression of endodermal (AFP), mesodermal (TBXT), and ectodermal (MSI1) markers (Fig. 1H) assessed by qRT-PCR. Transcript levels were normalized to GAPDH expression and plotted relative to expression levels in respective hiPSC lines. Directed neural differentiation confirmed ectodermal potential through expression of human-specific Nestin (hNestin), expressed by human neural progenitors, and the neuronal-specific intermediate filament, Neurofilament, medium-chain polypeptide (NF-M) (Fig. 1I). Karyotypic analysis detected no chromosomal abnormalities (Fig. 1G) and hiPSCs were mycoplasma free (Fig. S3). Authentication of CRMi001-A-1 hiPSCs was verified by short tandem repeat analysis in comparison to parental CRMi001-A hiPSCs at 17 distinct loci (submitted in archive with journal).
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal hiPSC morphology | Fig. 1 Panel C |
| Phenotype | Qualitative analysis | Immunohistochemistry of pluripotency markers: Oct4A, Nanog, Sox2, TRA-1-60, TRA-1-81 | Fig. 1 Panel E |
| Quantitative analysis | Expression of pluripotency marker Oct4 by flow cytometry | Fig. 1 Panel D | |
| Genotype | Karyotype (G-banding) and resolution | Normal Karyotype: 46,XY, resolution: 480 band level | Fig. 1 Panel G |
| Identity | STR analysis | 17 loci tested, 100% match | Submitted in archive with journal |
| Mutation analysis (IF | PCR and sequencing | Heterozygous targeted integration | Fig. 1 Panel F |
| APPLICABLE) | Southern Blot OR WGS | Not performed. Top 10 predicted coding and non-coding off-targets were sequenced, detecting no indel formation. | Available by request |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by PCR: Negative | Supplementary Fig. 4 |
| Differentiation potential | Embryoid body formation and Directed neural differentiation | Expression of genes from three germ layers following spontaneous differentiation: AFP, TBXT, MSI1 Directed neural differentiation immunohistochemistry: Human Nestin, Medium-length neurofilament polypeptide |
Fig. 1 Panel H and I |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | N/A |
| Genotype additional info | Blood group genotyping | Not performed | N/A |
| (OPTIONAL) | HLA tissue typing | Not performed | N/A |
Materials and methods
Cell culture
hiPSCs were cultured in StemMACS iPS-Brew XF medium (Miltenyi Biotec) on Corning™ Matrigel™ hESC-qualified-coated plates. Cells were maintained at 37 °C in humidified 5% CO2 and 21% O2. hiPSCs were passaged every 5 days with 0.5 mM EDTA.
Targeting of the CLTA locus
sgRNAs targeting the CLTA locus were designed at crispr.mit.edu. An oligo pair, 5’-CACCGCAGATGTAGTGTTTCCACA-3′ and 3’-CGTCTACA TCACAAAGGTGTCAAA-5′, was annealed and subcloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 vector (kind gift from Feng Zheng). sgRNA activity was validated by T7 endonuclease assay.
1kB homology arms flanking the Turquoise2-P2A-puromyocin insert (Fig. 1A) were amplified from genomic DNA and inserted into the pDONOR3 vector by Golden gate assembly (Scott et al., 2018). To prevent Cas9 recognition after integration, the insert's PAM sequence was mutated (Table 1).
1 × 106 CRMi001-A hiPSCs were transfected using the Nucleofector 2b system, Nucleofector Kit V, 4 μg donor vector and 2.5 μg sgRNA/Cas9 vector. hiPSCs were plated onto Matrigel-coated plates in iPSC-Brew XF. Tq2 positive cells were isolated with a BD FACSJazz Cell Sorter.
Targeted integration analysis
Homology-directed repair was validated by PCR utilizing primers listed in Table 2. CRMi001-A hiPSCs transfected with donor vector alone were used to control for non-selective integration. The knock-in was sequenced entirely (3′ and 5′ targeting in Fig. 1B).
Table 2.
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency markers | Rabbit anti-OCT4 | 1:400 | Cell Signaling Technology Cat# 2840, RRID:AB_2167691 |
| Mouse anti-SSEA4 | 1:400 | Cell Signaling Technology Cat# 4755, RRID:AB_1264259 | |
| Rabbit anti-NANOG | 1:200 | Cell Signaling Technology Cat# 4903, RRID:AB_10559205 | |
| Mouse anti-TRA-1-60 | 1:400 | Cell Signaling Technology Cat# 4746, RRID:AB_2119059 | |
| Rabbit anti-SOX2 | 1:200 | Cell Signaling Technology Cat# 3579, RRID:AB_2195767 | |
| Mouse anti-TRA-1-81 | 1:400 | Cell Signaling Technology Cat# 4745P, RRID:AB_10829904 | |
| Differentiation markers | Mouse anti-human Nestin | 1:1000 | Molecular Probes Cat# A-31572, RRID:AB_162543 |
| Rabbit anti-Neurofilament M, CT | 1:1000 | Millipore Cat# MAB5326, RRID:AB_11211837 | |
| Secondary antibodies | Alexa Fluor 555 donkey anti-rabbit IgG (H + L) | 1:1000 | Molecular Probes Cat# A-31572, RRID:AB_162543 |
| Alexa Fluor 488 goat anti-mouse IgG (H + L) | 1:1000 | Thermo Fisher Scientific Cat# A-11001, RRID:AB_2534069 | |
| Alexa Fluor 647 goat anti-rabbit IgG (H + L) | 1:1000 | Thermo Fisher Scientific Cat# A-21244, RRID:AB_2535812 | |
| Flow cytometry | Oct-4A (C30A3) Rabbit mAb Alexa Fluor 488 conjugate | 1:50 | Cell Signaling Technology Cat# 5177S, RRID:AB_10693303 |
| Rabbit (DA1E) mAb IgG XP Isotype Control Alexa Fluor 488 conjugate | 1:50 | Cell Signaling Technology Cat# 2975S, RRID:AB_10699151 | |
| Primers | ||||
| Target | Forward/Reverse primer (5′-3′) | |||
|---|---|---|---|---|
| Pluripotency Markers (qPCR) | POU5F1 | CCAAGGAATAGTCTGTAGAAGTGC/ TGCATGAGTCAGTGAACAGG |
||
| Differentiation Markers (qPCR) | AFP | TCTGCATGAATTATACATTGACCAC/ AGGAGATGTGCTGGATTGTC |
||
| MSI1 | TCGTTCGAGTCACCATCTTG/ GGCTTCGTCACTTTCATGGA |
|||
| TBXT | CTATGTGGATTCGAGGCTCATAC/ CGTCTCCTTCAGCAAAGTCAA |
|||
| House-Keeping Genes (qPCR) | GAPDH | GCGCCCAATACGACCAA/ CTCTCTGCTCCTCCTGTTC |
||
| Targeted integration analysis/sequencing | PCR analysis | |||
| 1. Template control | TTGCTTGCCAGTGTCCCTCAGTTTA/ GCTTTAATAATTGCTTGGAACATCACCT |
|||
| 2. Tq2-Puro Integration | TACGAGCGGCTCGGCTTCA GCTTTAATAATTGCTTGGAACATCACCT |
|||
| 3. Mono-allelic integration | ATCTTGGGAAAGCCAGAATGTCATT / CCTAAACTGAGGGACACTGGCAA |
|||
|
Sequencing 5′ targeting |
ATCTTGGGAAAGCCAGAATGTCATT TACCAGCAGAACACCCCCAT |
|||
| 3′ targeting | TCACCGAGCTGCAAGAACTCTT CCTAAACTGAGGGACACTGGCAA |
|||
| T7 endonuclease assay | 1 kB amplicon | ATCTTGGGAAAGCCAGAATGTCATT CCTAAACTGAGGGACACTGGCAA |
||
Karyotyping, STR analysis, and mycoplasma detection
G-banding karyotype analysis (450 band resolution) of eight cells was performed by the Sanford Cytogenetics Laboratory. STR analysis was performed using the ATCC Cell Line Authentication Service and compared to the CRMi001-A line. Mycoplasma assessment was performed using a PCR Detection Kit (Applied Biological Materials).
Immunocytochemistry
hiPSCs were grown on Matrigel-coated chamberslides, fixed with 4% paraformaldehyde, permeabilized with 0.2% Trition X-100, and incubated in blocking buffer (0.1% Triton X-100, 5% donkey serum). Primary antibodies were incubated overnight at 4 °C in blocking buffer. Alexa Fluor conjugated secondary antibodies and Hoechst counterstain were incubated for 1 hr at room temperature. Images were taken using a Nikon NiE fluorescent microscope and processed using NIS Elements software and Adobe Photoshop.
Quantitative analysis of pluripotency
hiPSCs were dissociated with accutase and fixed in 2% paraformaldehyde overnight at 4 °C. Cells were incubated with Alexa Fluor 488-conjugated Oct4A and analyzed on a BD LSRFortessa.
Germ layer differentiation
hiPSCs were incubated with 10 μM Y27632 prior to accutase dissociation. 1 × 106 cells were plated into the AggreWell™800 system (Stem Cell Technologies). After 5 days in differentiation media (DMEM/F12, 20% Knockout Serum Replacement, 2 mM l-glutamine, 1% NEAA, 1000 units/mL Penicillin-streptomycin, 100 μM β-mercaptoethanol), aggregates were transferred to gelatinized plates and spontaneously differentiated for 10 days.
Directed neural differentiation
Neural rosettes were isolated from EBs, plated onto laminin/PLO coated coverslips and differentiated for 7 days toward neural lineages.
Real-time PCR analysis
RNA was isolated using the E.Z.N.A RNA Kit (Omega Biotek). 1 μg of RNA was reverse transcribed using the Superscript III First-Strand Synthesis System (Invitrogen). Quantitative PCR was performed with Absolute Blue qPCR Master Mix on an Applied Biosystems 7500 instrument. Samples were analyzed in triplicate from independent experiments. Transcript abundance was normalized to GAPDH and quantified using the 2−ΔΔCt method. Primers used are listed in Table 2.
Supplementary Material
Acknowledgements
This research was supported by the National Institutes of Health (grant numbers P20GM103620, P20GM103548, and F30NS106788), the National Science Foundation/EPSCoR Cooperative Agreement #IIA-1355423, the South Dakota Research and Innovation Center, BioSNTR, and the State of South Dakota BOR CRGP.
Footnotes
Declarations of interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2018.10.001.
References
- Scott BL, et al. , 2018. Membrane bending occurs at all stages of clathrin-coat assembly and defines endocytic dynamics. Nat. Commun 9, 419. 10.1038/s41467-018-02818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.