INTRODUCTION
Meningiomas are the most common primary tumor of the CNS; there are approximately 28,000 new diagnoses annually in the United States.1 Currently, there are no approved systemic therapies for meningiomas that recur after local treatment: chemotherapy and hormonal agents have demonstrated minimal benefit in numerous clinical trials.2-4
Meningiomas comprise a heterogeneous group of neoplasms driven by mutations in a wide array of tumor suppressor genes and oncogenes.5-17 Characterization of these mutations has revealed opportunities for rational therapy.18-20 For example, a durable therapeutic response has been reported for a metastatic AKT1 E17K–mutant meningioma treated with a pan-AKT inhibitor.21
Studies also suggest the potential for treating meningioma with immune checkpoint modulators22-24: programmed death ligand 1 (PD-L1) is expressed in a subset of meningiomas, and the tumor microenvironment is immunosuppressive.22-28 Higher-grade meningiomas also harbor mutations predicted to generate neoantigens, which may foster susceptibility to immunotherapies.29
On the basis of these data, we initiated a phase II study of nivolumab, a humanized IgG4 programmed death 1 (PD-1)–blocking monoclonal antibody, in patients with higher-grade meningiomas that recurred after surgery and radiotherapy (ClinicalTrials.gov identifier: NCT02648997). We report here a patient with an atypical meningioma that was not controlled by repeated surgery and radiation but that was highly response to nivolumab.
CASE REPORT
A 50-year-old woman with progressive headaches underwent a gross total resection (surgery 1) of a gadolinium-enhancing right frontal convexity atypical meningioma (WHO grade 2; Fig 1A). The tumor recurred 10 months later and was treated with stereotactic radiosurgery (SRS; SRS 1, 25 Gy/five fractions). Recurrent atypical meningioma was debulked 17 months (surgery 2) and 21 months (surgery 3) after the original diagnosis. The patient then underwent conventional radiotherapy (RT; conformal RT, 54 Gy/27 fractions). Three months later, she again underwent SRS (SRS 2, 14 Gy/one fraction) because of tumor spread to the sphenoid ridge and infratemporal fossa. Additional debulking surgeries for recurrent atypical meningioma were performed at 46 months (surgery 4) and 49 months (surgery 5). She then received bevacizumab for 11 months, mifepristone for 7 months, and temozolomide for 4 months. Each therapy was discontinued after disease progression occurred. A sixth debulking surgery (surgery 6) at 70 months confirmed recurrent atypical meningioma.
Fig 1.
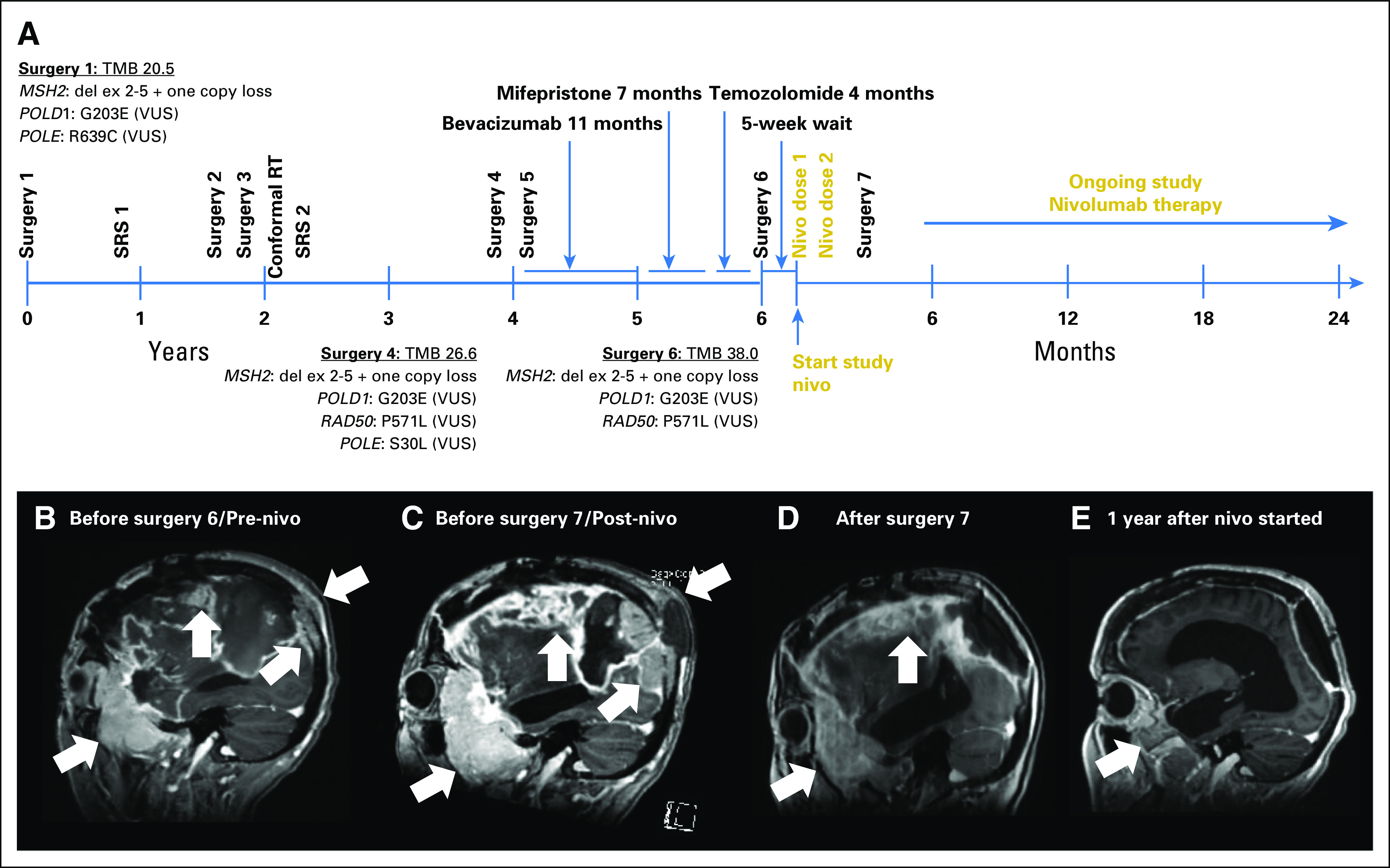
Patient treatment and radiographic assessment. (A) Timeline of patient therapy. Nivo, nivolumab; RT, radiation therapy; SRS, stereotactic radiosurgery; TMB, tumor mutational burden; VUS, variant of unknown significance. Sagittal T1 gadolinium-enhanced images (B) before initiation of study nivolumab therapy (before surgery 6), (C) after two doses of nivolumab therapy (before surgery 7), and (D) after subtotal resection (surgery 7) that demonstrate necrosis and no viable tumor; (E) ongoing response 1 year after initiation of nivolumab therapy. White arrows indicate bulky tumor burden, including occipital lesion growing into soft tissue external to skull. MSH2: del ex 2-5' indicates homozygous deletion of exons 2 to 5 of the DNA mismatch repair (MMR) gene MSH2.
Five weeks later, the patient enrolled in our phase II nivolumab clinical trial. At that time, 75 months after the original diagnosis, painful subcutaneous masses overlaid the right convexity. The patient required oxycodone for scalp pain and dexamethasone (4 mg/day) for worsening headache. Pretreatment magnetic resonance imaging (MRI) showed an enlarging-enhancing right sphenoid wing mass, plaque-like dural enhancement over the right convexity, tumor erosion through the occipital skull, and an increased fluid-attenuated inversion recovery (FLAIR) signal abnormality that involved the right hemisphere (Fig 1B).
The patient received nivolumab 3 mg/kg on days 1 (dose 1) and 15 (dose 2) of cycle 1 without incident, but her headaches and scalp pain worsened. The scalp masses were notably larger, erythematous, and more tender by day 28. Brain MRI showed significantly enlarged dural-based enhancing lesions, increased T2 and FLAIR signal abnormalities, and enlarged scalp masses (Fig 1C). Her physical examination and MRI appeared consistent with progressive disease, and we withheld additional nivolumab. She underwent palliative debulking surgery 3 weeks later (surgery 7). Immediately before surgery, her scalp masses were modestly smaller and less tender. Postoperative MRI showed a subtotal resection (Fig 1D).
MATERIALS AND METHODS
Patient Consent
Our patient provided informed consent for our institutional review board–approved study and consented to this publication. We obtained all permissions required by law and by the Dana-Farber Cancer Institute (DFCI)/Harvard Cancer Center to allow the publication of images from the patient.
Immunohistochemistry
We used immunohistochemistry (IHC) to evaluate protein expression using Envision Plus detection (Dako, Carpinteria, CA; Data Supplement) and published protocols.30,31
Multiplexed Immunofluorescence
The protocol for tissue-based multiplexed cyclic immunofluorescence (t-CyCIF) of conventionally prepared formalin-fixed, paraffin-embedded specimens and image analysis are described elsewhere.32,33 Antibodies are listed in the Data Supplement.
Oncopanel Sequencing
We used targeted next-generation exome sequencing (Oncopanel version 3; DFCI/Brigham and Women's Hospital [BWH]) to detect mutations and copy number variations in 447 cancer genes. We processed and annotated data as previously described.9,34,35 We calculated tumor mutational burden (TMB) by determining the number of nonsynonymous mutations per megabase (Mb) of exonic sequence data across sequenced genes.35,36
RESULTS
Histopathologic examination of the tissue resected 5 weeks before nivolumab treatment (sample 6) revealed a highly proliferative atypical meningioma (Fig 2A). In contrast, tissue resected 3 weeks after nivolumab dose 2 (sample 7) had dense fibrosis with extensive immune cell infiltration and necrosis (Fig 2C). Tumor cells were absent in the hematoxylin and eosin–stained slides, but IHC with a marker of meningioma, SSTR2a30 (Figs 2B and 2D), revealed one small cluster (approximately 1,000 cells/µm2) of possible residual tumor cells (which represented < 0.0001% of resected tissue; image and data not shown).
Fig 2.
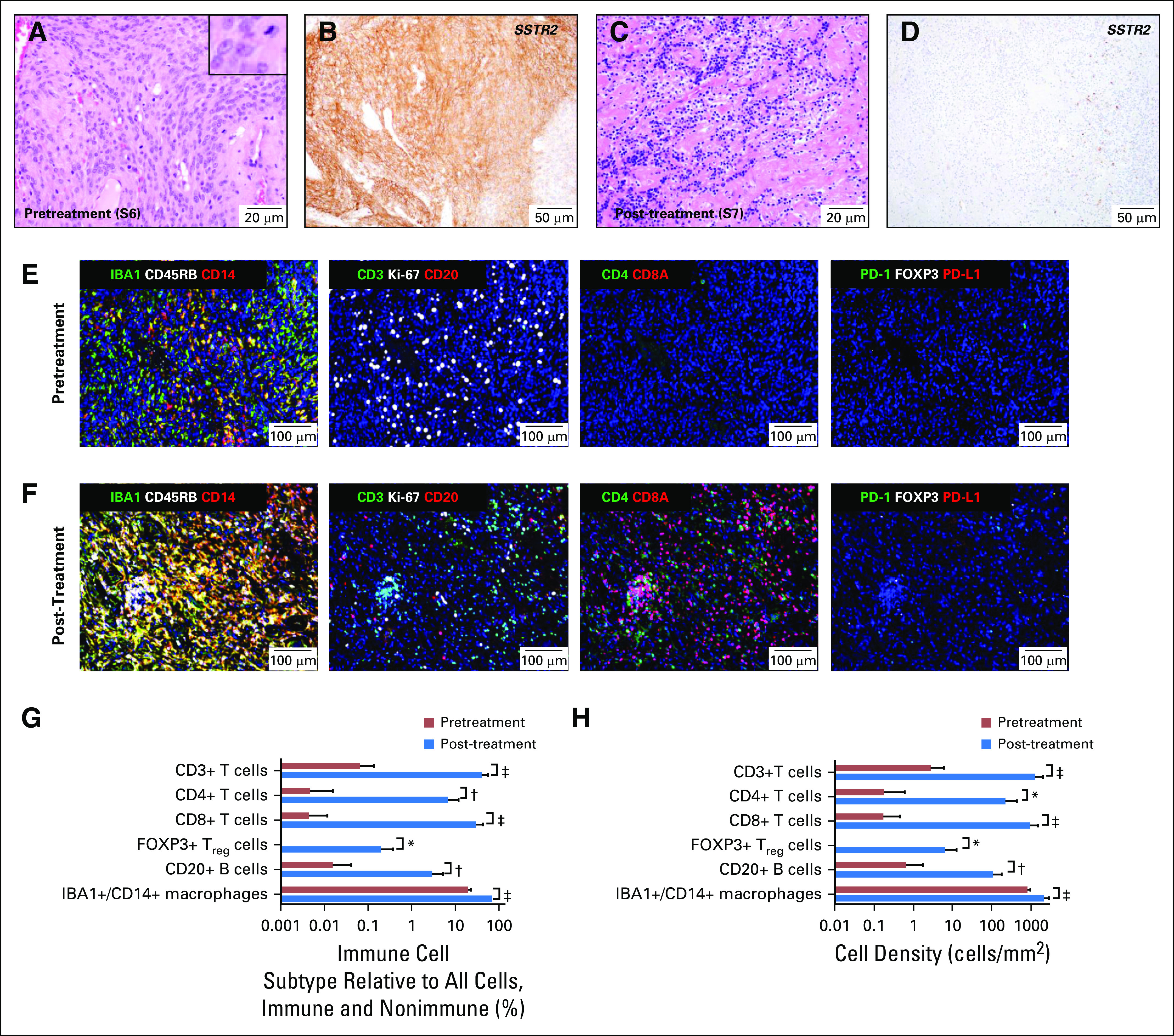
Histologic, immunohistochemical, and tissue-based multiplexed cyclic immunofluorescence (t-CyCIF) characterization of meningioma resection samples. Representative images of hematoxylin and eosin–stained section of meningioma resection (A) before treatment with nivolumab (surgery 6/sample 6; S6) and (C) after two doses of nivolumab (surgery 7/sample 7; S7). Representative images of immunohistochemistry for meningioma marker SSTR2a on resections (B) before treatment with nivolumab (surgery 6/sample 6; S6) and (D) after two nivolumab doses (surgery 7/sample 7; S7). The inset in (A) shows a mitotic figure and cells with prominent nucleoli; an elevated mitotic index of more than four mitoses per 10 high-powered fields was used for grading. (E) A representative field of view of t-CyCIF imaging data of 11 biomarkers (ionized calcium-binding adapter molecule 1 [IBA], CD45RB, CD14, CD3, marker of proliferation Ki-67 [Ki-67], CD20, CD4, CD8A, programmed cell death 1 [PD-1], forkhead box P3 [FOXP3], programmed cell death ligand 1 [PD-L1]) generated from a single section of pretreatment meningioma (surgery 6/sample 6) and (F) from a single section of the post-treatment meningioma (surgery 7/sample 7). Comparison of the images in (E) and (F) showed a marked increase in macrophages; T lymphocytes (CD3+), including CD4+ T cells and CD8A+ T cells; and B lymphocytes (CD20+) after nivolumab treatment. The majority of T cells after treatment were CD8A+. The antibodies used for this characterization and a detailed analysis of the absolute number, relative number and density of various immune subtypes are provided in Table 1 and the Data Supplement. (G) Bar graph of the percentage of immune cell subtypes relative to all cells (immune and nonimmune) before (red bars) and after (blue bars) nivolumab treatment. (H) Bar graph of cell density before (red bars) and after (blue bars) nivolumab treatment. The analysis was performed on 10 representative views (tiles) from the t-CyCIF data collected from samples 6 and 7. t test statistical analysis was performed. *, P < .01, †, P < .001, ‡, P < .0001. The bar graphs represent the most pertinent data derived from the immune profile of the pre-nivolumab (sample 6) and post-nivolumab (sample 7)–treated meningioma samples using t-CyCIF. Detailed data are listed in Table 1 and the Data Supplement.
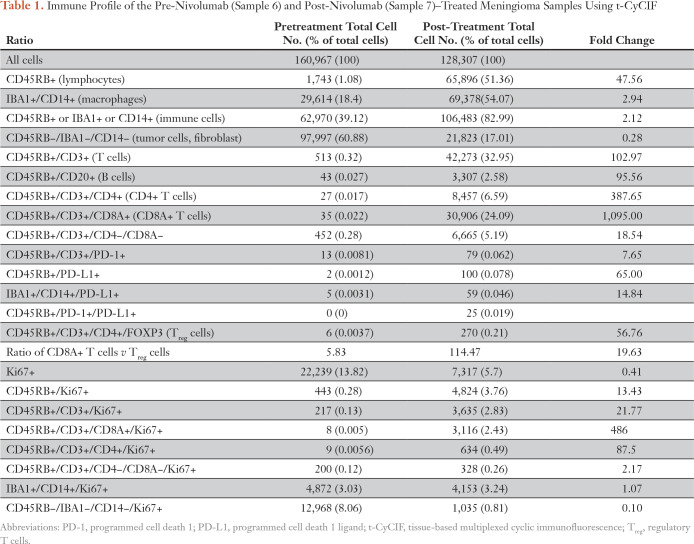
To characterize the effects of nivolumab, we profiled the tumor and its microenvironment using t-CyCIF, a method for highly multiplexed immunofluorescence imaging of formalin-fixed, paraffin-embedded specimens.32 We imaged 11 markers (Data Supplement) on pre- and post-treatment samples to measure changes in immune cell types and the relative density of the immune infiltrate (Table 1; Figs 2E-2H; Appendix Fig A1; Data Supplement). We have previously shown that PD-L1 is overexpressed in tumor cells of some higher-grade meningiomas,22 but, in this pretreatment specimen (sample 6), we found PD-L1 restricted to immune cells. Post-treatment (in sample 7), we observed a marked increase in IBA1+/CD14+ macrophages, CD4+ and CD8+ T cells, CD20+ B cells, and FOXP3+ regulatory T cells (Treg cells; Table 1; Figs 2E-2H; Appendix Fig A1). The fraction of CD8+ T cells increased from 7% to 73% (Table 1) and CD8+ T-cell density increased from 0.5 to 304 cells/mm2; the CD8+ T cell–to–Treg ratio increased 20-fold (from 5.8 to 115; Figs 2G-2H; Data Supplement). These data are consistent with a highly active antitumor immune response after treatment.
Table 1.
Immune Profile of the Pre-Nivolumab (Sample 6) and Post-Nivolumab (Sample 7)–Treated Meningioma Samples Using t-CyCIF
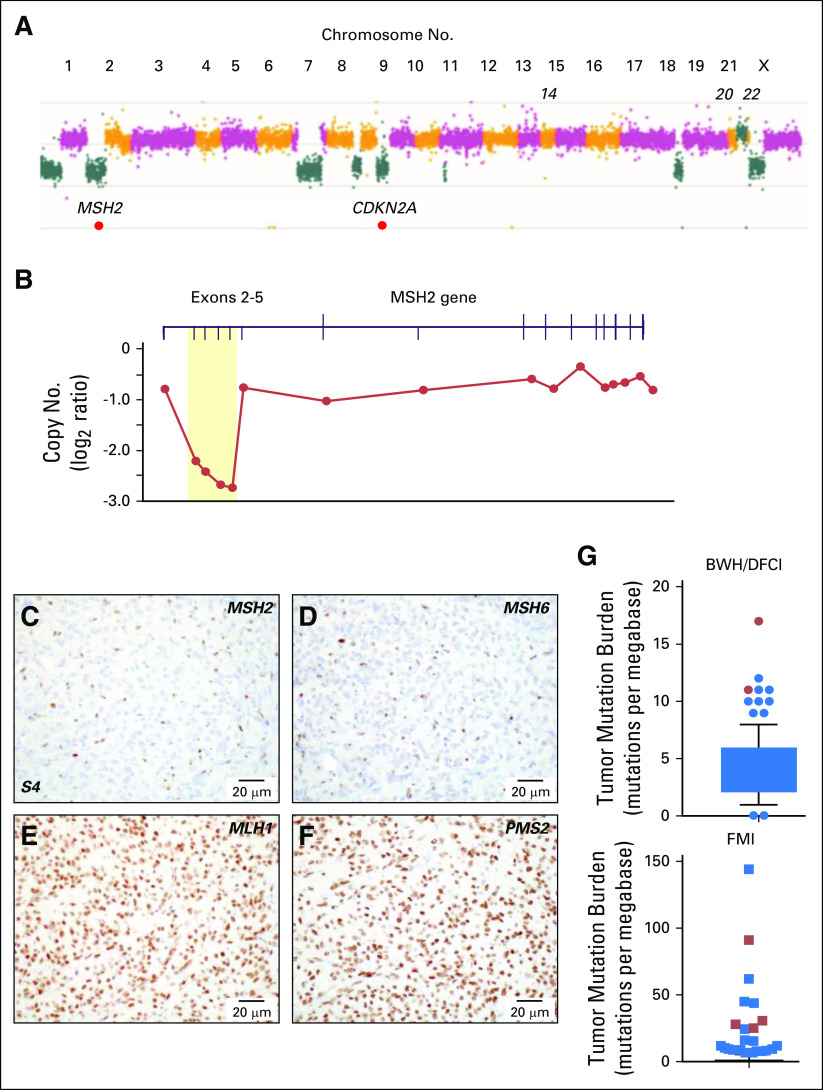
We used targeted-exome sequencing to characterize genomic aberrations in specimens from the original resection (sample 1); the resection before treatment with bevacizumab, mifepristone, and temozolomide (sample 4); and the resection before treatment with nivolumab (sample 6; Fig 1A; Data Supplement). This analysis revealed a significantly elevated TMB of 20.5, 26.6, and 38.0 mutations/Mb in the samples, respectively (Data Supplement).37 These levels were greater than those of 228 other meningiomas sequenced as part of the BWH/DFCI profile initiative.35 Copy number variation in the three samples was characteristic of higher-grade aggressive meningioma38-41 (Fig 3A; Data Supplement). Notably, homozygous deletion of exons 2 to 5 of the DNA mismatch repair (MMR) gene MSH2 was present in all three samples (Figs 3A-3B) but was absent in a peripheral-blood specimen.
Fig 3.
Genomic and immunohistochemical characterization of meningioma samples. (A) Copy number analysis from targeted next-generation sequencing data from sample 6 identified a genome-wide profile characteristic of a high-grade meningioma, including loss of 1p, 9p and monosomy of chromosomes 18 and 22. Focal homozygous deletion of CDKN2A/CDKN2B and intragenic deletion of MSH2 were present. (B) Gene-level view of MSH2 showed a homozygous intragenic deletion of exons 2 through 5 of MSH2 (NM_000251; log2 ratio from −2.21 to −2.73) in the setting of 2p arm-level single copy loss. Copy number is depicted as a log2 ratio value. Immunohistochemistry on pretreatment meningioma resection (surgery 4/sample 4; S4) for (C) MSH2, (D) MSH6, (E) MLH1, and (F) PMS2. MSH2 and MSH6 staining was present only in nontumor cells. (G) Box and whiskers plot (5th to 95th percentiles) of tumor mutational burden (TMB; mutations per megabase) for Brigham and Women’s Hospital (BWH)/Dana-Farber Cancer Institute (DFCI) profile initiative cohort (n = 228 sequenced cases; Data Supplement) plus sequencing data for the mismatch repair (MMR)–deficient case BWH/DFCI 2 and for the Foundation Medicine cohort (FMI; n = 615 sequenced cases; Data Supplement). Patient cases with loss of function changes in MMR and MMR-related genes (from Table 2) are highlighted in red (dots and squares). For the BWH/DFCI profile initiative cohort, the mean TMB was 4.25 mutations/megabase (standard deviation, 2.55; standard error of the mean, 0.17; lower 95% CI of mean, 3.91; upper 95% CI of mean, 4.58). For the Foundation Medicine cohort, the mean TMB was 2.77 mutations/megabase (standard deviation, 8.08; standard error of the mean, 0.33; lower 95% CI of mean, 2.14; upper 95% CI of mean, 3.41).
IHC showed that tumor cells were negative for the MSH2 protein and its heterodimeric partner, MSH6, but that expression of MLH1 and PMS2 protein was retained (Figs 3C-3F). Loss of MSH2/MSH6 in sample 1 demonstrated that MSH2 had been inactivated before any therapy and independent of temozolomide exposure, a known driver of acquired MMR deficiency in gliomas.42,43 Thus, the tumor was MMR deficient at the original diagnosis, and there was a progressive increase in TMB (Data Supplement).
The patient resumed nivolumab after surgery 7 and continued biweekly therapy for more than 2 years. MRI scans have shown gradual and continued regression of enhancing lesions and associated T2/FLAIR signal abnormalities (Fig 1E). Scalp lesions have disappeared, and narcotics and dexamethasone are no longer required for pain control and cerebral edema.
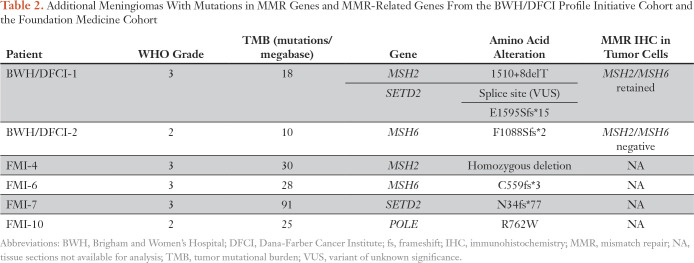
Given the dramatic response of this MSH2-deficient tumor to nivolumab, we investigated the prevalence of elevated TMB and MMR deficiency in meningioma (Fig 3G; Data Supplement). We used sequencing data from the BWH/DFCI profile initiative (n = 228 patient cases)35 or BWH/DFCI patient cases screened only by MMR-protein IHC (n = 237 patient cases)22,44 to study specimens from 465 patients (n = 288 with grade 1, n = 132 with grade 2, and n = 45 with grade 3 disease). Among the 228 sequenced specimens, the cohort mean TMB was 4.2 mutations/Mb (grade 1, 4.0; grade 2, 4.4; grade 3, 6.5; Data Supplement). Seven of the 228 specimens had TMBs of 10 or more mutations/Mb, a commonly used threshold for hypermutation36; one of these meningiomas (TMB, 18 mutations/Mb) had a truncating mutation in the DNA MMR regulator SETD2 as well as a splice site mutation in MSH2 of unclear functional significance. We discovered one occurrence with MMR protein loss discovered by IHC (one of 237 occurrences), and sequencing confirmed biallelic inactivation of MSH6 and a TMB of 10 mutations/Mb (Table 2; Fig 3G; Data Supplement). Among the sequenced meningiomas, elevated TMB was significantly positively associated with anaplastic histology, Ki-67 proliferative index, and chromosomal instability38 but not with prior radiotherapy, radiation induced meningioma, or recurrent tumor (Data Supplement).
Table 2.
Additional Meningiomas With Mutations in MMR Genes and MMR-Related Genes From the BWH/DFCI Profile Initiative Cohort and the Foundation Medicine Cohort
In a second cohort of 615 sequenced meningiomas (Foundation Medicine cohort), 14 tumors had a TMB of more than 10 mutations/Mb. Among these 14 specimens, two grade 3 meningiomas had inactivating mutations in MSH2 and MSH6; one grade 2 meningioma had a substitution in the DNA polymerase domain of the POLE subunit of DNA polymerase epsilon, and another grade 3 meningioma had a truncating mutation in SETD2 (Table 2; Fig 3G; Data Supplement). Thus, across two cohorts of sequenced meningioma samples (n = 843), 21 (2.5%) had high TMBs.36 Three of 1,080 meningiomas screened by sequencing or IHC (0.3%) had detectable inactivating mutations in MSH2 or MSH6. The number of notable tumors increased to six (0.6%) when MMR-related genes (eg, SETD2, POLE) were considered.
DISCUSSION
In our patient with an MSH2-deficient meningioma, nivolumab treatment generated a robust anticancer immune response, as evidenced by dramatically increased infiltrating CD8+ T cells and a durable therapeutic response. Notably, the enlargement of lesions and increased signal abnormalities seen on the MRI after nivolumab reflected pseudoprogression, not tumor growth. In addition to inactivation of MSH2, sequencing of the patient’s tumors revealed missense mutations in POLD1 (R639C), RAD50 (P571L), and POLE (G203E, S30L), but these changes are of unclear significance and are predicted to be nonpathogenic.45 An increase in TMB from 26.6 mutations/MB in sample 4 (before experimental therapies) to 38.0 in sample 6 raises the possibility that temozolomide can augment TMB in meningioma. However, this neoplasm was MMR deficient at the time of original diagnosis; thus, temozolomide did not drive acquired MMR deficiency.
This work also shows that approximately 2.5% of meningiomas have a high mutation burden, a phenotype that has been linked with neoantigen expression in other tumor types.46,47 In a subset of mutation-rich meningiomas, loss of function alterations in MMR and MMR-related genes can be detected. It is possible that other, as yet unknown or undetected, mutations contribute to high TMB in the remaining tumors. In all, this first report, to our knowledge, of a dramatic response of an MMR-deficient meningioma to immunotherapy and our characterization of meningioma cohorts from two different institutions indicate that screening meningiomas is warranted to identify a molecularly defined subtype that is likely responsive to immunotherapy.
ACKNOWLEDGMENT
We thank George Zanazzi, MD, for assistance with sample procurement, the staff at the Brigham and Women’s Hospital Center for Advanced Molecular Diagnostics for sequencing, Terri Woo for assistance with immunohistochemistry, and members of the Lab for Systems Pharmacology for helpful comments on the manuscript.
Appendix
Fig A1.
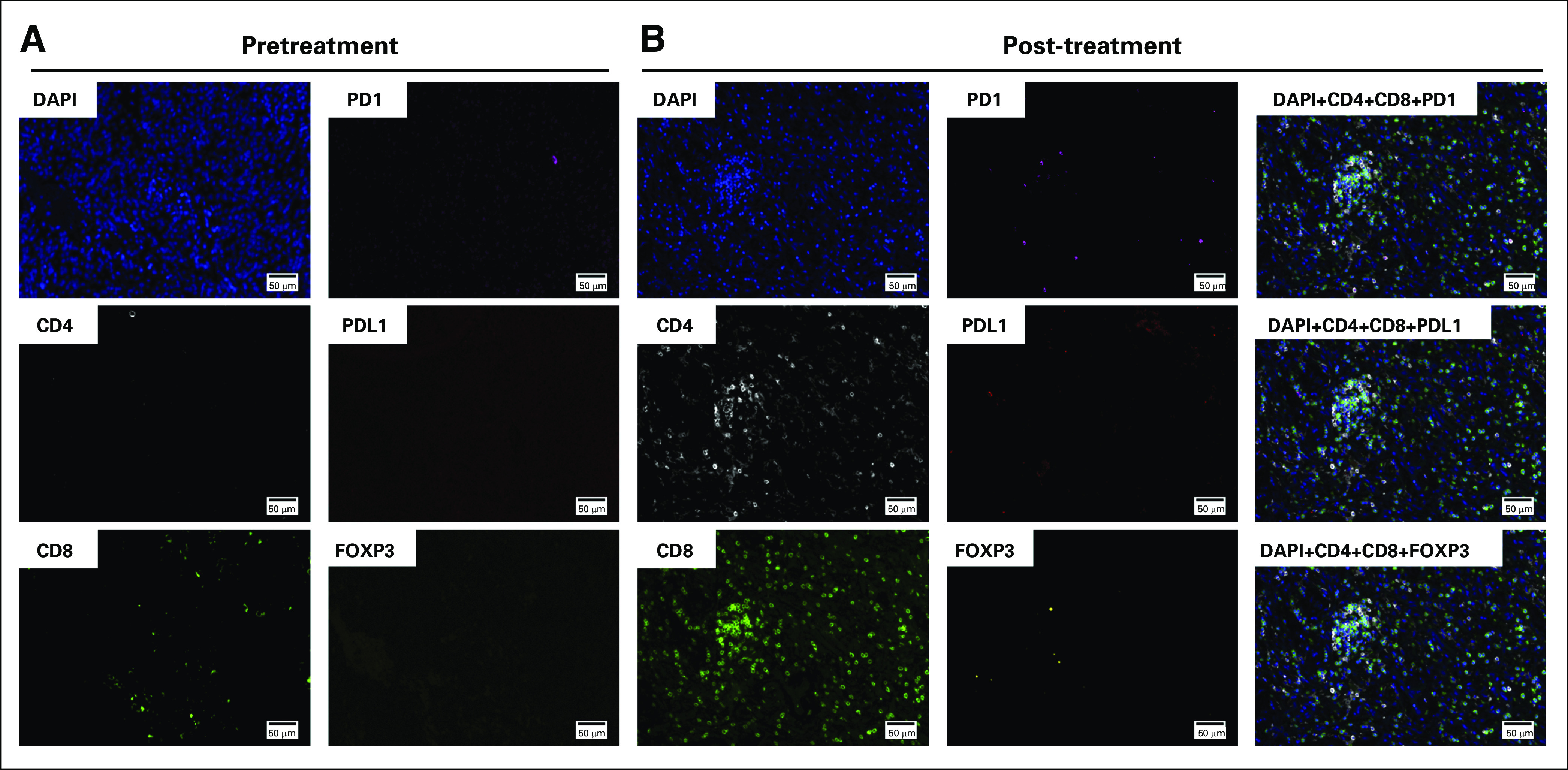
Tissue-based multiplexed cyclic immunofluorescence (t-CyCIF) characterization of resection tissues before and after nivolumab treatment. (A) A representative field of view of t-CyCIF imaging data of selected biomarkers generated from a single section of pretreatment meningioma (surgery 6/sample 6). (B) A representative field of view of t-CyCIF imaging data of selected biomarkers generated from a single section of the post-treatment meningioma (surgery 7/sample 7). FOXP3, forkhead box P3; PD-1, programmed death 1; PD-L1, programmed death ligand 1.
Footnotes
Supported by the Ludwig Center at Harvard Medical School (P.K.S. and S.S.) and by National Cancer Institute Grant No. U54- CA225088 (to P.K.S. and S.S.).
AUTHOR CONTRIBUTIONS
Conception and design: Ian F. Dunn, Mehdi Touat, Peter K. Sorger, Keith L. Ligon, Sandro Santagata, David A. Reardon
Provision of study material or patients: Ian F. Dunn, Michael B. Sisti, Peter D. Canoll, Keith L. Ligon, Sandro Santagata, David A. Reardon
Administrative support:Eric Severson, Brenda Acevedo
Financial support: Peter K. Sorger, Sandro Santagata
Data analysis and interpretation: Mehdi Touat, Patrick Y. Wen, Renato Umeton, Adrian M. Dubuc, Matthew Ducar, Peter D. Canoll, Eric Severson, Shakti H. Ramkissoon, Jia-Ren Lin, Peter K. Sorger, Keith L. Ligon, Sandro Santagata, David A. Reardon
Collection and assembly of data: Ziming Du, Mehdi Touat, Michael B. Sisti, Julia A. Elvin, Shakti H. Ramkissoon, Lais Cabrera, Brenda Acevedo, Keith L. Ligon, Sandro Santagata, David A. Reardon
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Ian F. Dunn
No relationship to disclose
Ziming Du
No relationship to disclose
Mehdi Touat
Consulting or Advisory Role: Agios, Taiho PharmaceuticalTravel, Accommodations, Expenses: Merck Sharp & Dohme
Michael B. Sisti
No relationship to disclose
Patrick Y. Wen
Consulting or Advisory Role: AbbVie, Genentech, Roche, Agios, AstraZeneca, Karyopharm Therapeutics, Vivus, Monteris Medical, Aurora Biopharma, Vascular Biogenics, Kadmon, Ziopharm Oncology, GW Pharmaceuticals, Lilly, Immunomic Therapeutics
Speakers' Bureau: Merck
Research Funding: Agios (Inst), Abbvie (Inst), Angiochem (Inst), Genentech (Inst), Roche (Inst), GlaxoSmithKline (Inst), AstraZeneca (Inst), ImmunoCellular Therapeutics (Inst), Karyopharm Therapeutics (Inst), Merck (Inst), Novartis (Inst), Oncoceutics (Inst), Sanofi (Inst), ARIAD (Inst), Vascular Biogenics (Inst), Lilly
Research Funding: Karyopharm Therapeutics
Renato Umeton
No relationship to disclose
Adrian M. Dubuc
No relationship to disclose
Matthew Ducar
No relationship to disclose
Peter D. Canoll
No relationship to disclose
Eric Severson
Employment: Foundation Medicine, Partners Healthcare
Julia A. Elvin
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Shakti H. Ramkissoon
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Jia-Ren Lin
No relationship to disclose
Lais Cabrera
No relationship to disclose
Brenda Acevedo
No relationship to disclose
Peter K. Sorger
Leadership: RareCyte, Merrimack, Applied BioMath
Stock and Other Ownership Interests: RareCyte, Merrimack, Applied BioMath, Glencoe Software
Honoraria: Novartis
Research Funding: Merck (Inst)
Keith L. Ligon
Stock and Other Ownership Interests: Travera
Consulting or Advisory Role: Midatech
Research Funding: Plexxikon (Inst), Amgen (Inst), X4 Pharma (Inst), Tragara (Inst), Bristol-Myers Squibb (Inst)
Patents, Royalties, Other Intellectual Property: Molecular Diagnostics Assay patent
Sandro Santagata
Consulting or Advisory Role: RareCyte
David A. Reardon
Honoraria: AbbVie, Cavion, Genentech, Roche, Merck, Midatech, Momenta Pharmaceuticals, Novartis, Novocure, Regeneron, Stemline Therapeutics, Celldex, Oxigene, Monteris Medical, Bristol-Myers Squibb, Juno Therapeutics, Inovio Pharmaceuticals, Oncorus
Consulting or Advisory Role: Cavion, Genentech, Roche, Merck, Momenta Pharmaceuticals, Novartis, Novocure, Regeneron, Stemline Therapeutics, Bristol-Myers Squibb, Inovio Pharmaceuticals, Juno Therapeutics, Celldex, Oxigene, Monteris Medical, Midatech, Oncorus
Research Funding: Celldex (Inst), Incyte (Inst), Midatech (Inst), Tragara (Inst), Inovio Pharmaceuticals (Inst), Agenus (Inst), EMD Serono (Inst), Acerta Pharma (Inst)
REFERENCES
- 1.Zhang AS, Ostrom QT, Kruchko C, et al. Complete prevalence of malignant primary brain tumors registry data in the United States compared with other common cancers, 2010. Neuro-oncol. 2017;19:726–735. doi: 10.1093/neuonc/now252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaley T, Barani I, Chamberlain M, et al. Historical benchmarks for medical therapy trials in surgery- and radiation-refractory meningioma: A RANO review. Neuro-oncol. 2014;16:829–840. doi: 10.1093/neuonc/not330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldbrunner R, Minniti G, Preusser M, et al. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17:e383–e391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- 4.Furtner J, Schöpf V, Seystahl K, et al. Kinetics of tumor size and peritumoral brain edema before, during, and after systemic therapy in recurrent WHO grade II or III meningioma. Neuro-oncol. 2016;18:401–407. doi: 10.1093/neuonc/nov183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brastianos PK, Horowitz PM, Santagata S, et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet. 2013;45:285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark VE, Erson-Omay EZ, Serin A, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339:1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuss DE, Piro RM, Jones DTW, et al. Secretory meningiomas are defined by combined KLF4 K409Q and TRAF7 mutations. Acta Neuropathol. 2013;125:351–358. doi: 10.1007/s00401-013-1093-x. [DOI] [PubMed] [Google Scholar]
- 8.Clark VE, Harmancı AS, Bai H, et al. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48:1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abedalthagafi MS, Merrill PH, Bi WL, et al. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 2014;5:10596–10606. doi: 10.18632/oncotarget.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abedalthagafi MS, Bi WL, Merrill PH, et al. ARID1A and TERT promoter mutations in dedifferentiated meningioma. Cancer Genet. 2015;208:345–350. doi: 10.1016/j.cancergen.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abedalthagafi M, Bi WL, Aizer AA, et al. Oncogenic PI3K mutations are as common as AKT1 and SMO mutations in meningioma. Neuro-oncol. 2016;18:649–655. doi: 10.1093/neuonc/nov316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahm F, Schrimpf D, Olar A, et al. TERT promoter mutations and risk of recurrence in meningioma. J Natl Cancer Inst. 2015;108:108. doi: 10.1093/jnci/djv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peyre M, Gaillard S, de Marcellus C, et al. Progestin-associated shift of meningioma mutational landscape. Ann Oncol. 2017;29:681–686. doi: 10.1093/annonc/mdx763. [DOI] [PubMed] [Google Scholar]
- 14.Shankar GM, Abedalthagafi M, Vaubel RA, et al. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro-oncol. 2017;19:535–545. doi: 10.1093/neuonc/now235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peyre M, Gauchotte G, Giry M, et al. De novo and secondary anaplastic meningiomas: A study of clinical and histomolecular prognostic factors. Neuro-oncol. 2018;20:1113–1121. doi: 10.1093/neuonc/nox231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerrini-Rousseau L, Dufour C, Varlet P, et al. Germline SUFU mutation carriers and medulloblastoma: Clinical characteristics, cancer risk and prognosis. Neuro-oncol. 2017;20:1122–1132. doi: 10.1093/neuonc/nox228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juratli TA, McCabe D, Nayyar N, et al. DMD genomic deletions characterize a subset of progressive/higher-grade meningiomas with poor outcome. Acta Neuropathol. 2018;136:779–792. doi: 10.1007/s00401-018-1899-7. [DOI] [PubMed] [Google Scholar]
- 18.Nowosielski M, Galldiks N, Iglseder S, et al. Diagnostic challenges in meningioma. Neuro-oncol. 2017;19:1588–1598. doi: 10.1093/neuonc/nox101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yesilöz Ü, Kirches E, Hartmann C, et al. Frequent AKT1 E17K mutations in skull base meningiomas are associated with mTOR and ERK1/2 activation and reduced time to tumor recurrence. Neuro-oncol. 2017;19:1088–1096. doi: 10.1093/neuonc/nox018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boetto J, Bielle F, Sanson M, et al. SMO mutation status defines a distinct and frequent molecular subgroup in olfactory groove meningiomas. Neuro-oncol. 2017;19:345–351. doi: 10.1093/neuonc/now276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weller M, Roth P, Sahm F, et al. Durable control of metastatic AKT1-mutant WHO grade 1 meningothelial meningioma by the AKT inhibitor, AZD5363. J Natl Cancer Inst. 2017;109:1–4. doi: 10.1093/jnci/djw320. [DOI] [PubMed] [Google Scholar]
- 22.Du Z, Abedalthagafi M, Aizer AA, et al. Increased expression of the immune modulatory molecule PD-L1 (CD274) in anaplastic meningioma. Oncotarget. 2015;6:4704–4716. doi: 10.18632/oncotarget.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SJ, Reis G, Kohanbash G, et al. Expression and prognostic impact of immune modulatory molecule PD-L1 in meningioma. J Neurooncol. 2016;130:543–552. doi: 10.1007/s11060-016-2256-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Liechty B, Patel S, et al. Programmed death ligand 1 expression and tumor infiltrating lymphocytes in neurofibromatosis type 1 and 2 associated tumors. J Neurooncol. 2018;138:183–190. doi: 10.1007/s11060-018-2788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang L, Lowther DE, Meizlish ML, et al. The immune cell infiltrate populating meningiomas is composed of mature, antigen-experienced T and B cells. Neuro-oncol. 2013;15:1479–1490. doi: 10.1093/neuonc/not110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingues PH, Teodósio C, Otero Á, et al. Association between inflammatory infiltrates and isolated monosomy 22/del(22q) in meningiomas. PLoS One. 2013;8:e74798. doi: 10.1371/journal.pone.0074798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domingues PH, Teodósio C, Ortiz J, et al. Immunophenotypic identification and characterization of tumor cells and infiltrating cell populations in meningiomas. Am J Pathol. 2012;181:1749–1761. doi: 10.1016/j.ajpath.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 28.Talari NK, Panigrahi M, Madigubba S, et al. Altered tryptophan metabolism in human meningioma. J Neurooncol. 2016;130:69–77. doi: 10.1007/s11060-016-2225-7. [DOI] [PubMed] [Google Scholar]
- 29.Bi WL, Greenwald NF, Abedalthagafi M, et al. Genomic landscape of high-grade meningiomas. NPJ Genom Med. 2017;2:pii:15. doi: 10.1038/s41525-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menke JR, Raleigh DR, Gown AM, et al. Somatostatin receptor 2a is a more sensitive diagnostic marker of meningioma than epithelial membrane antigen. Acta Neuropathol. 2015;130:441–443. doi: 10.1007/s00401-015-1459-3. [DOI] [PubMed] [Google Scholar]
- 31.Watkins JC, Yang EJ, Muto MG, et al. Universal screening for mismatch-repair deficiency in endometrial cancers to identify patients with Lynch syndrome and Lynch-like syndrome. Int J Gynecol Pathol. 2017;36:115–127. doi: 10.1097/PGP.0000000000000312. [DOI] [PubMed] [Google Scholar]
- 32.Lin J-R, Izar B, Wang S, et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife. 2018;7:7. doi: 10.7554/eLife.31657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coy S, Rashid R, Lin J-R, et al. Multiplexed immunofluorescence reveals potential PD-1/PD-L1 pathway vulnerabilities in craniopharyngioma. Neuro-oncol. 2018;20:1101–1112. doi: 10.1093/neuonc/noy035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramkissoon SH, Bandopadhayay P, Hwang J, et al. Clinical targeted exome-based sequencing in combination with genome-wide copy number profiling: precision medicine analysis of 203 pediatric brain tumors. Neuro-oncol. 2017;19:986–996. doi: 10.1093/neuonc/now294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1:e87062. doi: 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell BB, Light N, Fabrizio D, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171:1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowak JA, Yurgelun MB, Bruce JL, et al. Detection of mismatch repair deficiency and microsatellite instability in colorectal adenocarcinoma by targeted next-generation sequencing. J Mol Diagn. 2017;19:84–91. doi: 10.1016/j.jmoldx.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aizer AA, Abedalthagafi M, Bi WL, et al. A prognostic cytogenetic scoring system to guide the adjuvant management of patients with atypical meningioma. Neuro-oncol. 2016;18:269–274. doi: 10.1093/neuonc/nov177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahm F, Schrimpf D, Stichel D, et al. DNA methylation-based classification and grading system for meningioma: A multicentre, retrospective analysis. Lancet Oncol. 2017;18:682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 40.Olar A, Wani KM, Wilson CD, et al. Global epigenetic profiling identifies methylation subgroups associated with recurrence-free survival in meningioma. Acta Neuropathol. 2017;133:431–444. doi: 10.1007/s00401-017-1678-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 42.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Z, Brewster R, Merrill PH, et al. Meningioma transcription factors link cell lineage with systemic metabolic cues. Neuro-oncol. 2018;20:1331–1343. doi: 10.1093/neuonc/noy057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116–120. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 47.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology: Mutational landscape determines sensitivity to PD-1 blockade in non–small-cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]