Abstract
Rodents shift their nose from side to side when they actively explore and lateralize odors in the space. This motor action is driven by a pair of muscles, the deflector nasi. We studied the premotor control of this motion. We used replication-competent rabies virus to transsynaptically label inputs to the deflector nasi muscle and find putative premotor labeling throughout the parvocellular, intermediate, and gigantocellular reticular formations, as well as the trigeminal nuclei, pontine reticular formation, midbrain reticular formation, red nucleus, and superior colliculus. Two areas with extensive labeling were analyzed for their impact on nose movement. One area is in the reticular formation caudal to the facial motor nucleus and is denoted the nose retrofacial area. The second is in the caudal part of the intermediate reticular region near the oscillator for whisking (the nose IRt). Functionally, we find that optogenetic activation of glutamatergic cells in both areas drives deflection of the nose. Ablation of cells in the nose retrofacial area, but not the nose IRt, impairs movement of the nose in response to the presentation of odorants but otherwise leaves movement unaffected. These data suggest that the nose retrofacial area is a conduit for a sensory-driven orofacial motor action. Furthermore, we find labeling of neurons that are immediately upstream of premotor neurons in the preBötzinger complex that presumably synchronizes a small, rhythmic component of nose motion to breathing.
NEW & NOTEWORTHY We identify two previously undescribed premotor areas in the medulla that control deflection of the nose. This includes a pathway for directed motion of the nose in response to an odorant.
Keywords: medulla, nose, orofacial, premotor, sensorimotor
INTRODUCTION
Movement of the nose is a surprisingly multifaceted behavior that is controlled by a single set of muscles, the deflector nasi (Deschênes et al. 2015; Kurnikova et al. 2017). The nose exhibits slow deflections, such as orienting toward an odor (Esquivelzeta Rabell et al. 2017; Kurnikova et al. 2017), as well as small rhythmic movements locked to breathing. The slow, orienting movements of the nose likely play a role in olfactory navigation since lateral deflections affect air flow between sides of the nasal cavity (Deschênes et al. 2016a) and bilateral comparisons improve trail-tracking (Duistermars et al. 2009; Louis et al. 2008; Martin 1965; Porter et al. 2007; Steck et al. 2010). Synchronization of movement on a breath-by-breath cycle is seen in many types of orofacial movement and may play a role in binding multimodal sensory inputs (Heck et al. 2017; Kepecs et al. 2006; Kleinfeld et al. 2014a, 2014b; Moore et al. 2013; Ranade et al. 2013; Sirotin et al. 2014; Wachowiak 2011; Welker 1964).
Nose movement can be evoked by microstimulation in the motor cortex (Brecht et al. 2004). Lesions in the anterior olfactory nucleus, which is an area of the olfactory cortex that supports commissural projections (Brunjes et al. 2005; Yan et al. 2008) and compares odor inputs between two sides (Kikuta et al. 2010), have been found to disrupt nose orienting toward an odor (Esquivelzeta Rabell et al. 2017). However, the downstream brain stem motor circuit responsible for these motor actions, and its candidate anatomical connections to the motor and olfactory systems, have not been identified. In this report we seek putative premotor nuclei for movement of the nose. We take a combined structural and functional approach. We perform retrograde tracing with the use of rabies virus to identify pre-motor areas, optogenetic labeling of excitatory neurons in these regions to confer sufficiency, and lesion studies to confer necessity that control movement of the nose occurs via the identified premotor pathways.
We identify two premotor areas that control movement of the nose: an area caudal to the facial motor nucleus that we call the “retrofacial area” and an area near the vibrissa intermediate reticular region (IRt) oscillator that we call the “nose IRt.” We confirm functional relevance of the identified areas by showing that stimulation evokes nose movement, whereas ablation affects movement to the lesioned side.
MATERIALS AND METHODS
Subjects
Our data were collected from 30 Long-Evans adult female rats, 200–350 g in mass, and 23 adult Vglut2-ires-cre (JAX 028863) mice. All experimental procedures on our animals were accordance with the Guide for the Care and Use of Laboratory Animals and have been approved by the Institutional Animal Care and Use Committee at University of California, San Diego.
Tracing with Replication-Competent Rabies Virus
We set up an isolated biosafety level 2 enhanced (BSL2+) facility to perform replication-competent rabies virus injections and to house injected animals. The facility and our experimental protocol follow all recommendations described in Kelly and Strick (2000). To perform the injections, rats were anesthetized using ketamine-xylazine and placed in a stereotaxic apparatus. We exposed the deflector nasi muscle and checked that the correct muscle was identified by stimulating with pulses of current up to 0.1 mA to evoke movement of the nose (Deschênes et al. 2015). Finally, we used a pressure injector (Drummond Scientific NANOJECT II) to inject a total of 2 μl of CVS-N2c strain rabies virus (Peter Schnell Laboratory) in five spots along the muscle. Rats were perfused at select time points between 48 and 96 h after the injection.
A total of 19 rats were injected with rabies virus (Wirblich and Schnell 2011). Of these, the five brains collected at time points of 50, 67, 72, 61.5, and 79.5 h had no labeled cells and thus were excluded from analysis. Additionally, two brains with time points of 96 h were excluded from analysis because labeling in the brain stem was too dense for processing. Finally, a brain collected at a time point of 72 h was excluded from analysis because midline sections were missing in histology; the labeling in this brain was consistent with observed labeling patterns at primary labeled time points.
After perfusion, brains were extracted and placed in 30% (wt/vol) sucrose solution. Brains were cut on a freezing sliding microtome at 60 μm and stained for cytochrome oxidase. Rabies-labeled cells were stained using anti-rabies-L mouse antibody (Schnell Laboratory) and revealed as dark product using a biotinylated secondary antibody, the ABC and SG kits (Vector Laboratories). For seven of the brains, every third section was cut at 40 μm and stained with NeuroTrace blue, whereas rabies-labeled cells were stained using anti-rabies-L mouse antibody and revealed in fluorescence with a secondary anti-mouse antibody conjugated to Alexa 488 (Invitrogen A11029). In addition, in three brains the sections were also stained for somatostatin (T-4103; Peninsula Laboratories) to identify the location of the preBötzinger complex (Tan et al. 2010).
Reconstruction and Alignment
We used Neurolucida software (MBF Bioscience) to create full three-dimensional reconstructions of labeled cell body locations in all brains that had labeled cells. Brains were annotated by outlining key brain stem structures visible in cytochrome staining. In brains with alternating NeuroTrace sections, nucleus ambiguus and the inferior colliculus were outlined based on the cytoarchitecture. Finally, in brains with somatostatin labeling, the preBötzinger complex was identified as the area with higher somatostatin-positive cell density. One brain was fully annotated with all midbrain and hindbrain structures described in Paxinos and Watson (1986). All outlined structures are listed in Table 1.
Table 1.
Structure and fiber names and abbreviations
| Abbreviation | Structure Name |
|---|---|
| 5N | Trigeminal motor nucleus |
| 7N | Facial motor nucleus |
| AP | Anterior pretectal nucleus |
| Amb | Nucleus ambiguus |
| DMSp5 | Dorsomedial spinal trigeminal nucleus |
| Dk | Nucleus of Darkschewitz |
| DR | Dorsal raphe nucleus |
| Forel | Fields of Forel |
| IO | Inferior olive |
| Gi | Gigantocellular reticular nucleus |
| GiA | Gigantocellular reticular nucleus, alpha part |
| GiV | Gigantocellular reticular nucleus, ventral part |
| GP | Globus pallidus |
| HDB | Nucleus of the horizontal limb of the diagonal band |
| Hipp | Hippocampus |
| IC | Inferior colliculus |
| IMLF | Interstitial nucleus of the mediolateral formation |
| iTg | Intertrigeminal nucleus |
| IRt | Intermediate reticular nucleus |
| KF | Kölliker-Fuse nucleus |
| LDT | Laterodorsal tegmental nucleus |
| LH | Lateral hypothalamic area |
| LPB | Lateral parabrachial nucleus |
| LPBE | Lateral parabrachial nucleus, pars externa |
| LPGi | Lateral paragigantocellular nucleus |
| LRt | Lateral reticular nucleus |
| MCPO | Magnocellular preoptic nucleus |
| MPB | Medial parabrachial nucleus |
| MdD | Medullary reticular nucleus, dorsal part |
| MdV | Medullary reticular nucleus, ventral part |
| MiTg | Microcellular tegmental nucleus |
| mRT | Midbrain reticular formation |
| Mx | Matric region of the medulla |
| NLOT | Nucleus of the lateral olfactory tract |
| NPCom | Nucleus of the posterior commissure |
| PAG | Periaqueductal gray |
| PCRt | Parvicellular reticular nucleus |
| PCRtA | Parvicellular reticular nucleus, alpha part |
| PH | Posterior hypothalamic area |
| PMnR | Paramedian reticular nucleus |
| PPTg | Posterior pretectal nucleus |
| PR | Prerubral field |
| PR5VL | Principal sensory trigeminal nucleus, dorsomedial part |
| PaR | Pararubral area |
| PnC | Pontine reticular nucleus, pars caudalis |
| PnO | Pontine reticular nucleus, pars oralis |
| Pr5 | Principal sensory trigeminal nucleus |
| PreBötC | PreBötzinger complex |
| pCom | Precommissural nucleus |
| RIP | Raphe interpositus nucleus |
| RMg | Raphe magnus nucleus |
| RN | Red nucleus |
| ROB | Raphe obscurus nucleus |
| RPa | Raphe pallidus |
| RtTg | Reticulotegmental nucleus of the pons |
| SC_InG | Superior colliculus, inferior gray |
| SC_SuG | Superior colliculus, superior gray |
| SNC | Substantia nigra, pars compacta |
| SNL | Substantia nigra, pars lateralis |
| SNR | Substantia nigra, pars reticularis |
| SPTg | Subpeduncular tegmental nucleus |
| SPVmu | Spinal trigeminal nucleus, pars muralis |
| Sol | Nucleus of the solitary tract |
| SpVI | Spinal trigeminal nucleus, pars interpolaris |
| SpVC | Spinal trigeminal nucleus, pars caudalis |
| SpVO | Spinal trigeminal nucleus, pars oralis |
| Su5 | Supratrigeminal nucleus |
| SubC | Subcoeruleus nucleus |
| Tu | Olfactory tubercle |
| VLL | Ventral nucleus of the lateral lemniscus |
| VP | Ventral pallidum |
| VTA | Ventral tegmental nucleus |
| Ve | Vestibular nuclei |
| ZI | Zona incerta |
| Abbreviation | Fiber tract name |
|---|---|
| 7n | Facial tract |
| ac | Anterior commissure |
| f | Fornix |
| fr | Fasciculus retroflexus |
| ml | Medial lemniscus |
| mlf | Medial longitudinal fasciculus |
| pc | Posterior commissure |
| scp | Superior cerebellar peduncle |
First, a reference atlas was created by averaging six reconstructed brains (Chen et al. in press). The facial motor nucleus (7N), facial motor tract (7n), lateral reticular nucleus (LRt), trigeminal motor nucleus (5N), and inferior olive (IO) were used for aligning the stacks to a single ‘reference’ stack using an affine transform. Average center positions for all traced structures across stacks were then calculated. A midline plane was determined as a fit to the midpoints between only the alignment structures, and structure positions for the atlas were rotated such that they were set symmetrically about the midline. Finally, structure shapes were calculated by aligning and averaging each structure individually. The final symmetrical reference atlas was constructed by placing each averaged structure at its average position bilaterally. In a final step, each of the reconstructed brains was aligned to the “alignment” structures in the reference atlas using an affine transform with a scaling range between 99% and 101%.
Data Analysis
Retrograde tracing cell counts were calculated from the aligned reconstructions. Cell counts were done by reslicing the aligned data sets at a thickness of 15 μm and assigning each marked cell to the traced and reconstructed region. To account for gaps in traced regions that come from smoothing the reconstruction, cells that did not fall within any traced boundary were assigned to the boundary to which they were nearest. If a cell was within an overlap of two boundaries, it was assigned to the boundary where it was more deeply embedded. An example of a single section with cells colored by assigned region is shown in Supplemental Fig. S1B. (Supplemental Material for this article is available online at the Journal of Neurophysiology website.) Cell count analysis was performed using custom scripts written in Python.
Cell density was evaluated by computing a kernel density estimate with a 200-µm bandwidth Gaussian kernel on each full volume. Maximum contours in steps of 10% of labeling were defined by density areas greater than a threshold corresponding to the 10th percentile of the density individually by stack. To evaluate maximally overlapping areas across different labeled brains, volumes in which at least four stacks had a overlap of at least 15% maximum density were constructed. A Gaussian mixture model with two components was fit to all cells within this maximum density volume, with weights assigned as the inverse of the number of labeled cells per brain to ensure equal contribution of each labeled brain. Aikake and Bayesian information criteria for the Gaussian mixture model were evaluated for models of one, two, and three components to determine optimal fit.
To compare mouse functional experiments with the tracing results from rat data, the rat atlas was aligned to mouse volumes with an affine transform and allowable scaling up to 40%. An identical transform was applied to the results of the Gaussian fit to obtain an estimate of the location of the two putative premotor areas.
Lesions in Rats
Eight rats were injected with concentrated Sindbis-152-GFP virus (G. Patrick laboratory, UCSD) to drive expression virus at the injection site and cause cell death after 4–5 days (Moore et al. 2013). When tested in culture, the virus infected >90% of cells, and thus we expect that a majority of cells at the injection site take up the virus. An additional three rats were electrolytically lesioned by lowering a stainless steel electrode into the caudal IRt area and passing current of 300 µA for 5 s.
Rats were implanted with a head bar for head fixation and a thermocouple (model 5TC-TT-K-36-36; Omega Engineering) to monitor breathing 2 days after virus injection. Rats were placed in the setup for acclimation once before testing and were tested for evoked nose movement in response to odor 5 days after virus injection. For testing, bedding odor was presented alternately on either side of the snout (Ganguly and Kleinfeld 2004; Kurnikova et al. 2017).
Video data were collected via a high-speed camera (Basler A602f) using custom code written in MATLAB and standard LabChart functions. Rats were perfused and brains sectioned on a freezing microtome. For Sindbis virus lesions, 30-μm horizontal sections were stained for NeuN (MAB377; Millipore), and the lesion site was identified by the area of cell death as evidenced by a lack of NeuN-stained cells. For electrolytic lesions, 60-μm sections were stained with Neutral red, and the lesion site was identified as a hole or obvious damage to the tissue. Three-dimensional reconstructions of the lesion site in every third section were made using Neurolucida and were further processed using custom code in Python.
Lesions in Mice
Ten Vglut2-ires-cre mice (JAX 028863) were injected with AAV8-mCherry-flex-dTA (UNC Vector Core) virus to drive expression of diphtheria toxin subunit A in glutamatergic cells at the injection site. Mice were implanted with a head bar for head fixation and a fast-time NTC thermistor (MEAS-G22K7MCD419; Measurement Specialties) to monitor breathing (McAfee et al. 2016) 3–4 wk after virus injection. Mice were placed in the setup for acclimation at least once before testing and were tested for evoked nose movement 5–6 wk after virus injection. For testing, bedding odor was presented alternately on either side of the snout Ganguly and Kleinfeld 2004; (Kurnikova et al. 2017).
Video data were collected via a high-speed camera (Basler A602f) using custom code written in MATLAB and standard LabChart functions. Mice were perfused and brains sectioned at 25 μm. Sections were stained for NeuroTrace blue (N21479; Thermo-Fischer), and the size and location of the injection site were estimated by the tdTomato labeling in nonglutamatergic cells. Three-dimensional reconstructions of labeled cells and key structures in every third section were made using Neurolucida. All reconstructions were aligned to a single selected mouse brain using procedures from Chen et al. (in press) and were displayed and compared using custom code in Python.
We performed a cell count in the retrofacial area in all lesioned animals, and in five contralateral side sections as control, using a cell-counting algorithm in Neurolucida. We counted all NeuroTrace blue-labeled cells in a single section ~1.0 mm lateral of midline, manually adjusting counting parameters to ensure reasonable results in each instance. Cell-counted regions of interest (ROI) were aligned to each other by applying translation parameters from the three-dimensional alignment, with no rotation to avoid affecting the cell density.
Optogenetic Stimulation in Mice
Thirteen Vglut2-ires-cre mice (JAX 028863) were injected with AAV8-flex-ReaChR-citrine (Addgene no. 50955; Salk Vector Core) virus to drive expression of red-shifted channelrhodopsin (ReaChR) in glutamatergic cells at the injection site. An additional five control mice, i.e., negative for Cre, were injected with the same virus as a sham injection. Sham-injected mice were tested as blind controls along with the positive mice, to avoid bias in LED placement during comparison of results. Mice were implanted with a head bar for head fixation and a fast-time NTC thermistor (MEAS-G22K7MCD419; Measurement Specialties) to monitor breathing (McAfee et al. 2016) 2 wk after virus injection. Mice were placed in the setup for acclimation at least once before testing and were tested for evoked nose movement 3 wk after virus injection. For optogenetic stimulation, a Mightex 2-channel LED controller (SLC-MA02-U; Mightex Systems) was used to drive a 617-nm Luxeon LED (Lumileds LXM2-PH01-0070), placed in the ear, at 355 mA with 10-ms pulses. Video data were collected via a high-speed camera (Basler A602f) using custom code written in MATLAB and standard LabChart functions.
Mice were perfused and brains sectioned at 30 μm. Sections were stained for NeuroTrace far red (N21483; Thermo-Fischer), and the injection site was identified by the citrine label in cell membranes. Three-dimensional reconstructions of labeled cells and key structures in every third section were made using Neurolucida and were further processed using custom code in Python. All reconstructions were aligned to a single selected mouse brain using procedures from Chen et al. (in press) and were displayed and compared using custom code in Python.
RESULTS
Tracing with Rabies Virus
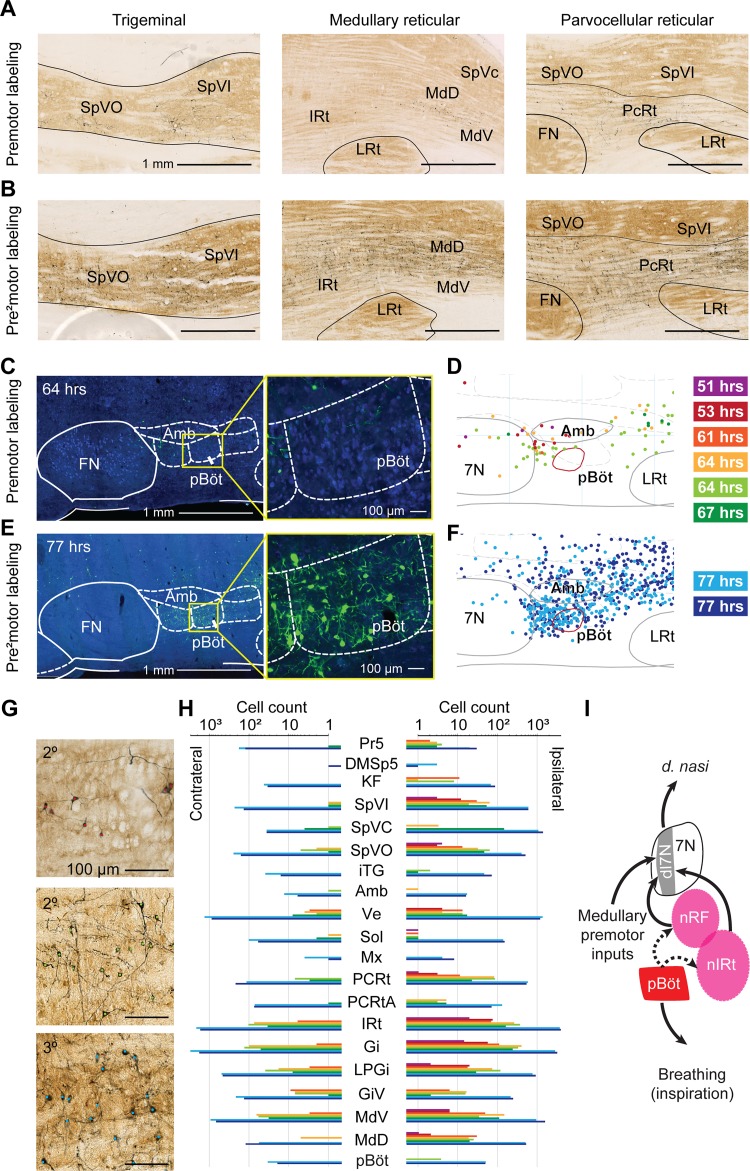
We identified putative premotor areas and pre2motor areas, i.e., immediately upstream of premotor areas, for nose motion by retrograde tracing using replication-competent rabies virus from the deflector nasi muscle (Fig. 1A). At all time points, corresponding to different levels of retrograde transport, cells in the facial motor nucleus appeared intact (Fig. 1B). In addition, motor neurons and their dendritic fields were found in the dorsal lateral portion of the facial motor nucleus (Fig. 1C), which is consistent with the reported location of motoneurons for the deflector nasi (Deschênes et al. 2016a). Thus we are confident that all observed labeling is from the injected muscle, with no contamination from virus spillover.
Fig. 1.
Key premotor areas identified by retrograde tracing from the deflector nasi (d. nasi). A: diagram of the experimental procedure. The deflector nasi was injected with N2c replication-competent rabies virus (CVS-N2c RV), and rats were perfused at primary, secondary, or tertiary time points. B: examples of labeling in the facial motor nucleus (7N) at primary, secondary, and tertiary time points. Motoneurons remain intact at all time points. C: reconstructions of labeled motoneurons and motoneuron dendrites in the facial motor nucleus. Cell bodies and dendritic fields were found in the dorsolateral part of 7N. Colors correspond to individual time points (colors in A). Top, primary labeled time points (grayscale); middle, secondary labeled time points (red-green); bottom, tertiary labeled time points (blues). D, dorsal; V, ventral; M, medial; L, lateral. D: example of premotor labeling in the retrofacial area at a 64-h time point in a sagittal section ~1.7 mm lateral of midline. Labeled cells are revealed in dark product, whereas structures are identified in a cytochrome oxidase (CO) stain. IRt, intermediate reticular nucleus; LRt, lateral reticular nucleus; 7n, facial tract. E: reconstructions of premotor labeling in the retrofacial area in 6 rats at secondary labeled time points. Left, reconstructions of labeled cells in a 200-µm-thick coronal slice at 10.7 mm caudal of bregma. Right, 10% maximum density contours in the same coronal slice. Secondary time points shown are 51 h (purple), 53 h (red), 61 h (orange), 64 h (yellow), 64 h (light green), and 67 h (green). Four of six contours (53, 64, 64, and 67) overlap in the nose retrofacial (nRF) region. Amb, nucleus ambiguus; IO, inferior olive; SpVI, Spinal trigeminal nucleus, pars interpolaris; Ve, vestibular nucleus. F: example of premotor labeling in the nose intermediate reticular (nIRt) area at a 64-h time point ~1.5 mm lateral of midline. Labeled cells are revealed in dark product, whereas structures are identified in a CO stain. Gi, gigantocellular reticular nucleus. G: reconstructions of premotor labeling in the caudal IRt/Gi area in 6 rats at secondary labeled time points. Left, reconstructions of labeled cells in a 200-µm-thick coronal slice at 11.5 mm caudal of bregma. Right, 10% maximum density contours in the same coronal slice. Secondary time points shown are 51 h (purple), 53 h (red), 61 h (orange), 64 h (yellow), 64 h (light green), and 67 h (green). Four of six contours overlap for 51, 61, 64, and 67 h in the caudal IRt/Gi area (nIRt). H: 3-dimensional display of a 2-component Gaussian mixture model fit to the high-density labeled area in 6 rats. Reconstructed cells are shown as small spheres. Centroids of the 2 labeled areas are shown as large black spheres. Three-sigma radius ellipsoids are shown in magenta. Two areas emerge from the fit: a more rostral area that we call the nRF area and a more caudal nose area on the border of Gi and IRt that we call the nIRt area. Centroids are located (rostral/caudal, M/L, D/V) at (−10.7 mm, 1.7 mm, 7.8 mm) for the retrofacial area and at (−11.5 mm, 1.5 mm, 7.7 mm) for the nIRt. Forty-five degree (left) and sagittal (right) projections of the data are displayed. 5N, trigeminal motor nucleus; I: diagram of proposed nose motion circuit based on the results of premotor labeling from the d. nasi muscle. Motoneurons located in the dorsolateral facial motor nucleus (7Ndl) send projections to the ipsilateral deflector nasi muscle and receive input from the ipsilateral nRF and nIRt premotor areas.
Key Premotor Areas for the Deflector Nasi
We aligned all brains to a common reference atlas of the brain stem (Chen et al. in press). Retrograde labeling was found across extensive regions of the hindbrain and midbrain. We consider first two regions with particularly dense secondary labeling across all brains. An example of labeled cells in an area just caudal to the facial motor nucleus, denoted the nose retrofacial (nRF) region, is shown in Fig. 1D, whereas an example of labeling in a more caudal region, denoted the nose intermediate reticular formation (nIRt), near the location of the vibrissa IRt (Moore et al. 2013), is shown in Fig. 1F. Reconstructions across all animals showed consistent labeling in each of these two regions across all rats at the secondary time point. A subset of four of the six rats at secondary time points had high-density labeling in the retrofacial region, as revealed by overlapping reconstructed cells and 10% density contours (Fig. 1E and Supplemental Fig. S2, A–C). A different subset of four of six rats had high-density labeling in the nIRt region (Fig. 1G and Supplemental Fig. S2, D–F). In summary we found a stretch of labeling in the medulla on the border of the IRt and gigantocellular reticular formation that has high density among at least four of six injected rats. To quantify the region of labeling, we fit a Gaussian mixture model to the high-density labeled area across all rats (Supplemental Fig. S2I), which implies that the labeled premotor areas can best be described as two loci of high-density labeling (Fig. 1H). The two overlapping areas are entered on the coordinates: (rostral-caudal = −10.7 mm, medial-lateral = 1.7 mm, dorsal-ventral = −7.8 mm) for the nRF and (−11.5 mm, 1.5 mm, −7.7 mm) for the nIRt. When scaled to mice, these coordinates are approximately (−6.9 mm, 1.0 mm, −5.4 mm) for the nose retrofacial area and (−7.3 mm, 0.9 mm, −5.1 mm) for the nIRt.
To confirm the labeling centroids, we repeated the clustering analysis using a point-by-point density-based clustering algorithm, DBSCAN (Ester et al. 1996). We found that a broad region of parameter space yields two clusters in the labeling. At the biologically plausible agglomeration parameter ε = 200 µm and minimum neighborhood size N = 50, we obtained clusters centered at (−10.5 mm, 1.8 mm, −7.9 mm) and (−11.4 mm, 1.6 mm, −7.7 mm), close to the clustering obtained by the Gaussian mixture model (Supplemental Fig. S2, G–I).
Although not all animals had labeling in each of the two key areas, the emergence of the labeling is not a matter of delayed transport. Critically, of the two earliest time points in which labeling was very sparse, the 53-h time point showed labeling in the nRF area, whereas the 51-h time point showed labeling in the nIRt. We propose that both the nRF and nIRt are likely to be premotor areas, although it is possible that they project to disparate subsets of motoneurons in the facial motor nucleus.
Extensive Premotor and Pre2motor Labeling in Other Brain Areas
Hindbrain.
Beyond the dense labeling in the nRF and nIRt, labeling from the deflector nasi was found in multiple areas of the medulla at both secondary and tertiary time points (Fig. 2). At secondary time points, brain stem reticular formation had dense ipsilateral labeling in the parvocellular, gigantocellular, intermediate, and dorsal and ventral medullary subdivisions, as well as weaker contralateral labeling in all these areas (Fig. 2, A, G and H). Only sparse labeling is observed in the contralateral nRF and nIRt. At tertiary time points, both ipsilateral and contralateral labeling increased dramatically (Fig. 2, B, G and H, and Supplemental Fig. S3).
Fig. 2.
Medullary labeling from the deflector nasi. A: example putative premotor areas labeled at secondary time points. Left, labeling in the ipsilateral trigeminal nuclei is most dense in the dorsal part of the spinal trigeminal nucleus pars oralis (SpVO) and pars interpolaris (SpVI). The section displayed is 2.9 mm lateral to midline, ipsilateral to the injection. Middle, labeling in the medullary reticular nuclei (MdD and MdV). The section displayed is 1.8 mm lateral to midline, ipsilateral to the injection. IRt, intermediate reticular nucleus; LRt, lateral reticular nucleus; SpVc, spinal trigeminal nucleus, pars caudalis). Right, labeling in the parvocellular reticular nucleus (PcRt). The section displayed is 2.5 mm lateral to midline, ipsilateral to the injection. Labeling density increases at the tertiary time points, suggesting additional circuits involved in pre2motor control. Structures are outlined from cytochrome oxidase (CO) stain; rabies-labeled cells are revealed in dark product. FN, facial motor nucleus. B: example putative pre2motor areas labeled at tertiary time points, as in A. Left, the section displayed is 2.9 mm lateral to midline, ipsilateral to the injection. Middle, the section displayed is 1.8 mm lateral to midline, ipsilateral to the injection. Right, the section displayed is 2.5 mm lateral to midline, ipsilateral to the injection. C: example sections showing no labeling in preBötzinger complex (pBöt) at secondary time point (64 h). Cells are stained with NeuroTrace blue, and rabies-labeled cells are revealed in green. Structures are annotated from the atlas of Paxinos and Watson (1986). Amb, nucleus ambiguus. D: sections, cut at 200 µm, from reconstruction of premotor (secondary) labeling in the preBötzinger complex (red circled area). Location of preBötzinger complex is identified from somatostatin stain. No labeling is observed at secondary time points. 7N, facial motor nucleus. E: example sections showing dense labeling in preBötzinger complex at a tertiary time point (77 h). Cells are stained with NeuroTrace blue, and rabies-labeled cells are revealed in green. Structures are annotated from the atlas of Paxinos and Watson (1986). F: sections from reconstruction of pre2motor (tertiary) labeling in the preBötzinger complex (red). Location of preBötzinger complex is identified from somatostatin stain. Dense labeling is shown at tertiary time points. G: example data of labeled neurons with somata indicated by colored dots, asigned as in Fig. 1 D, F, and H: bar chart of labeled cell count (logarithmic scale) in the medulla. Counts for ipsilateral (top) and contralateral (bottom) sides are shown. No labeling outside of the facial motor nucleus was observed at the primary time points. Labeling across all areas increases dramatically at the tertiary time points (77 h). Structure abbreviations are defined in Table 1; nose retrofacial area (nRF) and nose intermediate reticular formation (nIRt) cell counts are included in the IRt. I: proposed circuit for control of nose motion. Motoneurons in the dorsal lateral facial nucleus (7Ndl) send input to the d. nasi and receive input from premotor areas. We hypothesize that premotor areas nRF and nIRt receive respiratory input from the preBötzinger complex.
The trigeminal nuclei also had ipsilateral labeling at secondary time points, most notably in the ventral parts of spinal trigeminal nuclei interpolaris and oralis (Fig. 2A). Some animals at the secondary time points also showed labeling in subnucleus caudalis. At tertiary time points, the trigeminal nuclei were densely labeled both ipsilaterally and contralaterally (Fig. 2H).
At secondary time points, we observed sparse pre2motor labeling bilaterally in the pontine reticular formation oralis and caudalis, subcoeruleus nucleus, Kölliker-Fuse nucleus, and supratrigeminal nucleus. At tertiary time points, labeling density increased throughout the pontine reticular formation oralis and caudalis, subcoeruleus nucleus, and Kölliker-Fuse nucleus. In addition, dense labeling was observed bilaterally in the deep cerebellar nuclei, the lateral and medial parabrachial nuclei, the pedunculopontine tegmental nucleus, and the intertrigeminal nucleus (Supplemental Figs. S3 and S4).
PreBötzinger complex.
We single out this region in light of past data showing that the preBötzinger complex drives the oscillator for whisking (Moore et al. 2013). We observed no labeling at the secondary time points and dense labeling at tertiary time points (Fig. 2, C–F). This pattern of labeling is consistent with the circuit hypothesis that the preBötzinger complex provides inputs to a variety of premotor areas to drive orofacial motion coordination to breathing. Based on their locations, both the retrofacial and caudal IRt are likely candidates to receive input from the preBötzinger complex (Tan et al. 2010). We thus propose that the nRF and nIRt provide premotor input to the facial motor nucleus and that one or both regions receive input from the preBötzinger complex to synchronize nose motion to breathing (Fig. 2I).
Midbrain.
We found premotor labeling in the contralateral superior colliculus (Supplemental Fig. S4, A and B) and red nucleus (Supplemental Fig. S4, E and F), as well as in the midbrain reticular formation and periaqueductal gray at secondary time points (Supplemental Fig. S3C). We also found premotor labeling bilaterally in the interstitial nucleus of the medial lemniscus and ipsilaterally in the nucleus of Darkschewitz (Supplemental Figs. S3C and S4I). This labeling pattern is similar to tracing results from the facial motor nucleus (Hattox et al. 2002; Isokawa-Akesson and Komisaruk 1987), which labeled inputs to the vibrissa system, consistent with a similar circuit structure for control of whisking and nose motion.
At tertiary time points, we observed increased labeling across the midbrain, including bilaterally in superior colliculus (Supplemental Fig. S4, C and D), red nucleus (Supplemental Fig. S4, G and H), midbrain reticular formation, raphe magnus nucleus, paralemniscal nucleus, periaqueductal gray (Supplemental Fig. S3C). We also observe the appearance of labeled cells in the substantia nigra reticular part and lateral part, zona incerta, fields of Forel, prerubral fields, nuclei of the posterior commissure, and ventral tegmental area (Supplemental Fig. S3C). Tertiary midbrain labeling was denser contralaterally but present on both sides.
Forebrain.
At tertiary time points, we observed labeling in the forebrain, including bilateral labeling in motor, prefrontal, and sensory cortical areas (Supplemental Fig. S5, A and B), lateral hypothalamic area (Supplemental Fig. S5C), posterior hypothalamic area, and sparse labeling in the ipsilateral ventral pallidum, the ipsilateral globus pallidus, ipsilateral nucleus of the horizontal limb of the diagonal band, and in the ipsilateral lateral habenula (Supplemental Fig. S5D). In one example we observed sparse labeling in the ipsilateral nucleus of the lateral olfactory tract and the ipsilateral magnocellular preoptic nucleus, and dense labeling in the ipsilateral olfactory tubercle (Supplemental Fig. S5, C and D). These putative pre2motor projections are candidate areas for descending motor pathways from the olfactory system and motor cortex. Anterograde tracing could be used to determine specific pathways by which these areas send inputs to the nose motion control.
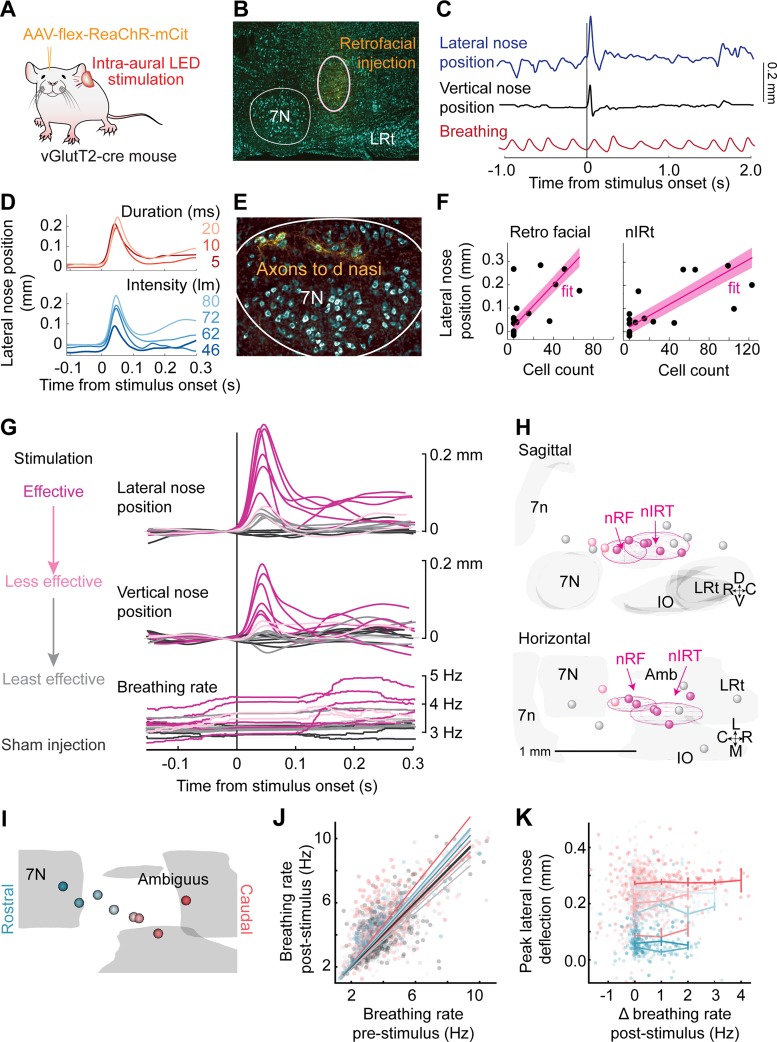
Optogenetic Stimulation Evokes Nose Deflection Independent of Sniffing
Cells in the nRF area send primarily glutamatergic projections to the facial motor nucleus, whereas motoneurons themselves do not express vesicular glutamate transporter 2 (vGluT2) in perinatal mice (Takatoh et al. 2013) or in adult mice (Deschênes et. al. 2016b). We optogenetically stimulate the glutamatergic population of cells in the nRF and nIRt areas of mice to determine the effects of their stimulation on nose motion We inject AAV-flex-ReaChR, a red-shifted channelrhodopsin (Lin et al. 2013), into Vglut-cre mice to drive expression of ReaChR at the injection site (Fig. 3A). For an injection site in the retrofacial area (Fig. 3B), labeled axons were found in the dorsolateral part of the facial motor nucleus (Fig. 3E), and light stimulation through the ear evoked lateral and vertical movement of the nose (Fig. 3C). No change in the profile of evoked movement was observed for changing stimulus duration at constant power, and an increase in peak amplitude of movement was observed for increasing stimulus intensity (Fig. 3D).
Fig. 3.
Optogenetic stimulation in nose retrofacial (nRF) and nose intermediate reticular (nIRt) areas evoke nose movement. A: diagram of the experimental setup. Transgenic mice were injected with an AAV virus (AAV-flex-ReaChR-mCit) to drive expression of red-shifted channelrhodopsin (ReaChR) in glutamatergic cells at the injection site. Nose movement was monitored by high-speed video in head-fixed mice, while breathing was monitored with a thermistor implanted in the nasal cavity. Stimulation was done by ReaChR stimulation with an LED through the ear canal. B: example histological identification of an injection site in the nRF area. Cell bodies were stained with NeuroTrace blue (cyan) and monomeric citrine (mCit)-labeled cells in yellow. C: example trace of lateral nose motion (blue), vertical nose motion (black), and breathing (red). The nose deflects laterally and upward after stimulation with a 10-ms LED pulse. Injection site is shown in B. D, top: example average lateral nose motion response to stimulation at 5 (dark red), 10 (red), and 20 ms (light red), with current adjusted such that the power remains constant across parameters. Bottom, example average lateral nose motion response to stimulation with 10-ms pulses at stimulation values of 46 (dark blue), 62 (medium blue), 72 (blue), and 80 lumens (light blue). E: axons (yellow) labeled in the dorsolateral facial motor nucleus after AAV-flex-ReaChR-citrine injection into the nRF area. Cell bodies were stained with NeuroTrace Blue (cyan). Injection site is shown in B. F: average lateral nose peak position after stimulation with a 10-ms pulse as a function of cell counts in the nRF and nIRt areas. Each point is the average from a single mouse. Linear fits are shown in magenta. G: average traces of lateral nose movement, vertical nose movement, and breathing rate after stimulus onset. Trials were selected by movement variance before stimulation onset and <0.5-Hz breathing rate change as compared before and 100 ms after stimulation onset. Effective stimulation sites (magenta), less effective stimulation sites (orange), and least effective stimulation sites (gray) are defined from the functional data. Sham injections (black) were done in Cre-negative mice. H: 3-dimensional reconstructions of ReaChR injection centroids. Sagittal (top) and horizontal (bottom) views are shown. Colors are drawn from the functional results in G. Centroids of effective stimulation sites (magenta) tend to overlap with the nRF and nIRt regions. I: color coding of all effective stimulation sites from rostral (blue) to caudal (red). J: breathing rate pre- vs. poststimulation. Stimulation of some injection sites showed a slight increase in breathing rate, but on most trials the breathing rate remained constant. K: peak lateral nose deflection as a function of change in breathing rate poststimulation. Flat profile of the graph indicates that lateral nose movement evoked by stimulation does not depend on eliciting a change (Δ) in breathing rate.
Lateral and upward nose movement, a signature of activity of the deflector nasi, was evoked after light stimulation in 8 of 13 mice, whereas no movement at all was evoked in the non-cre control mice (Fig. 3, E–H). In off-target injection sites, we observed a small lateral nose movement but no consistent upward nose movement (Fig. 3G). Thus the movement of the nose could be an effect of pulling other muscles in the snout. We find that effective stimulation sites have centroids in the nRF and caudal nIRt regions (Fig. 3, H and I). We also considered the lateral evoked nose movement as a function of number of cells counted within the nRF and nIRt regions in each mouse. We found that both cell count in the nRF region and cell count in the nIRt region predict lateral nose movement response (F statistic = 4.5, P < 0.05 and F statistic = 8.3, P < 0.02, respectively), and the combination of the two cell counts predicts the lateral nose response well (F statistic = 13.6, P = 0.0004), with coefficients of 0.0019 mm/cell (P = 0.03) in the nRF and 0.0014 mm/cell (P = 0.004) in the nIRt (Fig. 3F).
Movement of the nose occurs during sniffing (Kurnikova et al. 2017), and thus we examined whether nose movement evoked by stimulation is a result of triggering a sniffing bout. We found that although at some injection sites an increase in breathing rate is evoked by stimulation (Fig. 3J), the amplitude of nose movement is independent of change in breathing rate (Fig. 3K). We conclude that optogenetic stimulation of glutamatergic cells in both the nRF and in the nIRt can evoke nose movement independently of a change in breathing rate.
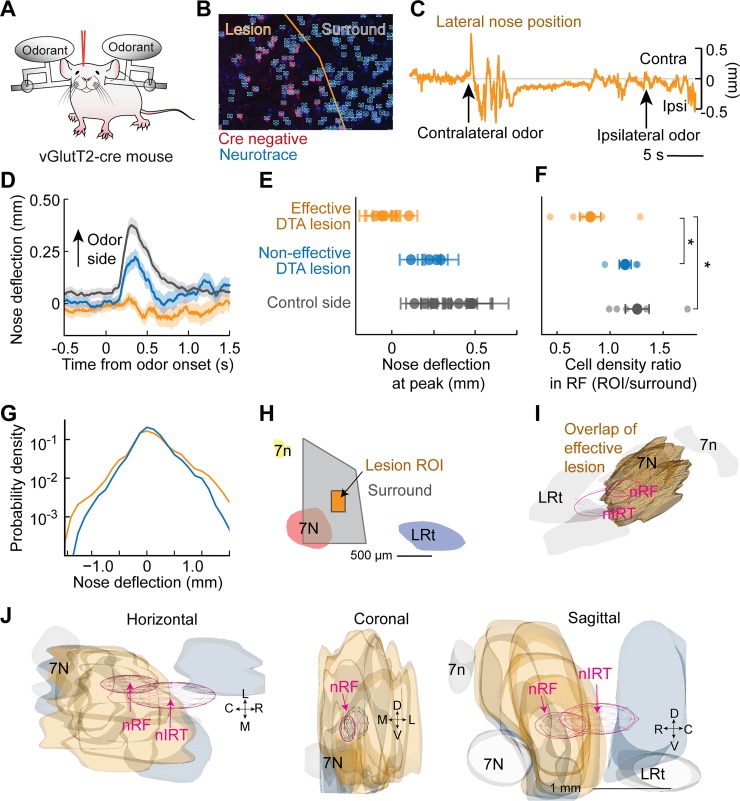
Retrofacial Area Lesion, But Not nIRt Lesion, Affects Nose Odor Response
To establish the functional significance of the identified areas, we aimed to test whether ablating the nRF area can affect the nose movement response to odor presentation. Because the nRF area is dense with fiber tracts, we chose Sindbis virus as a lesioning method to kill cells without damaging the passing fibers (Fig. 4, A–D). We also include analysis from electrolytic lesions in the nIRt region. We found that three of the Sindbis virus-lesioned rats had a reduced average peak nose deflection compared with the control averages (Fig. 4, E and F). Three-dimensional reconstructions of effective lesion positions show that all effective lesions were in the nRF area, whereas noneffective lesions flanked the region (Fig. 4, G and H). The average traces for each condition show that there was no change in the shape of the nose response for the noneffective lesions, whereas the nose motion response for nRF-lesioned animals was greatly reduced (Fig. 4D). Finally, we considered the distribution of all nose positions for the lesioned vs. control animals. We found that on a longer timescale, the lesioned animals were still able to move their nose both to the control side and lesioned side (Fig. 4F). As a note, it is possible that lesions with a larger extent would cause an asymmetry of motion of the nose; however, we cannot increase the size of the Sindbis virus lesions without risk of damaging the facial motor nucleus. Our present data show that lesions of the nRF area can disrupt nose movement response to odor without completely removing the ability of the rat to move the nose to the affected side.
Fig. 4.
Lesions in nose retrofacial area (nRF) but not nose caudal intermediate reticular formation (nIRt) disrupt nose odor response. A: diagram of the rat testing setup. Bedding odor was presented on alternate sides, and nose position was monitored using high-speed video in a head-restrained rat. Rats were tested 5 days after Sindbis virus (Sindbis-152-GFP) injection or electrolytic lesion. B: example histological section to confirm location of Sindbis virus injection (green). NeuN stain (red) used to establish region of cell death. 7N, facial motor nucleus; Amb, nucleus ambiguus. C: example trace of lateral nose position response to odor presentation in a rat lesioned with Sindbis virus in the retrofacial area. The nose deflects toward the nonlesioned side when an odor is presented on the nonlesioned side but does not deflect toward the lesioned side when an odor is presented on that side. D: average trace of lateral nose position toward presented bedding odor. Averages are shown for 4 conditions: contralateral side from all lesions (gray), ipsilateral side for noneffective electrolytic lesion (green), ipsilateral side for noneffective Sindbis lesion (blue), and ipsilateral side for effective Sindbis lesion (gold). Error bars are SE. E: average peak nose position of ipsilateral (rectified) nose motion averaged between 350 and 450 ms post-odor stimulus presentation. Average values for individual rats are shown for 4 conditions: contralateral side from all lesions (gray), ipsilateral side for noneffective electrolytic lesion (green), ipsilateral side for noneffective Sindbis lesion (blue), and ipsilateral side for effective Sindbis lesion (gold). F: average histograms of the lateral nose positions for noneffective electrolytic lesion (green), noneffective Sindbis lesion (blue), and effective Sindbis lesion (gold). These histograms were computed over all ongoing motions to show that movement in general is not impaired by the lesion. G: 3-dimensional reconstructions of lesion locations. Slices cut at 400 μm are shown in the horizontal, coronal, and sagittal planes. C, caudal; R, rostral; D, dorsal; V, ventral; M, medial; L, lateral. Locations of the nRF and nIRt are outlined in magenta. Effective lesion sites overlap with the nRF region and not the nIRt region. H: 3-dimensional reconstruction of the location of the overlap of all effective lesions (gold). Locations of the nRF and nIRt are outlined in magenta. All effective lesions overlap in the nRF area. 7n, facial tract; IO, inferior olive; LRt, lateral reticular nucleus; SpV, spinal trigeminal nucleus; Ve, vestibular nucleus.
As a further control for lesion to the retrofacial area, we aimed to confirm that ablating the glutamatergic cells in the nRF area can affect the nose movement response to odor presentation. We chose to use adult mice for these experiments given the robust expression achieved with Vglut-cre mice and our need to label exclusively excitatory cells. A cre-dependent diphtheria toxin subunit a (DTA) virus, AAV-tdTom-flex-DTA, was injected into the vicinity of the nRF to ablate vGluT2-expressing cells (Fig. 5, A and B). Critically, we observed a reduced nose motion response to odor presentation in five of the nine lesioned mice (Fig. 5, C–G). All effective injections were centered in the nRF area (Fig. 5, H–J). In addition, we found that the cell count was reduced in a region of interest (Fig. 5, F and H) in the retrofacial area compared with both the control side and with animals with no change in nose movement.
Fig. 5.
Glutamatergic cell lesions in retrofacial area disrupt nose odor response. A: diagram of the mouse testing setup. Bedding odor was presented on alternate sides and nose position monitored using high-speed video in a head-restrained mouse. vGluT2-ires-cre mice were tested 5–6 wk after AAV-tdTom-flex-DTA injection. B: example histological section to confirm location of virus injection based on the locations of Cre-negative cells (red). To confirm cell death in a region of interest in the nose retrofacial (nRF) area, NeuroTrace blue-stained cells (blue) were counted using an automated algorithm (cyan). C: example trace of lateral nose position response to odor presentation in a mouse lesioned with DTA in the nRF area. The nose deflects toward the nonlesioned side when an odor is presented on the nonlesioned side but does not deflect toward the lesioned side when an odor is presented on that side. D: average trace of lateral nose position toward presented bedding odor. Averages are shown for 3 conditions: contralateral side from all lesions (gray), ipsilateral side for noneffective lesion (blue), and ipsilateral side for effective lesion (gold). E: average peak nose position of ipsilateral (rectified) nose motion averaged between 250 and 300 ms post-odor stimulus presentation. Effective lesions (gold) were defined as mice in which the average peak position was less than all control side (black) averages. Noneffective DTA lesions (blue) had an average peak position similar to control side averages. F: ratio of cell density in the region of interest (ROI) in the nRF area vs. cell density in the surrounding area (gray). Mice with a behavioral reduction in nose response (gold) showed a reduced cell count compared with the control side (black) (T statistic = −2.5, P = 0.035) and compared with noneffective lesions (blue) (T statistic = −2.4, P = 0.044), whereas noneffective lesions showed no change compared with the control side (T statistic = −0.8, P = 0.47). G: average histograms of the lateral nose positions for noneffective Sindbis lesion (blue) and effective Sindbis lesion (gold). H: definition of region of interest for cell count in nRF and surround region displayed in a slice reconstruction 1.0 mm lateral of midline. 7n, facial tract; 7N, facial motor nucleus; LRt, lateral reticular nucleus. I: 3-dimensional reconstruction of the location of the overlap of all effective lesions (gold). Locations of the nRF and nose intermediate reticular formation (nIRt) are outlined in magenta. All effective lesions overlap in the nRF area. J: 3-dimensional reconstructions of lesion locations. Slices cut at 400 μm are shown in the horizontal, coronal, and sagittal directions. C, caudal; R, rostral; D, dorsal; V, ventral; M, medial; L, lateral. Locations of the nRF and nIRt are outlined in magenta.
DISCUSSION
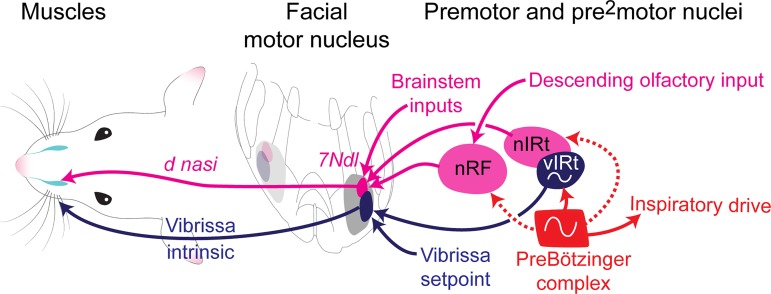
We have identified two key premotor areas for controlling nose motion, an area caudal to the facial motor nucleus that we call the retrofacial area (nRF) and an area near the vibrissa IRt oscillator that we call the nose IRt (nIRt). Anatomically, the nRF and nIRt have the densest labeling at secondary time points and are consistently labeled across different animals (Fig. 1, E–H, and Supplemental Fig. S2). We have established that glutamatergic inputs from both of these areas can drive motion of the nose (Fig. 3); however, only the nRF area is involved in the nose response to odor (Figs. 4 and 5). The proposed nose motion circuit based on this work is summarized in Fig. 6.
Fig. 6.
Circuits for orofacial motor control. Diagram shows the hypothesized circuit for control of nose motions via the deflector nasi (d nasi; magenta) and the known circuit for vibrissa motor control (blue). Motoneurons in the facial motor nucleus send projections to the muscles and receive inputs from premotor areas. Premotor areas receive inputs from the preBötzinger complex to drive synchronization to breathing. 7Ndl, dorsal lateral facial nucleus; nRF, nose retrofacial area; nIRT, nose intermediate reticular formation; vIRt, vibrissa intermediate reticular formation.
Our data show that although the nose moves in general after lesion of the nRF, it has reduced response to ipsilateral odorant presentation. This suggests that nRF is part of a pathway from the olfactory bulb that mediates bilateral odor comparisons, presumably involving the anterior olfactory nucleus (AON) (Esquivelzeta Rabell et al. 2017). The AON is part of a network that mediates complex odor processing and sends and receives projections from areas including the olfactory bulb, motor and piriform cortex, hippocampus, olfactory tubercle, and lateral hypothalamus (Brunjes et al. 2005; Davis and Macrides 1981; Haberly and Price 1978; Moyano and Molina 1980; Price et al. 1991; Scott et al. 1980). In our labeling results we saw no evidence of pre- or pre2motor projections directly from the AON; however, several known targets of the AON, including the olfactory tubercle and lateral hypothalamic area, had tertiary labeling. We propose that pathways through the lateral hypothalamic area or the olfactory tubercle are potential candidates to relay olfactory information to the nRF area.
We have also provided evidence that the preBötzinger complex sends putative premotor input to the nose premotor neurons. The nIRt is located near the vibrissa IRt (vIRt) (Moore et al. 2013), whereas the nRF is located in a similar location previously described as a respiratory area (Anderson et al. 2016); thus both areas are positioned such that they may receive respiratory inputs. Past work on orofacial and respiratory circuits in the medulla has pointed to the hypothesis that orofacial movement is linked to breathing via a signal from the preBötzinger complex to a premotor area (Del Negro et al. 2018; Kleinfeld et al. 2014b; Tan et al. 2010). Thus a circuit motif might involve a premotor area that drives motor neurons in the facial motor nucleus and receives input from the preBötzinger complex that drives the synchronization of movement to breathing. In our work we have characterized the contribution of glutamatergic inputs from the nRF and nIRt areas to nose motion. However, whisking generation is known to occur via an inhibitory oscillator that is positioned similarly to the nose caudal IRt.
The action of the nose deflector muscle consists of pulling the nasal cartilage in the caudal direction (Deschênes et al. 2015). There is no antagonistic muscle that pulls the cartilage rostrally. When the right and left muscles are equally activated in a synchronous manner, the resulting motion is an upward deflection of the nose. Unequal activation of these muscles produces upward and lateral deviation of the nose. Thus lateral nose deflection to the right, which is associated with odor delivery to the right side of the nose, likely results from activation of nose deflector motoneurons in the right facial nucleus and inhibition or disfacilitation of the contralateral pool of nose deflector motoneurons. It follows that there should exist inhibitory connections between the left and right pools of premotor neurons in the nRF or the nIRt, or between these premotor regions and the contralateral pool of nose deflector motoneurons. Determination of the cell types involved in control of nose movement and potential contribution of inhibitory drive to nose motion would allow integration of this medullary circuit to the ones that control sniffing and whisking (Moore et al. 2013, 2014).
GRANTS
We thank Matthias Schnell for supplying the rabies virus under National Institutes of Health (NIH) Grant P40 OD010996.
This study was supported by Canadian Institutes of Health Research Grant MT-5877 (to M. Deschênes), NIH Grants U01 NS0905905 and U19 NS107466 (to M. Deschênes and D. Kleinfeld), and Ruth L. Kirschstein National Research Service Award F31 NS089316 (to A. Kurnikova).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.D., A.K., and D.K. conceived and designed research; A.K. and D.K. performed experiments; A.K. analyzed data; A.K., M.D., and D.K. interpreted results of experiments; A.K. and D.K. prepared figures; A.K. drafted manuscript; A.K., M.D., and D.K. edited and revised manuscript; A.K., M.D., and D.K. approved final version of manuscript.
Supplemental Data
ACKNOWLEDGMENTS
We thank Agnieszka Brzozowska-Prechtl and Timothy Tran for assistance with histology and Celine Mateo, Jeffrey Moore, Peter Strick, and Fan Wang for helpful discussions.
REFERENCES
- Anderson TM, Garcia AJ 3rd, Baertsch NA, Pollak J, Bloom JC, Wei AD, Rai KG, Ramirez JM. A novel excitatory network for the control of breathing. Nature 536: 76–80, 2016. doi: 10.1038/nature18944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M, Krauss A, Muhammad S, Sinai-Esfahani L, Bellanca S, Margrie TW. Organization of rat vibrissa motor cortex and adjacent areas according to cytoarchitectonics, microstimulation, and intracellular stimulation of identified cells. J Comp Neurol 479: 360–373, 2004. doi: 10.1002/cne.20306. [DOI] [PubMed] [Google Scholar]
- Brunjes PC, Illig KR, Meyer EA. A field guide to the anterior olfactory nucleus (cortex). Brain Res Brain Res Rev 50: 305–335, 2005. doi: 10.1016/j.brainresrev.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Chen Y, McElvain LE, Tolpygo A, Ferrant D, Friedman B, Mitra PP, Karten HJ, Freund Y, Kleinfeld D. An active texture-based digital atlas enables automated mapping of structure and markers across brains. Nat Methods. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BJ, Macrides F. The organization of centrifugal projections from the anterior olfactory nucleus, ventral hippocampal rudiment, and piriform cortex to the main olfactory bulb in the hamster: an autoradiographic study. J Comp Neurol 203: 475–493, 1981. doi: 10.1002/cne.902030310. [DOI] [PubMed] [Google Scholar]
- Del Negro CA, Funk GD, Feldman JL. Breathing matters. Nat Rev Neurosci 19: 351–367, 2018. doi: 10.1038/s41583-018-0003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes M, Haidarliu S, Demers M, Moore J, Kleinfeld D, Ahissar E. Muscles involved in naris dilation and nose motion in rat. Anat Rec (Hoboken) 298: 546–553, 2015. doi: 10.1002/ar.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes M, Kurnikova A, Elbaz M, Kleinfeld D. Circuits in the ventral medulla that phase-lock motoneurons for coordinated sniffing and whisking. Neural Plast 2016: 1–9, 2016a. doi: 10.1155/2016/7493048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschênes M, Takatoh J, Kurnikova A, Moore JD, Demers M, Elbaz M, Furuta T, Wang F, Kleinfeld D. Inhibition, not excitation, drives rhythmic whisking. Neuron 90: 374–387, 2016b. doi: 10.1016/j.neuron.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duistermars BJ, Chow DM, Frye MA. Flies require bilateral sensory input to track odor gradients in flight. Curr Biol 19: 1301–1307, 2009. doi: 10.1016/j.cub.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivelzeta Rabell J, Mutlu K, Noutel J, Martin Del Olmo P, Haesler S. Spontaneous rapid odor source localization behavior requires interhemispheric communication. Curr Biol 27: 1542–1548.e4, 2017. doi: 10.1016/j.cub.2017.04.027. [DOI] [PubMed] [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining Portland, OR, August 2–4, 1996, p. 226–231. [Google Scholar]
- Ganguly K, Kleinfeld D. Goal-directed whisking behavior increases phase-locking between vibrissa movement and electrical activity in primary sensory cortex in rat. Proc Natl Acad Sci USA 101: 12348–12353, 2004. doi: 10.1073/pnas.0308470101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberly LB, Price JL. Association and commissural fiber systems of the olfactory cortex of the rat. II. Systems originating in the olfactory peduncle. J Comp Neurol 181: 781–807, 1978. doi: 10.1002/cne.901810407. [DOI] [PubMed] [Google Scholar]
- Hattox AM, Priest CA, Keller A. Functional circuitry involved in the regulation of whisker movements. J Comp Neurol 442: 266–276, 2002. doi: 10.1002/cne.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck DH, McAfee SS, Liu Y, Babajani-Feremi A, Rezaie R, Freeman WJ, Wheless JW, Papanicolaou AC, Ruszinkó M, Sokolov Y, Kozma R. Breathing as a fundamental rhythm of brain function. Front Neural Circuits 10: 115, 2017. doi: 10.3389/fncir.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isokawa-Akesson M, Komisaruk BR. Difference in projections to the lateral and medial facial nucleus: anatomically separate pathways for rhythmical vibrissa movement in rats. Exp Brain Res 65: 385–398, 1987. doi: 10.1007/BF00236312. [DOI] [PubMed] [Google Scholar]
- Kelly RM, Strick PL. Rabies as a transneuronal tracer of circuits in the central nervous system. J Neurosci Methods 103: 63–71, 2000. doi: 10.1016/S0165-0270(00)00296-X. [DOI] [PubMed] [Google Scholar]
- Kepecs A, Uchida N, Mainen ZF. The sniff as a unit of olfactory processing. Chem Senses 31: 167–179, 2006. doi: 10.1093/chemse/bjj016. [DOI] [PubMed] [Google Scholar]
- Kikuta S, Sato K, Kashiwadani H, Tsunoda K, Yamasoba T, Mori K. From the Cover: Neurons in the anterior olfactory nucleus pars externa detect right or left localization of odor sources. Proc Natl Acad Sci USA 107: 12363–12368, 2010. doi: 10.1073/pnas.1003999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D, Deschênes M, Wang F, Moore JD. More than a rhythm of life: breathing as a binder of orofacial sensation. Nat Neurosci 17: 647–651, 2014a. doi: 10.1038/nn.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D, Moore JD, Wang F, Deschênes M. The brainstem oscillator for whisking and the case for breathing as the master clock for orofacial motor actions. Cold Spring Harb Symp Quant Biol 79: 29–39, 2014b. doi: 10.1101/sqb.2014.79.024794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurnikova A, Moore JD, Liao S-M, Deschênes M, Kleinfeld D. Coordination of orofacial motor actions into exploratory behavior by rat. Curr Biol 27: 688–696, 2017. doi: 10.1016/j.cub.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables neuronal activation through the intact skull. Nat Neurosci 16: 1499–1508, 2013. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis M, Huber T, Benton R, Sakmar TP, Vosshall LB. Bilateral olfactory sensory input enhances chemotaxis behavior. Nat Neurosci 11: 187–199, 2008. doi: 10.1038/nn2031. [DOI] [PubMed] [Google Scholar]
- Martin H. Osmotropotaxis in the honey-bee. Nature 208: 59–63, 1965. doi: 10.1038/208059a0. [DOI] [Google Scholar]
- McAfee SS, Ogg MC, Ross JM, Liu Y, Fletcher ML, Heck DH. Minimally invasive highly precise monitoring of respiratory rhythm in the mouse using an epithelial temperature probe. J Neurosci Methods 283: 89–94, 2016. doi: 10.1016/j.jneumeth.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Deschênes M, Furuta T, Huber D, Smear MC, Demers M, Kleinfeld D. Hierarchy of orofacial rhythms revealed through whisking and breathing. Nature 497: 205–210, 2013. doi: 10.1038/nature12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Kleinfeld D, Wang F. How the brainstem controls orofacial behaviors comprised of rhythmic actions. Trends Neurosci 37: 370–380, 2014. doi: 10.1016/j.tins.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyano HF, Molina JC. Axonal projections and conduction properties of olfactory peduncle neurons in the rat. Exp Brain Res 39: 241–248, 1980. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. San Diego, CA: Academic, 1986. [Google Scholar]
- Porter J, Craven B, Khan RM, Chang S-J, Kang I, Judkewitz B, Volpe J, Settles G, Sobel N. Mechanisms of scent-tracking in humans. Nat Neurosci 10: 27–29, 2007. [Erratum in Nat Neurosci 10: 263, 2007.] doi: 10.1038/nn1819. [DOI] [PubMed] [Google Scholar]
- Price JL, Slotnick BM, Revial MF. Olfactory projections to the hypothalamus. J Comp Neurol 306: 447–461, 1991. doi: 10.1002/cne.903060309. [DOI] [PubMed] [Google Scholar]
- Ranade S, Hangya B, Kepecs A. Multiple modes of phase locking between sniffing and whisking during active exploration. J Neurosci 33: 8250–8256, 2013. doi: 10.1523/JNEUROSCI.3874-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, McBride RL, Schneider SP. The organization of projections from the olfactory bulb to the piriform cortex and olfactory tubercle in the rat. J Comp Neurol 194: 519–534, 1980. doi: 10.1002/cne.901940304. [DOI] [PubMed] [Google Scholar]
- Sirotin YB, Costa ME, Laplagne DA. Rodent ultrasonic vocalizations are bound to active sniffing behavior. Front Behav Neurosci 8: 399, 2014. doi: 10.3389/fnbeh.2014.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck K, Knaden M, Hansson BS. Do desert ants smell the scenery in stereo? Anim Behav 79: 939–945, 2010. doi: 10.1016/j.anbehav.2010.01.011. [DOI] [Google Scholar]
- Takatoh J, Nelson A, Zhou X, Bolton MM, Ehlers MD, Arenkiel BR, Mooney R, Wang F. New modules are added to vibrissal premotor circuitry with the emergence of exploratory whisking. Neuron 77: 346–360, 2013. doi: 10.1016/j.neuron.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Pagliardini S, Yang P, Janczewski WA, Feldman JL. Projections of preBötzinger complex neurons in adult rats. J Comp Neurol 518: 1862–1878, 2010. doi: 10.1002/cne.22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachowiak M. All in a sniff: olfaction as a model for active sensing. Neuron 71: 962–973, 2011. doi: 10.1016/j.neuron.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker WI. Analysis of sniffing of the albino rat. Behaviour 12: 223–244, 1964. doi: 10.1163/156853964X00030 [DOI] [Google Scholar]
- Wirblich C, Schnell MJ. Rabies virus (RV) glycoprotein expression levels are not critical for pathogenicity of RV. J Virol 85: 697–704, 2011. doi: 10.1128/JVI.01309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Tan J, Qin C, Lu Y, Ding C, Luo M. Precise circuitry links bilaterally symmetric olfactory maps. Neuron 58: 613–624, 2008. doi: 10.1016/j.neuron.2008.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.